Abstract
Cell cycle progression and cell proliferation are tightly controlled processes physiologically; however, in cancerous cells, uncontrolled cell proliferation may be attributed to abnormal expression of the cyclin genes. Therefore, analysis of the expression of the cyclin genes may result in the discovery of biomarkers that can be used to predict a prognosis and help to evaluate the therapeutic efficacy more accurately in several types of cancer, including breast cancer. In this study, 15 subtypes of the cyclin genes in breast cancer from public databases were selected using bioinformatics analysis, the correlation between their transcriptional expression levels and survival rates were analyzed, and the results were further confirmed using reverse transcription-quantitative PCR in vitro in various breast cancer cell lines. The expression of the majority of the cyclin genes in SK-BR-3, a HER2 overexpressing breast cancer cell line, was lower than that in MCF-10A cells. CCNC mRNA expression was higher and CCNH mRNA expression was lower in tumor and tumor-adjacent tissues compared with that in normal tissues; however, CCNC expression was lower and CCNH expression was higher in breast cancer cell lines compared with that in MCF-10A cells. The expression of the 13 other cyclin genes in breast cancer cell lines was generally consistent with the data from the bioinformatics analyses of breast cancer tissue samples, tumor-adjacent tissues, and normal tissues. Low expression of CCNA2, CCNB1/2, CCNC, CCND1, CCNE1/2 and CCNF, and high expression of CCNA1, CCNB3, CCND2/3, CCNG1/2 and CCNH genes was correlated with a higher survival rate for breast cancer patients (P<0.05). In conclusion, CCNA2, CCNB1/2, CCND1/2 and CCNE1/2 may serve as relatively mature and accurate biomarkers, and CCNG1/2 may be used to evaluate the prognosis and therapeutic efficacy of hormone receptor-positive breast cancer. Furthermore, CCNA1, CCNB3, CCNC, CCND3, CCNF and CCNH may serve as promising targets for the management of breast cancer.
Keywords: biomarker, gene target, cyclin, bioinformatics, breast cancer
Introduction
Physiologically, cell cycle progression and cell proliferation are under precise and coordinated control, whereas uncontrolled cell proliferation caused by abnormal cell cycle progression is a key feature of cancer development/progression. Understanding the progression and regulation of the cell cycle is of significant importance for improving cancer treatments (1). The cell cycle consists of a G1 phase, S phase (DNA synthesis), G2 phase, and M phase (mitosis), and each step is jointly regulated by cyclin proteins and related cyclin-dependent kinases (CDKs) (2). To date, eight types of cyclin proteins, cyclin A to H, have been identified in mammalian cells (2,3), and can be further divided into multiple sub-types depending on their functions. It has been widely shown that cyclin genes play regulatory roles in a variety of cancers (4), including urinary malignant tumors (5,6), digestive tract malignant tumors (7,8), reproductive malignant tumors (9,10), and respiratory malignant tumors (11), amongst others.
Breast cancer is caused by the uncontrolled proliferation of breast epithelial tissue cells and is affected by various carcinogenic factors, the environment, genetics, and other factors (12). Based on the presence or absence of marker proteins including estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor 2 (HER-2), and Ki-67, breast cancer can be classified into four subtypes, namely luminal A, luminal B, HER-2 enriched, and triple-negative (13). According to the 2020 Global Cancer Statistics (14), breast cancer not only ranks highest amongst the most common malignant tumors in women worldwide, but also surpasses lung cancer as the most commonly diagnosed cancer. Breast cancer is an extremely heterogeneous malignant tumor with inter-tumor and intra-tumor variability (15). Therefore, novel molecular mechanisms and biomarkers for improving the detection of early-stage breast cancer and management of breast cancer are needed, with a long-term goal of improving individualized therapy.
Although there are several causes underlying the development of breast cancer, the cyclin family of genes has consistently been shown to play a pivotal role in aberrant cell cycle progression. CyclinA genes are divided into CCNA1 and CCNA2. CCNA1 exerts differential effects in different diseases (10,16), whereas there are fewer studies on the role of CCNA1 in breast cancer (17). CyclinA2 is widely upregulated in a variety of cancers (18) and plays a significant role in regulating the cell cycle (19,20).
CCNB1 is expressed in almost all tissues in humans and is highly expressed in a variety of cancers (21). It binds to CDK1 to form a complex, a key factor regulating the G2-to-M transition and mitotic progression (22). In solid tumors, the expression of CCNB1 is considered a substantial prognostic parameter (4,23). CyclinB2 also binds to CDK1 to form a complex, which inhibits the G2-to-M transition, thereby inducing cell cycle arrest (24). The expression of CCNB2, an oncogene, is upregulated in several types of malignant tumors (7,24–26), and its upregulated expression is associated with a poor prognosis. CyclinB3 possesses homology with cyclinA and cyclinB1/2, and is expressed in animals, insects, and human tissue (27). CCNB3 is more interrelated with BCOR, which encodes the BCL6 co-repressor for co-transcriptional expression, and does not appear to exhibit any obvious specificity in its upregulation regarding cancer type (28–30).
In contrast with other the cyclin genes, the function of CCNC-encoding cyclinC remains largely unknown. It binds to CDK3 and regulates the cell cycle in the G1 and G2 phases, and stimulates the reactivation of the cell cycle from a resting state (31,32). CCNC was shown to be upregulated (33) and increased cell proliferation in 82.6% of breast cancer cases (34).
D-type cyclins can bind to CDK4/6 and phosphorylate various substrates involved in the G1-to-S phase transition (35). In >50% of breast cancer subtypes, cyclinD1 protein expression is upregulated (36–38) and this in turn reduces the efficacy of treatments (39). The specificity of CCND1 expression for the differential diagnosis of benign and malignant mesothelial hyperplasia has been shown to approach 100% (40). CCND2 is one of the essential factors affecting endocrine resistance in breast cancer (41). Hypermethylation of CCND2 significantly increases the risk of death and is deemed an independent factor of a poor prognosis in triple-negative breast cancer (42,43). A meta-analysis showed that the upregulated expression of CCND3 was related to poorer overall survival (OS) in breast cancer and bladder cancer patients (44).
Overexpression of cyclinE1 and cyclinE2 accelerates cell cycle progression by shortening the G1-to-S phase transition period, and promoting cell proliferation and tumorigenesis, resulting in poorer survival rates (45–48). CCNF is expressed in all stages of the cell cycle, and accumulates in the S phase, reaching a peak in the G2 phase and gradually declining in the M phase (3). Its expression in different types of cancer varies (49), for example, its expression is low in liver cancer (50), high in gastric cancer (51), and unknown in breast cancer (52).
CCNG1-encoding cyclinG1 was identified as a target of p53-regulated transcription (53). Several studies have shown that CCNG1, which is hypothesized to be an estrogen regulatory gene (54), is downregulated in breast cancer (55–57). CCNH typically binds to CDK7 and promotes cancer cell migration during carcinogenesis (58). It has been shown that high expression of CCNH is correlated with a poor prognosis in patients with lung cancer (58) and gastrointestinal cancer (8). However, the related role of CCNH in breast cancer has been difficult to determine.
All types of cyclin genes are associated in some manner with the prognosis and/or drug tolerance in breast cancer. Thus far, certain cyclin genes have been verified as biomarkers in the prognostic prediction, evaluation of curative effects, and exploration of drug tolerance mechanisms. Yet, the role/effects of the remainder of the cyclin genes in breast cancer are incompletely understood. Therefore, in this study, bioinformatics methods were used to analyze data from publicly available databases to investigate the expression and mutation of different cyclin genes in breast cancer, attempting to excavate novel biomarkers.
Material and methods
Cell culture
All cell lines used in the present study were obtained from ATCC, including MCF-10A (normal breast tissue cells), MCF-7 (hormone receptor-positive breast cancer cells), MDA-MB-231, MDA-MB-468, and BT-549 (triple negative breast cancer cell lines), and SK-BR-3 (HER2 positive breast cancer cells). All breast cancer cell lines were maintained in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (NEWZERUM, Ltd.), and 100 U/ml penicillin and 100 mg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.). MCF-10A cells were cultured in DMEM/F12 supplemented with 5% (vol/vol) horse serum, 20 ng/ml EGF, 100 ng/ml cholera toxin, 0.01 mg/ml insulin, and 500 ng/ml hydrocortisone (MCF-10A specific medium; CM-0525-125; Procell Life Science & Technology Co., Ltd.). All cell lines were maintained at 37°C in a humidified incubator supplied with 5% CO2.
