Abstract
Vibrio vulnificus contains homologues of the V. harveyi luxR and luxS genes. A null mutation in smcR (luxR) resulted in a defect in starvation survival, inhibition of starvation-induced maintenance of culturability that occurs when V. vulnificus is starved prior to low-temperature incubation, and increased expression of stationary-phase phenotypes.
The opportunistic pathogen Vibrio vulnificus is a common isolate of marine and estuarine waters. At water temperatures below 10°C, V. vulnificus enters a viable but nonculturable (VBNC) state (20, 25). Starvation of V. vulnificus prior to shifting cells to low temperatures induces the starvation-induced maintenance of culturability (SIMC) response which delays the induction of the VBNC response (24, 27), suggesting that starvation-induced or stationary-phase genes are important for the adaptation of this organism to low-temperature survival, in addition to other stress conditions.
In many bacteria, the regulation of phenotypes is controlled via signaling pathways where extracellular factors are used to coordinate the expression of phenotypes at the population level, many of which are induced during stationary phase. For example, Rhizobium leguminosarum uses signal molecules to induce stationary phase (11), and conditioned supernatants have been shown to induce carbon starvation proteins in Vibrio angustum (32). Signaling molecules have also been demonstrated or suggested to regulate the expression of virulence factors in a variety of organisms (4, 23, 28, 31), several of which are induced during stationary phase in V. vulnificus.
Vibrio harveyi possesses genes (luxR and luxS) which encode regulatory proteins that are members of a signaling system recently identified in a broad range of organisms (2); however, the genes regulated by this system are generally unknown. We report here the characterization of a V. vulnificus smcR mutant, which is a homologue of the V. harveyi luxR gene (21). This regulatory gene appears to play an important role in starvation adaptation and in the regulation of many stationary-phase-regulated genes, including some virulence factors. Furthermore, we report that V. vulnificus produces extracellular signals. The role of signals in the expression of these stationary-phase proteases and in the development of starvation adaptation is supported by the inhibition of these phenotypes upon addition of a signal antagonist that represses autoinducer system 2 (AI-2) phenotypes.
The plasmids and bacterial strains used in this study and their genotypes are listed in Table 1. The V. harveyi strains were a gift from Bonnie Bassler. Where specified, glucose was added to a final concentration of 0.5% for Luria-Bertani medium (LB) and 0.4% for 2M minimal medium (27). The antibiotics ampicillin, streptomycin, chloramphenicol, and colistin were used at concentrations of 50, 200, 34, and 100 μg ml−1, respectively. General chemicals were purchased from Sigma Chemical Co., St. Louis, Mo. Genomic DNA was isolated by the method of Tillett and Neilan (37). Restriction enzymes, molecular weight markers, shrimp alkaline phosphatase, ligase, Pwo polymerase, and T4 DNA ligase were purchased from Boehringer Mannheim (Indianapolis, Ind.).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| V. vulnificus | ||
| C7184 | Human wound isolate | 26 |
| DM7 | smcR::Sm, derived from C7184 | This study |
| UTHS-1 | 39 | |
| V. harveyi | ||
| BB170 | luxN::Tn5 (sensor 1− sensor 2+) | 3 |
| BB152 | luxL::Tn5 (AI-1− AI-2+) | 3 |
| E. coli | ||
| BW20767 | Smr (RP4-2 tet:Mu-1 kan::Tn7 integrant) tra+ leu-63::IS10 recA1 creC510 hsdR17 endA1 zbf-5 uidA(Mlu1):pir+ thi | 22 |
| DH5α | supE44 ΔlacU169 (Φ80lacZΔM15)hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 12 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZDM15 Tn10(Tetr)] | 5 |
| Plasmids | ||
| pBluescript II SK(+) | Apr, multiple cloning site flanked by T3 and T7 promoters, lac promoter fused to the α peptide of ′LacZ, derived from pUC19 | Stratagene |
| pCAM140 | Smr, Apr, mini-Tn5 gusA in pUT mini-Tn5 Sm/Sp | 38 |
| pCVD442 | Apr, positive selection vector, pGP704 with sacB inserted in multiple cloning site | 7 |
| pLG401 | Cmr, pACYC Ori, Mob, promoterless gfp with multiple cloning site | Lynn Gilson, University of Hawaii |
| pMacSB | Cmr, sacB from pCVD442 inserted as PstI-EcoRV fragment into cloning site of pLG401 | This study |
| pMacSmcRK | pMacSB with disrupted smcR fragment from pSmcR.SM inserted into EcoRV site of pMacSB | This study |
| pUC19 | Apr, multiple cloning site, lac promoter fused to the α peptide of ′LacZ | 40 |
| pSmcR8 | Apr, pUC19 with 720-bp insert containing V. vulnificus smcR | This study |
| pSmcR8.18 | Apr, pBluescript with ∼770-bp insert containing the EcoRI-HindIII smcR fragment from pSmcR8 | This study |
| pSmcR.SM | Apr, Smr, pSmcR8.18 with ∼2.0-kb Sm fragment from pCAM140 inserted into the BglII site of the smcR insert | This study |
