Abstract
The MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa was purified as a C-terminal histidine-tagged protein by metal chelate affinity chromatography. The purified protein was shown to bind ca. 200 bp upstream of mexA, at two sites, each of which contains a repeat of the nucleotide sequence GTTGA in inverse orientation. DNA sequence analysis identified mexA and mexR promoters within the MexR binding regions, consistent with the previously observed negative regulation of mexR and mexAB-oprM expression by MexR. Transcription of mexA from the promoter originating within the MexR binding site II was confirmed and shown to be markedly enhanced in a nalB (i.e., mexR) mutant of P. aeruginosa. A second mexA promoter was also identified, ca. 70 bp upstream of mexAB-oprM, and transcription from this promoter appeared to occur in both the wild type and a nalB mutant. Production of MexAB-OprM in wild-type cells may be due to expression from a constitutively expressed proximal promoter, while MexAB-OprM hyperexpression in nalB mutants is due to the additional expression from a MexR-regulated distal promoter.
In Pseudomonas aeruginosa, the combination of multidrug efflux systems and low outer membrane permeability produces an innate resistance to a wide array of antimicrobial agents (10, 17, 25, 27). Several multidrug efflux systems have been identified in P. aeruginosa, including MexAB-OprM (9, 31, 32), MexCD-OprJ (30), MexEF-OprN (16), and MexXY-OprM (1, 24). The Mex efflux systems are members of a family of three component multidrug resistance efflux system comprised of an inner membrane drug-proton antiporter of the resistance-nodulation-division (RND) family (e.g., MexB), an outer membrane efflux protein (e.g., OprM), and a membrane fusion protein which couples the activities of the two membrane proteins (e.g., MexA) (26).
Expression of MexCD-OprJ and MexEF-OprN appears to be lacking in wild-type cells, at least under normal laboratory growth conditions, occurring instead in nfxB (11, 22, 30) and nfxC (8, 16, 22) multidrug-resistant mutants, respectively. In contrast, both MexAB-OprM and MexXY-OprM are expressed to some extent in wild-type cells, where they contribute to intrinsic resistance to aminoglycosides (MexXY-OprM) (1) and β-lactams, quinolones, chloramphenicol, tetracycline, trimethoprim, sulfamethoxazole, and novobiocin (MexAB-OprM) (15, 18, 33, 37, 38). Hyperexpression of MexAB-OprM and an attendant increase in resistance to substrate antibiotics have also been described for nalB (22, 38) and nalC (39) multidrug-resistant strains. Although the nalB mutation is now known to occur in a gene, mexR, encoding a repressor of mexAB-oprM expression (33, 35, 39), the nature of the nalC mutation is not known. The mexR gene is located 274 bp upstream of mexA and transcribed divergently from the efflux genes (33). The association of mutations in mexR with increased drug resistance of clinical strains (13, 41) highlights the importance of studying the regulation of the mexAB-oprM operon by MexR.
MexR belongs to the MarR family of regulatory proteins (23), which includes PecS (34), CinR (5), and Hpr (29). MarR, repressor of the Escherichia coli multiple antibiotic resistance (marRAB) operon, binds the mar operator as a dimer at two locations, recognizing a 5-bp sequence which occurs as an inverted repeat (20). CinR, repressor of the Butyrivibrio fibrisolvens E14 cinnamoyl ester hydrolase-encoding gene (cinB), binds the cinR-cinB intergenic region, which is known to contain two 16-bp inverted repeats, although binding to this sequence has yet to be demonstrated (5). Hpr, a repressor of subtilisin (aprE), neural protease (nprE), and the oligopeptide permease (opp and app) operons of Bacillus subtilis, binds at a 4-bp inverted repeat (14). PecS regulates several genes associated with pectin degradation in Erwinia chrysanthemi, although a consensus binding sequence has not been identified (34). Members of the MarR family are proposed to bind DNA through a conserved helix-turn-helix motif or motifs (2, 5). In this study, purified histidine-tagged MexR was used to confirm that MexR binds the mexA-mexR intergenic region at two sites upstream of mexR, near promoters for both mexR and mexA.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli strain BL21/DE3(pLysE) (40) was host to the mexR expression vector pKLE1, a pET23a derivative (see below). P. aeruginosa wild-type strain K870 (33) and nalB strain OCR1 (21) were also used. Bacteria were grown in Miller's Luria broth base (Difco) containing 2 g of NaCl per liter of H2O (L broth) supplemented with ampicillin (100 μg/ml) and/or chloramphenicol (30 μg/ml) as indicated. Cultures were typically grown at 37°C with vigorous shaking (200 rpm).
Plasmids.
