Abstract
Major depressive disorder (MDD) is one of the most common mental health conditions that has been intensively investigated for its association with brain atrophy and mortality. Recent studies suggest that the deviation between the predicted and the chronological age can be a marker of accelerated brain aging to characterize MDD. However, current conclusions are usually drawn based on structural MRI information collected from Caucasian participants. The universality of this biomarker needs to be further validated by subjects with different ethnic/racial backgrounds and by different types of data. Here we make use of the REST-meta-MDD, a large scale resting-state fMRI dataset collected from multiple cohort participants in China. We develop a stacking machine learning model based on 1101 healthy controls, which estimates a subject’s chronological age from fMRI with promising accuracy. The trained model is then applied to 1276 MDD patients from 24 sites. We observe that MDD patients exhibit a +4.43 years (p < 0.0001, Cohen’s d = 0.31, 95% CI: 2.23–3.88) higher brain-predicted age difference (brain-PAD) compared to controls. In the MDD subgroup, we observe a statistically significant +2.09 years (p < 0.05, Cohen’s d = 0.134525) brain-PAD in antidepressant users compared to medication-free patients. The statistical relationship observed is further checked by three different machine learning algorithms. The positive brain-PAD observed in participants in China confirms the presence of accelerated brain aging in MDD patients. The utilization of functional brain connectivity for age estimation verifies existing findings from a new dimension.
Subject terms: Depression, Diagnostic markers
Introduction
Global population aging is expected to be one of the prominent social changes of the 21st century [1]. The resulting burden of age-related functional decline and disease would challenge all sectors of society, especially healthcare [2]. Therefore, understanding the biological link between aging and disease risk becomes increasingly important to provide effective care and treatment [3]. Aging can be regarded as a dynamic process in which an individual gradually losses her function as cumulative age-related damage accumulates. The brain structure and function are also significantly changed during this process [4]. As the central nervous system may age dissimilarly to the rest of the body [5], brain-specific aging markers may be of particular importance in assessing the risk of cognitive decline and propensity to neurodegenerative diseases [6].
Accelerated aging of the brain refers to the phenomenon that an individual’s brain appears older compared with the expected chronological age. Brain predicted age difference (brain-PAD), calculated as the difference between the estimated brain age from neuroimaging and the chronological age, is predisposed to be associated with the risk of cognitive aging or age-related brain disorders [7]. This neuroimaging-based biomarker is observed in several neurological disorders [8, 9], including schizophrenia [10–12], Alzheimer’s disease [13–15], epilepsy [16], multiple sclerosis [17], and traumatic brain injury [18]. Furthermore, an association between brain-PAD and mortality [19] has also been reported.
Recent research set out to explore the relationship between accelerated brain aging and major depressive disorder (MDD), a widespread, debilitating, and disabling psychiatric disorder [20] associated with cellular senescence [21, 22] and cognitive decline [23]. Despite the positive association reported [11, 24–29], current studies still present several limitations. First, the size of samples can have a great influence on the stability of the relationship discovered. While subjects with diverse sizes are investigated [8, 30], only one study analyzes data from more than 1000 individuals [31], to the best of our knowledge. Moreover, most of the current studies are based on Caucasian subjects. The generalizability of the association needs to be further verified in subjects from different ethnic and cultural backgrounds. In the meanwhile, the brain age is usually estimated from the structural MRI, with the gray or white matter volume and cortical thickness as the key feature [32]. But the brain age estimated from other types of neuroimaging data is needed for verifications. Finally, current studies often rely on only one machine learning algorithm to estimate brain age. Since different algorithms yield different estimations, it is reasonable to suspect that the conclusion drawn is algorithm sensitive. The statistically significant association originally reported may vanish when a new algorithm is applied.
To cope with these limitations, we make use of the resting-state functional magnetic resonance imaging (rsfMRI) data [33] collected by the REST-meta-MDD [34, 35], which is a coordinated multisite project from China containing over 1000 MDD patients and normal controls. We utilize three different machine learning algorithms to estimate brain age from resting-state functional connectivity [36–38]. We further propose a stacking model to combine results from the three algorithms to reach a more optimal age estimation. We conduct separate analyses on results obtained from each algorithm to check the robustness of the conclusion drawn. We confirm the existence of the positive association between accelerated brain aging and MDD based on subjects in China. The brain-PAD is significantly higher in MDD patients compared to controls and the conclusion is not affected by the machine learning algorithm applied. We separately analyze MDD patients with different depression severity, illness duration, episode status, and medication status to investigate the association between brain-PAD with demographic (age, sex) and clinical characteristics. We find a significant correlation between brain-PAD and illness duration in MDD patients as well as a higher brain-PAD in antidepressant users than in medication-free patients.
Methods
Samples
We conduct this study through rsfMRI indices of MDD patients and matched controls (aged 12–82 years) from the REST-meta-MDD consortium, which consists of 25 research groups from 17 hospitals in China. All MDD patients are hospital diagnosed and conducted at least a T1-weighted structural scan and a rsfMRI scan. All subjects agree to provide diagnosis, age, gender, and education years. The 17-item Hamilton Depression Rating Scale (HDRS) is administered to some patients with other tabular data provided, including episode status (if the patient’s prior and current episodes are diagnosed as MDD according to ICD10 or DSM-IV), medication status (whether antidepressants are used), and illness duration. After quality control, we reach a sample set of 1276 MDD patients and 1101 controls from 24 sites. The sample size and scanning parameters for each site are provided in Supplementary Table S1 and Table S2. Written informed consent is signed by participants at each local site, and all data are de-identified and anonymized. Besides, approvals from the local institutional review board and ethics committee are granted at all sites.
rsfMRI data preprocessing and functional brain network construction
The Data Processing Assistant for Resting State fMRI (DPARSF) [39] is used as a standardized preprocessing pipeline. To obtain functional connectivity, we first extract 116 averaged blood oxygen level-dependent (BOLD) signals based on the Automated Anatomical Labeling (AAL) atlas. Next, we calculate the Pearson correlation coefficients between the BOLD activity time series. We use the Z-Score method [40] to normalize the functional connectivity of each subject to reduce the effect of the imaging sites. We also test the Combat method [41] whose results are presented in Supplementary Information Tables S3 and S4. In general, the Z-Score method performs better in this study. More details of the data preprocessing process are presented in Supplementary Information S2.
