Abstract
A limitation for recombinant adeno-associated virus (rAAV)-mediated gene transfer into the central nervous system (CNS) is the low penetration of vectors across the human blood-brain barrier (BBB). High doses of intravenously delivered vector are required to reach the CNS, which has resulted in varying adverse effects. Moreover, selective transduction of various cell types might be important depending on the disorder being treated. To enhance BBB penetration and improve CNS cell selectivity, we screened an AAV capsid-shuffled library using an in vitro transwell BBB system with separate layers of human endothelial cells, primary astrocytes and/or human induced pluripotent stem cell-derived cortical neurons. After multiple passages through the transwell, we identified chimeric AAV capsids with enhanced penetration and improved transduction of astrocytes and/or neurons compared with wild-type capsids. We identified the amino acids (aa) from regions 451–470 of AAV2 associated with the capsids selected for neurons, and a combination of aa from regions 413–496 of AAV-rh10 and 538–598 of AAV3B/LK03 associated with capsids selected for astrocytes. A small interfering RNA screen identified several genes that affect transcytosis of AAV across the BBB. Our work supports the use of a human transwell system for selecting enhanced AAV capsids targeting the CNS and may allow for unraveling the underlying molecular mechanisms of BBB penetration.
Keywords: blood brain barrier, CNS capsids, transwell, human BBB, central nervous system, chimeric AAV vectors
Graphical abstract
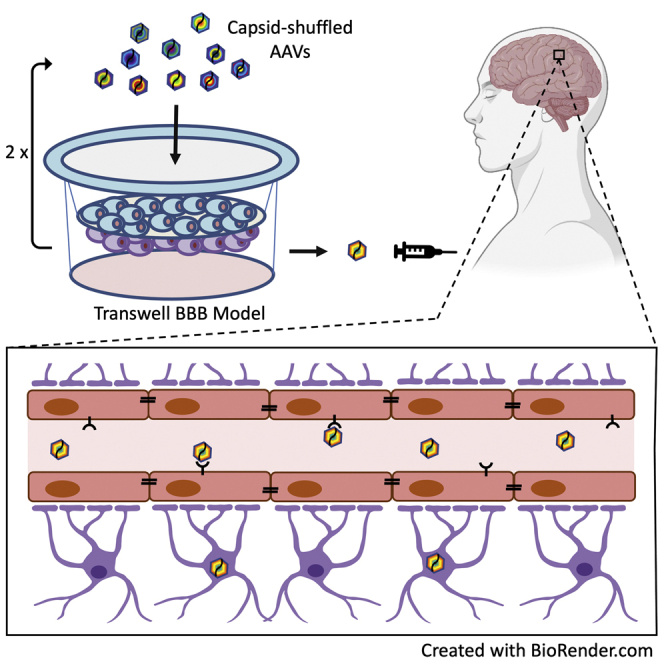
Many genetic and acquired CNS disorders could be treated using rAAV gene therapy if widespread dissemination of vector throughout the brain can be achieved. We have used a human transwell BBB model to achieve selection of AAV vectors with improved transport across the BBB and transduction of cells of CNS.
Introduction
Recombinant adeno-associated viral (rAAV) vectors have become one of the most popular gene therapy delivery tools for the clinical treatment of genetic disorders. Zolgensma is an US Food and Drug Administration-approved gene therapy product that uses an rAAV9 vector to deliver the SMN1 gene into motor neurons for the treatment of patients with lethal spinal muscular atrophy.1 Both AAV9 and AAV-rh10 are the current gold standard capsids known to cross the blood-brain barrier (BBB) and most often used for delivery into the central nervous system (CNS).2,3 Nonetheless, AAV9 and AAV-rh10 still have limited BBB penetration and CNS transduction. Owing to these limitations, high doses of AAV-based therapeutics are required to reach the target cells, while a large amount of the vector are taken up by other tissues leading to varying degrees of adverse effects, including death.4 Therefore, efficient delivery of cell type-specific rAAV vectors across the BBB represents an important goal for the field.
The BBB is mainly composed of endothelial cells that line the walls of the brain capillaries, which account for 85% of vessel length in the brain.5 Tight junctions formed between these endothelial cells limit paracellular transport of molecules between the blood and the brain. This ensures that molecular exchange between the circulating blood and the brain is tightly regulated to achieve a homeostatic supply of nutrients to the brain, while also fending off harmful molecules to prevent unwanted neuronal excitation. On the neural tissue side of the BBB, pericytes and astrocytes directly associate with the endothelial cells to further strengthen the integrity and functional maintenance of the BBB. Delivery across the BBB remains one of the most efficient way to achieve widespread disseminated gene transfer into the cells in the CNS.6
A number of laboratories have been working toward developing rAAV vectors that can efficiently cross the BBB in rodent or non-human primate (NHP) models.7 However, one of the major limitations is the lack of predictability of AAV transduction efficiency between species. A notable example is AAV-PHP.B, which was selected from a screen in C57BL/6 mice and showed significant improvement over AAV9.8 However, this improvement was not observed when AAV-PHP.B was tested in the NHP models.9,10 Further investigations found AAV-PHP.B specifically engages the LY6A receptor that is functional only in certain strains of mice.11,12 Another challenge is that capsids selected for improved BBB penetration do not necessarily transduce the target cell types with high efficiency. Many rAAV vectors that are able to successfully pass the BBB have a low preference toward neurons, while the transduction of supporting cells that directly associate with the BBB, such as astrocytes, is relatively more efficient.2,13 Methods in using transgenic rodent models carrying various cell type-specific promoters have facilitated the selection of AAVs that specialize in transduction of certain cells.14 However, these selection methods remain difficult to adapt to human models. More recently, parallel screens of AAV capsids have been performed in various human induced pluripotent stem cell (hiPSC)-derived neural cell types, but the BBB penetration abilities of these AAVs remain untested.15
To overcome the unpredictable differences in the BBB between species and select for cell type specific AAVs, we have employed an in vitro model of the human BBB that consists of a co-culture of human cerebral microvasculature endothelial cells (hCMEC/D3) and primary astrocytes in a transwell system.16 We also used hiPSC neurons derived in three-dimensional cortical organoids to select for enhanced transduction efficiency.17 We validated this BBB model by demonstrating that AAV9 and AAV-rh10 capsids pass the BBB more efficiently than other wild-type AAV capsids. Using a barcoded and capsid-shuffled AAV library18 to select for improved capsids in this human BBB transwell system, we were able to isolate a repertoire of capsids with an enhanced ability to penetrate the BBB and transduce hiPSC-derived cortical neurons and/or primary astrocytes. These novel AAV capsids not only provide new development opportunities for human gene therapeutics for the CNS, but also serve as tools to identify molecular mechanisms that facilitate BBB penetration and determine CNS cell type specificity.
Results
Establish and validate a transwell model of human BBB for selection of AAVs
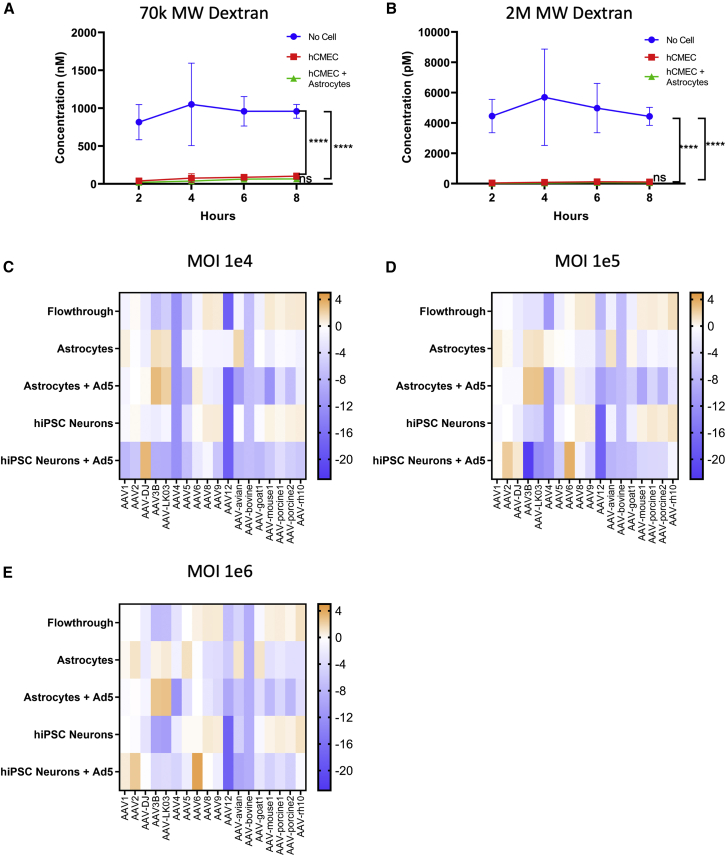
To overcome the species and cell-type limits of the current AAV vectors, we decided to select AAV vectors in a transwell culture model of the human BBB that is commonly used to study BBB structure and function.16 A key component of the BBB is the layer of endothelial cells that form tight junctions to prevent large molecules crossing from the blood into the brain by paracellular transport.5 We cultured a confluent layer of human cerebral microvascular endothelial cells (hCMEC/D3) on the top layer of the transwell system (Figure S1A). The transwell consists of a polyester membrane with 0.4 μm pores, which keeps the endothelial cells (10–20 μm) on the upper side of the transwell, while allowing AAV (approximately 20 nm) to cross as long as they are able to penetrate the endothelial cell layer. A culture time of 7 days was sufficient to prevent the diffusion of dextran molecules of 70,000 MW (regularly used to test cell layer integrity) and 2,000,000 MW (approximately one-half the size of AAV) from crossing the transwell (Figures 1A and 1B). Furthermore, a higher number of AAV9 penetrated the layer of hCMEC/D3 cells into the media below (flowthrough) as compared with AAV2 (Figure S1B). This observation aligns well with previously published data comparing AAV9 and AAV2 in the same transwell model using primary human brain microvascular endothelial cell cultures.19 As astrocytes are directly associated with endothelial cells in the BBB, we added a layer of primary human astrocytes to the bottom side of the porous membrane (Figure S1A). The addition of the astrocytes did not interfere with the dextran permeability of the hCEMC/D3 cells (Figure 1A) and allowed us to perform selection of AAVs that would enter the astrocytes after crossing the hCMEC/D3 cells.
