Abstract
The plasmid-borne cfr gene, mediating multiple drug resistance (MDR), has been observed in many Gram-positive bacteria. The prevalence of cfr and its co-occurrence with additional antimicrobial resistance (AMR) determinants in Escherichia coli is an ongoing issue. Additionally, the prevalence and transfer mechanism of the cfr gene remain partially investigated. Here, eight cfr-positive E. coli strains were screened using PCR from an extensive collection of E. coli (n = 2,165) strains isolated from pigs and chickens in 2021 in China, with a prevalence rate of 0.37%. All of them were MDR and resistant to florfenicol and tetracycline. These strains can transfer the cfr gene to E. coli J53 by conjugation (1.05 × 10−1 – 1.01 × 10−6). Moreover, the IncX4 plasmid p727A3-62 K-cfr (62,717 bp) harboring cfr in strain EC727A3 was confirmed using Oxford Nanopore Technology. The unknown type plasmid p737A1-27K-cfr (27,742 bp) harboring cfr in strain EC737A1 was also identified. Notably, it was verified by PCR that three of the eight E. coli strains were able to form the cfr-IS26 circular intermediate. It was 2,365 bp in length in strains EC727A3 and ECJHZ21-173, and 2,022 bp in length in EC737A1. Collectively, this study demonstrated that IS26 plays a vital role in transmitting the MDR gene cfr in E. coli via conjugation and provided updated knowledge regarding cfr in E. coli in Eastern China.
Keywords: Escherichia coli, florfenicol, cfr, antimicrobial resistance, circular intermediate
Introduction
Antimicrobial resistance (AMR) is a serious threat to global public health. The capability of bacteria to acquire and transfer antibiotic resistance and virulence genes is dangerous and urgently crucial to both human and animal health. The multidrug-resistance (MDR) gene cfr encodes 23S rRNA methylase, which is resistant to five classes of antimicrobials, including phenols, lincosamides, oxazolidinones, pleuromutilin, and streptomycin A class antibiotics (PhLOPSA phenotype) (Kehrenberg et al., 2005; Long et al., 2006), and has decreased susceptibility to the 16-membered macrolides spiramycin, and josamycin (Smith and Mankin, 2008). For the first time, the discovery of multiple AMR gene cfr in Staphylococcus bovis isolates has attracted attention in a global sense (Schwarz et al., 2000). Insertion sequences and transposons are associated with the spread of cfr in Gram-negative and Gram-positive bacteria, including but not limited to, Enterococcus, Bacillus, Jeotgalicoccus, Macrococcus, Pasteurella multocida, Vibrio diabolicus, Escherichia coli, Streptococcus, and Proteus vulgaris (Dai et al., 2010; Wang et al., 2011, 2012a,b, 2013; Chen et al., 2020a,b; Liu et al., 2022), considering that cfr is usually located on plasmids containing related insertion sequences and transposons (Shen et al., 2013; Partridge et al., 2018).
Based on published articles to date, a total of 112 strains of E. coli containing the MDR gene cfr have been identified in various provinces of China; the primary source of these E. coli strains are pigs, which may be related to the overuse of florfenicol for disease prevention and treatment in pig farms (Wang et al., 2012a, 2018; Zhang et al., 2014, 2015, 2016; Liu et al., 2017; Ma et al., 2021; Tang et al., 2021). For example, it coexists with the extended-spectrum-β-lactamase gene blaCTX-M-14b, tigecycline resistance gene tet(X4), colistin resistance gene mcr-1, and florfenicol resistance gene floR (Zhang et al., 2015; Ma et al., 2021; Tang et al., 2021). These plasmids carrying the cfr gene in E. coli belong to the plasmid replicon type, including IncX4, IncA/C, IncF14: A-: B-, IncN-IncX1 (Zhang et al., 2014; Sun et al., 2015; Wang et al., 2018; Tang et al., 2021), of which, IncX4 plasmids are frequently detected in China (Wang et al., 2018). However, few studies have investigated the mechanisms of transmission of the MDR gene cfr in E. coli.
In this study, the prevalence and characteristics of E. coli cfr-positive strains in food animals were investigated. All cfr-positive strains were further sequenced by Illumina or Nanopore platforms, and the cfr-harboring plasmids were also identified and characterized. It was confirmed that circular intermediate and conjugation transfer promoted the transfer of the cfr gene. Our study highlights the severe threat posed by cfr-carrying E. coli to public health and provides new insight on its role in dissemination.
Materials and methods
Screening of the cfr gene
From May to December 2021, 2,103 E. coli strains were isolated from 11 cities in Zhejiang, including Hangzhou, Jinhua, Jiaxing, Qvzhou, Ningbo, Taizhou, Shaoxing, Zhoushan, Lishui, Wenzhou and Huzhou, including 1,186 strains from pigs, 904 from strains in chickens and 13 strains from ducks. Thirty-six E. coli strains were isolated from Jiangxi Province, 25 E. coli strains were isolated from Hunan Province, and one was isolated from Anhui Province (Table 1). PCR screening of isolated strains was performed to obtain the prevalence of the cfr gene in the above E. coli isolates with primer sequences (F: GTGAAGCTCTAGCCAACCGTC; R: GCAGCGTCAATATCAATCCC), as described previously (Osman et al., 2019).
Table 1.
Strain information for screening the cfr gene.
| Province | Animal | Number |
|---|---|---|
| Zhejiang | Pig | 1,186 |
| Chicken | 904 | |
| Duck | 13 | |
| Jiangxi | Duck | 36 |
| Hunan | Chicken | 25 |
| Anhui | Pig | 1 |
| Total | – | 2,165 |
Antimicrobial Susceptibility Test
Escherichia coli was inoculated on Luria-Bertani (LB) agar medium for pure culture, according to the micro-dilution method recommended in the M100-S31 document of the American Committee for Clinical Laboratory Standardization (CLSI) (Humphries et al., 2021; Tang et al., 2022b). The antimicrobial susceptibility of E. coli to 13 tested antibiotics were, ampicillin (2–128 μg/ml), amoxicillin-clavulanate acid (4/2–128/64 μg/ml), cefotaxime (0.06–64 μg/ml), meropenem (0.5–16 μg/ml), amikacin (2–64 μg/ml), gentamicin (0.25–32 μg/ml), colistin (0.125–8 μg/ml), ceftiofur (0.25–32 μg/ml), ciprofloxacin (0.06–8 μg/ml), trimethoprim-sulfamethoxazole (0.5/9.5–16/304 μg/ml), tetracycline (0.25–64 μg/ml), tigecycline (0.25–32 μg/ml), and florfenicol (2–128 μg/ml). E. coli ATCC 25922 served as quality control bacteria.