Model training and evaluation
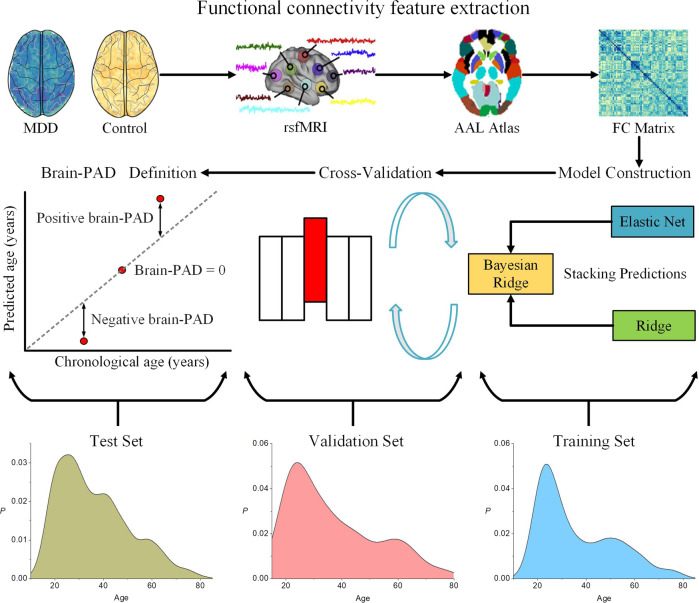
To obtain the input feature for the model, we reshape the upper triangle of the whole-brain correlation matrix into a one-dimensional vector with 6670 elements. To determine the brain aging pattern in healthy individuals, we first train a brain age prediction model on the training set containing 1101 normal controls. Next, we utilize the model to estimate the brain age of 1276 MDD patients on the test set. The brain age prediction is first carried out by three classical supervised learning algorithms: elastic net [42, 43], bayesian ridge [44], and ridge regression [31, 45, 46]. Furthermore, we introduce a stacking model [36] from ensemble learning [47] to combine results from the three algorithms, which gives the best estimation results. The flow is shown in Fig. 1 and Supplementary Fig. S1. The four models all come to consistent conclusions in subsequent experiments. To avoid switching between different methods and make the flow of the paper more concise, we use the results from the stacking method in the main text. Analyses based on the other three algorithms are included in Supplementary Table S5.
Fig. 1. The flow chart of the analysis.
Time series for each subject are extracted from rsfMRI according to the AAL atlas. The Pearson correlation is used to get the functional connectivity matrix. Next, the upper triangle of the functional connectivity matrix is stretched into a one-dimensional vector to get each subject’s feature vector. The feature vectors are concatenated to obtain input data matrices. A brain age prediction model is trained and five-fold cross-validated on the training set. Finally, brain-PAD scores are obtained by applying the model in the hold-out validation set and test set.
We evaluate our model performance in the control and MDD groups separately. We first evaluate the model on the entire training set with five-fold cross-validation. Then, the same model in each fold is used to predict the brain age of MDD patients on the entire test set. The performance of the four models is evaluated based on the following three metrics: mean absolute error (MAE), mean squared error (MSE), and mean coefficient of determination (R2). All models are implemented through the Python-based sklearn package with all parameters set as the default value.
Statistical analyses
To determine whether brain aging is accelerated in MDD patients relative to controls, we split the entire controls to get a fixed training set and a validation set using the hold-out method [48]. While modest in size, this hold-out validation set consists of normal controls from all sites with the entire age span, providing an unbiased age representation of Rest-meta-MDD. As the aim of this study is to explore potential brain age difference between normal controls and MDD patients, we separately estimate the brain age in the two groups. The model is trained and tested in the hold-out validation set composed of normal controls. The trained model is then applied to all MDD patients in the test set to estimate their brain ages. The chronological age is subtracted from the estimated age to get the brain-PAD as the outcome variable for statistical analysis. The five-fold cross-validation is used to compare the overall performance of different models. The hold-out validation set is used as a normal control group for brain-PAD comparison. Due to factors such as regression dilution and non-Gaussian age distribution [49], we need to perform an age-bias correction. We apply a post hoc correction for the residual age effect on the test set [45, 50–53]. Following Peng et al. [54], we train a linear regression model for brain age bias correction. We calculate the regression line between the chronological age and the estimated age on the hold-out validation set. Then the slope and intercept of the regression line are used to adjust brain-predicted age values in the testing set. The steps of this process are shown in Supplementary Information S4. The brain-PAD is independent of chronological age after the age-bias correction (Supplementary Figs. S2 and S3). We apply the univariate generalized linear model (GLM) with gender, diagnosis, age, and age as covariates to explore the relationship between brain-PAD and clinical characteristics [27]. Furthermore, the two-sample t-test is used to compare the brain-PAD in different subgroups. Multiple comparisons are corrected by false discovery rate correction. The threshold for statistical significance is set at p < 0.05.
Results
Model performance
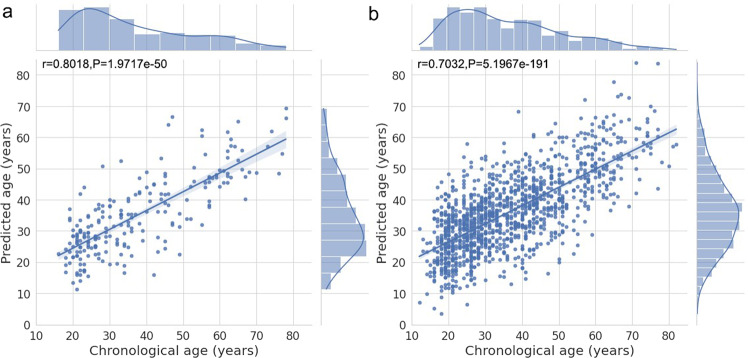
The models obtained from each fold of the training set are used to estimate the brain age of individuals for the rest of the controls in the validation set as well as the MDD patients in the test set. Table 1 shows the performance of four models with 882 training subjects, 219 validation subjects, and 1276 test subjects. Among the three classical machine learning algorithms, the bayesian ridge achieves the best performance. But the stacking model with ensemble learning outperforms all of them, giving rise to the lowest MAE and MSE in both the validation and test set. The performance of other widely used models, such as XGBoost [26], SVM [27], and MLP [13] is not as good as the three models applied under default parameters (Supplementary Table S6). The correlation between chronological age and predicted age on the validation set and test set are presented in Fig. 2a, b.
Table 1.
Performance of four models.
| Model | MAE | MSE | R2 |
|---|---|---|---|
| Validation set | |||
| Elastic net | 8.2327 ± 0.4608 | 103.7454 ± 10.9086 | 0.5670 ± 0.0486 |
| Ridge | 8.7749 ± 0.5662 | 127.4526 ± 17.7452 | 0.4691 ± 0.0665 |
| Bayesian ridge | 7.8057 ± 0.4420 | 97.4546 ± 10.5659 | 0.5934 ± 0.0440 |
| Stacking | 7.7287 ± 0.5547 | 95.3625 ± 11.8727 | 0.6026 ± 0.0456 |
| Test set | |||
| Elastic net | 8.4156 ± 0.0582 | 110.2473 ± 1.2672 | 0.4839 ± 0.0059 |
| Ridge | 9.4921 ± 0.2447 | 143.8590 ± 6.8188 | 0.3265 ± 0.0319 |
| Bayesian ridge | 8.3817 ± 0.0609 | 110.6582 ± 1.2202 | 0.4820 ± 0.0057 |
| Stacking | 8.3055 ± 0.0535 | 108.4852 ± 1.2633 | 0.4837 ± 0.0071 |
Fig. 2. Correlation between the chronological age and predicted age.
a The predicted age (y-axis)on the validation set. b The predicted age on the test set. The lines represent the regression results and the shades correspond to the 95% confidence interval.