V. vulnificus produces compounds that induce luminescence in V. harveyi.
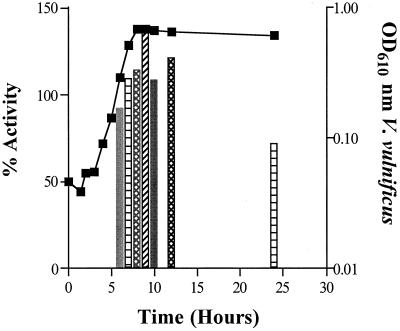
The ability of V. vulnificus supernatants to induce bioluminescence in the V. harveyi AI-2 reporter strain BB170 was determined as previously described (33). Cell-free supernatants were prepared from late-exponential-phase cells of V. vulnificus C7814 and UTHS-1 (optical dencity at 610 nm [OD610] = 0.796) grown in LB with aeration at 37°C and added to the reporter strain at a concentration of 10%. Induction by the V. vulnificus strain C7184 or UTHS-1 was 215 or 350%, respectively, of the positive control activity (data not shown). Maximal signal production in V. vulnificus occurred as cells enter the stationary phase of growth (Fig. 1). To assess the effect of nutrient starvation conditions on the induction of AI-2 activity in V. vulnificus, cells were grown to mid-exponential phase in LB containing NaCl (20 g liter−1), collected by centrifugation (8,000 × g, 10 min, 24°C), and washed and resuspended in 0.5× NSS (27). Cell-free supernatants taken immediately after the shift to starvation conditions (time zero) induced 0.4% of the luminescence observed in the V. harveyi reporter strain in the presence of the positive control supernatant. Induction of luminescence increased to 849% for supernatants taken from cells after 4 h of starvation; by 9 h of starvation, luminescence had dropped to 245% (data not shown). These data indicate that signal production is growth phase regulated and that starvation conditions are able to stimulate the production of AI-2-like activity in V. vulnificus.
FIG. 1.
Effect of growth phase on the production of substances able to induce luminescence in V. harveyi BB170. V. vulnificus C7184 was grown with aeration in 2M at 37°C, and OD610 was determined (squares). Cell-free supernatants were prepared at various times and assayed for the ability to induce luminescence in the V. harveyi reporter strain (bars). The activity of supernatants is presented as the percentage of activity obtained when V. harveyi BB152 cell-free spent supernatant is added to the reporter strain. Data presented are representative of results obtained in at least three independent experiments.
In contrast to Salmonella enterica serovar Typhimurium and Escherichia coli (34), the addition of 0.5% glucose to LB inhibits the production of AI-2 activity in V. vulnificus. Furthermore, AI-2 activity was produced by V. vulnificus cells grown in LB or the minimal medium, 2M (27), at room temperature and at 37°C (data not shown). Supernatants collected from the smcR mutant (see below) were also able to induce luminescence in V. harveyi to similar levels as the wild type (data not shown), indicating smcR is not required for signal production. The autoinducer activity of cell-free supernatants heated to 80°C for 10 min was reduced by 52.7%, while heat treatment at 100°C for 10 min abolished activity, indicating that the V. vulnificus AI-2-stimulating factor is a heat-labile compound.