Whole-genome sequencing
To further understand the genetic background of themultiple AMR gene cfr in E. coli, a genomic DNA extraction kit (Generay, Shanghai, China) was used to extract bacterial genomic DNA from all cfr positive strains for whole-genome sequencing (WGS). An Illumina sequencing library was generated using the NEXTflex DNA sequencing kit (Bioo Scientific, Austin, United States). Illumina paired-end sequencing was performed using the HiSeq-PE150 strategy, and the readings were filtered using fastp v0.12. Clean data were reconstructed using CLC Genomic Workbench 12.0. Prototypical strains were simultaneously whole-genome sequenced on the Oxford Nanopore GridION platform (Oxford, United Kingdom). The above genomic DNA library was prepared using the SQKLSK109 kit (Oxford Nanopore Technologies, Oxford, United Kingdom). Guppy v3.2.4 was used for base invocation and removal of adapter sequences. Sequences were assembled from scratch using a mixture of short and long reads from the Unicycler v0.4.4 pipeline (Wick et al., 2017). The reconstruction of plasmids from next generation sequence pair-end datasets was performed by PLACNETw (Vielva et al., 2017).
Antimicrobial resistance gene, virulence gene, phylogenetic tree and plasmid analysis
Acquired AMR genes and chromosomal mutations were predicted using ResFinder 4.11 with a percentage identification threshold of 90% and a minimum coverage length of 60%. The virulence genes were predicted using VirulenceFinder 2.0.2 Plasmid replicon type identification using PlasmidFinder 2.13 with a percentage identification threshold of 95% and percentage coverage length of 60%. Multilocus sequence typing (MLST) was performed using MLST 2.0.4 Phylogenetic analysis of genomes and plasmids based on maximum likelihood was performed using kSNP3 (Gardner et al., 2015). Easyfig 2.2.3 was used to compare the gene–environment (Sullivan et al., 2011). BRIG was used to plot circles of multiple plasmids for comparison (Alikhan et al., 2011).
Conjugation transfer assay
The E. coli strain J53 was selected as the recipient strain, and cfr-positive E. coli was selected as the donor strain. Florfenicol and sodium azide were added for the selection. First, we determined that cfr-positive E. coli could not be grown on LB plates containing 100 mg/l sodium azide, and J53 could no longer be grown on LB plates containing10 mg/l florfenicol. The method of conjugation transfer was mentioned in previous reports (Xu et al., 2021; Tang et al., 2022a). The donor bacteria and recipient bacteria were inoculated into LB broth and cultured on a shaker for 4–6 h. One milliliter of the bacterial solution was taken for centrifugation, and the donor and recipient bacteria were added to the LB plate overnight at 37°C. After gradient dilution with PBS, they were inoculated onto LB square plates containing 10 mg/l florfenicol and 100 mg/l sodium azide. Finally, single colonies that grew after mating were identified via PCR to exclude false-positive cases.
Detection of IS26-mediated circularization with a cfr-containing gene
To verify the circularization potential of the IS26 flanking fragments in a plasmid, a pair of primers were designed and amplified by PCR to observe whether they could form the circular intermediate of cfr-IS26. The primers used to identify the cfr-IS26 circular intermediate are shown (F: GTTGCCTGGTGTAAATGATTC; R: CTGCTAAGAGCTTGATATTC). The size of the cfr-IS26 circular intermediate was determined by Sanger sequencing.
Results
Antimicrobial susceptibility test of E. coli carrying the cfr gene
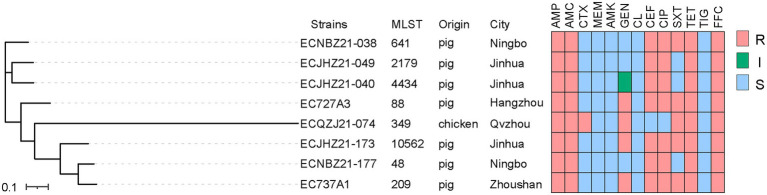
Eight cfr-positive isolates were identified from 2,165 E. coli isolates (1,187 from pigs, 929 from chickens, and 49 from ducks), and the prevalence was 0.37% (Table 2). Seven of the cfr-positive strains were isolated from pigs, and one strain was isolated from chicken. The AST results of eight positive E. coli isolates showed that all strains were resistant to ampicillin, amoxicillin-clavulanic acid, tetracycline, and florfenicol (Figure 1; Supplementary Table 1). All the strains were sensitive to colistin, meropenem, tigecycline, and amikacin.
Table 2.
cfr-positive E. coli isolates in this study.
| Strains | Source | Animal | City | Plasmid | Accession number |
|---|---|---|---|---|---|
| ECJHZ21-040 | Feces | Pig | Jinhua | – | JAMYDT000000000 |
| ECJHZ21-049 | Feces | Pig | Jinhua | – | JAMYDS000000000 |
| ECNBZ21-038 | Feces | Pig | Ningbo | – | JAMYDR000000000 |
| ECNBZ21-177 | Feces | Pig | Ningbo | – | JAMYDQ000000000 |
| ECJHZ21-173 | Feces | Pig | Jinhua | – | JAMYDP000000000 |
| ECQZJ21-074 | Feces | Chicken | Qvzhou | – | JAMYDO000000000 |
| EC727A3 | Feces | Pig | Hangzhou | p727A3-62K-cfr | CP100062-CP100071 |
| EC737A1 | Gut | Pig | Zhoushan | p737A1-27K-cfr | CP100005-CP100012 |
| ECJHZ21-058 | Feces | Pig | Jinhua | – | JAMYDT000000001 |
Figure 1.
