Abstract
The biosynthetic genes pchDCBA and pchEF, which are known to be required for the formation of the siderophore pyochelin and its precursors salicylate and dihydroaeruginoate (Dha), are clustered with the pchR regulatory gene on the chromosome of Pseudomonas aeruginosa. The 4.6-kb region located downstream of the pchEF genes was found to contain three additional, contiguous genes, pchG, pchH, and pchI, probably forming a pchEFGHI operon. The deduced amino acid sequences of PchH and PchI are similar to those of ATP binding cassette transport proteins with an export function. PchG is a homolog of the Yersinia pestis and Y. enterocolitica proteins YbtU and Irp3, which are involved in the biosynthesis of yersiniabactin. A null mutation in pchG abolished pyochelin formation, whereas mutations in pchH and pchI did not affect the amounts of salicylate, Dha, and pyochelin produced. The pyochelin biosynthetic genes were expressed from a vector promoter, uncoupling them from Fur-mediated repression by iron and PchR-dependent induction by pyochelin. In a P. aeruginosa mutant lacking the entire pyochelin biosynthetic gene cluster, the expressed pchDCBA and pchEFG genes were sufficient for salicylate, Dha, and pyochelin production. Pyochelin formation was also obtained in the heterologous host Escherichia coli expressing pchDCBA and pchEFG together with the E. coli entD gene, which provides a phosphopantetheinyl transferase necessary for PchE and PchF activation. The PchG protein was purified and used in combination with PchD and phosphopantetheinylated PchE and PchF in vitro to produce pyochelin from salicylate, l-cysteine, ATP, NADPH, and S-adenosylmethionine. Based on this assay, a reductase function was attributed to PchG. In summary, this study completes the identification of the biosynthetic genes required for pyochelin formation from chorismate in P. aeruginosa.
The opportunistic human pathogen Pseudomonas aeruginosa responds to iron-limiting growth conditions by the production of two major siderophores, pyoverdin (6, 19) and pyochelin (4, 28). These compounds are released to the extracellular environment, where they form a complex with iron and deliver it to the bacterial cell via their specific membrane receptors, FpvA (23) and FptA (1), respectively. Both siderophores, which contribute to the virulence of P. aeruginosa (5, 20, 36), are made by a thiotemplate mechanism (13, 18, 26, 27).
The genes required to make pyochelin and its precursors salicylate and dihydroaeruginoate (Dha) are clustered on the P. aeruginosa chromosome, next to the pyochelin receptor gene fptA (Fig. 1). Two biosynthetic operons, pchDCBA and pchEF, whose expression is repressed by the Fur protein in the presence of iron, have been identified (27, 33). Interestingly, pyochelin biosynthesis is autoregulated by a positive-feedback loop which requires the transcriptional regulator PchR. The pchR gene is located between the two biosynthetic operons (15, 27) (Fig. 1).
FIG. 1.
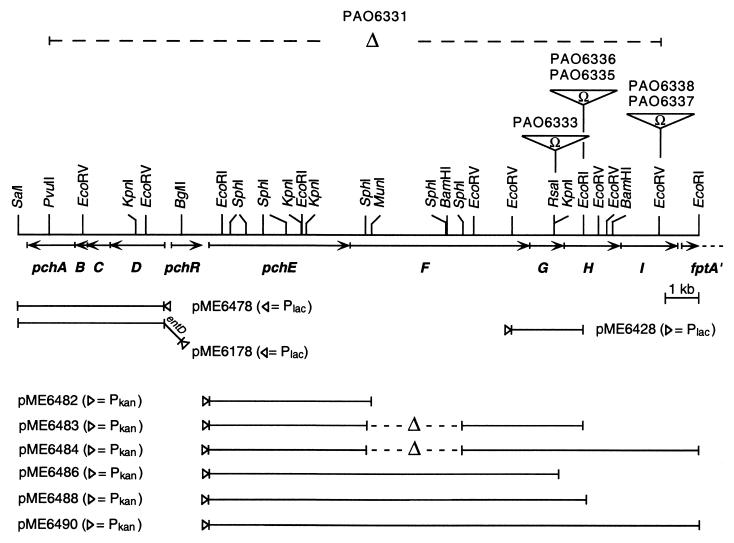
Physical map of the pchDCBA-pchR-pchEFGHI-fptA region of P. aeruginosa PAO1. The pchDCBA operon and the pchR, pchEF, and fptA genes have been described previously (1, 15, 27, 32). The ΩSm/Sp cassette present in mutants PAO6333, PAO6335, PAO6336, PAO6337, and PAO6338 is designated by Ω. Deletions are indicated by Δ. Subclones used for complementation experiments were constructed such that the respective vector promoter (indicated by a triangle) was located upstream of pchE, pchG, pchD, and entD. The restriction enzymes used for subcloning experiments are indicated. Note that for SalI, PvuII, MunI, and RsaI, only the locations relevant to this study are shown.
In the initial steps of pyochelin biosynthesis, salicylate is made from chorismate via isochorismate. These reactions are catalyzed by an isochorismate synthase and an isochorismate-pyruvate lyase encoded by the pchA and pchB genes, respectively (32; C. Gaille and D. Haas, unpublished results). Salicylate and l-cysteine, adenylated by PchD and PchE, respectively, are loaded onto the peptide synthetase PchE via covalent thioester linkages provided by two posttranslationally added phosphopantetheine prosthetic groups (26, 27). Subsequent condensation and cyclization reactions, catalyzed by PchE, generate an enzyme-bound hydroxyphenyl-thiazoline intermediate (26) which, when released by the action of a thioesterase, produces Dha (alternatively designated hydroxyphenyl-thiazolinyl-carboxylate). A domain of PchF displays such a thioesterase activity (26), and a putative external thioesterase is also encoded by the pchC gene (33). Pyochelin formation additionally requires the peptide synthetase PchF, which adenylates a second molecule of l-cysteine and anchors it, again via a phosphopantetheine prosthetic group, to a carrier domain in PchF (26, 27). PchF then catalyzes the condensation of the hydroxyphenyl-thiazoline-S-PchE intermediate with cysteinyl-S-PchF and generates the second thiazoline ring (26). At this stage, the basic skeleton of pyochelin is completed and the second thiazoline ring needs to undergo reduction and methylation before biosynthesis is terminated by pyochelin release from PchF due to cleavage of the remaining thioester bond.
In vitro reconstitution experiments with purified PchE and PchF, primed posttranslationally with phosphopantetheine, and purified PchD have shown that these proteins are able to make the pyochelin backbone from salicylate and l-cysteine but that they are not sufficient to catalyze the modification of the second thiazoline ring (26). A detailed genetic analysis of the pyochelin biosynthetic gene cluster indicates that additional genes located downstream of pchF may encode these missing functions (27).
