Abstract
Oxygen controls competence development in Streptococcus pneumoniae. Oxygen signaling involves the two-component signal transduction systems CiaRH and ComDE and the competence-stimulating peptide encoded by comC and processed by ComAB. We found that NADH oxidase (Nox) was required for optimal competence. Transcriptional analysis and genetic dissection showed that Nox was involved in post-transcriptional activation of the response regulator ComE and in the transcriptional control of ciaRH and comCDE. Thus, in S. pneumoniae, Nox, with O2 as its secondary substrate, is part of the O2-signaling pathway.
Competence development in Streptococcus pneumoniae is strongly dependent on the availability of oxygen in cultures. The comCDE operon, which encodes ComC, the precursor of the competence-stimulating peptide, and its dedicated two-component system (TCS), ComDE (4, 5), is the major target of this regulation. A correlation has been found between the level of comCDE transcripts and developmental transformability in culture. Another TCS, encoded by ciaRH, is involved in this regulation (3). Loss-of-function mutations in ciaRH derepress competence, suggesting that ciaRH negatively regulates the level of comCDE transcripts and competence development (2). The NADH oxidase (Nox) is required for optimal competence expression in aerobic cultures. Loss-of-function mutations in nox reduce bacterial transformability by 50% and alter its expression pattern (1). The role of Nox in signaling the O2 status of the cultures has been investigated. We assessed transformability and the levels of comCDE and ciaRH mRNA in strains carrying a nox loss-of-function mutation alone or in addition to the comE38KE gain-of-function missense mutation and the ciaR loss-of-function mutation. The results obtained show that Nox, whose enzymatic activity relies on oxygen availability, belongs to the O2 signaling network.
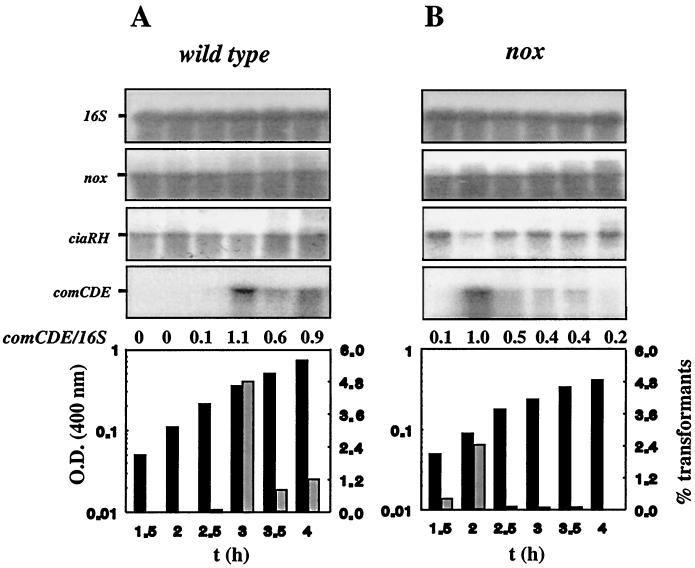
Influence of Nox on levels of ciaRH and comCDE mRNA.
It has been suggested that the O2 reductase/NADH oxidase, Nox, of S. pneumoniae is involved in O2 sensing (1). We therefore compared the levels of comCDE and ciaRH mRNA in Nox null mutant (Nox0) and wild-type bacteria to determine whether Nox affected the transcription of these TCSs involved in the O2 response (2).