Reverse transcription-quantitative (RT-q)PCR
Cells were plated in 6-well plates and cells in the logarithmic growth phase were used for RNA extraction. Total RNA was extracted using an ESscience RNA-Quick Purification Kit. Total RNA concentration and purity were analyzed in duplicate using a NanoDrop One (Thermo Fisher Scientific, Inc.; cat. no. AZY1705838). cDNA was synthesized from qualified 1,000 ng RNA using an RT-PCR reverse transcription kit (Hifair® III 1st Strand cDNA Synthesis SuperMix for qPCR (gDNA digester plus), Shanghai Yeasen Biotechnology Co., Ltd.). The reverse transcription temperature protocol was: 25°C for 5 min, 55°C for 15 min, and 85°C for 5 min, as per the manufacturer's protocol. cDNA was stored at −20°C until required or kept on ice if used immediately. qPCR was performed using SYBR Green PCR reagents (Hieff UNICON® qPCR SYBR Green Master Mix; Shanghai Yeasen Biotechnology Co., Ltd.) on a ROCHE LightCycler 480 II detection system. The thermocycling conditions were: 95°C for 10 min; followed by 40 cycles at 95°C for 10 sec and 60°C for 30 sec. GAPDH was used as the internal control, and relative mRNA levels were calculated using the 2−∆∆cq method (59). Primers (sequences provided in Table I) were designed using Primer Bank (https://pga.mgh.harvard.edu/primerbank/) and were synthesized by Guangzhou IGE Biotechnology, Ltd. Primer targets were confirmed using NCBI blast (https://www.ncbi.nlm.nih.gov/tools/primer-blast/primertool.cgi?ctg_time=1656555059&job_key=6OI3EQEuDIYruAm9BN0tj37GPL1T1SegUg).
Table I.
Sequences of the primers used.
| Gene symbol | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| CCNA1 | GAGGTCCCGATGCTTGTCAG | GTTAGCAGCCCTAGCACTGTC |
| CCNA2 | CGCTGGCGGTACTGAAGTC | GAGGAACGGTGACATGCTCAT |
| CCNB1 | AATAAGGCGAAGATCAACATGGC | TTTGTTACCAATGTCCCCAAGAG |
| CCNB2 | CCGACGGTGTCCAGTGATTT | TGTTGTTTTGGTGGGTTGAACT |
| CCNB3 | ATGAAGGCAGTATGCAAGAAGG | CATCCACACGAGGTGAGTTGT |
| CCNC | CCTTGCATGGAGGATAGTGAATG | AAGGAGGATACAGTAGGCAAAGA |
| CCND1 | GCTGCGAAGTGGAAACCATC | CCTCCTTCTGCACACATTTGAA |
| CCND2 | ACCTTCCGCAGTGCTCCTA | CCCAGCCAAGAAACGGTCC |
| CCND3 | TACCCGCCATCCATGATCG | AGGCAGTCCACTTCAGTGC |
| CCNE1 | GCCAGCCTTGGGACAATAATG | CTTGCACGTTGAGTTTGGGT |
| CCNE2 | TCAAGACGAAGTAGCCGTTTAC | TGACATCCTGGGTAGTTTTCCTC |
| CCNF | CCCCGAAGATGTGCTCTTTCA | GCCTTCATTGTAGAGGTAGGCT |
| CCNG1 | GAGTCTGCACACGATAATGGC | GTGCTTGGGCTGTACCTTCA |
| CCNG2 | TCTCGGGTTGTTGAACGTCTA | GTAGCCTCAATCAAACTCAGCC |
| CCNH | TGTTCGGTGTTTAAGCCAGCA | TCCTGGGGTGATATTCCATTACT |
| GAPDH | GGAGCGAGATCCCTCCAAAAT | GGCTGTTGTCATACTTCTCATGG |
Bioinformatics analysis
Oncomine (60), an online large-scale tumor gene chip database, can be used to analyze the transcriptional expression levels of cyclin genes in clinical breast cancer samples compared with normal breast tissues (Fold change=2, P<0.001, top 10% gene rank, for screening out samples with relatively significant differential expression in cyclin genes).
GEPIA (61) is primarily used for differential expression analysis between cancer and normal tissues, and for correlation analysis between gene expression and clinical pathological stage. The OS and risk-score analyses of breast cancer patients were assessed using Kaplan-Meier Plotter (62) and METABRIC using the auto-select best cutoff. cBioportal (63) was further used to analyze the genomic profiles of cyclin genes in TCGA, and analyze the relationship amongst genes through protein-protein interaction (PPI) analysis.
Database for annotation, visualization, and integrated discovery (DAVID) knowledgebase
DAVID contains species-specific gene/protein identifiers and their annotations from a variety of public genomic resources such as NCBI, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG), amongst others, to allow the incorporation of a diverse range of arrays of functional and sequencing annotations, greatly enriching the level of biological information available for a required gene (e.g. gene ID, pathways, etc.) (64). In this study, the GO and KEGG enrichment analysis of cyclin genes by DAVID were used to explore the functions and mechanisms of cyclin genes, R-4.0.4 (65,66) was used to draw figures.
Statistical analysis
A one-way ANOVA was used to compare the differences between multiple groups; a post-hoc Dunnett's multiple comparisons test was used to compare the mean of each column with the mean of the control group (MCF-10A). All statistical analyses were performed in GraphPad Prism version 9 (GraphPad Software, Inc.). For all bioinformatics analysis, all analyses were performed by the specific tools used in the corresponding website. P<0.05 was considered to indicate a statistically significant difference.
Results
Transcriptional expression of the cyclin genes in breast cancer patients
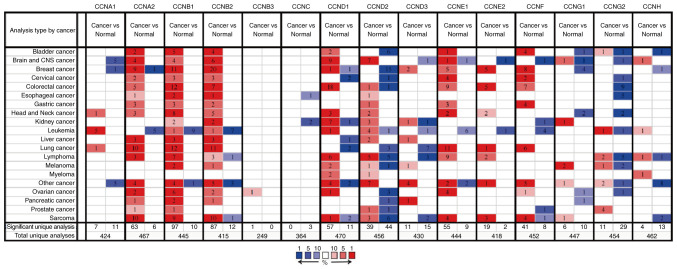
The transcriptional expression levels of the cyclin genes in breast cancer and normal breast tissues were compared using data obtained from Oncomine (Fig. 1). CCNA2, CCNB1, CCNB2, CCND1, CCND3, CCNE1, CCNE2, and CCNF expression was upregulated in the tumor tissues; CCND3 and CCNE1 expression was moderately upregulated, whereas the rest of the genes exhibited significant upregulation. Of note, CCNA2 mRNA expression in one of the datasets was significantly lower in the breast cancer tissues. Similarly, CCND1 mRNA expression was slightly downregulated in one of the datasets. The mRNA expression levels of CCNA1, CCND2, CCNG1, and CCNH were downregulated in the tumor tissues, particularly CCND2. Of note, the transcriptional expression of CCNB3, CCNC, and CCNG2 had not been collected in the Oncomine database.
Figure 1.
Transcriptional expression of the cyclin genes in different types of cancer compared with the respective normal tissue. CNS, central nervous system.
Relationship between the mRNA expression of cyclin genes and clinicopathological stages of breast cancer
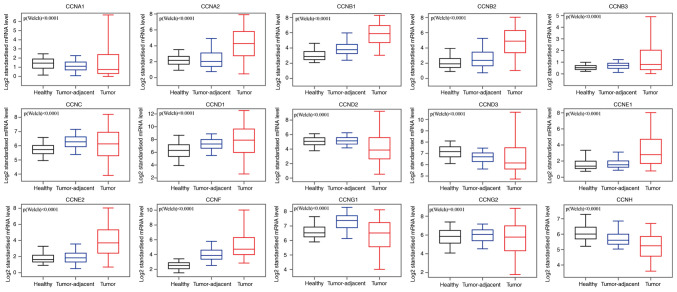
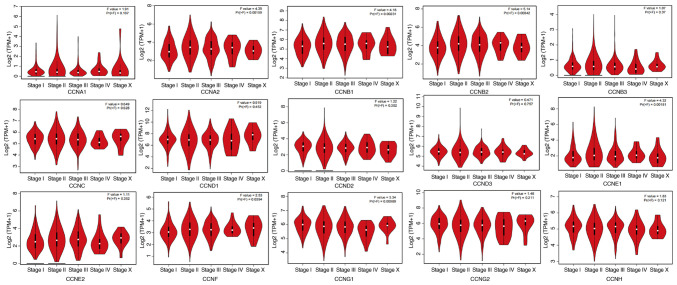
Comparative analysis of the transcriptional expression of cyclin genes in breast cancer tissues, tumor-adjacent tissues, and normal breast tissues in the GEPIA database demonstrated that the mRNA expression of CCNA2, CCNB1, CCNB2, CCNB3, CCNC, CCND1, CCNE1, CCNE2, and CCNF in breast cancer and tumor-adjacent tissues was higher than that in the normal breast tissues (Fig. 2), whereas CCNA1, CCND2, CCND3, CCNG1, and CCNH mRNA expression was higher in the normal tissues compared with the other tissues. CCNG2 transcriptional expression did not differ significantly between tumors and normal tissues, and in both, its expression was lower compared with the tumor-adjacent tissues. We also analyzed the transcriptional expression of cyclin genes in the different clinical stages of breast cancer (Fig. 3). CCNA2, CCNB1, CCNB2, CCNE1, CCNF and CCNG1 expression differed significantly, and the gene expression was highest in stage IV; however, the other genes did not exhibit differential expression based on stage.