Figure 1.
Establish and validate a transwell model of human BBB for selection of AAVs
(A and B) Permeability of transwells with no cells, hCMEC/D3 or hCMEC/D3 and human primary astrocytes were tested using 70k MW (A) and 2M MW (B) dextran. Significance determined by two-way ANOVA test, ∗∗∗∗p < 0.0001. (C–E) Eighteen uniquely barcoded AAVs were pooled and passaged through the transwell BBB model with hCMEC/D3, primary astrocytes and hiPSC-derived cortical neurons with MOI of 1e4 (C), 1e5 (D), or 1e6 (E). The log2-fold change of the percentage of each AAV in flowthrough media, primary astrocytes with or without Ad5, and in hiPSC-derived cortical neurons with or without Ad5 are graphed in comparison to the percentage in the input.
To further demonstrate that the transwell system indeed recapitulates the human BBB in allowing only certain known AAV vectors to cross efficiently, we tested a pool of 18 uniquely barcoded AAVs by passaging them through our model system. This pool of barcoded AAVs had been previously developed by our group to test other model systems and to optimize the AAV selection strategy.18,20 The pool included AAVs known to cross the BBB more efficiently, such as AAV9 and AAV-rh10, and those previously described not to cross the BBB (e.g., AAV-DJ).2,3 When tested in the transwell BBB model, we first compared the virus composition in the input and flowthrough of the transwell and found that AAV-rh10 and AAV9 are among the viruses whose proportions increased the most in the flowthrough compared with input (Figures 1C–1E and Table S1). Depending on the multiplicity of infection (MOI) used, the increase was 2.0- to 2.8-fold for AAV-rh10 and 1.8- to 1.9-fold for AAV9. AAV8, AAV-mouse1, AAV-porcine1, and AAV-porcine2 also showed increased proportions of up to 2.0-fold in the flowthrough. Comparatively, flowthrough proportions of AAV-DJ decreased more than four-fold in comparison with the input. The flowthrough from each transwell was also added to hiPSC-derived neurons, isolated by immunopanning from human cortical organoids, to evaluate the transduction efficiency of the respective AAVs (Figure S1A). We found that the proportion of viruses that transduced the hiPSC-derived cortical neurons were not significantly different from that in the flowthrough (Figures 1C–1E and Table S1). These results suggest that the pool of AAVs found in the flowthrough was a good indicator for neuron transduction efficiency.
However, the composition of the pool of viruses that entered the layer of primary human astrocytes was drastically different from that of the flowthrough and in hiPSC-derived neurons (Figures 1C–1E and Table S1). The AAVs that showed a consistent increase in astrocytes across of all MOIs tested include AAV1 (1.2- to 1.9-fold), AAV3B (1.5- to 2.8-fold), AAV-LK03 (1.8- to 2.5-fold), and AAV-avian (2.2- to 3.4-fold). These data suggest that the transwell BBB model will allow us to select for BBB-penetrating capsids with different preferences in transducing different types of cells in the CNS.
We then proceeded to evaluate how the addition of adenovirus 5 (Ad5) to the hiPSC-derived cortical neurons or primary astrocytes would affect the proportion of AAVs that transduced these cells. We found that Ad5 resulted in the dominance of variants that replicate well in the respective cell type, a phenomenon that had been previously described.20 We found that AAV-DJ, AAV2, or AAV6 had become the dominating species in hiPSC-derived cortical neurons, while AAV3B and AAV-LK03 had taken over more than 68% of the population in astrocytes (Figures 1C–1E and Table S1). Although removing Ad5 from the selection process may alleviate this bias, the capsids selected in the absence of Ad5 may not necessarily exhibit improved transduction efficiency, since multiple steps after cell entry are important for successful transgene expression. In addition, we observed that viral copy numbers were very low when Ad5 was omitted from the screen, making consecutive rounds of selection difficult. Thus, we decided to conduct hybrid schemes of selection of our AAV library with or without the addition of Ad5.
Selection of AAV vectors that efficiently cross the BBB and transduce hiPSC-derived cortical neurons and primary astrocytes
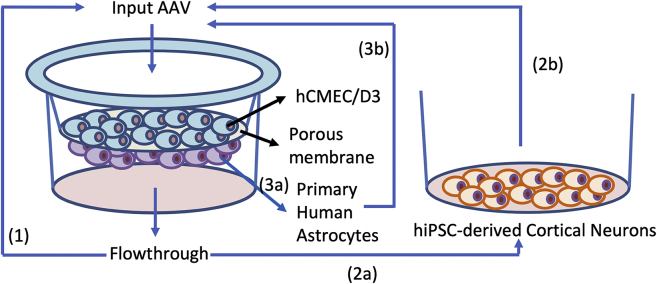
Over the past 20 years, our laboratory has established methods to make complex multispecies interbred AAV capsid libraries.18,21, 22, 23, 24 More recently, we have generated capsid-shuffled libraries where each AAV harbors a short and unique barcode sequence downstream of the cap polyA signal.18,25 This method allowed us to track AAV variant enrichment by high-throughput sequencing of the barcodes. We added an AAV library that contained chimeric capsids derived from 10 parents to the top of the transwell human BBB model and performed three separate selection experiments (Figure 2). The selections were carried out in two consecutive rounds in the flowthrough, hiPSC-derived cortical neurons and primary astrocytes. For the selection in flowthrough that enriched for AAV variants capable of BBB penetration, we did not use Ad5 to replicate viral variants throughout the entire process. We also did not include Ad5 during the selection process of capsids that efficiently cross the endothelial cell layer and transduce neurons or astrocytes. However, to generate sufficient input for the second round of selection, Ad5 was added to the neurons and astrocytes after the first round of selection. The barcode sequences of AAV from the input and output of each round of the three selections were identified by Illumina sequencing. Each selection was done in biological replicates and we identified barcodes that were present in both replicates and were continuously enriched in two rounds of selection. We found a total of 25, 90, and 250 such barcodes from the flowthrough, neuron, and astrocyte screens, respectively.
Figure 2.
Selection of AAV vectors in the human BBB transwell model
Three separate selection schemes were conducted using an barcoded and capsid-shuffled AAV library (MOI 1e6) as input added to the top of the transwell containing hCMEC/D3 and primary astrocytes. (1) AAVs collected from media in the flowthrough at 24 h were determined by sequencing and were added to a new transwell for a second round of enrichment. (2a) Media from flowthrough at 24 h was added to hiPSC-derived cortical neurons to select for AAVs that transduce this cell type. (2b) In a separate culture of hiPSC-derived neurons, media from flowthrough at 24 h were added along with Ad5 to generate input for the second round of selection in transwell and in hiPSC-derived neurons as in (2a). (3a) Astrocytes from the transwell were collected at 24 h to determine AAVs that transduce this cell type. (3b) In a separate transwell, Ad5 was added to the astrocytes at 24 h to generate input for the second round of selection in astrocytes as in (3a).
Validation of selected rAAV vectors in crossing the transwell BBB model
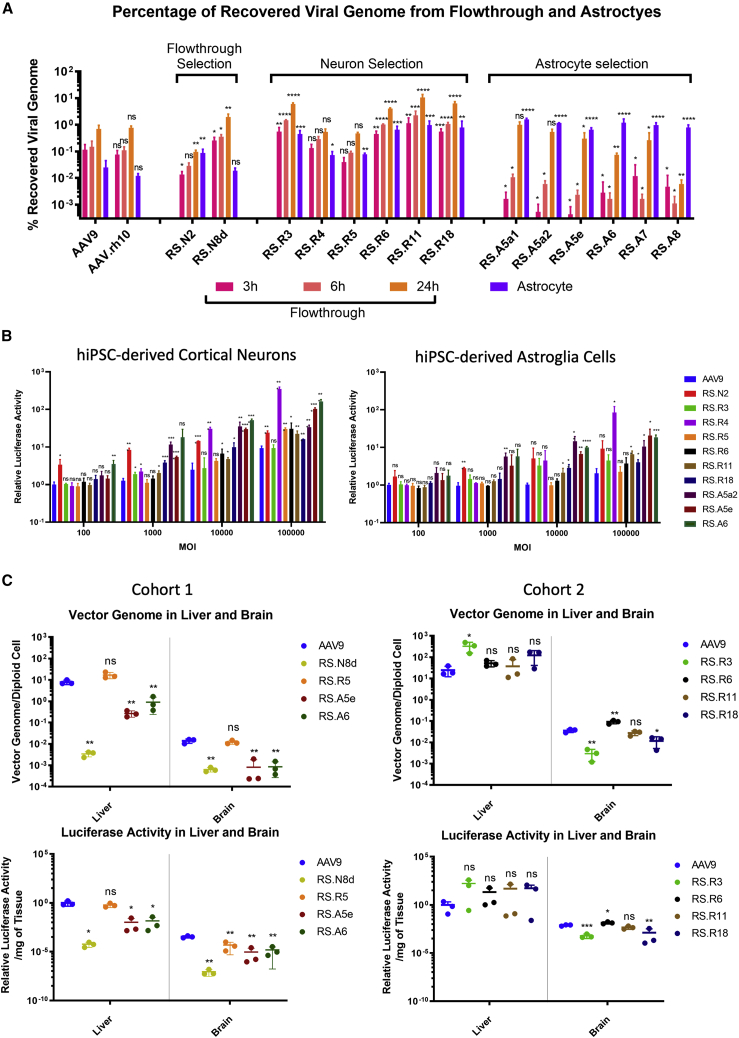
We vectorized a total of 14 AAV capsid sequences, 2 from the flowthrough selection, 6 from the neuron selection, and 6 from the astrocyte selection. Each rAAV capsid was used to package a firefly luciferase expression construct under transcriptional control of a CAG promoter (CAG-Fluc). We first tested each capsid variant separately for their ability to cross both the hCMEC/D3 and primary astrocyte cell layers in the transwell BBB model by measuring the abundance of vector genomes in the flowthrough (Figure 3A). The AAVs from the flowthrough and neuron selections crossed the transwell with high efficiency as early as 3 h post transduction (hpt) and continued to transcytose during the 24-h testing period. When compared with the control AAV9 at 24 hpt, we found 5 of the rAAV vectors were 2.8-fold (RS.N8d), 8.4-fold (RS.R3), 5.7-fold (RS.R6), 14.8-fold (RS.R11), and 8.9-fold (RS.R18) more efficient. Two of the rAAV vectors, RS.R4 and RS.R5, showed similar efficiency to the controls. RS.N2 was the only variant that was less efficient at crossing than AAV9 and AAV-rh10. The AAV capsids selected from the astrocyte screen, in contrast, were all less efficient at crossing the hCMEC/D3 and primary astrocyte cell layers especially at 3 and 6 hpt. Although RS.A5a1 and RS.A5a2 reached similar levels to the two control AAVs at 24 hpt, the four other AAVs remained less efficient when compared with AAV9 and AAV-rh10.