Here we investigate the role of three additional genes, pchG, pchH, and pchI, in pyochelin biosynthesis and translocation and show that PchG, together with PchD and the holoenzyme forms of PchE and PchF, is sufficient to catalyze pyochelin formation from salicylate, l-cysteine, ATP, NADPH, and S-adenosylmethionine (SAM) in vitro.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table 1. Bacteria were usually grown on nutrient agar and in nutrient yeast broth (35) at 37°C. To quantify salicylate, Dha, and pyochelin in culture supernatants of P. aeruginosa, strains were grown in GGP medium (3). Minimal medium M9 (29), supplemented with 20 mM sodium succinate and the weak iron chelator 2,2′-dipyridyl (500 μM), was used as an iron-depleted medium to test pyochelin-mediated iron uptake in pyoverdin-negative mutants of P. aeruginosa. Antibiotics, when required, were added to the growth media at the following concentrations: tetracycline, 25 μg/ml for Escherichia coli and 100 μg/ml for P. aeruginosa; ampicillin, 100 μg for E. coli; carbenicillin, 250 μg/ml for P. aeruginosa; spectinomycin, 50 μg/ml for E. coli and 1 mg/ml for P. aeruginosa; gentamicin, 10 μg/ml for E. coli and P. aeruginosa; and kanamycin, 25 μg/ml for E. coli. To counterselect E. coli donor cells in matings with P. aeruginosa, chloramphenicol was used at 10 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Bacterial strains | ||
| E. coli | ||
| DH5α | recA1 endA1 hsdR17 deoR thi-1 supE44 gyrA96 relA1 Δ(lacZYA-argF)U169 (φ80dlacZΔM15) | 29 |
| MT147 | thi trpE purE proC leuB lacY mtl xyl rpsL azi fhuA tsx supA entC::Kmr | 21 |
| S17-1 | thi pro hsdR recA; chromosomal RP4 (Tra+ Tcs Kms Aps) | 34 |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (λDE3) | Novagen |
| P. aeruginosa | ||
| PALS128 | pvdB | 37 |
| PALS128-17 | pvdB pchB or pchA | 32 |
| PAO1 | Wild type | ATCC 15692 |
| PAO6331 | Δpch | This study (Fig. 1) |
| PAO6333 | pchG::ΩSm/Sp | This study (Fig. 1) |
| PAO6335 | pchH::ΩSm/Sp | This study (Fig. 1) |
| PAO6336 | pvdB pchH::ΩSm/Sp | This study (Fig. 1) |
| PAO6337 | pchI::ΩSm/Sp | This study (Fig. 1) |
| PAO6338 | pvdB pchI::ΩSm/Sp | This study (Fig. 1) |
| Plasmids | ||
| pPchD | pchD overexpression construct; Kmr | 26 |
| pPchE | pchE overexpression construct; Kmr | 26 |
| pPchF | pchF overexpression construct; Kmr | 26 |
| pPchG | pchG overexpression construct derived from pET29b; Kmr | This study |
| pET28b-entD | entD overexpression construct; Kmr | 16 |
| pET29b | Expression vector; Kmr | Novagen |
| pHP45Ω | ΩSm/Sp-containing plasmid; Apr Smr Spr | 24 |
| pME3087 | Suicide vector; Tcr; ColE1 replicon; EcoRI KpnI BamHI XbaI PstI SphI HindIII polylinker | 38 |
| pME3088 | Suicide vector; Tcr; ColE1 replicon; EcoRI KpnI DraII XhoI HindIII polylinker | 38 |
| pME3300 | pLAFR3 carrying the structural genes of PAO1 for the biosynthesis of salicylate, Dha, and pyochelin | 27, 32, 33 |
| pME6001 | pBBR1-based cloning vector; Gmr | 2 |
| pME6012 | pVS1-p15A shuttle vector; Tcr | 14 |
| pME6176 | pME3088 carrying the 3′ part of pchA on a 0.95-kb SalI-PvuII fragment joined to the 3′ part of pchI on a 1.2-kb EcoRV-EcoRI fragment | This study |
| pME6178 | pME6001 carrying entD pchDCBA under Plac control | This study (Fig. 1) |
| pME6180 | pME3088 carrying a 4.2-kb EcoRV-EcoRI fragment containing pchG interrupted by an ΩSm/Sp cassette at RsaI | This study |
| pME6428 | pUCPKS carrying pchG under Plac control on a 2.2-kb EcoRV-EcoRI fragment | This study (Fig. 1) |
| pME6478 | pME6001 carrying pchDCBA under Plac control | This study (Fig. 1) |
| pME6482 | pME6012 carrying pchE under Pkan control | This study (Fig. 1) |
| pME6483 | pME6012 carrying pchEG under Pkan control | This study (Fig. 1) |
| pME6484 | pME6012 carrying pchEGHI under Pkan control | This study (Fig. 1) |
| pME6486 | pME6012 carrying pchEF under Pkan control | This study (Fig. 1) |
| pME6488 | pME6012 carrying pchEFG under Pkan control | This study (Fig. 1) |
| pME6490 | pME6012 carrying pchEFGHI under Pkan control | This study (Fig. 1) |
| pME6498 | pME3087 carrying a 3.8-kb KpnI-BamHI fragment containing part of pchH interrupted by an ΩSm/Sp cassette at EcoRI | This study |
| pME6499 | pME3087 carrying a 4.6-kb BamHI-EcoRI fragment containing pchI interrupted by an ΩSm/Sp cassette at EcoRV | This study |
| pUCPKS and pUCPSK | ColE1-pRO1600 shuttle vectors; Apr | 39 |
Construction of plasmids and gene replacement mutants.
All plasmids used in this study are listed in Table 1. To uncouple the expression of the pch genes from iron and pyochelin control, a two-plasmid system was developed to allow constitutive pyochelin formation in homologous and heterologous backgrounds. First, the pchDCBA operon was fused at the ATG start codon of pchD to the lac promoter (Plac) carried by vector pME6001 by using a linker region containing a ribosome binding site (5′-AAGCTTGATATCGAATTGTGAGCGGATAAC AATTTCACACAGAATTGATTAAAGAGGAGAAATTAAGCATG-3′; the HindIII site used for cloning into pME6001 is italicized; the ATG start codon of pchD is underlined). This procedure gave plasmid pME6478 (Fig. 1). Then, the entD gene, encoding a phosphopantetheinyl transferase from E. coli, was excised from pET28b-entD by using XbaI and HindIII, ligated to a ClaI-XbaI DNA linker (5′-ATCGATAAGCTCTAGA-3′), and cloned into pME6478 between ClaI and HindIII to give pME6178 (Fig. 1). Constitutive expression of the pchEFGHI operon or parts thereof was achieved by fusing the ATG start codon of pchE to the kanamycin promoter (Pkan) present on pME6012 by using a linker region with a ribosome binding site (5′-AGATCTATCGATGCATGCCATGGTACCCAACTTTAAGAAGGAGATATACCCATG-3′; the BglII site used for cloning into pME6012 is italicized; the ATG start codon of pchE is underlined). In subsequent cloning steps, a series of constructs carrying pchE only (pME6482), pchEG (pME6483), pchEGHI (pME6484), pchEF (pME6486), pchEFG (pME6488), and pchEFGHI (pME6490) were generated (Fig. 1).