To produce polyhistidine-tagged MexR (MexR-His), the mexR gene was cloned into plasmid pET23a (Novagen, Madison, Ws.). To achieve this, the mexR gene was amplified from P. aeruginosa strain K870 chromosomal DNA using primers K7 (5′-GGATTCATATGAACTACCCCGTGAATCCC-3′), which anneals at the 5′ end of mexR and incorporates an NdeI restriction site, and K6 (5′-ATCGCTCGAGAATATCCTCAAGCGGTTGC-3′), which anneals at the 3′ end of mexR and incorporates an XhoI restriction site. The reaction mixture (100 μl) included 2 U of Vent DNA polymerase (New England Biolabs, Mississauga, Ontario, Canada), 60 pmol of each primer, 0.2 mM each deoxynucleotide, 2.5 mM MgSO4, 10% (vol/vol) dimethyl sulfoxide, 1 μg of chromosomal DNA, and 1× ThermoPol buffer. The mixture was treated for 5 min at 94°C, followed by 30 cycles of 2 min at 56°C, 1.5 min at 72°C, and 1 min at 94°C and finally 2 min at 56°C and 5 min at 72°C. Products were examined on a 0.8% (wt/vol) agarose gel and purified with a QIAquick-spin PCR purification kit (Qiagen, Inc., Chatsworth, Calif.). The PCR product was digested with NdeI and XhoI and cloned into NdeI-XhoI-restricted pET23a, to produce pKLE1.
To isolate target DNA for the gel shift and DNase I footprinting assays, the mexA-mexR intergenic region was cloned into plasmid pCRII-TOPO (Invitrogen, Calif.). To achieve this, the mexA-mexR intergenic region was amplified using primers K9 (5′-CTGAAGATCTGTTGCATAGCGTTGTCCTCA), which anneals to the 5′ end of mexA, and K10 (5′-ACGGGGTACCCGGGGTAGTTCATTGGTTTG-3′), which anneals to the 5′ end of mexR. The reaction mixture (100 μl) included 2.5 U of Taq DNA polymerase (GibcoBRL, Burlington, Ontario, Canada), 60 pmol of each primer, 0.2 mM each deoxynucleotide, 1.5 mM MgCl2, 1 μg of P. aeruginosa strain K870 chromosomal DNA, and 1× Taq PCR buffer. PCR conditions were as described above, except that the last elongation step at 72°C was for 30 min instead of 5 min. After examination on a 0.8% (wt/vol) agarose gel, the PCR product was cloned into pCRII-TOPO according to the manufacturer's instructions for the TOPO TA Cloning kit (Invitrogen). DNA sequencing confirmed the absence of PCR-induced mutations in the resulting vector, pKLE2.
DNA techniques.
Plasmid transformations were carried out as previously described (36), using competent cells prepared by the method of Inoue et al. (12). Chromosomal DNA was prepared as previously described (3). Midi-preparations of plasmid DNA were prepared using Qiagen columns according to the method supplied by the manufacturer, whereas mini-preparations of plasmid DNA were prepared by the alkaline lysis procedure (36). Restriction endonucleases and T4 DNA ligase were obtained from New England Biolabs and used according to the manufacturer's instructions or as described previously (36). Restriction fragments were isolated, as required, from agarose gels (0.8% [wt/vol]) using the Prep-a-Gene glass matrix (Bio-Rad, Mississauga, Ontario, Canada) as recommended by the manufacturer. DNA sequencing was carried out by the Laboratory Services Division, University of Guelph, Guelph, Ontario, Canada.
Purification of MexR.
An overnight culture of E. coli BL21/DE3(pLysE) carrying plasmid pKLE1 was diluted 1:100 into 250 ml of L broth supplemented with ampillicin (100 μg/ml) and chloramphenicol (30 μg/ml) and incubated at 37°C until the culture reached an optical density at 600 of 0.3 to 0.5. Expression of MexR-His was then achieved by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 1 mM and incubation for a further 3 h. Bacteria were harvested by centrifugation at 6,000 × g at 4°C for 10 min, and the pellet was resuspended in 1 ml of lysis buffer (20 mM Tris-HCl [pH 8.0], 100mM NaCl) and stored at −20°C overnight. The cells were subsequently lysed by sonication (two sonic bursts of 45 s, power 40 with a VibraCell sonicator [Sonics & Material Inc., Danbury, Conn.T]). Unlysed cells and insoluble material were removed by centrifugation at 16,000 × g at 4°C for 10 min, and the supernatant was recovered. The volume of the lysate was brought up to 10 ml with lysis buffer, and 1 ml of Talon resin (Clontech, Palo Alto, Calif.), prepared according to the manufacturer's instructions, was added. The lysate and resin were gently agitated for 20 min at room temperature, at which time the resin was pelleted by centrifugation at 3,000 × g at 4°C for 5 min. Following removal of the supernatant, the resin was washed three times by adding 10 ml of lysis buffer, agitating gently for 10 min, and repeating the centrifugation. A more stringent wash was then carried out using lysis buffer containing 10 mM imidazole. The MexR-His protein was eluted from the Talon resin by adding 0.5 ml of elution buffer (lysis buffer containing 200 mM imidazole) and gently agitating for 10 min. Following centrifugation as for the wash steps, the MexR-His-containing supernatant was recovered. Protein concentration was determined by the Lowry assay (19), and purified protein was stored at 4°C.
Gel shift assay.