The relative feature importance for normal controls
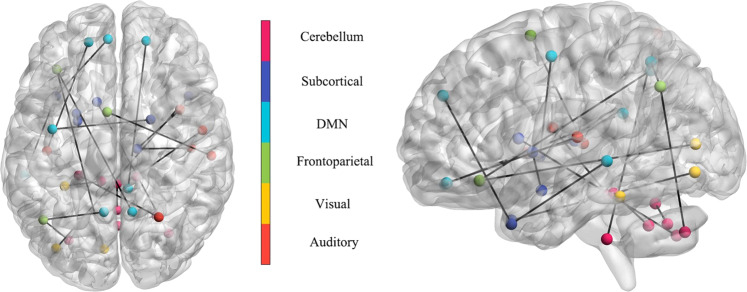
We calculate the correlation between the functional connectivity features and the chronological age (Supplementary Fig. S4). Among all the total 6670 functional connectivity features, 3196 features show positive correlations with age (mean correlation = 0.0645 ± 0.0495, range (6.1691e−05, 0.3017)). 3474 features show negative correlations with age (mean correlation = −0.0691 ± 0.0545, range (−0.3334, 3.7818e−05)). In particular, the most positive correlation is found for the precentral gyrus-Heschel gyrus [55, 56]. The most negative correlation is found for the median cingulate and paracingulate gyry-inferior parietal gyrus, excluding supramarginal and angular gyri [57, 58]. In addition, we identify brain regions that the machine learning algorithm considers to be significant in brain age estimation using the feature importance [59]. The feature importance values are normalized to give the top 20 functional connectivity features (Fig. 3). The main brain regions include the cerebellum [60] superior and vermis8, the medial superior frontal gyrus and middle temporal gyrus [61], the amygdala and lenticular nucleus putamen. These brain regions are associated with brain development and atrophy, which are consistent with previous studies.
Fig. 3. The top 20 most important functional connectivity features obtained from the bayesian ridge model.
These features consist of functional connectivity between the cerebellum superior and vermis8, medial superior frontal gyrus and middle temporal gyrus, amygdala and lenticular nucleus putamen, orbital inferior frontal gyrus and precuneus, angular gyrus and precuneus. Statistical values are obtained from Pearson correlations two-sided test.
Accelerated functional brain aging in MDD
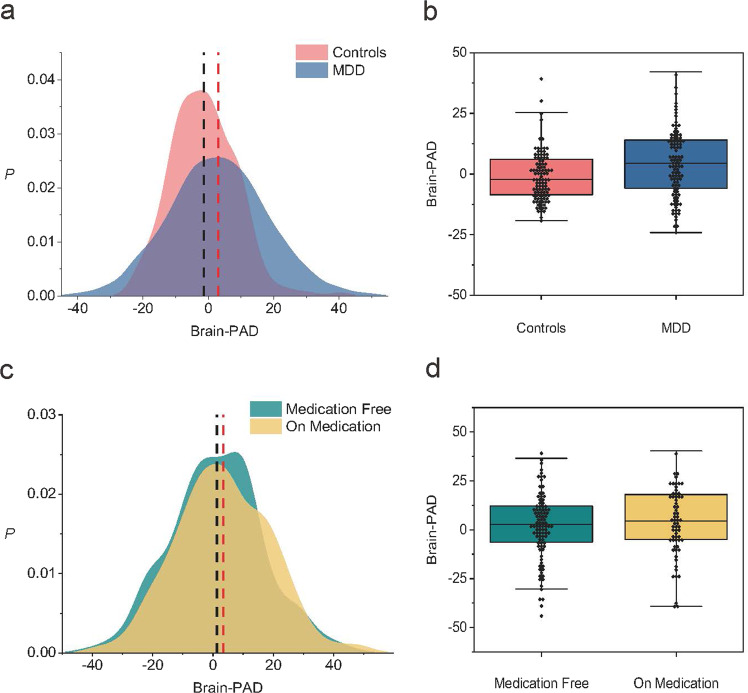
We compare the resulting brain-PAD scores of the MDD patients with the controls in the hold-out validation set to determine whether brain aging is accelerated in MDD patients. Overall, the brain-PAD score before age-bias correction is −1.3731 (SD 9.91) years in the control group and −0.0712 (SD 10.56) years in MDD patients. After applying an age-bias correction procedure, the brain-PAD is +4.43 years higher in MDD patients than in normal controls (p < 0.0001, Cohen’s d = 0.31, 95% CI: 2.23–3.88), which is shown in Fig. 4a, b. Although different estimations are obtained through different models, results from the other three models all demonstrate a consistent pattern that MDD patients have statistically significant higher brain-PAD scores compared to controls. In addition, GLM shows significant main effects for age (p < 0.001), age (p < 0.002) and diagnosis (p < 0.0001), but not for gender (Table 2A).
Fig. 4. The brain-PAD in different subgroups.
a The brain-PAD in controls and MDD patients. b Group analysis shows that the corrected brain-PAD is significantly higher in MDD patients than in controls (p < 0.0001, Cohen’s d = 0.31, 95% CI: 2.23–3.88). c The brain-PAD in antidepressant users and medication-free patients. d Group analysis shows that the brain-PAD is higher in MDD patients who are taking medication (+3.38 years, 95% CI: 1.86–4.90) than in those medication-free ones (+1.29 years, 95% CI: −0.14–2.72). The difference passes the statistically significant threshold (p = 0.0499).
Table 2.
Parameter estimates for all main effects and significant interactions in other clinical characteristics.
