Summary
Background
Osteoarthritis (OA) is the most common degenerative joint disease primarily characterized by cartilage destruction. The aim of this study was to investigate the role, molecular characteristics and potential therapeutic target of chondrocyte ferroptosis in the pathogenesis of OA.
Methods
The expression of ferroptotic hallmarks (iron and lipid peroxidation accumulation, glutathione deletion) were analyzed in paired intact and damaged cartilages from OA patients. Single cell RNA sequencing (scRNA-seq) analysis was performed on 17,638 chondrocytes to verify the presence, investigate the molecular signatures and unveil the potential therapeutic target of ferroptotic chondrocyte cluster in human OA cartilages. Destabilization of medial meniscus (DMM)-induced OA model and tert-butyl hydroperoxide (TBHP)-treated primary mouse chondrocytes and human cartilage explants were used to evaluate the protective effect of pharmacologically activated transient receptor potential vanilloid 1 (TRPV1). The downstream molecular mechanisms of TRPV1 was further investigated in glutathione peroxidase 4 (Gpx4) heterozygous genetic deletion mice (Gpx4+/−).
Findings
The concentrations of iron and lipid peroxidation and the expression of ferroptotic drivers in the damaged areas of human OA cartilages were significantly higher than those in the intact cartilage. scRNA-seq analysis revealed a chondrocyte cluster characterized by preferentially expressed ferroptotic hallmarks and genes, namely ferroptotic chondrocyte cluster. Comprehensive gene set variation analysis revealed TRPV1 as an anti-ferroptotic target in human OA cartilage. Pharmacological activation of TRPV1 significantly abrogated cartilage degeneration by protecting chondrocytes from ferroptosis. Mechanistically, TRPV1 promoted the expression of GPX4, and its anti-ferroptotic role was largely mitigated in the OA model of Gpx4+/− mice.
Interpretation
TRPV1 activation protects chondrocytes from ferroptosis and ameliorates OA progression by upregulating GPX4.
Funding
National Key R&D Program of China (2018YFC1105904), Key Program of NSFC (81730067), National Science Foundation of China (81772335, 81941009, 81802196), Natural Science Foundation of Jiangsu Province, China (BK20180127), Jiangsu Provincial Key Medical Talent Foundation, Six Talent Peaks Project of Jiangsu Province (WSW-079).
Keywords: Osteoarthritis, Ferroptosis, Single cell RNA sequencing, TRPV1, GPX4
Research in context.
Evidence before this study
Cartilage degeneration is the primary pathogenesis of OA. Ferroptosis is an iron-dependent regulated cell death that requires the accumulation of iron and lipid peroxides, both of which impair cartilage homeostasis. Although chondrocyte ferroptosis has been shown associated with OA progression, the molecular characteristics and potential therapeutic target of ferroptotic chondrocytes are largely unknown.
Added value of this study
We investigated the molecular signatures of ferroptotic chondrocytes and screened TRPV1 as its therapeutic target by the evidence that (i) Ferroptotic hallmarks accumulated in OA cartilage, especially in the damaged areas; (ii) Single cell RNA-seq analysis of human OA chondrocytes identified a ferroptotic chondrocyte cluster with preferentially expressed ferroptotic hallmarks and genes; (iii) TRPV1 was screened and verified as the anti-ferroptotic target both in vitro and in vivo; (iv) Mechanistically, TRPV1 protects chondrocytes from ferroptosis by promoting the expression of GPX4.
Implications of all the available evidence
This work identified the role and molecular characteristics of ferroptotic chondrocyte cluster in human OA cartilage. We further screened and verified TRPV1 as an anti-ferroptotic target, which provided potential therapeutic target to alleviate OA progression.
Alt-text: Unlabelled box
Introduction
Osteoarthritis (OA) is the most common form of joint degeneration and the leading cause of disability worldwide.1,2 The socioeconomic cost caused by OA accounts for 1–2.5% of gross national product, and with the growing prevalence of OA, its burden is substantially increasing.3 However, there is no disease-modifying therapy for OA because the precise molecular mechanisms that underlie its pathogenesis are poorly understood.
OA is primarily characterized by progressive cartilage degeneration, in which the survival crisis and metabolic disorder of chondrocytes, the sole cell type in cartilage, plays an indispensable role.4 Various types of chondrocyte death have recently been shown involved in OA development, among which, ferroptosis, driven by iron-dependent lipid peroxidation, attracted much attention.5, 6, 7 Different from other types of cell death, ferroptosis possesses distinctive morphological and mechanistical features; for example, unlike executioner protein-centered apoptosis and necroptosis, lipid peroxides accumulation is the cardinal feature of ferroptosis.8 Despite the essential role of iron for human life, iron overload has been reported to induce lipid reactive oxygen species (ROS) accumulation, changes of ferroptosis-related protein expression and chondrocyte death, leading to enhanced catabolism and weakened anabolism of chondrocytes.9 In addition, iron overload was also involved in cartilage destruction and was positively correlated with OA progression.10,11 The lipid repair enzyme glutathione peroxidase 4 (GPX4) plays an anti-ferroptotic role by directly reducing lipid peroxidation.12 A recent study showed that the decrease of GPX4 in OA cartilage resulted in aggravated oxidative stress and cartilage extracellular matrix degradation.13 These results indicate that chondrocyte ferroptosis participate in the pathogenesis of cartilage destruction in OA. However, the molecular characteristics and potential therapeutic targets of ferroptotic chondrocytes in human OA cartilage are largely unknown.
In this study, we investigated the role of ferroptotic chondrocytes in OA pathogenesis and explored potential therapeutic target for disease-modifying treatment of OA. Using single-cell RNA sequencing (scRNA-seq) analysis for human OA cartilages, we unveiled the existence and molecular characteristics of ferroptotic chondrocyte cluster. Further, we identified transient receptor potential vanilloid 1 (TRPV1) as an anti-ferroptotic target in chondrocytes, which abrogated ferroptosis by promoting GPX4 expression.
Materials and methods
Ethics statement
Human articular cartilage tissue and synovial fluid were obtained from patients with knee OA who underwent total knee arthroplasty, which was approved by the Ethical Committee of the Nanjing Drum Tower Hospital, the Affiliated Hospital of Nanjing University Medical School (2020-156-01). All experiments using mice were authorized and strictly performed in accordance with the Animal Care and Use Committee of Nanjing Drum Tower Hospital, The Affiliated Hospital of Nanjing University (2020AE01102).
Clinical specimen collection
Human articular cartilage samples from patients with knee OA (65–83 years old, Kellgren–Lawrence grade 4) undergoing knee arthroplasty surgery were obtained at the Nanjing Drum Tower Hospital and divided into intact (lateral condyle) and damaged (medial condyle) areas. Among the cartilage samples, 11 pairs were arranged for measurements of iron content, LPO concentration, glutathione (GSH) and GSH/GSSG (GSH/oxidized GSH) levels, and GPX4 activity, and eight pairs were used for extracting proteins. The synovial fluid from 52 knee OA patients (53–83 years old, Kellgren–Lawrence grade 4) were obtained before knee arthroplasty surgery and stored at −80 °C before the measurement of iron and matrix metallopeptidase 3 (MMP3) concentrations. The demographic characteristics of OA patients were shown in Supplementary Tables 1-3.
Single cell RNA-seq library construction and sequencing
After the acquirement, fresh cartilage tissues were washed by sterile PBS for three times. The cartilage tissues were minced into 1–2 mm pieces and digested by 0.2% collagenase II (#2195526, Gibco, USA) diluted in DMEM solution at 37 °C for 4 h. Then, the cell suspension was filtered with 70 μm strainer to remove the incompletely digested cartilage extracellular matrix (ECM). After centrifuged, chondrocytes were suspended in DPS buffer (#14190144, Thermo Fisher, USA) and their viability was assessed by Countess® II Automated Cell Counter (Thermo Fisher). According to the manufacturer's protocol, the Chromium Single Cell 3’ Library, Gel Bead Kit and the Chromium Single Cell A Chip Kit (10 × Genomics V2, USA) were used to construct scRNA-seq libraries. Briefly, cells were loaded in each channel of the 10 × chip and partitioned into Gel Beads in Emulsion in the Chromium instrument. Then, the RNA from cell lysis was barcoded for reverse transcription, followed by amplification, fragmentation and 5’adaptor and sample index attachment. An Illumina HiSeq Xten was used for the libraries sequencing.
Quality control and single cell RNA sequencing data pre-processing
The raw sequencing reads were first processed by the Cell Ranger toolkit (version 6.0.1) using the standard pipeline (10 × Genomics). To generate the FASTQ files, raw base call (BCL) files were used with the Cell Ranger “mkfastq” command. Next, we used Cell Ranger “count” to preform alignment. The reads were aligned to the GRCh38 reference genome using a pre-built annotation package download from the 10X Genomics website. From each sample, the output data was processed with the Seurat package (version 4.0.6) in R software (version 4.0.5). Low-quality cells were filtered by removing cells with expressed genes (>6000 and <200) or the percent of mitochondrial/ribosomal genes >10%. Cells identified as doublets were removed using DoubletFinder package (version 2.0.2). We detected 36,183 genes in a total of 17,638 cells for further bioinformatic analysis.
Data integration, dimension reduction and cell clustering
The Seurat object with gene expression data was processed to normalize and scale by using the functions in the Seurat package. We normalized the objects by ‘‘NormalizeData’’ function with setting normalization method as “LogNormalize’’. The top 2000 highly variable genes from the normalized expression matrix were identified. Dimensional reduction was performed via principal components analysis (PCA). The batch effects were removed by the integrated logarithm on the top 30 PCA components identified. The clustering analysis was performed on the integrated objects by using “FindClusters” function in the Seurat package. The identified clusters were visualized produced with the t-distributed stochastic neighbor embedding (t-SNE) method. For a sub-clustering analysis, we applied a similar procedure, including the variable genes identification, dimension reduction and the clustering identification.
Annotating cell clusters
We identified the differentiation marker genes in the specific cluster when compared to remaining clusters with the Wilcoxon Rank-Sum Test using the “FindAllMarkers” function in Seurat (adjusted P-value <0.05, only.pos = T and logfc.threshold = 0.1). Top 100 marker genes were used to enrich Gene Ontology (GO) terms by clusterProfiler packages (version 3.14.3). The previously reported cellular biomarkers and GO were utilized to annotate the cell clusters.
