Abstract
Extended periods of bed rest and limb immobilization are required for healing post-injury or disease, yet disuse can result in significant muscle atrophy and decreased quality of life in older adults. Physical rehabilitation is commonly prescribed to recover these deficits, yet accumulation of reactive oxygen species (ROS) and sustained rates of protein degradation persist during the rehabilitation period that can significantly delay or prevent recovery. Pericytes, considered the primary mesenchymal and vascular stromal cell in skeletal muscle, secrete beneficial factors that maintain baseline muscle mass, yet minimal information exists regarding the pericyte response to disuse and recovery. In the current study, single-cell RNA sequencing (scRNA-Seq) and functional assays were performed to demonstrate that pericytes in mouse skeletal muscle lose the capacity to synthesize antioxidants during disuse and recovery. This information was used to guide the design of a strategy in which healthy donor pericytes were stimulated with hydrogen peroxide (H2O2) to produce small extracellular vesicles (sEVs) that effectively restored myofiber size in adult and aged muscle after disuse. Proteomic assessment detected 11 differentially regulated proteins in primed sEVs that may account for recovery of muscle, including proteins associated with extracellular matrix composition and anti-inflammatory and antioxidant processes. This study demonstrates that healthy H2O2-primed pericyte-derived sEVs effectively improve skeletal muscle recovery after immobilization, representing a novel acellular approach to rebuild muscle mass in older adults after a period of disuse.
Keywords: skeletal muscle, disuse atrophy, pericyte, extracellular vesicles, antioxidant
Graphical Abstract
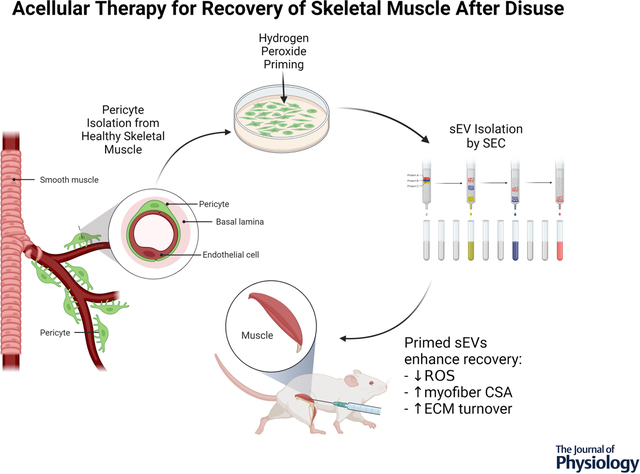
Hydrogen peroxide treatment of healthy, muscle-resident pericytes in vitro stimulates the release of small extracellular vesicles (sEVs) that effectively reduce oxidative stress and recover skeletal muscle fiber size and collagen remodeling in mice after disuse.
Introduction
Disuse-induced skeletal muscle atrophy can result in metabolic disorders and physical disabilities which are associated with loss of independence and premature death in older adults (Atherton et al., 2016). Resistance exercise is the most effective approach to recover loss of muscle mass following periods of bedrest or limb immobilization; however, capacity to engage in rehabilitation is often limited in older adults due to pain, impairments in balance, and decreased mobility (Peeters et al., 2016). In addition, older adults do not always elicit an appropriate growth response to resistance training (Suetta et al., 2009; Hvid et al., 2010; Suetta et al., 2013; Pisot et al., 2016). Alternative strategies are highly desired to improve recovery of muscle mass after a period of disuse.
Conditions of disuse and unloading significantly decrease the rate of muscle protein synthesis (MPS), as well as enhance muscle protein breakdown (MPB) (Baehr et al., 2016; Roberson et al., 2020). The onset of remobilization can rapidly recover MPS in both young and aged rodents (White et al., 2015; Baehr et al., 2016; Roberson et al., 2020), yet MPB may persist and impede restoration of muscle mass, particularly with aging (Baehr et al., 2016; Roberson et al., 2020). The mechanistic basis for the enhancement in MPB during reloading is not clear, but the onset of muscle contraction results in the accumulation of mitochondria-derived reactive oxygen species (ROS; superoxide, O2− and hydrogen peroxide, H2O2) due to devastating losses in mitochondria density, morphology and function during disuse (Kang & Ji, 2013; Kang et al., 2015). ROS can directly disrupt the sarcolemma and increase calcium influx, subsequently activating calpains necessary for Z-band degradation and the ubiquitination and proteasomal degradation of myofibrillar proteins [(Talbert et al., 2013); reviewed by Powers (Powers, 2014)]. Antioxidants are naturally synthesized to defend against ROS-mediated stress in most mammalian cells, yet antioxidant content and activity (catalase (CAT) and glutathione peroxidase (GPX)) has been reported to be deficient in skeletal muscle after disuse (Lawler et al., 2003). Thus, redox imbalance and prolonged ubiquitin proteasomal degradation are key events that may underlie delays in recovery following disuse.
Mesenchymal stem/stromal cells (MSCs) secrete anti-fibrotic and anti-inflammatory factors and are highly resistant to oxidative stress due to their ability to produce antioxidants, such as superoxide dismutase 1 and 2 (SOD1, SOD2), CAT, GPX, glutathione (GSH), and heme oxygenase-1 (HO-1) [(Caplan & Dennis, 2006; Fisher, 2009; Pan et al., 2016; Stavely & Nurgali, 2020); extensively reviewed by Stavely et al. in (Stavely & Nurgali, 2020)]. MSC-derived small extracellular vesicles (sEVs) similarly encapsulate antioxidants and contribute to MSC-mediated suppression of oxidative stress [(Eirin et al., 2018; Baek et al., 2019; Wiklander et al., 2019; Yao et al., 2019); extensively reviewed in (Stavely & Nurgali, 2020)]. Perivascular stem cells, or pericytes (CD146+CD45−CD31−), are considered the primary MSC in skeletal muscle and secrete beneficial immunomodulatory, anti-fibrotic, and pro-angiogenic factors in response to contraction that promote skeletal muscle remodeling (Dvoretskiy et al., 2019). Despite these intriguing results, minimal information exists regarding the impact of disuse and recovery on pericyte function.
In the current study, single-cell RNA sequencing (scRNA-Seq) and functional assays were performed to demonstrate that pericytes in mouse skeletal muscle lose the capacity to synthesize antioxidants during disuse and recovery. This information was used to guide the design of a strategy in which healthy donor pericytes were stimulated with H2O2 to produce small extracellular vesicles (sEVs) that effectively enhanced recovery of mouse skeletal muscle after disuse. The results from this study represent an important first step in developing an alternative approach to rebuild muscle mass and prevent disability in older adults.
Results
Single-Cell Analysis of Adult Skeletal Muscle After Disuse
scRNA-Seq was performed on pooled mononuclear cell suspensions obtained from adult mouse tibialis anterior (TA) muscles after 2 weeks of immobilization (Fig. 1a). After cell filtering, as well as normalization and quality control of scRNA-Seq data, 4,224 cells from Mobile (control) limbs and 5,404 cells from Immobile limbs were captured. The data were analyzed using the R package of Seurat with unsupervised graph-based clustering. Cells were then identified based on expression of established markers (Vanlandewijck et al., 2018) and were visualized using t-distributed stochastic neighborhood embedding (t-SNE; Fig. 1b). 23 cell clusters were identified, including 9 clusters of endothelial cells (Pecam1, Cdh5, Tek, Kdr), 2 clusters of pericytes (Rgs5, Cspg4, Mcam, Pdgfrb, Vtn, Abcc9, Kcnj8), 2 clusters of glial cells (Plp1, Kcna1, S100b, Mbp), muscle stem cells (MuSCs; Pax7, Myod1, Vcam1, Itga7), fibroadipogenic progenitor cells (FAPs; Pdgfra, Cd34, Dcn, Col1a1, Ly6a), smooth muscle cells (Acta2, Tagln, Cnn1, Myl9), and tendon cells (Scx, Tnc, Tnmd). Immune cells (macrophage, neutrophil, T and B cell) and unknown clusters were also present in small quantities (Fig. 1b). Analysis of differentially expressed genes provided additional molecular and surface markers for proper identification (Table 1 in 10.6084/m9.figshare.16778053).
Figure 1. Single-cell Analysis of Skeletal Muscle After 2 Weeks of Immobilization.
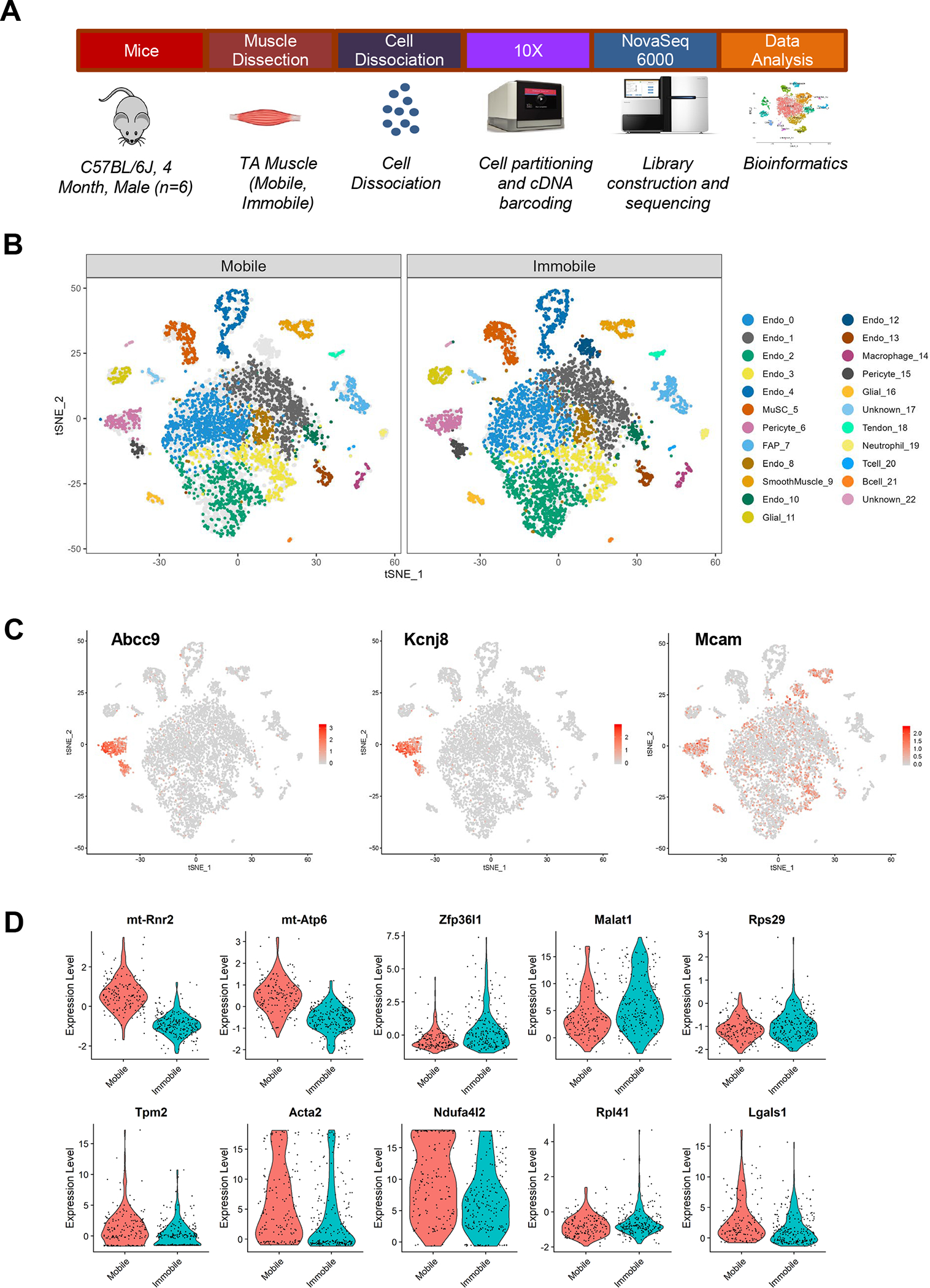
(a) Schematic diagram of experimental design for scRNA-Seq. Mice (C57BL/6J, 4 month-old, male; n=6) were subjected to 2 weeks of unilateral limb stapling and immobilization. Tibialis anterior muscles were collected from mobile and immobile limbs and separately pooled prior to preparation of single-cell suspensions by mechanical and enzymatic dissociation. After exclusion of debris and doublets from single-cell suspensions, cells were loaded into the 10X Chromium system. Library preparation and sequencing was performed on the NovaSeq 6000 platform and raw data were processed by Cell Ranger (v. 3.0.2) to generate a gene-cell expression matrix. Further data analysis was carried out using Seurat (v. 3.1.4). After gene and cell filtering, 17,927 genes were detected in 4,224 Mobile cells and 5,404 Immobile cells. (b) t-Stochastic neighbor embedding (tSNE) plots showing the distribution of the 23 primary cell populations identified in TA muscle in the mobile control limbs versus the immobilized limbs. Each dot represents a single cell; light gray dots show relative positions of cells from the other sample for comparison. Endothelial cluster 12 was only present after immobilization. (c) Marker expression illustrated by cluster. Pericyte clusters 6 and 15 were identified by expression of Abcc9 and Kcnj8. (d) Top 10 genes differentially expressed in pericyte cluster 6.