| Coef | SE | z | p | 0.025 | 0.975 | |
|---|---|---|---|---|---|---|
| (A) | ||||||
| Intercept | 7.64 | 2.57 | 2.972 | 0.003 | 2.602 | 12.678 |
| Gender | −0.6052 | 0.771 | −0.785 | 0.432 | −2.116 | 0.905 |
| Diagnosis | 4.7237 | 1.045 | 4.519 | 0 | 2.675 | 6.772 |
| Age | −0.4421 | 0.129 | −3.433 | 0.001 | −0.695 | −0.19 |
| Age2 | 0.0046 | 0.002 | 3.048 | 0.002 | 0.002 | 0.008 |
| (B) | ||||||
| Intercept | 1.8688 | 5.087 | 0.367 | 0.713 | −8.102 | 11.839 |
| Gender | −1.2863 | 1.255 | −1.025 | 0.305 | −3.746 | 1.173 |
| Episode | 2.2518 | 1.532 | 1.47 | 0.141 | −0.75 | 5.254 |
| Medication | 2.9454 | 1.291 | 2.282 | 0.023 | 0.415 | 5.475 |
| Age | −0.0285 | 0.252 | −0.113 | 0.91 | −0.522 | 0.465 |
| Age2 | −0.0007 | 0.003 | −0.219 | 0.827 | −0.007 | 0.006 |
| Education | −0.0022 | 0.156 | −0.014 | 0.989 | −0.307 | 0.303 |
| Month | 0.006 | 0.012 | 0.511 | 0.609 | −0.017 | 0.029 |
Brain-PAD comparison for clinical characteristics
To explore the association between brain-PAD scores and clinical characteristics, we use the GLM to fit the brain-PAD of MDD patients with the following explanatory variables: sex, medication status, episode status, education years, and illness duration months (Table 2B). The medication status (p = 0.023) has a main effect on the brain-PAD scores of MDD patients. We further apply a two-sample t-test to determine whether the brain-PAD mean value in antidepressant users and medication-free patients are significantly different from each other (Fig. 4c, d). Brain-PAD is +2.09 years higher (p = 0.0499, Cohen’s d = 0.13452) in antidepressant users than in medication-free ones. Comparisons of other subgroups (sex, episode status) with controls can be found in Supplementary Table S7. While significant differences are observed in all MDD subgroups compared to normal controls, posthoc comparisons of brain-PAD in other clinical characteristics do not demonstrate any significant differences between MDD subgroups except for the medication status. For the two continuous-type clinical characteristics (education years and illness months), we divide the subgroups according to their medians (both are 12 in this study) for brain-PAD comparisons (Supplementary Table S8). Overall, MDD patients with fewer than 12 years of education have a 2.28 years higher brain-PAD than those with greater than or equal to 12 years of education (p = 0.00679). Brain-PAD of MDD patients with fewer than 12 months of illness is 1.69 years higher than that in patients with greater than or equal to 12 months of illness. We also calculate correlations between brain-PAD scores and illness months, education years, and HDRS scores separately. Only illness duration is found to be significantly correlated with brain-PAD scores (Spearman R = −0.067, p < 0.05, Supplementary Fig. S5).
Discussion
Biological aging can be defined as a progressive process of decline involving multiple organ systems. While all individuals age chronologically at the same rate, the rate of their biological aging varies from one to the other [62]. Resting-state functional MRI is developed as a common approach to interrogate the myriad of functional systems in the brain without the constraints of any prior assumptions [63]. Machine learning algorithms based on functional connectivity and the availability of large-scale reliable samples allow us to develop generalized models to estimate the brain age of individual subjects [64]. Here, we make use of the Rest-Meta-MDD consortium from China to verify the accelerated brain aging in MDD patients, which is previously observed in Caucasian participants using structural MRI information. We apply four machine learning algorithms based on functional connectivity features to estimate the brain age of individuals with the entire adult lifespan (12–82 years). We observe manifestly accelerated brain aging in 1276 MDD patients. Furthermore, we compare brain-PAD scores between MDD subgroups divided according to clinical characteristics such as medication status and episode status. We confirm that the conclusion drawn in this paper is not algorithm sensitive as results from different algorithms lead to the same conclusion.
Our study benefits from a reliable experimental design. The dataset contains 24 cohorts so the potential site effect is effectively avoided. Instead of using samples from some independent sites as the fixed validation set, we randomly select samples from all sites to constitute the training set and validation set. In this way, the generalizability of the model is improved and the model outcomes are evaluated more objectively [65, 66]. Moreover, we split the normal controls into a fixed training set and a hold-out validation set. We compare the brain-PAD scores of the controls in this hold-out validation set to the MDD patients in the test set. As the validation set is not involved in the development of the brain age prediction model, the risk of overfitting is effectively prevented [67]. The application of four different machine learning algorithms allows us to further validate the consistency of the patterns observed.
The accuracy of our model (Stacking, MAE = 8.3055, R2 = 0.4837) is better than some previous studies, such as Peter et al. [32] (GPR, MAE = 8.587 years), and Gonneaud et al. [13] (DNN, MAE = 11.90 years). We acknowledge that algorithms used in some other studies yield more accurate age estimation than ours. But multiple factors can affect the performance of the models, such as different features considered and the age distribution of the subjects. The model tends to yield a better prediction using sMRI features than fMRI features [36]. In the meanwhile, the prediction error tends to be smaller when the age distribution of the subjects is narrower. To the best of our knowledge, our work provides a very accurate brain age estimation based on functional connectivity features in a date covering a wide age span (12–82 years).
Although multiple studies are carried out on relatively small samples, conclusions drawn from larger samples tend to be more reliable. First, machine learning algorithms are sensitive to sample size [68–71]. The small size of samples brings a bigger prediction error and a higher risk of overfitting [72]. Moreover, larger samples tend to contain subjects with a wider age distribution. While a wide range of ages makes the estimation challenging, it effectively increases the generalizability of the conclusion [73]. In this study, we make use of the Rest-meta-MDD consortium, which is the largest rsfMRI database of MDD patients. To the best of our knowledge, only one study from ENIGMA uses more subjects than ours, which contains a total of 6989 subjects aged from 18 to 75 years old [31]. But compared with ENIGMA, subjects in Rest-meta-MDD have a bigger life span, ranging from 12 to 82 years old (see the age distribution of the two datasets in Supplementary Figs. S6 and S7). The sufficiently large samples with a wider age span lead to similar conclusions drawn in ENIGMA.
Our results show a +4.43 years gap in terms of the brain-PAD between MDD patients and normal controls (with a Cohen’s d effect sizes size of 0.31) at the group level. Compared to previous studies on accelerated brain aging in MDD patients, such as the one by Koutsouleris et al. [11] (+4.0 years, N = 104), Han et al. [31] (+1.16 years, N = 2675), Dunlop et al. [27] (+2.11 years, N = 112), Han et al. [24] (+0.586 years, N = 195), our results demonstrate a higher brain-PAD. We speculate that this may be related to the stigmatization of depression in traditional Chinese culture [74]. Some studies suggest that compared to Caucasians, the Chinese tend to deny the existence of depression [75]. Consequently, the level of depression tends to be significantly elevated when MDD is diagnosed [76–78].
We find the significant main effects of age, age, and diagnosis in our regression analysis. In particular, the main effect of medication status on brian-PAD is observed after the inclusion of other clinical characteristics, which is in line with the finding by Sacchet et al. [79]. The comparisons between subgroups of MDD patients show that antidepressant users have a statistically significant higher brain-PAD than medication-free patients (+2.09 years, p = 0.0499, Cohen’s d = 0.13452). Explanations for this phenomenon are discussed by Han [31]. The antidepressant users are likely to have a more severe or chronic course of the disorder at the time of scanning. Therefore, the larger brain-PAD scores in antidepressant users may be confounded by clinical standards recommending antidepressant use mainly for severe or chronic MDD [80]. In other words, patients with milder symptoms tend not to take antidepressants [81]. To fully understand the adaptation of brain-PAD in response to pharmacotherapy, randomized controlled intervention studies are needed which require more information on the clinical use of antidepressants, such as the dosage and duration. It is also noteworthy that the p-value in the study by Han [31] is slightly above 0.05 whereas in our study it is slightly below the threshold of statistical significance. But it is still near the boundary of the threshold line. We honestly report the result, and we also admit that it is far early to draw any conclusion based on this statistic.