Gene set enrichment analysis
The differential GO and pathways in the cell clusters were identified by using the gene set enrichment analysis (GSEA) in GSEA package (version 4.1.0) with the curated gene set. The hallmark gene sets in the MSigDB databases (https://www.gsea-msigdb.org/gsea/msigdb) were used for the GSEA analysis. The mean gene expression level was calculated and the log-two fold change between the specific cell cluster and the other cells was applied as the test statistic, the functions with larger normalized enrichment score value and p-value <0.05 were marked as the significant differences. Next, the significant differential GO or pathways were combined into specific data sets. We applied the non-parametric and unsupervised algorithm named gene set variation analysis (GSVA) in the GSVA package (version 1.38.2) to assign functional differences activity estimates to subpopulations.
Cell-cell communication analysis
To investigate potential interactions across different cell clusters, a cell–cell communication analysis was performed using CellPhoneDB Python package (version 2.1.7), which is a publicly available repository of curated receptors and ligands and their interactions. We first randomly permuted the cluster labels of all cells 1000 times to determine the mean of the average receptor and ligand expression levels of the interacting clusters. Then, based on the expression of a receptor by one cell type and the expression of the corresponding ligand by another cell type, we enriched receptor-ligand interactions between two cell types. Next, the most relevant cell type-specific interactions between ligands and receptors were identified, and receptors and ligands expressed in less than 10% in the corresponding subcluster of the cells were removed. By statistical interactions of ligand and receptor in each cell cluster, we summarized the inferred intercellular communication network of each ligand-receptor pair and each signaling pathway and visualized by circle plots.
Animal study
Wild type C57BL/6 mice and Gpx4+/− mice, generated by CRISPR/Cas9 technology, were purchased from Model Animal Research Center of Nanjing University. All mice were kept under pathogen-free and 12 h light/dark cycle environment with free access to food and water. To determine the expression of Trpv1 and Gpx4 during OA progression, 10-week-old male C57BL/6 mice were subjected to DMM surgery14,15 on the right knee joint and sacrificed 2, 4 and 8 weeks after surgery. In addition, 10-week-old male C57BL/6 mice, Gpx4+/− mice and their littermate controls were randomly subjected to: (1) Sham surgery (Sham); (2) CPS (Capsaicin, #ab141000, Abcam, Cambridge, UK) treatment (intra-articular injection, 50 μM, 8 μL per time, 3 times per week); (3) DMM (destabilization of medial meniscus), and (4) DMM with CPS treatment. Mice in the Sham and DMM groups were injected with 8 μL normal saline. The number of mice in each group was calculated according to the formula reported by Charan and Kantharia.16 All animal studies complied with the ARRIVE guidelines.
DMM surgery-induced OA mouse model
The surgically induced OA by DMM was created as previously reported.15 Briefly, after mice were anaesthetized by isoflurane, DMM surgery was performed on the right knee joint by sectioning the medial menisco-tibial ligament, which anchored medial meniscus to tibial plateau. A sham operation followed the same procedure without ligament sectioning was performed on the control mice. Mice were sacrificed at 2, 4, or 8 weeks after surgery.
Iron, LPO, GSH, GSH/GSSG, GPX4 activity detection in human OA cartilage and MMP3 detection in synovial fluide
After morphological and histological division of intact and damaged areas, human OA cartilage were cut into small pieces and ground into powder under liquid nitrogen condition. Then, we homogenate the cartilage powder with cold normal saline and extracted the supernatant after centrifugation (12,000 rpm, 20 min, 4 °C). The iron contents (#E-BC-K139-S, Elabscience, Wuhan, China), LPO levels (#A106-1-1, Jiancheng, Nanjing, China), GSH levels and GSH/GSSG ratios (#S0053, Beyotime, Shanghai, China), and GPX4 activity (#A005, Jiancheng) were detected according to the manufacturer's instructions. In addition, the iron detection kit (#A039-1-1, Jiancheng) and MMP3 enzyme-linked immunosorbent assay kit (#KE00160, Proteintech, Wuhan, China) were used to detect iron and MMP3 levels in the synovial fluid, respectively.
Histological analysis
After decalcified by 10% EDTA (#1340, Biofroxx, Germany) solution, human cartilage and mouse knee joints were embedded in paraffin blocks and cut into continuous coronal slides (5 μm thick) by a microtome (Thermo, Germany). Following a previously reported procedure,15 every fifth section of two slides were selected and stained with Safranin-O/fast green (S.O.) (#G1371, Solarbio, Beijing, China) and hematoxylin and eosin (H&E) (#C0105S, Beyotime) to evaluate the cartilage lesions and synovitis, respectively. Cartilage destruction was assessed by blinded observers using the Osteoarthritis Research Society International (OARSI) grading system (0-6) and synovitis was scored by a 0-3 scoring system as previously reported.17 The highest OARSI score and synovitis score for every section were recorded and the means of all scores were calculated.
Immunofluorescence and immunohistochemical staining
The deparaffinized and hydrated histological slides of human cartilage or mouse knee joint were blocked by 5% bovine serum albumin (BSA) (1 h, room temperature) and incubated overnight (4 °C) with primary antibodies against Collagen II (#BA0533, Boster, Wuhan, China), MMP3 (#14351, Cell Signaling Technology, USA, RRID:AB_2798459), 4-HNE (#ab46545, Abcam), COX2 (#66351-1-Ig, Proteintech, RRID:AB_2881731), TRPV1 (#66983-1-Ig, Proteintech, RRID:AB_2882302), FTH1 (#4393, Cell Signaling Technology, RRID:AB_11217441), GPX4 (#ab125066, Abcam, RRID:AB_10973901), NCOA4 (#66849, Cell Signaling Technology), Tf (#17435-1-AP, Proteintech, RRID:AB_2035023), MMP13 (#ab219620, Abcam) and p-CaMKII (#ab124880, Abcam, RRID:AB_305050). After washed by TBST, the slides were incubated with tetramethylrhodamine-5-(and 6)-isothiocyanate- or fluorescein isothiocyanate (FITC)-conjugated secondary antibody (Abcam) for immunofluorescence staining, which was imaged with a fluorescence microscope (Zeiss, Germany). For immunohistochemical staining, 3% (v/v) H2O2 was used to quench the endogenous peroxidase activity and a horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG) (#BL003A, Biosharp, Shanghai, China) was used as the secondary antibody. An ultra-sensitive DAB Kit (#1205250, Typing, Nanjing, China) was used to visualize the immunohistochemical staining. Instead of the primary antibody, nonimmune IgG was used as a negative control of immunofluorescence and immunohistochemical staining. The proportions of positive cells in human and mouse cartilage were quantified by two observers under blind condition.
Micro-computed tomography (micro-CT) analysis
Before the histological analysis, mice knee joints were evaluated via micro-CT using a VivaCT 80 scanner (Scanco Medical AG, Switzerland) with a 70 kVp source as previously described.18 Joints were scanned at a resolution of 18.38 μm. The reconstructed 3-dimensional (3D) images were acquired by Scanco Medical software at a Scanco threshold of 220. The number of osteophytes, identified as protrusions, were calculated using the 3D reconstructive images. The subchondral plate thickness was calculated, as previously reported,19 by multiplying the number of cross-sections forming the subchondral plate and section thickness. The subchondral bone analysis of medial femoral condyle begins by starting at the distal edge of the epiphyseal line until 20 layers were analyzed by blinded observers. All non-cortical bone is included.19
The culture and treatment of human OA cartilage explants
The culture of human articular cartilage from OA patients undergone total knee arthroplasty was performed as previously reported.19 Briefly, full thickness cartilages were obtained and flushed with sterile saline solution. The cartilages were cut into small pieces with a side length of 0.5 cm and cultured overnight in low-glucose Dulbecco's modified Eagle's medium at 37 °C. Then, the cartilage pieces were randomly divided into five groups as: (1) Control (Con); (2) 50 μM capsaicin (CPS) treatment; (3) 50 μM tert-butyl hydroperoxide (TBHP); (4) 50 μM TBHP + 50 μM CPS; (5) 50 μM TBHP + 5 μM Fer-1. The drug-containing culture medium was replenished every 72 h. After 7 days’ treatment, the cartilage pieces were fixed with 4 % paraformaldehyde, embedded in paraffin, and cut into 5-μm sections for histological analysis.
Terminal deoxynucleotidyl transferase dUTP nick-end labeling (Tunel) assay
The live and dead cells measurement of treated human OA cartilage explants was performed using a CF488-Tunel Detection Kit (#G1504-50T, Servicebio) according to the manufacturer's instructions.
Cell culture
The primary mouse chondrocytes were collected and cultured as previously described.20 Briefly, tibial plateaus and femoral heads from 3-days-old C57BL/6 mice were dissected and digested with 4 ml 0.2% Collagenase 2 (Gibco, USA) for 4 hours at 37 °C. After soft tissue removal, the chondrocytes were cultured in Dulbecco's modified Eagle's medium contained 1 g/L glucose (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin and streptomycin (Gibco) at 37°C and 5% CO2 condition. The cell culture medium was replenished every 72 h. Chondrocytes were treated with TBHP (#MKCH9944, Sigma, USA), 50 μM capsaicin (CPS, #ab141000, Abcam), 10 μM JNJ-17203212 (JNJ, #HY-100129, MCE-MedChemExpress, New Jersey, USA), 10 μM RSL3 (#S8155, Selleck USA), 10 μM DFO (#HY-B0988, MCE), 5 μM Fer1 (#S7243, Selleck), 10 μM ZVAD-FMK (#HY-16658B, MCE), or 10 μM Necrosulfonamide (#HY-100573, MCE) as indicated.
Short interfering RNA (siRNA) transfection of mouse primary chondrocytes
The transfection of siRNAs was processed essentially as previously described.21 siRNAs were designed and synthesized as the following sequences: mouse Trpv1: 5’-GCUACACAGACAGCUACUACATT-3’; mouse Gpx4: 5’-GGAGCCAGGAAGUAAUCAAGATT-3’ (Hippobio, Huzhou, China).