V. vulnificus possesses the AI-2 synthase gene, luxS.
The presence of a putative luxS homologue in V. vulnificus was previously suggested by Southern hybridization (21). A 320-bp fragment was amplified and cloned from V. vulnificus using primers based on the AI-2 synthase gene, luxS, of V. harveyi. Sequence analysis determined that the fragment, luxSVv, showed >80, 79, and 68% nucleotide identities to the luxS gene from V. harveyi, a putative luxS in the Vibrio cholerae genome database, and the ygaG gene of E. coli (data not shown). The high degree of nucleotide identity and the presence of AI-2 activity in the supernatants of V. vulnificus confirms the presence of a luxS gene in V. vulnificus.
Characterization of a mutant in V. vulnificus of the luxR transcriptional regulator homologue.
A potential rho-independent terminator lies 19 nucleotides downstream of the smcR stop codon, and the smcR coding region is followed by a convergently transcribed homologue of lpd (dihydrolipoamide dehydrogenase) (21). pUC19 (40) and pBluescript II SK were used as shuttle vectors for the cloning of smcR. A null mutation in smcR was generated by disruption with the insertion of a streptomycin resistance cassette from pCAM140 (38) (pSmcR.SM) 183 bp downstream from the ATG codon. The vector used for delivery and homologous recombination, pMacSB, was constructed by the insertion of the sacB gene derived from pCVD442 (7) into pLG401 (constructed by Lynn Gilson, University of Hawaii). The smcR gene, containing the streptomycin disruption was inserted into pMacSB to generate pMacSmcRK. The null mutant was generated by conjugation of E. coli BW20767(pMacSmcRK) with V. vulnificus C7184 and selection for streptomycin and sucrose resistance. This disruption was confirmed by Southern hybridization and PCR.
SmcR is involved in the regulation of starvation survival and the SIMC response.
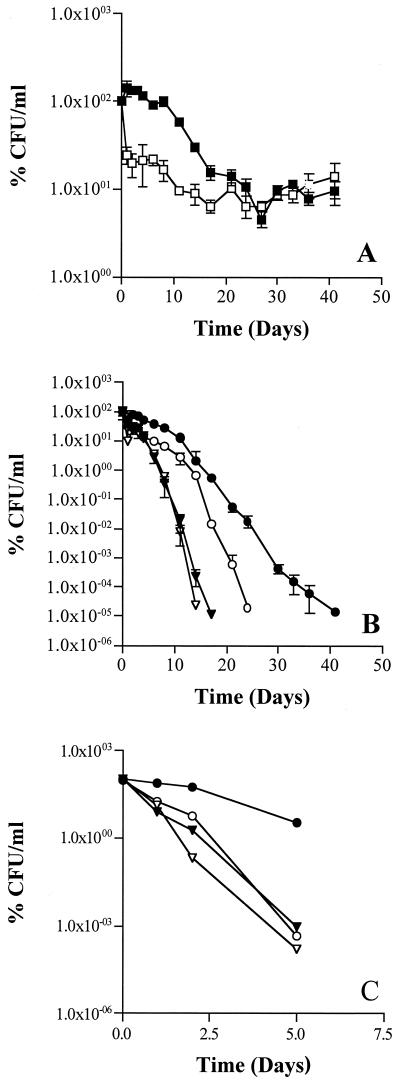
The effect of smcR on stationary-phase survival was determined by growth of cells of V. vulnificus C7184 and the smcR mutant (DM7) to early exponential phase in LB with 20 g of NaCl per liter (OD610 nm = 0.22; 4.0 × 108 CFU ml−1), followed by resuspension in 2 M lacking glucose (2M-C) (27) at 1:100 dilution. During room temperature starvation, there was an initial decrease of 76% in the CFU for the mutant strain and no decrease for the wild type after 1 day (Fig. 2A). After 14 days, the smcR mutant strain exhibited a decrease of 91% of CFU whereas the wild type had a loss of 70%. The loss in CFU for the two strains was not significantly different after the first 14 to 20 days of starvation.
FIG. 2.
SmcR affects starvation survival and SIMC at low temperature. V. vulnificus C7184 (filled symbols) and DM7 (smcR::Sm) (open symbols) were grown to mid-exponential phase in LB with NaCl (20 g liter−1), the cells were collected by centrifugation (10,000 × g, 10 min), washed in 2M-C and resuspended in 2M-C. Cultures were held statically at 24°C (A) or were allowed to starve for 0 (B; ▿, ▾) or 4 (B; ○, ●) h before being shifted to 4°C. (C) C7184 starved in the presence (open symbols) or absence (closed symbols) of C2. Determination of CFU was performed on DVNSS agar plates. Data are presented as percentages of the initial count (1.1 × 105 to 2.9 × 105 CFU ml−11) and are representative of three independent experiments. Error bars represent the 95% confidence interval.