Figure 2.
mRNA expression levels of the cyclin genes in healthy, tumor-adjacent, and tumor tissues.
Figure 3.
Relationship between mRNA expression levels of the cyclin genes and the clinicopathological stages in breast cancer patients. TPM, transcripts per million.
Correlation between the transcriptional expression of the cyclin genes and the survival period of patients with breast cancer
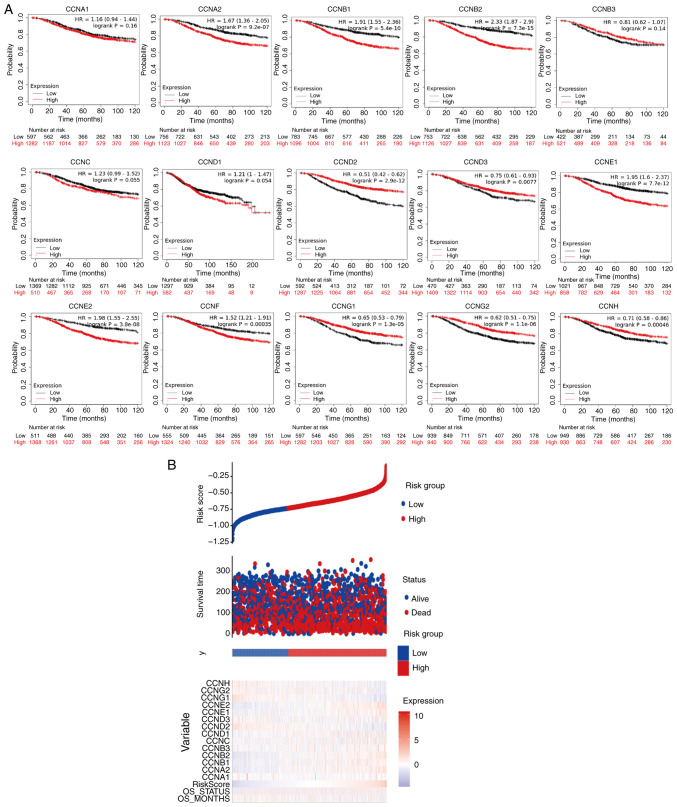
To further explore the role of cyclin genes in the survival of breast cancer patients, Kaplan-Meier Plotter was used (Fig. 4A). The results showed that lower expression of CCNA2, CCNB1, CCNB2, CCNE1, CCNE2, and CCNF and higher expression of CCND2, CCND3, CCNG1, CCNG2, and CCNH was significantly correlated with a better OS (P<0.05). However, high expression of CCNB3 mRNA was not associated with OS in patients (P=0.13). The curve trend showed that OS was longer if the transcriptional expression of CCNC was further reduced (P=0.064). The risk-score analysis provided basic evidence for the above survival analysis (Fig. 4B), the only thing lacking was the analysis of CCNF, but from the results, the relationship between low CCNF mRNA expression and high survival was unexpected.
Figure 4.
Association between cyclin genes and survival rate. (A) Correlation analysis between the transcriptional levels of the cyclin genes and the survival of patients with breast cancer. (B) Risk-score analysis of cyclin gene expression. HR, hazard ratio.
Alterations of expression of the cyclin genes in breast cancer and correlation analysis
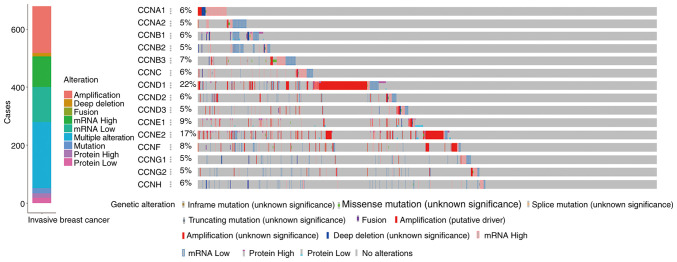
The online cBioportal tools were used to analyze the alterations of cyclin genes and the related correlation in invasive breast cancer from TCGA. Among the samples from 1,084 breast cancer patients, various alterations were detected in 679 samples (Fig. 5). Mutations including inframe, missense, splicing, and truncations occurred in 18 samples, fusion-mutations occurred only in 1 sample, amplification occurred in 161 samples, deep deletions occurred in 12 samples, upregulated mRNA expression was observed in 105 samples, downregulated mRNA expression was observed in 121 samples, upregulated protein expression was observed in 15 samples, downregulated protein expression was observed in 18 samples, and multiple alterations were observed in 228 samples.
Figure 5.
Alteration analysis of cyclin gene expression in breast cancer.
As shown in the specific variation analysis, the mRNA expression of CCNA1, CCNC, CCND3, CCNE1, and CCNB3 was upregulated, and that of CCNA2, CCNB1, CCNB2, CCND2, CCNG1, CCNG2, and CCNH was downregulated. Gene amplifications were more pronounced with CCND1, CCNE2, and CCNF. Furthermore, a fusion mutation was observed in the CCNF gene.
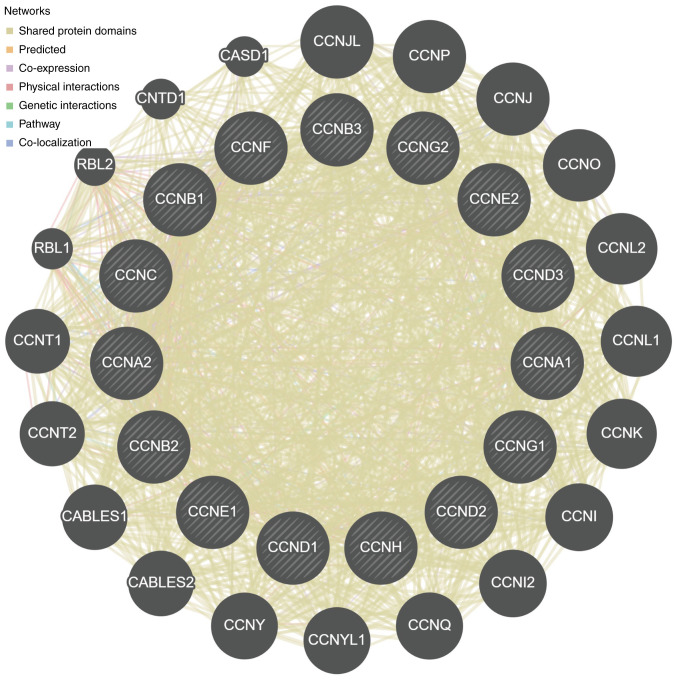
Based on the correlation analysis among the cyclin genes (Fig. 6), for cyclin genes and other interacting genes, the majority of shared protein domains accounted for 69.48% of the total interacting network nodes, predicted and co-expression accounted for 14.21 and 9.12%, respectively, and the remaining accounted for <10%.
Figure 6.
PPI network based on the cyclin genes.
Cyclin gene functions and molecular signaling pathways based on GO and KEGG enrichment analysis
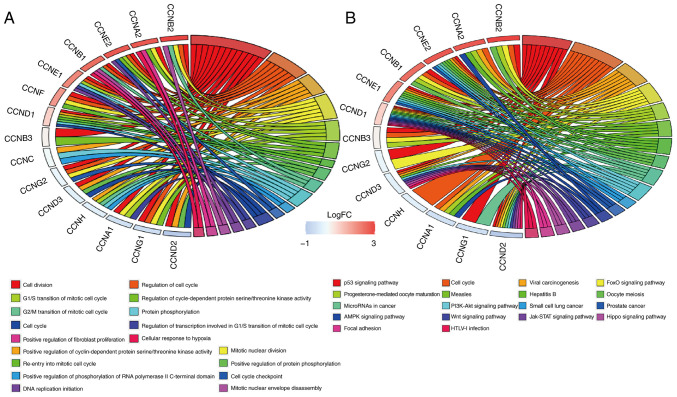
Functional involvement of cyclin genes such as biological processes (BP) was predicted using GO enrichment analysis. The results showed that cell division (GO: 0051301), regulation of cell cycle (GO: 0051726), positive regulation of cyclin-dependent protein serine/threonine kinase activity (GO: 0045737), G1/S transition of mitotic cell cycle (GO: 0000082), and regulation of cyclin-dependent protein serine/threonine kinase activity (GO: 0000079) were significantly affected by cyclin genes in BP (Fig. 7A).
Figure 7.
Relative molecular pathway analysis. (A) GO-BP and (B) KEGG enrichment analysis.
KEGG enrichment analysis indicated that there were 18 molecular signaling pathways that cyclin genes participated in (Fig. 7B). Signaling pathways closely associated with breast cancer included the Wnt signaling pathway (map04310), PI3K-Akt signaling pathway (map04151), p53 signaling pathway (map04115), microRNAs in cancer (map05206), JAK-STAT signaling pathway (map04630), hippo signaling pathway (map04390), cell cycle (map04110), and AMPK signaling pathway (map04152); the cyclin genes were mostly involved in cell cycle regulation and p53 signaling pathway.