Pericytes were identified in Clusters 6 and 15, which uniquely expressed Abcc9 and Kcnj8 (Fig. 1c). Canonical pericyte markers such as Mcam (CD146), Cspg4 (NG2), Rgs5, and Pdgfrb were also expressed, yet Cspg4 (NG2) was also expressed in smooth muscle cells and FAPs (Fig. 1c). Mcam (CD146) and Rgs5 were also expressed in endothelial cells and/or smooth muscle cells, but not FAPs (Fig. 1c). Itga7, which is commonly used to identify MuSCs along with Vcam1, was unexpectedly low in MuSCs and high in pericyte cluster 6. Thus, pericytes may represent the Itga7+Vcam1− cluster identified in skeletal muscle by Giordani et al. (Giordani et al., 2019). MSC markers CD105 and CD29, but not CD90, were expressed in pericyte cluster 6. Endothelial-specific genes were higher in pericyte cluster 15 compared to 6, suggesting the potential for differentiation. Finally, cluster 12 was only present after immobilization, and although classified as endothelial cells based on high expression of canonical endothelial markers, the cells also expressed Mcam, Cd34, and Ly6a.
Differentially expressed genes between Mobile and Immobile groups were analyzed. Several genes associated with mitochondrial complex proteins were significantly decreased in nearly every cluster, including mitochondrially encoded 16S RNA (mt-Rnr2; mitochondrial large subunit ribosomal RNA), subunits 2 and 4 of NADH dehydrogenase (ubiquinone or Complex I) (mt-Nd2, mt-Nd4), and mitochondrially encoded ATP synthase membrane 6 (mt-Atp6; ATP synthase Fo subunit 6) (Table 2 in 10.6084/m9.figshare.16778053). Specific to pericyte cluster 6, NADH dehydrogenase 1 alpha subcomplex, 4-like2 (Ndufa4l2) and several genes important for contractility (Tpm2, Acta2, Tnnc2, Tagln, Myl9) were decreased after immobilization (Fig. 1d and Table 2 in 10.6084/m9.figshare.16778053). Metastasis associated lung adenocarcinoma transcript 1 (Malat1), a long noncoding RNA that acts as a transrepressor for nuclear factor erythroid 2-related factor 2 (Nrf2) and antioxidant production (Chen et al., 2018), was significantly upregulated only in pericyte cluster 6 (Fig. 1d). In MuSCs, the signal transducer and activator of transcription 3 (Stat3) was significantly increased, which can inhibit self-renewal and induce muscle atrophy (Sala et al., 2019) (Table 2 in 10.6084/m9.figshare.16778053).
Antioxidant Response is Impaired in Pericytes in Response to H2O2 In Vivo and In Vitro
The significant decrease in mitochondrial-related gene expression and the elevation in Malat1 suggests that pericytes may experience redox imbalance during disuse. Despite confirmation of an increase in muscle oxidative stress three days following remobilization (Fig. 2b) (Kang & Ji, 2013), pericyte antioxidant gene expression was minimal (except Sod1, p=0.030) (Fig. 2d). Interestingly, a trend toward an increase in Malat1 gene expression was still detected at this time point (p=0.068) (Fig. 2c).
Figure 2. Antioxidant Response is Impaired in Pericytes After Immobilization.
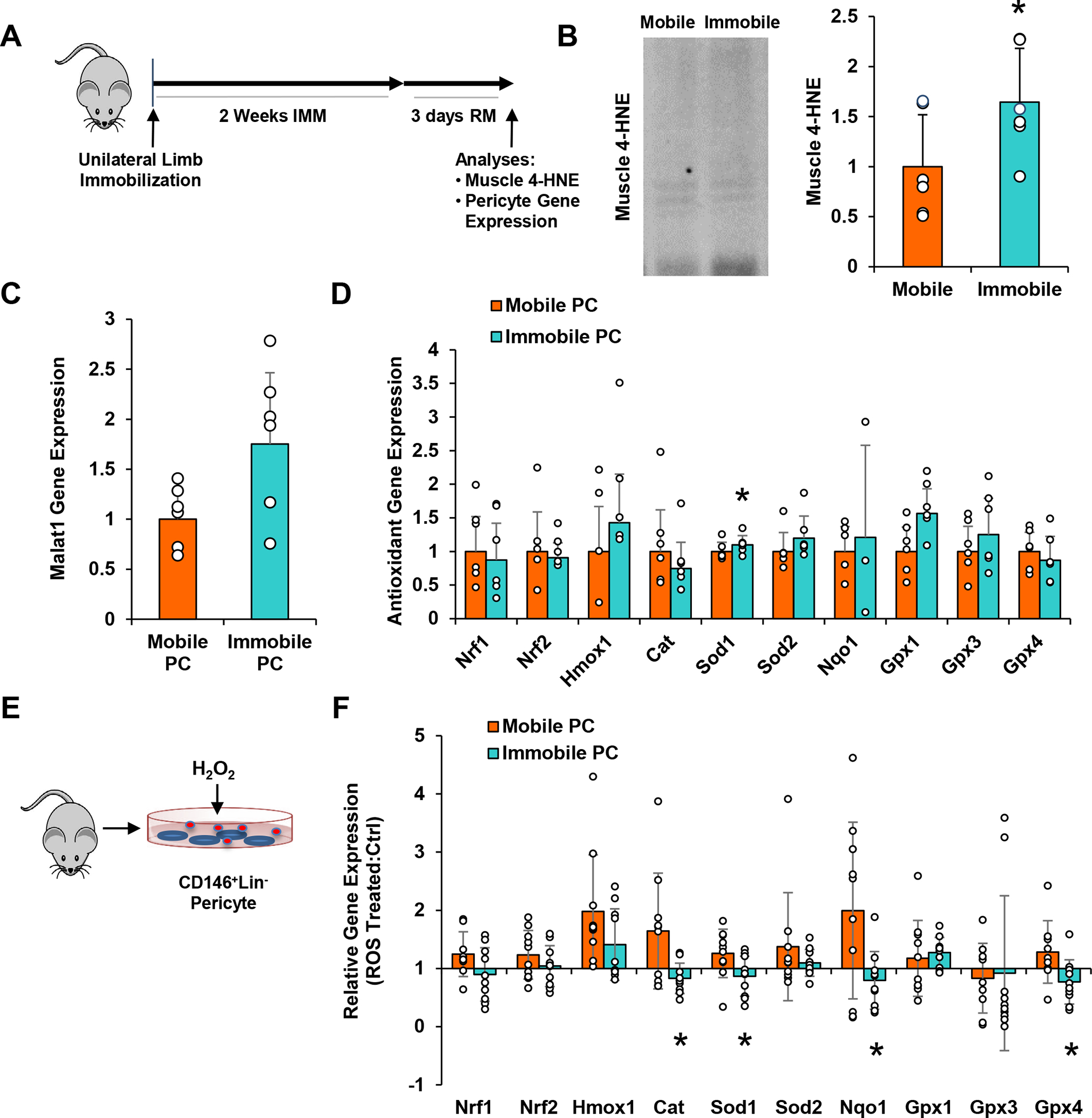
(a) Schematic diagram of experimental design for in vivo pericyte gene analysis. Three days after remobilization, (b) muscle was evaluated for reactive oxygen species based on 4-hydroxynonenal (4-HNE), and pericytes were isolated and qPCR analysis was performed to assess (c) Malat1 and (d) antioxidant defense system-associated gene expression. n=6/group. (e) Schematic diagram of experimental design for in vitro pericyte gene analysis. Pericytes were isolated from mobile and immobilized TA muscles, exposed to H2O2 for 3 hours, and then qPCR analysis was performed to assess (f) antioxidant defense system-associated gene expression. Data are expressed as a ratio of ROS-treated/non-treated groups. Values of non-treated groups were set at 1. n=10/group. All values expressed as mean ± SEM. *p<0.05 vs. Mobile.
To compare the capacity for healthy versus immobilized pericytes to mount an antioxidant response to ROS, pericytes were isolated from Mobile and Immobile TA muscles, expanded in vitro for 10 days, then exposed to H2O2 (Fig. 2e). Mobile pericytes demonstrated high capacity to respond to H2O2 stimulation compared to non-stimulated mobile pericytes; however, significantly less antioxidant gene expression was observed for Immobile pericytes compared to Mobile pericytes, including Cat (p=0.031), Sod1 (p=0.034), Nqo1 (p=0.037), and Gpx4 (p=0.027) (Fig. 2f). A trend was also observed for Nrf1 (p=0.093).
Immobilized Pericytes are Defective in Ability to Ameliorate Oxidative Stress and Suppress MPB
Prolonged oxidative stress is a barrier to muscle recovery following immobilization (Kang & Ji, 2013). Based on the results in Figure 2, suggesting that pericytes are deficient in the ability to mount an antioxidant response to H2O2, Mobile and Immobile pericytes were compared for their ability to reduce oxidative stress in muscle after disuse. Pericytes were transplanted into TA muscles immediately after immobilization, and oxidative stress was analyzed 3 days later (Fig. 3a). Mobile pericyte treatment decreased muscle 4-HNE protein compared to PBS treatment (p=0.012), but Immobile pericyte treatment did not (Fig. 3b). Similarly, Mobile pericyte treatment decreased 8-hydroxydeoxyguanosine-positive (8-OHdG+) nuclei to a greater extent than PBS treatment (p=0.013), whereas Immobile pericyte treatment did not (Fig. 3c). 8-OHdG+ nuclei quantity was significantly decreased with Mobile pericyte treatment compared to Immobile pericyte treatment (p=0.040) (Fig. 3c).
Figure 3. Pericytes Isolated from Muscle After Disuse Fail to Ameliorate Oxidative Stress or Suppress Muscle Protein Breakdown during Recovery.