Cp8056 (nox71K stop codon mutation) and Cp1015 (nox+) (Table 1) were subjected to a transformation test, and total RNA from these strains was subjected to Northern blotting with comE- and ciaR-specific DNA probes, as previously described (2). In Cp8056, we observed an early and narrow transformability window, not exceeding an optical density at 400 nm (OD400) of 0.1 to 0.2, typical of Nox-defective strains, with an early competence peak and a rapid loss of transformability during exponential growth (1). The amount of comCDE mRNA paralleled transformability. The inverse pattern was observed with ciaRH transcripts. Four to five times more ciaRH mRNA was detected in cultures in which competence was abolished than in the wild-type cultures. In cultures grown for 2 h, in which competence was maximal, the level of comCDE transcripts increased as the level of ciaRH decreased to wild-type levels (Fig. 1B). This suggests that opposite regulatory events control comCDE and ciaRH transcript levels in nox strains. Therefore, Nox influences the expression of both ciaRH and comCDE. It should be stressed that nox expression was not controlled by Nox activity, regardless of genetic background and growth status (Fig. 1), as previously suggested by Western blot analysis and specific activity measurements (1).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Phenotype | Source or reference |
|---|---|---|---|
| Strains | |||
| Cp1015 | Rx derivative, str1 hexA | Smr | 7 |
| Cp1800 | ciaR::pPt4 | Spr | 2 |
| Cp1856 | nox71K stop codon; ciaR::pPT4; this strain was obtained by transformation of Cp8056 with pPT4 as described previously (2) | Spr | This work |
| Cp6600 | comE38KE | Rifr | 2 |
| Cp6656 | comE38KE nox71K stop codon; this strain was obtained by transformation of Cp6600 with pN1 as described previously (1) | Rifr | This work |
| Cp8056 | nox71K stop codon | 1 | |
| Plasmids | |||
| pN1 | Mutagenic plasmid containing a stop codon at position 71K of nox | Apr | 1 |
| pNOX2 | The nox gene inserted into pMOS-Blue; a 1.1-kb EcoRI-BamHI fragment was used as the nox probe | Apr | 1 |
| pPS16 | The 16S rRNA gene inserted into pAM239; this DNA fragment was used as the 16S rRNA probe | Spr | 2 |
| pPT4 | An internal fragment of ciaR inserted into pAM239; this DNA fragment was used as the ciaR probe | Spr | 2 |
| pPT18 | The comCDE operon inserted into pWSK29; a 0.58-kb SpeI-PstI fragment was used as the comE probe | Apr | 2 |
FIG. 1.
Effect of the nox71K stop codon mutation on levels of ciaRH and comCDE mRNA and competence development. Aliquots of cultures of Cp1015 (A) and Cp8056 (nox) (B) growing under aerobiosis in CTM medium (1) were withdrawn at 30-min intervals. The change in biomass was measured as OD400 (black bars), and cultures were subjected to the transformation test (grey bars) or Northern blotting (2). For transformation tests, chromosomal DNA (1 μg/ml) carrying the rif-23 allele, conferring resistance to rifampin (2 μg/ml), was used. Rifr transformants were selected on plates, and their frequency in the population was calculated as described by Auzat et al. (1). For Northern blotting, probes specific for comE, nox, and ciaR were used to detect the corresponding 2.4-, 1.4-, and 2.1-kb mRNAs, with 16S rRNA used as a qualitative and quantitative internal control. Quantitative densitometry of 16S rRNA showed differences of <18% between samples taken throughout the period of exponential growth from a given culture. The experiment was repeated with independent cultures to test reproducibility. The columns are aligned with the corresponding signal in the Northern blots.
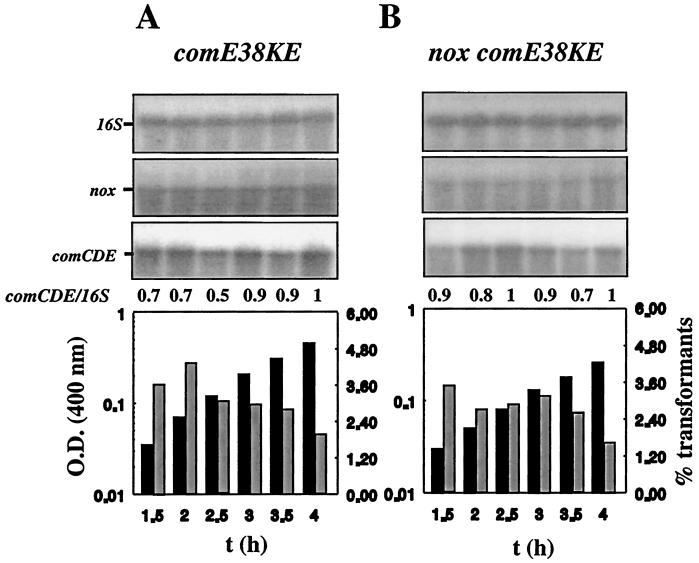
Role of Nox in signal transduction.
We used genetic dissection to investigate the correlation between the specific regulation of comCDE and ciaRH mRNA levels and the transformability of nox strains. Strain Cp6656 (nox71K stop codon, comE38KE) had a transformability profile and level of competence development similar to those of strain Cp6600 (comE38KE) (Fig. 2), indicating that the gain-of-function mutation comE38KE is epistatic to nox. Therefore, there was enough active ComE38KE in this Nox-defective strain for full transformability throughout the growth cycle. This demonstrates that Nox activity intervenes in maintaining threshold cellular levels of active ComE for competence or in the ComE activation step.