To overexpress and purify the PchG protein, its coding region was PCR amplified from genomic DNA of P. aeruginosa strain PAO1 by using the forward primer 5′-ATGCCAGAGGAGGCGAGCATATGAGCGACGTTCGTTCCG- 3′ and the reverse primer 5′-AGCAGGCGCCACAGCACCGCTCGAGCGAGGCTTGCTCC-3′; the forward primer introduces a silent mutation in the fourth codon to replace the low-frequency valine codon GTA with a GTT valine codon (italicized). The amplified product was digested with NdeI and XhoI and ligated to expression vector pET29b, digested with the same enzymes, to generate plasmid pPchG.
To generate chromosomal insertion and deletion mutations in pch genes (Fig. 1), derivatives of the suicide plasmids pME3087 and pME3088 (Table 1) were mobilized from E. coli S17-1 to strain PAO1 and chromosomally integrated, with selection for tetracycline resistance. Excision of the vector via a second crossing over was obtained by enrichment for tetracycline-sensitive cells (40). All mutants were checked by Southern analysis.
DNA manipulation and sequencing.
DNA manipulations were carried out as described by Sambrook et al. (29). Small-scale preparation of plasmid DNA was carried out by the cetyltrimethylammonium bromide (CTAB) method (7); large-scale preparation was performed with Qiagen-tip 100 columns (Qiagen Inc.). Chromosomal DNA was prepared as described by Gamper et al. (11). Restriction and DNA-modifying enzymes were used by following the instructions of the manufacturers. DNA fragments were purified from agarose gels with a Gene Clean DNA extraction kit (Bio 101, La Jolla, Calif.). E. coli and P. aeruginosa were transformed by the standard CaCl2 procedure (29) or by electroporation (8). Nucleotide sequences were determined on both strands with a dye terminator kit (Perkin-Elmer product no. 402080) and an ABI PRISM 373 sequencer and, in part, by the Euro Sequence Gene Service. Sequences were compared to the DNA sequence available from the P. aeruginosa genome sequencing project (http://www.pseudomonas.com) and found to be identical except for a single mismatch at position 1555 (T instead of G) of the pchI coding sequence. Nucleotide sequences were analyzed with programs from the University of Wisconsin Genetics Computer Group package (version 9.1).
Identification of salicylate, Dha, and pyochelin in culture supernatants of P. aeruginosa and E. coli.
P. aeruginosa strains were grown in GGP medium to stationary phase (optical density at 600 nm, ≈7 to 11, as measured with a Pharmacia Ultrospec III spectrophotometer), and recombinant E. coli strains were grown in nutrient yeast broth for 42 h to reach a final optical density at 600 nm of ≈2 to 2.5. For high-pressure liquid chromatography (HPLC) analysis, ethyl acetate extracts of acidified culture supernatants were dried by evaporation, dissolved in 60% (vol/vol) methanol–10 mM H3PO4, and injected into an HPLC system as described previously (27). Compounds were identified by their retention time and UV spectra. Dha, salicylate, and pyochelins I and II were quantified at 256, 237, 258, and 254 nm, respectively.
Overproduction and purification of Sfp and Pch proteins in E. coli.
Overproduction and purification of Sfp, PchD, PchE, and PchF proteins have been described previously (25, 26). For production in E. coli BL21(DE3) and purification of hexahistidine-tagged PchG, all techniques were identical to the methods used for the purification of PchD, PchE, and PchF, except that cultures were grown at 25°C for 24 h without induction.
Production and detection of salicylate-containing compounds.
In vitro reconstitution reactions contained 75 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 5 mM ATP, 0.1 mM coenzyme A, 1 mM salicylate, 5 mM l-cysteine, 2.5 μM PchE, 2.5 μM PchF, 4 μM PchD, and 0.5 μM Sfp. When needed, NADPH (2.0 mM), SAM (2.0 mM), and/or PchG (6 μM) was added. Reaction mixtures were incubated at 25°C for 1 h to allow phosphopantetheinylation, prior to initiation of the reaction by the addition of ATP. Incubation was continued for 2 h before the addition of HCl (100 mM final concentration) and extraction with ethyl acetate (1 ml) as reported previously (26). Dried samples were dissolved in 10% acetonitrile and analyzed by HPLC using a Vydac C18 reverse-phase column (4.6 mm by 25 cm) on a Beckman Gold HPLC system with monitoring at 254 nm. The mobile phases used were as follows: mobile phase A, 0.1 ml of formic acid and 0.2 ml of triethylamine in 1 liter of water; and mobile phase B, acetonitrile. Samples were eluted at 1 ml/min using a linear gradient of mobile phase B from 8 to 100% over 30 min. Mass spectrometry was carried out as described previously (26).
Nucleotide sequence accession number.
The nucleotide sequence of the pchG, pchH, and pchI genes of P. aeruginosa PAO1 has been assigned GenBank accession no. AF074705. The nucleotide sequence of pchG from strain PAO1 is almost identical to that from strain PA103 determined by De Voss and coworkers (GenBank accession no. AF116557).
RESULTS
The peptide synthetase genes pchE and pchF are followed by three additional genes, pchG, pchH, and pchI, in P. aeruginosa.
Previous work has indicated that the pchEF transcriptional unit contains additional genes downstream of pchF which may be necessary for pyochelin formation (27). To characterize these genes, the DNA region between pchF and the pyochelin receptor gene fptA was subcloned from cosmid pME3300 (32) and sequenced. Three open reading frames (ORFs) were identified. The first ORF, designated pchG, extends for 1,047 bp. The predicted ATG start codon overlaps with the stop codon of pchF and is preceded by a plausible ribosome binding site (AGGA). The second ORF, pchH, consists of 1,710 bp. The predicted GTG start codon overlaps with the TGA stop codon of pchG. A putative ribosome binding site (GGAG) is located at the 3′ end of pchG. The stop codon of pchH overlaps with the presumed ATG start codon of the third ORF, pchI, which consists of 1,722 bp and is also preceded by a plausible Shine-Dalgarno sequence (GGAG). The structural organization of the pchEFGHI genes strongly indicates that they belong to the same transcriptional unit, whereas the downstream fptA gene has its own iron-regulated promoter, as shown previously (1).