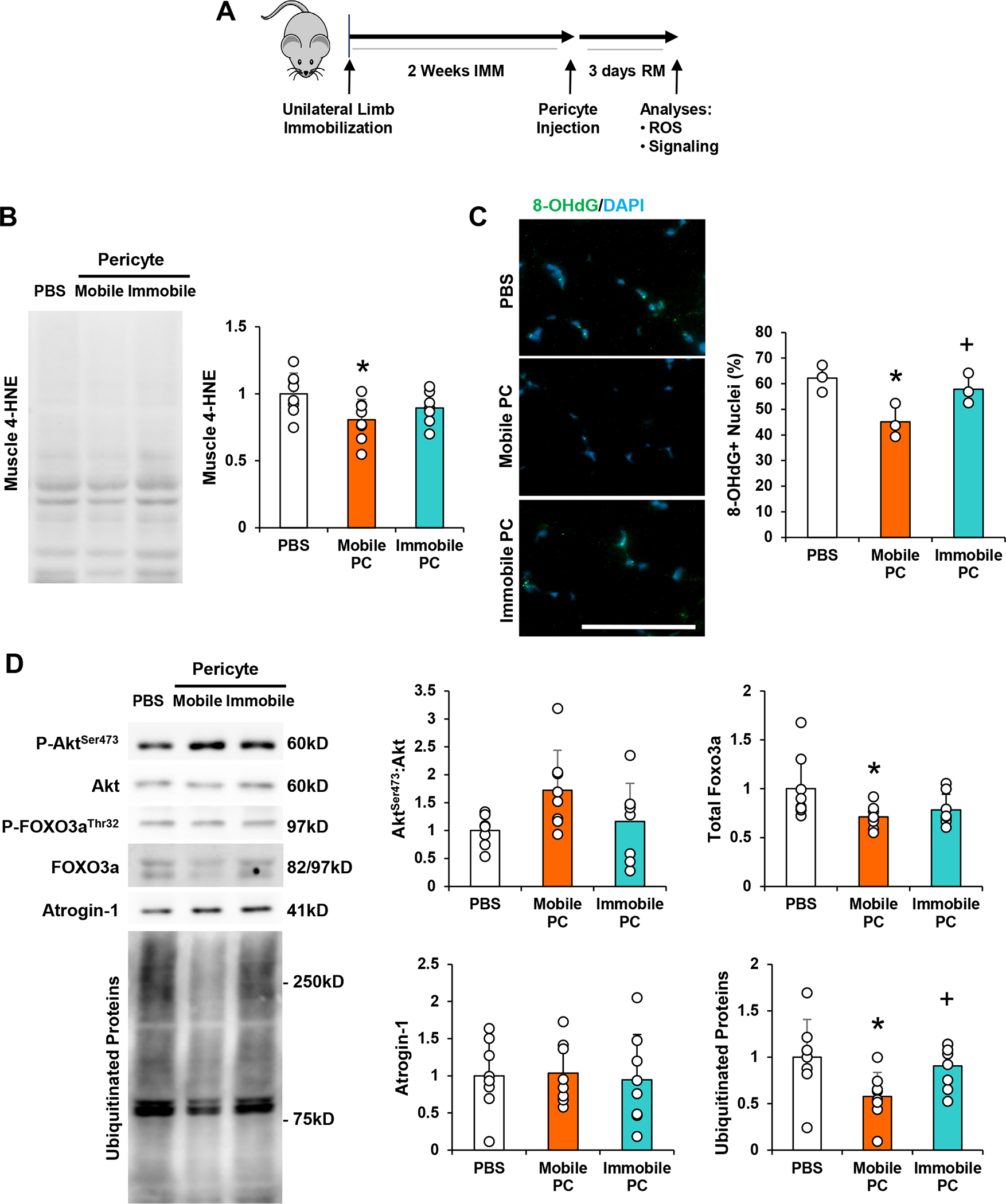
(a) Schematic diagram of experimental design for pericyte injection. Young adult (4 month) mice were subjected to unilateral limb stapling for 2 weeks. Immediately after removal of the staple, pericytes isolated from mobile limbs of donor mice (Mobile Pericyte/PC) or derived from stapled and immobilized limbs of donor mice (Immobile Pericyte/PC) were injected into the TA muscle. A group of mice also received PBS as a control. TA muscles were then dissected and evaluated from indices of recovery after 3 days of remobilization. (b) Assessment of 4-hydroxynonenal (4-HNE), in skeletal muscle after 3 days of remobilization with PBS, Mobile PC, and Immobile PC treatment. (c) Assessment of 8-Oxo-2’-deoxyguanosine (8-OHdG), a marker of oxidative stress, in skeletal muscle after 3 days of remobilization with PBS, Mobile PC, and Immobile PC treatment. (d) Assessment of AktSer473:Akt, and FOXO3a, Atrogin-1, and ubiquitinated protein in skeletal muscle after 3 days of remobilization. n=8/group. All values expressed as mean ± SEM. *p<0.05 vs. PBS; +p<0.05 vs. Mobile Pericyte. Scale bar: 50μm.
HO-1 can reduce oxidative stress and increase the phosphorylation of Akt (Pachori et al., 2007). One of the downstream substrates of Akt is the forkhead box O transcripton factor 3a (FOXO3a), an inducer of atrophy-related genes, which upon phosphorylation is sequestered in the cytoplasm for degradation (Sandri et al., 2004). In the current study, a trend toward a difference in AktSer473:Akt was observed between treatment groups (one-way ANOVA, p=0.072) (Fig. 3d). Direct comparison of the mobile pericyte group with PBS revealed a significant increase in AktSer473:Akt (unpaired t-test, p=0.027). Mobile pericytes did not impact FOXO3aThr32 or FOXO3aThr32:FOXO3a, but total FOXO3a protein was significantly reduced compared to PBS (p=0.017). No significant change in Atrogin-1 protein was detected among the three treatment groups (one-way ANOVA, p=0.938) (Fig. 3d). However, Immobile pericytes significantly decreased the amount of ubiquitinated proteins compared to both PBS (p=0.013) and Immobile pericytes (p=0.046) (Fig. 3d). Overall, these results suggest that immobilization reduces the capacity for endogenous pericytes to secrete antioxidants and defend against oxidative stress during reload, and transplantation of healthy donor pericytes can effectively restore redox balance and prevent skeletal muscle degradation during recovery.
Transplantation of H2O2-Primed Pericyte-Derived EVs Improves the Recovery of Young Adult and Aged Muscles Following Disuse
We previously demonstrated that donor pericyte transplantation can effectively improve muscle recovery following immobilization in young adult mice (Munroe et al., 2019). Given the results obtained in Figure 3 and the fact that MSCs secrete small EVs (sEVs) that encapsulate antioxidants (Eirin et al., 2018; Baek et al., 2019; Wiklander et al., 2019; Yao et al., 2019), we hypothesized that pericyte-derived sEVs may represent a novel strategy to reduce oxidative stress and improve recovery after disuse. Thus, CD146+Lin− pericytes were evaluated for the capacity to secrete sEVs that may provide an alternative cell-free approach to recover muscle mass post-disuse. Muscle-derived pericytes were conditioned in EV-depleted growth media for 48 hours, then EVs were isolated from conditioned media via SEC. Particle concentration, protein concentration, and EV marker expression of SEC fractions were analyzed (Fig. 4a). Small EVs (average size = 126.8 ± 0.5 nm) were present in Fractions 5 and 6, and slightly in 4, as verified by tetraspanin EV markers CD9, CD63, and CD81, and these fractions possessed low amounts of protein (Fig. 4b). In contrast, Fractions 7–10 were enriched with soluble proteins that did not express the tetraspanins (Fig. 4b). sEVs in Fractions 4–6 were obtained from pericyte media after 48 hours of DMEM (Ctrl sEV) or H2O2 stimulation (Primed sEV), were then intramuscularly injected into donor mice (young adult and aged) after 14 days of immobilization (Fig. 4c). Primed sEVs significantly decreased muscle 4-HNE protein 3 days after remobilization in young adult muscle compared to Ctrl sEVs (p=0.031) or PBS controls (p=0.006) (Fig. 4d). Variable results were observed in aged muscle, particularly in the PBS treatment group.
Figure 4. Transplantation of H2O2-Primed Pericyte-Derived EVs Improves the Recovery of Young Adult and Aged Muscles Following Disuse.
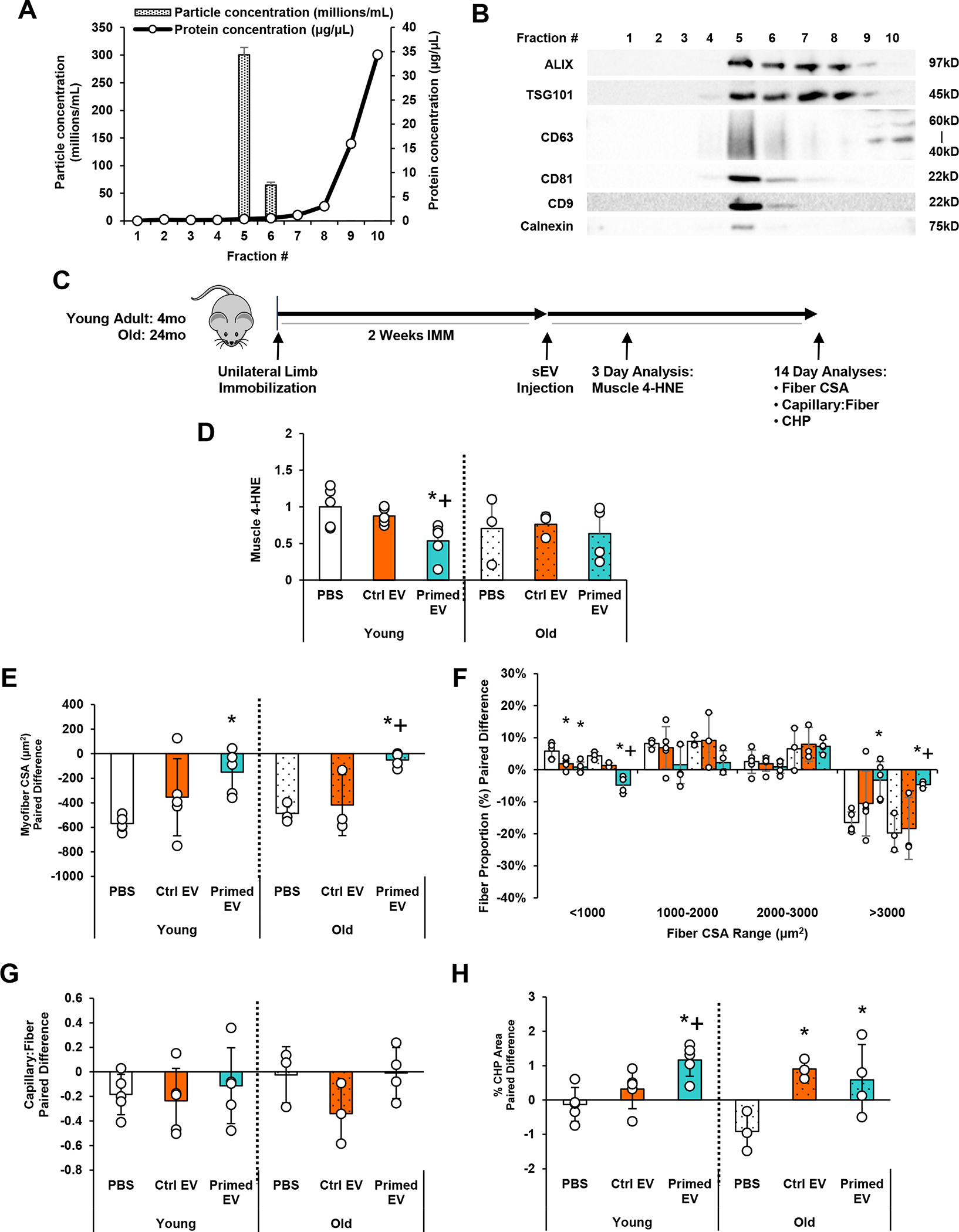
(a) Small EVs (sEVs) were isolated from pericyte conditioned media using SEC and verified in Fractions 4–6 using nanoparticle tracking analysis (NTA) (size and concentration) and bicinchoninic acid (BCA) assay (protein content) and (b) Western blotting (EV markers). (c) Schematic diagram of experimental design for sEV transplantation. Young adult (4 month) and aged (24 month) mice were subjected to unilateral limb stapling for 2 weeks. Immediately after removal of the staple, sEVs isolated from unprimed pericytes (Ctrl EV) or H2O2-primed pericytes (Primed EV) were injected into the TA muscle. A group of mice also received PBS as a control. (d) TA muscles were then dissected and evaluated for 4-HNE after 3 days of remobilization. A group of mice also received PBS as a control. (e) Separate TA muscles were evaluated for indices of recovery after 2 weeks of remobilization. Differences between mobile and immobilized limbs are reported for (e) mean myofiber CSA, (f) myofiber size distribution, (g) capillarization, and (h) collagen turnover as assessed by the collagen hybridizing peptide (CHP) assay. n=3–5/group. All values expressed as mean ± SEM. *p<0.05 vs. PBS; +p<0.05 vs. Ctrl EV.