The defect in survival during the first 2 weeks of starvation exhibited by DM7 prompted us to investigate the effect of the smcR null mutation on the SIMC response. This SIMC effect possibly allows cells to synthesize proteins that will be important in survival and recovery when conditions are again favorable. Cells of C7184 and DM7 prepared as above were starved for 0 or 4 h at 24°C and then shifted to 4°C. The results in Fig. 2B clearly indicate that the smcR mutant strain is defective in mounting the SIMC response. The cultures that were shifted to 4°C without prestarvation showed very little difference in the rate of loss of culturability, in contrast to those starved at room temperature before cold incubation (Fig. 2B). By the third day of cold incubation, DM7 had lost 77% of total CFU, while the wild-type strain showed a decrease of only 28%. This trend continued throughout the cold incubation.
These data indicate that SmcR affects the prestarvation response. Given that signals regulate starvation in some bacteria (14, 16, 32, 36), we tested the effect of a signal antagonist on the starvation response. The marine red alga Delisea pulchra has been shown to produce a range of halogenated furanones that specifically inhibit signaling phenotypes regulated by the acylated homoserine lactone and AI-2 systems in bacterial species (9, 10, 17, 18, 32). Cells of C7184 were collected during early exponential phase, washed and resuspended in 2M-C with or without furanone compound 2 (C2; 2 μg ml−1) and shifted to 4°C at time zero and after 4 h of starvation at room temperature (Fig. 2C). Room temperature starvation of cells in the presence of C2 does not allow V. vulnificus to mount the SIMC response which occurs in the absence of C2. C2 was added at 10 μg ml−1 to growing cultures of V. vulnificus C7184 during exponential phase (OD610 = 0.4) to assess whether signal transduction is required for the production of autoinducer activity. Supernatants collected from V. vulnificus during growth with C2 induced V. harveyi 152%.
DM7 exhibits increased exoenzyme production.
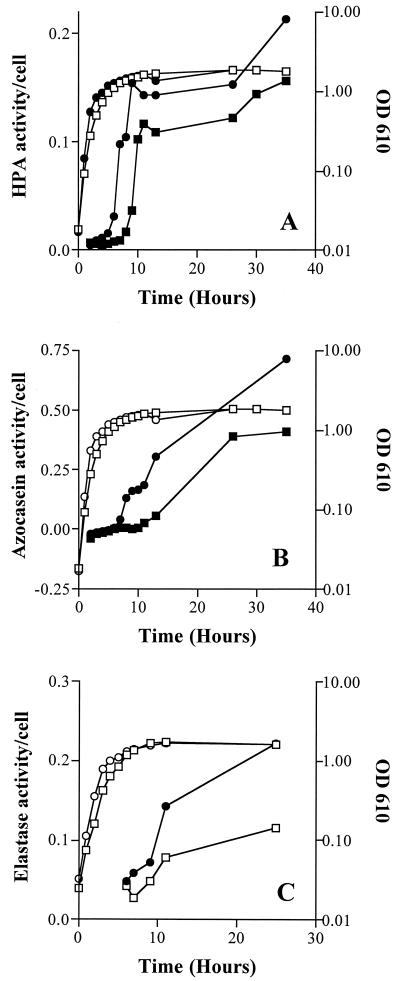
Typical results for exoprotease expression of cells grown in LB at 37°C as determined by HPA (1), azocasein (35), and elastin-Congo red (8) substrate degradation by cell-free supernatants are represented in Fig. 3. Similar trends in exoenzyme activity were obtained from supernatants collected from cultures grown at 24°C (data not shown) and for expression of alkaline phosphatase activity (Table 2). In all cases, exoenzyme expression of the smcR mutant occurred earlier and the final activity was higher than for the wild-type strain. Growth of the wild-type strain in the presence of C2 inhibited protease production (Table 2), indicating that the signaling pathway is important for protease production. In V. cholerae, a mutation in the luxR homologue, hapR, resulted in a loss of expression of the hemagglutinin/protease metalloenzyme (15). Interestingly, our results indicate that unlike hapR in V. cholerae, smcR is involved in the repression of protease expression during exponential growth rather than its induction.