The mexA-mexR intergenic region cloned into pCRII-TOPO in producing pKLE2 is bracketed by NotI and BglII cleavage sites present on pCRII-TOPO. Owing to the existence of a NcoI cleavage site in the middle of the mexA-mexR intergenic region, then, target DNA for gel shift experiments was recovered from pKLE2 by digestion with NotI and NcoI (for the mexR upstream region) or with BglII and NcoI (for the mexA upstream region), yielding fragments of 168 and 183 bp, respectively. The digested fragments were end labeled with [α-32P]dGTP (3,000 Ci mmol−1) using Klenow fragment (36) and purified from an 8% (wt/vol) polyacrylamide gel by the crush-and-soak method (36). The labeled DNA probes (3,000 cpm) were incubated with purified MexR-His at concentrations ranging from 3.7 nM to 15 μM for 15 min at room temperature in 20 μl of binding buffer [20 mM HEPES (pH 7.6), 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM dithiothreitol (DTT), 0.2% (wt/vol) Tween 20, 30 mM KC1] containing 1 μg of poly(dI-dC) and 0.1 μg of poly-l-lysine. The reaction mixtures were then submitted to electrophoresis on a nondenaturing 8% (wt/vol) polyacrylamide gel in 0.25 × TBE (22 mM Tris, 22 mM boric acid, 0.5 mM EDTA [pH 8.0]), and the labeled DNA was visualized by autoradiography. For competitor experiments, 0.5 μg of unlabeled DNA was added to the reaction mixture prior to the addition of MexR-His.
DNase I footprinting assay.
DNase I footprinting was carried out using a Sure Track footprinting kit (Amersham Pharmacia Biotech, Baie d'Urfé, Quebec, Canada). Target DNA (mexR-mexA intergenic region) was again recovered from pKLE2 by digestion with EcoRV and BglII (for labeling of the mexA coding strand) or with NotI and SacI (for labeling the mexR coding strand), yielding a fragment of 328 or 407 bp, respectively. The digested fragments were end labeled with [α-32P]dGTP (3,000 Ci mmol−1) using Klenow fragment (36) and purified from an 8% (wt/vol) polyacrylamide gel by the crush-and-soak method (36). Approximately 100,000 cpm of target DNA was incubated for 15 min at room temperature with purified MexR-His (5 to 30 mM) in 50 μl of gel shift assay buffer containing 1 μg of poly(dI-dC) and 0.1 μg of poly-l-lysine. The reaction mixtures were adjusted to 1 mM MgCl2 and 0.5 mM CaCl2 before the addition of 3 U of DNase I. Digestion was performed at room temperature for 1 min and stopped by the addition of 140 μl of stop solution (192 mM sodium acetate, 32 mM EDTA, 0.14% [wt/vol] sodium dodecyl sulfate [SDS], 64 μg of yeast RNA ml−1). After extraction with 200 μl of phenol-chloroform (1:1, vol/vol), DNA fragments were ethanol precipitated and separated by electrophoresis on an 8% (wt/vol) polyacrylamide–7 M urea sequencing gel (36). The DNase I digestion profile was subsequently revealed by autoradiography. A Maxam-Gilbert G+A sequencing reaction was performed on 100,000 cpm of the appropriate target DNA according to instructions provided with the SureTrack footprinting kit.
Mapping the mexA transcription start site.
The start of transcription of the mexA gene was determined by the 5′ rapid amplification of cDNA ends (RACE) protocol (7) using a 5′/3′ RACE kit essentially as recommended by the manufacturer (Roche Diagnostics, Laval, Quebec, Canada). Total RNA was prepared from P. aeruginosa strains K767 (wild type) and K766 (nalB) using a Qiagen RNeasy Mini kit. DNA contamination was removed by digestion with 10 U of RQ1 RNase-free DNase (Promega) for 2 h at 37°C. Total RNA (2 μg) in a 20-μl reaction volume was reverse transcribed at 55°C with avian myeloblastosis virus reverse transcriptase and mexA-specific primer JT-9 (5′-GGCGGGGTCGATCTGGTAGAGCTGCTG-3′), which anneals 264 bp downstream of the mexA translational start site. A homopolymeric tail was appended to the 3′ end of the synthesized first-strand cDNA (corresponds to the 5′ end of any mexA mRNA that was reverse transcribed in the above reaction) using terminal transferase and dATP by incubation at 37°C for 20 min as described in the RACE kit protocol. The dA-tailed cDNA was PCR amplified using an oligo (dT)-anchor primer (5′-GACCACGCGTATC-GATGTCGACTTTTTTTTTTTTTTTTV-3′ and another mexA-specific primer (LA21 [5′ GAACAGGCGCTTGAGGAT-3′]) which anneals 218 bp downstream of the mexA translational start site. The PCR product obtained was again PCR amplified with the nested mexA-specific primer LA20 (5′AGGATGATGCCGTTCACCTG-3′) and a kit-provided PCR anchor primer (5′ GACCACGCGTATCGATGTCGAC-3′) in order to eliminate any nonspecific PCR products from the first reaction. The amplified products were purified using a High Pure PCR product purification kit (Roche Diagnostics) and cloned into the pCR-Blunt II TOPO vector (Invitrogen). Automated DNA sequencing of the RACE inserts was performed by Cortec DNA Service Laboratories, Inc. (Queen's University, Kingston, Ontario, Canada).
RT-PCR.