Primed sEVs (Fractions 4–6) were then intramuscularly injected into donor mice (young adult and aged) after 2 weeks of immobilization to examine tissue recovery 2 weeks later (Fig. 4a). Consistent with prior results (Munroe et al., 2019), no significant changes were detected in TA muscle weight with treatment in both young adult and old mice. However, primed sEVs significantly restored myofiber CSA in both young adult (p=0.009) and old (p=0.006) mice compared to PBS groups (Fig. 4e). The degree of loss and restoration was similar in young adult and old likely due to the severe deficits induced by staple-based immobilization which fixes the TA in full dorsiflexion. Primed sEVs specifically decreased the quantity of myofibers below 1000 μm2 (young adult: p=0.002; old: p<0.001) and increased those over 3000 μm2 in both age groups (young adult: p=0.011; old: p=0.014) (Fig. 4f). Ctrl sEVs similarly decreased the quantity of myofibers below 1000 μm2 in young adult (p=0.007) but not old (p=0.093) mice. Interestingly, primed sEVs improved myofiber CSA compared to the Ctrl sEV group only in aged mice (mean myofiber CSA, p=0.004; quantity of myofibers over 3000 μm2, p=0.021) (Fig. 4f).
Deficits in the capillary:fiber ratio and collagen degradation were observed in young adult (capillarization) and aged (collagen degradation) mice after disuse. sEV injection did not significantly impact TA muscle capillarization in both young adult and old mice (one-way ANOVA, p=0.746 and 0.186, respectively) (Fig. 4g). However, primed sEVs significantly increased collagen remodeling in young adult mice compared to PBS and control sEVs (p=0.002; p=0.022), and both primed and control sEVs increased collagen remodeling in old mice (p=0.036; p=0.022) compared to PBS groups (Fig. 4h).
Proteomic Analysis of sEVs
Proteomic analysis of sEVs isolated from H2O2-primed and unprimed pericytes (Fractions 4–6) was then performed to reveal content that may facilitate muscle recovery. Across all samples, there were 802 unique proteins identified by the untargeted LC-MS analysis (Table 3 in 10.6084/m9.figshare.16778053). Among the proteins identified in this study, 23 were classified by the GO term ‘extracellular exosome,’ which included CD9, CD81, and CD63, verifying the presence of EVs in the samples. To assess overlap with previously identified EV proteins, the proteins in this study were compared with the EV protein repository ExoCarta. To perform this comparison, the protein accession numbers obtained from the LC-MS analysis were converted to gene IDs to facilitate comparison between the lists of proteins in this study and ExoCarta. This conversion resulted in 827 proteins, as multiple gene IDs corresponded to the same UniProt accession number. Of the proteins in the current study, there were 337 proteins shared with ExoCarta, indicating an overlap of 41% between the proteins in this dataset with ExoCarta (Fig. 5a). 490 proteins were identified in the current dataset that were not present in the ExoCarta set, corresponding to 59% of the proteins.
Figure 5. Proteomic Analysis of H2O2-Primed Pericyte-Derived EVs.
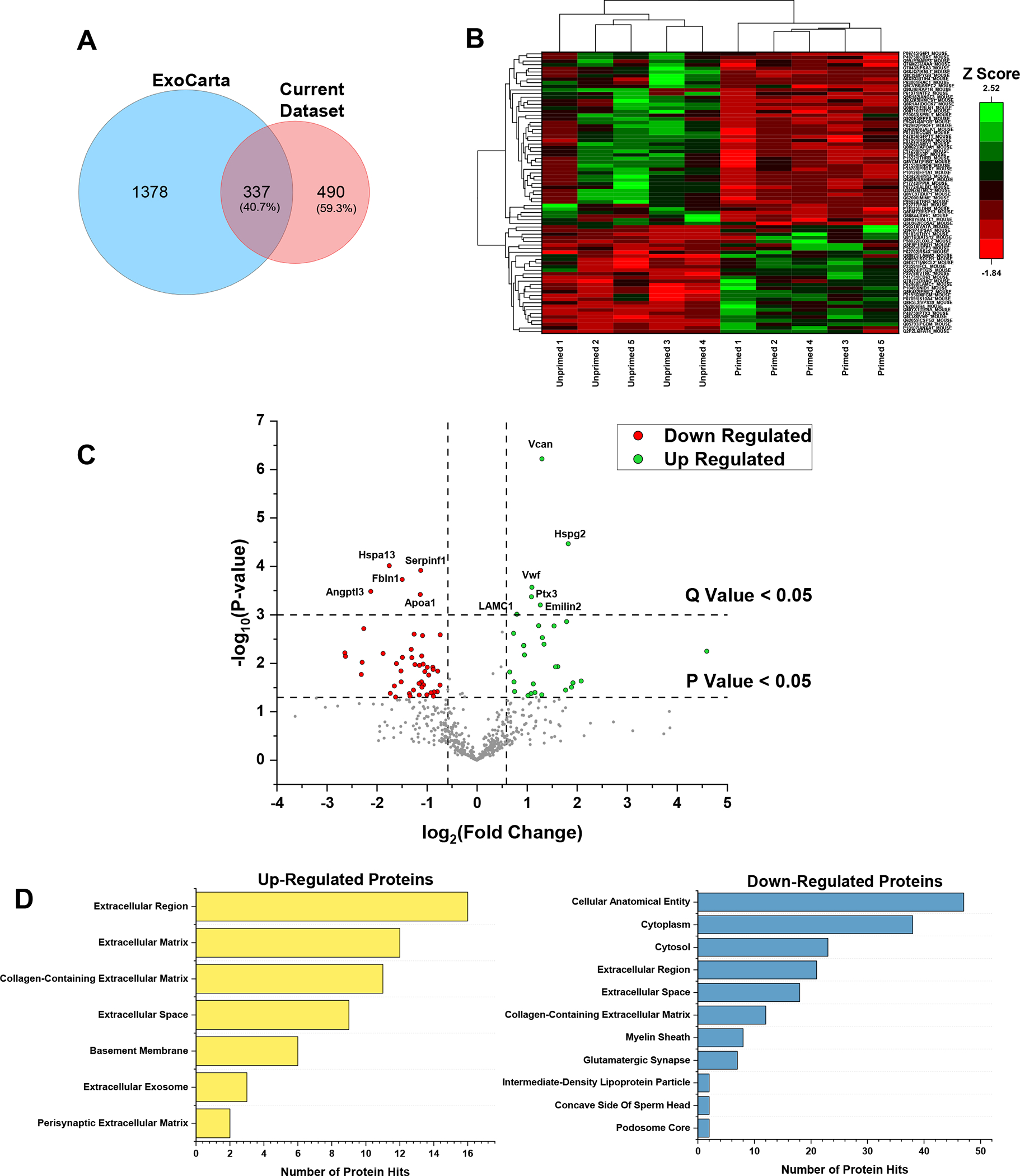
(a) Venn diagram showing the unique number of proteins in ExoCarta and in the current dataset. (b) Heat map representing the abundance in terms of Z scores for the up- and down-regulated proteins in H2O2-primed pericyte-derived EVs (Primed EV) relative to the unprimed pericyte-derived EVs (Ctrl EV). (c) Volcano plot showing the up-regulated (green) and down-regulated (red) proteins based on having a Q value less than 0.05. (d) GO analysis for the up- and down-regulated proteins depicting the enriched GO terms in the cellular component category. n=5/group.
For label-free quantitation, 531 unique proteins that passed the PEAKS Q filters, as described in the Methods section, were quantified by averaging the peak areas for peptides with the top 3 highest peak areas for each protein as previously described by Ning et al. (Ning et al., 2012) (Table 3 in 10.6084/m9.figshare.16778053). Student t-tests were performed to compare protein abundance between H2O2-primed and unprimed sEVs, and 78 proteins had both a P value less than 0.05 and a fold-change greater than 1.5 or less than −1.5, and these were considered up- or down-regulated relative to the unprimed samples, respectively (Table 3 in 10.6084/m9.figshare.16778053). Hierarchical cluster analysis was applied between the up- and down-regulated proteins to visualize protein abundance differences between both sample groups. Z-scores calculated row-wise were used to represent protein abundances based on the averaged top 3 peptide peak areas for each sample (Fig. 5b). A Benjamini-Hochberg FDR correction was performed on the quantified proteins to minimize the number of falsely significant proteins. After performing the FDR correction, 11 proteins were identified with a Q value less than 0.05, and these proteins were considered significantly different (Fig. 5c). Finally, GO analysis was performed to assess which cellular components were most highly represented. Upon analysis of the different categories for the up-regulated proteins, it is interesting to note that in the cellular component category, the ‘extracellular exosome’ term is among the enriched GO terms, in which the proteins Annexin A1, syntenin-1, and CD63 were mapped (Fig. 5d). In this same category, the most enriched GO term was ‘Extracellular Region’ to which various extracellular matrix proteins were found including laminin subunit beta-1, laminin subunit gamma-1, and versican core protein, as well as various basement membrane associated proteins such as tenascin, nidogen-1 and lysyl oxidase homolog 2. For the down-regulated proteins, the most enriched GO term in the cellular component category was ‘cellular anatomical entity’ which included various proteins involved in glycolysis including glyceraldehyde-3-phosphate dehydrogenase, glucose-6-phosphate isomerase and beta-enolase, and proteins involved in metabolite synthesis and catabolism including 4-hydroxyphenylpyruvate dioxygenase, L-lactate dehydrogenase B chain, and farnesyl pyrophosphate synthase. These results demonstrate that the majority of up-regulated proteins are involved in extracellular matrix processes, while most of the down-regulated proteins play a role in glucose metabolism and metabolite interconversion.
Discussion
This study outlines the development of a pericyte-based, cell-free strategy to recover muscle mass after disuse in aged mice. The significant increase in Malat1 gene expression in pericytes provided a clue that pericytes may be deficient in their capacity to mount an antioxidant response to ROS after a period of unloading, which was confirmed. The extent to which pericyte-mediated release of antioxidants is essential for recovery of muscle mass after disuse is not known, but here we demonstrate that EVs retrieved from H2O2-primed healthy pericytes are able to successfully recover collagen remodeling and myofiber size in both young adult and aged mice after immobilization. Thus, pericyte-derived EVs may provide a cell-free approach to revitalize aged skeletal muscle after atrophy.
Increased oxidative stress in muscle after disuse is the result of an imbalance between ROS production and antioxidant defense (Lawler et al., 2003; Kang & Ji, 2013). Mitochondria residing in myofibers, particularly in the subsarcolemma region, have been regarded as a main source of ROS production after muscle unloading (Krieger et al., 1980; Min et al., 2011). In the current study, scRNA-Seq data revealed that mitochondria-associated gene expression was significantly decreased following muscle disuse in several cell clusters, including pericytes, endothelial cells, MuSCs, FAPs, and smooth muscle cells. Prior studies have demonstrated that mitochondrial gene expression is compromised, and cellular ROS production is enhanced, in aged muscle stem cells (Bortoli et al., 2003; Fulle et al., 2005). However, this study is the first to suggest that nearly all muscle-resident mononuclear cells, particularly pericytes, are potential sources of ROS production during muscle unloading and reloading.
Cells naturally mount an adaptive response to ROS via initiation of Nrf2 binding to the antioxidant response element (ARE) in the upstream promoter region of many antioxidant and cytoprotective genes, including Sod1/2, Cat, Gpx, Hmox1, Nqo1 and glutathione S-transferases (Gst) (Nguyen et al., 2003; Valle-Prieto & Conget, 2010; Raghunath et al., 2018; Laporte et al., 2019). Whereas SODs catalyze superoxides to hydrogen peroxide, CAT and GPX reduce hydrogen peroxide to oxygen and water. Additionally, Nqo1 can catalyze the reduction and detoxification of quinones that cause oxidative stress, and HO-1 catalyzes the breakdown of heme into the antioxidant biliverdin (Stocker et al., 1987). Here we demonstrate that an acute H2O2 stimulus can induce antioxidant gene expression in healthy pericytes in vitro, similar to MSCs reported elsewhere (Gorbunov et al., 2013; Park et al., 2016; Stavely & Nurgali, 2020), and that transplantation of healthy pericytes can mitigate oxidative stress, increase Akt phosphorylation and reduce ubiquitination in adult mice 3 days after remobilization. Previous studies have reported similar increases in Hmox1 and other antioxidants within one week of disuse that are no longer present with prolonged unloading (Hunter et al., 2001; Matsushima et al., 2006; Agostini et al., 2010; Reich et al., 2010). The extent to which long-term disuse can desensitize cells to ROS stress and suppress antioxidant gene expression in muscle-resident perivascular stem/stromal cells has not been reported.