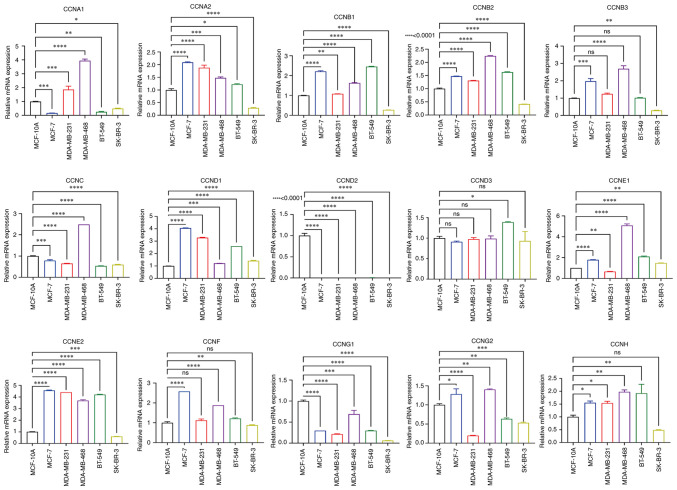
Expression of the cyclin genes in the breast cancer cell lines
To verify cyclin gene expression in vitro, RT-qPCR analysis of the breast cancer cell lines was performed using MCF-10A cells as the standard control. The results demonstrated that the mRNA expression levels of most of the cyclin genes in SK-BR-3 were lower than that in control, except for CCND1 and CCNE1 mRNA expression, whose expression was slightly higher (Fig. 8). The mRNA expression levels of CCNA2, CCNB1, CCNB2, CCNB3, CCND1, CCNE1, CCNE2, and CCNF in the cancer cell lines were significantly higher than that in MCF-10A cells, whereas CCND3 and CCNG2 exhibited lower expression. CCNA1 and CCNG2 mRNA expression varied based on the breast cancer cell line, and CCND2 was barely expressed in any of the cell lines. In summary, these results are generally consistent with the gene expression analysis of the data from the databases. However, the mRNA expression levels of CCNC and CCNH in cell lines were lower and higher than that in MCF-10A cells, respectively; their expression in tissues showed the opposite trend compared with that in the cell lines.
Figure 8.
Relative mRNA expression levels of the cyclin genes in different breast cancer cell lines *P<0.05, **P<0.01, ***P<0.001, ****P<0.001.
Discussion
The cyclin genes play significant roles in a variety of malignant tumors (4–11), and evaluation of the transcriptional expression levels of the different cyclin genes may be used to predict the prognosis and assess drug efficacy in malignant tumors. However, studies assessing this are lacking due to the numerous sub-types of cyclin genes and the high degree of heterogeneity in breast cancer. Thus, the present study aimed to analyze and discuss the value of the analysis of the diverse range of cyclin genes in breast cancer at the transcriptional level, using online public databases and bioinformatics methods to identify potential biomarkers to improve survival and prognostic prediction, and/or serve as therapeutic targets.
The functions of different cyclin genes vary in enrichment analysis, and regulation, in general, is associated with the phase of the cell cycle (67–72). However, in the enrichment analysis of breast cancer-related molecular signaling pathways, the regulation of CCNC and CCNF was not involved. Therefore, certain cyclin genes, especially CCNC and CCNF, may have greater value as potential biomarkers based on our results.
First, the median transcriptional expression of CCNA1 in breast cancer tissue was lower than that in tumor-adjacent and normal breast tissues. The protein encoded by CCNA1 is involved in the process of anthracycline resistance, and the methylation status of CCNA1 and CCND2 in normal tissues was lower in normal breast tissues than that in tumors, and its expression in breast cancer was downregulated after treatment (5,73,74). These data indicated that high transcriptional expression of CCNA1 and CCND2 may slow down the process of acquisition of resistance to anthracyclines and prolong the effects of the drug in breast cancer cells. Therefore, they may be used to predict anthracycline/filomycin sensitivity. Secondly, there were no significant differences in the mRNA expression levels of CCNA1 between the clinical tumor stages of breast cancer. The results showed that its high expression could significantly increase the patients' RFS, while low expression increased the patients' OS, although the difference was not significant. From the analysis of TCGA data, CCNA1 mRNA was mainly highly expressed in breast cancer caused by mutations. The contradictory results may be due to the fact that mutations may frequently occur due to alterations in breast cancer heterogeneity during disease development or treatment (15). Previous studies on the investigation of CCNA1 expression in breast cancer are lacking; however, based on these results, CCNA1 may exhibit potential as a biomarker for evaluating drug tolerance in breast cancer.
CCNA2 is a relatively more reliable biomarker for predicting the prognosis and assessing drug resistance. Gao et al (18) confirmed that CCNA2 was powerful in predicting the survival prognosis of ER-positive breast cancer patients. Overexpression of CCNA2 mRNA could lead to Tamoxifen resistance. However, additional evidence is required to understand the mechanism by which tamoxifen resistance is reversed. In this study, the mRNA expression levels of CCNA2 in breast cancer were significantly higher than that in normal tissues, highlighting the potential of CCNA2 as a biomarker. Additionally, its expression in different clinical stages of breast cancer varied significantly. Based on survival length, lower transcriptional expression of CCNA2 was significantly correlated with a better OS and RFS in cancer patients, a result consistent with a previous study (75). Additionally, the mRNA expression levels of CCNA2 in one dataset were significantly lower than that in normal tissues, and analysis of data from TCGA also showed that CCNA2 mRNA was lower in normal tissues. Furthermore, CCNA2 mRNA expression was lower in SK-BR-3 cells than in MCF-10A cells. Together, the results not only reflect the heterogeneity of breast cancer (15), but also provide a novel direction for the in-depth investigation of breast cancer. Thus, future studies should analyze CCNA2 expression for predicting prognosis and evaluating its clinical efficacy.
Similar to CCNA2, CCNB1, and CCNB2 show significant potential for predicting the prognosis and assessing drug resistance. In both tumor tissues and cancer cell lines, the mRNA expression levels of CCNB1 and CCNB2 were significantly higher than that in normal tissues and cells. The low transcriptional expression of the two genes may indicate a better OS in patients, consistent with previous studies (22,75–77). It has also been found that their high transcriptional expression can result in tamoxifen resistance and is positively correlated with endocrine resistance (22,78).
Unlike CCNB1 and CCNB2, CCNB3 in breast cancer has not been extensively studied and thus could not be validated. Corresponding mRNA expression data was not available in the Oncomine database. Transcriptional expression of CCNB3 in breast cancer was slightly higher than that in the normal tissues, and the in vitro analysis showed the same trend. In TCGA analysis, the number of samples that showed upregulated or downregulated expression of CCNB3 mRNA was approximately the same. Although the P-value was >0.05, the survival curve showed a trend such that the upregulated expression of CCNB3 mRNA may predict an improved OS for patients. At present, whether CCNB3 is suitable for future use to judge the prognosis in patients with breast cancer and the efficacy of drug response is still open for discussion, a definite correlation between CCNB3 and breast cancer has been confirmed. Additional studies are required to explore the biological function of CCNB3.
Analysis of the GEPIA dataset showed that CCNC mRNA expression in breast cancer was significantly higher than that in normal tissues, similar to the primary alteration in TCGA data. There were also a small number of mutations with lower transcriptional expression. In the breast cancer cell lines, CCNC mRNA expression was lower than that in MCF-10A cells. Li et al (79) showed that CCNC regulates the expression of the NOTCH1 oncogene to exert a tumor suppressor effect. Thus far, a more direct explanation for the low expression of CCNC mRNA is lacking. Tumor cells may adopt various means to circumvent the inhibitory effects of CCNC to grow. Additionally, CCNC is also involved in the transcriptional regulation of SRC2 in human breast cancer, and their expression was positively correlated (80). SRC is upregulated in the early stages of breast intraductal carcinoma compared with normal breast tissue. Decreased expression of SRC2 was associated with the progression of the disease and the formation of invasive ductal carcinoma (81), thus it is highly likely that CCNC mRNA expression may be similarly altered with SRC2. It has been suggested that CCNC participates in the RB-E2F pathway during the progression of breast cancer (31), and its expression may also vary as the disease progresses. Therefore, it may serve as a potential therapeutic target for repressing the cell cycle and a novel prognostic predictor.
CCND1, an oncogene, is currently the most extensively studied gene related to endocrine resistance (39,82). Its expression was significantly increased in hormone receptor-positive breast cancer (38) and was closely related to mechanisms of endocrine resistance. CCND1 has been used in HER-2-positive breast cancer as a continuous variable marker to predict the therapeutic benefit of trastuzumab (83). In addition, CCND1 had a different relationship with the aggressiveness of tumors and its low transcriptional expression may enhance the migration of breast cancer subgroups (43,84). In this study, CCND1 mRNA expression in breast cancer was significantly increased. Its low expression was shown to correlate with a better RFS in patients with breast cancer, consistent with previous studies (36,37). Analysis of the Oncomine dataset showed that the expression of CCND1 mRNA in breast cancer was lower than that in normal tissues, and the same trend of expression in breast cancer samples was found in TCGA, providing evidence for why the low expression of CCND1 can enhance the migration of breast cancer sub-types. Based on the above findings, it is understandable that CCND1 may serve as a biomarker for breast cancer diagnosis, prognostic prediction, drug efficacy, and resistance.