The phylogenetic tree and AMR of E. coli isolates. ‘S’ represents susceptibility, ‘I’ represents intermediate, and ‘R’ represents resistance. Penicillins: AMP, ampicillin; AMC, amoxicillin-clavulanic acid; Cephalosporins: CTX, cefotaxime; CEF, ceftiofur; Carbapenems: MEM, meropenem; Aminoglycosides: AMK, amikacin; GEN, gentamicin; Polypeptides: CL, colistin; Quinolones: CIP, ciprofloxacin; Sulfonamides: SXT, trimethoprim-sulfamethoxazole; Tetracyclines: TET, tetracycline; TIG, tigecycline; Chloramphenicol: FFC, florfenicol.
Molecular characterization and conjugative transfer of cfr-positive isolates
The contigs carrying cfr gene assembled by the second generation sequence are between 1 and 3 Kb in length (Supplementary Figure 1). The cfr-harboring E. coli strains isolated from chicken and pig belonged to different branches. Among the strains ECJHZ21-040, ECJHZ21-049, and ECNBZ21-038 were clustered together. Additionally, ECQZJ21-074 belonged to independent lineages, and there were differences between them and in the seven strains mentioned above (Figure 1). The eight E. coli isolates had distinct sequence types (STs) with ST641, ST2179, ST4434, ST88, ST349, ST10562, ST48, and ST209, indicating that cfr was widely distributed in E. coli with different genetic backgrounds.
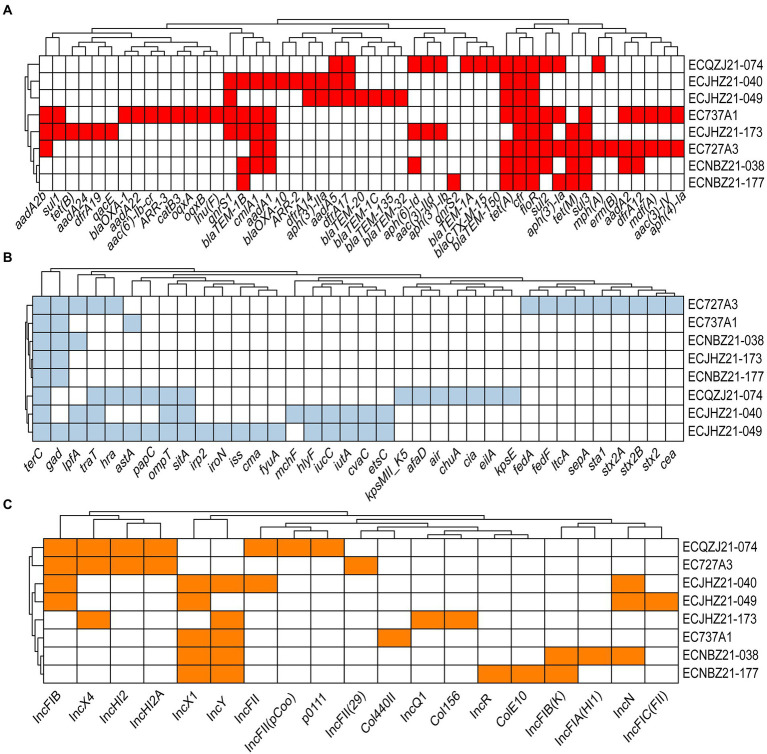
A total of 49 types of AMR determinants within 10 classes of antibiotics were detected (Figure 2A). In addition, there were two florfenicol genes (cfr, floR), three tetracycline genes (tet(A), tet(B), and tet(M)), 10 β-lactam genes (blaCTX-M-15, blaTEM-150, blaTEM-1A, blaTEM-1B, blaOXA-10, blaTEM-1C, blaOXA-20, blaOXA-135, blaTEM-32, blaOXA-1), two quinolone genes (qnrS1, qnrS2), two rifamycin genes (ARR-2, ARR-3), three macrolide genes (mph(A), mdf(A), and erm(B)), one lincosamide gene (Inu(F)), six folate pathway antagonist genes (sul1, sul2, sul3, dfrA12, dfrA17, dfrA19), 14 aminoglycoside genes (aadA2b, aph(4)-Ia, aac(3)-IV, aadA2, aph(3′)-Ia, aph(3″)-Ib, aac(3)-IId, aph(6)-Id, aadA5, aph(3′)-IIa, aadA1, aac(6′)-Ib-cr, aadA22, aadA24) and some additional AMR determinants (Figure 2A). The virulence genes of the strains included terC, traT, gad, lpfA, ompT, sitA, astA, hra, etc. (Figure 2B). Among them, astA is a virulence gene encoding heat-stable enterotoxin of enteroaggregative E. coli, which may produce related toxins with the possibility of pathogenicity. Importantly, strain EC727A3 contains the virulence genes stx2A and stx2B that produce Shiga toxin, which may cause self-limiting diarrhoeal disease and sometimes bloody diarrhea as well as complications such as hemorrhagic colitis and hemolytic uremic syndrome (HUS) (Fitzpatrick, 1999; Launders et al., 2016; Mcfarland et al., 2017). Plasmid replicons include 19 types such as IncFIC(FII), IncN, IncFIA(HI1), IncFIB(K), ColE10, IncR, Col156, IncQ1, Col440II, IncFII(29), p0111, IncFII(pCoo), IncFII, IncY, IncX1, IncHI2A, IncHI2, IncX4, and IncFIB. The plasmid types of the eight isolates remained genetically diverse (Figure 2C).
Figure 2.