Protein extraction and western blot
The total protein from human cartilages and chondrocytes with various treatment were extracted using RIPA lysis buffer (#R0010, Solarbio) containing 1 mM phosphatase inhibitor cocktail (#B15002, Bimake, USA) and 1 mM phenylmethanesulfonyl fluoride (#329-98-6, Solarbio). Western blot was performed essentially as previously described.18 The primary antibodies used were: NCOA4 (#66849, Cell Signaling Technology), Tf (#17435-1-AP, Proteintech, RRID:AB_2035023), COX2 (#66351-1-Ig, Proteintech, RRID:AB_2881731), MMP3 (#14351, Cell Signaling Technology, RRID:AB_2798459), Collagen II (#BA0533, Boster), FTH1 (#4393, Cell Signaling Technology, RRID:AB_11217441), MMP13 (#ab219620, Abcam), TRPV1 (#66983-1-Ig, Proteintech, RRID:AB_2882302), GPX4 (#ab125066, Abcam, RRID:AB_10973901) and GAPDH (#5174, Cell Signaling Technology, RRID:AB_10622025) (Supplementary Table 4). After horseradish peroxidase-conjugated goat anti-rabbit/mouse secondary antibodies (#BL003A or #BL001A, Biosharp) incubation, western blots were imaged by the ChemiDocXRS + Imaging System (Tanon, Shanghai, China). Image J (version 1.8.0) was used for the quantitative analysis of proteins. All antibodies used in this study were commercial antibodies.
Quantitative real-time polymerase chain reaction (qPCR)
The mRNA isolation and qPCR were essentially performed as previously described.18 Cellular mRNA was isolated using the RNA-quick Purification Kit (#RN001, ES Science, Shanghai, China) and qPCR was conducted using SYBR Green Q-PCR Kit (#Q411-02, Vazyme, Nanjing, China) on a LightCycler 480 PCR System (Roche, Switzerland). The primer sequences used are listed in Supplementary Table 5.
Intracellular ROS, lipid ROS and Fe2+ detection
Intracellular levels of ROS, lipid peroxidation and ferrous iron (Fe2+) were measured by 10 μM ROS probe DCFH-DA (#S0033S, Beyotime), 5 μM lipid ROS probe C11-BODIPY 581/591 (#GC40165, GLPBIO, USA) and 1 μM Fe2+ probe FerroOrange (#F374, Dojindo, Japan) as previously reported.22 Briefly, after overnight starvation, chondrocytes were incubated with probes for 30 min at 37 °C and then treated with 50 μM TBHP with or without 50 μM CPS and 10 μM JNJ for 4 h. The cells were suspended in flow buffer and run through a BD AccuriC6 Plus Flow CytoMeter (BD Biosciences, USA). FlowJo software (version 10) was used to analyze the data.
Cell Counting Kit-8 (CCK8) assay and Live/Dead assay
The cell viability and live/dead staining of treated chondrocytes were assessed by CCK8 (#CK04, Dojindo) assay and AM/PI staining (#41260A, BestBio, Shanghai, China), respectively, according to the manufacturer's instructions.
Statistical analysis
Statistical analysis was conducted using the SPSS software (version 25.0) and GraphPad Prism software (version 8.0). Quantitative data represents at least three independent experiments. During the analysis, no samples or animals were excluded. Shapiro-Wilk test and Levene method were used for the estimation of the data normal distributions and homogeneity of variance, respectively. For the comparation of mean values between two groups, paired or unpaired two-tailed Student's t test was used. One-way or two-way analysis of variance (ANOVA) followed by Tukey's post-hoc tests were used to assess the statistical significance of the mean values of more than two groups. Data were presented as mean values ± SD. The statistical significance was set at P < 0.05.
Role of funders
The funders played no role in the design, experiment conduction, data analysis and preparation of the manuscript of this work.
Results
Ferroptotic hallmarks accumulate in OA cartilage
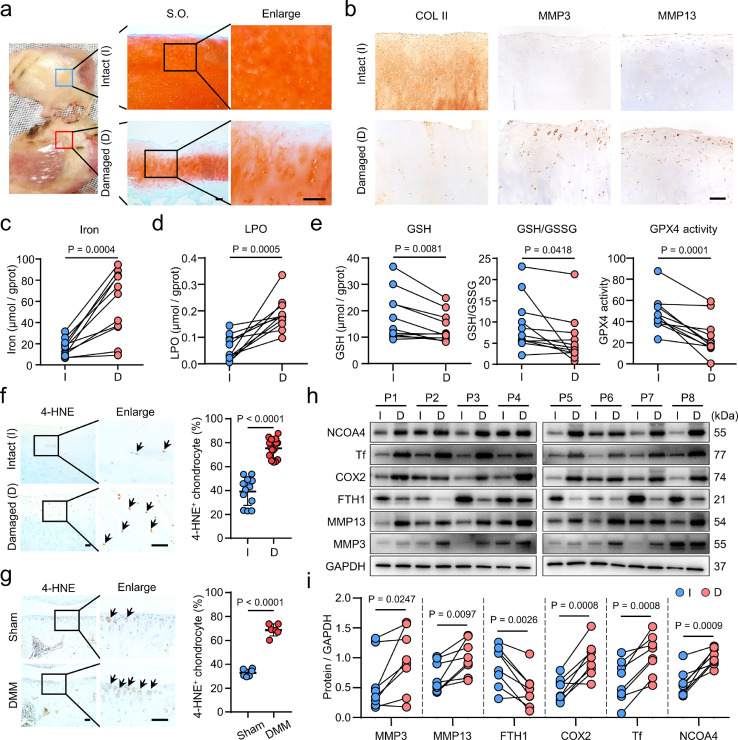
To address the association between ferroptosis and OA, we analyzed the expression of ferroptotic hallmarks23 in morphologically and histologically defined intact and damaged human OA cartilages (Figure 1a, b). Compared to the intact cartilages, the contents of iron and LPO were significantly elevated in damaged cartilages (Figure 1c, d), suggesting that OA chondrocytes may encounter ferroptosis. We then measured the levels of GSH and the activity of GPX4 to evaluate the anti-ferroptotic ability in the cartilages. The damaged cartilage showed lower levels of GSH, GSH/GSSG ratio and GPX4 activity compared with that of intact cartilage (Figure 1e). To further clarify the link between lipid peroxidation and OA pathogenesis, we examined the expression of 4-hydroxynonenal (4-HNE), a primary lipid peroxidation product.24 Coincident with the increased levels of LPO (Figure 1d), both damaged human OA cartilages and destabilization of medial meniscus (DMM) surgery-induced mice cartilages exhibited substantial increases of 4-HNE levels (Figure 1f, g). The ferroptotic hallmarks, including GSH depletion, iron and LPO accumulation25 suggest ferroptosis could occur in OA cartilage.
Figure 1.
Ferroptotic hallmarks accumulate in osteoarthritis (OA) cartilage. (a) Safranin O-Fast Green (S.O) staining of intact (I) and damaged (D) articular cartilages from OA patients. (b) Immunohistochemical (IHC) staining of Collagen II (COL II), matrix metallopeptidase 3 (MMP3) and MMP13 in human OA cartilages. (c-e) The levels of total iron (c), lipid peroxidation (LPO) (d), glutathione (GSH), GSH/GSSG (oxidized GSH) ratios and glutathione peroxidase 4 (GPX4) activities (e) in paired intact (I) and damaged (D) (n = 11) human OA cartilages. gprot, gram of protein. (f, g) IHC staining of 4-hydroxynonenal (4-HNE) and its quantification in the intact (n=12) and damaged (n=16) human OA cartilages (f), and in Sham or destabilization of medial meniscus (DMM)-induced mice (n=6) for 8 weeks (g). (h, i) Western blot analysis (h) and quantification (i) of indicated proteins in paired intact (I) and damaged (D) (n=8) human OA cartilages. P, patient. Scale bars, 50 μm (b, f, g), 200 μm (a). Two-tailed paired (c-e, i) or unpaired (f, g) t-test. Data are shown as mean ± SD.
Next, we examined the expression of canonical ferroptotic markers23,26 in the human OA cartilages. We found the ferroptotic drivers, nuclear receptor coactivator 4 (NCOA4), transferrin (Tf) and cyclooxygenase 2 (COX2) were substantially increased in damaged cartilages, as well as matrix metallopeptidase 3 (MMP3) and MMP13, two enzymes responsible for cartilage degradation (Figure 1h, i). Whereas, the ferroptotic suppressor, ferritin heavy chain 1 (FTH1), was significantly decreased in the damaged human OA cartilages (Figure 1h, i). Immunofluorescence staining confirmed that COX2 was significantly increased in the damaged human OA cartilage and OA mice model, and was gradually increased with the growth of mouse (Supplementary Figure 1a-c). Furthermore, the iron content in synovial fluid from OA patients showed a positive correlation with the MMP3 levels (Supplementary Figure 1d). Taken together, these results suggest that ferroptosis occurs in OA cartilage.
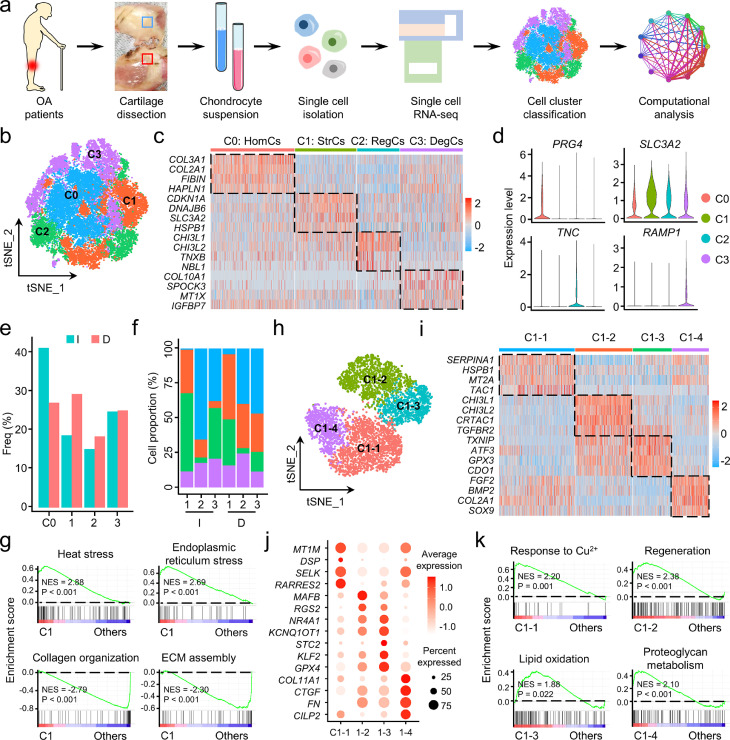
Transcriptional heterogeneity of human OA chondrocytes
To decipher the characteristics of ferroptotic chondrocyte cluster in OA, scRNA-seq analysis on three pairs of intact and damaged human OA cartilages was performed (Figure 2a). After initial stringent quality control and doublet removal, a total of 17,638 individual human chondrocytes were profiled, including 8,292 cells from the intact area and 9,346 cells from the damaged area (Supplementary Table 6). The cells were sequenced in a depth of UMIs (unique molecular indexes) ranged from 8,740 to 11,540 per cell, with the mean of 2,051 to 2,388 detected genes per cell (Supplementary Figure 2).