CCND2 mRNA expression was significantly lower in breast cancer samples compared with the normal tissues in several databases including Oncomine, GEPIA, and TCGA. Its high expression was typically associated with a better OS and RFS, consistent with previous reports (42,85). Further studies showed that CCND2 methylation was negatively correlated with its gene expression (43,86). The integrated analysis of the correlation between CCND2 methylation and gene expression may be used to improve the predictive performance of breast cancer outcomes, which may be conducive to the expansion of the potential clinical applications of CCND2.
The Oncomine dataset showed that CCND3 expression was highly upregulated in breast cancer and its mRNA expression was upregulated in the TCGA dataset. Analysis of the GEPIA dataset illustrated that CCND3 mRNA expression was high in normal breast tissues, and the RT-qPCR results showed that CCND3 mRNA expression in MCF-10A cells was higher than that in other cancer cell lines. Based on the survival analysis it was shown that upregulated expression of CCND3 mRNA was associated with a better OS. Conversely, Justenhoven et al (87) found that the low expression of E2F2, CCND1, and CCND3 genes reduced the levels of factors that downregulate HER-2, thereby allowing upregulated expression of HER-2 in tumors, and it is hypothesized that these three gene sub-types are potential indicators of HER-2 status in breast cancer. Combining all the results of the databases together, it can be suggested that CCND3 plays various functions in different breast cancer sub-types. Since the current exploration of CCND3 is still limited, further research is required to better understand the potential of CCND3 as a possible biomarker for prognostic prediction and improving drug efficacy.
The expression of CCNE1 and CCNE2, two subtypes of the CCNE gene, was approximately the same. The expression of both of these genes was upregulated in breast cancer, a result that was confirmed by RT-qPCR. Patients with low transcriptional levels of both genes in breast cancer had a significantly better OS. One difference between the two subtypes was that the expression of CCNE1 was correlated with the clinical tumor stage. The clinical trials PALPMA-3 and POP (88) demonstrated that the efficacy of Palbociclib decreased when CCNE1 mRNA expression was increased. Therefore, CCNE1 mRNA is predictive of metastatic tumors. Previously it has been shown that the amplification of CCNE1 was associated with a poorer OS of metastatic triple-negative breast cancer, and it was speculated that this phenomenon may be caused by CCNE1 upregulation-induced chemotherapeutic resistance (89). Moreover, both CCNE1 and CCNE2 could be used as independent prognostic markers for patients with lymph node-negative breast cancer (90). Thus, as typical oncogenes, CCNE1 and CCNE2 may be employed as factors for the prognostic prediction and for countering drug resistance mechanisms in breast cancer.
The functional involvement of CCNF in breast cancer remains unclear. Studies have used CCNF as a target gene for RNAi targeting in breast cancer (91,92). CCNF may be involved in the non-KEGG-enriched molecular signaling pathways to regulate drug resistance in breast cancer. Additionally, the CCNF protein possesses a unique characteristic, an F-BOX homologous sequence, which is suggestive of CCNF possessing a similar function to that of F-BOX. As for F-BOX, high mRNA expression of its subtypes (FBXO1, FBXO31, and FBXO5) may serve as biomarkers for predicting prognosis, and their expression was significantly correlated with a poor prognosis in patients with breast cancer (93). Therefore, it was hypothesized that CCNF may be used as a biomarker similar to F-BOX. In this study, CCNF mRNA expression was upregulated in breast cancer, and it was primarily observed as gene amplification and the only example of a fusion alteration found in TCGA in the present study. The clinical stage of breast cancer was significantly related to the differential expression of CCNF, and patients with low CCNF mRNA expression had a significantly better OS. Therefore, CCNF has potential as an advanced biomarker for prognostic prediction and as a gene target for counteracting therapeutic tolerance.
CCNG1 is the only cyclin gene that has both positive and negative effects on cellular growth (94,95) as the cyclinG1 protein encoded by CCNG1 is highly unstable. Tian et al (96) found out that the administration of estrogen and progesterone promoted cell proliferation and upregulated the expression of cyclinG1 in MCF-7 cells. Estradiol and progesterone-mediated cell viability and clonal ability were restricted after CCNG1 was knocked down using shRNA, suggesting that estradiol and progesterone promote cell proliferation in breast cancer partially by inducing cyclinG1 expression. In the present study, CCNG1 mRNA expression was low in breast cancer, and cancer patients with upregulated CCNG1 expression had a better OS. CCNG1 expression was also significantly associated with the clinicopathological stage. Taken together, despite the instability of the protein encoded by CCNG1, it may still serve as a biomarker for evaluating the prognosis of breast cancer and as a therapeutic target in certain subtypes of breast cancer.
As an estrogen-regulated gene, CCNG2 has previously been used as a target gene for judging the efficacy of endocrine-based medicine, as low CCNG2 mRNA expression is indicative of a poorer prognosis. In the present study, CCNG2 mRNA expression in breast cancer tissues was higher than that in normal tissues although the difference was not statistically significant. RT-qPCR analysis showed that CCNG2 mRNA expression was higher in MCF-7 cells and lower in MDA-MB-231 and BT-549 cells, and patients with high transcriptional expression of CCNG2 had a better OS. Consistent with a previous study (57), CCNG2 expression was downregulated in highly aggressive breast cancer subtypes with poor prognoses. Consequently, CCNG2 may exhibit potential for the prognostic prediction of ER-positive breast cancer and as a therapeutic target. However, additional studies on CCNG2 in other subtypes of breast cancer are required to examine its value as a biomarker.
Studies on the expression or the function of CCNH in breast cancer are limited. Shahi et al (97) performed whole-exome sequencing of 492 cancer-related genes as well as high-risk genes and mutation analysis of BRCA1 and BRCA2 negative breast cancer patients. In total, 31 gene variants/genes were identified for breast cancer susceptibility predisposition including CCNH. However, CCNH has not been extensively studied with regard to familial breast cancer. In the present study, CCNH expression was low in breast cancer, and upregulated expression was predictive of a better OS. It has been indirectly shown that the increased expression of CCNH in ER-positive breast cancer was associated with indicators of good prognosis (98). Yet, RT-qPCR analysis of breast cancer cell lines showed higher CCNH mRNA expression than that in MCF-10A. At present, there is currently no sufficient evidence to explain this phenomenon. Thus, further studies are required to determine the value of CCNH as a biomarker for evaluating the prognosis and exploring the mechanisms of drug action.
In conclusion, we systematically analyzed the expression of the cyclin genes in breast cancer based on its value in predicting prognosis and evaluating drug efficacy, enabling a deeper understanding of the heterogeneity of breast cancer. Our results indicated that CCNA2, CCNB1, CCNB2, CCND1, CCND2, CCNE1, and CCNE2 may be mature and valid biomarkers in predicting prognosis and judging efficacy. CCNG1 and CCNG2 may be used as biological markers in specific types of breast cancer. At present, there are few relevant studies on CCNA1, CCNB3, CCNC, CCND3, CCNF, and CCNH in breast cancer; however, the results of the present and previous studies showed their potential as unique predictors of prognosis and potential targets for countering therapeutic resistance. Together, the discovery of novel biomarkers and gene targets will facilitate the exploration of more personalized therapeutic strategies; however, further studies are required to verify their biological functions.
Acknowledgements
Not applicable.
