Abstract
Background
The genome of SARS-CoV-2, is mutating rapidly and continuously challenging the management and preventive measures adopted and recommended by healthcare agencies. The spike protein is the main antigenic site that binds to the host receptor hACE-2 and is recognised by antibodies. Hence, the mutations in this site were analysed to assess their role in differential infectivity of lineages having these mutations, rendering the characterisation of these lineages as variants of concern (VOC) and variants of interest (VOI).
Methods
In this work, we examined the genome sequence of SARS-CoV-2 VOCs and their phylogenetic relationships with the other PANGOLIN lineages. The mutational landscape of WHO characterized variants was determined and mutational diversity was compared amongst the different severity groups. We then computationally studied the structural impact of the mutations in receptor binding domain of the VOCs. The binding affinity was quantitatively determined by molecular dynamics simulations and free energy calculations.
Results
The mutational frequency, as well as phylogenetic distance, was maximum in the case of omicron followed by the delta variant. The maximum binding affinity was for delta variant followed by the Omicron variant. The increased binding affinity of delta strain followed by omicron as compared to other variants and wild type advocates high transmissibility and quick spread of these two variants and high severity of delta variant.
Conclusion
This study delivers a foundation for discovering the improved binding knacks and structural features of SARS-CoV-2 variants to plan novel therapeutics and vaccine candidates against the virus.
Keywords: SARS-CoV-2, Molecular modelling, Mutation landscape, Omicron, Delta, hACE2
Abbreviations
- SARS-CoV-2
Severe Acute Respiratory Syndrome Corona Virus-2
- hACE-2
Human Angiotensin-Converting Enzyme 2
- WHO
World Health Organization
- VOC
Variants of Concern
- VOI
Variants of Interest
- VUM
Variants under Monitoring
- GISAID
Global Initiative on Sharing All Influenza Data
- PANGO
Phylogenetic Assignment of Named Global Outbreak
- RBD
Receptor Binding Domain
- RBM
Receptor Binding Motif
- MD
Molecular Dynamics
- MMGBSA
Molecular Mechanics Generalized Born Surface Area
1. Introduction
Severe acute respiratory syndrome corona virus-2 (SARS-CoV-2) emerged in November 2019 and caused a worldwide pandemic, leading to public health emergencies by affecting social and economic conditions globally due to imposed social distancing and lockdowns [1,2]. Subsequently, the world faced an unprecedented situation due to the emergence of novel variants of SARS-CoV-2 that ignited significant health concerns [3]. The first confirmed case was reported in Wuhan, the capital of Central China's Hubei Province [4]. The emergence of COVID-19 as a pandemic has significantly affected the research perspective of healthcare workers, as fast and instantaneous management measures are required to deal with the rapidly evolving pathogen [5,6]. The advancements in technology have aided the strengthening of sequence and structure databases which can be strategically utilized to improve the diagnosis, treatment and prevention of this life-threatening infection [[7], [8], [9]]. The causative agent, SARS-CoV-2, is continuously evolving into novel variants, which makes the management of COVID-19 extremely challenging [10,11].
The SARS-CoV-2 belongs to the Coronaviridae family of viruses comprising a single-stranded positive-sense RNA genome. RNA viruses have a lower fidelity rate, making it easy to accumulate mutations for enhanced transmissibility and improved host adaptability [12]. The aggressive mutational landscape of SARS-CoV-2 has led to the parallel development of various nomenclature systems to differentiate the variants and lineages significantly [13]. GISAID (Global Initiative on Sharing All Influenza Data) segregated the variants into eight different clades termed as S, O, L, V, G, GH, GR, and GV; Nextstrain categorized them into 13 major clades including 19A to 19B, 20A to 20J and 21A whereas PANGO (Phylogenetic Assignment of Named Global Outbreak) divided these into 1517 lineages and sub lineages [[14], [15], [16]]. Whole-genome sequencing provides a complete insight into the mutations responsible for differential pathogenicity in these variants. More than 12.2 million sequences available on GISAID as on July 29, 2022, reveal that the mutations in spike protein critically influence both pathogenicity and immunity evasion and hence need to be evaluated extensively to understand the evolution of the novel variants [11,17]. The variants with a significant impact on transmissibility and severity were classified as Variants of concern (VOC), Variants of interest (VOI) and Variants under monitoring (VUM) by the World Health Organization (WHO) [18]. These VOC, VOI and VUM were designated as Greek alphabetical letters such as Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2) etc. on May 31, 2021 [19].
SARS-CoV-2 contains four structural proteins: Spike protein (S), envelope protein (E), membrane protein (M), and nucleocapsid protein (N) [20]. The Spike protein, an outward-positioned trimeric glycoprotein of coronaviruses, aids the attachment of the virus to the host cells by binding to human angiotensin-converting enzyme 2 (hACE2) and facilitates virus-cell membrane fusion in the course of viral infection [21]. Hence, this glycoprotein was considered a key target for developing vaccines and therapeutics against SARS-CoV-2 [22]. Each monomer of spike glycoprotein comprises 1273 residues and contains two subunits; S1, which interacts with the host receptor and S2 subunit, which facilitates the fusion of viral and host cell membranes [23]. S1 subunit harbours the main hACE2 interacting domain called the receptor binding domain (RBD) [24]. The S2 subunit contains five domains namely, cytoplasmic domain, two heptapeptide repeats (HR1 and HR2), transmembrane domain (TM) and a fusion peptide (FP) which collectively trigger the conformational changes that mediate the fusion of spike protein with the host cell membrane [24]. The receptor binding domain of S1 subunit not only interacts with the hACE2 receptor but also binds to a wide range of antibodies. Hence, the mutations occurring in this region exert selective pressure, forcing the development of novel variants [11,25]. The receptor binding motif (RBM) is the key motif of RBD, which directly binds to hACE2 and harbours the majority of mutational hotspot residues determining the severity of concerned variants [26].
There exist five SARS-CoV-2 lineages that are characterized as VOC by WHO [19]. The first VOC, Alpha variant, contains 7 mutations and two deletions in the spike protein, and these mutations are responsible for increased pathogenicity. Hence, it was included in the list of VOCs [27]. Similarly, the second and third VOCs, Beta (7 mutations and 1 deletion) and Gamma (12 mutations), also harbor substitutions in their antigenic and hACE2 interacting spike region, and are responsible for higher infectivity [28]. The number of substitutions in spike protein of the fourth VOC, Delta variant, when it posed a significant health emergency across the globe, were only five [28]. The emergence of the super-mutated variant, omicron possessing as many as 32 mutations in spike protein, with high transmissibility and immunity bypassing capability has necessitated a relook into the prospective preventive measures [29].
Omicron (Clade GRA, Lineage B.1.1.529.1), first reported in Botswana (November 11, 2021), was designated as a variant of concern [30]. The omicron variant speedily spread to neighboring countries and has now become the most prevalent variant worldwide [31]. Nevertheless, WHO documented the spread of this variant at an unexpectedly high rate with ability to evade immunity developed through vaccination or previous infections due to the presence of 32 mutations in the spike region [32]. Among the 32 mutations, 15 are present on the spike protein receptor-binding domain (RBD) [29]. Furthermore, about 10 mutations occur at the binding interface of RBD with hACE2 receptor protein [33].
A deeper insight is required to assess the impact of these mutations in different variants of concern which will aid the development of novel treatment modalities and vaccination candidates to protect against the variants with mutated versions of the spike protein [34]. Hence, it is important to pinpoint the mutational correlation amongst different VOCs, VOIs and VUMs to speculate on other probable key mutations that are central to the evolution of dangerous variants and their overall effect on the binding affinity with the hACE2 receptor [35].
The present study analyzes the phylogenetic relatedness and mutational diversity of different VOCs, VOIs and VUMs in comparison to other PANGOLIN lineages while estimating the functional impact of the spike mutations in VOCs. The molecular modelling approach adopted in this study, affords structural insights into the impact of RBD mutations on the three-dimensional conformation of the protein and interaction with the hACE2 receptors.
2. Methods
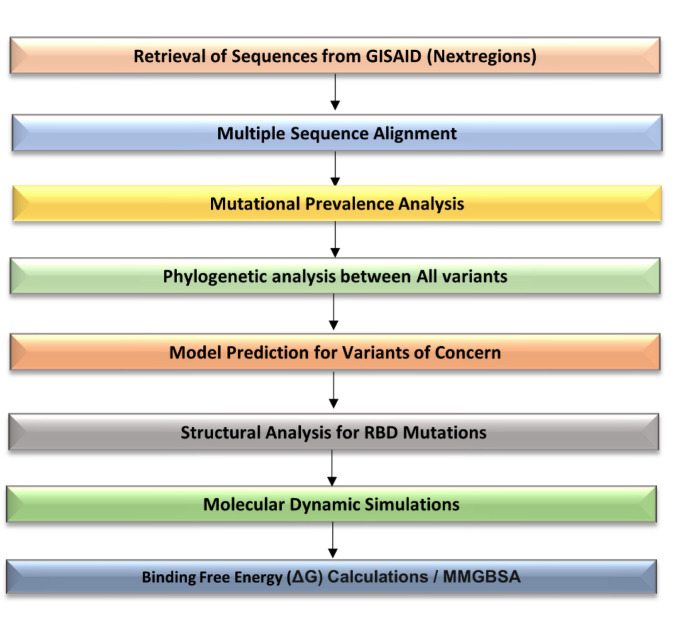
This study comprises two aspects; the first concerns the detailed whole-genome sequence analysis and characterization of mutations in VOCs, while the second involves molecular modelling studies to gain structural insights into mutant RBDs and the impact of the mutations on their binding affinity with the hACE2 receptor. The detailed plan of the study undertaken is given in Fig. 1 .