The AMR genes, plasmid replicons, and virulence genes in cfr-positive E. coli. (A) Acquired AMR gene in cfr-positive E. coli. Red indicates the AMR gene. (B) Virulence gene in cfr-positive E. coli. Light blue represents the presence of the virulence gene, and white represents the absence of the virulence gene. terC, tellurium ion resistance; gad, glutamate decarboxylase; lpfA, long polar fimbriae; traT, outer membrane protein complement resistance; hra, heat-resistant agglutinin; astA, EAST-1 heat-stable toxin; papC, outer membrane usher P fimbriae; ompT, outer membrane protease; sitA, iron transport; irp2, high molecular weight protein 2 non-ribosomal peptide synthetase; iroN, enterobactin siderophore receptor; iss, increased serum survival; cma, colicin M; fyuA, siderophore receptor; mchF, ABC transporter; hlyF, hemolysin F; iucC, aerobactin synthetase; iutA, ferric aerobactin receptor; cvaC, microcin C; etsC, putative type I secretion outer membrane; kpsMII_K5, polysialic acid transport; afaD, afimbrial adhesion; air, enteroaggregative immunoglobulin; chuA, outer membrane hemin receptor; cia, colicin ia; eilA, salmonella HilA homolog; kpsE, capsule polysaccharide export inner-membrane; fedA, fimbrial protein F107 subunit A; fedF, fimbrial adhesin AC precursor; ltcA, heat-labile enterotoxin A subunit; sepA, shigella extracellular protein A; sta1, Heat-stabile enterotoxin ST-Ia; stx2A, shiga toxin 2, subunit A; stx2B, shiga toxin 2, subunit B; stx2, O139 S1191, variant e; cea, colicin E1. (C) Plasmid replicon type in cfr-positive E. coli. Orange represents the plasmid replicon type; white represents none of the genes predicted.
The conjugation transfer assay demonstrated that all transconjugants from cfr-positive E. coli strains and E. coli J53 could grow normally on LB plates containing 100 mg/l sodium azide and 10 mg/l florfenicol. Further, PCR confirmed that the transconjugant contained the cfr gene, which indicated that the conjugative transfer experiment was successful, with a transfer frequency of 1.05 × 10−1–1.01 × 10−6.
Genetic environment of the cfr-positive isolates.
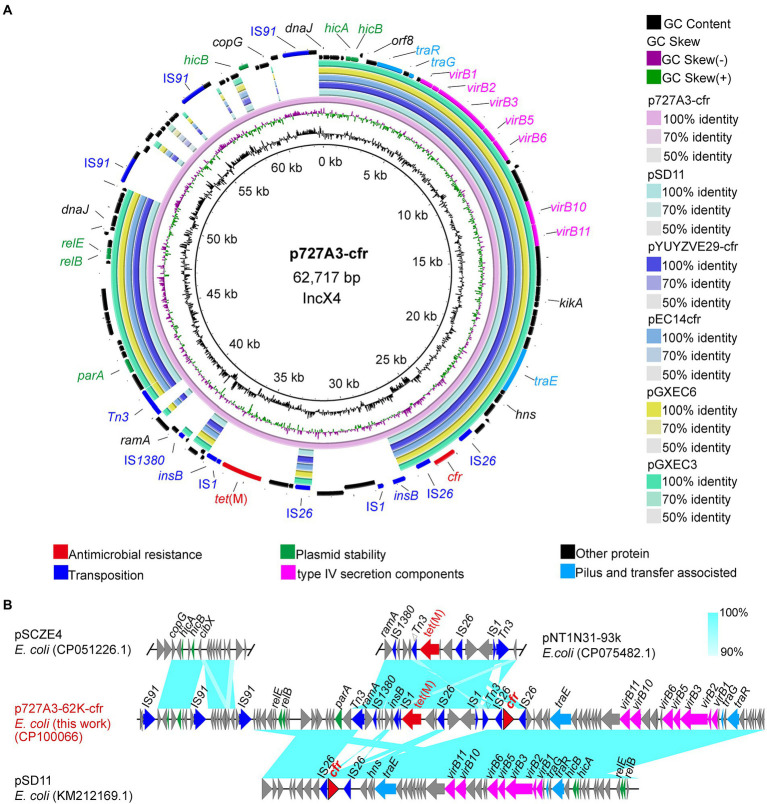
Two isolates were randomly selected from the eight cfr-positive strains for nanopore sequencing to obtain their complete genome sequences. To understand how cfr is transmitted, the genetic background of the cfr gene was further investigated. The cfr gene was located on the IncX4-type plasmid p727A3-62K-cfr (CP100066) in strain EC727A3. The length of p727A3-62K-cfr was 62,717 bp, and the GC content was 44% (Figure 3A). Moreover, p727A3-62K-cfr had high similarity with another cfr-carrying plasmid and had the highest homology with plasmid pSD11 (KM212169.1, 37,672 bp) from porcine E. coli strain 8ZG6D (65% query coverage and 99.99% identity). The collinear comparison showed that p727A3-62K-cfr and pSD11 had two different gene arrangements. The 12,647 bp region had high homology with the sequence containing the tet(M) gene in pNT1N31-93k (CP075482, 93,332 bp), and there was an insertion sequence IS1 upstream of tet(M) compared with pNT1N31-93k (Figure 3B). The other 10,831 bp region had a higher homology to a part of pSCZE4 (CP051226, 60,732 bp), and this sequence had three more IS91 insertion sequences in the same direction than pSCZE4 (Figure 3B).
Figure 3.
Sequence alignment of plasmid p727A3-62K-cfr and the gene–environment of cfr. (A) Comparison of the plasmid sequences between p727A3-62K-cfr, pSD11, pYUYZVE29-cfr, pEC14cfr, pGXEC6, and pGXEC3 of E. coli strains. (B) Linear comparison of the plasmid sequences of E. coli p727A3-62K-cfr and E. coli pSCZE4, pSD11, pNT1N21-93k. Open arrows indicate coding sequences (red arrows, AMR genes; green arrows, plasmid replication; blue arrows, transfer and transfer-related sequences; gray arrows, hypothetical, and unclassified) and indicate the direction of transcription.