Figure 3.
Validation of Selected rAAV vectors in transwell, hiPSC-derived cortical neurons and astroglia cells, and mice
(A) Fourteen AAV capsids from the three selections were vectorized and tested in the transwell BBB system with hCMEC/D3 and primary astrocytes, each rAAV was tested in separate transwells and viral genome copy numbers as a percentage of input virus were determined in flowthrough media at 3, 6, and 24 h and in astrocytes at 24 h. (B) Transduction efficiency of 12 AAV capsids from the three selections packaged with firefly luciferase expression cassette were tested in hiPSC-derived cortical neurons and astroglia cells. Relative luciferase activity as compared with AAV9 in each cell type were determined after 72 h. (C) Delivery of vector genomes and expression of luciferase in mouse liver and brain after retro-orbital injection was determined for RS.N8d, RS.R5, RS.A5e, and RS.A6 at 21 days and for RS.R3, RS.R6, RS.R11, and RS.R18 at 28 days. n ≥ 3 for all conditions, significance determined by the Student t test; ns, not significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
We selected the representative rAAV vectors from each selection (RS.R5, RS.R6, RS.R11, RS.R18, RS.A5e, and RS.A6), and package each capsid with a CAG human frataxin (CAG-hFXN) expression construct with a unique barcode sequence between the stop codon and polyA signal. This allowed us to pool the AAV capsids together with AAV9 and AAV-DJ at equimolar ratios, and tested their ability to cross the same transwell BBB when subjected to competition (Figure S2). We found that the data from this pooled approach correlated well with the data obtained when each virus was tested separately. RS.R6, RS.R11 and RS.R18 remained the most efficient variants at crossing into flowthrough, RS.R5 performed similar to AAV9, while RS.A5e, RS.A6, and DJ were the least efficient capsid variants. Taken together, these data suggest that our flowthrough and neuron selection schemes in the transwell BBB model were highly effective at enriching for AAVs that cross the endothelial and astrocyte layers efficiently.
Validation of selected rAAV vectors in transducing primary astrocytes in the transwell BBB model
Using the same transwell experiments as described above, we examined the efficiency of each rAAV capsid variant in transducing the primary astrocytes after crossing the endothelial cells. We found that all rAAV variants from the astrocyte selection delivered higher amounts of vector genome DNA to astrocytes than the two control serotypes AAV9 and AAV-rh10 (Figure 3A). These rAAV vectors also delivered higher proportions of vector DNA to astrocytes compared with the flowthrough. Apart from RS.N8d, the rAAV capsids derived from the flowthrough and neuron screens were also capable of delivering higher amounts of vector DNA to astrocytes when compared with the control AAVs. However, in contrast with the variants derived from the astrocyte screen, delivery of vector genomes into astrocytes was less efficient than the flowthrough media. When we tested the rAAV vectors in the pooled approach, we found that RS.A5e and RS.A6, two capsid variants enriched in the astrocyte selection, were the most effective at delivering vector genome DNA to astrocytes (Figure S2). When the expression of vector RNA was tested, RS.A6 remained the most dominant in astrocytes, while the performance of RS.A5e dropped behind RS.R11 and RS.R18, but remained more efficient than the other vectors in the pool (Figure S2). These data suggest that our astrocyte selection scheme in the transwell BBB model can select for AAV capsid variants that cross the endothelial layer and transduce astrocytes.
Testing transduction efficiency of the selected rAAV vectors in hiPSC-derived cortical neurons and astroglia cells as well as hCMEC/D3
The transduction efficiency of the selected vectors was examined in hiPSC-derived neurons and astroglia that were isolated from cortical organoids using immunopanning for Thy1 and HepaCAM as previously shown.26 We transduced the two cell types with rAAV vectors carrying the CAG-Fluc construct at increasing MOI from 100 to 100,000 (Figure 3B). Apart from RS.R3, all rAAV vectors from the flowthrough and neuron selection were more efficient at transducing hiPSC-derived neurons than AAV9, especially at a higher MOI. Notably, while RS.N2 and RS.R4 were the most efficient at direct transduction of hiPSC-derived neurons, these two rAAV vectors were the least efficient at penetrating the endothelial and primary astrocyte cell layers (Figure 3A), suggesting a negative correlation between the ability to transduce and the efficiency to cross the BBB in this model system. RS.N2, RS.R4, RS.11, and RS.18 were also more efficient at transducing hiPSC-derived astroglia at a higher MOI. All three capsids derived from the astrocyte screen were more efficient at transducing both human cortical organoid neurons and astroglia as compared with AAV9. We also examined the transduction of hCMEC/D3 cells with the pooled rAAV vectors RS.R5, RS.R6, RS.R11, RS.R18, RS.A5e, RS.A6, AAV9, and AAV-DJ carrying uniquely barcoded CAG-hFXN construct (Figure S2). While AAV-DJ was the dominating species both in terms of vector DNA delivery and RNA expression, all representative rAAV vectors derived from our various screens also showed higher transduction of endothelial cells when compared with AAV9.
Evaluation of the selected rAAV vectors in a mouse model
Although mouse models may not be the most relevant for the prediction of AAV capsids that can cross the human BBB, we examined representative rAAVs selected in our screens for their ability to transduce the liver and brain in comparison with AAV9 (Figure 3C). We found that RS.R6 was 2.6- and 1.8-fold better than AAV9 at delivering vector genome and expressing luciferase in the mouse brain, respectively, while transduction efficiency for liver was similar between the two capsids. For all other rAAV variants, although in our in vitro human BBB transwell model they were predicted to be superior to AAV9, in the mouse model they were either similar or less efficient at transducing both the brain and liver. These findings further suggest that mouse models may not be the most relevant for evaluating AAVs in crossing the human BBB.
Xover analysis of AAV capsid sequences reveal specific amino acid patterns that may contribute to BBB crossing phenotype
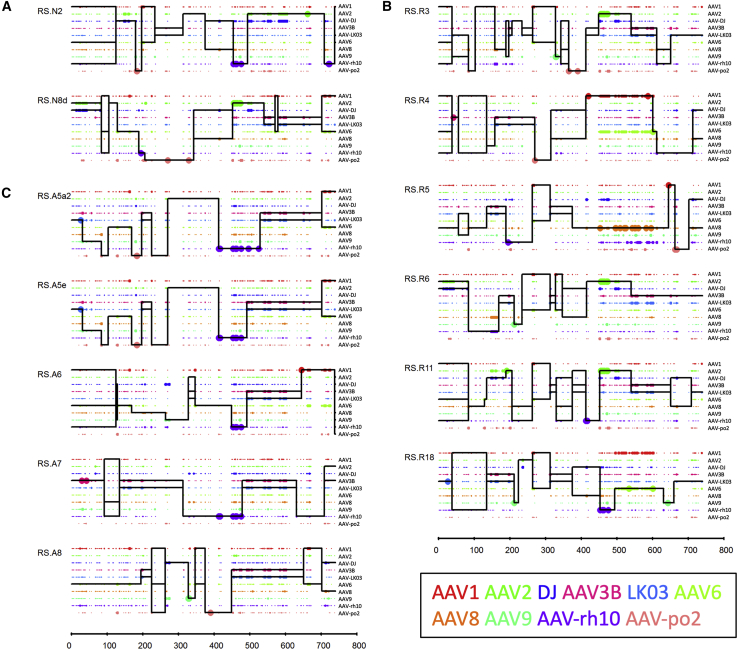
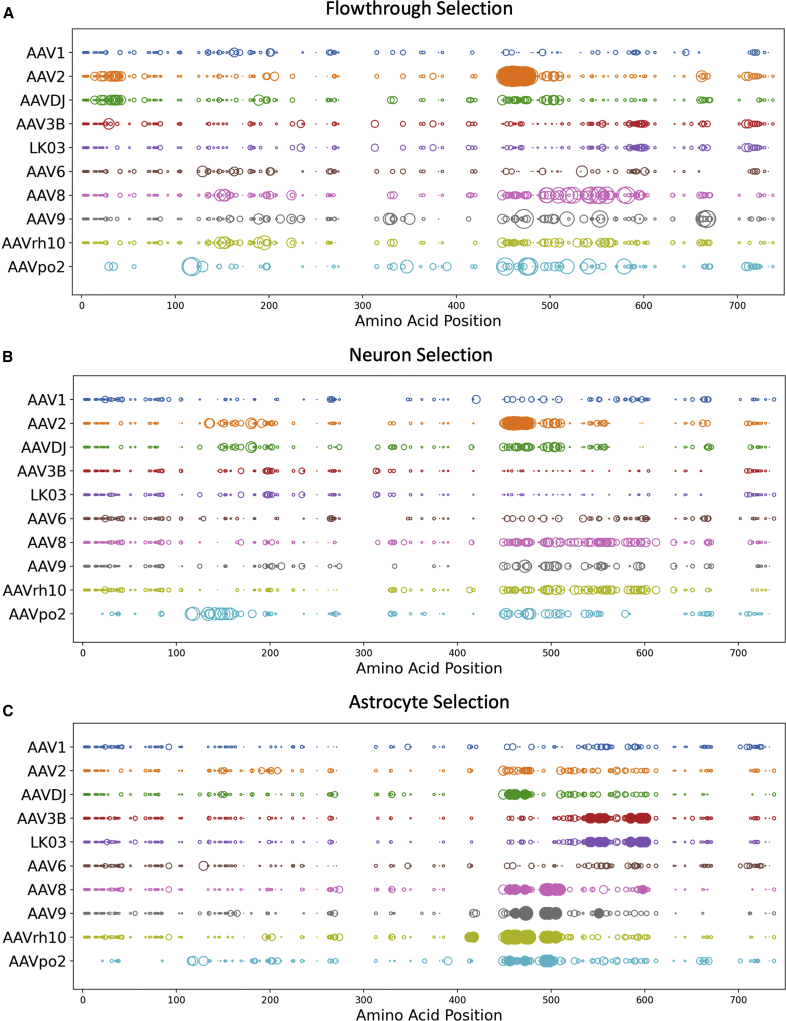
To understand whether the distribution of certain amino acids (aa) contributed to the observed phenotypes of the selected capsid variants, we performed an Xover analysis using the protein sequences of each capsid (Figure 4).27 This analysis reveals the crossover patterns of each chimeric vector, where the parental sequences aligning to the longest uninterrupted stretch in the shuffled capsid sequence are chosen as the most probable contributing source. By comparing the graphs generated from Xover analysis, we noticed two distinct patterns in the approximately 450–650 aa region (Figure 4).