Fig. 1.
Detailed work plan of the study: The whole genome sequences of SARS-CoV-2 global data were downloaded from GISAID and analysed for the prevalence of mutations, followed by the characterization of mutations in different WHO signified variants. The molecular modelling and dynamics studies were then performed to decipher the impact of these mutations on the binding affinity of VOCs with hACE2.
2.1. Sequence analysis
We performed the sequence analysis to delineate the differential mutational landscape in WHO characterized variants and other PANGOLIN lineages of SARS-CoV-2 [16,17]. A comprehensive analysis was carried out on the sequences retrieved from the public database, followed by multiple sequence alignment and phylogenetic analysis amongst various lineages.
2.1.1. Data retrieval
A representative global dataset (N = 3244) retrieved from GISAID (https://www.gisaid.org/) nextregions genomic epidemiology global database contained region-specific auspice source files from different geographical regions. Additionally, a collection of 2365 high-quality reference genomes comprising different lineages in Nextclade (https://clades.nextstrain.org/) were included as a reference for phylogenetic interpretation [36]. This complete dataset with 5609 sequences was used to perform phylogenetic inference using the Nextclade client interface (CLI) tool [36].
2.1.2. Proportionate prevalence of mutations
The pipeline from Nextstrain was employed to screen the presence of Single Nucleotide Polymorphisms (SNPs) and Insertions and Deletions (InDels) in the various lineages of SARS-CoV-2. A genome-wide mutation detection analysis was also performed from Nextstrain (https://github.com/nextstrain/ncov) [36]. The mutation frequencies amongst the VOCs were calculated. The spike protein sequences were separately aligned using MAFFT software (Version 7.467) to characterise the mutations present in spike proteins of all WHO signified variants [37].
2.1.3. Frequency of mutations in global data
The global mutation prevalence in the spike protein of all VOCs was calculated from the documented amino acid substitutions at SARS-CoV-2 mutations Situation Reports (https://outbreak.info/situation-reports). The data was analysed using the R package available for outbreak.info analysis [38].
All the genomic sequences available at GISAID (as on August 31, 2022) (N = 1,29,31,232) were included in the analysis of mutation prevalence [39].
2.1.4. Phylogenomic analyses
All the genomic sequences were aligned to the reference genome Wuhan-Hu-1 and the phylogenetic clade and lineage assignment was accomplished using Nextclade version 0.7.2 [36]. Phylogenomic analysis was also performed with the Nextstrain workflow available at https://github.com/nextstrain/ncov.
Nextclade operates on banded Smith–Waterman alignment and uses affine gap-penalty for sequence analysis [15]. The phylogenetic analysis was performed using Nextstrain pipelines, which contain an integrated Augur software for analysis and auspice viewer to visualise the data [40]. The Nextstrain uses its reference genomes for each lineage which are characterised by mapping with the reference genome Wuhan 2019-1 [41]. The phylogenetic relationship amongst each lineage was inferred by running a comparative approach using all the Nextstrain reference genomes as a representative of all PANGOLIN lineages [15]. IQ-TREE tool was employed to construct a maximum-likelihood phylogenetic tree while estimating the molecular clock branch lengths and reconstruction of nucleotide and amino acid changes via TreeTime [42,43]. The resulting tree was visualized using Nextstrain auspice and annotated with iTol [44].
2.2. Structural analysis of interactions with hACE2
The present study investigated the mechanisms of interactions and binding conformation of the receptor binding domain of wild type and different variants (alpha, beta, gamma, delta and omicron) with hACE2 receptor. The structural impact of the mutations in receptor binding domain (RBD) of all VOCs was assessed using the molecular modelling approach. The binding affinity of all VOCs were calculated using structural modelling, molecular docking, molecular dynamic simulations and molecular mechanics generalized Born surface area (MM/GBSA).
2.2.1. Protein structure modelling and validation
The spike glycoprotein sequence of SARS-CoV-2 was retrieved from the UniProt protein sequence database in FASTA format [45]. The 3D structure of the wild type spike protein was modelled using the Prime module of Schrodinger as the experimentally reported RBD structures contain several missing residues in our region of interest. Hence, we constructed these regions as per the reported template (PDB ID: 7KRS) to generate the complete three-dimensional structure of RBD with all residues [46]. The experimentally available structure of SARS-CoV-2 complexed with hACE2 (PDB ID: 6M0J) was considered for assessing the interactions of hACE2 with RBD [47].
The three-dimensional structures of receptor binding domains of different VOCs (alpha, beta, gamma, delta and omicron) were modelled by retrieving the information about the mutations from the GISAID database [14]. Win Coot was adopted to manually incorporate mutational changes and residues missing in the template structure by considering acceptable geometry and rotamer configuration. Apart from this, we also constructed three insertions and six deletions reported in the omicron variant manually using Win Coot [48].
The modelled 3D structures of different VOCs were validated using SAVES online server which incorporates ERRAT, VERIFY 3D and PROCHECK [49]. The Ramachandran plot evaluates the steric clashes and the reliability of the three-dimensional structure, whereas, the ERRAT plot denotes the overall error frequency rate of the modelled structures. No residues were observed in the disallowed region in Ramachandran plot (Supplementary File S1). The best-validated models of RBD of the SARS-CoV-2 spike glycoprotein of wild type and VOCs, obtained after geometry optimization and energy minimization were subsequently considered for calculation of interactions with hACE2 receptor.
2.2.2. Molecular docking
Molecular Docking was performed by using HDOCK online server to estimate the interactions of wildtype and VOC spike proteins with hACE2 receptor (http://hdock.phys.hust.edu.cn/, accessed on June 14, 2022) [20]. The best protein–protein complex model was selected from the top ten conformers based on their docking score.
2.2.3. Molecular dynamics (MD) simulations
The dynamic stability and flexibility of modelled RBD structures were assessed by molecular dynamics (MD) simulation method. The MD simulation study facilitated the understanding of the structural and conformation changes occurring due to mutations in the spike protein of VOCs. We used the Desmond module of Schrödinger suite for MD simulation study. Firstly, solvated the modelled three-dimensional protein systems with the TIP3P explicit solvent model in OPLS_2005 force field. Further, we neutralized the systems by adding 0.15 M NaCl into the solvent box and minimized by steepest descent and LBFGs algorithm with a maximum of 2000 iterations with convergence criteria of 1 kcal/mol/Å. Finally, the production run of 100 ns was performed for all the energy minimized complexes in eight discrete phases with specific limitations. The initial seven phases of molecular dynamics included the equilibration process, while the last step, constituted the extensive simulation production phase. The long-lasting production phase was carried out for 100 ns at a constant temperature (300 K) using the Nosé–Hoover chain coupling scheme. The reversible reference system propagator algorithms (RESPA) integrator was used throughout the molecular dynamics simulation. The bonding interactions for a time step of 2 fs were calculated in the final production run. The Particle Mesh Ewald (PME) algorithm was used to determine the long series electrostatic interactions during the simulation. The same protocol was employed for molecular dynamic studies for all the complexes (wild and mutational variants). After completion of the production run, simulation trajectories were analysed employing various parameters, such as root mean square deviation (RMSD), root mean square fluctuation (RMSF), and the number of hydrogen bonds formed during the simulation.
2.2.4. Prime MM-GBSA calculation
The post dynamics ΔGbinding free energy change of the simulated RBD-hACE2 complex of wild-type and VOCs were assessed using the molecular mechanics generalized Born surface area (MM/GBSA) approach in the Prime module of Schrodinger. The binding energies between the RBD/hACE2 of wild as well as all five VOCs were calculated as
| ΔGbinding = ΔGcomplex− (ΔGprotein+ΔGligand) |
Where, ΔGcomplex, ΔGprotein and ΔGligand represent the total free energies of the complex, the protein and the ligand respectively.
The binding energy was determined utilizing a total of 50 snapshots taken from the last 10 ns of the stable simulation trajectories and computed using the “thermal_mmgbsa.py” script. Per residue interaction energy was also calculated using the RBD domain as a ligand and hACE2 as receptor using the script “breakdown_MMGBSA_by_residue.py”.
3. Results and discussion
3.1. Sequence analysis
Whole genome sequences were analysed to evaluate the phylogenomic association amongst various lineages of SARS-CoV-2 and delineate the specific variations in significant groups. A comprehensive sequence analysis supports identifying the genomic regions presenting rapid evolution compared to others.
3.1.1. Data retrieval
The ‘nextregions database’ of GISAID contains the sequences subsampled to represent a global phylogeny by including the data from each geographical region. Thus, the 3244 sequences downloaded from GISAID nextregions genomic epidemiology global database represent a set of subsampled sequences from all reported SARS-CoV-2 lineages worldwide. The sequences analysed using Nextclade command-line interface (Nextclade CLI) contain a reference database of 2365 sequences representing the various lineages and use Nextalign tool to align the query sequences. The corresponding metadata for the analysed sequences is included as Supplementary file S2.