FIG. 3.
Exoprotease activity of V. vulnificus C7814 and DM7 (smcR::Sm). Cultures of V. vulnificus C7814 (□, ■) and DM7 (○, ●) were grown in LB at 37°C with shaking at 200 rpm on a rotary shaker. At various time points, aliquots were removed and cell-free supernatants were prepared by centrifugation (10,000 × g, 10 min). The supernatant was then fi through 0.2-pore-size μm filters. Exoprotease activity (closed symbols) was assayed by degradation of HPA (A), azocasein (B), and elastin-Congo red (C) at 37°C. Results are presented as the exoprotease activity per cell and are representative of at least three independent experiments.
TABLE 2.
Selected phenotypes of a V. vulnificus smcR mutant
| V. vulnificus strain | Signal productiona | Alkaline phosphataseb | Biofilm formationc | Protease activityd |
|---|---|---|---|---|
| DM7 (smcR) | + | 230 | 507.4 | 121.4 |
| C7184 (smcR+) | + | 100 | 100.0 | 100 |
| C7184 + C2e | + | NDf | ND | 0.001 |
Cell-free supernatants were collected from early-stationary-phase cultures, and their ability to induce luminescence in the V. harveyi reporter strain was tested.
Alkaline phosphatase production as percentage of wild-type activity.
Biofilm formation at 24 h in microtiter plates, presented as percentage of wild-type attachment.
Protease production as percentage of wild-type HPA activity.
C2 was added at a concentration of 10 μg ml−1 during mid-exponential phase, and the culture was incubated at 37°C for 4 h (stationary phase) prior to collection of supernatants
ND, not determined.
Recent intriguing discoveries suggest that signal molecules may regulate phenotypes that are not density dependent but are regulated in relation to growth phase or in response to local environmental conditions. For example, signal molecules have been shown to regulate the induction of stationary phase in R. leguminosarum (36) and Pseudomonas aeruginosa (41) and to induce the carbon starvation response in V. angustum (32). These reports suggest there are density-independent signaling systems in some bacteria that regulate starvation and/or stationary-phase phenotypes.
SmcR is important for starvation survival and SIMC. While the general features of the AI-2 systems appear to be highly conserved across a broad range of genera and species, some of the specific features of the system clearly differ and may reflect individual adaptation of the AI-2 system to the specific needs of particular bacteria. In V. vulnificus, the SmcR appears to act as an activator as well as a repressor, in contrast to data presented for similar phenotypes on other bacteria (15, 19).
Indeed, it has been previously suggested that LuxR may function as a repressor. For example, LuxR binds independently to two sites upstream of its own open reading frame (6) and represses transcription from the luxR promoter as a result of possibly interfering with and displacing RNA polymerase from the promoter (6). LuxR is a member of the TetR family of transcriptional regulators, which act as repressors (13). Taken together, these data indicate that the primary function of the LuxR regulator, at least in some organisms, may in fact be the repression, rather than activation, of gene transcription. We propose that SmcR in V. vulnificus appears to act as both an activator and a repressor, similarly to TyR (29) and nitrogen regulator I (30). It seems likely that signal production and recognition, which occurs at the transition into stationary phase or shortly after entry into starvation, when these phenotypes are normally expressed, may be the mediator of this relief of repression.
The loss of a functional smcR impairs starvation survival and prevents V. vulnificus from exhibiting the SIMC response upon starvation prior to low-temperature incubation. In addition to an increase in exoenzyme production, the mutant strain also exhibited increases in motility, fimbria production (data not shown), and biofilm formation (Table 2). We propose that the defect in starvation survival by the smcR mutant may be reflected by the altered regulation of the many stationary-phase phenotypes reported here. To our knowledge, this is the first report of the regulation of starvation adaptation by a V. harveyi luxR homologue. This discovery expands the role of signaling systems to include global regulation of nongrowth physiology.
Acknowledgments
We thank Bonnie Bassler for her gift of the V. harveyi reporter strains and Evi Fuary for performing biofilm experiments.
Funding for this project was provided by Centre for Marine Biofouling and Bio-Innovation.