Figure 2.
Single-cell transcriptomic analysis of intact and damaged human osteoarthritis (OA) cartilages. (a) Schematic workflow of the experimental strategy. (b) The t-distributed stochastic neighbor embedding (t-SNE) plot of the four identified main chondrocyte clusters in OA cartilage. (c) Heatmap revealing the scaled expression of preferentially and differentially expressed genes for each chondrocyte cluster. (d) Violin plots demonstrating the normalized gene expression levels of representative marker genes among the four main clusters. (e) The frequency of each chondrocyte cluster in the intact (I) and damaged (D) human OA cartilages. Freq, frequency. (f) Relative proportion of each chondrocyte cluster across three pairs of intact (I) and damaged (D) cartilages as indicated. (g) Gene set enrichment analysis (GSEA) revealing the enrichment of Gene Ontology (GO) terms in C1 and the other three clusters. NES, normalized enrichment score; ECM, extracellular matrix. (h) Visualization of t-SNE colored subclusters within C1. (i) Heatmap showing the highly expressed genes in the four subclusters within C1. (j) Dot plot showing the mean expression of preferentially expressed genes among the four subclusters. (k) GSEA showing the representative function among each subcluster. NES, normalized enrichment score.
Based on t-SNE analysis, unbiased clustering of the cells parallelly identified four main clusters according to their gene profiles and canonical markers (Figure 2b). Particularly, these cell clusters were: (C0) the homeostatic chondrocytes (HomCs) highly expressing COL2A1,27 COL3A1,28 HAPLN129 and PRG4;30 (C1) the stressed chondrocytes (StrCs) preferentially expressing CDKN1A,31 DNAJB6,32 SLC3A233 and HSPB1;34 (C2) the regulatory chondrocytes (RegCs) specifically expressing CHI3L1,35 CHI3L2,35 NBL136 and TNC;37 and (C3) the degenerative chondrocytes (DegCs) with high expression of COL10A1,27 MT1X,38 IGFB7,39 and RAMP140 (Figure 2c, d). We noticed that, in damaged cartilage, the frequency of HomCs was declined, while StrCs and RegCs were increased (Figure 2e), indicating loss of cartilage homeostasis and increased cellular stress in the damaged area than the intact area. Moreover, the varied proportion of the four chondrocyte clusters among the six samples suggests high heterogeneity of OA chondrocytes (Figure 2f). To decipher the specified characteristics of StrCs, we performed a GSEA and found cellular stress-related GO terms were significantly enriched in StrCs when compared to the other three clusters (Figure 2g, Supplementary Figure 3a). In contrast, the ECM-related GO terms were significantly enriched in cell clusters except for StrCs (Figure 2g, Supplementary Figure 3b).
Since stressed cells by various stimulus may acquire unequal features,41,42 we further divided StrCs into four subclusters (Figure 2h). Among them, C1-1 was identified as metal ion-related cluster with high expression of genes encoding proteins that transport and bind metal ions (Figure 2i, j). C1-2 and C1-4 were regulatory and homeostatic clusters with specifically expressed genes related to chondrogenesis (CHI3L1 and TGFBR2) and ECM production (COL2A1 and SOX9), respectively (Figure 2i, j). Furthermore, C1-3 was identified as an oxidative stress cluster because of preferentially expressed oxidation and antioxidant genes, including TXNIP, ATF3, GPX3, CDO1, and GPX4 (Figure 2i, j). GSEA also confirmed the characteristics of these four subclusters (Figure 2k). Since ferroptosis was a specific type of oxidative stress,43 to gain insight into whether ferroptotic chondrocyte cluster was contained in C1-3, we assessed the expression levels of genes involving classical types of cell death. C1-3 exhibited apparently higher levels of ferroptosis marker genes than apoptosis, necroptosis and pyroptosis (Supplementary Figure 4), suggesting ferroptosis was the most likely form of chondrocyte death within C1-3.
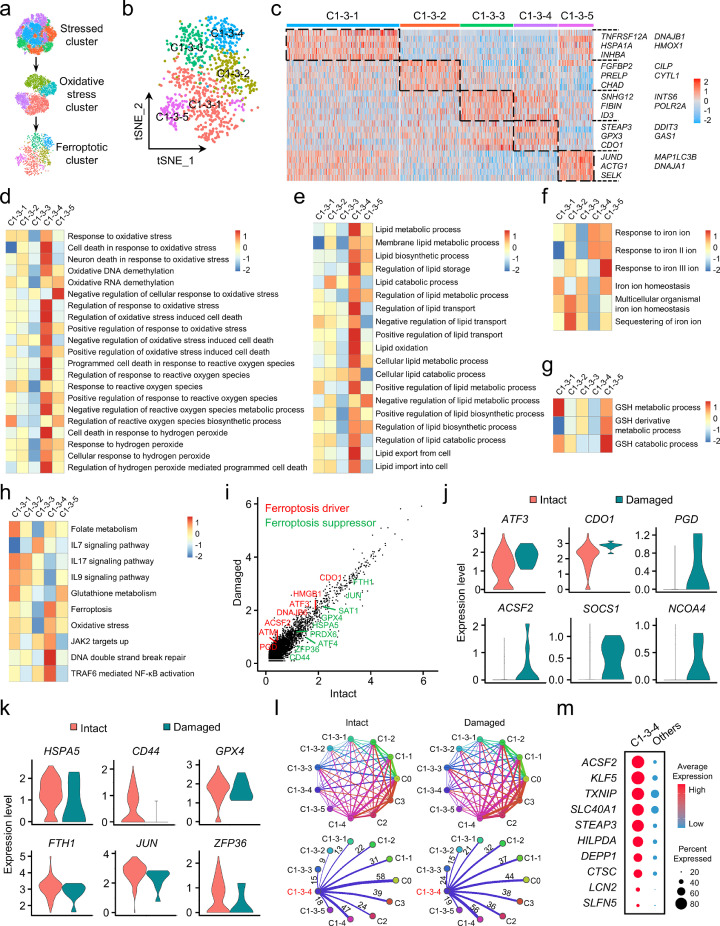
Identification and molecular characterization of the ferroptotic chondrocyte cluster in OA
To characterize the features of the ferroptotic chondrocyte cluster, we divided the oxidative stress cluster into five subclusters (Figure 3a, b) and found that C1-3-4 preferentially expressed the marker genes of ferroptosis (Figure 3c). Competitive GSEA showed that the GO terms related to oxidative stress, and, especially, oxidative stress-induced cell death were prominently enriched in C1-3-4 (Figure 3d, Supplementary Figure 5a). Moreover, C1-3-4 exhibited relatively high enrichment of lipid synthesis, transport and oxidation, as well as response to ferrous iron (Fe2+) and iron ion transmembrane transport (Figure 3e, f, Supplementary Figure 5a). However, GSH metabolism related GO terms were not apparently enriched in C1-3-4 (Figure 3g). More impressively, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed highly enriched “Ferroptosis” in C1-3-4 (Figure 3h). In addition, ferroptosis-related genes were relatively abundant in C1-3-4, while genes involved in other types of cell death were barely detectable (Supplementary Figure 5b). These results indicate that C1-3-4 is the ferroptotic chondrocyte cluster in OA.
Figure 3.
Identification and molecular characterization of the ferroptotic chondrocyte cluster in osteoarthritis (OA). (a) Graphic view of the analysis roadmap. (b) The t-distributed stochastic neighbor embedding (t-SNE) plot of the five main chondrocyte clusters within the oxidative stress cluster (C1-3). (c) Heatmap revealing the preferentially expressed genes among the subclusters. (d-g) The heatmap of gene set enrichment analysis (GSEA) of the hallmark gene sets in MSigDB database revealing the enrichment of oxidative stress (d), lipid (e), iron (f) and glutathione (g) related Gene Ontology (GO) terms among subclusters within C1-3. (h) The heatmap of Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis showing differentially enriched signaling pathways among the subclusters. (i) The scatter plot of the differentially expressed genes in C1-3-4 between chondrocytes from the intact and damaged cartilages. (j, k) Violin plots showing the normalized expression levels of ferroptosis drivers (j) and suppressors (k) in C1-3-4 across the intact and damaged cartilages. (l) Overview of the cell-cell crosstalk (upper panel) and the communications of ferroptotic chondrocyte cluster with the others (lower panel) in intact and damaged cartilages. (m) Dot plot showing the overexpressed genes in ferroptotic chondrocyte cluster (C1-3-4).
Next, we investigated the molecular characteristics of ferroptotic chondrocyte cluster. Among the differentially expressed genes, the ferroptotic drivers were higher expressed in the damaged cartilage, whereas ferroptotic suppressors were more abundant in the intact cartilage (Figure 3i-k, Supplementary Figure 5c, d), indicating more ferroptosis occurs in more severely damaged area. To evaluate the role of ferroptosis in OA, we investigated the intercellular crosstalk between C1-3-4 and other clusters. Compared to the intact cartilage, C1-3-4 showed more active signaling interactions with clusters possessing cartilage regeneration function (C2, C1-2 and C1-4) in the damaged cartilage (Figure 3l), suggesting chondrocyte ferroptosis might interfere the self-repair mechanisms of OA cartilage. Furthermore, by comparing the gene profiles in cells from other clusters, we also explored the top 10 substantially overexpressed genes in C1-3-4 as potential markers of ferroptotic chondrocytes (Figure 3m). Taken together, these results confirm the existence and illustrate the molecular characteristics of ferroptotic chondrocyte cluster in OA cartilage.