Funding Statement
This study was supported by funding from the National Nature Science Foundation of China, The Third Affiliated Hospital of Kunming Medical University: Yunnan Cancer Center, Kunming, Yunnan, China (grant nos. 81760480 and 81960479).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
NQL, WHC and JN confirm the authenticity of all the raw data. NQL participated in the design of study, and was responsible for collecting and interpreting data, performing the bioinformatics analysis, visualizing the presentation, and drafting and revising the original manuscript. WHC collected the data, performed the bioinformatics analysis and drafted the original manuscript. XW and JC participated in data curation, confirmed and analyzed the data, and assisted in drafting the revised manuscript. JN mainly provided the research conceptualization and funding support, supervised and assisted in designing the study, and participated in the revision of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cai Z, Liu Q. Cell cycle regulation in treatment of breast cancer. Adv Exp Med Biol. 2017;1026:251–270. doi: 10.1007/978-981-10-6020-5_12. [DOI] [PubMed] [Google Scholar]
- 2.Steeg PS, Zhou Q. Cyclins and breast cancer. Breast Cancer Res Treat. 1998;52:17–28. doi: 10.1023/A:1006102916060. [DOI] [PubMed] [Google Scholar]
- 3.Bai C, Richman R, Elledge SJ. Human cyclin F. EMBO J. 1994;13:6087–6098. doi: 10.1002/j.1460-2075.1994.tb06955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye C, Wang J, Wu P, Li X, Chai Y. Prognostic role of cyclin B1 in solid tumors: A meta-analysis. Oncotarget. 2017;8:2224–2232. doi: 10.18632/oncotarget.13653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee YS, Ryu SW, Bae SJ, Park TH, Kwon K, Noh YH, Kim SY. Cross-platform meta-analysis of multiple gene expression profiles identifies novel expression signatures in acquired anthracycline-resistant breast cancer. Oncol Rep. 2015;33:1985–1993. doi: 10.3892/or.2015.3810. [DOI] [PubMed] [Google Scholar]
- 6.Miftakhova R, Hedblom A, Semenas J, Robinson B, Simoulis A, Malm J, Rizvanov A, Heery DM, Mongan NP, Maitland NJ, et al. Cyclin A1 and P450 aromatase promote metastatic homing and growth of stem-like prostate cancer cells in the bone marrow. Cancer Res. 2016;76:2453–2464. doi: 10.1158/0008-5472.CAN-15-2340. [DOI] [PubMed] [Google Scholar]
- 7.Li R, Jiang X, Zhang Y, Wang S, Chen X, Yu X, Ma J, Huang X. Cyclin B2 overexpression in human hepatocellular carcinoma is associated with poor prognosis. Arch Med Res. 2019;50:10–17. doi: 10.1016/j.arcmed.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Dorn J, Spatz H, Schmieder M, Barth TF, Blatz A, Henne-Bruns D, Knippschild U, Kramer K. Cyclin H expression is increased in GIST with very-high risk of malignancy. BMC Cancer. 2010;10:350. doi: 10.1186/1471-2407-10-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang KC, Yang J, Ng MC, Ng SK, Welch WR, Muto MG, Berkowitz RS, Ng SW. Cyclin A1 expression and paclitaxel resistance in human ovarian cancer cells. Eur J Cancer. 2016;67:152–163. doi: 10.1016/j.ejca.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chujan S, Kitkumthorn N, Siriangkul S, Mutirangura A. CCNA1 promoter methylation: A potential marker for grading Papanicolaou smear cervical squamous intraepithelial lesions. Asian Pac J Cancer Prev. 2014;15:7971–7975. doi: 10.7314/APJCP.2014.15.18.7971. [DOI] [PubMed] [Google Scholar]
- 11.Takashima S, Saito H, Takahashi N, Imai K, Kudo S, Atari M, Saito Y, Motoyama S, Minamiya Y. Strong expression of cyclin B2 mRNA correlates with a poor prognosis in patients with non-small cell lung cancer. Tumour Biol. 2014;35:4257–4265. doi: 10.1007/s13277-013-1556-7. [DOI] [PubMed] [Google Scholar]
- 12.Wiseman M. The second world cancer research fund/American institute for cancer research expert report. Food, nutrition, physical activity, and the prevention of cancer: A global perspective. Proc Nutr Soc. 2008;67:253–256. doi: 10.1017/S002966510800712X. [DOI] [PubMed] [Google Scholar]
- 13.Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389:1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 14.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 15.Zeng X, Liu C, Yao J, Wan H, Wan G, Li Y, Chen N. Breast cancer stem cells, heterogeneity, targeting therapies and therapeutic implications. Pharmacol Res. 2021;163:105320. doi: 10.1016/j.phrs.2020.105320. [DOI] [PubMed] [Google Scholar]
- 16.Ochsenreither S, Majeti R, Schmitt T, Stirewalt D, Keilholz U, Loeb KR, Wood B, Choi YE, Bleakley M, Warren EH, et al. Cyclin-A1 represents a new immunogenic targetable antigen expressed in acute myeloid leukemia stem cells with characteristics of a cancer-testis antigen. Blood. 2012;119:5492–5501. doi: 10.1182/blood-2011-07-365890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang R, Nakamaki T, Lübbert M, Said J, Sakashita A, Freyaldenhoven BS, Spira S, Huynh V, Müller C, Koeffler HP. Cyclin A1 expression in leukemia and normal hematopoietic cells. Blood. 1999;93:2067–2074. [PubMed] [Google Scholar]
- 18.Gao T, Han Y, Yu L, Ao S, Li Z, Ji J. CCNA2 is a prognostic biomarker for ER+ breast cancer and tamoxifen resistance. PLoS One. 2014;9:e91771. doi: 10.1371/journal.pone.0091771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hein JB, Nilsson J. Interphase APC/C-Cdc20 inhibition by cyclin A2-Cdk2 ensures efficient mitotic entry. Nat Commun. 2016;7:10975. doi: 10.1038/ncomms10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pei J, Zhang J, Yang X, Wu Z, Sun C, Wang Z, Wang B. NEK5 promotes breast cancer cell proliferation through up-regulation of cyclin A2. Mol Carcinog. 2019;58:933–943. doi: 10.1002/mc.22982. [DOI] [PubMed] [Google Scholar]
- 21.Ding K, Li W, Zou Z, Zou X, Wang C. CCNB1 is a prognostic biomarker for ER+ breast cancer. Med Hypotheses. 2014;83:359–564. doi: 10.1016/j.mehy.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 22.Liu HY, Liu YY, Yang F, Zhang L, Zhang FL, Hu X, Shao ZM, Li DQ. Acetylation of MORC2 by NAT10 regulates cell-cycle checkpoint control and resistance to DNA-damaging chemotherapy and radiotherapy in breast cancer. Nucleic Acids Res. 2020;48:3638–3656. doi: 10.1093/nar/gkaa130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu AQ, Wang ZX, Wu W, Chen KY, Yan SR, Mao ZB. Circular RNA CircCCNB1 sponges micro RNA-449a to inhibit cellular senescence by targeting CCNE2. Aging (Albany NY) 2019;11:10220–10241. doi: 10.18632/aging.102449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shubbar E, Kovács A, Hajizadeh S, Parris TZ, Nemes S, Gunnarsdóttir K, Einbeigi Z, Karlsson P, Helou K. Elevated cyclin B2 expression in invasive breast carcinoma is associated with unfavorable clinical outcome. BMC Cancer. 2013;13:1. doi: 10.1186/1471-2407-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qian X, Song X, He Y, Yang Z, Sun T, Wang J, Zhu G, Xing W, You C. CCNB2 overexpression is a poor prognostic biomarker in Chinese NSCLC patients. Biomed Pharmacother. 2015;74:222–227. doi: 10.1016/j.biopha.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 26.Gao Z, Man X, Li Z, Bi J, Liu X, Li Z, Zhu Y, Zhang Z, Kong C. Expression profiles analysis identifies the values of carcinogenesis and the prognostic prediction of three genes in adrenocortical carcinoma. Oncol Rep. 2019;41:2440–2452. doi: 10.3892/or.2019.7021. [DOI] [PubMed] [Google Scholar]
- 27.Lozano JC, Perret E, Schatt P, Arnould C, Peaucellier G, Picard A. Molecular cloning, gene localization, and structure of human cyclin B3. Biochem Biophys Res Commun. 2002;291:406–413. doi: 10.1006/bbrc.2002.6458. [DOI] [PubMed] [Google Scholar]
- 28.Han H, Bertrand KC, Patel KR, Fisher KE, Roy A, Muscal JA, Venkatramani R. BCOR-CCNB3 fusion-positive clear cell sarcoma of the kidney. Pediatr Blood Cancer. 2020;67:e28151. doi: 10.1002/pbc.28151. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida A, Arai Y, Hama N, Chikuta H, Bando Y, Nakano S, Kobayashi E, Shibahara J, Fukuhara H, Komiyama M, et al. Expanding the clinicopathologic and molecular spectrum of BCOR-associated sarcomas in adults. Histopathology. 2020;76:509–520. doi: 10.1111/his.14023. [DOI] [PubMed] [Google Scholar]