3.1.2. Proportionate prevalence of mutations
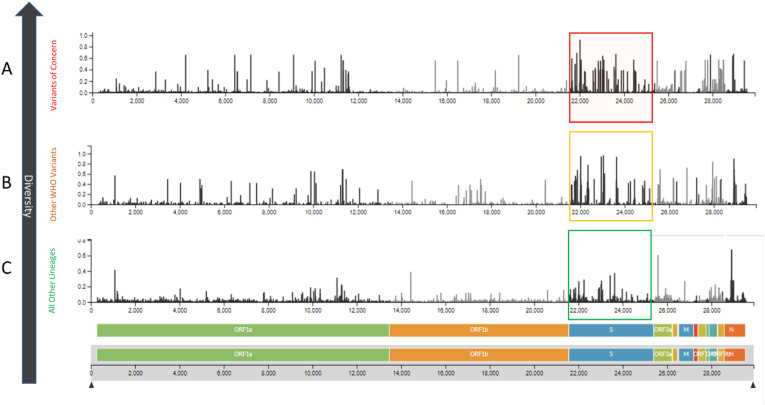
The mutations present in VOC v/s VOI v/s other lineages were analysed descriptively to reveal the prevalence of mutations across these clinically significant groups. The maximum number of mutations were observed in WHO signified variants (Alpha to Omicron). The Auspice tool measures the degree of genomic variations as entropy, which denotes the changes in amino acid residues during the evolution process. Additionally, the entropy of SNP variations across the genomes of various WHO variant classes indicated that the majority of the mutations were concentrated in the spike protein region. The spike protein region constitutes the main interacting site for binding to the host receptors and antibodies and hence contributes markedly to the evolution of SARS-CoV-2 lineages. Several studies state that the ratio of nonsynonymous to synonymous divergence is highest in Spike proteins [50,51]. It was reported that out of the 20 significant mutations contributing to the viral fitness in 1.6 million SARS-CoV-2 genomes, 14 were present within the spike protein region [52]. The number of overall mutations observed in VOCs (Fig. 2 A) was greater than the VOIs and VUMs (Fig. 2B). These numbers are comparatively higher than the mutations present in other lineages (Fig. 2C).
Fig. 2.
Prevalence of mutations in the analysed SARS-CoV-2 sequences. The abundance of SNPs in spike region of VOCs (red box), VOI & VUM (yellow box) and other PANGOLIN lineages (green box). The prevalence of mutations is highest in VOC followed by VOI/VUM and lowest in other lineages which are not a part of WHO variants nomenclature.
The spike protein is located at the virus's surface and aids interaction with the hACE-2 receptor leading to the fusion and entry into the host cells [47]. A total of 84 polymorphism sites were observed in the spike glycoprotein. The maximum number of polymorphisms were harboured by the omicron variant spike protein (32 mutations) followed by the delta variant (5 mutations).
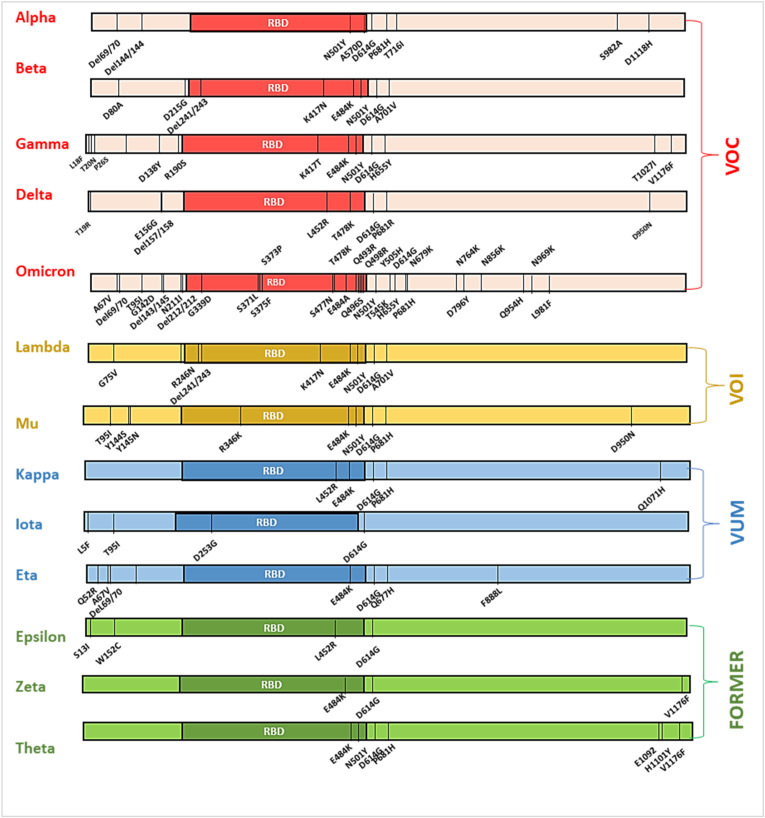
Descriptive analysis of mutation prevalence revealed that the most widely distributed key mutations involving A67V, T95I, G142D, K417 N, S477 N, T478K, N501Y, D614G and P681H, were observed to be collectively present in alpha, beta, gamma, delta, omicron, eta, iota, kappa and theta variants (Fig. 3 ). The majority of the spike glycoprotein variations were located in the receptor binding domain (RBD), which constitutes the main interacting domain with the hACE-2 receptor [53].
Fig. 3.
Mutational landscape of WHO characterised variants. The relative number and position of mutations in VOC, VOI, VUM and former variants now removed from the list of variants by WHO.
The most common mutation present in the spike protein was D614G, detected in 5321 out of the total 5609 sequences which is the most prevalent mutation reported in SARS-CoV-2 globally and known to confer fitness to the virus [54]. The prevalent mutations found in variants of concern were L452R, E484K, and N501Y, which are positioned within the spike protein's receptor binding motif (RBM) (Fig. 4 ). RBM is the interaction interface of the spike protein and hACE-2 receptor, and the mutations in this region affect the affinity with hACE-2. Hence these substitutions are significant in making lineages as variants of concern and interest [55].
Fig. 4.
Multiple sequence alignment of receptor binding domains (RBD: 239–519) of variants of concern indicating the location of mutations. The residues in the Receptor Binding Motif (RBM), which form the main interface interacting with the hACE2 receptor, are highlighted in the red box.
3.1.3. Frequency of mutations in global data
The SARS-CoV-2 genomic sequence data was retrieved from GISAID (N = 1,29,31,232). The lineages included for mutation analysis are given as supplementary file S3. Total 78 substitutions and indels (insertions and deletions) were having a prevalence of >50% in VOCs genomic sequences, out of which ten (T19R/I, Del 69/70, G142D, K417 T/N, T478K, E484 A/K, N501Y, D614G, H655Y and P681 H/R) were harboured by more than one variant. Of the 68 unique substitutions, maximum mutations were present in the omicron variant (N = 29). The percent prevalence of mutations in all VOCs is depicted as a sunburst plot (Fig. 5 ).
Fig. 5.
Sunburst diagram depicting the global percent incidence of mutations in variants of concern as per the genomic data available at GISAID (N = 1,29,31,232) (https://outbreak.info/situation-reports). The most common mutation was D614G (shared by all VOCs) followed by N501Y. (Present in four VOCs).
3.1.3.1. Phylogenetic analysis
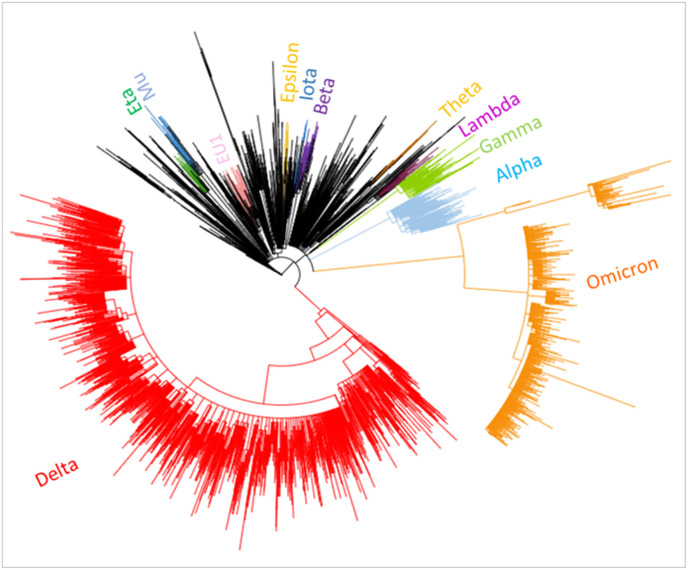
The phylogenetic analysis of the SARS-CoV-2 genomes revealed the parallel evolution of VOCs and VOIs, which are heterogeneously distributed throughout the phylogenetic tree. The phylogenetic distribution of clades across the tree was comparable with the other studies performed on a large number of strains advocating that the SARS-CoV-2 exhibits convergent evolution [[56], [57], [58]]. The graphical analysis of lineage diversity in all variants revealed that omicron is the most distant variant forming a separate clade from all other lineages (Fig. 6 ) as also documented in other reports [29]. In the global dataset, >40% of the sequences belong to the delta variant, which clustered into three separate groups belonging to 21A, 21I and 21J clades [59]. A study on phylogenomic analysis with a smaller number of genomes demonstrated that in the whole genome-based phylogeny (n = 478 genomes) including VOC, VOI and VUM, the omicron variant forms a separate phylogroup with alpha and lambda variants [60].
Fig. 6.