REFERENCES
- 1.Albertson N H, Nyström T, Kjelleberg S. Exoprotease activity of two marine bacteria during starvation. Appl Environ Microbiol. 1990;56:218–223. doi: 10.1128/aem.56.1.218-223.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler B L. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr Opin Microbiol. 1999;2:582–587. doi: 10.1016/s1369-5274(99)00025-9. [DOI] [PubMed] [Google Scholar]
- 3.Bassler B L, Wright M, Showalter R E, Silverman M R. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9:773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- 4.Beck von Bodman S, Farrand S K. Capsular polysaccharide biosynthesis and pathogenicity in Erwinia stewartii require induction by an N-acylhomoserine lactone autoinducer. J Bacteriol. 1995;177:5000–5008. doi: 10.1128/jb.177.17.5000-5008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullock W O, Fernandez J M, Short J M. XL1-blue: a high efficiency plasmid transforming recA Escherichia coli strain with β-galactosidase selection. BioTechniques. 1987;5:376–379. [Google Scholar]
- 6.Chatterjee J, Miyamoto C M, Meighen E A. Autoregulation of luxR: the Vibrio harveyi lux-operon activator functions as a repressor. Mol Microbiol. 1996;20:415–425. doi: 10.1111/j.1365-2958.1996.tb02628.x. [DOI] [PubMed] [Google Scholar]
- 7.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gambello M J, Iglewski B H. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Givskov M, deNys R, Manefield M, Gram L, Maximilien R, Eberl L, Molin S, Steinberg P D, Kjelleberg S. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J Bacteriol. 1996;178:6618–6622. doi: 10.1128/jb.178.22.6618-6622.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gram L, deNys R, Maximilien R, Givskov M, Steinberg P, Kjelleberg S. Inhibitory effects of secondary metabolites from the red alga Delisea pulchra on swarming motility of Proteus mirabilis. Appl Environ Microbiol. 1996;62:4284–4287. doi: 10.1128/aem.62.11.4284-4287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray K M, Pearson J P, Downie J A, Boboye B E A, Greenberg E P. Cell-to-cell signaling in the symbiotic nitrogen-fixing bacterium Rhizobium leguminosarum: autoinduction of a stationary phase and rhizosphere-expressed genes. J Bacteriol. 1996;178:372–376. doi: 10.1128/jb.178.2.372-376.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 13.Hinrichs W, Kiser C, Düvel M, Müller A, Tovar K, Hillen W, Saenger W. Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance. Science. 1994;264:418–420. doi: 10.1126/science.8153629. [DOI] [PubMed] [Google Scholar]
- 14.Huisman G W, Kolter R. Sensing starvation: a homoserine lactone-dependent signaling pathway in Escherichia coli. Science. 1994;265:537–539. doi: 10.1126/science.7545940. [DOI] [PubMed] [Google Scholar]
- 15.Jobling M G, Holmes R K. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol Microbiol. 1997;26:1023–1034. doi: 10.1046/j.1365-2958.1997.6402011.x. [DOI] [PubMed] [Google Scholar]
- 16.Lazazzera B A. Quorum sensing and starvation: signals for entry into stationary phase. Curr Opin Microbiol. 2000;3:177–182. doi: 10.1016/s1369-5274(00)00072-2. [DOI] [PubMed] [Google Scholar]
- 17.Manefield M, deNys R, Kumar N, Read R, Givskov M, Steinberg P, Kjelleberg S. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology. 1999;145:283–291. doi: 10.1099/13500872-145-2-283. [DOI] [PubMed] [Google Scholar]
- 18.Manefield M, Harris L, Rice S A, deNys R, Kjelleberg S. Halogenated furanones from Delisea pulchra inhibit bioluminescence and virulence in the Paneus monodon pathogen Vibrio harveyi. Appl Environ Microbiol. 2000;66:2079–2084. doi: 10.1128/aem.66.5.2079-2084.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCarter L L. OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio parahaemolyticus. J Bacteriol. 1998;180:3166–3173. doi: 10.1128/jb.180.12.3166-3173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDougald D, Rice S A, Kjelleberg S. New perspectives on the viable but nonculturable response. Biologia. 1999;54:617–623. [Google Scholar]
- 21.McDougald D, Rice S A, Kjelleberg S. Vibrio vulnificus encodes a putative homologue of the Vibrio harveyi regulatory gene, luxR: a genetic and phylogenetic comparison. Gene. 2000;248:213–221. doi: 10.1016/s0378-1119(00)00117-7. [DOI] [PubMed] [Google Scholar]
