Abstract
A yeast glc7-1 mutant expressing a variant of protein phosphatase type 1 fails to accumulate glycogen. This defect is associated with hyperphosphorylated and inactive glycogen synthase, consistent with Glc7p acting directly to dephosphorylate and activate glycogen synthase. To characterize the glycogen synthesis defect of this mutant in more detail, we isolated 26 pseudorevertants of the glc7-1 mutant. All pseudoreversion events were due to missense mutations in GSY2, the gene encoding the major isoform of glycogen synthase. A majority of the mutations responsible for the suppression were in the 3′ end of the gene, corresponding to the phosphorylated COOH terminus of Gsy2p. Phosphorylation of the mutant proteins was reduced, suggesting that they are poor substrates for glycogen synthase kinases. Suppressor mutations outside this domain did not decrease the phosphorylation of the resulting proteins, indicating that these proteins are immune to the regulatory effects of phosphorylation. Since no growth defect has been observed for strains with altered glycogen levels, the relative levels of fitness of GSY2 mutants that fail to accumulate glycogen and that hyperaccumulate glycogen were assayed by cocultivation experiments. A wild-type strain outcompeted both hypo- and hyperaccumulating strains, suggesting that glycogen levels contribute substantially to the fitness of yeast.
The wide distribution of the storage carbohydrate glycogen in nature suggests that it plays important physiological roles. Indeed, genetic, biochemical, and physiological analyses of mammals indicate that glycogen metabolism plays an essential role in energy homeostasis. As one might expect, the synthesis and degradation of glycogen are regulated by multiple signaling pathways. Glycogen also accumulates in fungi; in the budding yeast Saccharomyces cerevisiae, where the pathway has been studied in detail, the metabolism of glycogen is remarkably similar to that in mammals. Most of the enzymatic components (glycogenin, glycogen synthase, phosphorylase, and branching enzyme) are conserved in yeast and mammals (5, 7, 8, 22, 31, 37), but the signaling pathways that regulate glycogen synthase have diverged. In mammals, glycogen metabolism is under hormonal control, whereas in yeast, it is controlled largely by nutrients. Yeast cells normally accumulate glycogen near the end of log-phase growth, due to a decrease in the levels of nutrients such as glucose, phosphate, nitrogen, and sulfate (23). This accumulation is due in part to the increased expression of genes whose products act in the pathway (15, 24, 25, 31) and in part to post translational regulatory mechanisms (10, 16, 21, 27, 30). At least for glucose, the accumulation of glycogen does not seem dependent on the absolute amount present; rather, glycogen accumulates when approximately half the glucose is consumed (23). Although many of the key regulators have been identified, the basic mechanism that controls this accumulation is unknown.
Glycogen synthase is regulated at the level of gene expression and by posttranscriptional mechanisms. mRNA levels for the major isoform of glycogen synthase (GSY2 mRNA) increase as cells approach stationary phase when grown in glucose-containing medium (15, 24, 25). Gsy2p is also phosphorylated and negatively regulated by phosphorylation at its COOH terminus (16). Glycogen synthase can be fully activated by glucose-6-phosphate (G6P), even in the fully phosphorylated state (26). Therefore, the ratio of glycogen synthase activities assayed in the absence and presence of G6P (activity ratio) provides a convenient means of assessing the phosphorylation state of the protein. Mammalian glycogen synthase is phosphorylated at its NH2 and COOH termini (28). In contrast, yeast glycogen synthase is phosphorylated only at its COOH terminus. A deletion variant of Gsy2p lacking amino acid residues after amino acid (aa) 643 has a very high activity ratio and is not phosphorylated in vivo (16). Other Gsy2p variants that contain alanine substitutions for Ser-650 (S650A), Ser-654 (S654A), or Thr-667 (T667A) also have high activity ratios, suggesting that these residues are the major sites of phosphorylation (16).
Biochemical characterization of glycogen synthase kinase in yeast has revealed at least two distinct activities (18), one of which is cyclin-dependent kinase Pho85p activity. Null mutations in the genes encoding the catalytic subunit, PHO85 (18, 38), or two of its many cyclin subunits, PCL8 and PCL10 (19), result in the hyperaccumulation of glycogen. Pho85p-Pcl10p can phosphorylate Gsy2p in vitro (19, 39). However, Gsy2p variants that lack Ser-654 and Thr-667 are no longer Pho85p-Pcl10p substrates, suggesting that Pho85p phosphorylates Gsy2p at these two sites while another, as-yet-unidentified kinase phosphorylates Gsy2p at Ser-650. Further evidence indicates that at least one other kinase is involved in down-regulating Gsy2p. Yang et al. (40) have recently found that a block to respiration inhibits glycogen accumulation by increasing the phosphorylation state of glycogen synthase. The activity ratio of glycogen synthase is very low in respiration-deficient mutants, and GSY2 mutants that lack any one of the three key phosphorylation sites (Ser-650, Ser-654, or Thr-667) accumulate glycogen in a respiration-deficient background. However, the loss of Pho85 kinase does not restore glycogen accumulation to respiration-deficient cells, indicating that another protein kinase is responsible for the effect.
On the reverse side of the regulatory pathway, protein phosphatase type 1 is responsible for dephosphorylating and activating glycogen synthase. Some mutations in GLC7, which encodes the catalytic subunit of protein phosphatase type 1, result in a failure to accumulate glycogen due to retention of glycogen synthase in the inactive, phosphorylated form (11, 27). The phosphatase directly responsible for activating glycogen synthase is thought to be composed of Glc7p and the regulatory subunit Gac1p. Gac1p binds to both Glc7p and glycogen synthase through separate domains and appears to act as a scaffold to target the substrate to the phosphatase (X. Wu, H. Hart, C. Cheng, P. J. Roach, and K. Tatchell, submitted for publication). The related protein Pig1p, first identified in a screen for glycogen synthase binding proteins, may have a role similar to that of Gac1p. The low levels of glycogen in a gac1 mutant are reduced further by deletion of PIG1 (4). The glycogen deficiency of glc7-1 or gac1 mutants can be suppressed either by elimination of the phosphorylation sites in Gsy2p (16) or by inactivation of Pho85p-Pcl8p and Pho85p-Pcl10p kinase (19), furthering the idea that the Glc7-Gac1p phosphatase reverses the phosphorylation and inactivation of Gsy2p by Pho85p-Pcl8p or Pho85p-Pcl10p.
To further the understanding of glycogen metabolism, we have isolated revertants of glc7-1 (glycogen defect) mutants. All mutants isolated in this screen are defective in the major isoform of glycogen synthase, Gsy2p, and all suppress the glc7-1 mutation by increasing the activity of glycogen synthase. The mutations in some suppressors are located in the region known to encode the phosphorylated COOH terminus. However, other mutants alter portions of the protein not previously known to have a regulatory role. In addition, we demonstrate for the first time the importance of glycogen accumulation by showing that levels of glycogen play an important role in overall fitness. Strains that accumulate too much or too little glycogen are rapidly outcompeted by wild-type strains.
MATERIALS AND METHODS
Strains and media.