DNA-free total RNAs from P. aeruginosa strains K767 (wild type) and K766 (nalB) prepared as described above were reverse transcribed and subsequently PCR amplified using primers K14 (5′-CGTCGCTGCCTTCCTTGAACA-3′; anneals 229 bp downstream of the mexA start codon) and LA19 (5′-GACCTTATCAACCTTGTTTCAGG-3′; anneals 185 bp upstream of mexA coding region). The reaction was carried out using a OneStep reverse transcription-PCR (RT-PCR) kit (Qiagen) according to the manufacturer's instructions.
RESULTS
Purification of MexR.
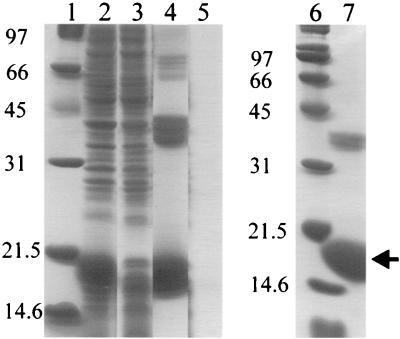
The mexR gene was cloned into the pET23a vector such that six histidine residues were added to the C terminus of MexR. Overexpression of MexR-His was achieved by IPTG induction of E. coli BL21/DE3 (pLysE) carrying plasmid pKLE1 (Fig. 1, lane 2). Metal affinity chromatography permitted ready purification of MexR-His at a concentration of 5 mg/ml in the eluted fraction. When this fraction was run on an SDS-polyacrylamide gel, we observed two major bands of approximately 19 kDa, in good agreement with the predicted size of 17.9 kDa (molecular mass of MexR plus six histidines, although higher-molecular-mass bands in multiples of 19 kDa were also present in pKLE1 containing E. coli BL21/DE3(pLysE) (lane 4). These bands were all absent in the control strain E. coli BL21/DE3(pLysE) carrying the pET23a vector control (lane 5), indicating that these high-molecular-mass proteins were derived from MexR-His. Treatment of the MexR-His-containing fraction with DTT reduced the two major bands to a single 19-kDa band and eliminated most of the higher-molecular-mass species (lane 7). The latter, therefore, were likely multimers of disulfide-bonded MexR-His, while the 19-kDa protein that was eliminated by DTT treatment was probably a intramolecular disulfide-bonded form of MexR (lane 7). These disulfide-bonded forms of MexR-His were undoubtedly artifacts of the obvious hyperexpression of the protein, such forms having been observed upon hyperexpression of other regulatory proteins (6). The in vivo significance, if any, of this disulfide bonding is unclear, although DTT-treated MexR-His was active in gel shift and DNase I footprinting experiments (see below).
FIG. 1.
Overexpression and purification of MexR-His. E. coli BL21/DE3(pLysE) carrying pKLE1 (lanes 2 and 4) or pET23a (lanes 3 and 5) was cultured as described in the text, and MexR-His was purified from pKLE1-containing E. coli BL21/DE3(pLysE) by metal chelate affinity chromatography (Talon). Lanes 1 and 6, molecular mass markers; lanes 2 and 3, clarified lysate; lanes 4 and 5, eluant from Talon column; lane 7, DTT (1.5% [wt/vol])-treated sample from lane 4. The arrow indicates the location of the MexR-His monomer.
MexR binds to the mexA-mexR intergenic region.
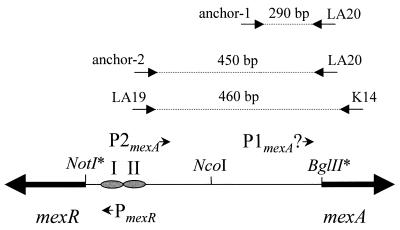
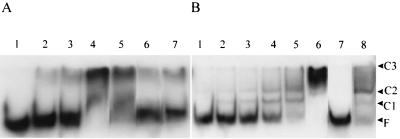
DNA from the mexA-mexR intergenic region was recovered from pKLE2 by digestion with NotI and NcoI (for the fragment upstream of the mexR translational start) or BglII and NcoI (for the fragment upstream of the mexA translation start) (see Fig. 4) and used to assess MexR binding in a gel retardation assay (Fig. 2). MexR-dependent gel shifts were observed for both fragments, although 1,000-fold more MexR-His was needed to shift the mexA upstream region compared to the mexR upstream region (lane 4 in Fig. 2A compared to lane 4 in Fig. 2B). Moreover, the interaction of MexR-His with the mexA upstream region was shown to be nonspecific, since MexR-His binding to this fragment was lost in the presence of the unrelated calf thymus DNA (Fig. 2A, lane 7). At least three MexR-DNA complexes were observed for the mexR upstream region fragment with increasing amounts of protein (Fig. 2B, lanes 2 to 6), which is consistent with the presence of multiple binding sites (see below). The smaller C1 and C2 complexes occurred at the lowest MexR-His concentration (3.7 nM), while the largest, C3, occurred only at a very high protein concentration (7.4 μM). Binding of MexR-His to the mexR upstream region was shown to be specific, since MexR-His binding to the labeled mexR upstream fragment was abrogated in the presence of mexR upstream DNA (Fig. 2B, lane 7) but not in the presence of unrelated calf thymus DNA (Fig. 2B, lane 8). Consistent with the preferred binding of MexR-His to the mexR upstream region, the unlabeled mexR upstream fragment obviated the MexR shift of the mexA upstream fragment (Fig. 2A, lane 6).