Figure 4.
Xover analysis of vectorized AAV capsid aa sequences
Xover analysis were performed for each of the vectorized AAV capsids from flowthrough (A), neuron (B) and astrocyte (C) selection. RS.A5a1 was excluded as it only has one aa (L464Q) difference from RS.A5a2. Parental AAVs are depicted in different colors. Large dots represent 100% match to the parental AAV aa and small dots represent more than one parental match at the position. The horizontal black lines represent the AAV parents contributing to each rAAV capsid chimera. The vertical black lines are predicted crossover positions between parents.
Specifically, all AAV capsids from the astrocyte selection with the exception of RS.A8 harbor the stretch between aa 453 and 477 from AAV-rh10 and a stretch between aa 526 and 627 from AAV3B/LK03 (Figure 4C). The 453–477 aa region of AAV-rh10 includes three unique aa (453S, 459G and 462Q) in the variable region (VR) IV,28 and three additional aa (468A, 472N, and 475A) in close proximity of VR IV that are suggested to confer the ability to cross the BBB.29 All AAV capsids from the astrocyte screen contains the aa 526–627 from AAV3B/LK03. This aa region covers many unique aa within or in proximity to VR VI (538H and 540N), VII (549T and 554E), and VIII (582N, 592T, 594R, and 598D). Interestingly, RS.N2 and RS.R18, which were derived from the flowthrough and neuron selection screens, respectively, also contain AAV-rh10 aa sequence from 453 to 477. However, neither of these capsids share the aa 526–627 region of AAV3B/LK03. Thus, the unique combination of AAV-rh10 aa 453–477 and AAV3B/LK03 aa 526–627 may contribute to the phenotype of efficiently crossing the BBB endothelial cells and transducing astrocytes.
Another pattern that is less prominent is the combination of AAV2 aa 449–535 and AAV3B/LK03 aa 537–608. Three of the AAV capsids enriched in the neuron screen (RS.R3, RS.R6, and RS.R11), as well as RS.N8d from the flowthrough screen, harbor this pattern (Figures 4A and 4B). The aa 449–535 region derived from AAV2 encompasses a total of six unique aa, four (451P, 456T, 461Q, and 467A) within and two (469D and 470I) in close proximity to VR IV. There are also three additional aa (492S, 493A and 500Y) that are part of VR V and are shared only between AAV2 and AAV-DJ. Although the AAV3B/LK03-derived aa stretch between 537 and 608 is a shorter region compared with that observed in the AAV capsids selected in the astrocyte screen, both segments encompassed the same unique aa. The unique combination of AAV2 aa 449–535 and AAV3B/LK03 aa 537–608 may contribute to the improved endothelial cell and astrocyte layer penetration and the enhanced transduction of hiPSC-derived cortical neurons in our BBB model system.
Long read sequencing analysis confirms specific aa patterns in AAV capsid sequences from different selections
The aa patterns identified in the of vectorized AAV capsid sequences were intriguing, but the number of sequences analyzed was very limited. This prompted us to examine a broader number of capsid sequences by submitting our round one and round two selections to Pacific Biosciences SMRT single DNA molecule long read sequencing to obtain full length capsid sequences along with its barcode.20 From the 25, 90, and 250 barcodes that were continuously enriched in two rounds of duplicated selection in the flowthrough, neuron, and astrocyte, we were able to pull out 6, 28, and 43 associated full capsid sequences, respectively. For every identified AAV capsid sequence, we calculated the probability of the contribution of each parental AAV at every aa position. We then averaged the parent contribution at each aa position for all 6, 28, and 43 capsid sequences from the flowthrough, neuron and astrocyte screens, respectively, and calculated fold change and statistical significance comparing to the average of 11,613 sequences from the unselected input AAV library.
For the capsids from the flowthrough selection screen, four aa from AAV2 (451P, 456T, 461Q, and 469D) were identified as significantly enriched (Figures 5A, S3A and Table S2). The enriched aa in capsids from the neuron selection screen share the same four aa, with two additional (467A and 470D) also from AAV2 (Figures 5B, S3B and Table S2). These positions were also identified in the Xover analysis of the vectorized capsid sequences of RS.R3, RS.R6, and RS.R11 from the neuron selection and RS.N8d from the flowthrough selection. However, unlike the Xover analysis, we did not find any aa between 537 and 608 region AAV3B/LK03 in the flowthrough or neuron selection screen-derived capsids to be significantly different from the input sequences.
Figure 5.
Long read sequencing analysis of AAV capsid aa sequences from different selections
Fold change of parental AAV contribution at each aa position comparing 6, 28, and 43 full capsid sequences from flowthrough (A), neuron (B), and astrocyte (C) selection, respectively, with 11,613 sequences from the unselected input AAV library is plotted as circle size. Fold changes of more than 1 are plotted, and larger circles indicate higher parental contribution at given aa position. Parental AAVs are depicted in different colors. Filled circles indicate statistical significant (Fisher test, p < 0.05) change compared with unselected input AAV, empty circles indicate non-statistical significant change.
An analysis of the 43 capsid sequences from the astrocyte selection identified significant contribution of aa from seven of the ten AAV parents, out of which AAV-rh10, AAV8, and AAV3B/LK03 contributed unique aa (Figures 5C, S3C and Table S2). Similar to the Xover analysis of the vectorized capsid sequences from astrocyte selection, we found AAV-rh10 contributing residues in the 413–496 region, with unique aa 413E, 417Q, 453S, 459G, 462Q, 475A, 495L, and 496S significantly enriched compared with sequences from input. We also found AAV3B/LK03 contributing to the 538–598 region with the exact same unique aa identified by Xover analysis (538H, 540N, 549T, 554E, 582N, 592T, 594R, and 598D) showing significant enrichment compared with input. These data confirm that a combination of AAV-rh10 aa in the 413–496 region and AAV3B/LK03 aa in the 538–598 stretch may confer ability to penetrate endothelial cells and transducing astrocytes. In contrast with the Xover analysis, we also found AAV8 contributing three unique aa 495T, 496G, and 508G. Further validation will be required to confirm whether these aa from AAV8 also contribute to penetrating endothelial cells and transducing astrocytes.