Crosses were performed and analyzed by standard yeast genetic practices (14). Petite strains were generated by treatment with ethidium bromide as described previously (14). Hyperactive GSY2 mutants were crossed to strains containing snf1, bcy1, or gac1 mutations (Table 1). The resultant diploids were allowed to sporulate, and double mutants were identified by tetrad analysis. Isolates were confirmed by backcrossing to the parental strain and examining meiotic progeny from this cross. Strains CV182 and CV218 were derived from a strain kindly provided by Peter Roach (8) by three serial backcrosses to KT1113. All other strains were derivatives of JC482 (3). Ethyl methanesulfonate mutagenesis of strain KT1118 was performed as described previously (14). Since spontaneous suppressors were infrequent, cells were mutagenized to approximately 99% killing and plated on either 1% yeast extract–2% peptone–2% dextrose medium (YPD) or synthetic medium (2% glucose and 0.67% yeast nitrogen base without amino acids, supplemented with required amino acids) at 30 °C for screening. All strains accumulated more glycogen on synthetic medium, and all subsequent screens were performed with this medium. Suppression of the glc7-1 glycogen defect was scored qualitatively by inverting freshly grown plates over iodine crystals for 30 to 60 s. Increased glycogen accumulation results in darker brown staining. Putative suppressor mutants were backcrossed three times to the parental strain (glc7-1) and retested for glycogen accumulation. Mutants with suppressors that segregated in a Mendelian fashion were then backcrossed three times to the wild-type strain (KT1113). These suppressor mutants in the wild-type strain background were crossed to a gsy2::HIS3 strain (CV218), and suppressors were scored for linkage of the hyperaccumulation of glycogen to HIS3. All suppressors were linked, indicating that the suppressors were located in or near GSY2.
TABLE 1.
Yeast strains
| Strain | Genotypea | Source or reference |
|---|---|---|
| CV73 | MATaura3 leu2 | This study |
| CV74 | MATα ura3 leu2 glc7-1 | This study |
| CV75 | MATaura3 leu2 glc7-1 | This study |
| KT1112 | MATaura3 leu2 his3 | 35 |
| KT1113 | MATα ura3 leu2 his3 | 12 |
| KT1124 | MATaura3 leu2 bcy1-13 | 1 |
| KT1155 | MATaura3 leu2 snf1::URA3 | This study |
| KT1141 | MATα ura3 leu2 gac1::URA3 CTT1::lacZ | This study |
| CV83 | MATaura3 leu2 his3 GSY2-E590K | This study |
| CV129 | MATaura3 leu2 his3 glc7-1 | This study |
| CV130 | MATα ura3 leu2 his3 glc7-1 | This study |
| CV182 | MATα ura3 leu2 his3 trp1-1 gsy2::HIS3 | This study |
| CV218 | MATaMATα ura3 leu2 his3 trp1-1 gsy2::HIS3 | This study |
| CV317 | MATaura3 leu2 his3 gsy2::HIS3 | This study |
a All strains are congenic relative to KT1112 (35).
Reduction of glucose and/or other medium constituents due to autoclaving resulted in a significant increase in glycogen accumulation. For this reason, assays to compare the effects of GLC7, glc7-1, gac1, snf1, and rho− were performed with filter-sterilized media. Solid media contained 2% agar. Strains were typically grown at 30°C. Sporulation was performed on 1% yeast extract–2% peptone–2% potassium acetate medium (YPA) for 3 days at 24°C or longer if required. Although strains homozygous for glc7-1 will not sporulate on traditional sporulation medium, it was found that sufficient, although poor, sporulation could be achieved on YPA after 5 days.
Quantitative analysis.
Glycogen assays were performed as described previously (9). Cell numbers were determined by direct counts with a hemacytometer. Glycogen synthase activity assays were performed as described previously (36). Yeast strains were inoculated at 2 × 106 cells per ml and grown at 30°C until late-log or early-stationary phase, approximately 12 h. Protein extracts were made as described below. Yeast extract (30 μl) was combined with 60 μl of synthase assay mixture (36) in duplicate and incubated for 10 min at 30°C. A 75-μl sample of the reaction mixture was spotted on Whatman 31 ET paper, which was immediately placed in 66% ethanol kept at 4°C. The paper samples were washed twice in cold ethanol, twice in ethanol at room temperature, and once in acetone; then, they were dried and placed into scintillation vials, and counts were determined. Background counts were determined by spotting 60 μl of assay mixture (no protein) on Whatman paper, which was then treated with experimental samples.
Plasmid construction and sequencing.
Standard methods (32) were used for plasmid generation, identification, and amplification. Unless otherwise stated, all digestions and ligations were performed using enzymes obtained from New England Biolabs. pBluescript with a 3.8-kb SalI fragment containing GSY2 was kindly provided by Peter Roach (8). The 3.8-kb SalI fragment from this plasmid was subcloned into the SalI site of YEp351 (pCV58). pCV58 was digested with NdeI to excise a 2.8-kb fragment of GSY2 and religated to produce pCV59. This plasmid was digested with NdeI, and the resulting linearized plasmid was transformed into the suppressor strains to gap repair the mutant GSY2 genes. The gap-repaired plasmids were isolated from yeast (29), transformed into CV130, and assayed for the ability to suppress the glycogen accumulation defect of glc7-1. To localize the mutation responsible for suppression, pCV58 was digested with XhoI and KpnI (excision of a 1.7-kb fragment), KpnI and SacI (excision of a 0.7-kb fragment), or XhoI and SacI (excision of a 2.4-kb fragment); the restriction fragments containing portions of GSY2 were replaced with the same fragments from the suppressor-containing GSY2 genes. These plasmids were transformed into CV130 and assayed for glc7-1 suppression. Sequence analysis was performed using a Sequenase 2.0 kit (U.S. Biochemicals) to identify the mutation or mutations present in this region.
To construct a six-histidine-tagged wild-type GSY2 gene, two PCR products were generated. The first consisted of the GSY2 promoter, the ATG, and a sequence encoding a nine-amino acid tag (RGSHHHHHH) directly after the ATG and followed by an XmaI/EcoRI site at the 3′ end (primers 5′ GTCGACCTGCAGGTCAACGGATCACAAA 3′ and 5′ AAGTTTTGACTACCTCAGAGAAAAATTTTGATGAGAGGTTCGCATCATCATCATCATCATTTCCCGGGAATTCTG 3′). The other product consisted of the entire coding region of GSY2 with an XmaI site added at the 5′ and 3′ ends by PCR (primers 5′ ATTTTCCCGGGGATGTCCCGTGACCTAC 3′ and 5′ TCCCCCGGGGGACGCCTCGAAATGTCGTATGTC 3′). These products were cloned into pBluescript (0.8-kb Sau3A/EcoRI promoter fragment into BamHI/EcoRI and 2.2-kb XmaI coding fragment into XmaI). The promoter fragment and part of the multicloning site of pBluescript were excised with EagI and XhoI and ligated into the EagI/XhoI sites of pRS316 (33). The 2.2-kb XmaI fragment containing the coding region was then ligated into this plasmid to create pCV116. The 2.6-kb EcoRI/SalI fragment of wild-type GSY2 and the gap-repaired mutants was then inserted into an EcoRI/XhoI fragment of pCV116, replacing all but 0.4 kb of the PCR-generated coding region to create plasmids pCV117 GSY2, pCV117 GSY2-G264R, and so forth, which were then transformed into CV218 (gsy2::HIS3).
Protein extracts.
