Abstract
Saccharomyces cerevisiae normally will not take up sterols from the environment under aerobic conditions. A specific mutant, upc2-1, of the predicted transcriptional activator UPC2 (YDR213w) has been recognized as a strain that allows a high level of aerobic sterol uptake. Another predicted transcriptional activator, the YLR228c gene product, is highly homologous to Upc2p. In fact, at the carboxy terminus 130 of the last 139 amino acids are similar between the two proteins. Since these proteins are very similar, the effect of mutations in the YLR228c open reading frame (ORF) was compared with like alterations in UPC2. First, the YLR228c ORF was insertionally inactivated and crossed with various UPC2 constructs. Deletion of YLR228c and UPC2 in combination resulted in nonviability, suggesting that the two proteins have some essential overlapping function. The upc2-1 point mutation responsible for aerobic sterol uptake was duplicated in the homologous carboxy region of the YLR228c ORF using site-directed mutagenesis. This mutation on a high-copy vector resulted in an increase in sterol uptake compared to an isogenic wild-type strain. The combination of both point mutations resulted in the greatest level of aerobic sterol uptake. When the YLR228c point mutation was expressed from a low-copy vector there was little if any effect on sterol uptake. Gas chromatographic analysis of the nonsaponifiable fractions of the various strains showed that the major sterol for all YLR228c and UPC2 combinations was ergosterol, the consensus yeast sterol.
The consensus sterol in Saccharomyces cerevisiae is ergosterol, which is capable of fulfilling the multiple functions described for sterols in this organism (17). The production of ergosterol is very costly for the cell. One enzymatic step (C24 transmethylation) requires 12 to 14 ATP equivalents (14). Several other enzymatic reactions in the ergosterol biosynthetic pathway require oxygen for activity. Molecular oxygen is a substrate in the squalene epoxidase reaction, catalyzed by ERG1, and it is required for the demethylation and desaturation reactions that occur subsequently (15). Therefore, in the absence of oxygen, sterols cannot be produced and must be acquired from the environment for survival. Under aerobic conditions S. cerevisiae is unable to take sterols from the medium (1), an effect termed aerobic sterol exclusion (12). It has been demonstrated that heme competency prohibits uptake under aerobic conditions, but the mechanism of exclusion has not been determined (8, 22).
Bourot and Karst (3) identified a gene involved in sterol uptake, while they were seeking strains that were resistant to the morpholine antifungal agent, fenpropimorph. The overexpression of SUT1 (YGL162w) resulted in fenpropimorph resistance. Fenpropimorph acts by inhibiting two enzymes in the ergosterol biosynthetic pathway, C14 reductase activity (ERG24) and Δ8-7 isomerase (ERG2) (2, 13). Drug resistance via SUT1 overexpression could stem from the aerobic uptake of ergosterol from the environment, in effect bypassing the need for the inhibited enzymes. In comparison to the wild type, an isogenic strain overexpressing SUT1 mediates a 2.6-fold increase in sterol uptake (3). This gene is hypoxically controlled (3, 20). SUT1 hypoxic regulation is not surprising because the mechanism of sterol uptake is needed for survival under anaerobic conditions.
Lewis et al. (11) isolated a yeast mutant, upc2-1, which resulted in aerobic sterol uptake. Under native conditions the level of sterol uptake was 10- to 20-fold greater than with the isogenic wild type. The mutant was heme and ergosterol competent and in fact resulted in increased ergosterol production. Crowley et al. (4) showed that the mutation was a single transition from guanine to adenine resulting in a predicted switch from glycine to aspartate at amino acid 888 near the carboxy terminus of the YDR213w open reading frame (ORF). This ORF, designated UPC2, belongs to a group of more than 80 fungal proteins that contain a Zn(II)2Cys6 binuclear cluster DNA-binding domain (19, 21). The domain is unique to fungal proteins, most of which are transcriptional regulators. The transcriptional activators Gal4p and Hap1p are the most widely studied examples of this group.
Another ORF, containing the unique DNA-binding domain and having a high predicted amino acid sequence homology with Upc2p (45%), is YLR228c. In fact, at the carboxy terminus where the upc2-1 point mutation occurs, 36 consecutive amino acids are identical, and 130 out of the last 139 amino acids are similar (Fig. 1). Because of the high homology, we characterized this ORF to determine if it had some of the same phenotypes as Upc2p. A YLR228c knockout strain was constructed and mated with strains containing different UPC2 alleles (Table 1). The resulting strains were tested for sensitivity to cations, sterol species present, and sterol uptake levels. Additionally, the upc2-1 point mutation was duplicated in the homologous carboxy region of YLR228c. The resulting strain was also phenotypically analyzed.