Small interfering RNA screening to identify receptors that mediate transcytosis of AAV across the BBB
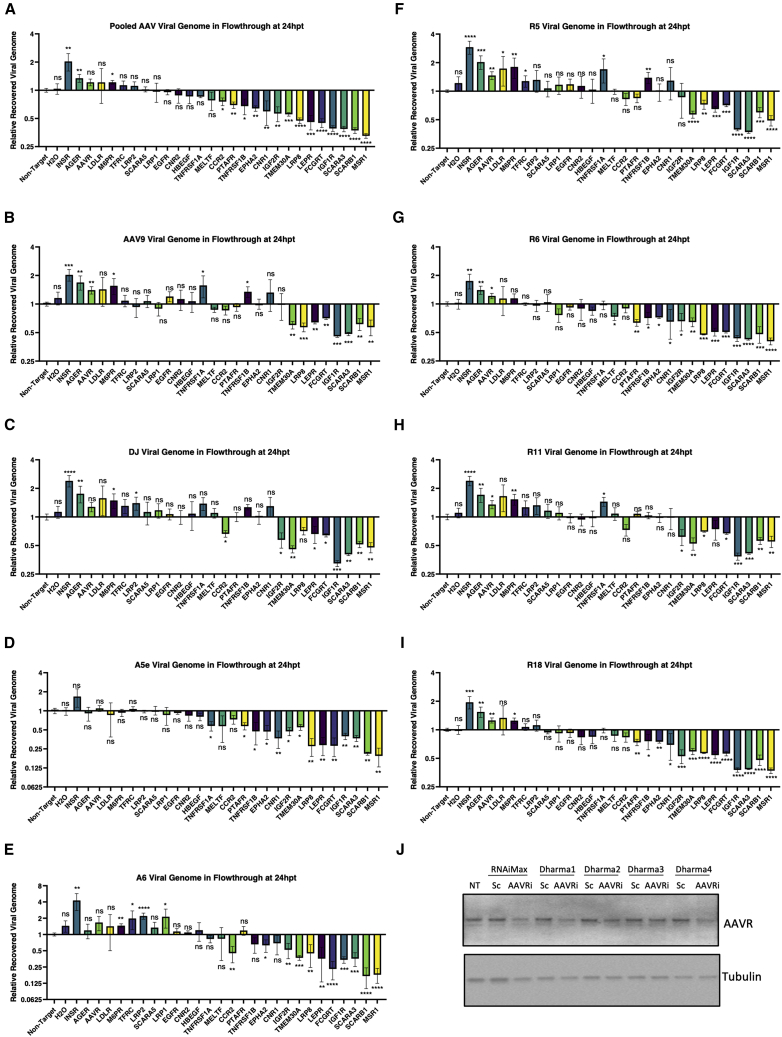
Transport of macromolecules from the blood to the brain across the BBB is tightly regulated by receptor-mediated transcytosis. Several receptors expressed on endothelial cells have been reported to mediate transcytosis of various types of cargo (Table S3), and we hypothesize that these receptors may be used by AAVs for transcytosis across the BBB. Crossing the BBB is likely a complex process involving multiple independent or interacting pathways. Depending on the receptor engaged, endocytosed AAV capsids may be targeted for transduction of endothelial cells, transduction of astrocytes, or transcytosis across the BBB. To determine which of these receptors mediate the transcytosis of AAV capsids, we used a small interfering RNA (siRNA) approach to knock down the receptors in hCMEC/D3 cells grown in transwells. We then added the pool of rAAV vectors consisting of equimolar of AAV9, AAV-DJ, and representative rAAV vectors from each selection (RS.R5, RS.R6, RS.R11, RS.R18, RS.A5e, and RS.A6) and tested their ability to enter or transcytose the hCMEC/D3 and primary astrocyte cell layers in the transwell system.
Knocking down of several scavenger receptors (MSR1, SCARB1, and SCARA3), insulin-like growth factor receptors (IGF1R and IGF2R), antibody transporting receptors (FCGRT and TMEM30A), as well as LEPR, LRP8, CNR1, EPHA2, PTAFR, and CCR2, resulted in reduced transcytosis of two or more rAAVs (Figure 6 and Table S4). This observation suggests that these receptors may be important mediators of rAAV transcytosis. Knocking down of MSR1, SCARB1, SCARA3, IGF1R, IGF2R, FCGRT, LEPR, LRP8, CCR2, and MELTF also led to an increased amount of rAAV genomes in the endothelial cells and/or astrocytes, suggesting these receptors may function in the exocytosis of rAAV vectors from endothelial cells and/or directing them to astrocytes (Figures S4, S5 and Table S4). Knocking down CNR1, EPHA2, and PTAFR did not significant change the level of rAAV genomes in the endothelial cells, but decreased the levels in astrocytes for some capsid variants, suggesting that these receptors likely mediate the transfer of AAV from endothelial cells to astrocytes. Unlike the other receptors, knocking down the TMEM30A also decreased the number of rAAV genomes in endothelial cells, suggesting that this receptor may affect global endocytosis of rAAV vectors.
Figure 6.
siRNA screen to identify receptors that mediate transcytosis of AAV across the BBB
A-I siRNA was used to knock down 27 receptors in hCMEC/D3 cells grown in transwells containing both hCMEC/D3 and primary astrocytes. The individually barcoded rAAV vectors of AAV9, AAV-DJ, RS.R5, RS.R6, RS.R11, RS.R18, RS.A5e, and RS.A6 were pooled and passaged through these transwell BBB model with siRNA treatment. The number of vector genomes of each virus in the flowthrough of every siRNA condition were normalized to that of non-target siRNA. n = 3 for all conditions, significance determined by Student’s t test, ns, not significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. J. siRNA knock down of AAVR protein expression in hCMEC/D3 cells.
INSR, AGER, AAVR, M6PR, TFRC, and LRP2 may participate in the inhibition of rAAV transcytosis across the endothelial and astrocyte cell layers, as siRNA knock down led to increased amount of two or more rAAVs in the flowthrough portion of the transwell (Figure 6 and Table S4). Apart from AAVR, knocking down of the receptors in this category also increased the level of rAAV genomes in endothelial cells, suggesting that these receptors and the pathways they are involved in may compete for resources needed for transcytosis of rAAV (Figure S5 and Table S4). AAVR is the receptor mediating cell entry for several AAVs including AAV9 and AAV-DJ and we show successful knock-down of AAVR as confirmed by western blot (Figure 6J). Knocking down AAVR may have decreased the AAV directed for transduction of endothelial cells, thus increasing the amount of rAAV for transcytosis. Knockdown of INSR and M6PR also decreased the level of rAAV in astrocytes, suggesting that these receptors may play a role in AAV astrocyte transduction (Figure S4 and Table S4).
The tumor necrosis factor (TNF) receptors TNFRSF1A and TNFRSF1B have differential effects on different rAAV (Figures 6, S4, S5 and Table S4). Knocking down these receptors increased the levels of AAV9 and RS.R5 in the flowthrough, but decreased their levels in endothelial cells and/or astrocytes, suggesting that these receptors likely mediate their transduction of endothelial cells and astrocytes. TNFRSF1A also increased the level of RS.R11 and reduced the level of RS.A5e in the flowthrough, while TNFRSF1B decreased the levels of RS.A5e, RS.R6, and RS.R18. These results suggest that the TNF receptors may involve in multiple pathways to mediate different process for different rAAVs.
There are also several receptors that do not affect the transcytosis of rAAV across the BBB, but may impact other processes (Figures 6, S4, S5, and Table S4). According to our data, the receptors SCARA5 and MELTF seemed to play a role in endocytosis and the transduction of endothelial cells for all rAAVs tested. Receptors LDLR, CNR, and HBEGF are likely involved in the transduction of astrocytes for several rAAVs, as the knocking down LDLR increased vector genome copies while knocking down CNR or HBEGF decreased vector genome copies in astrocytes.
Discussion
This study has focused on selecting AAV vectors that efficiently cross the human BBB in an in vitro transwell cell culture model, and suggest this approach is an effective alternative model to identify AAV capsids with improved human CNS properties. The fact that our transwell model allowed us to predict relative enhancement or reduction in BBB penetration of a number of known capsids already tested in NHP pre-clinical studies and/or human trials suggests that this is an approach worth further consideration. Using this model, we have expanded the repertoire of AAV vectors that are not only efficient at transcytosing endothelial cells, but also exhibit improved transduction of hiPSC-derived cortical neurons and/or primary astrocytes compared with the current clinical gold standards, AAV9 and AAV-rh10. We also used this model to explore the molecular mechanisms that allow these AAV capsids to exemplify such improved performance. Specifically, using long-read high-throughput PacBio sequencing to analyze a large repertoire of capsid sequences, we were able to highlight distinctive capsid features that possibly contribute to the improved performance of our selected capsids. To elucidate some of the cellular factors that play a role in transcytosis, we knocked down a wide range of receptors known to mediate transcytosis and evaluated their effects on AAV transcytosis. We showed that several receptors may play distinct roles in targeting the virus to its destination. Overall, our work demonstrates that the transwell culture model of the human BBB is not only effective for selecting AAV capsids that may be useful for CNS-based gene therapy applications, but is also a valuable tool for studying the biological pathways that allow AAV to cross and target the different cell layers in the human BBB.
We opted to use the transwell culture model system of the human BBB for our studies owing to its ability to overcome the limitations of using non-human models for AAV capsid selection. We also prefer this system as it is very easy to set up and is very versatile in the cell types that can be used to satisfy different selection purposes. This model has been commonly used for study of the structure and functions and BBB in supporting the understanding of how homeostasis is achieved with the transport of molecules between the blood and the brain.16 Previous studies have used simplified versions of the transwell BBB model to evaluate the ability of AAV in transcytosing the endothelial cell layer. In a transwell model consisting of primary human microvascular endothelial cells, Merkel et al.19 were the first to show that AAV9 penetrates the endothelial cell barrier more effectively than AAV2. More recently, Zhang et al.30 adopted the transwell BBB model consisting of a hCMEC/D3 cell barrier and showed that shuttle peptides bound to AAV8 enhanced its ability to cross the barrier.
In our current study, we have taken the use of the transwell model to the next level in various ways. By incorporating primary astrocytes to the bottom of the transwell that allow direct interaction with hCEMC/D3 cells, our model not only provides a closer resemblance to the BBB, but also enabled AAV selection and evaluation in astrocytes. Furthermore, we extended our AAV selection and evaluation in hiPSC-derived cortical neurons, eliminating capsids that may have high efficiency in penetration of the cell layers but not efficient at transducing neurons. With a model of improved complexity in cell types and closer resemblance of the BBB, we evaluated the performance of 18 AAVs with different capsid sequences and performed several selection regimens using a highly complex capsid-shuffled AAV library. Our results show that the transwell BBB model consistently enriched for those capsids that confer enhanced efficiency in crossing the cell layers and in transducing various cell types. Our transwell BBB model was also able to capture capsids such as AAV-DJ, which is particularly poor at crossing the BBB, but efficient at transducing human hiPSC-derived cortical neurons and astroglia cells shown here and in organoid models.8 Interestingly, we also found AAV-DJ highly enriched and expressed in the endothelial cells in our transwell model, suggesting it might be useful in disorders caused by dysfunctional BBB, such as 22q11.2 deletion syndrome.31
The transwell BBB model remains a static model where the top compartment does not flow and thus may not fully represent the interactions AAVs will have with the endothelial cells in a microvasculature wall. For future selections and evaluations of AAVs that cross the BBB, the use of the novel microfluidic models may prove to be more relevant.32 Furthermore, selection on BBB models alone does not guarantee the output AAV capsids are specific for CNS targeting. As exemplified in our 18-AAV validation assay in the transwell BBB model, AAV8 showed similar efficiency at crossing the BBB as AAV9 and AAV-rh10. Although intravascular administration of AAV8 has been previously shown to result in efficient brain cell transduction, this serotype remains a robust vector targeting the liver.33 In future screens, to eliminate selection of AAVs that may potentially target tissues other than the CNS, it may be beneficial to perform parallel on-chip selection schemes for various organs, such as liver, pancreas, kidney, and choose mutually exclusive AAV vectors.