Yeast cells were inoculated into selective minimal media and grown at 30°C with shaking until the cultures reached late log phase. Cells (7.5 × 108) were harvested, washed with breaking buffer (100 mM Tris, 200 mM NaCl, 1 mM EDTA, 5% glycerol [pH 7.0]), and resuspended in 0.25 ml of cold breaking buffer containing 1 mM phenylmethylsulfonyl fluoride and a 1/150 dilution of a protease inhibitor cocktail consisting of 5 mg each of chymostatin, leupeptin, antipain, and pepstatin in 20 ml of 50% ethanol. Cells were broken using a Mini-BeadBeater (Biospec Products) (four repetitions at a setting of 50 for 20 s). Samples were placed in an ice bath for a minimum of 1 min between repetitions. Breaking buffer (0.25 ml) with protease inhibitors (see above) was added to each sample, samples were centrifuged at 4°C for 2 min at 3,800 × g, and the supernatant was used immediately or stored at −80°C.
Immunoblot analysis.
Protein concentrations of cell extracts were determined using the bicinchoninic acid protein assay (Pierce) with bovine serum albumin as a standard. Protein loading dye (0.6 M Tris [pH 6.8], 10% glycerol, 2% sodium dodecyl sulfate [SDS], 5% 2-mercaptoethanol, 0.05% bromphenol blue) was added, and samples were heated to 80°C for 4 min. Equal concentrations of total protein were loaded in each well of an SDS–7.5% polyacrylamide gel, which was run at 200 V for approximately 45 min. The gel was rinsed in transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol) and transferred to nitrocellulose for 1 h at 100 V using a Bio-Rad Mini Trans-Blot apparatus. The membrane was blocked for 2 h in 5% nonfat milk in TBS (10 mM Tris-HCl [pH 7.5], 150 mM NaCl) and then incubated with a 1/1,000 dilution of primary antibody (RGS αHis antibody; Qiagen) in antibody binding solution (TBS with 1.5% bovine serum albumin and 1.5% nonfat milk) for 2 h. The membrane was washed twice with 20 mM Tris-HCl [pH 7.5]–500 mM NaCl–0.1% Tween 20– 0.4% Triton X-100 and once with TBS, 10 min each wash. The membrane was incubated with a 1/8,000 dilution of goat anti-mouse peroxidase-conjugated secondary antibody (Sigma) in antibody binding solution for 1 h and then washed as described above. Detection was performed using an Amersham ECL kit in accordance with the manufacturer's instructions.
Purification of His-tagged proteins.
His-tagged proteins were purified with Ni-nitrilotriacetic acid (NTA)– agarose (Qiagen) using a batch procedure. Cell extracts were added to Ni-NTA–agarose beads, incubated at room temperature for 30 min, and then washed three times with buffer C (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris [pH 6.3]). Purified protein was eluted with buffer C (pH 6.03) containing 100 mM EDTA. Immunoblotting was performed as described above, except that equal volumes of eluate rather than equal amounts of protein were loaded. As the amount of purified protein did not approach the binding capacity of the resin, the amount of purified protein in the eluate reflects the amount of the His-tagged protein in the cell extract.
32P labeling.
Strains from fresh overnight cultures were inoculated at 2 × 106 cells/ml into 50 ml of selective medium. After 12 h of incubation at 140 rpm and 30°C, 109 cells were pelleted, washed with water, and resuspended in 10 ml of low-phosphate medium. Low-phosphate medium was prepared by combining 3.3 g of yeast nitrogen base without amino acids, 5 ml of 1 M MgSO4, and 5 ml of concentrated NH4OH in 450 ml of distilled H2O. Precipitated phosphate was allowed to settle, the resultant supernatant was filtered through a 0.22-μm- pore-size cellulose acetate filter, and the pH of the medium was adjusted to 5.8 with concentrated HCl. Appropriate amino acids were added to sustain growth. After 3 h of incubation, 0.1 mCi of 32P (as orthophosphate) was added. The culture was incubated for 1 h, at which point cells were harvested and broken as described above with the following exception: phosphatase inhibitors were added to the breaking buffer-protease inhibitor mixture. After cells were broken, purification was performed as described above and the purified protein was electrophoresed by SDS-polyacrylamide gel electrophoresis (PAGE). Protein was transferred to nitrocellulose as for the immunoblot protocol, and the blot was exposed to film overnight to detect 32P incorporation. Phosphorimaging was performed to quantitate radioactivity in the Gsy2p bands. To quantitate protein, the blot was incubated in 7 ml of phosphatase buffer (50 mM Tris-HCl [pH 8.0], 1 mM MgCl2, 0.1 mM ZnCl2) containing 70 U of alkaline phosphatase. This step removed 32P from the nitrocellulose and allowed detection by immunoblotting, which was performed as described above.
RESULTS
Isolation and genetic analysis of suppressors of glc7-1 mutants.
Strains containing glc7-1 (R73C) accumulate only 10% of the glycogen levels found in wild-type strains (2). Glycogen synthase from these strains is in the phosphorylated, G6P-dependent form (11, 27). To further dissect the regulatory pathway required for the activation of glycogen synthase, we mutagenized a MATa glc7-1 strain (KT1118) with ethyl methanesulfonate as described in Materials and Methods and screened 30,000 colonies for revertants that accumulated glycogen by using the iodine staining method (6). Twenty-nine revertants were backcrossed three times to a MATα glc7-1 strain (KT1119), and suppressors which did not mate or sporulate, which stained a bluish-purplish color with iodine (indicating a defect typically found in mutants with mutations in the glycogen branching enzyme) (31, 37), or which had phenotypes that did not segregate 2:2 in meiotic progeny were discarded. Twenty-six of the 29 mutants survived this preliminary screen and were characterized in greater detail.
Tetrad analysis of crosses between the revertants and a wild-type GLC7 strain revealed that each suppressor segregated independently of GLC7 and induced a hyperglycogen phenotype in a wild-type GLC7 background. Diploid strains constructed by mating the revertants to a glc7-1 strain accumulated more glycogen than a homozygous glc7-1 strain but accumulated less a than a diploid strain homozygous for a suppressor, indicating that each revertant was semidominant. To determine if the suppressors were genetically linked, we performed tetrad analysis of glc7-1/glc7-1 diploid strains constructed by mating two different suppressor strains. If suppressors are in different loci, the low-glycogen glc7-1 phenotype should reappear in some spore clones; if they are tightly linked, all spores should display the hyperglycogen phenotype of one of the parent strains. All spore clones from crosses between different revertants retained the high-glycogen phenotype, indicating that all suppressors are tightly linked. To help determine which locus was mutated in the revertants, each suppressor in a wild-type GLC7 background was crossed to gac1::URA3 (KT1141) and gsy2::HIS3 (CV218) strains. These mutants represented two of the most logical suppressor classes. No linkage was observed with gac1::URA3. However, linkage was observed between each suppressor and the gsy2::HIS3 disruption. In every case, the two histidine auxotrophic spore clones from each tetrad accumulated higher-than-normal levels of glycogen and the histidine prototrophs failed to accumulate glycogen.
Cloning and sequencing of suppressors.