Chen et al. (Chen et al., 2018) recently demonstrated that Malat1 binds to Nrf2 and serves as a repressor of antioxidant gene expression, including Cat and Nqo1. Interestingly, Malat1 gene expression was enhanced in pericytes in our scRNA-Seq dataset in response to two weeks of hindlimb immobilization and continued to be elevated three days after remobilization. We confirmed that pericytes isolated from immobilized limbs were deficient in their ability to increase antioxidant gene expression (Cat, Sod1, Nqo1, Gpx4) in response to H2O2 in culture, and further demonstrated that these cells were incapable of suppressing oxidative stress and ubiquitination upon transplantation. The extent to which Malat1 serves as a direct regulator of Nrf2 binding and antioxidant gene expression in MSCs and pericytes remains to be determined. Regardless, the loss of antioxidant gene expression in pericytes may underlie the prolonged lack of recovery in muscle mass commonly observed for several days to weeks after disuse.
We previously demonstrated that donor pericyte transplantation can effectively improve muscle recovery following immobilization in young adult mice (Munroe et al., 2019). Given the results obtained in the current study and the fact that MSCs secrete sEVs that encapsulate antioxidants (Eirin et al., 2018; Baek et al., 2019; Wiklander et al., 2019; Yao et al., 2019), we hypothesized that pericyte-derived sEVs may represent a novel strategy to reduce oxidative stress and improve recovery after disuse. sEVs secreted from pericytes after H2O2 exposure in vitro effectively reduced muscle oxidative stress in young adult mice 3 days after transplantation and recovered myofiber size in both young adult and aged mice two weeks after disuse. Direct targets of Nrf2 were not differentially expressed in primed sEVs compared to unprimed, perhaps due to low expression or the inability to detect with our method. Pentraxin-3 (PTX3), a long chain member of the pentraxin family, was upregulated in primed sEVs along with several ECM proteins, including versican, basement membrane-specific heparan sulfate proteoglycan core protein (HSPG; perlecan), and laminins (α2, γ1). PTX3 and laminins are synthesized by multiple cell types in response to inflammatory stimuli, including pericytes (Dvoretskiy et al., 2019), and play an important role in tissue repair following injury (Ji & Tsirka, 2012; Zhu et al., 2018; Doni et al., 2021). In addition, HSPG has been found to be upregulated in sEVs under inflammatory and hypoxic conditions (Wang et al., 2017) and its cell surface localization on sEVs may participate in either cell targeting and fusion events or remodeling of the microenvironment when associated with matrix degrading enzymes (Cerezo-Magana et al., 2020). Thus, PTX3, laminins and HSPG may have facilitated some of the beneficial effects of primed sEVs on muscle recovery. On the other hand, heat shock 70kDa protein 13 (HSPA13), which can participate in protein folding and protect against oxidative stress (Reeg et al., 2016), was found to be significantly decreased in H2O2-primed sEVs. One possible explanation is that oxidative stress may have elicited increased need for HSPA13 in pericytes, resulting in less packaging and release. Overall, these proteomic results provide important insight, yet further studies are required to elucidate precise mechanisms by which H2O2-primed sEVs restore muscle mass in young adult and aged mice.
A discrepancy was noted between our prior study (Munroe et al., 2019) and the ability for pericyte-derived sEVs to promote capillarization after disuse. Specifically, no recovery or promotion of capillary content was noted with sEV injection in the current study. The presence of endothelial cell gene expression in one of our two pericyte clusters (cluster 15) suggests that pericytes may directly differentiate into endothelial cells (Hayes et al., 2018), and that EV transplantation alone is insufficient to confer this benefit. However, pericytes in cluster 15 may also be contaminated with endothelial cells due to the inability for pericytes to detach from endothelial cells during digestion (Vanlandewijck et al., 2018). Pericyte lineage tracing or fate mapping experiments will be required to address this important question. The fact that pericyte-derived EVs can recover muscle mass without improving capillarization in aged muscle suggests that vascular improvements are not required for pericyte-mediated benefits observed here and in our prior study (Munroe et al., 2019).
In conclusion, this study is the first to demonstrate that muscle-resident pericytes are deficient in the ability to produce antioxidants during and after a period of disuse. To overcome this deficiency, H2O2-primed pericyte-derived sEVs were injected into mice immediately following limb immobilization to demonstrate superior capacity to reduce oxidative stress, restore ECM remodeling and/or fiber size during recovery in adult and aged mice. The precise molecular cargo responsible for sEV-mediated revitalization of muscle growth after disuse is not yet known, but likely includes factors that enhance membrane stability and indirectly reduce oxidative stress. The success of this strategy outlined here may represent a viable approach to maintaining muscle mass and improving health span in older adults.
Materials and Methods
Ethical Approval
Protocols for animal use were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Illinois at Urbana-Champaign (#19120, 19122), and National Institutes of Health guidelines for the care and use of laboratory animals were strictly followed. The methods used in this study and the manner in which the results are reported are in compliance with the principles and standards outlined by The Journal of Physiology (Grundy, 2015).
Animals
Young adult (4-month-old) and old (24-month-old), male C57BL/6J mice were ordered from Charles River Laboratories (Wilmington, MA) and the National Institute on Aging (NIA), respectively. Mice were housed at the Beckman Institute for Advance Science and Technology at the University of Illinois at Urbana-Champaign, IL, USA. Mice were kept under a 12 hr light/dark cycle (lights on 07:00 to 19:00 h) in a pathogen free, temperature-controlled facility and given ad libitum access to food, standard laboratory chow, and water. Mouse strain and age were selected to reproduce results from Munroe et al. (Munroe et al., 2019).
Hindlimb Immobilization
Mice were subjected to unilateral hindlimb immobilization for 2 weeks as previously described (Munroe et al., 2019). Mice were weighed, anesthetized with isoflurane (~2.5%), and subcutaneously injected with buprenorphine SR (1.0 mg/kg mouse; ZooPharm Pharmacy, Laramie, WY) prior to immobilization. The right hindlimb was shaved, then immobilized using Covidien Royal AutoSuture 35W skin stapler (eSutures, Mokena, IL, USA). One tine of the staple inserted into the plantar of the foot, and the other tine inserted into the distal gastrocnemius. This restrained the foot in full dorsiflexion and immobilized the tibialis anterior (TA) muscle at a shortened position. Surgical sites were monitored daily for bleeding or injury for the following five days. On day 14 of immobilization, mice were anesthetized with isoflurane, and the surgical staple was removed. For pericyte or sEV transplantation experiments, the TA muscle of the immobilized limb then received an injection of either 1X PBS as control (30 μL), pericytes (5×104 pericytes/30 μL), or pericyte-derived EVs (5×108 vesicles/30 μL) via 29g needle (n=8–9 per group for pericytes; n=3–5 per group for sEVs). Mice resumed normal movement in cages and were sacrificed at 3 days or 2 weeks post-remobilization via carbon dioxide asphyxiation, which was confirmed by cervical dislocation. The TA muscles from mobile (“Mobile”; control) and previously immobilized (“Immobile”) limbs were excised and preserved in precooled 2-methyl-butane and stored at −80°C until analysis. TA muscles were also removed immediately after immobilization for scRNA-Seq analysis (no pericyte or EV injection) and 3 days after remobilization for evaluation of pericyte antioxidant gene expression (no pericyte or sEV injection).
Single-Cell RNA Sequencing
TA muscles were excised from both limbs of six mice (Mobile, n=6; and Immobile, n=6) immediately after two weeks of unilateral hindlimb immobilization. Mobile and immobile TA muscles were separately pooled and washed in cold 1X PBS with penicillin-streptomycin antibiotic (P/S) (Thermo Fisher Scientific, Grand Island, NY, USA). The cleaned muscles were minced and then enzymatically digested for ~60 minutes at 37°C. The enzyme solution contained 250 U/mL collagenase Type 2 (Worthington-Biochemical Corp., Lakewood, NJ, USA), 2.4 U/mL neutral protease (Dispase) (Worthington-Biochemical Corp.), 60 U/mL DNase I (Sigma-Aldrich, St. Louis, MO, USA), and 2.5 mM CaCl2 in 1X PBS. Enzyme digestion was inhibited with inhibition media containing DMEM (Corning; #10013CV), 20% FBS, 1 mM EDTA (UltraPure 0.5M EDTA, pH 8.0; Thermo Fisher Scientific; #15575–038) and P/S. The tissue solution was filtered (70 μm and 40 μm) to obtain mononuclear cell suspension, and the filtrate was centrifuged at 300 RCF for 5 minutes. Supernatant was decanted, and cell pellet was resuspended in PBS with 5% FBS.
Two single-cell libraries were constructed with the Chromium Single-Cell Gene Expression Kit Version 3 and sequenced on two 2×150nt SP lanes in a NovaSeq 6000. Fastq.gz files were generated and demultiplexed with 10X Genomic’s Cell Ranger 3.0.2. Each sample’s R1 and R2 fastq files were run through cellranger count using a custom reference made from Gencode M21 gene set and --expect-cells=5000 to call cells and de-duplicate individual reads to unique UMI counts per gene. The cells had not been washed prior to library construction so the ambient RNA in the supernatant lead to Cell Ranger calling 11,055 and 11,676 cells in Mobile and Immobile samples, respectively, even though only 5,000 cells had been targeted. The UMI counts were then read into R (v 3.6.3) using Seurat (v. 3.1.4)(Stuart et al., 2019). Genes were filtered out if they did not have at least 1 UMI in at least 10 cells combined between the two samples. Further cell filtering was performed first based on percentage of UMIs that mapped to mitochondrial (MT) genes. Both samples showed extreme bi-model distributions with ~55% of cells showing > 50% MT UMIs. These cells also had low total UMI counts and low total number of genes detected and were likely not true cells but ambient RNA that had been sequenced. Cells with ≥ 50% MT were filtered out and then additional QC was performed based on the number of UMIs and genes detected. Cells that had > 4 median absolute deviations of the total UMI counts were excluded as possible doublets. There were also a sub-group of cells with many fewer genes detected than the rest. These were determined to be red blood cells and so cells were filtered out if they had fewer than 250 genes detected. After gene and cell filtering, 17,927 genes were detected in 4,224 cells from the mobile limb and 5,404 cells from the immobile limb.
Cluster analysis was performed with the Seurat package. First, normalization was performed using the SCTransform method (Hafemeister & Satija, 2019; Stuart et al., 2019). Then linear dimensional reduction was done using Principal Components Analysis and the first 40 PCs were used as input to find clusters using a k-nearest neighbor graph and also for non-linear dimensional reduction using tSNE. Cell type identification was completed using candidate marker genes curated from the literature and the top unique genes for each cluster. Within each cluster, differential expression testing was evaluated between Mobile and Immobile cells using the non-parametric Wilcoxon Rank Sum test.
Pericyte Isolation from Skeletal Muscle via Fluorescent-Activated Cell Sorting (FACS)
To obtain primary CD146+Lin− (Lin- = lineage negative = CD31−CD45−) pericytes, muscles were washed in cold 1X PBS with penicillin-streptomycin antibiotic (P/S). The cleaned muscles were minced, enzymatically digested for ~60 minutes at 37°C prior to addition of inhibition media, as described in the scRNA-Seq section above. The tissue solution was filtered (70 μm and 40 μm) to obtain mononuclear cell suspension, and the filtrate was centrifuged at 300 RCF for 5 minutes. Supernatant was decanted, and cell pellet was resuspended in 2% FBS solution. Cells were incubated with anti-mouse CD16/CD32 (Thermo Fisher Scientific; #14–0161-81) for 10 minutes at 4°C to block Fc-mediated nonspecific binding, then were washed, centrifuged at 300 RCF for 5 minutes, supernatant removed, and cells resuspended with 2% FBS. A small portion of cells was aliquoted for unstained and single-stained controls, and all other cells were incubated with an antibody cocktail designed for FACS.