Comparison of the deduced amino acid sequence of pchG with the current sequence databases did not convincingly indicate the function for this protein. Some similarity was found only to proteins Irp3 (GenBank accession number Y12527; 28% identity) and YbtU (accession number AL031866; 27% identity), which are required for yersiniabactin formation in Yersinia enterocolitica and Y. pestis, respectively (12, 22). NrpU, a protein from Proteus mirabilis (accession number U46488), which shares 41% identical amino acids with Irp3, is also related to PchG (24% identity). However, no biochemical function has yet been attributed to Irp3, YbtU, and NrpU (12, 22).
The deduced amino acid sequences of PchH and PchI showed similarities to those of various ATP binding cassette (ABC) transport proteins with export function (30 to 35% identical amino acids). All four conserved sequence motifs characteristic of ABC proteins (17) are present in PchH and PchI; the Walker A motif (GPSGSGKST in PchH and GPSGAGKSS in PchI), resembling the consensus core sequence GXXGXGKST, and the Walker B motif (LLLLDEPT in PchH and PchI), resembling the consensus core sequence hhhhDEPT (h being a hydophobic residue), are necessary for ATP binding and hydrolysis. The ABC signature sequence LSGG, which is likely to be involved in energy transduction, is entirely conserved in PchH and PchI. An additional new motif consisting of a conserved histidine preceded by four hydrophobic residues and followed by a charged residue is present in PchH (VIVAHR) and PchI (LVLTHR) about 30 amino acids downstream of the conserved aspartic acid residue of the Walker B motif. PchH and PchI have membrane-spanning domains with six plausible transmembrane helices in their amino-terminal portions, whereas the ATPase domains are located in their carboxy-terminal portions.
Effects of pchG, pchH, and pchI mutations in P. aeruginosa on the formation of salicylate, Dha, and pyochelin.
To study the role of PchG, PchH, and PchI in pyochelin biosynthesis and translocation, several P. aeruginosa mutants were constructed. First, a pchI mutation was created by marker exchange using plasmid pME6499, which carries an ΩSm/Sp cassette at the EcoRV site in pchI. This mutation was made in the chromosome of the wild-type strain PAO1, giving strain PAO6337 (Fig. 1). Similar pyochelin and Dha concentrations were measured in culture supernatants of PAO6337 and PAO1 grown under iron-limiting conditions (Table 2), indicating that PchI is not involved in pyochelin or Dha export. An ΩSm/Sp insertion was also created in pchH (at the EcoRI site) using the suicide construct pME6498. Again, the mutation was transferred to wild-type strain PAO1, giving strain PAO6335 (Fig. 1). This insertion not only interrupts pchH but also has a polar effect on the expression of the downstream pchI gene. Salicylate, Dha, and pyochelin concentrations in culture supernatants of PAO6335 were determined and were found to be similar to those in the wild type and in the pchI mutant (Table 2). Thus, neither PchH nor PchI appears to be essential for pyochelin or Dha export.
TABLE 2.
Effects of pchG, pchH, and pchI mutations on the concentrations of salicylate, Dha, and pyochelin in culture supernatants of P. aeruginosaa
| Strain | Mutation | Plasmid | Gene carried | Culture supernatant concn (nmol/ml) of:
|
||
|---|---|---|---|---|---|---|
| Salicylate | Dha | Pyochelin | ||||
| PAO1 | None (wild type) | <8.0 | 69.3 ± 0.6 | 878.1 ± 59.0 | ||
| PAO6333 | pchG::ΩSm/Sp | 28.4 ± 3.4 | 5.5 ± 0.2 | <2.0 | ||
| PAO6333 | pchG::ΩSm/Sp | pME6428 | pchG | 27.7 ± 5.2 | 99.5 ± 1.4 | 619.2 ± 14.9 |
| PAO6335 | pchH::ΩSm/Sp | <8.0 | 75.2 ± 7.3 | 905.7 ± 71.8 | ||
| PAO6337 | pchI::ΩSm/Sp | <8.0 | 61.7 ± 3.0 | 824.1 ± 10.5 | ||
GGP medium (30 ml) containing, when required, carbenicillin (250 μg/ml) was inoculated with 0.3-ml samples of cultures grown in the same medium. After incubation at 37°C and 220 rpm for 33 h, supernatants were extracted and analyzed for salicylate, Dha, and pyochelin concentrations by HPLC (see Materials and Methods). The values given represent the means and standard deviations for three independent experiments.
The pchH and pchI ΩSm/Sp insertion mutations were also introduced into the pyoverdin-negative mutant PALS128, resulting in strains PAO6336 and PAO6338, respectively (Fig. 1). These strains grew well in minimal medium supplemented with the weak iron chelator dipyridyl, suggesting that they could utilize their own, stronger iron chelator pyochelin for iron uptake. In a control experiment, the double pyoverdin- and pyochelin-negative mutant PALS128-17 (32) grew very poorly in dipyridyl minimal medium. Therefore, an involvement of PchH and PchI in pyochelin-mediated iron uptake seems unlikely.
An ΩSm/Sp cassette was also inserted into the pchG gene of strain PAO1 using the suicide construct pME6180, resulting in strain PAO6333. As shown in Table 2, no pyochelin was detected in the supernatant of PAO6333 grown under iron-limiting conditions, implying that pchG is required for pyochelin biosynthesis. Dha was detected in small amounts, a result which could be explained by the fact that pyochelin is required to induce the transcription of the entire pchE operon (27). Although the mutation in PAO6333 is polar, the observed phenotype can be attributed solely to the mutation in pchG since, as shown above, insertions of Ω cassettes in pchH or pchI do not affect Dha and pyochelin production. Moreover, PAO6333 could be fully complemented by introducing plasmid pME6428, which carries pchG under the control of the constitutive lac promoter (Fig. 1 and Table 2). These results show that PchG is essential for pyochelin production.
Uncoupling salicylate, Dha, and pyochelin biosynthesis from Fur-mediated repression by iron and PchR-dependent induction by pyochelin.
We have shown previously that the expression of the two pyochelin biosynthetic operons is positively autoregulated by pyochelin and PchR (27). As a consequence, it can be difficult to study the importance of each gene individually in the biosynthetic pathway. Thus, although pchG is clearly needed for pyochelin production, a possible role in Dha formation cannot be excluded. We therefore decided to uncouple the expression of pchDCBA and pchEFGHI from positive autoregulation by pyochelin as well as from negative regulation by iron and Fur. To this end, the pchDCBA genes, together with the entD gene from E. coli, were subcloned into vector pME6001 and expressed under the control of the constitutive lac promoter in pME6178 (Fig. 1). The entD gene, which encodes a phosphopantetheinyl transferase with relaxed specificity, was added to ensure the conversion of the peptide synthetases PchE and PchF to their holoenzyme forms, in case the gene for phosphopantetheinyl transferase of P. aeruginosa were under iron and/or pyochelin control as well. This gene, which is not part of the pchDCBA and pchEFGHI operons, has not yet been identified in P. aeruginosa.