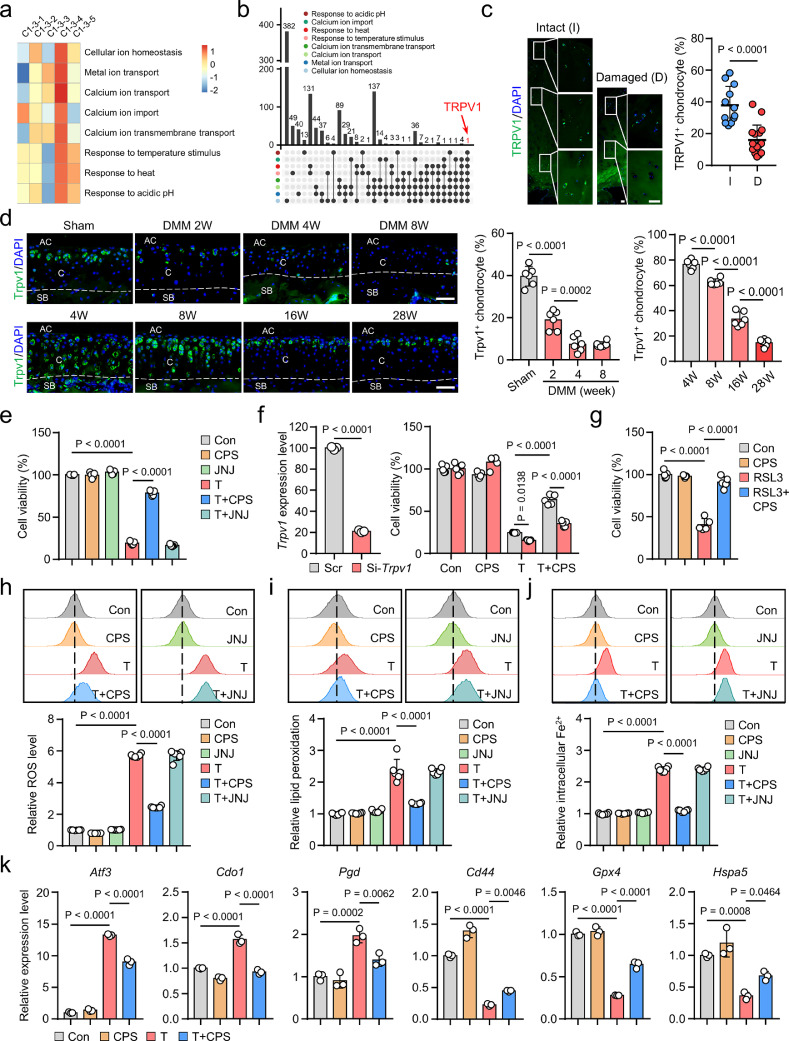
TRPV1 plays an anti-ferroptotic role in chondrocytes
Given the above results, we sought to explore the potential therapeutic target for chondrocyte ferroptosis. We performed a comprehensive GSVA to unveil the biological features of C1-3-4 (Figure 4a, Supplementary Figure 6). We noticed that calcium ion transport, temperature and acidic pH associated GO terms were highly enriched in C1-3-4 (Figure 4a), and, intriguingly, TRPV1 was the only overlapped gene among these biological processes (Figure 4b), suggesting that TRPV1, the heat- and acid-sensitive cation channel,44,45 may potentially regulate chondrocyte ferroptosis. Moreover, we found that the expression of TRPV1 was substantially decreased in damaged human OA cartilage (Figure 4c), as well as in DMM-induced and relatively older mice cartilage (Figure 4d), which promoted us to further explore whether TRPV1 alleviated chondrocyte ferroptosis in OA.
Figure 4.
Transient receptor potential vanilloid 1 (TRPV1) prevents chondrocyte ferroptosis. (a) The heatmap of gene set variation analysis revealing functional Gene Ontology (GO) terms highly enriched in C1-3-4. (b) The gene overlap analysis of the GO terms that revealed in (a). (c) Immunofluorescence (IF) staining and quantitative analysis for TRPV1 in intact (I) (n=12) and damaged (D) (n=16) human OA cartilages. (d) IF staining and quantitative analysis for Trpv1 expression in the cartilage of indicated mice (n=6). AC, articular cavity; C, cartilage; SB, subchondral bone. (e-g) Cell viability of mouse primary chondrocytes treated as indicated for 24 h (n=5). (h-j) Flow cytometry analysis and quantitative analysis of intracellular total reactive oxygen species (ROS) (h), lipid peroxidation (i) and ferrous iron (Fe2+) (j) levels in chondrocytes induced by 50 μM TBHP with or without 50 μM CPS or 10 μM JNJ for 4 h (n=6). (k) Q-PCR analysis of indicated genes in chondrocytes treated with 50 μM TBHP and/or 50 μM CPS for 6 hours (n=3). Scale bars, 50 μm (d), 100 μm (c). Two-tailed unpaired t-test (c, left panel of f), one-way (d, e, g-k) or two-way (right panel of f) ANOVA with Tukey's post-hoc test. Data are shown as mean ± SD.
Next, we tested the effect of TBHP, a widely used oxidative stress inducer,12 on chondrocyte ferroptosis in vitro. The TBHP treatment resulted in significantly increases of total ROS, lipid peroxidation and cell death in chondrocytes, which was significantly abrogated by ferroptosis inhibitors deferoxamine (DFO) and ferrostatin-1 (Fer-1) (Supplementary Figure 7a-d). In contrast, as previously reported,13 the apoptosis inhibitor (ZVAD) and necroptosis inhibitor (Necro) showed no significant effect on TBHP-induced cell death (Supplementary Figure 7d). These data suggest that TBHP could induce chondrocyte ferroptosis.
Then, we treated the cultured primary chondrocytes with a specific TRPV1 agonist, CPS, or a selective TRPV1 antagonist, JNJ, under the condition of TBHP. TRPV1 activation by CPS substantially recovered cell viability, while TRPV1 inhibition by JNJ showed no obvious effect (Figure 4e, Supplementary Figure 8a-c). Trpv1 knockdown significantly weakened the protection of pharmacologically activated Trpv1, excluding the off-target effect of CPS (Figure 4f). Moreover, TRPV1 activation significantly inhibited RSL3, a ferroptosis inducer,46 induced chondrocyte ferroptosis (Figure 4g). Furthermore, TRPV1-activated chondrocytes exhibited significant inhibition of the hallmarks of ferroptosis, including decrease of TBHP-induced total ROS, lipid peroxidation and Fe2+ accumulation (Figure 4h-j), as well as mitochondrial dysfunction (Supplementary Figure 8d). Q-PCR validated the anti-ferroptotic role of Trpv1 by downregulated ferroptotic drivers (Atf3, Cdo1, Pgd, Ptgs2) and upregulated ferroptotic suppressors (Cd44, Gpx4, Hspa5, Atf4, Lamp2) (Figure 4k, Supplementary Figure 8e). Moreover, the expression of Trpv1 itself and six of the marker genes screened in the ferroptotic chondrocyte cluster were also regulated by CPS-induced Trpv1 activation (Supplementary Figure 8e, f). These results indicate the vital role of TRPV1 as an anti-ferroptotic target in chondrocytes.
TRPV1 protects chondrocytes from ferroptosis in OA
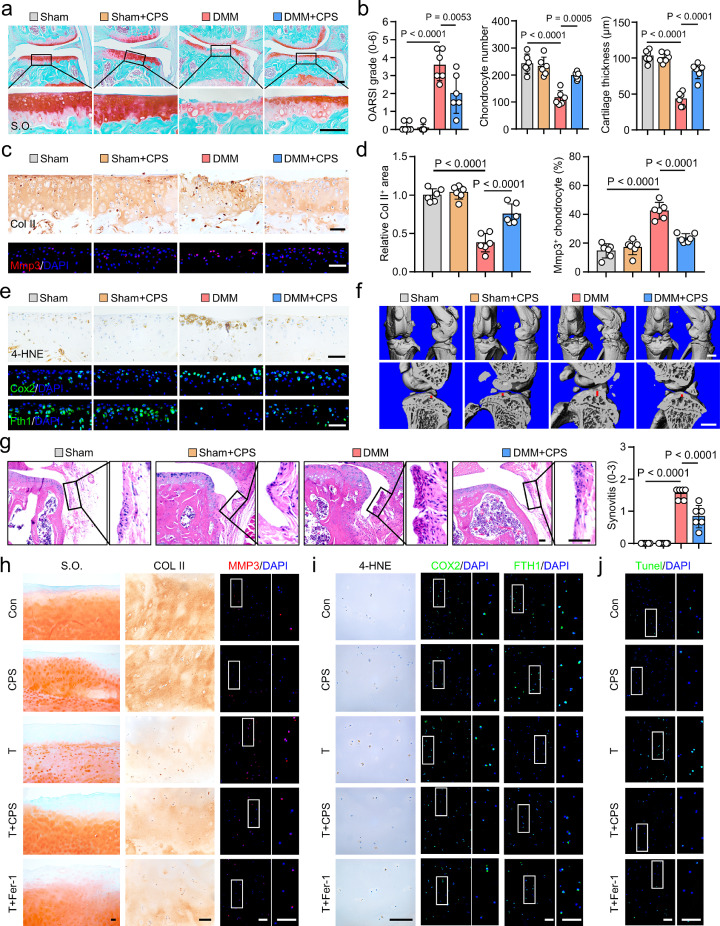
To gain further insight into the anti-ferroptotic role of TRPV1 in OA, we performed intra-articular injection of CPS in the OA mouse model. The phosphorylation of calcium/calmodulin-dependent protein kinase II (CaMKII) is dependent on TRPV1 activation.18 Firstly, the CPS injection caused significant upregulation of phosphorylated CaMKII (p-CaMKII) (Supplementary Figure 9a), indicating the effective activation of TRPV1 in the mice articular cartilage. The CPS treatment showed significant cartilage-protective effects, including reduced cartilage erosion and Osteoarthritis Research Society International (OARSI) score (Figure 5a, b), increased total chondrocyte number (Figure 5b), improved total cartilage thickness (Figure 5b) and uncalcified cartilage thickness (Supplementary Figure 9b). In addition, increased Col II+ areas, suggestive of cartilage anabolism, and decreased Mmp3+ chondrocytes, indicative of cartilage catabolism, were observed in CPS-treated DMM surgery group (Figure 5c, d).
Figure 5.
Transient receptor potential vanilloid 1 (TRPV1) plays an anti-ferroptotic role in osteoarthritis (OA). (a, b) Safranin-O/fast green (S.O.) staining (a) and quantification (b) of mice induced by Sham or destabilization of medial meniscus (DMM) surgery with or without intraarticular injection of 50 μM capsaicin (CPS) (n=6). (c, d) Immunohistochemical (IHC) staining of Col II, immunofluorescence (IF) staining of Mmp3 (c),and corresponding quantitative analysis (d) in the articular cartilage of mice treated as in (a) (n=6). (e) IHC staining of 4-hydroxynonenal (4-HNE), IF staining of cyclooxygenase 2 (Cox2) and ferritin heavy chain 1 (Fth1) of indicated mice knee articular cartilage. (f) 3D reconstructed images of mice knee joints and the sagittal view of the medial joint compartment revealing the changes to femoral and tibial surfaces and subchondral bone plate (SBP) thickness, respectively. Red line indicates the thickness of SBP. (g) Hematoxylin & eosin (H&E) staining of mice knee and relative quantification of synovitis scores as indicated (n=6). (h) Representative images of S.O. staining, IHC staining for COL II, and IF staining for MMP3 of the cultured human OA cartilage explants treated with 50 μM TBHP and/or 50 μM CPS, or 5 μM Fer-1 for 7 days. (i, j) Representative images of IHC staining for 4-HNE, IF staining for FTH1, COX2 (i) and Tunel (j) of human OA cartilage explants as treated in (h). Scale bars, 50 μm (c, e, g), 100 μm (a, h-j) and 1mm (f). One-way ANOVA with Tukey's post-hoc test. Data are shown as mean ± SD.