FIG. 4.
Schematic showing the positions of promoters and MexR binding sites within the mexA-mexR intergenic region. The relative positions and orientations of PmexR, P1mexA, P2mexA, and MexR binding sites I and II (from Fig. 3) are highlighted. Restriction sites marked with an asterisk do not occur within the mexA-mexR coding coding or intergenic sequence but instead flank the cloned intergenic region in plasmid pKLE2. They are indicated here to illustrate the specific portion of the mexA-mexR intergenic region that was encompassed by the restriction fragments used in the gel shift experiments in Fig. 2. The relative locations of RT-PCR and RACE primers (Fig. 5) as well as the predicted amplification products are also highlighted. Placement of the RACE anchor primers corresponds with the predicted transcription starts sites for mexA.
FIG. 2.
Interaction of MexR with the mexA-mexR intergenic region. (A) The 183-bp NcoI-BglII fragment of pKLE2 containing the mexA upstream region was incubated without MexR (lane 1) or with 3.7 μM (lane 2), 7.4 μM (lane 3), or 15 μM (lane 4) MexR-His. This fragment was incubated with 15 μM MexR and 0.5 μg of either unlabeled NcoI-BglII fragment (lane 5), NcoI-NotI fragment (lane 6), or calf thymus (lane 7) DNA as competitor DNA. (B) The 168-bp NcoI-NotI fragment containing the mexR upstream region was incubated without MexR (lane 1) or with 3.7 nM (lane 2), 7.4 nM (lane 3), 15 nM (lane 4), 30 nM (lane 5), or 7.4 μM (lane 6) MexR-His. This fragment was incubated with 30 nM MexR-His and 0.5 μg of either unlabeled NcoI-NotI fragment (lane 7) or calf thymus (lane 8) DNA as competitor DNA. MexR-DNA complexes (C1 to C3) and free DNA (F) are highlighted.
Identification of the MexR binding site.
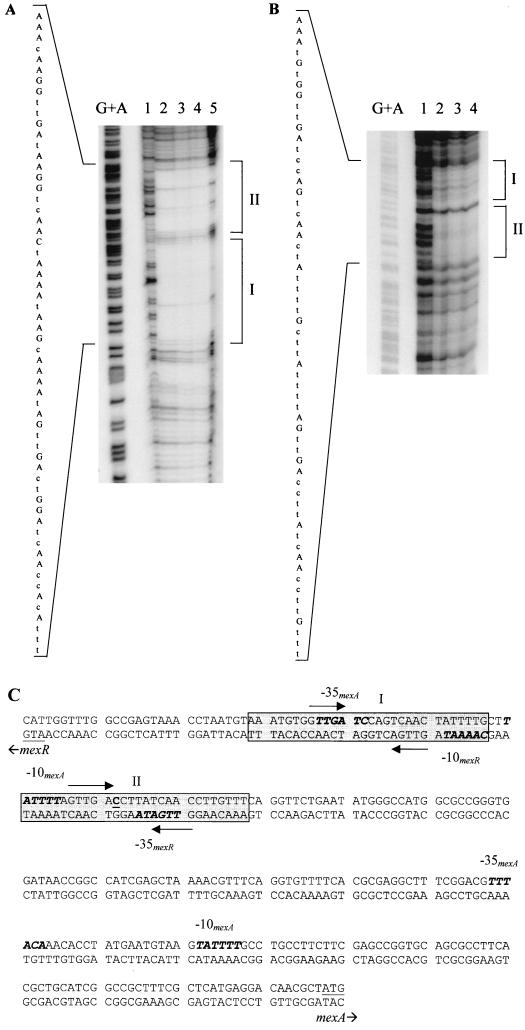
The mexA-mexR intergenic region was end labeled on either the mexA or mexR coding strand and then subjected to DNase I footprinting in an effort to ascertain the MexR binding site(s). DNase I footprinting revealed two protected areas, dubbed sites I and II, on both strands (Fig. 3A and B). The protected areas, some 28 bp in size, were separated by 3 bp and occurred 189 bp upstream of mexA (from the 3′ end of site I to the translational start of mexA) and 25 bp upstream of mexR (from the 3′ end of site II to the translational start of mexR). No binding of MexR-His was observed more proximal to mexA. The locations of the MexR binding sites were therefore consistent with the gel shift data. Within each binding site, the sequence 5′-GTTGA-3′ was repeated in inverse orientation after a space of 5 bp in the same position within each site. Maxam-Gilbert sequencing of the end-labeled DNA placed MexR binding site I overlapping the -35 region of a second putative promoter for mexA (the originally predicted mexA promoter, here dubbed P1mexA, was proximal to mexA [33]) and the -10 region of the putative mexR promoter (Fig. 3C). MexR binding site II overlaps the -35 region of the mexR promoter and the -10 region of the second putative mexA promoter (Fig. 3C).
FIG. 3.