Similarly, consistent with the finding obtained by Han et al. [24] using structural brain MRI, we observe a higher brain-PAD in first-episode patients (+4.19 years, N = 538) than in recurrent patients (+2.56 years, N = 282). We believe the same explanation can be applied. First, as pointed out in previous studies and observed in our work, there is a negative correlation between brain-PAD and illness duration (Spearman R = −0.067, p < 0.05). Furthermore, recurrent patients have a longer illness duration than first-episode patients. The median illness duration in recurrent patients (60 months) is 10 times greater than in first-episode patients (6 months) in our data. The corresponding median of brain-PAD score in recurrent patients (0.58 years) is 3.08 years smaller than in first-episode patients (3.66 years). The combination of the two effects gives rise to a higher brain-PAD in first-episode patients than that in recurrent patients. It is implied that there may be a clinically unstable period in first-episode patients. As more treatment is given, patients may become more stable in brain functioning. Hence the brain-PAD decreases with the illness duration [24]. But such a hypothesis needs more clinical information to be further verified through longitudinal studies.
Our results extend the generalizability of accelerated brain aging in MDD patients using the rsfMRI feature of Chinese participants. But several limitations should be considered. Although a standardized preprocessing pipeline is employed at all sites before the aggregation group analysis, some subjects still show measurement bias and missing values in the scan. We address this problem by applying various standardization methods to the features. Although the prediction error is within control, these operations may still bring impact on the final results. Next, multiple brain atlas could be considered to obtain the functional connectivity features. Different functional connectivity will have an impact on the subsequent analysis. Furthermore, different features and models could also have a dramatic effect on the final results. Several studies report the great potential of the multimodal features [82–84] and deep learning algorithms [85–87] in neuroimaging research. More comparisons of neuroimaging features and models are needed in the future to produce more convincing conclusions. Besides, all participants in Rest-meta-MDD are Chinese, the generalizability of our model to other ethnic/racial and cultural backgrounds remained to be explored. Finally, aging is a continuous process, yet few current studies address longitudinal investigations of brain aging, including stage-by-stage analyses of MDD to explore trends in brain-PAD with age to understand the progressive effects of the aging process. More clinical features are still desired in the future to determine the clinical significance of measuring brain-PAD and whether it can be considered as a clinically essential biomarker.
Supplementary information
Acknowledgements
We thank the other collaborative members of the REST-meta-MDD consortium for sharing the data.
Author contributions
YSL: design of the work, data analysis and interpretation, and drafting the manuscript. WYC: data analysis and interpretation. JQ: revising the manuscript. TJ: conception and design of the work, drafting the manuscript, and revising the manuscript.
Funding
This work is supported by Industry-University-Research Innovation Fund for Chinese Universities (No. 2021ALA03016), University Innovation Research Group of Chongqing (No. CXQT21005).
Code availability
Multi-site MDD dataset is available through a reasonable request to the Rest-meta-MDD consortium (http://rfmri.org/REST-meta-MDD). The code of brain age estimation is conditionally available upon request from the corresponding author.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41398-022-02162-y.
References
- 1.Vos T, Flaxman AD, Naghavi M, Lozano R, Michaud C, Ezzati M, et al. Years lived with disability (ylds) for 1160 sequelae of 289 diseases and injuries 1990–2010: A systematic analysis for the global burden of disease study 2010. Lancet. 2012;380:2163–96. doi: 10.1016/S0140-6736(12)61729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dinsdale NK, Bluemke E, Smith SM, Arya Z, Vidaurre D, Jenkinson M, et al. Learning patterns of the ageing brain in MRI using deep convolutional networks. Neuroimage. 2021;224:117401. doi: 10.1016/j.neuroimage.2020.117401. [DOI] [PubMed] [Google Scholar]
- 3.Cole JH, Franke K. Predicting age using neuroimaging: Innovative brain ageing biomarkers. Trends Neurosci. 2017;40:681–90. doi: 10.1016/j.tins.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Harman D. Aging: Overview. Ann NY Acad Sci. 2001;928:1–21. doi: 10.1111/j.1749-6632.2001.tb05631.x. [DOI] [PubMed] [Google Scholar]
- 5.Isaev NK, Genrikhs EE, Oborina MV, Stelmashook EV. Accelerated aging and aging process in the brain. Rev Neurosci. 2018;29:233–40. doi: 10.1515/revneuro-2017-0051. [DOI] [PubMed] [Google Scholar]
- 6.Dafflon J, Pinaya WH, Turkheimer F, Cole JH, Leech R, Harris MA, et al. An automated machine learning approach to predict brain age from cortical anatomical measures. Hum Brain Mapp. 2020;41:3555–66. doi: 10.1002/hbm.25028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott ML, Belsky DW, Knodt AR, Ireland D, Melzer TR, Poulton R, et al. Brain-age in midlife is associated with accelerated biological aging and cognitive decline in a longitudinal birth cohort. Mol Psychiatry. 2021;26:3829–38. doi: 10.1038/s41380-019-0626-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuo CY, Lee PL, Hung SC, Liu LK, Lee WJ, Chung CP, et al. Large-scale structural covariance networks predict age in middle-to-late adulthood: A novel brain aging biomarker. Cereb Cortex. 2020;30:5844–62. doi: 10.1093/cercor/bhaa161. [DOI] [PubMed] [Google Scholar]
- 9.Mishra S, Beheshti I, Khanna P. A review of neuroimaging-driven brain age estimation for identification of brain disorders and health conditions. IEEE Rev Biomed Eng. 2021;1:1–16. doi: 10.1109/RBME.2021.3107372. [DOI] [PubMed] [Google Scholar]
- 10.Shahab S, Mulsant BH, Levesque ML, Calarco N, Nazeri A, Wheeler AL, et al. Brain structure, cognition, and brain age in schizophrenia, bipolar disorder, and healthy controls. Neuropsychopharmacology. 2019;44:898–906. doi: 10.1038/s41386-018-0298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koutsouleris N, Davatzikos C, Borgwardt S, Gaser C, Bottlender R, Frodl T, et al. Accelerated brain aging in schizophrenia and beyond: A neuroanatomical marker of psychiatric disorders. Schizophrenia Bull. 2014;40:1140–53. doi: 10.1093/schbul/sbt142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schnack HG, Van Haren NE, Nieuwenhuis M, Hulshoff Pol HE, Cahn W, Kahn RS. Accelerated brain aging in schizophrenia: A longitudinal pattern recognition study. Am J Psychiatry. 2016;173:607–16. doi: 10.1176/appi.ajp.2015.15070922. [DOI] [PubMed] [Google Scholar]