Phylogenetic tree depicting the relationships among SARS-CoV-2 genomes. A total of 5609 sequences were used for the analysis. Variants of Concern and variants of interest are labelled and highlighted in distinct colours. Genome sequences from Omicron variant formed a separate clade suggesting that it evolved parallelly rather than diverging from Delta variant.
3.2. Structure analysis
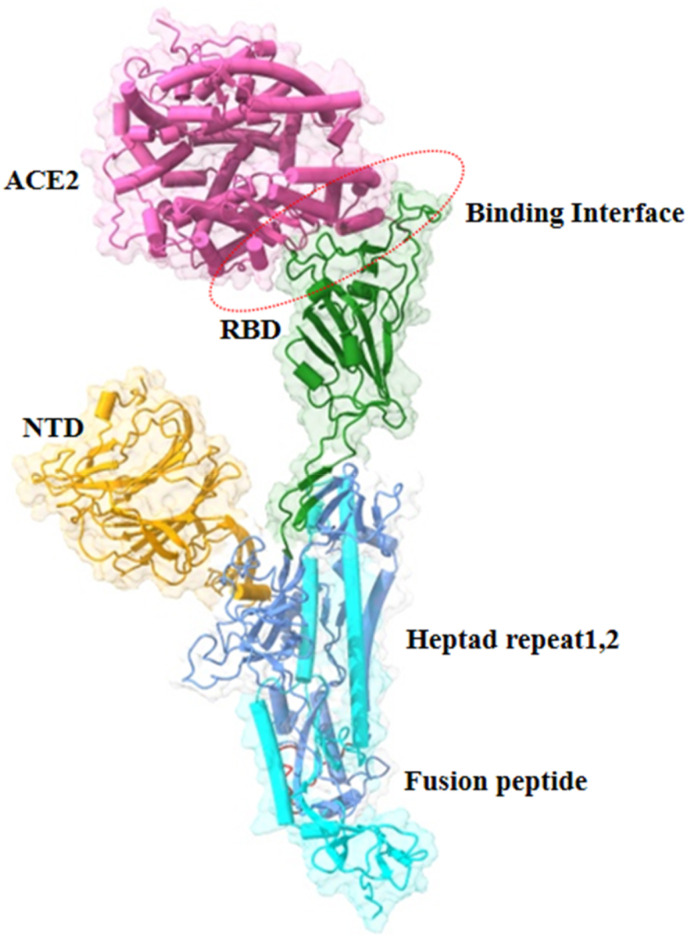
The SARS-CoV-2 variants have discrete mutations at their binding interface with the hACE2 receptor [61]. The coronavirus spike protein exists as a trimer, and each subunit possesses a Y shaped conformation containing two interacting domains (S1 and S2). The first supersite is situated at the N-terminal domain, which is the target for anti-NTD antibodies [22]. The second site constitutes the receptor-binding domain (RBD), which interacts with the hACE2 receptor and the neutralising antibodies [24]. The mutations present in the RBD region play a key role in influencing its attachment with the hACE2 receptor (Fig. 7 ). The present computational study emphasizes on identifying the interaction occurring between the different RBD variants and the hACE2 receptor.
Fig. 7.
Overall structure of spike protein complex with hACE2 receptor. The receptor binding domain is shown in green colour interacting with the hACE-2 receptor (purple) at the binding interface.
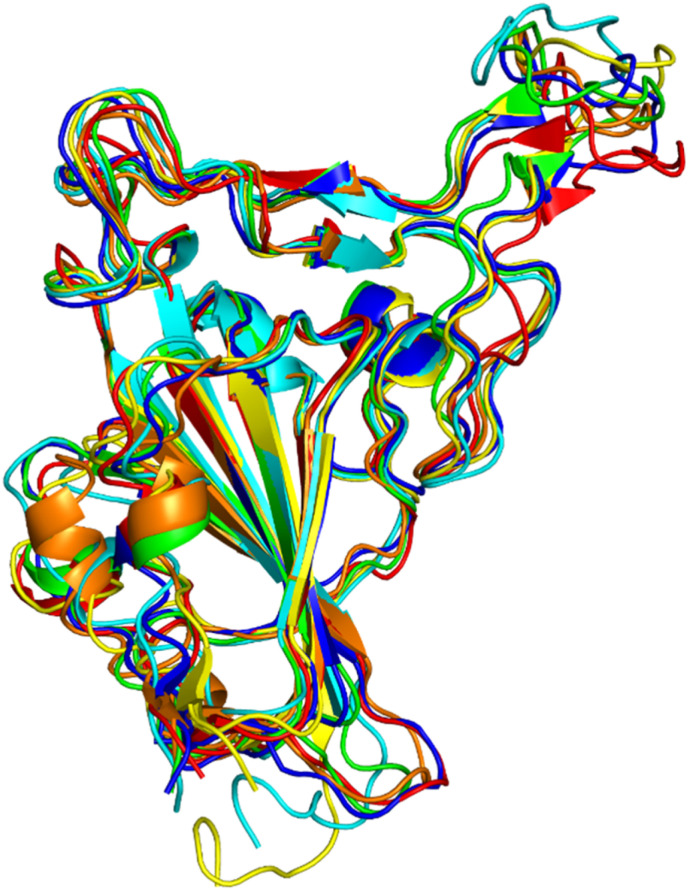
3.2.1. Molecular modelling and structural analysis
The models with the lowest energy were selected from all the generated models for further study. The structures of all the various modelled RBDs were superimposed to decipher the conformational alterations in the protein structure occurring as a result of the mutations (Fig. 8 ). The RMSD values so calculated for the RBD wild type when overlaid on the VOC RBDs, were 0.92 Å (alpha), 1.08 Å (beta), 1.13 Å (gamma), 0.78 Å (delta) and 1.01 Å (omicron).
Fig. 8.
Superimposed structures of receptor-binding domains (RBDs) of alpha (cyan), Beta (blue), Gamma (yellow), Delta (red) and Omicron (orange) with wild-type RBD protein structure (green).
3.2.2. Molecular docking
The protein-protein docking yielded in silico interaction energy between hACE-2 receptor and the RBDs of VOCs and the wild-type (WT) isolate from Wuhan to predict the binding scores. The predicted docking score from HDOCK for alpha, beta, gamma, delta, omicron and wild type with hACE2 was −307.44, −343.55, −341.54, −343.88, −344.76 and −345.14 kcal/mol respectively. Previous studies have also reported that WT, delta and omicron variants have better docking scores as compared to other variants [62,63]. In a wide-scale analysis of the effects of mutations on RBD–hACE2 affinity, it was reported that approximately 84.3% of mutations did not alter binding affinity, whereas only 3.8% of mutations decreased the binding strength [64]. This indicates that the docking score alone is not sufficient to assess the molecular binding. Hence, to explore the binding affinity; molecular dynamics, free energy analysis, interaction analysis and per residue contribution of mutated residues was performed.
3.2.3. Dynamic stability and flexibility analysis
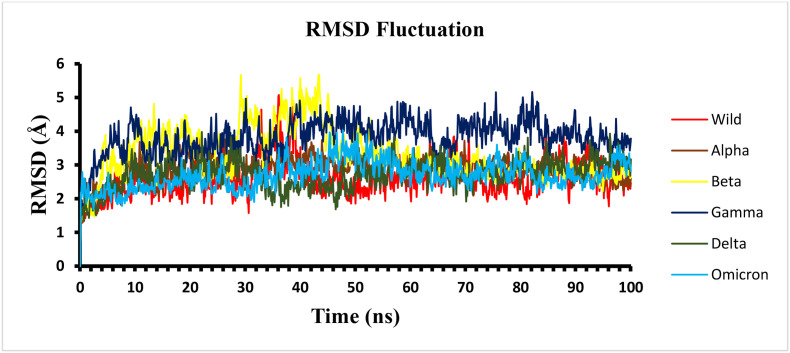
Untangling the structural and conformational dynamics of SARS-CoV-2 spike protein and its variants is an effective method to estimate the overall stability of complex. It also enables to predict the effect of mutations on the structure, function and overall binding of the protein. A 100 ns simulation run was performed for wild type protein and the different variants (alpha, beta, gamma, delta and omicron) with hACE2 receptor to monitor their dynamic behaviour. The observed changes enabled the prediction of the effects these mutations had on the binding affinity with host receptor in different RBD variants. Molecular dynamics studies facilitate in examining its implication in the virus fitness for humans regarding attachment and infection. The simulation trajectory was examined by evaluating the root mean square deviation (RMSD) to observe structural changes in the RBD-hACE2 complex under dynamic environment. Initially, all the complex systems displayed pre-equilibration phases up to 30 ns time. Subsequently, their trajectory assessment revealed lesser fluctuation, indicating a comparatively stable behaviour. The RMSD analysis from trajectory revealed that the stable structures were well-maintained with the least deviation up to 3 Å for all the variants as compared to the wild type (Fig. 9 ).
Fig. 9.
RMSD plot of the wild and variant RBD–hACE2 complexes (backbone) during 100 ns MD simulation.