Further, CPS-treated mice cartilage exhibited substantially decreased proportion of 4-HNE+, Cox2+, Ncoa4+ and Tf+ chondrocytes (Figure 5e, Supplementary Figure 9c-e), demonstrating declined ferroptosis in Trpv1-activated cartilage. Ferritin heavy chain 1 (Fth1) is the main intracellular iron storage protein and its autophagic degradation results in increased cellular labile iron content and sensitizes cells to ferroptosis.23 Our results showed that TRPV1 activated cartilage exhibited evidently increased Fth1 expression in DMM-induced mice (Figure 5e, Supplementary Figure 9c), suggesting the reduced free iron overload. CPS injection also preserved the expression of Trpv1 in the cartilage of OA mice (Supplementary Figure 9f).
We also evaluated the bone status of mice knee joints by micro-computed tomography analysis and three-dimensional reconstruction, and observed significant decrease of osteophyte formation and subchondral bone sclerosis in CPS-injection group (Figure 5f, Supplementary Figure 10). Moreover, the synovitis score was also reduced by Trpv1 activation (Figure 5g).
We further verified the relevance of our findings to humans by examining the role of TRPV1 in ex vivo TBHP-treated cartilage explants from OA patients, in which Fer-1 was used as a positive control. Similar to the results in mice cartilage, TRPV1 activation by CPS or Fer-1 treatment markedly abated cartilage ECM loss, preserved cartilage anabolism and inhibited cartilage catabolism (Figure 5h, Supplementary Figure 11a). Furthermore, CPS exerted similar anti-ferroptotic effect to Fer-1 by mitigating the TBHP-induced 4-HNE and COX2 expression and restoring FTH1 (Figure 5i, Supplementary Figure 11b). Then, we used Tunel assay to detect cell death47 and revealed significantly reduced Tunel+ chondrocytes in the TBHP with CPS or Fer-1 treated groups (Figure 5j, Supplementary Figure 11c). Taken together, these results indicate that TRPV1 exerts anti-ferroptotic and chondroprotective effects in both DMM-induced mice cartilage and human OA cartilage.
GPX4 mediates the anti-ferroptotic effect of TRPV1
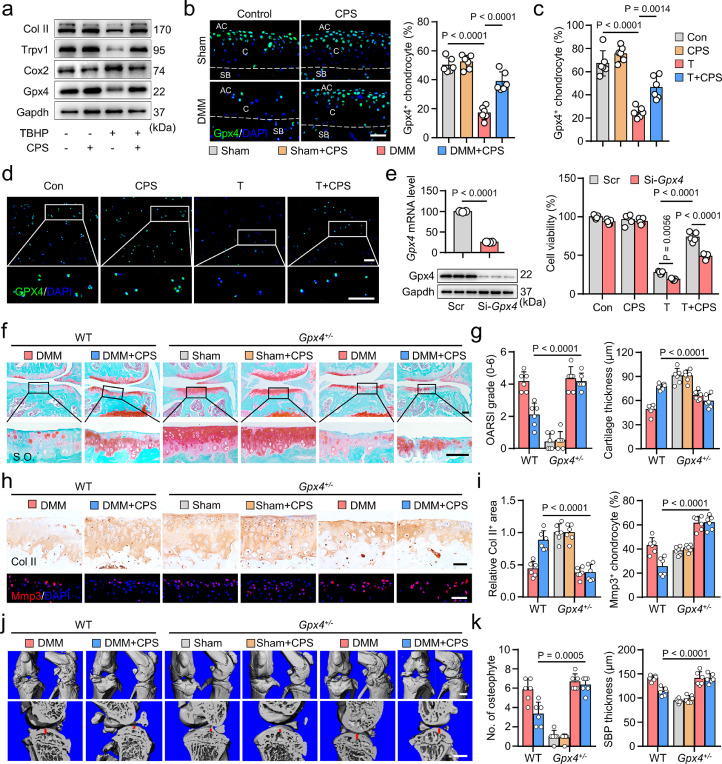
To clarify the downstream molecular mechanisms of TRPV1 resistance to chondrocyte ferroptosis, we analyzed the correlation between TRPV1 and ferroptosis-related genes in our scRNA-seq database. Our results showed that TRPV1 exhibited relatively strong relationship with GPX4 (Supplementary Figure 12a). The expression of Gpx4 in mouse cartilage was also positively correlated with Trpv1 levels (Supplementary Figure 12b), which promoted us to investigate whether Trpv1 regulated the expression of Gpx4. Under the TBHP treatment, mRNA and protein levels of Gpx4 were markedly decreased in the primary chondrocytes, while they were substantially restored by Trpv1 activation (Figure 4k, 6a, Supplementary Figure 13a). TRPV1 activation by CPS resulted in significant increase of GPX4 in both DMM-induced mice and TBHP-treated human OA cartilages (Figure 6b-d). Furthermore, coincident with the decreased Trpv1 (Figure 4d), Gpx4 was significantly decreased in DMM-induced and relatively older mice cartilages (Supplementary Figure 13b, c).
Figure 6.
Glutathione peroxidase 4 (GPX4) mediates the anti-ferroptotic effect of Transient receptor potential vanilloid 1 (TRPV1). (a) Western blot analysis of indicated proteins in mouse primary chondrocytes induced by 50 μM TBHP and/or 50 μM CPS for 8 h. (b) IF staining and quantitative analysis of Gpx4 in the indicated mice cartilage (n=6). AC, articular cavity; C, cartilage; SB, subchondral bone. (c, d) Quantification (c) and IF staining (d) of GPX4 in human OA cartilage explants treated as indicated for 7 days (n=6). (e) Cell viability (n=5) of mouse primary chondrocytes induced by 50 μM TBHP and/or 50 μM CPS for 24 h with or without pretreated Gpx4 siRNA. (f, g) S.O. staining (f) and quantitation of Osteoarthritis Research Society International (OARSI) scores and cartilage thickness (g) in Gpx4+/− mice and their littermate control mice treated as indicated (n=6). (h, i) IF staining of Col II and Mmp3 (h) and their quantification (i) in Gpx4+/− mice and their littermate control mice as treated in (f). (j, k) 3D reconstructed images and sagittal view of the medial joint compartment (j), as well as their relative quantitative analysis (k) of Gpx4+/− mice and their littermate control mice knee joints treated as in (f) (n=6). Scale bars, 50 μm (b, f, h), 200 μm (d), 1 mm (j). Two-tailed unpaired t-test (left panel of e), one-way (b, c) or two-way (right panel of e, g, i, k) ANOVA with Tukey's post-hoc test. Data are shown as mean ± SD.
To determine the casual relationship between Trpv1 and Gpx4, we knocked down Gpx4 by siRNA and demonstrated substantially decreased anti-ferroptotic effect of Trpv1 activation (Figure 6e). We further examined the role of Gpx4 in the downstream of Trpv1 by using Gpx4 knockdown mice (Gpx4+/−), which exhibited robust decrease of Gpx4 protein in articular cartilage (Supplementary Figure 13d). Trpv1 activation by intra-articular injection of CPS in DMM-induced Gpx4+/− mice did not significantly alter the cartilage damage (Figure 6f, g, Supplementary Figure 13e), cartilage metabolism (Figure 6h, i), synovitis (Supplementary Figure 13e), osteophyte formation and subchondral bone sclerosis (Figure 6j, k, Supplementary Figure 13f). Whereas, Trpv1 exhibited cartilage protective effects in the littermate control mice of Gpx4+/− mice. These results indicate that Gpx4 knockdown resulted in the loss of anti-ferroptotic effect of Trpv1 in OA cartilage. Thus, GPX4 mediates the anti-ferroptotic role of TRPV1 in OA.
Discussion
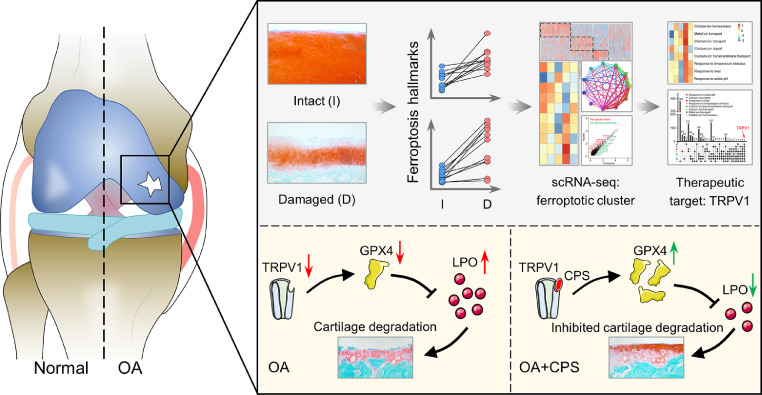
Chondrocyte ferroptosis has recently been proposed as risk factor for OA progression.5,9 However, the molecular characteristics and potential therapeutic targets of ferroptotic chondrocytes remain enigmatic. This study directly identified the ferroptotic chondrocyte cluster in human OA cartilage and deciphered its molecular characteristics and potential markers. We screened TRPV1 as a potential therapeutic target of chondrocyte ferroptosis and verified its anti-ferroptotic role in primary chondrocytes, human OA cartilage explants and DMM-induced OA mice model. Mechanistically, GPX4 is an essential mediator for the anti-ferroptotic effect of TRPV1. Activation of TRPV1 is thus a potential therapeutic target for protecting chondrocytes from ferroptosis in OA (Figure 7).
Figure 7.
A graphical scheme of revealing the ferroptotic chondrocyte cluster and the anti-ferroptotic role of TRPV1-GPX4 pathway in osteoarthritis (OA). TRPV1, transient receptor potential vanilloid 1; GPX4, Glutathione peroxidase 4; CPS, capsaicin.