FIG. 2.
Effect of the nox71K stop codon mutation on competence development in the comE38KE (Cp6656) genetic background. Strains Cp6600 (comE38KE) (A) and Cp6656 (nox comE38KE) (B) were grown under aerobiosis and analyzed as described in the legend to Fig. 1. Blots were hybridized separately with probes for 16S rRNA, comE, and nox. 16S rRNA signals differed by <20% between wells. The experiment was performed three times with independent cultures to test reproducibility.
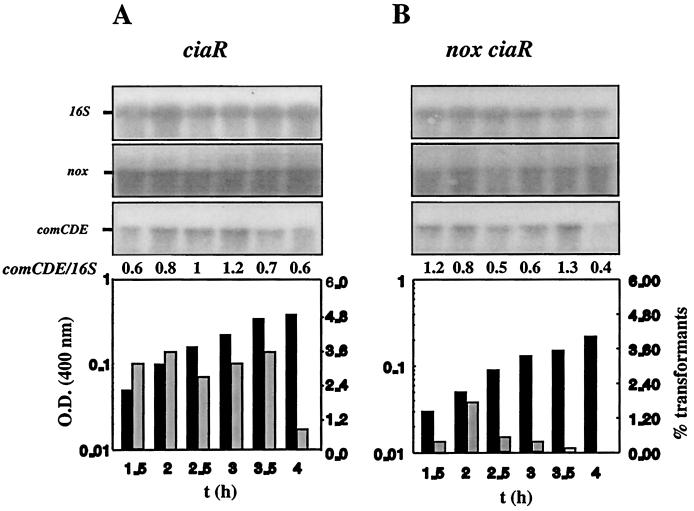
Cultures of strain Cp1856 (nox71K stop codon, ciaR) were also subjected to the competence test, and RNA from this strain was subjected to Northern blotting with comE-specific DNA. The results obtained were compared to those for Cp1800 (ciaR). In contrast to Cp1800, which is transformable throughout the growth cycle (Fig. 3A), the transformability pattern of Cp1856 (Fig. 3B) was similar to that of Cp8056 (Fig. 1B), with a peak early in exponential growth and a rapid decrease. However, the ciaR insertion mutation resulted in high levels of comCDE transcripts being present throughout the period of exponential growth, whatever the genetic background (nox+ and nox). This suggests that the absence of CiaR is sufficient for high levels of comCDE mRNA to be maintained in cells. It is possible that both CiaR and ComE control comCDE transcription independently. A negative control of CiaR in addition to the positive control of ComE might adjust the cellular levels of comCDE mRNA to cell needs. In line with this proposal it was shown that a ciaR loss-of-function mutation and a comE38KE gain-of-function mutation exhibit additive effects on competence derepression, leading to transformant yields of 25 to 30% in strains carrying both mutations (data not shown). For strain Cp8456 (ciaR nox), in which comCDE transcription was increased due to a plasmid insertion mutation in ciaR, the level of comCDE mRNA was not related to transformability in cultures at ODs higher than 0.1. Nox deficiency prevented further steps of signal transduction, resulting in the abortive transcription of comCDE in bacteria from cultures with an OD400 greater than 0.1. This effect of Nox was abolished by the comE38KE mutation (Fig. 2). Thus, Nox is specifically involved in the control of active ComE, which is essential for the expression of late competence genes.
FIG. 3.
Effect of the nox71K stop codon mutation on competence development in the ciaR genetic background. Strains Cp1800 (ciaR) (A) and Cp 1856 (nox ciaR) (B) were grown under aerobiosis and analyzed as described in the legend to Fig. 1. Blots were hybridized separately with probes for 16S rRNA, comE, and nox. 16S rRNA signals differed by <20% between wells. The experiment was performed three times with independent cultures to test reproducibility.
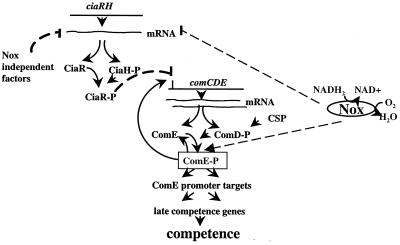
These results suggest that Nox exerts transcriptional control over ciaRH and comCDE and is required for the maintenance of threshold levels of active ComE during exponential growth for the positive regulation of late competence genes. Both effects culminate in the high yield and specific pattern of competence expression in cultures growing exponentially (see Fig. 4 for a possible regulatory scheme).