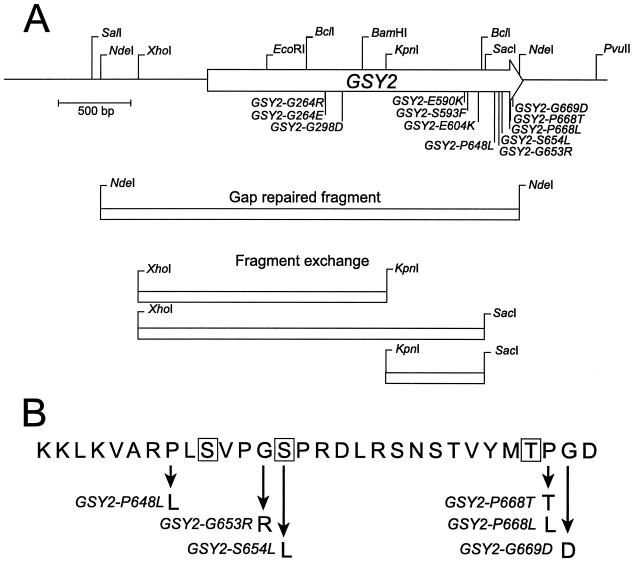
Since the suppressors resided in or near GSY2, the NdeI fragment of GSY2, which encodes all but the final four amino acid residues of Gsy2p, was cloned by gap repair from each mutant as described in Materials and Methods. Plasmids were transformed into the glc7-1 strain (CV129) to determine if the GSY2 gene recovered from the mutant strain would suppress the glc7-1 phenotype. Transformants containing each of the recovered GSY2 genes accumulated glycogen, indicating that the mutation responsible for the suppression resided within the NdeI fragment. Wild-type GSY2 in YEp351 is unable to restore glycogen accumulation to the glc7-1 strain. To more accurately localize the suppressor mutations, internal XhoI/SacI, XhoI/KpnI, and KpnI/SacI fragments of wild-type GSY2 in YEp351 (pCV58) were exchanged with the identical fragments from each of the mutant plasmids (Fig. 1). The presence of the suppressor in each of these subclones was identified by assaying glycogen levels in CV129 transformants. The DNA sequence was determined for each restriction fragment from the gap-repaired plasmids that complemented the glc7-1 defect. The sequence was also determined for the SacI/NdeI fragment from every suppressor clone because we were unable to test for the presence of the suppressor in this fragment.
FIG. 1.
Locations of mutations in GSY2 that suppress the glycogen defect of glc7-1. (A) Overall locations of the suppressor mutations in GSY2. The arrow corresponds to the GSY2 coding sequence. The arrowhead corresponds to the 3′ end of GSY2. The NdeI/NdeI DNA fragment used to gap repair each GSY2 allele and the DNA restriction endonuclease fragments used to locate each allele are indicated below the restriction map. (B) Locations of suppressor mutations in the COOH-terminal region of Gsy2p. The boxed residues, S650, S654, and T667, correspond to proposed sites of phosphorylation (16).
The 12 unique mutations identified from the 26 suppressors are listed in Table 2, and the locations of these mutations in the GSY2 gene are presented in Fig. 1A. Only a single mutation was found in each of the suppressors. Each mutant was named for the predicted change in the sequence of the gene product. GSY2-G264R, GSY2-G264E, GSY2-G298D, GSY2-P648L, and GSY2-P668T mutations were all recovered multiple times. For each of these five mutations, at least one of the duplicates was isolated independently from a separate mutagenesis experiment. The 12 mutations were located in three regions of GSY2. Three were located near the middle of the gene, three were located more distal, and the remaining six were located in the known COOH-terminal regulatory region. The six NH2-terminal mutations lay in the portion of Gsy2p that is closely related to mammalian glycogen synthase. Residues G298, E590, and S593, which are altered in GSY2-G298D, GSY2-E590K, and GSY2-S593F mutants, correspond to identical residues in human liver and muscle glycogen synthase. In contrast, the residues altered in the six most COOH-terminal mutants are not similar to residues found in this region of mammalian glycogen synthase. This region contains the amino acid residues that are phosphorylated in vivo (16).
TABLE 2.
GSY2 mutants
| Mutant | Amino acid residue alteration | Nucleotide alteration | No. of isolates | No. of mutagenic pools |
|---|---|---|---|---|
| GSY2-G264R | G265R | G1613A | 3 | 2 |
| GSY2-G264E | G265E | G1614A | 3 | 3 |
| GSY2-G298D | G265D | G1716A | 3 | 3 |
| GSY2-E590K | G591K | G2591A | 1 | 1 |
| GSY2-S593F | S494F | C2601T | 1 | 1 |
| GSY2-E604K | E605K | G2633A | 1 | 1 |
| GSY2-P648L | P649L | C2766A | 3 | 3 |
| GSY2-G653R | G654R | G2780C | 1 | 1 |
| GSY2-S654L | S655L | C2784T | 1 | 1 |
| GSY2-P668L | P669T | C2825A | 1 | 1 |
| GSY2-P668T | P669L | C2826T | 7 | 4 |
| GSY2-P669D | G670D | G2829A | 1 | 1 |
Glycogen accumulation in suppressor strains.
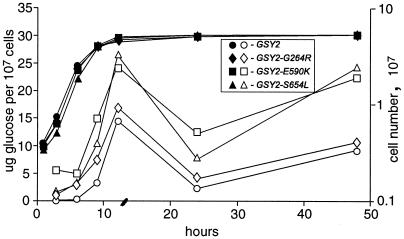
To investigate how the GSY2 mutations alter glycogen synthesis, we first assayed glycogen levels throughout the growth curve of the wild type and selected GSY2 mutants that were grown in batch culture. Glycogen stores are normally low in exponential phase, begin to increase as the cultures approach stationary phase, peak during late-log/early-stationary phase, and drop after 12 h in stationary phase. The three suppressors that we tested accumulated higher levels of glycogen at each time point but showed basically the same overall pattern of accumulation (Fig. 2). This similarity is not unexpected because genes involved in glycogen metabolism are known to be under transcriptional control (15, 25), which we would predict would not be altered in our GSY2 mutants. We note that glycogen levels are very sensitive to the method used to prepare the media (see Materials and Methods). All subsequent data were gathered from cells grown in filter-sterilized media.
FIG. 2.
Glycogen accumulation in yeast strains grown in batch cultures. Strains were inoculated into synthetic medium at 2 × 106 cells/ml (see Materials and Methods). At the indicated times, cell numbers were determined with a hemacytometer (filled symbols), and glycogen levels per 107 cells were assayed and expressed as micrograms of glucose (open symbols).
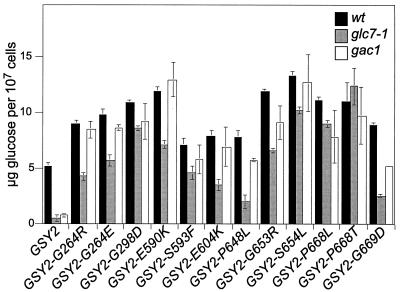
We next assayed glycogen levels of each suppressor mutant in glc7-1, GLC7, and gac1::URA3 backgrounds at cell densities (2.3 × 108 to 3 × 108 cells/ml) at which strains accumulate maximum levels of glycogen. As shown in Fig. 3, glc7-1 mutants accumulated less than 10% the glycogen accumulated by wild-type strains (5̃ μg of glucose per 107 cells). For a given GSY2 mutant, glycogen levels were usually higher in a wild-type background than in a glc7-1 background. The exceptions were GSY2-P668L and GSY2-P688T mutants, whose mutations caused the accumulation of the same levels in both backgrounds. This result suggests that most mutant proteins are not completely deregulated and require Glc7p to become fully active.