- 13.Gonneaud J, Baria AT, Pichet Binette A, Gordon BA, Chhatwal JP, Cruchaga C, et al. Accelerated functional brain aging in pre-clinical familial Alzheimer’s disease. Nat Commun. 2021;12:1–17. doi: 10.1038/s41467-021-25492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moradi E, Pepe A, Gaser C, Huttunen H, Tohka J, Initiative A D N. et al. Machine learning framework for early MRI-based Alzheimer’s conversion prediction in MCI subjects. Neuroimage. 2015;104:398–412. doi: 10.1016/j.neuroimage.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iturria-Medina Y, Sotero RC, Toussaint PJ, Mateos-P´erez JM, Evans AC. Early role of vascular dysregulation on late-onset Alzheimer’s disease based on multifactorial data-driven analysis. Nat Commun. 2016;7:1–14. doi: 10.1038/ncomms11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sone D, Beheshti I, Maikusa N, Ota M, Kimura Y, Sato N, et al. Neuroimaging-based brain-age prediction in diverse forms of epilepsy: A signature of psychosis and beyond. Mol Psychiatry. 2021;26:825–34. doi: 10.1038/s41380-019-0446-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole JH, Raffel J, Friede T, Eshaghi A, Brownlee WJ, Chard D, et al. Longitudinal assessment of multiple sclerosis with the brain-age paradigm. Ann Neurol. 2020;88:93–105. doi: 10.1002/ana.25746. [DOI] [PubMed] [Google Scholar]
- 18.Cole JH, Leech R, Sharp DJ, Initiative A D N. Prediction of brain age suggests accelerated atrophy after traumatic brain injury. Ann Neurol. 2015;77:571–81. doi: 10.1002/ana.24367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole JH, Ritchie SJ, Bastin ME, Hern´andez MV, Maniega SM, Royle N, et al. Brain age predicts mortality. Mol Psychiatry. 2018;23:1385–92. doi: 10.1038/mp.2017.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrari AJ, Charlson FJ, Norman RE, Patten SB, Freedman G, Murray CJ, et al. Burden of depressive disorders by country, sex, age, and year: Findings from the global burden of disease study 2010. PLoS Med. 2013;10:e1001547. doi: 10.1371/journal.pmed.1001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verhoeven J, R´ev´esz D, Picard M, Epel E, Wolkowitz O, Matthews K, et al. Depression, telomeres and mitochondrial DNA: Between-and within-person associations from a 10-year longitudinal study. Mol Psychiatry. 2018;23:850–7. doi: 10.1038/mp.2017.48. [DOI] [PubMed] [Google Scholar]
- 22.Verhoeven JE, R´ev´esz D, Epel ES, Lin J, Wolkowitz OM, Penninx BW. Major depressive disorder and accelerated cellular aging: Results from a large psychiatric cohort study. Mol Psychiatry. 2014;19:895–901. doi: 10.1038/mp.2013.151. [DOI] [PubMed] [Google Scholar]
- 23.John A, Patel U, Rusted J, Richards M, Gaysina D. Affective problems and decline in cognitive state in older adults: A systematic review and meta-analysis. Psychol. Med. 2019;49:353–65. doi: 10.1017/S0033291718001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han S, Chen Y, Zheng R, Li S, Jiang Y, Wang C, et al. The stage-specifically accelerated brain aging in never-treated first-episode patients with depression. Hum Brain Mapp. 2021;42:3656–66. doi: 10.1002/hbm.25460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christman S, Bermudez C, Hao L, Landman BA, Boyd B, Albert K, et al. Accelerated brain aging predicts impaired cognitive performance and greater disability in geriatric but not midlife adult depression. Transl Psychiatry. 2020;10:1–11. doi: 10.1038/s41398-020-01004-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaufmann T, van der Meer D, Doan NT, Schwarz E, Lund MJ, Agartz I, et al. Common brain disorders are associated with heritable patterns of apparent aging of the brain. Nat Neurosci. 2019;22:1617–23. doi: 10.1038/s41593-019-0471-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunlop K, Victoria LW, Downar J, Gunning FM, Liston C. Accelerated brain aging predicts impulsivity and symptom severity in depression. Neuropsychopharmacology. 2021;46:911–9. doi: 10.1038/s41386-021-00967-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drobinin V, Van Gestel H, Helmick CA, Schmidt MH, Bowen CV, Uher R. The developmental brain age is associated with adversity, depression, and functional outcomes among adolescents. Biol Psychiatry: Cogn Neurosci Neuroimaging. 2022;7:406–14. doi: 10.1016/j.bpsc.2021.09.004. [DOI] [PubMed] [Google Scholar]
- 29.Ballester PL, Suh JS, Nogovitsyn N, Hassel S, Strother SC, Arnott SR, et al. Accelerated brain aging in major depressive disorder and antidepressant treatment response: A can-bind report. NeuroImage: Clin. 2021;32:102864. doi: 10.1016/j.nicl.2021.102864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Besteher B, Gaser C, Nenadi´c I. Machine-learning based brain age estimation in major depression showing no evidence of accelerated aging. Psychiatry Res.: Neuroimaging. 2019;290:1–4. doi: 10.1016/j.pscychresns.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 31.Han LK, Dinga R, Hahn T, Ching CR, Eyler LT, Aftanas L, et al. Brain aging in major depressive disorder: Results from the enigma major depressive disorder working group. Mol Psychiatry. 2021;26:5124–39. doi: 10.1038/s41380-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Millar PR, Luckett PH, Gordon BA, Benzinger TL, Schindler SE, Fagan AM, et al. Predicting brain age from functional connectivity in symptomatic and preclinical Alzheimer disease. NeuroImage. 2022;256:119228. doi: 10.1016/j.neuroimage.2022.119228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vergun S, Deshpande A, Meier TB, Song J, Tudorascu DL, Nair VA, et al. Characterizing functional connectivity differences in aging adults using machine learning on resting state fMRI data. Front Comput. Neurosci. 2013;7:38. doi: 10.3389/fncom.2013.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan CG, Chen X, Li L, Castellanos FX, Bai TJ, Bo QJ, et al. Reduced default mode network functional connectivity in patients with recurrent major depressive disorder. Proc Natl Acad Sci USA. 2019;116:9078–83. doi: 10.1073/pnas.1900390116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang H, Chen X, Chen ZB, Li L, Li XY, Castellanos FX, et al. Disrupted intrinsic functional brain topology in patients with major depressive disorder. Mol Psychiatry. 2021;26:7363–71. doi: 10.1038/s41380-021-01247-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liem F, Varoquaux G, Kynast J, Beyer F, Masouleh SK, Huntenburg JM, et al. Predicting brain-age from multimodal imaging data captures cognitive impairment. Neuroimage. 2017;148:179–88. doi: 10.1016/j.neuroimage.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Zhai J, Li K. Predicting brain age based on spatial and temporal features of human brain functional networks. Front Hum Neurosci. 2019;13:62. doi: 10.3389/fnhum.2019.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Satterthwaite TD, Fan Y. Brain age prediction based on resting-state functional connectivity patterns using convolutional neural networks. 2018 IEEE 15th international symposium on biomedical imaging (isbi 2018). IEEE, Washington, USA, 2018: p. 101–4. [DOI] [PMC free article] [PubMed]