As with any human cellular model, the value of the model in prediction of human application will take more effort and time to justify. Until both NHP and human clinical trials engage additional naturally occurring and chimeric AAV capsids that have shown enhancement in a model such as the one used here, the true value of the model may not be appreciated. An example is AAV-LK03, the first chimeric capsid isolated from a library screen in a humanized liver model to be used in a clinical trial.22,34,35 This capsid has been shown to result in similar if not higher levels of human factor VIII when compared with a trial utilizing AAV5 at 30–50 times lower doses. Thus, confirming the effectiveness of the AAV capsids in crossing the BBB in NHP model and clinical trials is of crucial importance for supporting the use of the transwell in vitro model for prediction of capsids with improved properties for treatment of CNS diseases.
Understanding the molecular mechanism of how AAV vectors cross the BBB is also an important aspect for the development of future vectors. Previous studies have shown that transcytosis of AAV is a complex process that likely involves multiple cross-reacting pathways. Merkel et al.19 have shown that AAV2 tends to accumulate in oval shaped vesicular structures and nucleus, whereas AAV9 accumulates in tubular vesicular formations and basolateral plane of endothelial cells. This observation correlates well with the finding that AAV2 has a higher transduction rate for endothelial cells while AAV9 has a higher rate of penetration through endothelial barriers. Thus, we hypothesize that the interaction of the AAV capsid with specific receptors at the endothelial cell surface may determine the destiny of this particular vector, resulting in the transduction of the endothelial cells, the transport to astrocytes in direct contact with the endothelial cells, or the trafficking to the basolateral surface followed by dissemination to adjacent tissue.
Through Xover analysis of the vectorized AAV capsids and long read sequencing analysis of selected pools, we identified specific aa signatures that are associated with distinct phenotypes. In AAV capsids selected for transduction of astrocytes, the aa 413–496 of AAV-rh10 and aa 538–598 of AAV3B/LK03 have a significantly high frequency of occurrence. While many residues within the 413–496 aa region of AAV-rh10 have been suggested to confer the ability to cross the BBB in previous studies,29 the combination with a stretch derived from AAV3B/LK03 may be crucial for the association with cellular factors that result in astrocyte transduction. The capsids selected from the flowthrough and neuron screens showed a high contribution rate of AAV2 residues from aa 451 to 470. This is an unexpected finding; no previous studies have shown these AAV2 residues confer transcytosis capabilities. Further characterization and evaluation will be needed to confirm that these aa patterns are truly responsible for the observed phenotypes. Whether these aa residues interact with specific receptors that target AAV capsids to distinct cellular pathways will be another line of study worth pursuing.
Using an siRNA approach to knockdown the expression of various receptors that are known to mediate transcytosis of macromolecules across the BBB, we found different receptors may play a role in directing AAV capsids to different destinations. Based on our data, SCARA5 and MELTF are likely involved in the transduction of endothelial cells. Receptors INSR, M6PR, LDLR, CNR, and HBEGF may regulate the transport of AAV vectors to astrocytes that are in contact with endothelial cells. We also found several receptors that may enhance AAV transcytosis (MSR1, SCARB1, SCARA3, IGF1R, IGF2R, FCGRT, TMEM30A, LEPR, LRP8, CNR1, EPHA2, PTAFR, and CCR2), while others seemed to be negatively regulate the process (INSR, AGER, AAVR, M6PR, TFRC, and LRP2). The TNF receptors TNFRSF1A and TNFRSF1B were shown to have differential effects on transcytosis depending on the AAV capsid, suggesting that these receptors may be involved in multiple pathways. Further in-depth assessment will be needed to characterize if these receptors directly interact with AAV capsids to regulate the targeting process or play an indirect role.
Taken together, our studies show that the in vitro transwell BBB cell culture model is suited for selection of AAV vectors that efficiently transcytose and transduce certain cell types in the BBB. We hope to evaluate the efficacy and safety of these vectors in the NHP model and move these capsids to clinical use in gene therapy for CNS diseases. We also elucidated molecular mechanisms that may be responsible for the improved phenotype by detailed sequencing analysis and receptor knock-down experiments. We hope that the further characterization of these mechanisms will support future endeavors in rational design of better AAV vectors for clinical applications.
Materials and methods
Cell cultures
hCMEC/D3 cells (Cat #SCC066) and media (Cat #SCME004), dish coating material type I rat collagen (Cat #08–115) were purchased from MilliporeSigma. Primary human astrocytes isolated from cerebral cortex (Cat # 1800), astrocyte medium (Cat # 1801), trypsin neutralization solution (Cat # 0113) and dish coating material poly-L-lysine (Cat # 0403) were purchased from ScienCell Research Laboratories. All cells were maintained according to manufacturer instructions. Generation and culture of hiPSC-derived neurons and astroglia from human cortical spheroids (hCS) were described previously.17,26,36 Briefly, hiPSC derived from fibroblasts and maintained on MEFs were used to generate hCS for more than 150 days, after which they were enzymatically dissociated and immunopanned into purified neurons (Mouse anti-Thy1 [CD90], BD Biosciences, Cat. 550402) and astroglia cells (Mouse anti-HepaCAM, R&D, Cat # MAB4108) as previously described.26 The isolated cells were seeded on poly-d-lysine coated plates and maintained in a Neurobasal/DMEM based serum-free medium.26 All cell cultures were maintained in a humidified incubator at 37°C with 5% CO2. All human stem cell work was performed with approval from the Stanford Human Stem Cell Research Oversight committee.
Culture of transwell human BBB model
For construction of human BBB cultures in transwell, we followed a previously published protocol with some modifications.37 Briefly, for cultures containing hCMEC/D3 cells only, cells were seeded at 2 × 105 cells/cm2 on the top of a 0.4-μM PET membrane (Cat # 3450 or 3470, Corning) pre-coated with collagen. The 0.4-μM pore size was selected as it prevents cells from crossing between top and bottom sides, but allows AAVs of 20 nM to cross. Cultures containing both hCMEC/D3 cells and human primary astrocytes were set up as follows: astrocytes were seeded at 6 × 104 cells/cm2 on the bottom side of the 0.4-μM PET membrane pre-coated with poly-l-lysine. After 24 h of incubation, hCMEC/D3 cells were seeded at 2 × 105 cells/cm2 on the top membrane after coating with collagen. For all transwell BBB cultures, heparin sulfate included in the hCMEC/D3 media kit was omitted as it interferes with binding of several AAV serotypes to heparin sulfate proteoglycans. Full media changes were performed every 2–3 days, and cultures were maintained for 11 days after seeding of hCMEC/D3 cells before use for selection of AAV for BBB penetration.
Testing permeability in transwells using fluorophore-conjugated dextran
Transwells with hCEMC/D3 culture only or with both hCMEC/D3 and primary astrocytes were tested for permeability by adding to the top of the transwell: 70k MW Dextran-TRITC (Invitrogen) (ex 555 nm/em 580 nm) at 1 μM and 2M MW Dextran-FITC (Invitrogen) (ex 494 nm/em 521 nm) at 5 nM. Media from the bottom of the transwell were collected at 2, 4, 6, and 8 h after addition of the dextran. Fluorescent intensity of the collected media were measured using Tecan Infinite M1000Pro.
Production, purification, and titer of AAV libraries
The generation of 18 uniquely barcoded AAV pool and 10 parent capsid-shuffled and barcoded AAV library were described previously.18 Briefly, the 18 AAV pool contains serotypes (AAV1, AAV2, AAV3B, AAV4, AAV5, AAV6, AAV8, AAV9.hu14, AAV12, AAV-porcine1, AAV-porcine2, AAV-bovine, AAV-goat1, AAV-mouse1, AAV-avian, AAV-rh10, AAV-DJ, and AAV-LK03), which were separately cloned into a vector containing unique barcode sequence. AAVs were pooled at equimolar before purification. For the 10 parent capsid-shuffled AAV library, capsid sequences from AAV1, AAV2, AAV-DJ, AAV3B, AAV-LK03,AAV6, AAV8, AAV9.hu14, AAV-porcine2, and AAV-rh10 were used as parental sequences for the shuffling and resulted fragments were cloned into a BC library vector. The 18 AAV pool and 10 parent capsid-shuffled AAV library were purified by two consecutive CsCl gradient centrifugation, and genome titers were determined by quantitative polymerase chain reaction (qPCR) using an AAV2 REP gene-specific primer/probe set described in Ref.18
Selection of 18 AAV pool and 10 parent capsid-shuffled AAV library in a transwell BBB model
The AAV input was added to the top side of transwell BBB culture model containing both hCMEC/D3 cells and human primary astrocytes. After 24 h, astrocytes and flowthrough media on the bottom of the transwell culture were collected. In a separate transwell BBB culture, hCMEC/D3 and flowthrough media were removed at 24 h and Ad5 was added to the astrocytes for 72 h before collection. The flowthrough media were also added to hiPSC-derived cortical neuron cultures with or without Ad5 and collected after 72 h. DNA from input and all collected samples were tested for AAV types by next-generation sequencing (NGS) of barcodes previously described in Refs.18,20 For the 10 parent capsid-shuffled AAV library, second round of selections were performed with three different inputs from round one. In the first, flowthrough media from round one was the input added to a new transwell BBB culture, and flowthrough media from round two was collected after 24 h. In the second, astrocytes with Ad5 from round one were frozen and thaw three times, heat inactivated at 65°C and used as input on a new transwell BBB culture. Astrocytes from the new transwell was collected after 24 h. For the third, hiPSC-derived cortical neurons with Ad5 from round one were frozen and thaw three times, heat inactivated at 65°C and used as input on a new transwell BBB culture. Flowthrough media from the new transwell was collected after 24 h and added to fresh hiPSC-derived cortical neurons for 72 h before collection. Two biological replicates were performed at every selection step. Barcodes that were present in both replicates and were enriched in both rounds of selection were identified.