FIG. 3.
Glycogen accumulation for GSY2 and 12 glc7-1 suppressors in wild-type (wt), glc7-1, and gac1▵ backgrounds. Cultures were inoculated into synthetic medium at 2 × 106 cells/ml, cells were harvested 12 h later at densities of 2 × 107 to 3 × 107 cells/ml.
GAC1 encodes a regulatory subunit of Glc7p necessary for the activation of glycogen synthase (11). The subunit contains separate binding domains for Glc7p and glycogen synthase and appears to act in a manner similar to that of the mammalian glycogen targeting subunits (Wu et al., submitted). We assayed the ability of the GSY2 mutations to suppress the glycogen defect found in gacI strains. As shown in Fig. 3, all GSY2 suppressor mutations increased glycogen levels in a gac1 mutant, in many cases to the level found for the suppressor in a wild-type background. Most GSY2 mutations suppressed the glycogen defect of a gac1 mutant better than that of a glc7-1 mutant. This result is consistent with the observation that glc7-1 strains are usually more deficient in the ability to accumulate glycogen than are gac1 strains, possibly because of the partially redundant activity of Pig1p (4).
Effects of GSY2 mutations on regulatory mutations affecting glycogen accumulation.
In addition to Pho85p, two additional protein kinases have been shown to regulate the activity of glycogen synthase. Snf1p, closely related to the mammalian AMP-dependent protein kinase, is a positive regulator of glycogen synthesis. snf1 mutants fail to accumulate glycogen, in part because of a defect in the accumulation of GSY2 mRNA and in part because of a defect in the activation of glycogen synthase (15). This latter defect may be due to reduced levels of G6P accumulating in snf1 mutants (20). In contrast, cyclic AMP (cAMP)-dependent protein kinase has a negative role in the regulation of glycogen synthase. Cells with constitutively active cAMP-dependent protein kinase activity, due to defects in the cAMP binding inhibitory subunit Bcy1p, express very low levels of GSY2 mRNA and may have an altered Gsy2p phosphorylation state (15).
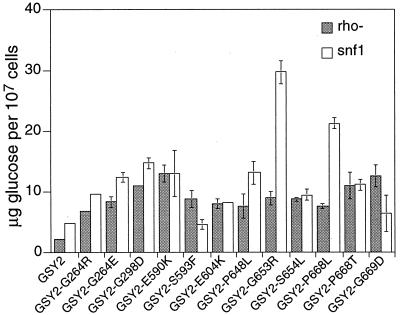
As shown in Fig. 4, our GSY2 mutations were able to suppress the glycogen defect of snf1 mutants, with the exception of GSY2-S593F and G669D. Surprisingly, snf1 GSY2-G653L and snf1 GSY2-P668L mutants accumulated much higher levels of glycogen than any other mutant we tested. In contrast to the results for snf1 mutants, none of our GSY2 mutations was able to suppress the glycogen defect of bcy1 mutants (data not shown), consistent with previous work indicating that cAMP-dependent protein kinase acts on the glycogen biosynthetic pathway, largely at the level of mRNA synthesis (11, 15).
FIG. 4.
Glycogen accumulation for GSY2 and 12 glc7-1 suppressors in snf1 and rho− backgrounds. Cultures were inoculated into synthetic medium at 2 × 106 cells/ml, and cells were harvested 12 h later at densities of 2 × 107 to 3 × 107 cells/ml.
Chester (6) noted that respiration-deficient yeast mutants fail to accumulate glycogen. More recently, Yang et al. (40) found that this defect can be suppressed in GSY2 mutants whose products cannot be phosphorylated. Surprisingly, mutation of PHO85, which encodes one of the kinases responsible for phosphorylating and inactivating glycogen synthase, does not rescue the defect, suggesting that another kinase may be responsible for the inhibition of glycogen synthase in respiration-deficient strains. We tested our collection of GSY2 mutants for suppression of this defect. As shown in Fig. 4, all GSY2 suppressors at least partially alleviated the glycogen defect. Interestingly, several of the suppressors produced a “ring” phenotype in qualitative plate assays, where the outer edge of the culture stained much darker than the center (data not shown). This result suggests that for these mutants, only rapidly growing cells or cells with access to the highest levels of nutrients accumulated excess glycogen. Hyperaccumulation was also observed in qualitative assays when the suppressors in a Rho+ background were grown anaerobically, indicating that these mutants do not require oxygen for increased glycogen accumulation (data not shown).
Abundance, phosphate incorporation, and activity of mutant Gsy2p enzymes.
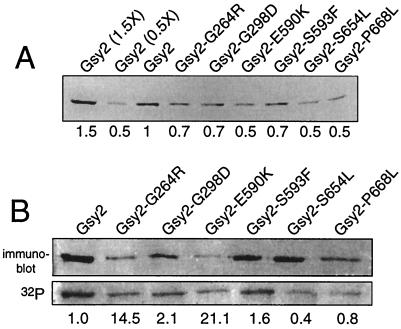
Although the overexpression of wild-type Gsy2p does not raise glycogen levels in either a wild-type or a glc7-1 mutant background, we nevertheless assayed the levels of Gsy2 proteins in our mutants to eliminate the possibility that protein abundance was affecting the glycogen phenotype. To facilitate the assay of Gsy2 protein levels, the sequence encoding RSH6 was inserted after the NH2-terminal methionine codon for efficient protein purification and detection. The six-histidine-tagged genes were cloned into a CEN plasmid that allowed expression from the native GSY2 promoter, and plasmids containing six different GSY2 mutations were transformed into a gsy2::HIS3 strain (CV218) to assay for the ability of the tagged genes to complement the gsy2::HIS3 disruption. Transformants containing six-histidine-tagged and untagged GSY2 genes accumulated similar levels of glycogen, indicating that the introduction of the six-histidine- tag at the NH2 terminus does not reduce activity. Protein extracts were collected from late-log cultures, at the point at which glycogen levels were at a maximum. Six-histidine-tagged Gsy2 proteins were purified on Ni-NTA–agarose from equal amounts of total protein, separated by SDS-PAGE, transferred to nitrocellulose, and detected using an antibody against the six-histidine tag. As shown in Fig. 5A, mutant protein concentrations were equal to or lower than wild-type protein levels. Ni-NTA-purified protein levels were assessed because of the relatively low level of enzyme in total extracts. However, immunoblotting was performed using total extracts, and similar results were obtained (data not shown). Thus, increased glycogen levels in the mutants were not due to increased Gsy2p abundance.
FIG. 5.