- 39.Yan C, Zang Y. Dparsf: A matlab toolbox for” pipeline” data analysis of resting-state fMRI. Front Syst Neurosci. 2010;4:13. doi: 10.3389/fnsys.2010.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wachinger C, Rieckmann A, P¨olsterl S, Initiative A D N. et al. Detect and correct bias in multi-site neuroimaging datasets. Med Image Anal. 2021;67:101879. doi: 10.1016/j.media.2020.101879. [DOI] [PubMed] [Google Scholar]
- 41.Beer JC, Tustison NJ, Cook PA, Davatzikos C, Sheline YI, Shinohara RT, et al. Longitudinal combat: A method for harmonizing longitudinal multi-scanner imaging data. Neuroimage. 2020;220:117129. doi: 10.1016/j.neuroimage.2020.117129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ball G, Kelly CE, Beare R, Seal ML. Individual variation underlying brain age estimates in typical development. NeuroImage. 2021;235:118036. doi: 10.1016/j.neuroimage.2021.118036. [DOI] [PubMed] [Google Scholar]
- 43.Khundrakpam BS, Tohka J, Evans AC, Group B D C. et al. Prediction of brain maturity based on cortical thickness at different spatial resolutions. Neuroimage. 2015;111:350–9. doi: 10.1016/j.neuroimage.2015.02.046. [DOI] [PubMed] [Google Scholar]
- 44.Xu Y, Li X, Yang Y, Li C, Shao X. Human age prediction based on DNA methylation of non-blood tissues. Comput Methods Prog Biomed. 2019;171:11–18. doi: 10.1016/j.cmpb.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 45.Niu X, Zhang F, Kounios J, Liang H. Improved prediction of brain age using multimodal neuroimaging data. Hum Brain Mapp. 2020;41:1626–43. doi: 10.1002/hbm.24899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chung Y, Addington J, Bearden CE, Cadenhead K, Cornblatt B, Mathalon DH, et al. Use of machine learning to determine deviance in neuroanatomical maturity associated with future psychosis in youths at clinically high risk. JAMA Psychiatry. 2018;75:960–8. doi: 10.1001/jamapsychiatry.2018.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Couvy-Duchesne B, Faouzi J, Martin B, Thibeau-Sutre E, Wild A, Ansart M, et al. Ensemble learning of convolutional neural network, support vector machine, and best linear unbiased predictor for brain age prediction: Aramis contribution to the predictive analytics competition 2019 challenge. Front Psychiatry. 2020;11:593336. doi: 10.3389/fpsyt.2020.593336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levman J, Jennings M, Kabaria P, Rouse E, Nangaku M, Berger D, et al. Hold-out validation for the assessment of stability and reliability of multivariable regression demonstrated with magnetic resonance imaging of patients with schizophrenia. Int J Dev Neurosci. 2021;81:655–62. doi: 10.1002/jdn.10144. [DOI] [PubMed] [Google Scholar]
- 49.Smith SM, Vidaurre D, Alfaro-Almagro F, Nichols TE, Miller KL. Estimation of brain age delta from brain imaging. Neuroimage. 2019;200:528–39. doi: 10.1016/j.neuroimage.2019.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le TT, Kuplicki RT, McKinney BA, Yeh HW, Thompson WK, Paulus MP, et al. A nonlinear simulation framework supports adjusting for age when analyzing brain age. Front Aging Neurosci. 2018;10:317. doi: 10.3389/fnagi.2018.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang H, Zhang F, Niu X. Investigating systematic bias in brain age estimation with application to post-traumatic stress disorders. Technical report. Wiley Online Libr. 2019;40:3143–52. doi: 10.1002/hbm.24588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cole JH. Multimodality neuroimaging brain-age in UK biobank: Relationship to biomedical, lifestyle, and cognitive factors. Neurobiol Aging. 2020;92:34–42. doi: 10.1016/j.neurobiolaging.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aycheh HM, Seong JK, Shin JH, Na DL, Kang B, Seo SW, et al. Biological brain age prediction using cortical thickness data: A large scale cohort study. Front Aging Neurosci. 2018;10:252. doi: 10.3389/fnagi.2018.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng H, Gong W, Beckmann CF, Vedaldi A, Smith SM. Accurate brain age prediction with lightweight deep neural networks. Med Image Anal. 2021;68:101871. doi: 10.1016/j.media.2020.101871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kovalev VA, Kruggel F, von Cramon DY. Gender and age effects in structural brain asymmetry as measured by MRI texture analysis. NeuroImage. 2003;19:895–905. doi: 10.1016/S1053-8119(03)00140-X. [DOI] [PubMed] [Google Scholar]
- 56.Amoroso N, La Rocca M, Bellantuono L, Diacono D, Fanizzi A, Lella E, et al. Deep learning and multiplex networks for accurate modeling of brain age. Front Aging Neurosci. 2019;11:115. doi: 10.3389/fnagi.2019.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MacDonald SW, Nyberg L, Sandblom J, Fischer H, B¨ackman L. Increased response-time variability is associated with reduced inferior parietal activation during episodic recognition in aging. J Cogn Neurosci. 2008;20:779–86. doi: 10.1162/jocn.2008.20502. [DOI] [PubMed] [Google Scholar]
- 58.Rivera SM, Reiss A, Eckert MA, Menon V. Developmental changes in mental arithmetic: Evidence for increased functional specialization in the left inferior parietal cortex. Cereb Cortex. 2005;15:1779–90. doi: 10.1093/cercor/bhi055. [DOI] [PubMed] [Google Scholar]