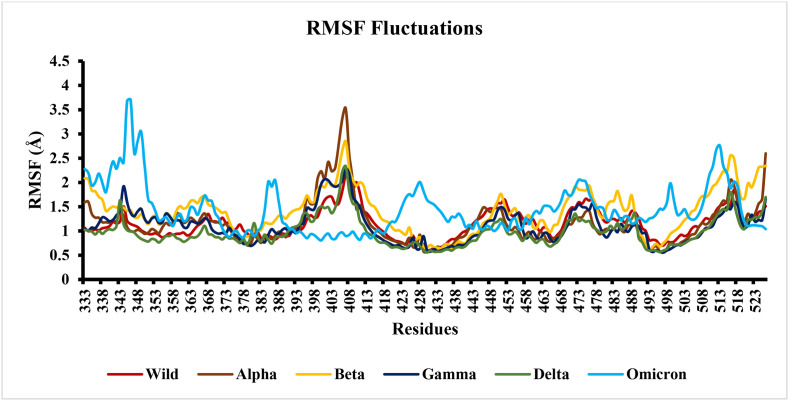
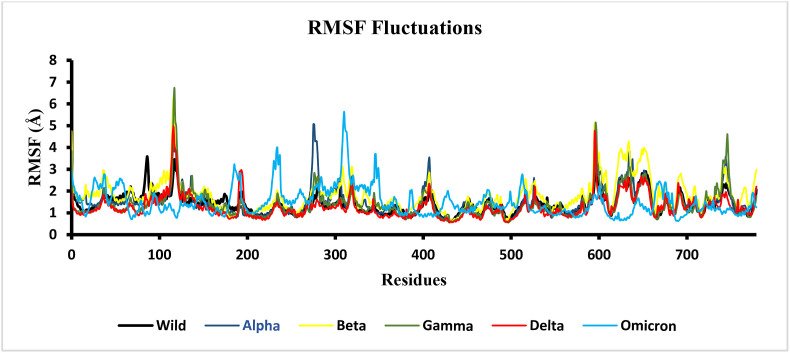
The conformation of a particular amino acid and nature of interactions with other residues in protein constitutes a vital role in various processes such as molecular rigidity, protein-protein complex interactions, and protein structure stability. The dynamic behaviour of the various amino acid residues in the wild type and mutant proteins during molecular dynamics simulation was also assessed by evaluating the root mean square fluctuation (RMSF). Since, the RBD of spike protein represents a significant mutational region, the residual fluctuations of the RBD in wild type and VOCs were plotted to observe the effect of the various mutations (Fig. 11). The RMSF plots permit the visualization of the flexibility for the different regions of proteins, such as the flexible loop region and terminal portions of the proteins. A lower RMSF value reflects a less flexible region, whereas a higher RMSF value indicates maximal movements in its average position during simulation. Generally, loop regions were observed to be highly flexible due to the absence of a stable secondary structure and, therefore, correspond to higher fluctuations and high RMSF values. The RBD of the spike protein consists of three loop regions, γ1 (474–485), γ2 (488–490) and γ3 (494–505), which contribute to the differential interaction of various variants to the human ACE2 receptor [49]. Previous studies have documented higher fluctuation in loop regions comprising residues 380 to 420 in wild type and mutants (Fig. 11) [62,65].
Fig. 11.
RMSF of RBD in the spike protein throughout the 100-ns MD simulation.
In the omicron complex, a sharp fluctuation was observed between residues 200–300 compared to the other variants (Fig. 10 ). The dynamic behaviour of conformational variations in the mutants revealed that the amino acid substitutions in the beta and omicron mutants caused greater fluctuations than other variants. Moreover, the loop region of RBD comprising residues 469–505 exhibited higher fluctuations in the delta and omicron variants (Fig. 11). These findings suggest that the spike protein undergoes structural modification differentially to enable their binding with hACE2 receptor and consequently increases the affinity for the host-receptor for entry into the host cells.
Fig. 10.
RMSF plot of hACE2 and RBD residues in wild type and VOCs throughout the 100 ns MD simulation.
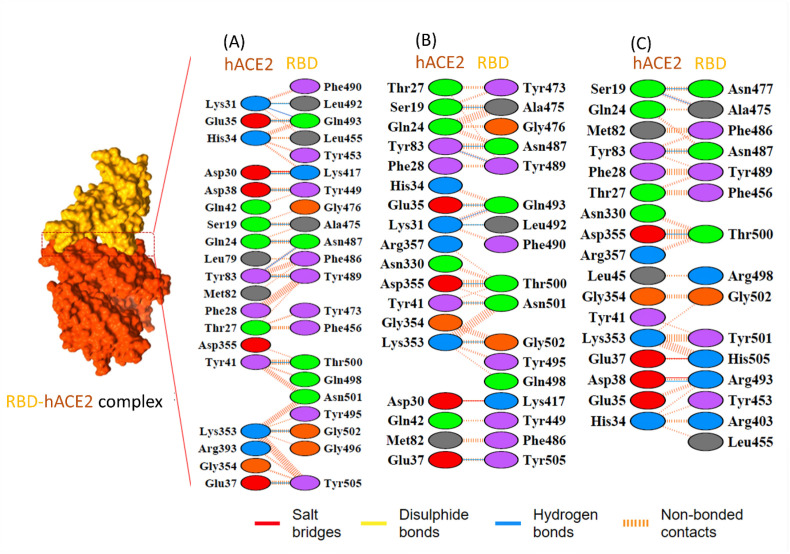
The stability of RBD-hACE2 complex can be judged through various parameters, amongst them hydrogen bonds, ionic interactions and hydrophobic interactions significantly impact the stability of the structure. During the entire simulation time, the studied systems exhibited a high proportion of hydrogen bonds that were continuously forming and breaking. The interaction analysis for wild type protein reflects that this complex is stabilized by 12 hydrogen bonds, one salt bridge and several hydrophobic interactions (Fig. 12 ). The crystal structures of the RBD complex (PDB Ids: 6M0J and 6LZG) reveal that residues K417, G446, Y449, Y453, L455, F456, A475, F486, N487, Y489, Q493, G496, Q498, T500, N501, G502 and Y505 interact directly with human ACE2 receptor. The residue N501 plays an important role in binding to the receptor and hence N501Y mutation is reported to be associated with lower free energy values, implying better binding affinity [17]. The substitutions at N501 and D614 were previously reported to enhance the infectivity in various in vitro and in vivo studies [66,67]. MD simulation indicated the stability of the complex wherein additional hydrophobic interaction with residues G476, F490, L492 and Y495 of RBD to hACE2 receptor were detected. The interacting regions in the hACE2 receptor majorly encompassing residues, 19–45, 82–83, and 352–357, formed close contacts with RBD (Fig. 13 ).
Fig. 12.
Representation of different interactions formed in the interface of hACE2 and RBD domain of wild type (A), Delta (B) and Omicron (C) variants. The amino acid residues are colored as per their properties. The positively charged residues (His,Lys,Arg) are shown in blue color, negatively charged amino acids (Asp, Glu) are shown in red, neutral amino acids (Ser, Thr, Asn, Gln) in green, aliphatic amino acid (Ala, Val, Leu, Ile, Met) in grey, aromatic amino acids (Phe, Tyr, Trp) in violet color, Pro and Gly are in orange color whereas Cys is represented in yellow color.
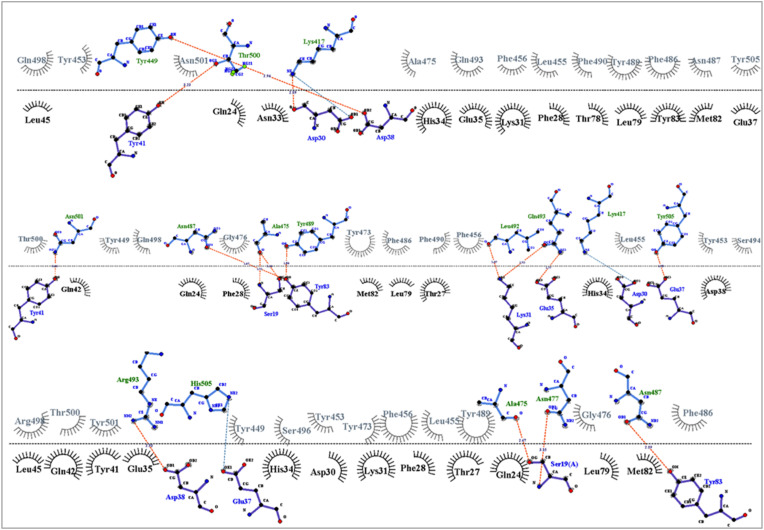
Fig. 13.
Ligplot analysis of the interactions between hACE2 and RBD of wild type (A), Delta (B) and Omicron (C) variant. The hACE2 amino acids are represented in black and RBD amino acids in grey color. The hydrogen bonds and salt bridges are shown in red and green color, respectively.
Furthermore, in Delta variant the interaction analysis between RBD and hACE2 receptor revealed the presence of 10 hydrogen bonded interactions, one ionic interaction, and several hydrophobic interactions (Table 1 ). Here, the 9 residues of RBD involved in hydrogen bond formation were A475, N487, Y489, L492, Q493, T500, Q501, G502 and Y505. The hACE2 receptor residues from 19 to 45, 82–83, N330 and 352–357 participate in close contact with RBD of delta variant similar to wild type. This complex revealed the presence of a salt bridge formed between K417 of RBD and D30 of hACE2. Similar interactions have also been reported in the experimentally determined structure [62,68].
Table 1.