To express different combinations of genes from the pchEFGHI operon constitutively, a series of plasmids (Fig. 1 and Table 3) were generated from vector pME6012, in which gene expression is driven from the kanamycin promoter. The individual roles of pchE, pchF, pchG, pchH, and pchI in Dha and pyochelin formation were then studied with PAO6331, a mutant of PAO1 in which the whole pyochelin region encompassing pchDCBA, pchR, and pchEFGHI had been removed by gene exchange using the suicide construct pME6176 (Fig. 1). As expected, salicylate, Dha, and pyochelin were not detectable in supernatants of PAO6331 grown in the presence or in the absence of iron (Table 3). The introduction of pME6178 (Fig. 1) resulted in iron-independent production of salicylate. In agreement with our previous findings (26, 27), constitutive production of Dha required, in addition, the presence of pchE on pME6482 (Fig. 1). PchG was not involved in Dha production, as similar amounts were made when pME6482 was replaced by plasmid pME6483, carrying pchEG. To test whether PchH and PchI would contribute to Dha export, we measured the content of Dha in PAO6331 carrying pME6178 and pME6484. Surprisingly, the presence of the pchHI genes strongly reduced the amounts of Dha and weakly reduced those of salicylate detected in culture supernatants (Table 3). Thus, a function of PchH and PchI in Dha export is unlikely, but an import activity for Dha (and perhaps salicylate) cannot be excluded. Constitutive Dha and pyochelin formation in PAO6331 was achieved with pME6178 and pME6488, carrying pchEFG. The requirement for pchG was further corroborated by the fact that no pyochelin was made when pME6488 was replaced by pME6486, carrying the pchEF genes only. Adding the pchHI genes downstream of pchEFG on pME6490 did not result in larger amounts of Dha and pyochelin being produced (Table 3), showing again that this ABC transport system is not necessary for Dha or pyochelin export.
TABLE 3.
Effects of the pchEFGHI genes on salicylate, Dha, and pyochelin formation in P. aeruginosaa
| Strain | Chromosomal mutation | Plasmid(s) | Genes carried | Culture supernatant concn (nmol/ml) of the following in the absence (−) or presenceb (+) of Fe:
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Salicylate
|
Dha
|
Pyochelin
|
|||||||
| − | + | − | + | − | + | ||||
| PAO1 | None (wild type) | <8 | <8.0 | 79 ± 10.0 | <1.2 | 824 ± 26 | <2.0 | ||
| PAO6331 | Δpch | <8 | <8.0 | <1.2 | <1.2 | <2.0 | <2.0 | ||
| PAO6331 | Δpch | pME6178 | entDpchDCBA | 688 ± 38 | 428 ± 11 | <1.2 | <1.2 | <2.0 | <2.0 |
| PAO6331 | Δpch | pME6178 + pME6482 | entDpchDCBA + pchE | 1,178 ± 93 | 1,044 ± 118 | 140 ± 11.7 | 145 ± 20 | <2.0 | <2.0 |
| PAO6331 | Δpch | pME6178 + pME6483 | entDpchDCBA + pchEG | 969 ± 118 | 1,075 ± 107 | 130 ± 15.6 | 148 ± 13.9 | <2.0 | <2.0 |
| PAO6331 | Δpch | pME6178 + pME6484 | entDpchDCBA + pchEGHI | 720 ± 146 | 621 ± 72 | 4 ± 1.7 | <1.2 | <2.0 | <2.0 |
| PAO6331 | Δpch | pME6178 + pME6486 | entDpchDCBA + pchEF | 773 ± 129 | 1,059 ± 14 | 104 ± 11.8 | 101 ± 12.1 | <2.0 | <2.0 |
| PAO6331 | Δpch | pME6178 + pME6488 | entDpchDCBA + pchEFG | 225 ± 114 | 477 ± 109 | 21 ± 3.5 | 25 ± 2.5 | 494 ± 60 | 424 ± 52 |
| PAO6331 | Δpch | pME6178 + pME6490 | entDpchDCBA + pchEFGHI | 233 ± 49 | 387 ± 13 | 19 ± 1.0 | 20 ± 1.2 | 437 ± 12 | 349 ± 29 |
| PAO6331 | Δpch | pME6478 + pME6488 | pchDCBA + pchEFG | 57 ± 19 | 88 ± 26 | 9 ± 1.7 | 16 ± 1.4 | 308 ± 52 | 296 ± 11 |
GGP medium (30 ml) containing, when required, gentamicin (10 μg/ml) and tetracycline (100 μg/ml) was inoculated with 0.3-ml samples of cultures grown in the same medium. After incubation at 37°C and 220 rpm for 33 h, supernatants were extracted and analyzed for salicylate, Dha, and pyochelin concentrations by HPLC (see Materials and Methods). The values given represent the means and standard deviations for three independent experiments.
The medium was supplemented with 100 μM FeCl3 (iron excess).
A control experiment showed that the deletion mutant PAO6331 was complemented by the pchDCBA and pchEFG genes alone, in the absence of the cloned entD gene; pyochelin, salicylate, and Dha were produced by strain PAO6331 carrying plasmids pME6478 and pME6488, and the addition of iron did not influence the amounts of these metabolites (Table 3, last entry). Thus, the expression of the chromosomal phosphopantetheinyl transferase gene of P. aeruginosa does not appear to restrict pyochelin synthesis when iron is present in the growth medium. We do not know at this stage whether this gene is expressed constitutively or is positively autoregulated by pyochelin, in parallel with the two pyochelin biosynthetic operons (27).
Pyochelin formation in E. coli expressing pyochelin biosynthetic genes from P. aeruginosa.
The results presented in Tables 2 and 3 indicate that the pchDCBA and pchEFG genes are sufficient to produce pyochelin from chorismate in P. aeruginosa. In an attempt to clarify the role of the pchHI genes, we also measured pyochelin formation in recombinant derivatives of E. coli, a host which is normally unable to make salicylate, Dha, and pyochelin. After 42 h of growth in nutrient yeast broth containing tetracycline and gentamicin for plasmid maintenance, pyochelin concentations were measured in supernatants from three independent cultures of strain MT147 carrying pME6178 and pME6488. Although the amount of pyochelin made by this strain (21 ± 1.5 nmol/ml) was considerably smaller than that measured in culture supernatants of P. aeruginosa, the result confirms that pchDCBA and pchEFG, together with entD, are sufficient for pyochelin biosynthesis in the heterologous host. Replacing pchEFG with pchEFGHI did not affect pyochelin production (data not shown); hence, the function of PchH and PchI remained elusive. The small amounts of pyochelin produced by E. coli recombinants may have been a result of low chorismate availability, since salicylate concentrations measured in culture supernatants of MT147 carrying pME6178 and pME6488 were also much lower (2.3 ± 0.3 nmol/ml) than those found in PAO6331 carrying the same two plasmids (Table 3).