- 30.Shibayama T, Okamoto T, Nakashima Y, Kato T, Sakurai T, Minamiguchi S, Kataoka TR, Shibuya S, Yoshizawa A, Toguchida J, Haga H. Screening of BCOR-CCNB3 sarcoma using immunohistochemistry for CCNB3: A clinicopathological report of three pediatric cases. Pathol Int. 2015;65:410–414. doi: 10.1111/pin.12319. [DOI] [PubMed] [Google Scholar]
- 31.Ren S, Rollins BJ. Cyclin C/cdk3 promotes Rb-dependent G0 exit. Cell. 2004;117:239–251. doi: 10.1016/S0092-8674(04)00300-9. [DOI] [PubMed] [Google Scholar]
- 32.Miyata Y, Liu Y, Jankovic V, Sashida G, Lee JM, Shieh JH, Naoe T, Moore M, Nimer SD. Cyclin C regulates human hematopoietic stem/progenitor cell quiescence. Stem Cells. 2010;28:308–317. doi: 10.1002/stem.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu W, Ji JY. Dysregulation of CDK8 and cyclin C in tumorigenesis. J Genet Genomics. 2011;38:439–452. doi: 10.1016/j.jgg.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu YN, Yip GW, Tan PH, Thike AA, Matsumoto K, Tsujimoto M, Bay BH. Y-box binding protein 1 is up-regulated in proliferative breast cancer and its inhibition deregulates the cell cycle. Int J Oncol. 2010;37:483–492. [PubMed] [Google Scholar]
- 35.Roy PG, Thompson AM. Cyclin D1 and breast cancer. Breast. 2006;15:718–127. doi: 10.1016/j.breast.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Elsheikh S, Green AR, Aleskandarany MA, Grainge M, Paish CE, Lambros MB, Reis-Filho JS, Ellis IO. CCND1 amplification and cyclin D1 expression in breast cancer and their relation with proteomic subgroups and patient outcome. Breast Cancer Res Treat. 2008;109:325–335. doi: 10.1007/s10549-007-9659-8. [DOI] [PubMed] [Google Scholar]
- 37.He Q, Wu J, Liu XL, Ma YH, Wu XT, Wang WY, An HX. Clinicopathological and prognostic significance of cyclin D1 amplification in patients with breast cancer: A meta-analysis. J BUON. 2017;22:1209–1216. [PubMed] [Google Scholar]
- 38.Villegas SL, Darb-Esfahani S, von Minckwitz G, Huober J, Weber K, Marmé F, Furlanetto J, Schem C, Pfitzner BM, Lederer B, et al. Expression of cyclin D1 protein in residual tumor after neoadjuvant chemotherapy for breast cancer. Breast Cancer Res Treat. 2018;168:179–187. doi: 10.1007/s10549-017-4581-1. [DOI] [PubMed] [Google Scholar]
- 39.Shi Q, Li Y, Li S, Jin L, Lai H, Wu Y, Cai Z, Zhu M, Li Q, Li Y, et al. LncRNA DILA1 inhibits cyclin D1 degradation and contributes to tamoxifen resistance in breast cancer. Nat Commun. 2020;11:5513. doi: 10.1038/s41467-020-19349-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pors J, Naso J, Berg K, Churg A. Cyclin D1 immunohistochemical staining to separate benign from malignant mesothelial proliferations. Mod Pathol. 2020;33:312–318. doi: 10.1038/s41379-019-0411-9. [DOI] [PubMed] [Google Scholar]
- 41.Kwapisz D. Cyclin-dependent kinase 4/6 inhibitors in breast cancer: Palbociclib, ribociclib, and abemaciclib. Breast Cancer Res Treat. 2017;166:41–54. doi: 10.1007/s10549-017-4385-3. [DOI] [PubMed] [Google Scholar]
- 42.Hung CS, Wang SC, Yen YT, Lee TH, Wen WC, Lin RK. Hypermethylation of CCND2 in lung and breast cancer is a potential biomarker and drug target. Int J Mol Sci. 2018;19:3096. doi: 10.3390/ijms19103096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Callahan CL, Wang Y, Marian C, Weng DY, Eng KH, Tao MH, Ambrosone CB, Nie J, Trevisan M, Smiraglia D, et al. DNA methylation and breast tumor clinicopathological features: The western New York exposures and breast cancer (WEB) study. Epigenetics. 2016;11:643–652. doi: 10.1080/15592294.2016.1192735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding ZY, Li R, Zhang QJ, Wang Y, Jiang Y, Meng QY, Xi QL, Wu GH. Prognostic role of cyclin D2/D3 in multiple human malignant neoplasms: A systematic review and meta-analysis. Cancer Med. 2019;8:2717–2729. doi: 10.1002/cam4.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luhtala S, Staff S, Tanner M, Isola J. Cyclin E amplification, over-expression, and relapse-free survival in HER-2-positive primary breast cancer. Tumour Biol. 2016;37:9813–9823. doi: 10.1007/s13277-016-4870-z. [DOI] [PubMed] [Google Scholar]
- 46.Keyomarsi K, Tucker SL, Buchholz TA, Callister M, Ding Y, Hortobagyi GN, Bedrosian I, Knickerbocker C, Toyofuku W, Lowe M, et al. Cyclin E and survival in patients with breast cancer. N Engl J Med. 2002;347:1566–1575. doi: 10.1056/NEJMoa021153. [DOI] [PubMed] [Google Scholar]
- 47.Lee C, Fernandez KJ, Alexandrou S, Sergio CM, Deng N, Rogers S, Burgess A, Caldon CE. Cyclin E2 promotes whole genome doubling in breast cancer. Cancers (Basel) 2020;12:2268. doi: 10.3390/cancers12082268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peek GW, Tollefsbol TO. Combinatorial PX-866 and raloxifene decrease Rb phosphorylation, cyclin E2 transcription, and proliferation of MCF-7 breast cancer cells. J Cell Biochem. 2016;117:1688–1696. doi: 10.1002/jcb.25462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindskog C. The potential clinical impact of the tissue-based map of the human proteome. Expert Rev Proteomics. 2015;12:213–215. doi: 10.1586/14789450.2015.1040771. [DOI] [PubMed] [Google Scholar]
- 50.Fu J, Qiu H, Cai M, Pan Y, Cao Y, Liu L, Yun J, Zhang CZ. Low cyclin F expression in hepatocellular carcinoma associates with poor differentiation and unfavorable prognosis. Cancer Sci. 2013;104:508–515. doi: 10.1111/cas.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao L, Jiang L, He L, Wei Q, Bi J, Wang Y, Yu L, He M, Zhao L, Wei M. Identification of a novel cell cycle-related gene signature predicting survival in patients with gastric cancer. J Cell Physiol. 2019;234:6350–6360. doi: 10.1002/jcp.27365. [DOI] [PubMed] [Google Scholar]
- 52.Noh JM, Kim J, Cho DY, Choi DH, Park W, Huh SJ. Exome sequencing in a breast cancer family without BRCA mutation. Radiat Oncol J. 2015;33:149–154. doi: 10.3857/roj.2015.33.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamura K, Kanaoka Y, Jinno S, Nagata A, Ogiso Y, Shimizu K, Hayakawa T, Nojima H, Okayama H. Cyclin G: A new mammalian cyclin with homology to fission yeast Cig1. Oncogene. 1993;8:2113–2118. [PubMed] [Google Scholar]
- 54.Zimmermann M, Arachchige-Don AP, Donaldson MS, Patriarchi T, Horne MC. Cyclin G2 promotes cell cycle arrest in breast cancer cells responding to fulvestrant and metformin and correlates with patient survival. Cell Cycle. 2016;15:3278–3295. doi: 10.1080/15384101.2016.1243189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu D, Han B, Guo L, Fan Z. Molecular mechanisms associated with breast cancer based on integrated gene expression profiling by bioinformatics analysis. J Obstet Gynaecol. 2016;36:615–621. doi: 10.3109/01443615.2015.1127902. [DOI] [PubMed] [Google Scholar]
- 56.Knowles LM, Smith JW. Genome-wide changes accompanying knockdown of fatty acid synthase in breast cancer. BMC Genomics. 2007;8:168. doi: 10.1186/1471-2164-8-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu ET, Bergh J. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mao L, Ling X, Chen J. Cyclin H regulates lung cancer progression as a carcinoma inducer. Comput Math Methods Med. 2021;2021:6646077. doi: 10.1155/2021/6646077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 60.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45((W1)):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagy Á, Munkácsy G, Győrffy B. Pancancer survival analysis of cancer hallmark genes. Sci Rep. 2021;11:6047. doi: 10.1038/s41598-021-84787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173:291–304.e6. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 65.R Core Team. R Foundation for Statistical Computing; Vienna: 2012. R: A language and environment for statistical computing. [Google Scholar]
- 66.RStudio Team, corp-author. Boston, MA: 2015. RStudio: Integrated Development for R. RStudio Inc. [Google Scholar]
- 67.Tang T, Guo C, Xia T, Zhang R, Zen K, Pan Y, Jin L. LncCCAT1 promotes breast cancer stem cell function through activating WNT/β-catenin signaling. Theranostics. 2019;9:7384–7402. doi: 10.7150/thno.37892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shao F, Pang X, Baeg GH. Targeting the JAK/STAT signaling pathway for breast cancer. Curr Med Chem. 2021;28:5137–5151. doi: 10.2174/0929867328666201207202012. [DOI] [PubMed] [Google Scholar]
- 69.Costa RLB, Han HS, Gradishar WJ. Targeting the PI3K/AKT/mTOR pathway in triple-negative breast cancer: A review. Breast Cancer Res Treat. 2018;169:397–406. doi: 10.1007/s10549-018-4697-y. [DOI] [PubMed] [Google Scholar]