Mononuclear cells were incubated for 60 minutes at 4°C with combinations of the following antibodies diluted in 2% FBS in 1X PBS: conjugated anti-mouse CD146 (melanoma cell adhesion molecule)-phyocerythrin (PE) (Clone ME-9F1) (1:50; BioLegend, San Diego, CA, USA; #134704), CD31 (PECAM-1)-allophycoctanin (APC) (Clone MEC 13.3) (1:100; BD Biosciences; #561814), and CD45-APC (Clone 30-F11) (1:100; BD Biosciences; #561018). Following antibody incubation, cells were washed, centrifuged at 300 RCF for 5 minutes at 4°C, supernatant removed, cells resuspended with 2% FBS, filtered through a 40 μm filter, and placed on ice.
CD146+Lin−, skeletal muscle pericytes were sorted using a BD FACS Aria II sorter by the Flow Cytometry Center at the UIUC Roy J. Carver Biotechnology Center (Urbana, IL, USA). Unstained and single stained control samples were analyzed to account for spectral overlap. For direct RNA or protein extraction, isolated cells were collected in lysis buffers. To obtain pericytes for injection into muscle, isolated cells were collected in growth media (10% FBS, 1% P/S in DMEM), centrifuged at 300 RCF for 5 minutes, supernatant removed, and cells resuspended in growth media. Cells were cultured in 60 mm dishes and allowed 10 days to expand prior to transplantation. The media was changed every 3–4 days.
H2O2 Stimulation of Pericyte Gene Expression In Vitro
CD146+Lin− primary pericytes were obtained from Mobile and Immobile TA muscles via FACS and were plated onto 96-well plates (approximately 5000 cells/well). Cells from each muscle were divided equally into two groups: Control (non-stimulated) and ROS-Stimulated, resulting in a total of 4 groups: Mobile (Control) Pericytes, Mobile ROS-Stimulated Pericytes, Immobile Pericytes, and Immobile ROS-Stimulated Pericytes (n=10 per group). Cells expanded in growth media (10% FBS, 1% P/S in DMEM) for 10 days prior to ROS stimulation. Cells were washed with 1X PBS twice, and then stimulated with hydrogen peroxide (H2O2) (50 μM) (Thermo Fisher Scientific) in growth media or growth media alone. Three hours later, cells were lysed and RNA was collected for qPCR analysis.
ROS Stimulation of Pericytes and Collection of EVs In Vitro
CD146+Lin− primary pericytes were cultured for 10 days to reach approximately 90% confluency, washed with 1X PBS twice, then stimulated with H2O2 (50 μM) in EV-depleted growth media (DMEM containing 10% EV-depleted FBS [#A2720803, Gibco] and 1X P/S) for 48 hours. Priming was reinforced at 24 hours by adding 20% more volume of corresponding media to the pericyte culture. At 48 hours post-priming, conditioned media was harvested, centrifuged at 5,000g for 10 minutes and filtered (0.22 μm) to remove cells and debris, then collected into ultrafilter tubes (Amicon® Ultra-15 Centrifugal Filter Unit – 30 kDa cutoff; Millipore; UFC903024) and centrifuged for 20 minutes at 4,000g. sEVs were isolated from the concentrate using size exclusion chromatography (SEC). SEC columns (qEV single/35 nm; Izon Science) were washed with 2 mL 1X PBS prior to fractioning. Fraction collection began immediately after 150 μL of the concentrate was loaded onto the column. 150 μL of 1X PBS was loaded at a time for elution, and every 300 μL of elute was collected as a fraction until fraction 10. Fractions 1–10 were then ultrafiltered (Amicon® Ultra-2 Centrifugal Filter Unit – 30 kDa cutoff; Millipore; UFC203024) at 4,000g for 20 minutes, and the concentrate was collected for further use or analysis. Fractions 4–6 were pooled to provide an EV-rich, low-soluble protein solution for transplantation. Particle size and concentration were assessed using Nanoparticle Tracking Analysis (NanoSight NS300, Malvern Panalytical).
Quantitative Real-Time Polymerase Chain Reaction (qPCR)
Total RNA was extracted from cell lysates using RNeasy Mini Kit (Qiagen), following the manufacturer’s instruction, and DNase I digestion was performed during the procedure to remove unwanted DNA. Quantity and quality of extracted RNA was assessed in duplicate on a Take-3 application plate using Synergy H1 Hybrid Multi-Mode Microplate Reader (BioTek, Winooski, VT, USA). 25 ng RNA was used to perform reverse transcription via the High Capacity cDNA Reverse Transcription Kit (Life Technologies, Grand Island, NY, USA), following manufacturer’s instructions. Reaction solution was prepared by mixing cDNA dilution, TaqMan primers (Applied Biosystems, Thermo Fisher Scientific), and Taqman Universal PCR Master Mix (Applied Biosystems, Thermo Fisher Scientific). qPCR was performed using the QuantStudio™ 7 Flex Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific). Relative gene expression level was calculated using the ΔΔCt (threshold cycle) method. All genes were normalized to glyceraldahyde-3-phosphate dehydrogenase (Gapdh), of which the Ct values remained unchanged among all experimental conditions. Primer information and gene expression assay ID numbers are listed in Table 4 in 10.6084/m9.figshare.16778053.
Western Blotting
The proximal half of frozen TA muscles were placed directly in 400 μL of ice-cold lysis buffer containing HEPES (20 mM), Triton X-100 (1%), glycerol (10%), protease inhibitor (Complete Mini; Roche) and phosphatase inhibitor (PhosSTOP; Roche) tablets. The tissue was homogenized using a handheld homogenizer and the homogenates were rotated at 4°C for 1 hr and centrifuged at 14,000g for 5 minutes at 4°C. Pericytes and EVs were lysed in 1X RIPA (dilute from 10X RIPA; Cell Signaling Technology) with 0.1% SDS and 1 mM PMSF. Lysates were rotated at 4°C for 1 hr and centrifuged at 14,000g for 5 minutes at 4°C. Supernatants were collected and the concentration of protein was determined with the BCA assay (Thermo Fisher Scientific) using bovine serum albumin (BSA) for the standard curve. Equal amounts of protein (30 μg) were subjected to SDS-PAGE using 8% or 10% acrylamide gels and transferred to nitrocellulose membranes. Equal protein loading was verified by Ponceau S staining. Membranes were blocked in Tris-buffered saline (pH 7.8) with 0.1% Tween 20 (TBS-T) containing 5% BSA for 1 hour at room temperature, then incubated overnight with primary antibodies of phospho-AktSer473 (1:1000, Cell Signaling #9271), Akt (1:1000, Cell Signaling #9272), phospho-Foxo3aThr32 (1:1000, Cell Signaling #9464), Foxo3a (1:1000, Cell Signaling #2497), Atrogin-1 (1:1000, ECM Biosciences #AP2041) or Ubiquitin (1:1000, Cell Signaling #3933) suspended in 2% BSA in TBS-T. Horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA; Cell Signaling, Danvers, MA) were applied for 1 hour at 1:5000 dilution for the 4-hydroxynonenal (4-HNE; R&D Systems, #MAB3249-SP) and 1:2000 for the other proteins in TBS-T containing containing 2.5% non-fat dry milk (NFDM). Bands were detected using SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific) and a Bio-Rad ChemiDoc Imaging System (Bio-Rad, Hercules, CA). Quantification was completed using Quantity One software (Bio-Rad). All target bands were normalized to all bands detected by Ponceau S. For phospho/total measures, the phospho and total were run on separate membranes, and each normalized to Ponceau S prior to calculating the ratio.
Immunofluorescence
The distal half of frozen TA muscles was transferred from −80°C to the cryostat (Leica CM3050S, Wetzlar, Germany) and embedded in optimum cutting temperature (OCT) (Tissue-Tek; Thermo Fisher Scientific). Sections (10 μm) were collected onto Superfrost microscope slides (Thermo Fisher Scientific) and stored at −80°C until staining.
Slides were thawed from −80°C to room temperature. Sections were fixed in −20°C acetone for 5 minutes, then hydrated for 5 minutes with 1X PBS for three times. Sections were blocked for 1 hour at room temperature with AffiniPure Fab Fragment goat anti-mouse IgG (H+L) (55 μg/mL; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA; #115–007-003) in 1X PBS with 5% bovine serum albumin (BSA) and 0.05% Tween-20 (Fisher Bioreagents; #BP337–500).
To assess myofiber cross-sectional area (CSA) and capillarization, sections were incubated with rat anti-mouse CD31 (clone 390) (1:100; Thermo Fisher Scientific; #14–0311-85) and rabbit anti-mouse dystrophin (1:100; Abcam, Cambridge, MA, USA; ab15277) in Wash Buffer (1X PBS with 1% BSA and 0.05% Tween-20) simultaneously for 1 hour at room temperature. Following Wash Buffer, Alexa Fluor 488 goat anti-rat IgG (1:200; Thermo Fisher Scientific; #A-11006) and Alexa Fluor 633 goat anti-rabbit IgG (1:100; Thermo Fisher, #A-21070) secondary antibodies were applied simultaneously for 1 hour at room temperature to visualize dystrophin and CD31. After final washes with Wash Buffer and then 1X PBS, slides were mounted with VECTASHIELD© HardSet with DAPI (Vector Laboratories, Burlingame, CA, USA; #H-1500–10) and stored at 4°C.
To assess collagen remodeling, slides were prepared as previously described, but without Fab fragment during the 1-hour block. Sections were washed in 1X PBS. 5μM CHP solution in 1X PBS (Collagen Hybridizing Peptide, 5-FAM Conjugate [F-CHP]; 3Helix, Salt Lake City, UT, USA; #FLU300) was prepared and heated for 5 minutes at 80°C to activate the compound. Solution was immediately placed on ice for 1 minute to cool, and then added to sections quickly. Slides were incubated overnight at 4°C. After slides were washed with 1X PBS, slides were sealed with VECTASHIELD© HardSet with DAPI and stored at 4°C.
To assess oxidative stress with 8-hydroxy-2’-deoxyguanosine (8-OHdG) stain, slides were prepared as previously described, but sections were blocked for 1 hour at room temperature with AffiniPure Fab Fragment goat anti-mouse IgG (H+L) in 1X PBS with 10% bovine serum albumin (BSA) and 0.05% Tween-20. Sections were washed with Wash Buffer, and then incubated with mouse anti-8-OHdG (1:1000; Santa Cruz; #sc-66036) in 1X PBS containing 3% BSA and 0.05% Tween-20 at 4°C overnight. Following washed with Wash Buffer, Alexa Fluor 488 goat anti-mouse IgG2b (1:500; Thermo Fisher; #A-21141) in 1X PBS containing 3% BSA and 0.05% Tween-20 for 1 hour at room temperature. After final washes with Wash Buffer and then 1X PBS, slides were mounted with VECTASHIELD© HardSet with DAPI and stored at 4°C.
Imaging and Analysis
Immunofluorescence stains were visualized using Zeiss Axiovert Inverted Fluorescent Microscope with Zeiss Axiocam digital camera and Axiovision software (Zeiss, Thornwood, NY, USA). Images were acquired at 10X magnification for CSA and collagen remodeling analysis, 20X magnification for capillary, and 63X magnification for 8-OHdG analysis. Images were acquired in separate color channels to allow for independent analyses of images. To assess total myofiber CSA, 10X dystrophin images of entire muscle sections were analyzed using Semi-automatic Muscle Analysis using Segmentation of Histology (SMASH; a MATLAB application by Smith and Barton, 2014). Images were uploaded to SMASH, and fibers were automatically outlined on the red channel (dystrophin-Alexa Fluor 633). Any incomplete fiber outlines were manually completed to ensure their inclusion. Improperly selected non-myofiber areas were manually excluded. Fibers above 0.98 eccentricity or below 0.65 convexity were excluded from analysis for illegitimate fiber morphology. Area values less than 250 μm2 were also excluded.
Muscle capillarization was assessed using 20X images taken at five random fields for each muscle section. Punctate CD31-AlexaFluor 488 dots were manually counted as capillary numbers. Myofiber number and CSA were quantitated using the SMASH method as described above. Incomplete myofibers cut by the edge of images were excluded, and the capillaries that made no close contact with any complete myofiber were excluded. Capillary-to-fiber ratio was obtained by dividing capillary number by myofiber number. Capillary density was obtained by dividing capillary number by the total myofiber CSA (mm2).