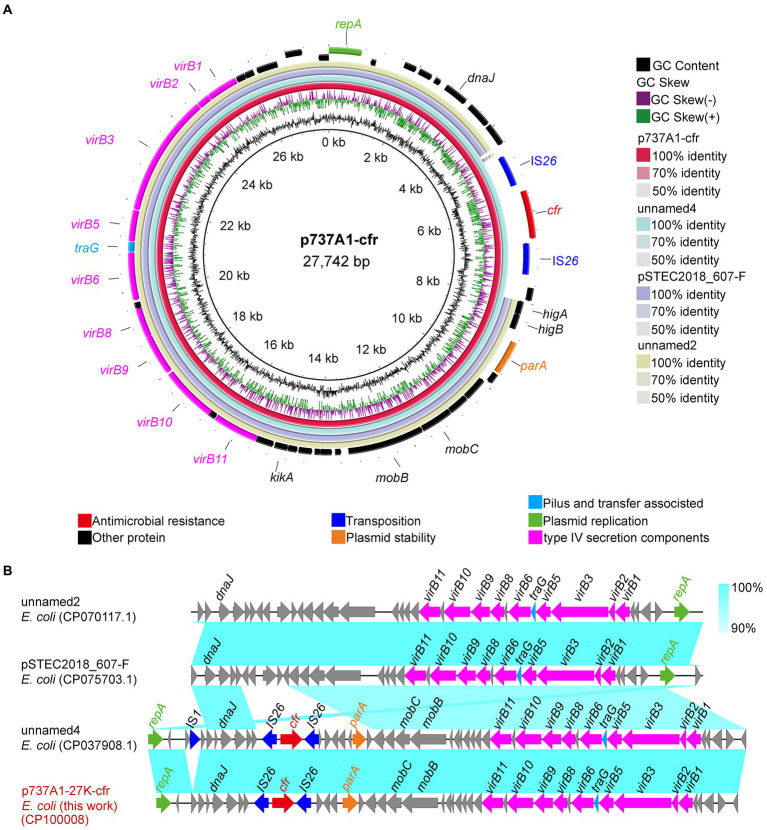
The cfr gene of strain EC737A1 was located on plasmid p737A1-27K-cfr (CP100008). The length of p737A1-27K-cfr was 27,742 bp, and the GC content was 43% (Figure 4A). Plasmid p737A1-27K-cfr had a high degree of homology (100% query coverage and 100% recognition) with plasmid unnamed4 (CP037908.1, 28,519 bp). The collinear comparison showed that a 777 bp region containing the IS1 mobile element was inserted into the plasmid p737A1-27K-cfr to form unnamed4. However, the type of plasmid had not yet been determined; it was only known that the backbone of plasmid p737A1-27K-cfr was derived from pSTEC2018_607-F (CP075703.1, 24,412 bp). The 4,270 bp construct containing the IS26-cfr-IS26-higA-higB-parK was inserted into the plasmid pSTEC2018_607-F (Figure 4B).
Figure 4.
Sequence alignment of plasmid p737A1-62K-cfr and the gene–environment of cfr. (A) Comparison of the plasmid sequences between E. coli p737A1-27K-cfr, E. coli plasmid unnamed4, pSTEC2018_607-F, unnamed2, FHI99-scaffold-18_conting-14, and pEC2547-KPC-2. (B) Linear comparison of the plasmid sequences of E. coli p737A1-27K-cfr and E. coli plasmid unnamed4, pSTEC2018_607-F, and plasmid unnamed2. Open arrows indicate coding sequences (red arrows, AMR genes; green arrows, plasmid replication; blue arrows, transfer, and transfer-related sequences; gray arrows, hypothetical, and unclassified) and indicate the direction of transcription.
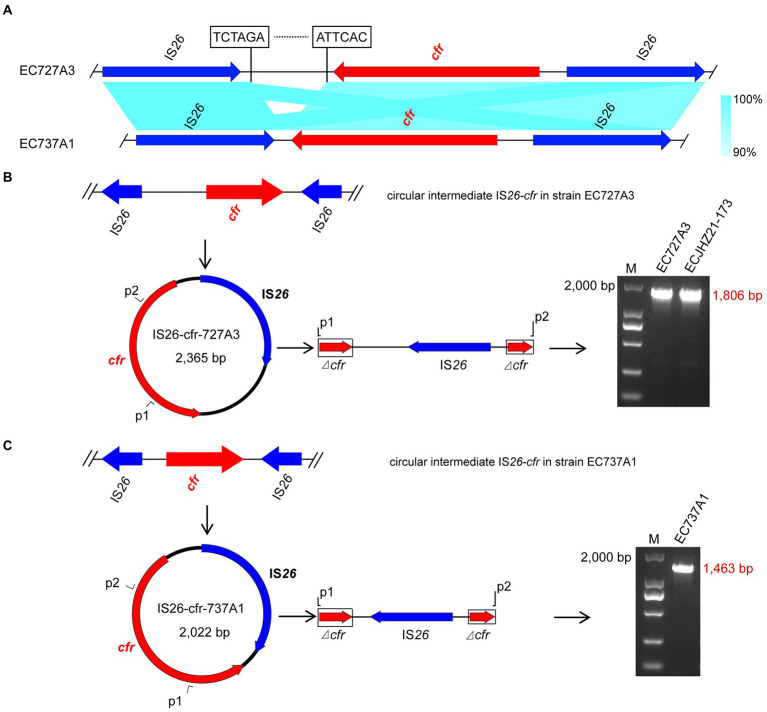
cfr-IS26 circular intermediate
Genome analysis found that both the upstream and downstream regions of the cfr gene in EC727A3 and EC737A1 had an IS26 element in the same direction, forming an IS26-cfr-IS26 structure (Figure 5A). However, there was a 343 bp size difference between the IS26-cfr-IS26 structures in EC727A3 and EC737A1. PCR determined that three out of eight E. coli strains could form cfr-IS26 cyclic intermediates of two different sizes. Among them, the size of the circular intermediate formed by ECJHZ21-173 and EC727A3 was the same, at 2,365 bp (Figure 5B). The size of the cfr-IS26 circular intermediate in EC737A1 was 2,022 bp (Figure 5C).