Gsy2p expression and phosphorylation. (A) Immunoblot of extracts prepared from wild-type and Gsy2 suppressor variants grown to late-log phase. Equal concentrations of total protein were loaded into each lane, except for the first and second lanes, which contained 1.5 and 0.5 times the amounts loaded in the other lanes, respectively. Numbers below each lane indicate relative Gsy2 protein levels, normalized to that of the wild-type protein, as determined by densitometry of the immunoblot. (B) Immunoblot and autoradiogram of cells labeled with 32P. Yeast strains were 32P labeled, cell extracts were prepared, and six-histidine-tagged, affinity-purified protein was electrophoresed by SDS-PAGE as described in Materials and Methods. Gsy2p present in each immunoprecipitate was assayed by immunoblotting, and the level of phosphorylation was determined by autoradiography. The numbers below each lane indicate the ratio of 32P incorporation in mutant proteins to that in total Gsy2 protein, normalized to wild-type Gsy2p levels.
Glycogen synthase assays were performed with the mutant proteins to determine if suppression ability correlated with increased protein activity. Assays were performed in the presence and absence of G6P to measure total and active glycogen synthase activities, respectively. All mutants examined exhibited an increased activity ratio (Table 3).
TABLE 3.
Glycogen synthase activity assaysa
| Strain | Glycogen synthase activity (mU/mg of protein) in the presence (+) or absence (−) of G6P
|
Ratio | |
|---|---|---|---|
| + | − | ||
| GSY2 | 4.22 ± 0.65 | 0.63 ± 0.08 | 0.15 ± 0.04 |
| GSY2-G264R | 4.79 ± 0.66 | 1.37 ± 0.11 | 0.43 ± 0.22 |
| GSY2-G298D | 6.10 ± 1.22 | 2.82 ± 0.39 | 0.37 ± 0.10 |
| GSY2-E590K | 9.14 ± 1.14 | 5.77 ± 1.01 | 0.63 ± 0.03 |
| GSY2-S593F | 4.13 ± 0.93 | 1.19 ± 0.30 | 0.29 ± 0.01 |
| GSY2-S654L | 6.88 ± 0.86 | 2.82 ± 0.90 | 0.33 ± 0.18 |
| GSY2-P668T | 6.66 ± 1.57 | 1.56 ± 0.02 | 0.24 ± 0.06 |
Values are reported as means and standard deviations. All strains were in a glc7-1 background.
To assay for phosphate incorporation into wild-type Gsy2p and mutant Gsy2p, a glc7-1 gsy2::HIS3 strain was transformed with plasmids containing the six-histidine-tagged GSY2 genes. Transformants were labeled with 32P (as orthophosphate) as described in Materials and Methods. The purified six-histidine-tagged Gsy2 proteins were subjected to SDS-PAGE and transferred to nitrocellulose, and 32P incorporation into Gsy2p was assayed by autoradiography (Fig. 5B). Gsy2p levels were assayed by immunoblot analysis after treatment with alkaline phosphatase. As shown by the relative incorporation of 32P into each mutant Gsy2 protein (Fig. 5B), each Gsy2p variant tested incorporated 32P. Variants with mutations in the phosphorylated domain, Gsy2-S654L and Gsy2-P668T, incorporated less 32P than wild-type Gsy2p, whereas Gsy2-G264R and Gsy2-E590K incorporated higher levels of 32P than the wild-type protein.
Relative fitness of glycogen synthase mutants.
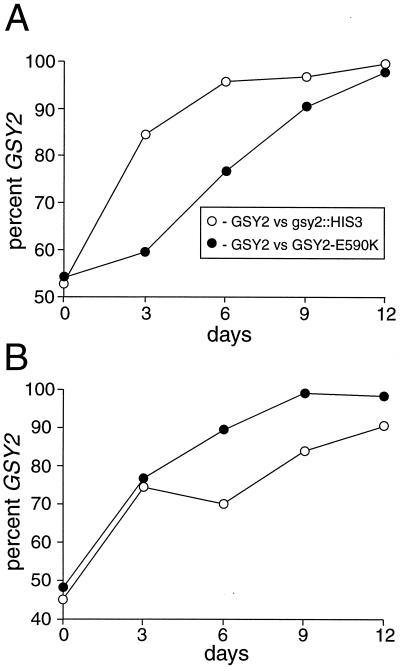
Although yeast cells have evolved a complex mechanism for the regulation of glycogen synthesis, little is known about the physiological role for this storage carbohydrate. The elimination of both GSY1 and GSY2 has been reported to have no effect on growth rate or heat shock resistance and to confer only a slight defect in sporulation (8). To provide a more sensitive assay for defects caused by altered levels of glycogen, we cocultivated a wild-type strain with either a gsy2::HIS3 mutant or a GSY2-E590K mutant. These congenic strains had identical nutritional requirements and showed no difference in exponential growth rates when cultured separately (data not shown). In each cocultivation experiment, the two strains were inoculated into fresh medium at a concentration of 2 × 104 cells/mI, and cultures were allowed to grow for 3 days before reinoculation into fresh medium. After 3 days, samples were diluted with fresh medium to a concentration of 2 × 104 cells/ml and incubated for an additional 3 days. This regimen was repeated twice. After each 3-day period, the ratio of the numbers of cells of the strains in the culture was determined using glycogen accumulation to score the two genotypes. As shown in Fig. 6A, after 12 days of growth in glucose-containing medium, greater than 95% of the cells in both cultures were wild type. In acetate-containing medium, gsy2::HIS3 and GSY2-E590K strains were also less fit than the wild type, although the gsy2::HIS3 strain showed higher fitness in acetate-containing medium than in glucose-containing medium.
FIG. 6.
Competition between wild-type and GSY2 mutants in synthetic complete medium and acetate-containing medium. Wild-type and either GSY2-E950K or gsy2::HIS3 strains were inoculated at 104 cells/ml into synthetic complete medium (A) or acetate-containing medium (B) and allowed to grow for 3 days at 30°C. Cultures were subcultured into fresh medium at 2 × 104 cells/ml and allowed to grow for another 3 days. This regimen was repeated twice. For each dilution, a culture sample was placed on solid YPD and colonies were allowed to grow. The ratio of wild-type to mutant GSY2 on the plate was determined by iodine staining. Each point represents of the average of at least two independent experiments.
The relatively rapid overgrowth of the wild-type strain in these experiments indicates that both decreased glycogen and increased glycogen levels lead to reduced fitness of the population. In principle, these differences in competition can be attributed to differences in growth rate, cell viability, or rate of outgrowth from stationary phase. We have not observed any differences in the doubling times of these strains in log phase or in the rate of outgrowth from stationary phase, but we have observed reduced viability of both the gsy2::HIS3 and the GSY2-E590K strains. The percentage of viable wild-type cells in cultures after 3 days of growth in synthetic glucose-containing medium was 93%, whereas those of the gsy2::HIS3 and GSY2-E590K strains were 71 and 79%, respectively. These differences could account for at least some of the competitive disadvantages of the GSY2 mutants and demonstrate for the first time the importance of the regulation of glycogen metabolism in yeast.
DISCUSSION
In a screen for revertants that restored glycogen accumulation to a glc7-1 strain, we identified 12 unique mutations in GSY2. All 12 probably act by altering the activity of glycogen synthase, because none of the GSY2 mutants tested showed an increase in protein levels, as assayed by immunoblot analysis. In addition, overexpression of the wild-type GSY2 gene does not suppress the glc7-1 phenotype, indicating that the concentration of glycogen synthase in the cell is not normally rate limiting for glycogen biosynthesis. Although there is no direct biochemical evidence in vitro that Glc7p is the phosphatase that activates glycogen synthase, the results presented here strengthen this hypothesis. It is surprising that no other genes were identified in our screen because mutations in REG1 (17) and PHO85 (18, 38) have also been shown to suppress the glc7-1 mutation. However, the exclusion of slowly growing colonies from our screen would have eliminated reg1 and pho85, since both show a slow-growth phenotype unrelated to their high level of glycogen accumulation.