Collagen remodeling was quantified with 10X CHP images using a threshold intensity program from ImageJ (NIH). The green channel was thresholded to remove background to determine the percent of CHP-stained area observed within each image. Four random 10x images were analyzed per sample.
8-OHdG-positive (8-OHdG+) nuclei was quantitated using ten 63X images taken from random fields for each muscle section. The green channel was thresholded to remove background signals with ImageJ. All DAPI-stained cell nuclei were counted, and numbers of nuclei co-localized with 8-OHdG were divided by the numbers of total nuclei to determine the percentage of 8-OHdG+ nuclei.
For all assessments, investigators were blinded to sample information until after data collection was complete.
Exosome Protein Extraction and Trypsin Digestion
Exosome samples (~100 μL each), control and H2O2-primed (n=5 per group) were lysed using the Perfect-FOCUS™ kit (G-Biosciences, St. Louis, MO) by following the manufacturer’s instructions. After the proteins were extracted, the samples were dried in a speed vac, and then reconstituted in 100 μL of 6M urea/50 mM Tris-HCl (pH 8), followed by the addition of 5 μL of 200 mM DTT/50 mM Tris-HCl (pH 8). Samples were incubated for 60 minutes at room temperature, and then 20 μL of 200 mM iodoacetamide/50 mM Tris-HCl (pH 8) was added to each sample. Samples were incubated again for 60 minutes at room temperature in the dark, and then 20 μL of 200 mM DTT/50 mM Tris-HCl (pH 8) was added to the samples. The samples were incubated with the DTT to neutralize unreacted iodoacetamide for 60 minutes at room temperature in the dark. Samples were then each diluted with 775 μL of 1 mM CaCl2/50 mM Tris-HCl (pH 7.6), and then Sequencing Grade Modified Trypsin (Promega, Madison, WI) was added to each sample at a trypsin:protein ratio of 1:20. The samples were digested for 20 hours in a 37°C water bath. After the incubation, the enzyme reaction was stopped by adding 1 μL of formic acid to reduce the pH to 3–4, and then the samples were dried down completely in the speed vac, and stored at −80°C until further use. The dried protein digests were reconstituted in 200 μL of 0.1% trifluoroacetic acid (TFA) in water, and they were desalted using Pierce™ Peptide Desalting Spin Columns (ThermoFisher Scientific) as per the manufacturer’s instructions. The tryptic peptides were eluted from the spin columns with 600 μL of 50/49.9/0.1 acetonitrile/water/TFA, and then the eluate was dried down completely in the speed vac and stored at −80°C.
Liquid Chromatography-Trapped Ion Mobility Spectrometry-Mass Spectrometry Analysis
Liquid chromatography (LC) separation of the tryptic peptides was performed on a nanoElute® LC system (Bruker Daltonics) with a pre-concentration setup. Mobile phases consisted of LC-MS grade water containing 0.1% formic acid (solvent A) and LC-MS grade acetonitrile containing 0.1% formic acid (solvent B). For each sample analysis, 200 ng of the peptide digest were loaded on Acclaim™ PepMap™ 100 C18 ( Thermo Scientific, 0.3 mm x 5mm, 100 Å pore size) using solvent A, for approximately 5 minutes, depending on the backpressure generated by the sample. The peptides were then separated on a nanoElue FIFTEEN column (Bruker Daltonics, 75 μm x 150 mm, C18, 1.9 μm particles, 120 Å pore size). The separation was carried out at 40°C with a flow rate of 400 nL/min using the following gradient: 2–10% B from 0–2 mins, 10–35% B from 2–90 mins, 35–45% B from 90 to 105 mins, followed by wash and equilibration steps during which data was not acquired.
The LC was coupled to a timsTOF Pro mass spectrometer (MS) (Bruker Daltonics) with a CaptiveSpray ion source. The MS was operated in parallel accumulation-serial fragmentation (PASEF) mode with TIMS on and dynamic exclusion, 10 PASEF MS/MS scans per 1.1 sec cycle. Active exclusion for precursor ions was selected to release after 0.40 minutes, and precursors were reconsidered for analysis if the current intensity/previous intensity was greater than 4.0. The mass range was set between 100 and 1700 m/z, and the ion mobility range was set between 0.60 and 1.60 V·s/cm2 with a ramp time of 100.0 milliseconds. The intensity threshold was set to 2,500 units and the target intensity was set to 20,000 units. The collision energy was ramped between 20.00 to 59.00 eV as a function of ion mobility.
Database Search and Label-Free Quantitation
The acquired raw data from each LC-TIMS-MS analysis were imported onto PEAKS Online X (Bioinformatics Solutions Inc., Waterloo, ON) for peptide sequencing and protein identification. The protein database for Mus musculus containing 17,058 reviewed proteins was downloaded from UniProt. The following parameters were employed for de novo peptide sequencing and protein identification via database search: precursor mass error tolerance = 15 ppm, fragment mass error tolerance = 0.05 Da, enzyme = trypsin, digest mode = specific, peptide length was set between 6 and 45 residues, and the maximum number of missed cleavages was set to 3. Carbamidomethylation of cysteine residues was set as a fixed modification and oxidation of methionine residues was set as a variable modification. Search result filters were set to a peptide false discovery rate (FDR) of 1% and a protein filter was set to a −10logP value ≥ 20.
For label-free quantitation, the PEAKS Q module was employed to extract the peak areas of the peptides in each sample (N = 5 per group, 10 samples total). The following parameters for the PEAKS Q quantitation were as follows: mass error tolerance = 15 ppm, retention time shift tolerance was set to “auto detect”, CCS error tolerance = 0.05, and feature intensity ≥ 0. The following peptide filters were selected: average area ≥ 0, peptide ID count ≥ 0, charge between 1 and 10, have at least 2 confident samples per group, and use in group coefficient of variation filter. No normalization was applied, since normalization was performed elsewhere, as described in the next section. Peptide ID transfer was enabled to reduce the number of missing peptides across samples.
Proteomic Evaluation Statistical Analysis
Peptide peak areas obtained from PEAKS Q were imported onto the Normalyzer (version 1.1.1) online tool for normalization, whereby peak areas were log2 transformed and then normalized by LOESS-G. This method of normalization has been shown to be well-suited for bottom-up proteomics in terms of minimizing random intragroup variation across samples during the MS acquisition for accurate quantitation. Missing peptide values were imputed with Perseus (version 1.6.15.0) from a normal distribution with the default settings using a Gaussian distribution width of 0.3 relative to the standard deviation of the measured values and a downward shift of 1.8 standard deviation units from the data. For protein abundance calculations, the top 3 highest peptide peak areas were averaged for each unique protein. Student’s unpaired T-tests were performed between the two sample groups, and a multiple hypothesis testing correction was also applied using Perseus, where a Benjamini-Hochberg FDR correction was applied. Hierarchical cluster analysis (HCA) was performed using OriginPro (version 2021b, OriginLab Corporation, Northampton, MA), where the standardization was set to “rows”, the cluster method for both columns and rows was set to “group average”, and the distance type was set to “Euclidean”. Gene ontology (GO) analysis was performed using the STRING database (version 11.0b). Protein class analysis was performed with Panther classification system (version 16.0). OriginPro was used to create the figures. For protein overlap comparison, proteins were downloaded from ExoCarta (version 5) and filtered to only contain proteins from Mus musculus.
Statistics
All data is presented as mean ± standard error of the mean (SEM). Independent t test was used to compare between two groups (Mobile vs. Immobile pericyte gene expression, H2O2-stimulated vs. unstimulated pericyte gene expression). One-way ANOVA was used to examine values among different treatments (PBS, Mobile Pericyte, and Immobile Pericyte; PBS, Ctrl sEV, and Primed sEV within each age cohort). LSD and Games-Howell post hoc analyses were applied when appropriate. Statistical analyses were completed using SPSS version 18 (IBM, Chicago, IL, USA). Differences were considered statistically significant at p≤0.05.
Supplementary Material
Key Points.
Prior studies suggest that prolonged oxidative stress represents a barrier to skeletal muscle recovery after a period of immobilization
In this study we demonstrate that muscle resident perivascular stromal cells (pericytes) become dysfunctional and lack the capacity to mount an antioxidant defense after disuse in mice
Hydrogen peroxide treatment of healthy pericytes in vitro simulates the release of small extracellular vesicles (sEVs) that effectively recover skeletal muscle fiber size and ECM remodeling in young adult and aged mice after disuse
Pericyte-derived sEVs represent a novel acellular strategy to recover skeletal muscle after disuse
Acknowledgements
The authors would like to thank Dr. Barbara Pilas and the Flow Cytometry Center at the Roy J. Carver Biotechnology Center (University of Illinois at Urbana-Champaign) for advice and assistance with FACS and flow cytometry, as well as Drs. Alvaro Hernandez and Chris Wright and the DNA Services Center at the Roy J. Carver Biotechology Center for completion of single cell sequencing. We would also like to thank Dr. Priscila Falagan Lotsch and Prof. Cathy Murphy, Department of Chemistry, for assistance with size exclusion chromatography. This study was supported by NIAMS of the National Institutes of Health under award number R01 AR072735 (to MDB) and NIH P30 DA018310 (to JVS). LD was supported by a University of Illinois Graduate College Fellowship.
Biography

Yu-Fu Wu received his PhD in Kinesiology in 2021 at the University of Illinois, USA. His doctoral study in the Molecular Muscle Physiology laboratory mainly focused on disuse atrophy of aging skeletal muscle. His scientific interests include aging, degeneration, and comorbidities in the human movement system. His future goal is to continue studying the molecular and cellular biology of muscular and nervous system.
Footnotes
Conflict of Interest
No conflicts of interest, financial or otherwise, are declared by the author(s).
Data Availability Statement
All data generated or analyzed during this study are included in this published article and found under GEO accession GSE185560 and Figshare file 10.6084/m9.figshare.16778053.