FIG. 4.
Possible regulatory scheme involving Nox and the CiaRH and ComDE TCSs, as deduced from genetic dissection.
Nox is involved in the recycling of NADH, thereby controlling the redox status of the cells. O2 is a secondary substrate of Nox, and the activity of Nox therefore depends directly on O2 availability. The development of competence in cultures probably involves metabolic signals produced by Nox and transduced via ciaRH and comDE, with the competence-stimulating peptide as an intercellular mediator. Loss-of-function mutation in the nox gene results in residual competence levels, while in wild-type bacteria grown under microaerobic conditions competence is below the detection level (2). Thus, another route in addition to that involving Nox must be involved in O2 signaling in S. pneumoniae. Nox activity also determines the virulence of this pathogen (1). It has recently been reported that the ciaR insertion mutation greatly reduces virulence (8) and facilitates competence development in bacteria grown under microaerobiosis (2). Steps in the cellular response to metabolic “messengers” produced by the O2-dependent Nox and transduced by the transfer of phosphate groups may be common to virulence and competence. This is the first time that such a mechanism, mediating O2 signaling via the activity of an O2-dependent Nox and related to TCS-mediated signal transduction, has been described. The high conservation of Nox among pathogens, notably Streptococcus faecalis and Streptococcus pyogenes (1, 6), raises the question as to whether similar mechanisms for O2 adaptation exist in other pathogenic species.
Acknowledgments
This work was supported by Université Paul Sabatier Toulouse and Rhône-Poulenc Rorer, France. J.R.E was supported by an RPR postdoctoral fellowship.
We thank Delphine Dos Santos and Suzanne Eychenne for technical assistance. We also thank the Technical Department of Institut Louis Bugnard/INSERM, Toulouse, for providing access to certain pieces of apparatus.
REFERENCES
- 1.Auzat I, Chapuy-Regaud S, Dos Santos D, Le Thomas I, Le Bras G, Onnuyighy D, Garel J-R, Paton J, Trombe M-C. The NADH oxidase of Streptococcus pneumoniae, its role in competence and virulence. Mol Microbiol. 1999;34:1018–1028. doi: 10.1046/j.1365-2958.1999.01663.x. [DOI] [PubMed] [Google Scholar]
- 2.Echenique J, Chapuy-Regaud S, Trombe M-C. Oxygen regulation of competence in Streptococcus pneumoniae: involvement of ciaRH and comCDE. Mol Microbiol. 2000;36:688–696. doi: 10.1046/j.1365-2958.2000.01891.x. [DOI] [PubMed] [Google Scholar]
- 3.Guenzi E, Gasc A M, Sicard M A, Hakenbeck R. A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol Microbiol. 1994;12:505–515. doi: 10.1111/j.1365-2958.1994.tb01038.x. [DOI] [PubMed] [Google Scholar]
- 4.Havarstein L S. Identification of a competence regulon in Streptococcus pneumoniae by genomic analysis. Trends Microbiol. 1998;6:297–299. doi: 10.1016/s0966-842x(98)01328-6. [DOI] [PubMed] [Google Scholar]
- 5.Havarstein L S, Coomaraswamy G, Morrison D A. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mallett T C, Claiborne A. Oxygen reactivity of an NADH oxidase C42S mutant: evidence for a C(4a)-peroxyflavin intermediate and a rate-limiting conformational change. Biochemistry. 1998;37:8790–8802. doi: 10.1021/bi9803630. [DOI] [PubMed] [Google Scholar]
- 7.Morrison D A, Trombe M-C, Hayden M K, Waszak G A, Chen J D. Isolation of transformation-deficient Streptococcus pneumoniae mutants defective in control of competence, using insertion-duplication mutagenesis with the erythromycin resistance determinant of pAM beta 1. J Bacteriol. 1984;159:870–876. doi: 10.1128/jb.159.3.870-876.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Throup J P, Koretke K K, Bryant A P, Ingraham K A, Chalker A F, Ge Y, Marra A, Wallis N G, Brown J R, Holmes D J, Rosenberg M, Burnham M K. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol Microbiol. 2000;35:566–576. doi: 10.1046/j.1365-2958.2000.01725.x. [DOI] [PubMed] [Google Scholar]