Recovery and evaluation of enriched AAV capsid in the transwell BBB model
Capsid sequences that have enriched barcode count in the two rounds of selection were PCR amplified using primer capF18 and a primer containing the specific barcode sequence. The capsids DNA sequences recovered from the selections were analyzed by Sanger sequencing and subjected to DNA shuffling pattern analysis using the Xover 3.0 online interface.27 The capsids were then used to package a single-stranded rAAV vector expressing firefly luciferase under control of the CAG promoter (pAAV-CAG-FLuc, Addgene, catalog 83281). rAAVs were purified using AAVpro Purification Kit for All Serotypes (Cat # 6666, Takara Bio USA) and each of the purified rAAVs were tested in separate transwell BBB cultures with hCMEC/D3 cells and primary astrocytes at MOI of 1e4. Flowthrough media was collected at 3, 6, and 24 h and astrocytes were collected at 24 h. Vector genome copies in input and collected samples were determined by qPCR using a FLuc-specific primer set described in Ref.18
Evaluation of rAAVs for transduction efficiency in hiPSC-derived cortical neurons and astroglia cells
Takara kit purified rAAVs capsids packaged with pAAV-CAG-FLuc were tested in separate cultures of hiPSC-derived cortical neurons and astroglia cells at MOI of 1e2, 1e3, 1e4, and 1e5. After 72 h, cells were lysed in plate and luciferase activity was measured using Luciferase 1000 Assay System (Cat #E4550, Promega).
Evaluation of rAAVs for transduction efficiency in mice
The rAAVs capsids packaged with pAAV-CAG-FLuc were purified by two consecutive CsCl gradient centrifugation. Each rAAV were test in separate B6 albino mouse (B6N-Tyrc-Brd/BrdCrCrl) through retro-orbital injection. Brain and liver tissues were collected 21 days or 28 days after injection. Vector genome copies in the collected samples were determined by qPCR using a FLuc-specific primer set described in Ref.18 Luciferase activity in the tissues was measured using Luciferase 1000 Assay System (Cat #E4550, Promega). All mouse procedures were approved by the Institutional Animal Care and Use Committee of Stanford University.
Construction and evaluation of pooled rAAVs in the transwell BBB model
A single-stranded rAAV vector expressing hFXN under control of the CAG promoter (pAAV-CAG-hFXN) was generated by replacing the FLuc sequence in the pAAV-CAG-FLuc plasmid with hFXN. For generation of the barcodes, oligonucleotides with 12 random nucleotides and restriction site sequences on both 5′- and -3′- ends (5′-CTTGAGAATTCGATATCNNNNNNNNNNNNAAGCTTATCGATAATCAACC-3′) were annealed to an oligonucleotide containing the antisense of the 3′ restriction site (5′-GGTTGATTATCGATAAGCTT-3′) and extended using Klenow polymerase devoid of exonuclease activity. Resultant fragments were inserted into the pAAV-CAG-hFXN plasmid between the stop codon of the hFXN and poly A signal. Individual clones were sequenced to assess the barcode identity. The rAAV were generated by packaging each capsid with a pAAV-CAG-hFXN carrying a unique barcode, purified using Takara kit and pooled at equimolar ratios. The pooled rAAVs were tested in the same transwell BBB culture with hCMEC/D3 cells and primary astrocytes at MOI of 1e4 and 1e5. Flowthrough media, hCMEC/D3 cells, astrocytes were collected at 24 h. DNA from input and all collected samples, RNA from the hCMEC/D3 cells and astrocytes were tested for AAV types by NGS of barcodes previously described in Refs.18,20
PacBio sequencing and analysis
All samples collected from the flowthrough, neuron and astrocyte selections of the 10 parent capsid-shuffled AAV library were used to amplify approximately 2.4-kb fragments containing the capsid as well as the BC sequences. Library preparation and Pacific Biosciences (PacBio) sequencing using Sequel Sequencing Kit 3.0 (Cat # 101-613-700) for 10 h movies were performed by the University of Washington PacBio Sequencing Services. Circular consensus sequence (CCS) reads were generated using the SMRT Link 6.0.0.47841 with filtering set at a minimum number of passes of 3 and a minimum predicted accuracy of 0.9. The barcodes were first extracted from the CCS reads using seeq, a DNA/RNA pattern-matching algorithm (https://github.com/ezorita/seeq). The barcodes with three or more CCS reads were included in downstream bioinformatics analyses to generate consensus DNA sequence, extract the open reading frame of a capsid sequence and translate into aa sequence. The translated capsid aa sequences along with 10 parent sequences were subjected to MAFFT v7.427, a multiple sequence alignment program for aa and nucleotide sequences. Based on the alignment, the aa contribution from each parent at every position were calculated. Barcodes that were previously identified enriched in two rounds of selection by barcode NGS sequencing were used to pulled out full length capsid sequences. The aa contribution from each parent at every position were averaged for capsid sequences from flowthrough, neuron or astrocyte selections. Then fold-change and statistical significance was calculated compared with capsid sequences in the input.
siRNA transfection and evaluation of the pooled rAAVs in the transwell BBB model
Pools of four siRNA targeting each receptor was purchased from Dharmacon, CO. siRNA transfection was performed on both day 6 and day 8 after seeding of hCMEC/D3 cells. Lipofectamine RNAiMax (Cat # 13778075, ThermoFisher Scientific) was used to transfect 60 nM of the siRNA pool on each of the 2 days transfection was performed. Knockdown efficiency for AAVR was assessed by western blot using an antibody against AAVR (Cat # ab105385, Abcam). The pooled rAAVs carrying uniquely barcoded CAG-hFXN were tested in the transwell BBB culture with hCMEC/D3 cells transfected with siRNA and primary astrocytes at MOI of 1e4. Flowthrough media, hCMEC/D3 cells, and astrocytes were collected at 24 h. DNA from input and all collected samples were tested for AAV types by NGS of barcodes.
Statistical analyses
Statistical calculations were conducted using Prism v8.4 or NumPy package in Python. Experimental values were assessed via two-way ANOVA test, Student t test, or Fisher’s exact test. A p value of less than 0.05 was considered statistically significant.
Acknowledgments
The authors acknowledge Shuyuan Zhang for writing the Python scripts used in sequence analysis, Se-Jin Yoon for assistance with immunopanning experiments, Xuhuai Ji from the Stanford Genomics Facility for performing the NGS, and Jeff Rubin for reviewing the manuscript.
R.S. was supported in part by a postdoctoral fellowship from Stanford Dean’s Fellowship. This work was supported by grants to M.A.K. NIH R01AI116698.
Author contributions
R.S. and M.A.K. designed the experiments. R.S., K.P., T.A.K., and F.Z. generated reagents and protocols, performed experiments and analyzed data. R.S., S.P.P., and M.A.K. acquired funding. R.S. wrote the manuscript and generated the figures. All authors reviewed, edited and commented on the manuscript.
Declaration of interests
R.S. is currently an employee and shareholder of Allogene Therapeutics. S.P.P. is an NYSCF Robertson Investigator, a CZI Ben Barres Investigator, and a CZ BioHub Investigator. Stanford University has submitted patents for the new AAV capsids described in this article, of which M.A.K. and R.S. are the inventors. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the various funding bodies or universities involved.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2022.09.002.
Supplemental information
Data availability
Packaging plasmids for any of the new capsids described herein must be obtained through a material transfer agreement (MTA) with Stanford University.