Figure 5.
Structures of IS26-cfr circular intermediates in EC727A3 and EC737A2. (A) Linear comparison of IS26-cfr-IS26 genomic sequences in cfr-positive E. coli EC727A3 and EC737A1. (B) Circular intermediates formed by the cfr gene in EC727A3 and the size of amplicons of cfr circular intermediates obtained by gel electrophoresis. (C) Circular intermediates formed by the cfr gene in EC737A1 and the size of amplicons of cfr circular intermediates obtained by gel electrophoresis.
Discussion
To date, the prevalence of the cfr gene in E. coli from animals has been reported to be 0.37% in Eastern China. In previous studies, most of the cfr genes in E. coli were isolated from pigs (Deng et al., 2014; Zhang et al., 2014). As far as we know, only four E. coli strains of chicken origin containing the cfr gene have been identified in Guangdong Province, Fujian Province and Heilongjiang Province (Zhao et al., 2016; Wang et al., 2018). No cfr gene has been found in human clinical E. coli isolates. In this study, we isolated the cfr gene from chicken sources in addition to pigs, and the prevalence of the cfr gene in E. coli isolates was higher than the initially reported at 0.08% (1/1230) (Wang et al., 2012a). This was similar to the previously reported 0.5% (2/398) (Liu et al., 2017) but much lower than the 13.7% (85/617) recently reported in Guangdong Province, China (Ma et al., 2021). According to the official, authoritative statement, in 2018 (P.R., 2019) and 2020 (P.R., 2021), the use of phenicols was 2,123 and 3,519 tons in animal breeding in China, respectively, and florfenicol was the primary antimicrobials in phenicols used in livestock and poultry breeding (Van Cuong et al., 2016). Previous global or national reports show that the florfenicol resistance gene is related to the long-term use of florfenicol (Li et al., 2020). Our study indicated that the cfr gene dissemination was significantly different in different provinces of China, and there was a possibility of rapid spread in a small area.
IS26 is a universal mobile element in various gram-negative bacteria, including E. coli, P. multocida, Acinetobacter baumannii, Klebsiella pneumoniae, V. diabolicus, and Proteus vulgaris (Post and Hall, 2009; Harmer et al., 2014; Chen et al., 2020a; Jin et al., 2021; Zhao et al., 2021). The presence of transfer elements plays a vital role in the transfer of the cfr gene. Previous studies confirmed the existence of different genetic environments for the cfr gene in E. coli, with one IS26 element on each side of the cfr gene being the most reported genetic environment in E. coli and the other two being one IS256 element on each side of cfr and one IS15 element on each side (Wang et al., 2012a, 2018; Zhang et al., 2016; Liu et al., 2017; Tang et al., 2021). In addition, IS26 was found to form circular intermediates mediating the transmission of cfr genes in V. diabolicus. Similarly, it was also found to form circular intermediates that mediate the transmission of other AMR genes in E. coli (Zhao et al., 2021; Liu et al., 2022). The current study results were inconsistent with previous studies verifying that cfr can form a circular intermediate of IS26-cfr during transmission and facilitate its transmission in E. coli.
Plasmid p727A3-62K-cfr obtained in the present study belonged to the IncX4 type. The IncX4 plasmids carrying the cfr gene have been found in E. coli isolated from Jiangsu, Guangdong, Guangxi, Liaoning, Jilin, and Heilongjiang Provinces in China (Deng et al., 2014; Mei et al., 2021). This result indicated that the IncX4-type plasmid might be a common plasmid carrying the MDR gene cfr. In addition, we also identified a plasmid p737A1-27K-cfr that had not yet been typed, which indicates that the types of plasmids carrying the cfr gene are gradually increasing, and it is necessary to pay close attention to the spread of the cfr gene in E. coli.
Conclusion
Eight strains containing the cfr gene were isolated from 2,165 strains of E. coli in 2021, seven strains were isolated from pig farms, and one strain was isolated from chicken farms, indicating that the cfr gene widely exists in a variety of food animals. An IncX4 type plasmid and an unknown type plasmid were found, and the IS26-cfr-IS26 structure was verified to form a cfr-IS26 circular intermediate for propagation. Since the widespread use of antibiotics, particularly florfenicol, may promote the spread of cfr genes among animals. It is necessary to strengthen the control of veterinary antibiotics and continuously monitor the spread of the E. coli multidrug resistance gene cfr to reduce the potential public health threat.
Data availability statement
The names of the repository/repositories and accession number(s) can be found in the article/Table 2.