PchG is required for pyochelin formation in vitro.
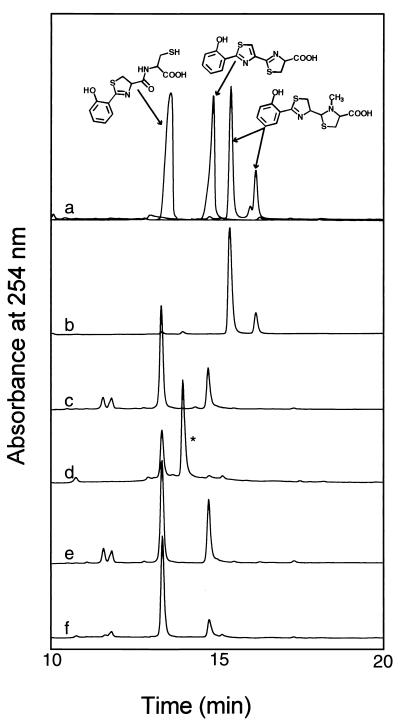
As reported previously (26), the in vitro reconstitution reaction containing PchD, PchE, PchF, salicylate, ATP, and l-cysteine results in the formation of hydroxyphenyl-thiazolinyl-cysteine (HPT-cys) and hydroxyphenyl-thiazolyl-thiazolinyl-carboxylic acid (HPTT-COOH) (Fig. 2, trace f). The addition of PchG alone did not alter the HPLC chromatogram (Fig. 2, trace e); however, the addition of PchG and NADPH produced a new product (indicated by an asterisk in trace d) that eluted between the standards for HPT-cys and HPTT-COOH. Mass spectrometric analysis showed this peak to have a mass of 311 Da. This mass corresponds to that of the predicted [M + H]+ ion for desmethyl-pyochelin (hydroxyphenyl-thiazolinyl-thiazolidinyl-carboxylic acid), in which the second thiazoline ring of the precursor hydroxyphenyl-bis-thiazolinyl-carboxylic acid has been reduced. The autoxidation of the middle ring, which is observed in the formation of HPTT-COOH, is not observed in this product.
FIG. 2.
HPLC elution profile showing intermediates and end products of pyochelin formation in vitro. Trace a, elution profile of the synthetic standards (from left to right) HPT-cys and HPTT-COOH (26) as well as a pyochelin standard purified from P. aeruginosa (as a 3:1 mixture of stereoisomers). Traces b to f, enzymatic products of in vitro reactions with mixtures containing 75 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 5 mM ATP, 0.1 mM coenzyme A, 1 mM salicylate, 5 mM l-cysteine, 2.5 μM PchE, 2.5 μM PchF, 4 μM PchD, and 0.5 μM Sfp. In addition, traces b to e also contained 6 μM PchG, traces b and c also contained 2.0 mM SAM, and traces b and d also contained 2.0 mM NADPH. The identity of the peak indicated by an asterisk was determined by mass spectrometry and corresponds to desmethyl-pyochelin (see Fig. 3). Note that peak sizes were normalized to show the identity of peaks and that they do not reflect relative amounts of products.
When SAM was added and NADPH was omitted, only HPT-cys and HPTT-COOH were produced (Fig. 2, trace c). Full reconstitution of pyochelin synthetase activity was observed upon inclusion of PchG, NADPH, and SAM (Fig. 2, trace b). The two peaks which had the same retention time as authentic pyochelin (Fig. 2, trace a) and which were observed in the full reconstitution were isolated and subjected to mass spectrometric analysis. The mass of each peak (325 Da) was in good agreement with that of the predicted [M + H]+ ion and was identical to the mass observed for the pyochelin standard purified from P. aeruginosa. Thus, both mass analysis and HPLC confirmed that the two peaks observed in Fig. 2, trace b, corresponded to the isomeric mixture (pyochelins I and II) of natural pyochelin (trace a), as previously reported (28).
DISCUSSION
We have shown in this study that the pchG gene, which is located downstream of pchEF, is essential for pyochelin formation in P. aeruginosa. Thus, we have identified all genes required to make pyochelin from chorismate, with the exception of a phosphopantetheinyl transferase gene, whose activity is necessary for priming the peptide synthetases PchE and PchF. This result was confirmed by the fact that transfer of the pchDCBA and pchEFG genes to E. coli resulted in pyochelin formation in this heterologous host.
The pchHI genes located immediately downstream of pchG encode proteins that resemble the subfamily of ABC transporters with export function. PchH and PchI each carry an ATP binding domain and a membrane-spanning domain on the same polypeptide, a configuration which is typical for bacterial export systems (9, 31). The absence of an adjacent gene coding for a periplasmic binding protein further supports a role for PchH and PchI in an export function rather than an import function. Based on these structural characteristics and on the likely coregulation of pchHI with the pchEFG genes, it was conceivable that PchH and PchI could be involved in pyochelin export. However, mutations in either pchH or pchI did not alter the amount of pyochelin (or Dha) present in culture supernatants. Thus, PchH and PchI may have another, as-yet-unknown transport function, or Dha and pyochelin may be exported by other transporters.
Two ABC transport genes, ybtP and ybtQ, have been identified in the yersiniabactin cluster in Y. pestis. Their corresponding proteins have the structural characteristics of ABC exporters. However, they are not necessary for yersiniabactin secretion but rather appear to play a role in iron uptake (10). We could not obtain evidence for an involvement of PchH and PchI in pyochelin-mediated iron uptake in P. aeruginosa. However, in the absence of pyochelin synthesis (due to the absence of pchF), the overexpressed pchHI genes significantly reduced the amount of Dha excreted (Table 3). One possible interpretation of this finding may be that PchH and PchI import Dha, thereby allowing the recycling of this compound. Unfortunately, there is no simple assay for such a function, in that free Dha does not act as a biosynthetic precursor of pyochelin (27).