DNase I footprinting of MexR-DNA interactions on the mexA-mexR intergenic region. DNase I footprinting was conducted with the 407-bp NotI-SacI fragment from pKLE2 (which labels the mexR coding strand) (A) or the 328-bp NcoI-BglII fragment from pKLE2 (which labels the mexA coding strand) (B) in the presence of no MexR (lane 1) or 30 μM (lane 2), 60 μM (lane 3), 120 μM (lane 4), or 180 μM (lane 5) MexR-His. The protected regions are bracketed at the right and labeled I and II. The nucleotide sequence of the protected region is indicated at the left, with bases identified in the G+A sequencing reaction (lane G+A) shown in uppercase. (C) Nucleotide sequence of the mexA-mexR intergenic region highlighting the MexR binding sites (shaded) and 5′-GTTGA-3′ inverted repeat sequences (arrows) within each binding site. Putative mexA and mexR -35/-10 promoter sequences are highlighted in bold italics, and the +1 site (as determined by the RACE method [Fig. 5]) for the more distal of the two mexA promoters is underlined and in boldface.
Identification of two transcriptional start sites for mexA.
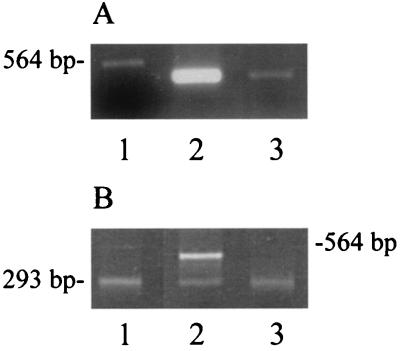
The lack of specific MexR binding proximal to mexA led to the recognition of a putative, more distal, second promoter (P2mexA [Fig. 3C and 4]), which is bound by MexR. Consensus −35/−10 hexamers were identified within the region protected by MexR, suggesting a mechanism by which MexR could negatively regulate mexA or mexAB-oprM expression. To confirm the activity of this distal mexAB-oprM promoter, RT-PCR was performed on total cellular RNA using primers that anneal at MexR binding site II (LA19) and within the mexA coding region (K14) (Fig. 4). Large amounts of the expected ca. 400-bp RT-PCR product (Fig. 4) were obtained from nalB strain OCR1 (Fig. 5A, lane 2), though barely detectable levels of this product were observed for wild-type strain PAO1 (K767) (Fig. 5A, lane 3). This was consistent both with the presence of mexA (i.e., mexAB-oprM) transcripts that extend at least as far as MexR binding site II and with the increased production of these transcripts in a nalB (i.e., mexR) mutant.
FIG. 5.
Identification and mapping of mexA transcripts. (A) RT-PCR of total RNA from P. aeruginosa strains OCR1 (nalB) (lane 2) and PAO1 (K767) (lane 3) using primers K14 and LA19 (Fig. 4). The 564-bp λ HindIII fragment is shown in lane 1. (B) 5′ RACE products from P. aeruginosa strains OCR1 (nalB) (lane 2) and PAO1 (K767) (lane 3) prepared with the mexA-specific LA20 primer and the PCR anchor primer provided with the RACE kit (Roche). A 293-bp RACE product produced using control template and primers provided with the RACE kit is shown in lane 1. The migration position of the 564-bp λ HindIII fragment is shown at the right.
To more precisely map the transcription start site for the putative P2mexA, the RACE procedure was used. The basis of this method is the cloning and nucleotide sequencing of cDNAs derived from the reverse transcription and subsequent amplification of, in this case, the 5′ end of each mexAB-oprM mRNA (see Materials and Methods). Using this procedure, an expected 450-bp mexA-derived cDNA (Fig. 4, anchor-2 and LA20 primers) was recovered from the nalB mutant OCR1 (Fig. 5B, lane 2, top band), and its sequencing confirmed that expression from P2mexA initiates at a cytosine residue appropriately placed downstream of the predicted -10 site for this promoter (Fig. 3C). No 430-bp cDNA was recovered from wild-type strain PAO1 (K767) (Fig. 5B, lane 3), consistent with the minimal expression expected from P2mexA in this MexR+ strain and in agreement with the RT-PCR results described above. Interestingly, a second mexA-derived cDNA was recovered from both the wild type and nalB mutant (Fig. 5B, lanes 2 and 3). The size of this DNA, ca. 300 bp, is consistent with the existence in these strains of a mexA transcript that initiates within the vicinity of the proximal P1mexA promoter (Fig. 4, anchor-1 and LA20 primers). Cloning and sequencing of these cDNAs failed, however, to consistently identify a single reside as the initiation site.
DISCUSSION
The binding site for MexR appears to involve an inverted repeat of the sequence GTTGA that by analogy with MarR likely constitutes the binding site for a MexR monomer (20) (i.e., the binding site itself accommodates a dimer). MarR (20) and other members of this family, including EmrR (4) and SlyA (28), are known to form oligomers; CinR, which also apparently recognizes an inverted repeat sequence, is also predicted to operate as a dimer (5). As for MarR (20), two closely linked binding sites were identified for MexR and dubbed sites I and II. MarR binding sites spanned both the −35/−10 hexamers of the marRAB promoter and the MarR ribosome binding site, but binding at the MarR ribosome binding site is not necessary either for MarR binding to the −35/−10 hexamers or for repression of the mar operon (20). Here, both MexR binding sites overlap components of the mexR and mexA promoters, and thus occupancy of either site would interfere with mexR and mexAB-oprM expression.