Author contributions
BT and MY: conceptualization. HY: funding acquisition. BT, JN, JL, HL, and YW: investigation. JN, JL, and BT: methodology. MY and HY: supervision. JN, HL, and BT: visualization. JN and BT: writing—original draft. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Key Research and Development Program of Zhejiang Province (2020C02031), the earmarked fund for China Agriculture Research System (CARS-42-27), the State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products (2010DS700124-ZZ2102), Collaborative Extension Plan of Major Agricultural Technologies in Zhejiang Province (2021XTTGXM03) and Major Special Project for the Construction of Agricultural Product Standardized Production Demonstration Counties (zjny2022001).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.999778/full#supplementary-material
Footnotes
References
- Alikhan N. F., Petty N. K., Ben Zakour N. L., Beatson S. A. (2011). BLAST ring image generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Deng H., Cheng L., Liu R., Fu G., Shi S., et al. (2020a). First report of the multiresistance gene cfr in Pasteurella multocida strains of avian origin from China. J. Glob. Antimicrob. Resist. 23, 251–255. doi: 10.1016/j.jgar.2020.09.018, PMID: [DOI] [PubMed] [Google Scholar]
- Chen L., Han D., Tang Z., Hao J., Xiong W., Zeng Z. (2020b). Co-existence of the oxazolidinone resistance genes cfr and optrA on two transferable multi-resistance plasmids in one enterococcus faecalis isolate from swine. Int. J. Antimicrob. Agents 56:105993. doi: 10.1016/j.ijantimicag.2020.105993, PMID: [DOI] [PubMed] [Google Scholar]
- Dai L., Wu C. M., Wang M. G., Wang Y., Wang Y., Huang S. Y., et al. (2010). First report of the multidrug resistance gene cfr and the phenicol resistance gene fexA in a Bacillus strain from swine feces. Antimicrob. Agents Chemother. 54, 3953–3955. doi: 10.1128/aac.00169-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H., Sun J., Ma J., Li L., Fang L. X., Zhang Q., et al. (2014). Identification of the multi-resistance gene cfr in Escherichia coli isolates of animal origin. PLoS One 9:e102378. doi: 10.1371/journal.pone.0102378, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick M. (1999). Haemolytic uraemic syndrome and E. coli O157. BMJ 318, 684–685. doi: 10.1136/bmj.318.7185.684, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner S. N., Slezak T., Hall B. G. (2015). kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31, 2877–2878. doi: 10.1093/bioinformatics/btv271 [DOI] [PubMed] [Google Scholar]
- Harmer C. J., Moran R. A., Hall R. M. (2014). Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. mBio 5, e01801–e01814. doi: 10.1128/mBio.01801-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries R., Bobenchik A. M., Hindler J. A., Schuetz A. N. (2021). Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100, 31st Edition. J. Clin. Microbiol. 59:e0021321. doi: 10.1128/jcm.00213-21, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Wang R., Gao H., Wang Q., Wang H. (2021). Identification of a novel hybrid plasmid encoding KPC-2 and virulence factors in Klebsiella pneumoniae sequence type 11. Antimicrob. Agents Chemother. 65, e02435–e02420. doi: 10.1128/aac.02435-20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrenberg C., Schwarz S., Jacobsen L., Hansen L. H., Vester B. (2005). A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 57, 1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x, PMID: [DOI] [PubMed] [Google Scholar]
- Launders N., Byrne L., Jenkins C., Harker K., Charlett A., Adak G. K. (2016). Disease severity of shiga toxin-producing E. coli O157 and factors influencing the development of typical haemolytic uraemic syndrome: a retrospective cohort study, 2009–2012. BMJ Open 6:e009933. doi: 10.1136/bmjopen-2015-009933, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Zhu T., Zhou D., Lu W., Liu H., Sun Z., et al. (2020). Analysis of resistance to florfenicol and the related mechanism of dissemination in different animal-derived bacteria. Front. Cell. Infect. Microbiol. 10:369. doi: 10.3389/fcimb.2020.00369, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. Q., Wang J., Li W., Zhao L. Q., Lu Y., Liu J. H., et al. (2017). Distribution of cfr in Staphylococcus spp. and Escherichia coli strains from pig farms in China and characterization of a novel cfr-carrying F43:A-:B-plasmid. Front. Microbiol. 8:329. doi: 10.3389/fmicb.2017.00329, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Zhang W., Hu Y., Chen P., Wang Z., Li R. (2022). Emergence of the cfr gene in Vibrio diabolicus of seafood origin. Antimicrob. Agents Chemother. 66:e0181921. doi: 10.1128/aac.01819-21, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long K. S., Poehlsgaard J., Kehrenberg C., Schwarz S., Vester B. (2006). The cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50, 2500–2505. doi: 10.1128/aac.00131-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Liu J., Chen L., Liu X., Xiong W., Liu J. H., et al. (2021). Rapid increase in the IS26-mediated cfr gene in E. coli isolates with IncP and IncX4 plasmids and co-existing cfr and mcr-1 genes in a swine farm. Pathogens 10:33. doi: 10.3390/pathogens10010033, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcfarland N., Bundle N., Jenkins C., Godbole G., Mikhail A., Dallman T., et al. (2017). Recurrent seasonal outbreak of an emerging serotype of shiga toxin-producing Escherichia coli (STEC O55:H7 Stx2a) in the south west of England, July 2014 to September 2015. Euro Surveill. 22:30610. doi: 10.2807/1560-7917.Es.2017.22.36.30610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei C. Y., Wu H., Wang Y., Wang Z. Y., Ma Q. C., Shen P. C., et al. (2021). First detection of the multiresistance gene cfr in Escherichia coli from retail vegetables. China. Int. J. Antimicrob. Agents. 57:106348. doi: 10.1016/j.ijantimicag.2021.106348, PMID: [DOI] [PubMed] [Google Scholar]
- Osman K. M., Badr J., Orabi A., Elbehiry A., Saad A., Ibrahim M. D. S., et al. (2019). Poultry as a vector for emerging multidrug resistant Enterococcus spp.: first report of vancomycin (van) and the chloramphenicol-florfenicol (cat-fex-cfr) resistance genes from pigeon and duck faeces. Microb. Pathog. 128, 195–205. doi: 10.1016/j.micpath.2019.01.006, PMID: [DOI] [PubMed] [Google Scholar]
- P.R. Ministry of Agriculture and Rural Affairs, China (2019). Report on the use of veterinary antibiotics of China in 2018. Off. Veterin. Bull. 21, 57–59. [Google Scholar]