Interactions formed in the interface of hACE2 and RBD domain of S protein complex in different mutants.
|
Variants |
Interface area | Number of interface residues | Number of Hydrogen Bond | Salt bridge |
|---|---|---|---|---|
| Wild | Chain A-941 Chain E−983 |
Chain A-19 Chain E−21 |
12 | 1 |
| Alpha | Chain A-941 Chain E−983 |
Chain A-19 Chain E−21 |
12 | 0 |
| Beta | Chain A-951 Chain E−963 |
Chain A-19 Chain E−18 |
9 | 0 |
| Gamma | Chain A-716 Chain E−774 |
Chain A-14 Chain E−13 |
3 | 0 |
| Delta | Chain A-606 Chain E−630 |
Chain A-9 Chain E−11 |
10 | 1 |
| Omicron | Chain A-822 Chain E−895 |
Chain A-17 Chain E−15 |
5 | 2 |
The omicron variant contains 15 mutational residues in the RBD segment of spike protein. Among these mutations, 11 substitutions (K417 N, N440K, G446S, S477 N, T478K, E484A, Q493K, G496S, Q498R, N501Y and Y505H) were observed at the interface of RBD-hACE2 complex. Due to these mutational variations, the dimeric interface attains different electrostatic surface potential that might have a supplementary effect on binding stability. Interaction analysis of this complex disclosed 5 hydrogen bonds, two salt bridges, and several hydrophobic interactions that contribute to the strength and stability of the complexed structure. The significant interactive residues from RBD (K417, N477, A475, N487 and R493 and T500) were involved in hydrogen bonded interactions while residues R493 and H505 formed salt bridges with D38 and E37 of hACE2, respectively. Our findings are consistent with previous reports stating that the main substitutions in amino acids occurring in omicron RBD (K417 N, G446S, Q493R, N501Y and Y505H) are linked with hACE2 through hydrogen bonding [62,69]. The mutations at Q493, Q498, and N501 of RBD are known to increase binding affinity as these residues are reported to form polar contacts [70]. Hence, the substitutions with polar residues at these three sites are well tolerated as minor variations in the polarity difference occurring due to the substitutions does not affect the binding affinity in SARS-CoV-2 mutants [70].
3.2.4. Binding energy analysis
Binding free energy using the MM/GBSA method is the most widely used approach to determine the binding affinity, structural stability and identification of binding hot spots. The binding free energy of the different variants was calculated using the MM/GBSA method implemented in the prime module of maestro. A total of 50 conformations that spanned the stable trajectories between 70 and 100 ns intervals of the MD production run were extracted from MM/GBSA calculations. The different energy components obtained after the calculation are presented in Table 2 . This binding energy enables the evaluation and prediction of the structure, function and interaction of the different mutants present in RBD of spike protein towards the hACE2 receptor. The maximum negative binding energy was observed for delta variant (−152.13 kcal/mol) as compared to wild type (−145.09 kcal/mol). Previous concordant findings indicated that the delta spike protein has better binding affinity than the wild type and causes severe illness due to its higher affinity towards the hACE2 receptor [59,71,72]. For other variants, namely alpha, beta, gamma and omicron, the binding energy was observed to be −132.20, −138.94, −137.89 and −140.06 kcal/mol, respectively.
Table 2.
Computed MM/GBSA binding free energies of wild type and different variants. All energy values are presented in kcal/mol.
| Model system | Binding energy | Coulomb energy | Covalent binding energy | van der Waals energy | Lipophilic energy | ΔG_Bind_Solv_GB |
|---|---|---|---|---|---|---|
| Wild | −145.09 | 1.49 | 1.27E-11 | −2083.07 | −27.49 | 23.71 |
| Alpha | −132.20 | −127.86 | −9.09E-12 | −1936.73 | −25.96 | 159.79 |
| Beta | −138.94 | −88.79 | −2.00E-11 | −1982.37 | −29.16 | 102.38 |
| Gamma | −137.90 | −181.41 | −2.73E-11 | −1888.27 | −26.04 | 203.34 |
| Delta | −152.13 | −122.33 | 1.64E-11 | −2045.12 | −29.11 | 141.90 |
| Omicron | −140.07 | −404.04 | −2.00E-11 | −2055.77 | −26.41 | 439.28 |
The RBD domain in omicron has a high proportion of hydrophobic amino acid residues located in the protein's core required for structural stability. In wild type, RBD residues 493, 498, and 501 were identified as key residues for interaction of SARS‐CoV‐2 with host receptors [63,65,67]. In omicron, mutations in these residues are likely to affect its interaction with hACE2 receptor. The calculated binding energy analysis also revealed the maximum negative binding energy for the delta variant (−152.13 kcal/mol) as compared to the omicron variant (−140.06 kcal/mol). Furthermore, interaction analysis also indicated a stable interaction for wild type, which formed 12 hydrogen bonds, one salt bridge and several hydrophobic interactions which enhanced its binding strength with the hACE2 receptor (Table 1), which correlates with our MMGBSA analysis. The binding strength for omicron for observed to be comparatively lesser, and during interaction analysis this variant made 5 hydrogen bonds and 2 salt bridges only with hACE2 receptor. Despite having comparatively better binding affinity than omicron, the replacement of delta variant by omicron can be attributed to the immune evasion of omicron in vaccinated and previously infected people as documented by other in silico, in vitro as well as in vivo studies [[73], [74], [75]].
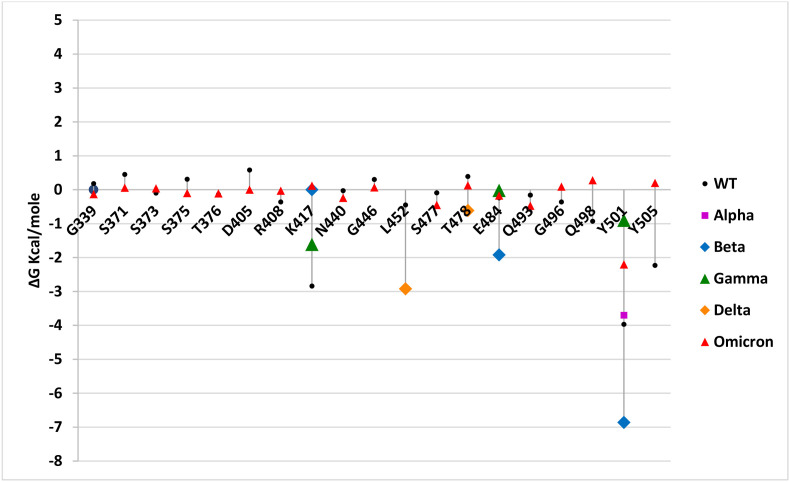
Additionally, the per residue contribution to the binding free energy was determined across the MD simulations for RBD-hACE2 complexes of wild-type and other VOCs. The ΔG of all wild-type RBD residues were plotted with respect to the mutated residues for alpha, beta, gamma, delta and omicron variants (Fig. 14 ). The significantly decreased value of ΔG at the amino acid positions 417, 501, 452, 478, 484 and 501 denotes the contribution of these mutated residues in the binding affinity in different variants. The per residue contribution of mutated residues in the omicron variant displayed positive and negative discrepancies in ΔG compared to the wild-type protein (Fig. 14 and File S3). The per residue contribution energy analysis suggested that the energy contribution of the residues 440, 498 and 505 are maximal in the omicron variant. Mutation at 478 residue in RBD of delta variant exists in a more solvent-oriented region that facilitates its interaction with hACE2 due to the resultant longer side chain length of the substituted residue [62]. The per residue contribution energy analysis suggests that all the mutations present in VOCs contribute significantly towards the free energy change (Fig. 14). Hence, may be responsible for its increased affinity towards the hACE2 receptor. The maximum negative binding energy perceived for the delta variant also correlates with the per residue contribution energy analysis as compared to other variants (Supplementary file S4).
Fig. 14.
Per residue free energy contribution of mutated residues in VOCs and wild type RBDs in simulated complexed structure with hACE2 receptor. Only the ΔG values for mutated residues in more than 50% of alpha, beta, gamma, delta and omicron RBDs are plotted to compare the variations in free energies of mutated residues and their contribution to overall binding affinity of these VOCs with hACE2.
Further analyses using structural bioinformatics and experimental biochemical methodologies are prerequisites to gain deeper insights into the structural modifications and fluctuations in specific pathogen–host receptor interactions triggering higher transmissibility of the SARS-CoV-2 omicron variant.
4. Conclusion
SARS-COV-2 exhibits an extensive genomic diversity pattern accompanied by major nonsynonymous mutations in the spike glycoprotein region. This mutational landscape is associated with differential interactions with the host receptor and antibodies and thus may induce higher infectivity and immunity evasion by the mutants. The increased binding affinity of delta strain followed by omicron compared to other variants and wild type hints to the opportunity for high transmissibility and quick spread of these two variants. Binding free energy analysis of the RBD-receptor complex exposed that the delta variant had the maximum negative score, specifying that it bound to hACE2 more strongly than the other. This can offer a clue for drug discovery endeavours by elucidating residues that could be targeted for disrupting this interface. Overall, this study provides molecular insights for the better affinity of the delta variant for human ACE2 receptor. Overall, this study provides a link between mutations in VOCs and their binding affinity to hACE2 receptor at the molecular level. Although a plethora of WGS data is available, only a few sequences are submitted with clinical metadata. Laboratories should be encouraged to submit clinical metadata along with the genome sequences so that the clinical correlation with the mutations can be carried out in time. This will help to warn the healthcare agencies about the future emerging variants. Due to repeated deadly surges caused by variants from time to time, there is a need to find prevention strategies by designing potent vaccines against the conserved domains.