The lack of MexR binding more proximal to mexA, in the vicinity of a previously proposed promoter sequence (shown here to indeed function as a promoter), was initially surprising, given the known MexR repression of mexAB-oprM expression (33). The presence of a second mexA promoter, which occurs substantially upstream of mexA and in the vicinity of a MexR binding site, provided a ready explanation. The observation that a mexA transcript is produced from P2mexA and that the levels of this transcript increase markedly in a MexR-deficient (nalB) strain clearly demonstrate that P2mexA is functional and that MexR regulates expression of mexAB-oprM via this promoter. Data provided here also suggests that P1mexA, the proximal promoter first identified as a mexAB-oprM promoter, is a functional promoter, possibly responsible for the modest mexAB-oprM expression and corresponding multidrug resistance of wild-type P. aeruginosa. Still, it is also possible that the RACE product that implicates P1mexA as an active promoter is a stable breakdown product of the P2mexA-derived RACE product or that the mRNA originating from P2mexA is itself unstable, yielding 5′-truncated RACE products during the reverse transcription and amplification steps of the RACE reaction. In this case, mexAB-oprM expression would initiate from a single MexR-regulated promoter located substantially upstream of mexA (i.e., P2mexA). In any case, hyperexpression of MexAB-OprM in nalB multidrug-resistant strains is due to enhanced mexAB-oprM transcription arising from P2mexA. The functional significance of the lengthy untranslated leader that results from expression from this promoter is unclear, although it may play a role in mexAB-oprM expression mediated by mutations in nalC (39).
ACKNOWLEDGMENTS
This work was supported by an operating grant from the Canadian Cystic Fibrosis Foundation (CCFF). K.E. holds a CCFF studentship. L.A. is supported by the Canadian Bacterial Diseases Network (a consortium of the Centres of Excellence Program). K.P. is a CCFF Scholar.
REFERENCES
- 1.Aires J R, Köhler T, Nikaido H, Plesiat P. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother. 1999;43:2624–2628. doi: 10.1128/aac.43.11.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alekshun M N, Kim Y S, Levy S B. Mutational analysis of MarR, the negative regulator of marRAB expression in Escherichia coli, suggests the presence of two regions required for DNA binding. Mol Microbiol. 2000;35:1394–1404. doi: 10.1046/j.1365-2958.2000.01802.x. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. 2nd ed. New York, N.Y: John Wiley & Sons, Inc.; 1992. [Google Scholar]
- 4.Brooun A, Tomashek J J, Lewis K. Purification and ligand binding of EmrR, a regulator of a multidrug transporter. J Bacteriol. 1999;181:5131–5133. doi: 10.1128/jb.181.16.5131-5133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalrymple B P, Swadling Y. Expression of a Butyrivibrio fibrisolvens E14 gene (cinB) encoding an enzyme with cinnamoyl ester hydrolase activity is negatively regulated by the product of an adjacent gene (cinR) Microbiology. 1997;143:1203–1210. doi: 10.1099/00221287-143-4-1203. [DOI] [PubMed] [Google Scholar]
- 6.Dean C R, Neshat S, Poole K. PfeR, an enterobactin-responsive activator of ferric enterobactin receptor gene expression in Pseudomonas aeruginosa. J Bacteriol. 1996;178:5361–5369. doi: 10.1128/jb.178.18.5361-5369.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frohman M A. Rapid amplification of complementary DNA ends for generation of full-length complementary DNAs: thermal RACE. Methods Enzymol. 1993;218:340–356. doi: 10.1016/0076-6879(93)18026-9. [DOI] [PubMed] [Google Scholar]
- 8.Fukuda H, Hosaka M, Hirai K, Iyobe S. New norfloxacin resistance gene in Pseudomonas aeruginosa PAO. Antimicrob Agents Chemother. 1990;34:1757–1761. doi: 10.1128/aac.34.9.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gotoh N, Tsujimoto H, Poole K, Yamagishi J-I, Nishino T. The outer membrane protein OprM of Pseudomonas aeruginosa is encoded by oprK of the mexA-mexB-oprK multidrug resistance operon. Antimicrob Agents Chemother. 1995;39:2567–2569. doi: 10.1128/aac.39.11.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock R E W. The bacterial outer membrane as a drug barrier. Trends Microbiol. 1997;5:37–42. doi: 10.1016/S0966-842X(97)81773-8. [DOI] [PubMed] [Google Scholar]
- 11.Hirai K, Suzue S, Irikura T, Iyobe S, Mitsuhashi S. Mutations producing resistance to norfloxacin in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1987;31:582–586. doi: 10.1128/aac.31.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1991;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 13.Jalal S, Wretlind B. Mechanisms of quinolone resistance in clinical strains of Pseudomonas aeruginosa. Microbiol Drug Resist. 1998;4:257–261. doi: 10.1089/mdr.1998.4.257. [DOI] [PubMed] [Google Scholar]
- 14.Kallio P T, Fagelson J E, Hoch J A, Strauch M A. The transition state regulator Hpr of Bacillus subtilis is a DNA-binding protein. J Biol Chem. 1999;266:13417. [PubMed] [Google Scholar]