Various types of chondrocyte death, including ferroptosis, apoptosis and necroptosis, have been investigated to participate in OA pathogenesis.48, 49, 50 Ferroptosis is a unique cell death mechanism, which is different from other types of cell death with distinctive morphological and mechanistical features.8 Morphologically, unlike the chromatin condensation in apoptosis, ferroptosis is characterized by shrunken mitochondria and reduced mitochondrial cristae.25 Mechanistically, distinct from executioner proteins-driven apoptosis (caspase 3) and necroptosis (mixed lineage kinase domain-like protein), ferroptosis occurs when cellular activities that promote ferroptosis overwhelming antioxidant capabilities of ferroptosis defense systems.12,51, 52, 53 However, there is still lack of distinct biomarkers or standard criteria to identify ferroptosis, especially in the disease.23 Therefore, we measured the indicators and genes of ferroptosis as much as possible in our scRNA-seq analysis to identify the ferroptotic chondrocyte cluster. We found that the C1-3-4 highly enriched oxidative stress, lipid oxidation, Fe2+ response related GO terms and preferentially expressed ferroptotic genes, which are consistent with the definition and molecular characteristics of ferroptosis.23 Further gene profile analysis revealed upregulated ferroptotic drivers and downregulated ferroptotic suppressors in damaged areas of OA cartilage, suggesting a relationship between chondrocyte ferroptosis and OA development. Recently, evidences showed that early and late ferroptotic cells could regulate other cells by their active secretion and passive release, respectively.54 Our cell–cell interaction analysis revealed enhanced signaling interactions between C1-3-4 and other clusters, particularly the cartilage regeneration clusters (C2, C1-2 and C1-4), in the damaged cartilage compared to the intact cartilage. We speculate this possibly due to the involvement of the secretory phenotype of ferroptotic chondrocytes, which needs further investigations. Moreover, it has been hypothesized that the crosstalk between ferroptosis and other types of cell death exists,55 which may drive a local auto-amplification loop and exaggerate cell death. Given the coexistence of various chondrocyte death types in OA pathogenesis, it is conceivable that ferroptosis may be entangled with other death pathways. Therefore, further investigations are warranted to unveil the exquisite interactions among different types of chondrocyte death in OA.
Given no golden standard for the measurement of ferroptosis, Stockwell et al. suggested that ferroptosis should be suppressed by both a lipophilic antioxidant and an iron chelator, and apoptosis and necroptosis should be ruled out when verifying ferroptosis.25 In our in vitro experiments, TBHP (50 μM)-induced chondrocytes showed total ROS, lipid ROS and Fe2+ accumulation with subsequent cell death, which was robustly abrogated by ferroptotic inhibitors (Fer-1 and DFO). Traditionally, TBHP was used to induce apoptosis and necroptosis.56 However, in our study, neither the apoptosis inhibitor (ZVAD) nor necroptosis inhibitor (Necro) showed obvious effect on TBHP (50 μM)-induced chondrocyte death. Recently, Miao et al. reported that ZVAD could suppress low concentration (30 μM), but showed no effect on high concentration (50 μM), of TBHP-induced chondrocyte death; therefore, they exclude the involvement of apoptosis in 50 μM TBHP-induced chondrocyte death.13 Furthermore, they also verified that the inhibitor of necroptosis showed no effects on TBHP (50 μM)-induced cellular ROS and lipid peroxidation accumulation, as well as the expression level of ferroptosis-related genes.13 Taken together, according to the suggestion of measuring ferroptosis,25 these results suggest that TBHP, at least in the concentration of 50 μM, could induce chondrocyte ferroptosis.
TRPV1 is primarily recognized as a nociceptive sensor of noxious stimuli, including heat, CPS, acidic pH, and various animal toxins.45,57 It has been reported that CPS exhibited pain management effects on OA by disrupting the entire terminal of TRPV1-expressing nociceptive fibers.58 Topical application of CPS has been included in the 2019 American College of Rheumatology/Arthritis Foundation Guideline for the pain management of knee OA.59 Moreover, we previously showed that TRPV1 was highly expressed by infiltrated M1 macrophages in OA synovium and its activation played an inhibitory role of M1 macrophage polarization by Ca2+/CaMKII/Nrf2 signaling pathway.18 Whereas, the role and mechanism of TRPV1 in chondrocytes are remain unclear. Herein, we surprisingly found that TRPV1-related GO terms were significantly enriched in the ferroptotic chondrocyte cluster. Pharmacological activation of TRPV1 markedly abrogated the ferroptotic hallmarks in TBHP-induced chondrocytes, DMM surgically induced OA mouse model and, especially, the human OA cartilage explants by promoting the expression of GPX4. Altogether, the anti-pain, anti-inflammation and anti-ferroptosis effect of TRPV1 suggest its potential as a disease-modifying target for the management of various pathogenesis of OA.
There are several limitations in this study: (1) The proportion of various types of chondrocyte death in human OA cartilage remains unclear. Future studies with large-scale human OA cartilage samples revealing the most critical type of chondrocyte death are expected. (2) We did not evaluate the effect of intra-articular injection of CPS on OA pain, let alone separately in male and female mice. (3) Although we screened the top 10 overexpressed genes in ferroptotic chondrocyte cluster, which may serve as ferroptotic biomarkers, the function and underlying mechanisms of them needs to be investigated.
In conclusion, despite the limitations, our investigation directly reveals that chondrocyte ferroptosis contributes to the progression of OA, and pharmacological activation of TRPV1 protects chondrocytes from ferroptosis, at least in part, by upregulating the expression of GPX4.
Contributors
D.S. and X.C. conceptualized and supervised the study. Z.Lv and J.L. conducted the most assays and acquired and analyzed data with the help of A.L., R.W., X.X. and W.Y.. Z.Lv and J.L. performed single cell RNA sequencing with the help of H.G., Y.F., Z.S. and M.W.. J.H. and C.F. performed bioinformatics analysis. Z.Lv, J.D., W.L., Y.X. and W.S. conducted the animal studies. J.C., Y.L., F.C. and Z.Liu collected and processed the samples. Z.Lv, J.H., X.C. and D.S. drafted the manuscript. Q.J. and S.I. revised the article for important intellectual content. Z.Lv, J.H., J.L., X.C. and D.S. have read and verified the underlying data. All authors have read and approved the final version of this manuscript.
Data sharing statement
All reagents and experimental protocols described in this work are available upon reasonable request. Single cell RNA-seq data reported in the present paper have been deposited in the National Genomics Data Center (accession number: HRA002569).
Declaration of interests
The authors have declared that no conflict of interest exists.
Acknowledgments
This work was supported by National Key R&D Program of China (2018YFC1105904), Key Program of NSFC (81730067), National Science Foundation of China (81772335, 81941009, 81802196), Natural Science Foundation of Jiangsu Province, China (BK20180127), Jiangsu Provincial Key Medical Talent Foundation, Six Talent Peaks Project of Jiangsu Province (WSW-079).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104258.
Contributor Information
Xiao Chen, Email: chenxiao-610@zju.edu.cn.
Dongquan Shi, Email: shidongquan@nju.edu.cn.
Appendix. Supplementary materials
References
- 1.Hunter DJ, March L, Chew M. Osteoarthritis in 2020 and beyond: a Lancet commission. The Lancet. 2020;396(10264):1711–1712. doi: 10.1016/S0140-6736(20)32230-3. [DOI] [PubMed] [Google Scholar]
- 2.Hunter DJ, Osteoarthritis B.-Z.S. Lancet. 2019;393(10182):1745–1759. doi: 10.1016/S0140-6736(19)30417-9. [DOI] [PubMed] [Google Scholar]
- 3.Hunter DJ, Schofield D, Callander E. The individual and socioeconomic impact of osteoarthritis. Nat Rev Rheumatol. 2014;10(7):437–441. doi: 10.1038/nrrheum.2014.44. [DOI] [PubMed] [Google Scholar]
- 4.Charlier E, Relic B, Deroyer C, et al. Insights on molecular mechanisms of chondrocytes death in osteoarthritis. Int J Mol Sci. 2016;17(12) doi: 10.3390/ijms17122146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J, Hu S, Bian Y, et al. Targeting cell death: pyroptosis, ferroptosis, apoptosis and necroptosis in osteoarthritis. Front Cell Dev Biol. 2022;9 doi: 10.3389/fcell.2021.789948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22(4):266–282. doi: 10.1038/s41580-020-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lei G, Zhuang L, Gan B. Targeting ferroptosis as a vulnerability in cancer. Nat Rev Cancer. 2022;22(7):381–396. doi: 10.1038/s41568-022-00459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao X, Sun K, Yu S, et al. Chondrocyte ferroptosis contribute to the progression of osteoarthritis. J Orthop Transl. 2021;27:33–43. doi: 10.1016/j.jot.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jing X, Lin J, Du T, et al. Iron overload is associated with accelerated progression of osteoarthritis: the role of DMT1 mediated iron homeostasis. Front Cell Dev Biol. 2021;8 doi: 10.3389/fcell.2020.594509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kennish L, Attur M, Oh C, et al. Age-dependent ferritin elevations and HFE C282Y mutation as risk factors for symptomatic knee osteoarthritis in males: a longitudinal cohort study. BMC Musculoskelet Disord. 2014;15:8. doi: 10.1186/1471-2474-15-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingold I, Berndt C, Schmitt S, et al. Selenium utilization by GPX4 is required to prevent hydroperoxide-induced ferroptosis. Cell. 2018;172(3):409–422. doi: 10.1016/j.cell.2017.11.048. [DOI] [PubMed] [Google Scholar]
- 13.Miao Y, Chen Y, Xue F, et al. Contribution of ferroptosis and GPX4′s dual functions to osteoarthritis progression. EBioMedicine. 2022;76 doi: 10.1016/j.ebiom.2022.103847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng Y, Lu J, Li W, et al. Reciprocal inhibition of YAP/TAZ and NF-κB regulates osteoarthritic cartilage degradation. Nat Commun. 2018;9(1):4564. doi: 10.1038/s41467-018-07022-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu X, Chen F, Lu K, Wei A, Jiang Q, Cao W. PPARγ preservation via promoter demethylation alleviates osteoarthritis in mice. Ann Rheum Dis. 2019;78(10):1420–1429. doi: 10.1136/annrheumdis-2018-214940. [DOI] [PubMed] [Google Scholar]