References
- 1.Al-Zaidy S.A., Mendell J.R. From clinical trials to clinical practice: practical considerations for gene replacement therapy in SMA type 1. Pediatr. Neurol. 2019;100:3–11. doi: 10.1016/j.pediatrneurol.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Foust K.D., Nurre E., Montgomery C.L., Hernandez A., Chan C.M., Kaspar B.K. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat. Biotechnol. 2009;27:59–65. doi: 10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanguy Y., Biferi M.G., Besse A., Astord S., Cohen-Tannoudji M., Marais T., Barkats M. Systemic AAVrh10 provides higher transgene expression than AAV9 in the brain and the spinal cord of neonatal mice. Front. Mol. Neurosci. 2015;8:36. doi: 10.3389/fnmol.2015.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Day J.W., Mendell J.R., Mercuri E., Finkel R.S., Strauss K.A., Kleyn A., Tauscher-Wisniewski S., Tukov F.F., Reyna S.P., Chand D.H. Clinical trial and postmarketing safety of onasemnogene abeparvovec therapy. Drug Saf. 2021;44:1109–1119. doi: 10.1007/s40264-021-01107-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greene C., Hanley N., Campbell M. Claudin-5: gatekeeper of neurological function. Fluids Barriers CNS. 2019;16:3. doi: 10.1186/s12987-019-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perez B.A., Shutterly A., Chan Y.K., Byrne B.J., Corti M. Management of neuroinflammatory responses to AAV-mediated gene therapies for neurodegenerative diseases. Brain Sci. 2020;10:E119. doi: 10.3390/brainsci10020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Challis R.C., Ravindra Kumar S., Chen X., Goertsen D., Coughlin G.M., Hori A.M., Chuapoco M.R., Otis T.S., Miles T.F., Gradinaru V. Adeno-associated virus toolkit to target diverse brain cells. Annu. Rev. Neurosci. 2022;45:447–469. doi: 10.1146/annurev-neuro-111020-100834. [DOI] [PubMed] [Google Scholar]
- 8.Deverman B.E., Pravdo P.L., Simpson B.P., Kumar S.R., Chan K.Y., Banerjee A., Wu W.L., Yang B., Huber N., Pasca S.P., et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 2016;34:204–209. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hordeaux J., Wang Q., Katz N., Buza E.L., Bell P., Wilson J.M. The neurotropic properties of AAV-PHP.B are limited to C57BL/6J mice. Mol. Ther. 2018;26:664–668. doi: 10.1016/j.ymthe.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuzaki Y., Konno A., Mochizuki R., Shinohara Y., Nitta K., Okada Y., Hirai H. Intravenous administration of the adeno-associated virus-PHP.B capsid fails to upregulate transduction efficiency in the marmoset brain. Neurosci. Lett. 2018;665:182–188. doi: 10.1016/j.neulet.2017.11.049. [DOI] [PubMed] [Google Scholar]
- 11.Hordeaux J., Yuan Y., Clark P.M., Wang Q., Martino R.A., Sims J.J., Bell P., Raymond A., Stanford W.L., Wilson J.M. The GPI-linked protein LY6A drives AAV-PHP.B transport across the blood-brain barrier. Mol. Ther. 2019;27:912–921. doi: 10.1016/j.ymthe.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Q., Chan K.Y., Tobey I.G., Chan Y.A., Poterba T., Boutros C.L., Balazs A.B., Daneman R., Bloom J.M., Seed C., et al. Delivering genes across the blood-brain barrier: LY6A, a novel cellular receptor for AAV-PHP.B capsids. PLoS One. 2019;14:e0225206. doi: 10.1371/journal.pone.0225206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gray S.J., Matagne V., Bachaboina L., Yadav S., Ojeda S.R., Samulski R.J. Preclinical differences of intravascular AAV9 delivery to neurons and glia: a comparative study of adult mice and nonhuman primates. Mol. Ther. 2011;19:1058–1069. doi: 10.1038/mt.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravindra Kumar S., Miles T.F., Chen X., Brown D., Dobreva T., Huang Q., Ding X., Luo Y., Einarsson P.H., Greenbaum A., et al. Multiplexed Cre-dependent selection yields systemic AAVs for targeting distinct brain cell types. Nat. Methods. 2020;17:541–550. doi: 10.1038/s41592-020-0799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flitsch L.J., Börner K., Stüllein C., Ziegler S., Sonntag-Buck V., Wiedtke E., Semkova V., Au Yeung S.W.C., Schlee J., Hajo M., et al. Identification of adeno-associated virus variants for gene transfer into human neural cell types by parallel capsid screening. Sci. Rep. 2022;12:8356. doi: 10.1038/s41598-022-12404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helms H.C., Abbott N.J., Burek M., Cecchelli R., Couraud P.O., Deli M.A., Förster C., Galla H.J., Romero I.A., Shusta E.V., et al. In vitro models of the blood-brain barrier: an overview of commonly used brain endothelial cell culture models and guidelines for their use. J. Cereb. Blood Flow Metab. 2016;36:862–890. doi: 10.1177/0271678X16630991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paşca A.M., Sloan S.A., Clarke L.E., Tian Y., Makinson C.D., Huber N., Kim C.H., Park J.Y., O'Rourke N.A., Nguyen K.D., et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pekrun K., De Alencastro G., Luo Q.J., Liu J., Kim Y., Nygaard S., Galivo F., Zhang F., Song R., Tiffany M.R., et al. Using a barcoded AAV capsid library to select for clinically relevant gene therapy vectors. JCI Insight. 2019;4:131610. doi: 10.1172/jci.insight.131610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merkel S.F., Andrews A.M., Lutton E.M., Mu D., Hudry E., Hyman B.T., Maguire C.A., Ramirez S.H. Trafficking of adeno-associated virus vectors across a model of the blood-brain barrier; a comparative study of transcytosis and transduction using primary human brain endothelial cells. J. Neurochem. 2017;140:216–230. doi: 10.1111/jnc.13861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Alencastro G., Pekrun K., Valdmanis P., Tiffany M., Xu J., Kay M.A. Tracking adeno-associated virus capsid evolution by high-throughput sequencing. Hum. Gene Ther. 2020;31:553–564. doi: 10.1089/hum.2019.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimm D., Lee J.S., Wang L., Desai T., Akache B., Storm T.A., Kay M.A. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lisowski L., Dane A.P., Chu K., Zhang Y., Cunningham S.C., Wilson E.M., Nygaard S., Grompe M., Alexander I.E., Kay M.A. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2014;506:382–386. doi: 10.1038/nature12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paulk N.K., Pekrun K., Charville G.W., Maguire-Nguyen K., Wosczyna M.N., Xu J., Zhang Y., Lisowski L., Yoo B., Vilches-Moure J.G., et al. Bioengineered viral platform for intramuscular passive vaccine delivery to human skeletal muscle. Mol. Ther. Methods Clin. Dev. 2018;10:144–155. doi: 10.1016/j.omtm.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paulk N.K., Pekrun K., Zhu E., Nygaard S., Li B., Xu J., Chu K., Leborgne C., Dane A.P., Haft A., et al. Bioengineered AAV capsids with combined high human liver transduction in vivo and unique humoral seroreactivity. Mol. Ther. 2018;26:289–303. doi: 10.1016/j.ymthe.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adachi K., Enoki T., Kawano Y., Veraz M., Nakai H. Drawing a high-resolution functional map of adeno-associated virus capsid by massively parallel sequencing. Nat. Commun. 2014;5:3075. doi: 10.1038/ncomms4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sloan S.A., Darmanis S., Huber N., Khan T.A., Birey F., Caneda C., Reimer R., Quake S.R., Barres B.A., Paşca S.P. Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron. 2017;95:779–790.e6. doi: 10.1016/j.neuron.2017.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang W., Johnston W.A., Boden M., Gillam E.M.J. ReX: a suite of computational tools for the design, visualization, and analysis of chimeric protein libraries. Biotechniques. 2016;60:91–94. doi: 10.2144/000114381. [DOI] [PubMed] [Google Scholar]
- 28.Govindasamy L., Padron E., McKenna R., Muzyczka N., Kaludov N., Chiorini J.A., Agbandje-McKenna M. Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J. Virol. 2006;80:11556–11570. doi: 10.1128/JVI.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mietzsch M., Barnes C., Hull J.A., Chipman P., Xie J., Bhattacharya N., Sousa D., McKenna R., Gao G., Agbandje-McKenna M. Comparative analysis of the capsid structures of AAVrh.10, AAVrh.39, and AAV8. J. Virol. 2020;94:e01769-19. doi: 10.1128/JVI.01769-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X., He T., Chai Z., Samulski R.J., Li C. Blood-brain barrier shuttle peptides enhance AAV transduction in the brain after systemic administration. Biomaterials. 2018;176:71–83. doi: 10.1016/j.biomaterials.2018.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greene C., Kealy J., Humphries M.M., Gong Y., Hou J., Hudson N., Cassidy L.M., Martiniano R., Shashi V., Hooper S.R., et al. Dose-dependent expression of claudin-5 is a modifying factor in schizophrenia. Mol. Psychiatry. 2018;23:2156–2166. doi: 10.1038/mp.2017.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X., Liu C., Muok L., Zeng C., Li Y. Dynamic 3D on-chip BBB model design, development, and applications in neurological diseases. Cells. 2021;10:3183. doi: 10.3390/cells10113183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakai H., Fuess S., Storm T.A., Muramatsu S.i., Nara Y., Kay M.A. Unrestricted hepatocyte transduction with adeno-associated virus serotype 8 vectors in mice. J. Virol. 2005;79:214–224. doi: 10.1128/JVI.79.1.214-224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.George L.A., Monahan P.E., Eyster M.E., Sullivan S.K., Ragni M.V., Croteau S.E., Rasko J.E.J., Recht M., Samelson-Jones B.J., MacDougall A., et al. Multiyear factor VIII expression after AAV gene transfer for hemophilia A. N. Engl. J. Med. 2021;385:1961–1973. doi: 10.1056/NEJMoa2104205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pasi K.J., Rangarajan S., Mitchell N., Lester W., Symington E., Madan B., Laffan M., Russell C.B., Li M., Pierce G.F., et al. Multiyear follow-up of AAV5-hFVIII-SQ gene therapy for hemophilia A. N. Engl. J. Med. 2020;382:29–40. doi: 10.1056/NEJMoa1908490. [DOI] [PubMed] [Google Scholar]
- 36.Khan T.A., Revah O., Gordon A., Yoon S.J., Krawisz A.K., Goold C., Sun Y., Kim C.H., Tian Y., Li M.Y., et al. Neuronal defects in a human cellular model of 22q11.2 deletion syndrome. Nat. Med. 2020;26:1888–1898. doi: 10.1038/s41591-020-1043-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatherell K., Couraud P.O., Romero I.A., Weksler B., Pilkington G.J. Development of a three-dimensional, all-human in vitro model of the blood-brain barrier using mono-co-and tri-cultivation Transwell models. J. Neurosci. Methods. 2011;199:223–229. doi: 10.1016/j.jneumeth.2011.05.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Packaging plasmids for any of the new capsids described herein must be obtained through a material transfer agreement (MTA) with Stanford University.