- 15.Koehler T, Kok M, Michea-Hamzehpour M, Plesiat P, Gotoh N, Nishino T, Kocjanici Curty L, Pechere J-C. Multidrug efflux in intrinsic resistance to trimethoprim and sulfamethoxazole in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1996;40:2288–2290. doi: 10.1128/aac.40.10.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koehler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J-C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 17.Li X-Z, Zhang L, Poole K. Interplay between the MexAB-OprM multidrug efflux system and the outer membrane barrier in the multiple antibiotic resistance of Pseudomonas aeruginosa. J Antimicrob Chemother. 2000;45:433–436. doi: 10.1093/jac/45.4.433. [DOI] [PubMed] [Google Scholar]
- 18.Li X-Z, Zhang L, Srikumar R, Poole K. β-Lactamase inhibitors are substrates of the multidrug efflux pumps of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:399–403. doi: 10.1128/aac.42.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowry O H, Rosenbrough A L, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 20.Martin R G, Rosner J L. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc Natl Acad Sci USA. 1995;92:5456–5460. doi: 10.1073/pnas.92.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuda N, Ohya S. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:1847–1851. doi: 10.1128/aac.36.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masuda N, Sakagawa E, Ohya S. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:645–649. doi: 10.1128/AAC.39.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller P F, Sulavik M C. Overlaps and parallels in the regulation of intrinsic multiple-antibiotic resistance in Escherichia coli. Mol Microbiol. 1996;21:441–448. doi: 10.1111/j.1365-2958.1996.tb02553.x. [DOI] [PubMed] [Google Scholar]
- 24.Mine T, Morita Y, Kataoka A, Mitzushima T, Tsuchiya T. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:415–417. doi: 10.1128/aac.43.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 26.Nikaido H. Multiple antibiotic resistance and efflux. Curr Opin Microbiol. 1998;1:516–523. doi: 10.1016/s1369-5274(98)80083-0. [DOI] [PubMed] [Google Scholar]
- 27.Nikaido H. The role of outer membrane and efflux pumps in the resistance of gram-negative bacteria. Can we improve drug access? Drug Res Updates. 1998;1:93–98. doi: 10.1016/s1368-7646(98)80023-x. [DOI] [PubMed] [Google Scholar]
- 28.Oscarsson J, Mizunoe Y, Uhlin B E, Haydon D J. Induction of haemolytic activity in Escherichia coli by the slyA gene product. Mol Microbiol. 1996;20:191–199. doi: 10.1111/j.1365-2958.1996.tb02500.x. [DOI] [PubMed] [Google Scholar]
- 29.Perego M, Hoch J A. Sequence analysis and regulation of the hpr locus, a regulatory gene for protease production and sporulation in Bacillus subtilis. J Bacteriol. 1988;170:2560–2567. doi: 10.1128/jb.170.6.2560-2567.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J-I, Li X-Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB multidrug resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 31.Poole K, Heinrichs D E, Neshat S. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol Microbiol. 1993;10:529–544. doi: 10.1111/j.1365-2958.1993.tb00925.x. [DOI] [PubMed] [Google Scholar]
- 32.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poole K, Tetro K, Zhao Q, Neshat S, Heinrichs D, Bianco N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob Agents Chemother. 1996;40:2021–2028. doi: 10.1128/aac.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reverchon S, Nasser W, Robert-Baudouy J. pecS; a locus controlling pectinase, cellulase and blue pigment production in Erwinia chrysanthemi. Mol Microbiol. 1994;11:1127–1139. doi: 10.1111/j.1365-2958.1994.tb00389.x. [DOI] [PubMed] [Google Scholar]
- 35.Saito K, Yoneyama H, Nakae T. nalB-type mutations causing the overexpression of the MexAB-OprM efflux pump are located in the mexR gene of the Pseudomonas aeruginosa chromosome. FEMS Microbiol Lett. 1999;179:67–72. doi: 10.1111/j.1574-6968.1999.tb08709.x. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Srikumar R, Kon T, Gotoh N, Poole K. Expression of Pseudomonas aeruginosa multidrug efflux pumps MexA-MexB-OprM and MexC-MexD-OprJ in a multidrug-sensitive Escherichia coli strain. Antimicrob Agents Chemother. 1998;42:65–71. doi: 10.1128/aac.42.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srikumar R, Li X-Z, Poole K. Inner membrane efflux components are responsible for the β-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J Bacteriol. 1997;179:7875–7881. doi: 10.1128/jb.179.24.7875-7881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srikumar R, Paul C J, Poole K. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J Bacteriol. 2000;182:1410–1414. doi: 10.1128/jb.182.5.1410-1414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 41.Ziha-Zarifi I, Llanes C, Koehler T, Pechere J-C, Plesiat P. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob Agents Chemother. 1999;43:287–291. doi: 10.1128/aac.43.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]