- 22.Metcalf W W, Jiang W, Daniels L L, Kim S-K, Haldimann A, Wanner B L. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 23.Novick R P. Regulation of pathogenicity in Staphylococcus aureus by a peptide-based density-sensing system. In: Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D. C.: American Society for Microbiology; 1999. pp. 129–146. [Google Scholar]
- 24.Oliver J D. Formation of nonculturable Vibrio vulnificus cells and its relationship to the starvation state. Appl Environ Microbiol. 1991;57:2640–2644. doi: 10.1128/aem.57.9.2640-2644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliver J D. The viable but non-culturable state in the human pathogen Vibrio vulnificus. FEMS Microbiol Lett. 1995;133:203–208. doi: 10.1111/j.1574-6968.1995.tb07885.x. [DOI] [PubMed] [Google Scholar]
- 26.Oliver J D, Warner R A, Cleland D R. Distribution and ecology of Vibrio vulnificus and other lactose-fermenting marine vibrios in coastal waters of the southeastern United States. Appl Environ Microbiol. 1982;44:1404–1414. doi: 10.1128/aem.44.6.1404-1414.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paludan-Müller C, Weichart D, McDougald D, Kjelleberg S. Analysis of starvation conditions that allow for prolonged culturability of Vibrio vulnificus at low temperature. Microbiology. 1996;142:1675–1684. doi: 10.1099/13500872-142-7-1675. [DOI] [PubMed] [Google Scholar]
- 28.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 29.Pittard A J, Davidson B E. TyrR protein of Escherichia coli and its role as repressor and activator. Mol Microbiol. 1991;5:1585–1592. doi: 10.1111/j.1365-2958.1991.tb01904.x. [DOI] [PubMed] [Google Scholar]
- 30.Reitzer L J, Magasanik B. Expression of glnA in Escherichia coli is regulated at tandem promoters. Proc Natl Acad Sci USA. 1985;82:1979–1983. doi: 10.1073/pnas.82.7.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sperandio V, Mellies J L, Nguyen W, Shin S, Kaper J B. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc Natl Acad Sci USA. 1999;96:15196–15201. doi: 10.1073/pnas.96.26.15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srinivasan S, Ostling J, Charlton T, deNys R, Takayama K, Kjelleberg S. Extracellular signal molecule(s) involved in the carbon starvation response of marine Vibrio sp. strain S14. J Bacteriol. 1998;180:201–209. doi: 10.1128/jb.180.2.201-209.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Surette M G, Bassler B L. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc Natl Acad Sci USA. 1998;95:7046–7050. doi: 10.1073/pnas.95.12.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Surette M G, Miller M B, Bassler B L. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci USA. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swift S, Lynch M J, Fish L, Kirke D F, Tomás J M, Stewart G S A B, Williams P. Quorum sensing-dependent regulation and blockade of exoprotease production in Aeromonas hydrophila. Infect Immun. 1999;67:5192–5199. doi: 10.1128/iai.67.10.5192-5199.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorne S H, Williams H D. Cell density-dependent starvation survival of Rhizobium leguminosarum bv. phaseoli: identification of the role of an N-acyl homoserine lactone in adaptation to stationary-phase survival. J Bacteriol. 1999;181:981–990. doi: 10.1128/jb.181.3.981-990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tillett D, Neilan B A. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J Phycol. 2000;36:1–8. [Google Scholar]
- 38.Wilson K J, Sessitsch A, Corbo J C, Giller K E, Akkermans A D L, Jefferson R A. β-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other Gram-negative bacteria. Microbiology. 1995;141:1691–1705. doi: 10.1099/13500872-141-7-1691. [DOI] [PubMed] [Google Scholar]
- 39.Wolf P W, Oliver J D. Temperature effects on the viable but nonculturable state of Vibrio vulnificus. FEMS Microbiol Ecol. 1992;101:33–39. [Google Scholar]
- 40.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 41.You Z, Fukushima J, Tanaka K, Kawamoto S, Okuda K. Induction of entry into stationary growth phase in Pseudomonas aeruginosa by N-acylhomoserine lactone. FEMS Microbiol Lett. 1998;164:99–106. doi: 10.1111/j.1574-6968.1998.tb13073.x. [DOI] [PubMed] [Google Scholar]