- 16.Charan J, Kantharia ND. How to calculate sample size in animal studies? J Pharmacol Pharmacother. 2013;4(4):303–306. doi: 10.4103/0976-500X.119726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi W-S, Lee G, Song W-H, et al. The CH25H–CYP7B1–RORα axis of cholesterol metabolism regulates osteoarthritis. Nature. 2019;566(7743):254–258. doi: 10.1038/s41586-019-0920-1. [DOI] [PubMed] [Google Scholar]
- 18.Lv Z, Xu X, Sun Z, et al. TRPV1 alleviates osteoarthritis by inhibiting M1 macrophage polarization via Ca2+/CaMKII/Nrf2 signaling pathway. Cell Death Dis. 2021;12(6):504. doi: 10.1038/s41419-021-03792-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlson EL, Karuppagounder V, Pinamont WJ, et al. Paroxetine-mediated GRK2 inhibition is a disease-modifying treatment for osteoarthritis. Sci Transl Med. 2021;13(580):eaau8491. doi: 10.1126/scitranslmed.aau8491. [DOI] [PubMed] [Google Scholar]
- 20.Gosset M, Berenbaum F, Thirion S, Jacques C. Primary culture and phenotyping of murine chondrocytes. Nat Protocols. 2008;3(8):1253–1260. doi: 10.1038/nprot.2008.95. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Sun Z, Lv Z, et al. Microtubule stabilization enhances the chondrogenesis of synovial mesenchymal stem cells. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.748804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun J, Zhou C, Zhao Y, et al. Quiescin sulfhydryl oxidase 1 promotes sorafenib-induced ferroptosis in hepatocellular carcinoma by driving EGFR endosomal trafficking and inhibiting NRF2 activation. Redox Biol. 2021;41 doi: 10.1016/j.redox.2021.101942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang D, Chen X, Kang R, Kroemer G. Ferroptosis: molecular mechanisms and health implications. Cell Res. 2021;31(2):107–125. doi: 10.1038/s41422-020-00441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong H, Yin H. Role of lipid peroxidation derived 4-hydroxynonenal (4-HNE) in cancer: focusing on mitochondria. Redox Biol. 2015;4:193–199. doi: 10.1016/j.redox.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stockwell BR, Friedmann Angeli JP, Bayir H, et al. Ferroptosis: a regulated cell death nexus linking metabolism. Redox Biol Dis Cell. 2017;171(2):273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng H, Schorpp K, Jin J, et al. Transferrin receptor is a specific ferroptosis marker. Cell Rep. 2020;30(10):3411–3423. doi: 10.1016/j.celrep.2020.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Q, Zhai L, Han M, et al. SHP2 inhibition attenuates osteoarthritis by maintaining homeostasis of cartilage metabolism via the DOK1/UPP1/uridine cascade. Arthr Rheumatol. 2022;74(3):462–474. doi: 10.1002/art.41988. [DOI] [PubMed] [Google Scholar]
- 28.Kreuser U, Buchert J, Haase A, Richter W, Diederichs S. Initial WNT/β-catenin activation enhanced mesoderm commitment, extracellular matrix expression, cell aggregation and cartilage tissue yield from induced pluripotent stem cells. Front Cell Dev Biol. 2020;8 doi: 10.3389/fcell.2020.581331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galeano-Garces C, Camilleri ET, Riester SM, et al. Molecular validation of chondrogenic differentiation and hypoxia responsiveness of platelet-lysate expanded adipose tissue-derived human mesenchymal stromal cells. Cartilage. 2017;8(3):283–299. doi: 10.1177/1947603516659344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuzaki T, Alvarez-Garcia O, Mokuda S, et al. FoxO transcription factors modulate autophagy and proteoglycan 4 in cartilage homeostasis and osteoarthritis. Sci Transl Med. 2018;10(428):eaan0746. doi: 10.1126/scitranslmed.aan0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ock S-A, Knott JG, Choi I. Involvement of CDKN1A (p21) in cellular senescence in response to heat and irradiation stress during preimplantation development. Cell Stress Chaperones. 2020;25(3):503–508. doi: 10.1007/s12192-020-01090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klaips CL, Gropp MHM, Hipp MS, Hartl FU. Sis1 potentiates the stress response to protein aggregation and elevated temperature. Nat Commun. 2020;11(1):6271. doi: 10.1038/s41467-020-20000-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu C, Li X, Li C, et al. SLC3A2 is a novel endoplasmic reticulum stress-related signaling protein that regulates the unfolded protein response and apoptosis. PLoS One. 2018;13(12) doi: 10.1371/journal.pone.0208993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kennedy D, Mnich K, Oommen D, et al. HSPB1 facilitates ERK-mediated phosphorylation and degradation of BIM to attenuate endoplasmic reticulum stress-induced apoptosis. Cell Death Dis. 2017;8(8):e3026. doi: 10.1038/cddis.2017.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji Q, Zheng Y, Zhang G, et al. Single-cell RNA-seq analysis reveals the progression of human osteoarthritis. Ann Rheum Dis. 2019;78(1):100–110. doi: 10.1136/annrheumdis-2017-212863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei T, Kulkarni NH, Zeng QQ, et al. Analysis of early changes in the articular cartilage transcriptisome in the rat meniscal tear model of osteoarthritis: pathway comparisons with the rat anterior cruciate transection model and with human osteoarthritic cartilage. Osteoarthr Cartil. 2010;18(7):992–1000. doi: 10.1016/j.joca.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 37.Matsui Y, Hasegawa M, Iino T, Imanaka-Yoshida K, Yoshida T, Sudo A. Tenascin-C prevents articular cartilage degeneration in murine osteoarthritis models. Cartilage. 2018;9(1):80–88. doi: 10.1177/1947603516681134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonaventura P, Lamboux A, Albarède F, Miossec P. Regulatory effects of zinc on cadmium-induced cytotoxicity in chronic inflammation. PLoS One. 2017;12(7) doi: 10.1371/journal.pone.0180879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawahara C, Forster T, Chapman K, Carr A, Loughlin J. Genetic association analysis of the IGFBP7, ADAMTS3, and IL8 genes as the potential osteoarthritis susceptibility that maps to chromosome 4q. Ann Rheum Dis. 2005;64(3):474–476. doi: 10.1136/ard.2004.027342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muschter D, Beiderbeck A-S, Späth T, Kirschneck C, Schröder A, Grässel S. Sensory neuropeptides and their receptors participate in mechano-regulation of murine macrophages. Int J Mol Sci. 2019;20(3):503. doi: 10.3390/ijms20030503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bakkenist CJ, Kastan MB. Initiating cellular stress responses. Cell. 2004;118(1):9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 42.Peters A, Nawrot TS, Baccarelli AA. Hallmarks of environmental insults. Cell. 2021;184(6):1455–1468. doi: 10.1016/j.cell.2021.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J, Kuang F, Kroemer G, Klionsky DJ, Kang R, Tang D. Autophagy-dependent ferroptosis: machinery and regulation. Cell Chem Biol. 2020;27(4):420–435. doi: 10.1016/j.chembiol.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dhaka A, Uzzell V, Dubin AE, et al. TRPV1 is activated by both acidic and basic pH. J Neurosci. 2009;29(1):153–158. doi: 10.1523/JNEUROSCI.4901-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang F, Zheng J. Understand spiciness: mechanism of TRPV1 channel activation by capsaicin. Prot Cell. 2017;8(3):169–177. doi: 10.1007/s13238-016-0353-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghoochani A, Hsu E-C, Aslan M, et al. Ferroptosis inducers are a novel therapeutic approach for advanced prostate cancer. Cancer Res. 2021;81(6):1583–1594. doi: 10.1158/0008-5472.CAN-20-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore CL, Savenka AV, Basnakian AG. TUNEL assay: a powerful tool for kidney injury evaluation. Int J Mol Sci. 2021;22(1):412. doi: 10.3390/ijms22010412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeon J, Noh H-J, Lee H, et al. TRIM24-RIP3 axis perturbation accelerates osteoarthritis pathogenesis. Ann Rheum Dis. 2020;79(12):1635–1643. doi: 10.1136/annrheumdis-2020-217904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hosseinzadeh A, Kamrava SK, Joghataei MT, et al. Apoptosis signaling pathways in osteoarthritis and possible protective role of melatonin. J Pineal Res. 2016;61(4):411–425. doi: 10.1111/jpi.12362. [DOI] [PubMed] [Google Scholar]
- 50.Zhou X, Zheng Y, Sun W, et al. D-mannose alleviates osteoarthritis progression by inhibiting chondrocyte ferroptosis in a HIF-2α-dependent manner. Cell Prolif. 2021;54(11):e13134. doi: 10.1111/cpr.13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doll S, Freitas FP, Shah R, et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575(7784):693–698. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 52.Soula M, Weber RA, Zilka O, et al. Metabolic determinants of cancer cell sensitivity to canonical ferroptosis inducers. Nat Chem Biol. 2020;16(12):1351–1360. doi: 10.1038/s41589-020-0613-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mishima E, Ito J, Wu Z, et al. A non-canonical vitamin K cycle is a potent ferroptosis suppressor. Nature. 2022;608(7924):778–783. doi: 10.1038/s41586-022-05022-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu J, Zhu S, Zeng L, et al. DCN released from ferroptotic cells ignites AGER-dependent immune responses. Autophagy. 2022;18(9):2036–2049. doi: 10.1080/15548627.2021.2008692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee Y-S, Lee D-H, Choudry HA, Bartlett DL, Lee YJ. Ferroptosis-induced endoplasmic reticulum stress: cross-talk between ferroptosis and apoptosis. Mol Cancer Res. 2018;16(7):1073–1076. doi: 10.1158/1541-7786.MCR-18-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao W, Feng H, Sun W, Liu K, Lu J-J, Chen X. Tert-butyl hydroperoxide (t-BHP) induced apoptosis and necroptosis in endothelial cells: roles of NOX4 and mitochondrion. Redox Biol. 2017;11:524–534. doi: 10.1016/j.redox.2016.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Julius D. TRP channels and pain. Annu Rev Cell Dev Biol. 2013;29:355–384. doi: 10.1146/annurev-cellbio-101011-155833. [DOI] [PubMed] [Google Scholar]
- 58.Campbell JN, Stevens R, Hanson P, et al. Injectable capsaicin for the management of pain due to osteoarthritis. Molecules. 2021;26(4):778. doi: 10.3390/molecules26040778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kolasinski SL, Neogi T, Hochberg MC, et al. 2019 American college of rheumatology/arthritis foundation guideline for the management of osteoarthritis of the hand, hip, and knee. Arthr Rheumatol. 2020;72(2):220–233. doi: 10.1002/art.41142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.