We envision three mechanisms by which our mutations in GSY2 could suppress the glc7-1 phenotype. One possibility is that the mutant proteins are resistant to negative regulation by phosphorylation, either directly due to reduced phosphorylation or indirectly due to changes in allosteric effects of phosphorylation on activity. Another possibility is that mutants could be more sensitive to positive regulation, either by becoming better substrates for phosphatase activity or by becoming more sensitive to the activating effect of G6P. A third possibility is that the suppressors act by increasing the activity of the encoded enzyme without changing its regulation. Six out of 12 GSY2 mutations identified in this screen probably act via the first mechanism. These are located at or near the proposed phosphorylation sites (Fig. 1B). One of the six directly alters phosphorylation site Ser-654, while the others alter residues one or two residues away from the phosphorylation sites.
Pho85p, together with its cyclin subunits Pcl8p and Pcl10p, comprises one of the kinases that phosphorylates Gsy2p at S654 and T667 (19, 39). The precise consensus site has not been determined for this activity, but the consensus sequence around these two sites is S/T-P-X-D-L. Two GSY2 mutations, GSY2-P668T and GSY2P688L, altered one of these prolines, supporting the above consensus recognition motif. The fact that GSY2-G669D also suppressed glc7-1 could indicate that the identity of the amino acid residue after the proline is also restricted for Pho85p-Pcl10p recognition.
The kinase responsible for phosphorylating S650 is not known, but the fact that GSY2-P648L suppresses glc7-1 leads to the prediction that the consensus recognition motif for this kinase contains a proline at the −2 position. It is worth noting that a proline is present at the −2 position in the sequence recognition motif for the mitogen-activated protein (MAP) kinase Erk1 (13, 34). François et al. (9) found that glycogen synthesis was inhibited by the mating pathway, a signaling pathway regulated by the MAP kinase Fus3p. However, to our knowledge there is no direct evidence that Fus3p can phosphorylate Gsy2p. We do not believe that the other six suppressor mutations alter the phosphorylation state of Gsy2p, but several of them could cause insensitivity to the effects of phosphorylation. These mutant proteins were phosphorylated to a greater extent than the wild-type protein, as judged by the relative levels of 32P incorporation in vivo.
A detailed biochemical characterization of 22 mutations of GSY2 revealed two mutants that were totally resistant to activation by G6P (26). One of these, Gsy2-R586A R588A R591A, had wild-type activity and was inactivated by phosphorylation. Another, Gsy2-R579A R580A R582A, showed reduced sensitivity to phosphorylation. Pederson et al. (26) proposed that these residues constitute a region of glycogen synthase that is important for G6P binding and/or changes between different activity states. It is noteworthy that two of our mutants, GSY2-E590K and GSY2-S593F, had mutations that lay in or near this region. Gsy2-E590K had the highest activity in the presence or absence of G6P, suggesting that this mutant might also be insensitive to the influence of G6P and C-terminal phosphorylation. A detailed biochemical characterization of these mutants would help distinguish between different possible mechanisms of suppression.
In both yeast and mammals, glycogen biosynthesis is under complex regulation. In mammals, where glycogen serves as a key energy store and for maintenance of blood glucose levels, the reasons for this regulation are obvious. In yeast, the accumulation of glycogen at the end of logarithmic growth would implicate the carbohydrate as having a role in survival in stationary phase or sporulation. However, glycogen synthase mutants sporulate well, have no growth defect, and survive heat shock stress (8). We revisited this issue using a cocultivation strategy, where wild-type and mutant strains with identical auxotrophies were cultured together in medium containing either glucose or acetate as a carbon source. Under both conditions, strains with low (gsy2::HIS3) and high (GSY2-E590K) levels of glycogen were rapidly overgrown by the wild-type strain (Fig. 6). This result provides a solid indication that glycogen levels must be tightly regulated, since as little as a 2-fold increase or a 10-fold decrease in glycogen is enough to dramatically reduce fitness levels. Although we do not know at present the physiological defects responsible for the reduced fitness of the GSY2 mutants, we have observed reduced viability of both gsy2::HIS3 and GSY2-E590K strains, a result which could account for the differences in fitness. Our results provide a means to study the basis for the requirement for the tightly regulated control of glycogen accumulation.
ACKNOWLEDGMENTS
We thank Lucy Robinson for comments on the manuscript and Peter Roach for strains, plasmids, and communication of results prior to publication.
This work was supported by National Institutes of Health grant GM-477899 and a predoctoral NSF fellowship to C.A.
REFERENCES
- 1.Cannon J F, Gitan R, Tatchell K. Yeast cAMP-dependent protein kinase regulatory subunit mutations display a variety of phenotypes. J Biol Chem. 1990;265:11897–11904. [PubMed] [Google Scholar]
- 2.Cannon J F, Pringle J R, Fiechter A, Khalil M. Characterization of glycogen-deficient glc mutants of Saccharomyces cerevisiae. Genetics. 1994;136:485–503. doi: 10.1093/genetics/136.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cannon J F, Tatchell K. Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol Cell Biol. 1987;7:2653–2663. doi: 10.1128/mcb.7.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng C, Huang D, Roach P J. Yeast PIG genes: PIG1 encodes a putative type 1 phosphatase subunit that interacts with the yeast glycogen synthase Gsy2p. Yeast. 1997;13:1–8. doi: 10.1002/(SICI)1097-0061(199701)13:1<1::AID-YEA49>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 5.Cheng C, Mu J, Farkas I, Huang D, Goebl M G, Roach P J. Requirement of the self-glucosylating initiator proteins Glg1p and Glg2p for glycogen accumulation in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6632–6640. doi: 10.1128/mcb.15.12.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chester V E. Heritable glycogen-storage deficiency in yeast and its induction by ultra-violet light. J Gen Microbiol. 1968;51:49–56. doi: 10.1099/00221287-51-1-49. [DOI] [PubMed] [Google Scholar]
- 7.Farkas I, Hardy T A, DePaoli-Roach A A, Roach P J. Isolation of the GSY1 gene encoding yeast glycogen synthase and evidence for the existence of a second gene. J Biol Chem. 1990;265:20879–20886. [PubMed] [Google Scholar]
- 8.Farkas I, Hardy T A, Goebl M G, Roach P J. Two glycogen synthase isoforms in Saccharomyces cerevisiae are coded by distinct genes that are differentially controlled. J Biol Chem. 1991;266:15602–15607. [PubMed] [Google Scholar]