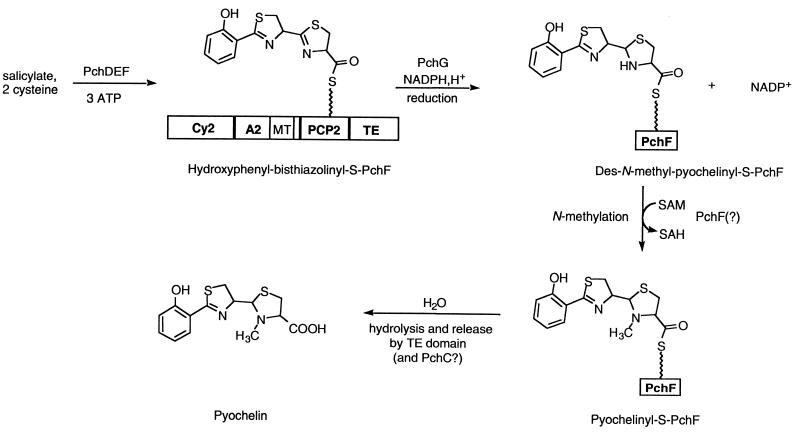
When the essential nature of PchG for pyochelin formation had been established by the genetic knockout results, the specific function of PchG as an enzyme was assayed in vitro in the presence of purified PchD, PchE, and PchF proteins. Only in the presence of both pure PchG protein and the cosubstrates NADPH and SAM was pyochelin produced in these incubations, starting from salicylate, cysteine, and ATP. Since we have previously shown that a tricyclic acyl enzyme intermediate (26) accumulates on PchF and is not released in the absence of PchG, we conclude that PchG is a catalytic protein whose action is required before the mature pyochelin acyl chain can be released from its covalent tether on PchF. PchG is weakly similar to biliverdin reductase (30), also an NADPH-dependent imine reductase, supporting the assignment in Fig. 3 of PchG as a thiazolinyl-S-enzyme-reductase converting bisthiazolinyl-S-PchF to the des-N-methyl–pyochelinyl-S-PchF acyl enzyme intermediate. SAM is additionally required for catalytic turnover; while it has not yet been proven whether the N-methyltransferase activity resides in PchG or PchF, we have argued (26) that a 35-kDa insert in the A domain of PchF is likely to be the methyltransferase. This question needs to be resolved by future mutagenesis experiments. Thus, in Fig. 3, N methylation produces mature pyochelinyl-S-PchF, which is presumed then to be a substrate for rapid hydrolysis by the C-terminal thioesterase domain of PchF or by PchC.
FIG. 3.
Action of PchG as a thiazolinyl reductase on an acyl-S-PchF intermediate in pyochelin biosynthesis. PchG reduces hydroxyphenyl-bisthiazolinyl-S-PchF to the des-N-methyl–pyochelinyl-S-acyl enzyme intermediate. The latter is then methylated by the putative methyltransferase (MT) domain of PchF and finally hydrolytically released from PchF. Domain structure of PchF: Cy2, cyclization; A2, adenylation; PCP2, peptidyl carrier protein. SAH, S-adenosyl-homocysteine; TE, thioesterase (26).
From these results, we conclude that PchG is an imine reductase, reducing thiazoline to a thiazolidine ring. There is mechanistic analogy between PchG and other NADPH-dependent imine reductases, for example, dihydrofolate reductase. Only after PchG action will the nitrogen in the thiazolidine ring be sufficiently basic and electron rich to attack the electron-deficient methyl group in SAM to yield the N-methylthiazolidine moiety characteristic of mature pyochelin. This initial elucidation of a thiazoline reductase activity of PchG is likely to presage equivalent reductase functions of Irp3 and YbtU, which are involved in the biosynthesis of the related but structurally more complex siderophore yersiniabactin (12, 22).
ACKNOWLEDGMENTS
We thank Thomas Keating for intellectual involvement with the in vitro reconstitution, Patrick Michaux for help with HPLC analysis, and SmithKline Beecham for the generous gift of carbenicillin.
This work was supported by the Swiss National Foundation for Scientific Research (project 31-56608.99; to D.H) and National Institutes of Health grant GM20011 (to C.T.W). H.M.P. is supported by a National Science Foundation fellowship.
REFERENCES
- 1.Ankenbauer R G, Quan H N. FptA, the Fe(III)-pyochelin receptor of Pseudomonas aeruginosa: a phenolate siderophore receptor homologous to hydroxamate siderophore receptors. J Bacteriol. 1994;176:307–319. doi: 10.1128/jb.176.2.307-319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blumer C, Heeb S, Pessi G, Haas D. Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc Natl Acad Sci USA. 1999;96:14073–14078. doi: 10.1073/pnas.96.24.14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carmi R, Carmeli S, Levy E, Gough F J. (+)-(S)-Dihydro-aeruginoic acid, an inhibitor of Septoria tritici and other phytopathogenic fungi and bacteria, produced by Pseudomonas fluorescens. J Nat Prod. 1994;57:1200–1205. doi: 10.1021/np50111a002. [DOI] [PubMed] [Google Scholar]
- 4.Cox C D. Iron uptake with ferripyochelin and ferric citrate by Pseudomonas aeruginosa. J Bacteriol. 1980;142:581–587. doi: 10.1128/jb.142.2.581-587.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox C D. Effect of pyochelin on the virulence of Pseudomonas aeruginosa. Infect Immun. 1982;36:17–23. doi: 10.1128/iai.36.1.17-23.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox C D, Adams P. Siderophore activity of pyoverdin in Pseudomonas aeruginosa. Infect Immun. 1985;48:130–138. doi: 10.1128/iai.48.1.130-138.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Del Sal G, Manfioletti G, Schneider C. A one-tube plasmid DNA mini-preparation suitable for sequencing. Nucleic Acids Res. 1988;16:9878. doi: 10.1093/nar/16.20.9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farinha M A, Kropinski A M. High efficiency electroporation of Pseudomonas aeruginosa using frozen cell suspensions. FEMS Microbiol Lett. 1990;58:221–225. doi: 10.1111/j.1574-6968.1990.tb13982.x. [DOI] [PubMed] [Google Scholar]
- 9.Fath M J, Kolter R. ABC transporters: bacterial exporters. Microbiol Rev. 1993;57:995–1017. doi: 10.1128/mr.57.4.995-1017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fetherston J D, Bertolino V J, Perry R D. YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol Microbiol. 1999;32:289–299. doi: 10.1046/j.1365-2958.1999.01348.x. [DOI] [PubMed] [Google Scholar]
- 11.Gamper M, Ganter B, Polito M, Haas D. RNA processing modulates the expression of the arcDABC operon in Pseudomonas aeruginosa. J Mol Biol. 1992;226:943–957. doi: 10.1016/0022-2836(92)91044-p. [DOI] [PubMed] [Google Scholar]
- 12.Gehring A M, DeMoll E, Fetherston J D, Mori I, Mayhew G F, Blattner F R, Walsh C T, Perry R D. Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem Biol. 1998;5:573–586. doi: 10.1016/s1074-5521(98)90115-6. [DOI] [PubMed] [Google Scholar]