- 70.Li X, Zeng Z, Wang J, Wu Y, Chen W, Zheng L, Xi T, Wang A, Lu Y. MicroRNA-9 and breast cancer. Biomed Pharmacother. 2020;122:109687. doi: 10.1016/j.biopha.2019.109687. [DOI] [PubMed] [Google Scholar]
- 71.Qiao K, Ning S, Wan L, Wu H, Wang Q, Zhang X, Xu S, Pang D. LINC00673 is activated by YY1 and promotes the proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo signaling pathway. J Exp Clin Cancer Res. 2019;38:418. doi: 10.1186/s13046-019-1421-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee MG, Kwon YS, Nam KS, Kim SY, Hwang IH, Kim S, Jang H. Chaga mushroom extract induces autophagy via the AMPK-mTOR signaling pathway in breast cancer cells. J Ethnopharmacol. 2021;274:114081. doi: 10.1016/j.jep.2021.114081. [DOI] [PubMed] [Google Scholar]
- 73.Woo SH, Seo SK, An S, Choe TB, Hong SI, Lee YH, Park IC. Implications of caspase-dependent proteolytic cleavage of cyclin A1 in DNA damage-induced cell death. Biochem Biophys Res Commun. 2014;453:438–442. doi: 10.1016/j.bbrc.2014.09.104. [DOI] [PubMed] [Google Scholar]
- 74.Klajic J, Busato F, Edvardsen H, Touleimat N, Fleischer T, Bukholm I, Børresen-Dale AL, Lønning PE, Tost J, Kristensen VN. DNA methylation status of key cell-cycle regulators such as CDKNA2/p16 and CCNA1 correlates with treatment response to doxorubicin and 5-fluorouracil in locally advanced breast tumors. Clin Cancer Res. 2014;20:6357–6366. doi: 10.1158/1078-0432.CCR-14-0297. [DOI] [PubMed] [Google Scholar]
- 75.Deng JL, Xu YH, Wang G. Identification of potential crucial genes and key pathways in breast cancer using bioinformatic analysis. Front Genet. 2019;10:695. doi: 10.3389/fgene.2019.00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tang J, Kong D, Cui Q, Wang K, Zhang D, Gong Y, Wu G. Prognostic genes of breast cancer identified by gene co-expression network analysis. Front Oncol. 2018;8:374. doi: 10.3389/fonc.2018.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jayanthi VSPKSA, Das AB, Saxena U. Grade-specific diagnostic and prognostic biomarkers in breast cancer. Genomics. 2020;112:388–396. doi: 10.1016/j.ygeno.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 78.Zhou H, Lv Q, Guo Z. Transcriptomic signature predicts the distant relapse in patients with ER+ breast cancer treated with tamoxifen for five years. Mol Med Rep. 2018;17:3152–3157. doi: 10.3892/mmr.2017.8234. [DOI] [PubMed] [Google Scholar]
- 79.Li N, Fassl A, Chick J, Inuzuka H, Li X, Mansour MR, Liu L, Wang H, King B, Shaik S, et al. Cyclin C is a haploinsufficient tumour suppressor. Nat Cell Biol. 2014;16:1080–1091. doi: 10.1038/ncb3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bozickovic O, Hoang T, Fenne IS, Helland T, Skartveit L, Ouchida M, Mellgren G, Sagen JV. Cyclin C interacts with steroid receptor coactivator 2 and upregulates cell cycle genes in MCF-7 cells. Biochim Biophys Acta. 2015;1853:2383–2391. doi: 10.1016/j.bbamcr.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 81.Kurebayashi J, Otsuki T, Kunisue H, Tanaka K, Yamamoto S, Sonoo H. Expression levels of estrogen receptor-alpha, estrogen receptor-beta, coactivators, and corepressors in breast cancer. Clin Cancer Res. 2000;6:512–518. [PubMed] [Google Scholar]
- 82.Butt AJ, McNeil CM, Musgrove EA, Sutherland RL. Downstream targets of growth factor and oestrogen signalling and endocrine resistance: The potential roles of c-Myc, cyclin D1 and cyclin E. Endocr Relat Cancer. 2005;12((Suppl 1)):S47–S59. doi: 10.1677/erc.1.00993. [DOI] [PubMed] [Google Scholar]
- 83.Filipits M, Dafni U, Gnant M, Polydoropoulou V, Hills M, Kiermaier A, de Azambuja E, Larsimont D, Rojo F, Viale G, et al. Association of p27 and cyclin D1 expression and benefit from adjuvant trastuzumab treatment in HER2-positive early breast cancer: A TransHERA study. Clin Cancer Res. 2018;24:3079–3086. doi: 10.1158/1078-0432.CCR-17-3473. [DOI] [PubMed] [Google Scholar]
- 84.Tobin NP, Sims AH, Lundgren KL, Lehn S, Landberg G. Cyclin D1, Id1 and EMT in breast cancer. BMC Cancer. 2011;11:417. doi: 10.1186/1471-2407-11-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fischer H, Chen J, Skoog L, Lindblom A. Cyclin D2 expression in familial and sporadic breast cancer. Oncol Rep. 2002;9:1157–1161. [PubMed] [Google Scholar]
- 86.Li Z, Heng J, Yan J, Guo X, Tang L, Chen M, Peng L, Wu Y, Wang S, Xiao Z, et al. Integrated analysis of gene expression and methylation profiles of 48 candidate genes in breast cancer patients. Breast Cancer Res Treat. 2016;160:371–383. doi: 10.1007/s10549-016-4004-8. [DOI] [PubMed] [Google Scholar]
- 87.Justenhoven C, Pierl CB, Haas S, Fischer HP, Hamann U, Baisch C, Harth V, Spickenheuer A, Rabstein S, Vollmert C, et al. Polymorphic loci of E2F2, CCND1 and CCND3 are associated with HER2 status of breast tumors. Int J Cancer. 2009;124:2077–2081. doi: 10.1002/ijc.24198. [DOI] [PubMed] [Google Scholar]
- 88.Turner NC, Liu Y, Zhu Z, Loi S, Colleoni M, Loibl S, DeMichele A, Harbeck N, André F, Bayar MA, et al. Cyclin E1 expression and palbociclib efficacy in previously treated hormone receptor-positive metastatic breast cancer. J Clin Oncol. 2019;37:1169–1178. doi: 10.1200/JCO.18.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao ZM, Yost SE, Hutchinson KE, Li SM, Yuan YC, Noorbakhsh J, Liu Z, Warden C, Johnson RM, Wu X, et al. CCNE1 amplification is associated with poor prognosis in patients with triple negative breast cancer. BMC Cancer. 2019;19:96. doi: 10.1186/s12885-019-5290-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sieuwerts AM, Look MP, Meijer-van Gelder ME, Timmermans M, Trapman AM, Garcia RR, Arnold M, Goedheer AJ, de Weerd V, Portengen H, et al. Which cyclin E prevails as prognostic marker for breast cancer? Results from a retrospective study involving 635 lymph node-negative breast cancer patients. Clin Cancer Res. 2006;12:3319–3328. doi: 10.1158/1078-0432.CCR-06-0225. [DOI] [PubMed] [Google Scholar]
- 91.Seyhan AA, Varadarajan U, Choe S, Liu W, Ryan TE. A genome-wide RNAi screen identifies novel targets of neratinib resistance leading to identification of potential drug resistant genetic markers. Mol Biosyst. 2012;8:1553–1570. doi: 10.1039/c2mb05512k. [DOI] [PubMed] [Google Scholar]
- 92.Gupta ED, Pachauri M, Ghosh PC, Rajam MV. Targeting polyamine biosynthetic pathway through RNAi causes the abrogation of MCF 7 breast cancer cell line. Tumour Biol. 2016;37:1159–1171. doi: 10.1007/s13277-015-3912-2. [DOI] [PubMed] [Google Scholar]
- 93.Wang X, Zhang T, Zhang S, Shan J. Prognostic values of F-box members in breast cancer: An online database analysis and literature review. Biosci Rep. 2019;39:BSR20180949. doi: 10.1042/BSR20180949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Piscopo DM, Hinds PW. A role for the cyclin box in the ubiquitin-mediated degradation of cyclin G1. Cancer Res. 2008;68:5581–5590. doi: 10.1158/0008-5472.CAN-07-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu F, Gao X, Yu H, Yuan D, Zhang J, He Y, Yue L. Effects of expression of exogenous cyclin G1 on proliferation of human endometrial carcinoma cells. Chin J Physiol. 2013;56:83–89. doi: 10.4077/CJP.2013.BAA079. [DOI] [PubMed] [Google Scholar]
- 96.Tian JM, Ran B, Zhang CL, Yan DM, Li XH. Estrogen and progesterone promote breast cancer cell proliferation by inducing cyclin G1 expression. Braz J Med Biol Res. 2018;51:1–7. doi: 10.1590/1414-431x20175612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shahi RB, De Brakeleer S, Caljon B, Pauwels I, Bonduelle M, Joris S, Fontaine C, Vanhoeij M, Van Dooren S, Teugels E, De Grève J. Identification of candidate cancer predisposing variants by performing whole-exome sequencing on index patients from BRCA1 and BRCA2-negative breast cancer families. BMC Cancer. 2019;19:313. doi: 10.1186/s12885-019-5494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Patel H, Abduljabbar R, Lai CF, Periyasamy M, Harrod A, Gemma C, Steel JH, Patel N, Busonero C, Jerjees D, et al. Expression of CDK7, cyclin H, and MAT1 is elevated in breast cancer and is prognostic in estrogen receptor-positive breast cancer. Clin Cancer Res. 2016;22:5929–5938. doi: 10.1158/1078-0432.CCR-15-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.