Data availability statement
The data as well as coordinates for the three-dimensional model structures used in the study will be made available on AIIMS website www.aiims.edu after the publication of the study.
Funding statement
The authors did not receive any grant for the present work.
Ethical approval
The study is ethically approved by institutional ethics committee (Ref no. IEC-46/14.01.2022).
Patient consent to participate
Not Applicable (There were no patients or any patient sample included in the present study).
Permission to reproduce materials from all the sources
The present study includes data retrieval from GISAID, Nextclade and RCSB which are freely available public repositories and permit free data access for analysis.
Consent for publication
All the authors have checked the final manuscript and given their consent for publication.
Summary
The emergence of COVID-19 as a pandemic has significantly affected the research perspective of healthcare workers, as fast and instantaneous management measures are required to deal with the rapidly evolving pathogen. The mutations in the receptor-binding domain of spike protein exert a selective pressure, forcing the development of novel variants. Receptor binding motif (RBM) is the key motif of RBD, which directly binds to hACE2 and harbours the majority of mutations determining the severity of concerned variants.
In the current research work, we have described the sequence variations in different lineages while focusing on the mutations characterised as variants of concern (VOCs), variants of interest (VOIs) and variants under monitoring (VUMs). Hence, a combination of sequence and structure analysis was performed to assess the effect of these variations on the interaction with host receptor (hACE-2) to understand the significance of these mutations.
The molecular modelling and dynamics studies were then performed to gain deeper insights into the impact of these mutations on the interaction of viruses with the human host by evaluating the binding affinity with hACE2. Per residue contribution was calculated to find the role of mutation hotspots in determining the severity of the disease. All the mutation hotspots including D614G, N501Y, E484 A/K, P681H, T478K and K417T play a crucial role in interaction with the host receptor and their comparative contribution revealed that the more number of mutations in omicron and delta variant were contributing to the lower binding energy as compared to other variants. The increased binding affinity of delta strain followed by omicron compared to other variants and wild type designated the opportunity for high transmissibility and quick spread of these two variants.
Declaration of competing interest
All the authors do not have any competing interest.
Acknowledgements
We hereby acknowledge GISAID, Nextclade and RCSB for public sharing of the data.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2022.106129.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91 doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicola M., Alsafi Z., Sohrabi C., Kerwan A., Al-Jabir A., Iosifidis C., Agha M., Agha R. The socio-economic implications of the coronavirus pandemic (COVID-19): a review. Int. J. Surg. 2020;78 doi: 10.1016/j.ijsu.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yi H., Wang J., Wang J., Lu Y., Zhang Y., Peng R., Lu J., Chen Z. The emergence and spread of novel SARS-CoV-2 variants. Front. Public Health. 2021;9 doi: 10.3389/fpubh.2021.696664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579 doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chilamakuri R., Agarwal S. Covid-19: characteristics and therapeutics. Cells. 2021;10 doi: 10.3390/cells10020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tripathi M.K., Singh P., Sharma S., Singh T.P., Ethayathulla A.S., Kaur P. Identification of bioactive molecule from Withania somnifera (Ashwagandha) as SARS-CoV-2 main protease inhibitor. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1790425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karn V., Ahmed S., Tsai L.-W., Dubey R., Ojha S., Singh H.N., Kumar M., Gupta P.K., Sadhu S., Jha N.K., Kumar A., Pandit S., Kumar S. Extracellular vesicle-based therapy for COVID-19: promises, challenges and future prospects. Biomedicine. 2021;9 doi: 10.3390/biomedicines9101373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar M., Roy A., Rawat R.S., Alok A., Tetala K.K.R., Biswas N.R., Kaur P., Kumar S. Identification and structural studies of natural inhibitors against SARS-CoV-2 viral RNA methyltransferase (NSP16) J. Biomol. Struct. Dyn. 2021:1–11. doi: 10.1080/07391102.2021.1997821. [DOI] [PubMed] [Google Scholar]
- 9.Tripathi M.K., Sharma S., Singh T.P., Ethayathulla A.S., Kaur P. Stud. Comput. Intell. 2021. Computational intelligence in drug repurposing for COVID-19. [DOI] [Google Scholar]
- 10.Tao K., Tzou P.L., Nouhin J., Gupta R.K., de Oliveira T., Kosakovsky Pond S.L., Fera D., Shafer R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021;22 doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta D., Sharma P., Singh M., Kumar M., Ethayathulla A.S., Kaur P. Structural and functional insights into the spike protein mutations of emerging SARS-CoV-2 variants. Cell. Mol. Life Sci. 2021;78 doi: 10.1007/s00018-021-04008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lauring A.S., Frydman J., Andino R. The role of mutational robustness in RNA virus evolution. Nat. Rev. Microbiol. 2013;11 doi: 10.1038/nrmicro3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X. Concerns on the multiple nomenclature systems for SARS-CoV-2. J. Med. Virol. 2022;94 doi: 10.1002/jmv.27406. [DOI] [PubMed] [Google Scholar]
- 14.GISAID. GISAID Initiative Adv. Virus Res. 2020;2008 [Google Scholar]
- 15.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. NextStrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34 doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rambaut A., Holmes E.C., O'Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5 doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data – from vision to reality. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konings F., Perkins M.D., Kuhn J.H., Pallen M.J., Alm E.J., Archer B.N., Barakat A., Bedford T., Bhiman J.N., Caly L., Carter L.L., Cullinane A., de Oliveira T., Druce J., El Masry I., Evans R., Gao G.F., Gorbalenya A.E., Hamblion E., Herring B.L., Hodcroft E., Holmes E.C., Kakkar M., Khare S., Koopmans M.P.G., Korber B., Leite J., MacCannell D., Marklewitz M., Maurer-Stroh S., Rico J.A.M., Munster V.J., Neher R., Munnink B.O., Pavlin B.I., Peiris M., Poon L., Pybus O., Rambaut A., Resende P., Subissi L., Thiel V., Tong S., van der Werf S., von Gottberg A., Ziebuhr J., Van Kerkhove M.D. SARS-CoV-2 Variants of Interest and Concern naming scheme conducive for global discourse. Nat. Microbiol. 2021;6 doi: 10.1038/s41564-021-00932-w. [DOI] [PubMed] [Google Scholar]
- 19.Parums D.V. Editorial: revised world health organization (who) terminology for variants of concern and variants of interest of sars-cov-2. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2021;27 doi: 10.12659/MSM.933622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395 doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papageorgiou A.C., Mohsin I. The SARS-CoV-2 spike glycoprotein as a drug and vaccine target: structural insights into its complexes with ACE2 and antibodies. Cells. 2020;9 doi: 10.3390/cells9112343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3 doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koley T., Madaan S., Chowdhury S.R., Kumar M., Kaur P., Singh T.P., Ethayathulla A.S. Structural analysis of COVID-19 spike protein in recognizing the ACE2 receptor of different mammalian species and its susceptibility to viral infection. 3 Biotech. 2021;11 doi: 10.1007/s13205-020-02599-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polydorides S., Archontis G. Computational optimization of the SARS-CoV-2 receptor-binding-motif affinity for human ACE2. Biophys. J. 2021;120 doi: 10.1016/j.bpj.2021.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Domingo P., de Benito N. Alpha variant SARS-CoV-2 infection: how it all starts. EBioMedicine. 2021;74 doi: 10.1016/j.ebiom.2021.103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duong D. Alpha, Beta, Delta, Gamma: what's important to know about SARS-CoV-2 variants of concern? CMAJ (Can. Med. Assoc. J.) 2021;193 doi: 10.1503/cmaj.1095949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin S., Cui M., Sun S., Zhou J., Du Z., Cui Y., Fan H. Genome characterization and potential risk assessment of the novel SARS-CoV-2 variant omicron (B.1.1.529) Zoon. 2021;1 doi: 10.15212/zoonoses-2021-0024. [DOI] [Google Scholar]
- 30.WHO . Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. Who; 2021. [Google Scholar]
- 31.Torjesen I. Covid-19: omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear. BMJ. 2021;375 doi: 10.1136/bmj.n2943. [DOI] [PubMed] [Google Scholar]
- 32.Chen J., Wang R., Gilby N.B., Wei G.W. Omicron variant (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. J. Chem. Inf. Model. 2022;62 doi: 10.1021/acs.jcim.1c01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar S., Thambiraja T.S., Karuppanan K., Subramaniam G. Omicron and Delta variant of SARS-CoV-2: a comparative computational study of spike protein. J. Med. Virol. 2022;94 doi: 10.1002/jmv.27526. [DOI] [PubMed] [Google Scholar]
- 34.Raman R., Patel K.J., Ranjan K. Covid-19: unmasking emerging sars-cov-2 variants, vaccines and therapeutic strategies. Biomolecules. 2021;11 doi: 10.3390/biom11070993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rehman M., Tauseef I., Aalia B., Shah S.H., Junaid M., Haleem K.S. Therapeutic and vaccine strategies against SARS-CoV-2: past, present and future. Future Virol. 2020;15 doi: 10.2217/fvl-2020-0137. [DOI] [Google Scholar]