- 13.Handfield M, Lehoux D E, Sanschagrin F, Mahan M J, Woods D E, Levesque R C. In vivo-induced genes in Pseudomonas aeruginosa. Infect Immun. 2000;68:2359–2362. doi: 10.1128/iai.68.4.2359-2362.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heeb S, Itoh Y, Nishijyo T, Schnider U, Keel C, Wade J, Walsh U, O'Gara F, Haas D. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in Gram-negative, plant-associated bacteria. Mol Plant Microbe Interact. 2000;13:232–237. doi: 10.1094/MPMI.2000.13.2.232. [DOI] [PubMed] [Google Scholar]
- 15.Heinrichs D E, Poole K. Cloning and sequence analysis of a gene (pchR) encoding an AraC family activator of pyochelin and ferripyochelin receptor synthesis in Pseudomonas aeruginosa. J Bacteriol. 1993;175:5882–5889. doi: 10.1128/jb.175.18.5882-5889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lambalot R H, Gehring A M, Flugel R S, Zuber P, LaCelle M, Marahiel M A, Reid R, Koshia C, Walsh C T. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 17.Linton K J, Higgins C F. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol Microbiol. 1998;28:5–13. doi: 10.1046/j.1365-2958.1998.00764.x. [DOI] [PubMed] [Google Scholar]
- 18.Merriman T R, Merriman M E, Lamont I L. Nucleotide sequence of pvdD, a pyoverdin biosynthetic gene from Pseudomonas aeruginosa: PvdD has similarity to peptide synthetases. J Bacteriol. 1995;177:252–258. doi: 10.1128/jb.177.1.252-258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer J M, Abdallah M A. The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physicochemical properties. J Gen Microbiol. 1978;107:319–328. [Google Scholar]
- 20.Meyer J M, Neely A, Stintzi A, Georges C, Holder I A. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect Immun. 1996;64:518–523. doi: 10.1128/iai.64.2.518-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozenberger B A, Brickman T J, McIntosh M A. Nucleotide sequence of Escherichia coli isochorismate synthetase gene entC and evolutionary relationship of isochorismate synthetase and other chorismate-utilizing enzymes. J Bacteriol. 1989;171:775–783. doi: 10.1128/jb.171.2.775-783.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pelludat C, Rakin A, Jacobi C A, Schubert S, Heesemann J. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J Bacteriol. 1998;180:538–546. doi: 10.1128/jb.180.3.538-546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poole K, Neshat S, Krebes K, Heinrichs D E. Cloning and nucleotide sequence analysis of the ferripyoverdin receptor gene fpvA of Pseudomonas aeruginosa. J Bacteriol. 1993;175:4597–4604. doi: 10.1128/jb.175.15.4597-4604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 25.Quadri L E N, Weinreb P H, Lei M, Nakano M M, Zuber P, Walsh C T. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 26.Quadri L E N, Keating T A, Patel H M, Walsh C T. Assembly of the Pseudomonas aeruginosa nonribosomal peptide siderophore pyochelin: in vitro reconstitution of aryl-4,2-bisthiazoline synthetase activity from PchD, PchE, and PchF. Biochemistry. 1999;38:14941–14954. doi: 10.1021/bi991787c. [DOI] [PubMed] [Google Scholar]
- 27.Reimmann C, Serino L, Beyeler M, Haas D. Dihydroaeruginoic acid synthetase and pyochelin synthetase, products of the pchEF genes, are induced by extracellular pyochelin in Pseudomonas aeruginosa. Microbiology. 1998;144:3135–3148. doi: 10.1099/00221287-144-11-3135. [DOI] [PubMed] [Google Scholar]
- 28.Rinehart K L, Staley A L, Wilson S R, Ankenbauer R G, Cox C D. Stereochemical assignment of the pyochelins. J Org Chem. 1995;60:2786–2791. [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Schluchter W M, Glazer A N. Characterization of cyanobacterial biliverdin reductase. J Biol Chem. 1997;272:13562–13569. doi: 10.1074/jbc.272.21.13562. [DOI] [PubMed] [Google Scholar]
- 31.Schneider E, Hunke S. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol Rev. 1998;22:1–12. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 32.Serino L, Reimmann C, Baur B, Beyeler M, Visca P, Haas D. Structural genes for salicylate biosynthesis from chorismate in Pseudomonas aeruginosa. Mol Gen Genet. 1995;249:217–228. doi: 10.1007/BF00290369. [DOI] [PubMed] [Google Scholar]
- 33.Serino L, Reimmann C, Visca P, Beyeler M, Della Chiesa V, Haas D. Biosynthesis of pyochelin and dihydroaeruginoic acid requires the iron-regulated pchDCBA operon in Pseudomonas aeruginosa. J Bacteriol. 1997;179:248–257. doi: 10.1128/jb.179.1.248-257.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–790. [Google Scholar]
- 35.Stanisich V A, Holloway B W. A mutant sex factor of Pseudomonas aeruginosa. Genet Res. 1972;19:91–108. doi: 10.1017/s0016672300014294. [DOI] [PubMed] [Google Scholar]
- 36.Takase H, Nitanai H, Hoshino K, Otani T. Impact of siderophore production on Pseudomonas aeruginosa infections in immunosuppressed mice. Infect Immun. 2000;68:1834–1839. doi: 10.1128/iai.68.4.1834-1839.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Visca P, Serino L, Orsi N. Isolation and characterization of Pseudomonas aeruginosa mutants blocked in the synthesis of pyoverdin. J Bacteriol. 1992;174:5727–5731. doi: 10.1128/jb.174.17.5727-5731.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voisard C, Bull C T, Keel C, Laville J, Maurhofer M, Schnider U, Défago G, Haas D. Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches. In: O'Gara F, Dowling D, Boesten B, editors. Molecular ecology of rhizosphere microorganisms. Weinheim, Germany: VCH Publishers; 1994. pp. 67–89. [Google Scholar]
- 39.Watson A, Alm R A, Mattick J S. Construction of improved vectors for protein production in Pseudomonas aeruginosa. Gene. 1996;172:163–164. doi: 10.1016/0378-1119(96)00026-1. [DOI] [PubMed] [Google Scholar]
- 40.Ye R W, Haas D, Ka J O, Krishnapillai V, Zimmermann A, Baird C, Tiedje J M. Anaerobic activation of the entire denitrification pathway in Pseudomonas aeruginosa requires Anr, an anolog of Fnr. J Bacteriol. 1995;177:3606–3609. doi: 10.1128/jb.177.12.3606-3609.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]