References
- Agostini F, Dalla Libera L, Rittweger J, Mazzucco S, Jurdana M, Mekjavic IB, Pisot R, Gorza L, Narici M & Biolo G. (2010). Effects of inactivity on human muscle glutathione synthesis by a double-tracer and single-biopsy approach. J Physiol 588, 5089–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton PJ, Greenhaff PL, Phillips SM, Bodine SC, Adams CM & Lang CH. (2016). Control of skeletal muscle atrophy in response to disuse: clinical/preclinical contentions and fallacies of evidence. Am J Physiol Endocrinol Metab 311, E594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehr LM, West DW, Marcotte G, Marshall AG, De Sousa LG, Baar K & Bodine SC. (2016). Age-related deficits in skeletal muscle recovery following disuse are associated with neuromuscular junction instability and ER stress, not impaired protein synthesis. Aging (Albany NY) 8, 127–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek G, Choi H, Kim Y, Lee HC & Choi C. (2019). Mesenchymal Stem Cell-Derived Extracellular Vesicles as Therapeutics and as a Drug Delivery Platform. Stem Cells Transl Med 8, 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortoli S, Renault V, Eveno E, Auffray C, Butler-Browne G & Pietu G. (2003). Gene expression profiling of human satellite cells during muscular aging using cDNA arrays. Gene 321, 145–154. [DOI] [PubMed] [Google Scholar]
- Caplan AI & Dennis JE. (2006). Mesenchymal stem cells as trophic mediators. J Cell Biochem 98, 1076–1084. [DOI] [PubMed] [Google Scholar]
- Cerezo-Magana M, Bang-Rudenstam A & Belting M. (2020). The pleiotropic role of proteoglycans in extracellular vesicle mediated communication in the tumor microenvironment. Semin Cancer Biol 62, 99–107. [DOI] [PubMed] [Google Scholar]
- Chen J, Ke S, Zhong L, Wu J, Tseng A, Morpurgo B, Golovko A, Wang G, Cai JJ, Ma X, Li D & Tian Y. (2018). Long noncoding RNA MALAT1 regulates generation of reactive oxygen species and the insulin responses in male mice. Biochem Pharmacol 152, 94–103. [DOI] [PubMed] [Google Scholar]
- Doni A, Mantovani A, Bottazzi B & Russo RC. (2021). PTX3 Regulation of Inflammation, Hemostatic Response, Tissue Repair, and Resolution of Fibrosis Favors a Role in Limiting Idiopathic Pulmonary Fibrosis. Front Immunol 12, 676702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvoretskiy S, Garg K, Munroe M, Pincu Y, Mahmassani ZS, Coombs C, Blackwell B, Garcia G, Waterstradt G, Lee I, Drnevich J, Rhodes JS & Boppart MD. (2019). The impact of skeletal muscle contraction on CD146(+)Lin(−) pericytes. Am J Physiol Cell Physiol 317, C1011–C1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirin A, Zhu XY, Jonnada S, Lerman A, van Wijnen AJ & Lerman LO. (2018). Mesenchymal Stem Cell-Derived Extracellular Vesicles Improve the Renal Microvasculature in Metabolic Renovascular Disease in Swine. Cell Transplant 27, 1080–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AB. (2009). Redox signaling across cell membranes. Antioxid Redox Signal 11, 1349–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulle S, Di Donna S, Puglielli C, Pietrangelo T, Beccafico S, Bellomo R, Protasi F & Fano G. (2005). Age-dependent imbalance of the antioxidative system in human satellite cells. Exp Gerontol 40, 189–197. [DOI] [PubMed] [Google Scholar]
- Giordani L, He GJ, Negroni E, Sakai H, Law JYC, Siu MM, Wan R, Corneau A, Tajbakhsh S, Cheung TH & Le Grand F. (2019). High-Dimensional Single-Cell Cartography Reveals Novel Skeletal Muscle-Resident Cell Populations. Mol Cell 74, 609–621 e606. [DOI] [PubMed] [Google Scholar]
- Gorbunov NV, Garrison BR, McDaniel DP, Zhai M, Liao PJ, Nurmemet D & Kiang JG. (2013). Adaptive redox response of mesenchymal stromal cells to stimulation with lipopolysaccharide inflammagen: mechanisms of remodeling of tissue barriers in sepsis. Oxid Med Cell Longev 2013, 186795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy D (2015). Principles and standards for reporting animal experiments in The Journal of Physiology and Experimental Physiology. J Physiol 593, 2547–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister C & Satija R. (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 20, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes KL, Messina LM, Schwartz LM, Yan J, Burnside AS & Witkowski S. (2018). Type 2 diabetes impairs the ability of skeletal muscle pericytes to augment postischemic neovascularization in db/db mice. Am J Physiol Cell Physiol 314, C534–C544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter RB, Mitchell-Felton H, Essig DA & Kandarian SC. (2001). Expression of endoplasmic reticulum stress proteins during skeletal muscle disuse atrophy. Am J Physiol Cell Physiol 281, C1285–1290. [DOI] [PubMed] [Google Scholar]
- Hvid L, Aagaard P, Justesen L, Bayer ML, Andersen JL, Ortenblad N, Kjaer M & Suetta C. (2010). Effects of aging on muscle mechanical function and muscle fiber morphology during short-term immobilization and subsequent retraining. J Appl Physiol (1985) 109, 1628–1634. [DOI] [PubMed] [Google Scholar]
- Ji K & Tsirka SE. (2012). Inflammation modulates expression of laminin in the central nervous system following ischemic injury. J Neuroinflammation 9, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C, Goodman CA, Hornberger TA & Ji LL. (2015). PGC-1alpha overexpression by in vivo transfection attenuates mitochondrial deterioration of skeletal muscle caused by immobilization. FASEB J 29, 4092–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C & Ji LL. (2013). Muscle immobilization and remobilization downregulates PGC-1alpha signaling and the mitochondrial biogenesis pathway. J Appl Physiol (1985) 115, 1618–1625. [DOI] [PubMed] [Google Scholar]
- Krieger DA, Tate CA, McMillin-Wood J & Booth FW. (1980). Populations of rat skeletal muscle mitochondria after exercise and immobilization. J Appl Physiol Respir Environ Exerc Physiol 48, 23–28. [DOI] [PubMed] [Google Scholar]
- Laporte C, Tubbs E, Cristante J, Gauchez AS, Pesenti S, Lamarche F, Cottet-Rousselle C, Garrel C, Moisan A, Moulis JM, Fontaine E, Benhamou PY & Lablanche S. (2019). Human mesenchymal stem cells improve rat islet functionality under cytokine stress with combined upregulation of heme oxygenase-1 and ferritin. Stem Cell Res Ther 10, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler JM, Song W & Demaree SR. (2003). Hindlimb unloading increases oxidative stress and disrupts antioxidant capacity in skeletal muscle. Free Radic Biol Med 35, 9–16. [DOI] [PubMed] [Google Scholar]
- Matsushima Y, Nanri H, Nara S, Okufuji T, Ohta M, Hachisuka K & Ikeda M. (2006). Hindlimb unloading decreases thioredoxin-related antioxidant proteins and increases thioredoxin-binding protein-2 in rat skeletal muscle. Free Radic Res 40, 715–722. [DOI] [PubMed] [Google Scholar]
- Min K, Smuder AJ, Kwon OS, Kavazis AN, Szeto HH & Powers SK. (2011). Mitochondrial-targeted antioxidants protect skeletal muscle against immobilization-induced muscle atrophy. J Appl Physiol (1985) 111, 1459–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe M, Dvoretskiy S, Lopez A, Leong J, Dyle MC, Kong H, Adams CM & Boppart MD. (2019). Pericyte transplantation improves skeletal muscle recovery following hindlimb immobilization. FASEB J 33, 7694–7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Sherratt PJ & Pickett CB. (2003). Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol 43, 233–260. [DOI] [PubMed] [Google Scholar]
- Ning K, Fermin D & Nesvizhskii AI. (2012). Comparative analysis of different label-free mass spectrometry based protein abundance estimates and their correlation with RNA-Seq gene expression data. J Proteome Res 11, 2261–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachori AS, Smith A, McDonald P, Zhang L, Dzau VJ & Melo LG. (2007). Heme-oxygenase-1-induced protection against hypoxia/reoxygenation is dependent on biliverdin reductase and its interaction with PI3K/Akt pathway. J Mol Cell Cardiol 43, 580–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Guan D, Liu X, Li J, Wang L, Wu J, Zhou J, Zhang W, Ren R, Zhang W, Li Y, Yang J, Hao Y, Yuan T, Yuan G, Wang H, Ju Z, Mao Z, Li J, Qu J, Tang F & Liu GH. (2016). SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res 26, 190–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CM, Kim MJ, Kim SM, Park JH, Kim ZH & Choi YS. (2016). Umbilical cord mesenchymal stem cell-conditioned media prevent muscle atrophy by suppressing muscle atrophy-related proteins and ROS generation. In Vitro Cell Dev Biol Anim 52, 68–76. [DOI] [PubMed] [Google Scholar]
- Peeters CM, Visser E, Van de Ree CL, Gosens T, Den Oudsten BL & De Vries J. (2016). Quality of life after hip fracture in the elderly: A systematic literature review. Injury 47, 1369–1382. [DOI] [PubMed] [Google Scholar]
- Pisot R, Marusic U, Biolo G, Mazzucco S, Lazzer S, Grassi B, Reggiani C, Toniolo L, di Prampero PE, Passaro A, Narici M, Mohammed S, Rittweger J, Gasparini M, Gabrijelcic Blenkus M & Simunic B. (2016). Greater loss in muscle mass and function but smaller metabolic alterations in older compared with younger men following 2 wk of bed rest and recovery. J Appl Physiol (1985) 120, 922–929. [DOI] [PubMed] [Google Scholar]
- Powers SK. (2014). Can antioxidants protect against disuse muscle atrophy? Sports Med 44 Suppl 2, S155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunath A, Sundarraj K, Nagarajan R, Arfuso F, Bian J, Kumar AP, Sethi G & Perumal E. (2018). Antioxidant response elements: Discovery, classes, regulation and potential applications. Redox Biol 17, 297–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeg S, Jung T, Castro JP, Davies KJA, Henze A & Grune T. (2016). The molecular chaperone Hsp70 promotes the proteolytic removal of oxidatively damaged proteins by the proteasome. Free Radic Biol Med 99, 153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich KA, Chen YW, Thompson PD, Hoffman EP & Clarkson PM. (2010). Forty-eight hours of unloading and 24 h of reloading lead to changes in global gene expression patterns related to ubiquitination and oxidative stress in humans. J Appl Physiol (1985) 109, 1404–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson PA, Shimkus KL, Welles JE, Xu D, Whitsell AL, Kimball EM, Jefferson LS & Kimball SR. (2020). A time course for markers of protein synthesis and degradation with hindlimb unloading and the accompanying anabolic resistance to refeeding. J Appl Physiol (1985) 129, 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala D, Cunningham TJ, Stec MJ, Etxaniz U, Nicoletti C, Dall’Agnese A, Puri PL, Duester G, Latella L & Sacco A. (2019). The Stat3-Fam3a axis promotes muscle stem cell myogenic lineage progression by inducing mitochondrial respiration. Nat Commun 10, 1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH & Goldberg AL. (2004). Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117, 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavely R & Nurgali K. (2020). The emerging antioxidant paradigm of mesenchymal stem cell therapy. Stem Cells Transl Med 9, 985–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker R, Yamamoto Y, McDonagh AF, Glazer AN & Ames BN. (1987). Bilirubin is an antioxidant of possible physiological importance. Science 235, 1043–1046. [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P & Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetta C, Frandsen U, Mackey AL, Jensen L, Hvid LG, Bayer ML, Petersson SJ, Schroder HD, Andersen JL, Aagaard P, Schjerling P & Kjaer M. (2013). Ageing is associated with diminished muscle re-growth and myogenic precursor cell expansion early after immobility-induced atrophy in human skeletal muscle. J Physiol 591, 3789–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetta C, Hvid LG, Justesen L, Christensen U, Neergaard K, Simonsen L, Ortenblad N, Magnusson SP, Kjaer M & Aagaard P. (2009). Effects of aging on human skeletal muscle after immobilization and retraining. J Appl Physiol (1985) 107, 1172–1180. [DOI] [PubMed] [Google Scholar]
- Talbert EE, Smuder AJ, Min K, Kwon OS, Szeto HH & Powers SK. (2013). Immobilization-induced activation of key proteolytic systems in skeletal muscles is prevented by a mitochondria-targeted antioxidant. J Appl Physiol (1985) 115, 529–538. [DOI] [PubMed] [Google Scholar]
- Valle-Prieto A & Conget PA. (2010). Human mesenchymal stem cells efficiently manage oxidative stress. Stem Cells Dev 19, 1885–1893. [DOI] [PubMed] [Google Scholar]
- Vanlandewijck M, He L, Mae MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Lavina B, Gouveia L, Sun Y, Raschperger E, Rasanen M, Zarb Y, Mochizuki N, Keller A, Lendahl U & Betsholtz C. (2018). A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480. [DOI] [PubMed] [Google Scholar]
- Wang X, Wilkinson R, Kildey K, Potriquet J, Mulvenna J, Lobb RJ, Moller A, Cloonan N, Mukhopadhyay P, Kassianos AJ & Healy H. (2017). Unique molecular profile of exosomes derived from primary human proximal tubular epithelial cells under diseased conditions. J Extracell Vesicles 6, 1314073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JR, Confides AL, Moore-Reed S, Hoch JM & Dupont-Versteegden EE. (2015). Regrowth after skeletal muscle atrophy is impaired in aged rats, despite similar responses in signaling pathways. Exp Gerontol 64, 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiklander OPB, Brennan MA, Lotvall J, Breakefield XO & El Andaloussi S. (2019). Advances in therapeutic applications of extracellular vesicles. Sci Transl Med 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Zheng J, Cai J, Zeng K, Zhou C, Zhang J, Li S, Li H, Chen L, He L, Chen H, Fu H, Zhang Q, Chen G, Yang Y & Zhang Y. (2019). Extracellular vesicles derived from human umbilical cord mesenchymal stem cells alleviate rat hepatic ischemia-reperfusion injury by suppressing oxidative stress and neutrophil inflammatory response. FASEB J 33, 1695–1710. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Cankova Z, Iwanaszko M, Lichtor S, Mrksich M & Ameer GA. (2018). Potent laminin-inspired antioxidant regenerative dressing accelerates wound healing in diabetes. Proc Natl Acad Sci U S A 115, 6816–6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and found under GEO accession GSE185560 and Figshare file 10.6084/m9.figshare.16778053.