- 36.Aksamentov I., Roemer C., Hodcroft E., Neher R. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 2021;6 doi: 10.21105/joss.03773. [DOI] [Google Scholar]
- 37.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30 doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsueng G., Mullen J., Alkuzweny M., Cano M., Rush B., Haag E., Curators O., Alaa Abdel L., Zhou X., Qian Z., Andersen K.G., Wu C., Su A.I., Gangavarapu K., Hughes L.D. Outbreak.info Research Library: a standardized, searchable platform to discover and explore COVID-19 resources and data. BioRxiv Prepr. Serv. Biol. 2022 doi: 10.1101/2022.01.20.477133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsueng G., Mullen J., Alkuzweny M., Cano M., Rush B., Haag E., Curators O., Alaa Abdel L., Zhou X., Qian Z., Andersen K.G., Wu C., Su A.I., Gangavarapu K., Hughes L.D. Outbreak.info Research Library: a standardized, searchable platform to discover and explore COVID-19 resources and data. BioRxiv Prepr. Serv. Biol. 2022 doi: 10.1101/2022.01.20.477133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huddleston J., Hadfield J., Sibley T., Lee J., Fay K., Ilcisin M., Harkins E., Bedford T., Neher R., Hodcroft E. Augur: a bioinformatics toolkit for phylogenetic analyses of human pathogens. J. Open Source Softw. 2021;6 doi: 10.21105/joss.02906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graham Carlos W., Dela Cruz C.S., Cao B., Pasnick S., Jamil S. Novel wuhan (2019-NCoV) coronavirus. Am. J. Respir. Crit. Care Med. 2020;201 doi: 10.1164/rccm.2014P7. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen L.T., Schmidt H.A., Von Haeseler A., Minh B.Q., Iq-Tree A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32 doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar S., Stecher G., Suleski M., Hedges S.B. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017;34 doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- 44.Letunic I., Bork P. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.UniProt The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jun Z., Yongfei C., Tianshu X., Jianming L., Hanqin P., M S.S., M W.R., Sophia R.-V., Haisun Z., N W.A., Wei Y., Piotr S., Bing C. Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science. 2021;372:525–530. doi: 10.1126/science.abf2303. 80-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 48.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66 doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laskowski R.A., Jabłońska J., Pravda L., Vařeková R.S., Thornton J.M. PDBsum: structural summaries of PDB entries. Protein Sci. 2018;27:129–134. doi: 10.1002/pro.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balloux F., Tan C., Swadling L., Richard D., Jenner C., Maini M., van Dorp L. Vol. 3. Oxford Open Immunol; 2022. (The Past, Current and Future Epidemiological Dynamic of SARS-CoV-2). iqac003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thye A.Y.K., Law J.W.F., Pusparajah P., Letchumanan V., Chan K.G., Lee L.H. Emerging sars-cov-2 variants of concern (Vocs): an impending global crisis. Biomedicines. 2021;9 doi: 10.3390/biomedicines9101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Obermeyer F., Jankowiak M., Barkas N., Schaffner S.F., Pyle J.D., Yurkovetskiy L., Bosso M., Park D.J., Babadi M., MacInnis B.L., Luban J., Sabeti P.C., Lemieux J.E. Analysis of 6.4 million SARS-CoV-2 genomes identifies mutations associated with fitness. Science. 2022;376:1327–1332. doi: 10.1126/science.abm1208. 80-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laffeber C., de Koning K., Kanaar R., Lebbink J.H.G. Experimental evidence for enhanced receptor binding by rapidly spreading SARS-CoV-2 variants. J. Mol. Biol. 2021;433 doi: 10.1016/j.jmb.2021.167058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., Mirchandani D., Scharton D., Bilello J.P., Ku Z., An Z., Kalveram B., Freiberg A.N., Menachery V.D., Xie X., Plante K.S., Weaver S.C., Shi P.Y. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592 doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nicholas K.B., Nicholas H.B.J. GeneDoc: a tool for editing and annotating multiple sequence alignments. Ieee Softw. 1997 [Google Scholar]
- 56.Hahn G., Lee S., Weiss S.T., Lange C. Unsupervised cluster analysis of SARS-CoV-2 genomes reflects its geographic progression and identifies distinct genetic subgroups of SARS-CoV-2 virus. Genet. Epidemiol. 2021;45 doi: 10.1002/gepi.22373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou Y., Zhang S., Chen J., Wan C., Zhao W., Zhang B. Vol. 40. Nan Fang Yi Ke Da Xue Xue Bao; 2020. (Analysis of Variation and Evolution of SARS-CoV-2 Genome). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rochman N.D., Wolf Y.I., Faure G., Mutz P., Zhang F., Koonin E.V. Ongoing global and regional adaptive evolution of SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 2021;118 doi: 10.1073/pnas.2104241118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stern A., Fleishon S., Kustin T., Dotan E., Mandelboim M., Erster O., Mendelson E., Mor O., Zuckerman N.S. MedRxiv; 2021. The Unique Evolutionary Dynamics of the SARS-CoV-2 Delta Variant. [Google Scholar]
- 60.Bansal K., Kumar S. Mutational cascade of SARS-CoV-2 leading to evolution and emergence of omicron variant. Virus Res. 2022;315 doi: 10.1016/j.virusres.2022.198765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen J., Wang R., Wang M., Wei G.W. Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 2020;432 doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Celik I., Khan A., Dwivany F.M., Fatimawali, Wei D.Q., Tallei T.E. Computational prediction of the effect of mutations in the receptor-binding domain on the interaction between SARS-CoV-2 and human ACE2. Mol. Divers. 2022 doi: 10.1007/s11030-022-10392-x. [DOI] [PubMed] [Google Scholar]
- 63.Shahhosseini N., Babuadze G., Wong G., Kobinger G.P. Mutation signatures and in silico docking of novel sars-cov-2 variants of concern. Microorganisms. 2021;9 doi: 10.3390/microorganisms9050926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Teng S., Sobitan A., Rhoades R., Liu D., Tang Q. Systemic effects of missense mutations on SARS-CoV-2 spike glycoprotein stability and receptor-binding affinity. Briefings Bioinf. 2021;22 doi: 10.1093/bib/bbaa233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suleman M., Yousafi Q., Ali J., Ali S.S., Hussain Z., Ali S., Waseem M., Iqbal A., Ahmad S., Khan A., Wang Y., Wei D.Q. Bioinformatics analysis of the differences in the binding profile of the wild-type and mutants of the SARS-CoV-2 spike protein variants with the ACE2 receptor. Comput. Biol. Med. 2021;138 doi: 10.1016/j.compbiomed.2021.104936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu H., Zhang Q., Wei P., Chen Z., Aviszus K., Yang J., Downing W., Jiang C., Liang B., Reynoso L., Downey G.P., Frankel S.K., Kappler J., Marrack P., Zhang G. The basis of a more contagious 501Y.V1 variant of SARS-CoV-2. Cell Res. 2021;31 doi: 10.1038/s41422-021-00496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Zhang L., Li X., Huang W., Wang Y. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182 doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gurung A.B., Ali M.A., Lee J., Farah M.A., Al-Anazi K.M., Al-Hemaid F., Sami H. Structural and functional insights into the major mutations of SARS-CoV-2 Spike RBD and its interaction with human ACE2 receptor. J. King Saud Univ. Sci. 2022;34 doi: 10.1016/j.jksus.2021.101773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumar S., Karuppanan K., Subramaniam G. Omicron (BA.1) and sub‐variants (BA.1.1, BA.2, and BA.3) of SARS‐CoV‐2 spike infectivity and pathogenicity: a comparative sequence and structural‐based computational assessment. J. Med. Virol. 2022 doi: 10.1002/jmv.27927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., King N.P., Veesler D., Bloom J.D. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182 doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee K.S., Wong T.Y., Russ B.P., Horspool A.M., Miller O., Rader N., Givi J.P., Winters M.T., Wong Z.Y., Cyphert H.A., Denvir J., Stoilov P.G., Barbier M., Roan N., Amin M.S., Martinez I., Bevere J.R., Damron F.H. SARS-CoV-2 Delta variant induces enhanced pathology and inflammatory responses in K18-hACE2 mice. bioRxiv. 2022 doi: 10.1371/journal.pone.0273430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Otto S.P., Day T., Arino J., Colijn C., Dushoff J., Li M., Mechai S., Van Domselaar G., Wu J., Earn D.J.D., Ogden N.H. The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr. Biol. 2021;31:R918–R929. doi: 10.1016/j.cub.2021.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang R., Zhang Q., Zhang R., Aw Z.Q., Chen P., Wong Y.H., Hong J., Ju B., Shi X., Ding Q., Zhang Z., Chu J.J.H., Zhang L. SARS-CoV-2 omicron variants reduce antibody neutralization and acquire usage of mouse ACE2. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.854952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Contractor D., Globisch C., Swaroop S., Jain A. Structural basis of Omicron immune evasion: a comparative computational study. Comput. Biol. Med. 2022;147 doi: 10.1016/j.compbiomed.2022.105758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.H., Porrot F., Staropoli I., Lemoine F., Péré H., Veyer D., Puech J., Rodary J., Baele G., Dellicour S., Raymenants J., Gorissen S., Geenen C., Vanmechelen B., Wawina -Bokalanga T., Martí-Carreras J., Cuypers L., Sève A., Hocqueloux L., Prazuck T., Rey F.A., Simon-Loriere E., Bruel T., Mouquet H., André E., Schwartz O. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602 doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data as well as coordinates for the three-dimensional model structures used in the study will be made available on AIIMS website www.aiims.edu after the publication of the study.