- 59.Nunes A, Schnack HG, Ching CR, Agartz I, Akudjedu TN, Alda M, et al. Using structural MRI to identify bipolar disorders-13 site machine learning study in 3020 individuals from the enigma bipolar disorders working group. Mol Psychiatry. 2020;25:2130–43. doi: 10.1038/s41380-018-0228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tiemeier H, Lenroot RK, Greenstein DK, Tran L, Pierson R, Giedd JN. Cerebellum development during childhood and adolescence: A longitudinal morphometric mri study. Neuroimage. 2010;49:63–70. doi: 10.1016/j.neuroimage.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tomoda A, Kinoshita S, Korenaga Y, Mabe H. Pseudohypacusis in childhood and adolescence is associated with increased gray matter volume in the medial frontal gyrus and superior temporal gyrus. Cortex. 2012;48:492–503. doi: 10.1016/j.cortex.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 62.Elliott ML, Caspi A, Houts RM, Ambler A, Broadbent JM, Hancox RJ, et al. Disparities in the pace of biological aging among midlife adults of the same chronological age have implications for future frailty risk and policy. Nat Aging. 2021;1:295–308. doi: 10.1038/s43587-021-00044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Biswal BB, Mennes M, Zuo XN, Gohel S, Kelly C, Smith SM, et al. Toward discovery science of human brain function. Proc Natl Acad Sci USA. 2010;107:4734–9. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Franke K, Gaser C. Ten years of brainage as a neuroimaging biomarker of brain aging: What insights have we gained? Front Neurol. 2019;10:789. doi: 10.3389/fneur.2019.00789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Orru‘ G, Monaro M, Conversano C, Gemignani A, Sartori G. Machine learning in psychometrics and psychological research. Front Psychol. 2020;10:2970. doi: 10.3389/fpsyg.2019.02970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cai XL, Xie DJ, Madsen KH, Wang YM, B¨ogemann SA, Cheung EF, et al. Generalizability of machine learning for classification of schizophrenia based on resting-state functional MRI data. Hum Brain Mapp. 2020;41:172–84. doi: 10.1002/hbm.24797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shim M, Lee SH, Hwang HJ. Inflated prediction accuracy of neuropsychiatric biomarkers caused by data leakage in feature selection. Sci Rep. 2021;11:1–7. doi: 10.1038/s41598-021-87157-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arbabshirani MR, Plis S, Sui J, Calhoun VD. Single subject prediction of brain disorders in neuroimaging: Promises and pitfalls. Neuroimage. 2017;145:137–65. doi: 10.1016/j.neuroimage.2016.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gianfrancesco MA, Tamang S, Yazdany J, Schmajuk G. Potential biases in machine learning algorithms using electronic health record data. JAMA Intern Med. 2018;178:1544–7. doi: 10.1001/jamainternmed.2018.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Varoquaux G. Cross-validation failure: Small sample sizes lead to large error bars. Neuroimage. 2018;180:68–77. doi: 10.1016/j.neuroimage.2017.06.061. [DOI] [PubMed] [Google Scholar]
- 71.Vabalas A, Gowen E, Poliakoff E, Casson AJ. Machine learning algorithm validation with a limited sample size. PLoS One. 2019;14:e0224365. doi: 10.1371/journal.pone.0224365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Flint C, Cearns M, Opel N, Redlich R, Mehler D, Emden D, et al. Systematic misestimation of machine learning performance in neuroimaging studies of depression. Neuropsychopharmacology. 2021;46:1510–7. doi: 10.1038/s41386-021-01020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cole JH, Franke K, Cherbuin N. Quantification of the biological age of the brain using neuroimaging. Biomarkers of human aging. Springer, Cham, 2019:293–328.
- 74.Georg Hsu L, Wan YM, Chang H, Summergrad P, Tsang BY, Chen H. Stigma of depression is more severe in Chinese Americans than Caucasian Americans. Psychiatry: Interpers Biol Process. 2008;71:210–8. doi: 10.1521/psyc.2008.71.3.210. [DOI] [PubMed] [Google Scholar]
- 75.Parker G, Gladstone G, Chee KT. Depression in the planet’s largest ethnic group: The Chinese. Am J Psychiatry. 2001;158:857–64. doi: 10.1176/appi.ajp.158.6.857. [DOI] [PubMed] [Google Scholar]
- 76.Young CB, Fang DZ, Zisook S. Depression in Asian-American and Caucasian undergraduate students. J Affect Disord. 2010;125:379–82. doi: 10.1016/j.jad.2010.02.124. [DOI] [PubMed] [Google Scholar]
- 77.Ai AL, Nicdao EG, Appel HB, Lee DH. Ethnic identity and major depression in Asian American subgroups nationwide: Differential findings in relation to subcultural contexts. J Clin Psychol. 2015;71:1225–44. doi: 10.1002/jclp.22214. [DOI] [PubMed] [Google Scholar]
- 78.Krieg A, Xu Y, Cicero DC. Comparing social anxiety between Asian Americans and European Americans: An examination of measurement invariance. Assessment. 2018;25:564–77. doi: 10.1177/1073191116656438. [DOI] [PubMed] [Google Scholar]
- 79.Sacchet MD, Camacho MC, Livermore EE, Thomas EA, Gotlib IH. Accelerated aging of the putamen in patients with major depressive disorder. J Psychiatry Neurosci: JPN. 2017;42:164–71. doi: 10.1503/jpn.160010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schmaal L, Hibar D, S¨amann PG, Hall G, Baune B, Jahanshad N, et al. Cortical abnormalities in adults and adolescents with major depression based on brain scans from 20 cohorts worldwide in the enigma major depressive disorder working group. Mol Psychiatry. 2017;22:900–9. doi: 10.1038/mp.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schmaal L, Veltman DJ, van Erp TG, S¨amann P, Frodl T, Jahanshad N, et al. Subcortical brain alterations in major depressive disorder: Findings from the enigma major depressive disorder working group. Mol Psychiatry. 2016;21:806–12. doi: 10.1038/mp.2015.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cherubini A, Caligiuri ME, P´eran P, Sabatini U, Cosentino C, Amato F. Importance of multimodal MRI in characterizing brain tissue and its potential application for individual age prediction. IEEE J Biomed Health Inform. 2016;20:1232–9. doi: 10.1109/JBHI.2016.2559938. [DOI] [PubMed] [Google Scholar]
- 83.De Lange AMG, Anatu¨rk M, Suri S, Kaufmann T, Cole JH, Griffanti L, et al. Multimodal brain-age prediction and cardiovascular risk: The Whitehall ii MRI sub-study. NeuroImage. 2020;222:117292. doi: 10.1016/j.neuroimage.2020.117292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rokicki J, Wolfers T, Nordhøy W, Tesli N, Quintana DS, Alnæs D, et al. Multimodal imaging improves brain age prediction and reveals distinct abnormalities in patients with psychiatric and neurological disorders. Hum Brain Mapp. 2021;42:1714–26. doi: 10.1002/hbm.25323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.J´onsson BA, Bjornsdottir G, Thorgeirsson T, Ellingsen LM, Walters GB, Gudbjartsson D, et al. Brain age prediction using deep learning uncovers associated sequence variants. Nat Commun. 2019;10:1–10. doi: 10.1038/s41467-019-13163-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koppe G, Meyer-Lindenberg A, Durstewitz D. Deep learning for small and big data in psychiatry. Neuropsychopharmacology. 2021;46:176–90. doi: 10.1038/s41386-020-0767-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abrol A, Fu Z, Salman M, Silva R, Du Y, Plis S, et al. Deep learning encodes robust discriminative neuroimaging representations to outperform standard machine learning. Nat Commun. 2021;12:1–17. doi: 10.1038/s41467-020-20655-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Multi-site MDD dataset is available through a reasonable request to the Rest-meta-MDD consortium (http://rfmri.org/REST-meta-MDD). The code of brain age estimation is conditionally available upon request from the corresponding author.