- 9.François J, Higgins D L, Chang F, Tatchell K. Inhibition of glycogen synthesis in Saccharomyces cerevisiae by the mating pheromone α-factor. J Biol Chem. 1991;266:6174–6180. [PubMed] [Google Scholar]
- 10.François J, Villanueva M E, Hers H-G. The control of glycogen metabolism in yeast. 1. Interconversion in vivo of glycogen synthase and glycogen phosphorylase induced by glucose, a nitrogen source, or uncouplers. Eur J Biochem. 1988;174:551–559. doi: 10.1111/j.1432-1033.1988.tb14134.x. [DOI] [PubMed] [Google Scholar]
- 11.François J M, Thompson-Jaeger S, Skroch J, Zellenka U, Spevak W, Tatchell K. GAC1 may encode a regulatory subunit for protein phosphatase type 1 in Saccharomyces cerevisiae. EMBO J. 1992;11:87–96. doi: 10.1002/j.1460-2075.1992.tb05031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frederick D L, Tatchell K. The REG2 gene of Saccharomyces cerevisiae encodes a type 1 protein phosphatase-binding protein that functions with Reg1p and the Snf1p protein kinase to regulate growth. Mol Cell Biol. 1996;16:2922–2931. doi: 10.1128/mcb.16.6.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez F A, Raden D L, Davis R J. Identification of substrate recognition determinants for human ERK1 and ERK2 protein kinases. J Biol Chem. 1991;266:22159–22163. [PubMed] [Google Scholar]
- 14.Guthrie C, Fink G R, editors. Methods in enzymology. 194. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press, Inc.; 1991. [PubMed] [Google Scholar]
- 15.Hardy T A, Huang D, Roach P J. Interactions between cAMP-dependent and SNF1 protein kinases in the control of glycogen accumulation in Saccharomyces cerevisiae. J Biol Chem. 1994;269:27907–27913. [PubMed] [Google Scholar]
- 16.Hardy T A, Roach P J. Control of yeast glycogen synthase-2 by COOH-terminal phosphorylation. J Biol Chem. 1993;268:23799–23805. [PubMed] [Google Scholar]
- 17.Huang D, Chun K T, Goebl M G, Roach P J. Genetic interactions between REG1/HEX2 and GLC7, the gene encoding the protein phosphatase type 1 catalytic subunit in Saccharomyces cerevisiae. Genetics. 1996;143:119–127. doi: 10.1093/genetics/143.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang D, Farkas I, Roach P J. Pho85p, a cyclin-dependent protein kinase, and the Snf1p protein kinase act antagonistically to control glycogen accumulation in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4357–4365. doi: 10.1128/mcb.16.8.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang D, Moffat J, Wilson W A, Moore L, Cheng C, Roach P J, Andrews B. Cyclin partners determine Pho85 protein kinase substrate specificity in vitro and in vivo: control of glycogen biosynthesis by Pcl8 and Pcl10. Mol Cell Biol. 1998;18:3289–3299. doi: 10.1128/mcb.18.6.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang D, Wilson W A, Roach P J. Glucose-6-P control of glycogen synthase phosphorylation in yeast. J Biol Chem. 1997;272:22495–22501. doi: 10.1074/jbc.272.36.22495. [DOI] [PubMed] [Google Scholar]
- 21.Huang K P, Cabib E. Yeast glycogen synthetase in the glucose 6-phosphate-dependent form. J Biol Chem. 1974;249:3851–3857. [PubMed] [Google Scholar]
- 22.Hwang P K, Tugendriech S, Fletterick R J. Molecular analysis of GPH1, the gene encoding glycogen phosphorylase in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:1659–1666. doi: 10.1128/mcb.9.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lillie S H, Pringle J R. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol. 1980;143:1384–1394. doi: 10.1128/jb.143.3.1384-1394.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ni H T, LaPorte D C. Response of a yeast glycogen synthase gene to stress. Mol Microbiol. 1995;16:1197–1205. doi: 10.1111/j.1365-2958.1995.tb02342.x. [DOI] [PubMed] [Google Scholar]
- 25.Parrou J L, Teste M A, Francois J. Effects of various types of stress on the metabolism of reserve carbohydrates in Saccharomyces cerevisiae: genetic evidence for a stress-induced recycling of glycogen and trehalose. Microbiology. 1997;143:1891–1900. doi: 10.1099/00221287-143-6-1891. [DOI] [PubMed] [Google Scholar]
- 26.Pederson B A, Cheng C, Wilson W A, Roach P J. Regulation of glycogen synthase: identification of residues involved in regulation by the allosteric ligand glucose-6-P and by phosphorylation. J Biol Chem. 2000;275:27753–27761. doi: 10.1074/jbc.M003342200. [DOI] [PubMed] [Google Scholar]
- 27.Peng Z-Y, Trumbly R J, Reimann E M. Purification and characterization of glycogen synthase from a glycogen-deficient strain of Saccharomyces cerevisiae. J Biol Chem. 1990;265:13871–13877. [PubMed] [Google Scholar]
- 28.Roach P J. Control of glycogen synthase by hierarchal protein phosphorylation. FASEB J. 1990;4:2961–2968. [PubMed] [Google Scholar]
- 29.Rose M D, Winston F, Hieter P. Methods in yeast genetics. A laboratory course manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 30.Rothman-Denes L, Cabib E. Two forms of yeast glycogen synthetase and their role in glycogen accumulation. Proc Natl Acad Sci USA. 1970;66:967–974. doi: 10.1073/pnas.66.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rowen D W, Meinke M, LaPorte D C. GLC3 and GHA1 of Saccharomyces cerevisiae are allelic and encode the glycogen branching enzyme. Mol Cell Biol. 1992;12:22–29. doi: 10.1128/mcb.12.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sikorksi R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Songyang Z, Lu K P, Kwon Y T, Tsai L H, Filhol O, Cochet C, Brickey D A, Soderling T R, Bartleson C, Graves D J, DeMaggio A J, Hoekstra M F, Blenis J, Hunter T, Cantley L C. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stuart J S, Frederick D L, Varner C M, Tatchell K. The mutant type 1 protein phosphatase encoded by glc7–1 from Saccharomyces cerevisiae fails to interact productively with the GAC1-encoded regulatory subunit. Mol Cell Biol. 1994;14:896–905. doi: 10.1128/mcb.14.2.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas J A, Schlender K K, Larner J. A rapid filter paper assay for UDPglucose-glycogen glucosyltransferase, including an improved biosynthesis of UDP-14C-glucose. Anal Biochem. 1968;25:486–499. doi: 10.1016/0003-2697(68)90127-9. [DOI] [PubMed] [Google Scholar]
- 37.Thon V J, Vigneron-Lesens C, Marianne-Pepin T, Montreuil J, Decq A, Rachez C, Ball S G, Cannon J F. Coordinate regulation of glycogen metabolism in the yeast Saccharomyces cerevisiae. J Biol Chem. 1992;267:15224–15228. [PubMed] [Google Scholar]
- 38.Timblin B K, Tatchell K, Bergman L W. Deletion of the gene encoding the cyclin-dependent protein kinase PHO85 alters glycogen metabolism in Saccharomyces cerevisiae. Genetics. 1996;143:57–66. doi: 10.1093/genetics/143.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson W A, Mahrenholz A M, Roach P J. Substrate targeting of the yeast cyclin-dependent kinase Pho85p by the cyclin Pcl10p. Mol Cell Biol. 1999;19:7020–7030. doi: 10.1128/mcb.19.10.7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang R, Chun K T, Wek R C. Mitochondrial respiratory mutants in yeast inhibit glycogen accumulation by blocking activation of glycogen synthase. J Biol Chem. 1998;273:31337–31344. doi: 10.1074/jbc.273.47.31337. [DOI] [PubMed] [Google Scholar]