- P.R. Ministry of Agriculture and Rural Affairs, China (2021). Report on the use of veterinary antibiotics of China in 2020. Off. Veterin. Bull. 23, 33–36. [Google Scholar]
- Partridge S. R., Kwong S. M., Firth N., Jensen S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31, e00088–e00017. doi: 10.1128/cmr.00088-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post V., Hall R. M. (2009). AbaR5, a large multiple-antibiotic resistance region found in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53, 2667–2671. doi: 10.1128/aac.01407-08, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S., Werckenthin C., Kehrenberg C. (2000). Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 44, 2530–2533. doi: 10.1128/aac.44.9.2530-2533.2000, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Wang Y., Schwarz S. (2013). Presence and dissemination of the multiresistance gene cfr in gram-positive and gram-negative bacteria. J. Antimicrob. Chemother. 68, 1697–1706. doi: 10.1093/jac/dkt092, PMID: [DOI] [PubMed] [Google Scholar]
- Smith L. K., Mankin A. S. (2008). Transcriptional and translational control of the mlr operon, which confers resistance to seven classes of protein synthesis inhibitors. Antimicrob. Agents Chemother. 52, 1703–1712. doi: 10.1128/aac.01583-07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Deng H., Li L., Chen M. Y., Fang L. X., Yang Q. E., et al. (2015). Complete nucleotide sequence of cfr-carrying IncX4 plasmid pSD11 from Escherichia coli. Antimicrob. Agents Chemother. 59, 738–741. doi: 10.1128/aac.04388-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B., Chang J., Chen Y., Lin J., Xiao X., Xia X., et al. (2022a). Escherichia fergusonii, an underrated repository for antimicrobial resistance in food animals. Microbiol. Spectr. 10:e0161721. doi: 10.1128/spectrum.01617-21, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B., Chang J., Luo Y., Jiang H., Liu C., Xiao X., et al. (2022b). Prevalence and characteristics of the mcr-1 gene in retail meat samples in Zhejiang Province. China. J. Microbiol. 60, 610–619. doi: 10.1007/s12275-022-1597-y, PMID: [DOI] [PubMed] [Google Scholar]
- Tang Y., Lai Y., Kong L., Wang X., Li C., Wang Y., et al. (2021). Characterization of three porcine Escherichia coli isolates co-harbouring tet(X4) and cfr. J. Antimicrob. Chemother. 76, 263–264. doi: 10.1093/jac/dkaa384, PMID: [DOI] [PubMed] [Google Scholar]
- Van Cuong N., Nhung N. T., Nghia N. H., Mai Hoa N. T., Trung N. V., Thwaites G., et al. (2016). Antimicrobial consumption in medicated feeds in Vietnamese pig and poultry production. EcoHealth 13, 490–498. doi: 10.1007/s10393-016-1130-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielva L., de Toro M., Lanza V. F., de la Cruz F. (2017). PLACNETw: a web-based tool for plasmid reconstruction from bacterial genomes. Bioinformatics 33, 3796–3798. doi: 10.1093/bioinformatics/btx462, PMID: [DOI] [PubMed] [Google Scholar]
- Wang X., Zhu Y., Hua X., Chen F., Wang C., Zhang Y., et al. (2018). F14:A-:B-and IncX4 Inc group cfr-positive plasmids circulating in Escherichia coli of animal origin in Northeast China. Vet. Microbiol. 217, 53–57. doi: 10.1016/j.vetmic.2018.02.003, PMID: [DOI] [PubMed] [Google Scholar]
- Wang Y., He T., Schwarz S., Zhou D., Shen Z., Wu C., et al. (2012a). Detection of the staphylococcal multiresistance gene cfr in Escherichia coli of domestic-animal origin. J. Antimicrob. Chemother. 67, 1094–1098. doi: 10.1093/jac/dks020, PMID: [DOI] [PubMed] [Google Scholar]
- Wang Y., Li D., Song L., Liu Y., He T., Liu H., et al. (2013). First report of the multiresistance gene cfr in Streptococcus suis. Antimicrob. Agents Chemother. 57, 4061–4063. doi: 10.1128/aac.00713-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang Y., Schwarz S., Shen Z., Zhou N., Lin J., et al. (2012b). Detection of the staphylococcal multiresistance gene cfr in Macrococcus caseolyticus and Jeotgalicoccus pinnipedialis. J. Antimicrob. Chemother. 67, 1824–1827. doi: 10.1093/jac/dks163, PMID: [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang Y., Wu C. M., Schwarz S., Shen Z., Zhang W., et al. (2011). Detection of the staphylococcal multiresistance gene cfr in Proteus vulgaris of food animal origin. J. Antimicrob. Chemother. 66, 2521–2526. doi: 10.1093/jac/dkr322, PMID: [DOI] [PubMed] [Google Scholar]
- Wick R. R., Judd L. M., Gorrie C. L., Holt K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Zhang J., Wang M., Liu M., Liu G., Qu H., et al. (2021). Mobilization of the nonconjugative virulence plasmid from hypervirulent Klebsiella pneumoniae. Genome Med. 13, 119. doi: 10.1186/s13073-021-00936-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Sun B., Wang Y., Lei L., Schwarz S., Wu C. (2016). Characterization of a cfr-carrying plasmid from porcine Escherichia coli that closely resembles plasmid pEA3 from the plant pathogen Erwinia amylovora. Antimicrob. Agents Chemother. 60, 658–661. doi: 10.1128/aac.02114-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. J., Wang X. M., Dai L., Hua X., Dong Z., Schwarz S., et al. (2015). Novel conjugative plasmid from Escherichia coli of swine origin that coharbors the multiresistance gene cfr and the extended-spectrum-β-lactamase gene blaCTX-M-14b. Antimicrob. Agents Chemother. 59, 1337–1340. doi: 10.1128/aac.04631-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. J., Xu X. R., Schwarz S., Wang X. M., Dai L., Zheng H. J., et al. (2014). Characterization of the IncA/C plasmid pSCEC2 from Escherichia coli of swine origin that harbours the multiresistance gene cfr. J. Antimicrob. Chemother. 69, 385–389. doi: 10.1093/jac/dkt355, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Hou H., Liu X., Li W., Zhang X., Liu J., et al. (2016). Investigation of epidemic characteristics of multi-resistance gene cfr in Escherichia coli isolates from food animal origins. Chin. Vet. Sci. 46, 1464–1468. doi: 10.16656/j.issn.1673-4696.2016.11.020 [DOI] [Google Scholar]
- Zhao Q. Y., Zhu J. H., Cai R. M., Zheng X. R., Zhang L. J., Chang M. X., et al. (2021). IS26 IS responsible for the evolution and transmission of blaNDM-harboring plasmids in Escherichia coli of poultry origin in China. mSystems 6:e0064621. doi: 10.1128/mSystems.00646-21, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The names of the repository/repositories and accession number(s) can be found in the article/Table 2.