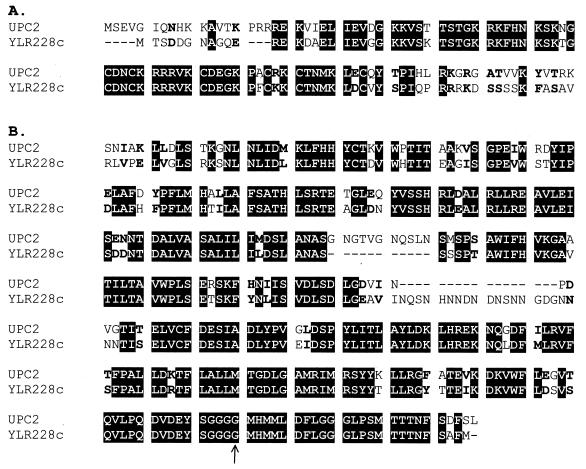
FIG. 1.
Alignment of Upc2p and the YLR228c ORF product. The BioEdit alignment program created by Tom Hall (North Carolina State University Department of Microbiology) was used to gap align the Upc2p and YLR228c amino acid sequences. The black boxes signify identity, and the boldface characters signify similarity. Only the homologous regions of the alignment are shown since the entire sequence is available in the Saccharomyces Genome Database. (A) The DNA-binding domain is present where the high homology occurs at the amino terminus. (B) The homologous region at the carboxy region is the proposed activation domain. The arrow shows where the upc2-1 point mutation occurs (4).
TABLE 1.
S. cerevisiae strains used in this study
| Straina | Genotypeb | Source or reference |
|---|---|---|
| 463-1C | MATaleu2 his3 trp1 ura3 | K. Tatchelc |
| 463-1D | MATα leu2 his3 trp1 ura3 | K. Tatchel |
| 463-1C/463-1D | MATa/MATα leu2/leu2 his3/his3 trp1/trp1 ura3/ura3 | K. Tatchel |
| CJ2-A | MATaupc2-1 leu2 his3 trp1 ura3 | 4 |
| CJ625 | MATaupc2::URA3 leu2 his3 trp1 ura3 | 4 |
| H1B | MATα ylr228c::LEU2 leu2 his3 trp1 ura3 | This study |
| H1B (+) | MATα ylr228c::LEU2 leu2 his3 trp1 ura3 | This study |
| KS93 | MATaupc2-1 ylr228c::LEU2 leu2 his3 trp1 ura3 | This study |
| KS93 (+) | MATaupc2-1 ylr228c::LEU2 leu2 his3 trp1 ura3 | This study |
| KS94 (+) | MATaupc2::URA3 ylr228c::LEU2 leu2 his3 trp1 ura3 | This study |
| H1B (++) | MATα ylr228c::LEU2 leu2 his3 trp1 ura3 | This study |
| KS122 (++) | MATaupc2-1 ylr228c::LEU2 leu2 his3 trp1 ura3 | This study |
| KS119 (++) | MATaupc2::URA3 ylr228c::LEU2 leu2 his3 trp1 ura3 | This study |
+, Strain is transformed with the plasmid YEp351 containing the YLR228c point mutation; ++, strain is transformed with the plasmid pRS313 containing the YLR228c point mutation.
upc2 = YDR213w.
Kelly Tatchel, Louisiana State University Medical Center, Shreveport, La.
MATERIALS AND METHODS
Materials.
Dextrose, sodium chloride, potassium hydroxide, potassium acetate, yeast extract, Bacto Agar, tryptone, peptone, and yeast nitrogen base (without amino acids) were purchased from Fisher Scientific for media preparation. The following media components were obtained from Sigma: succinic acid, ampicillin, nucleic acids, and all amino acids. Restriction enzymes and DNA modifying enzymes were purchased from New England Biolabs. The Taq PCR Core Kit by Qiagen was used for the PCR. [14C]cholesterol for the sterol uptake experiments was purchased from Amersham Life Sciences, Inc. The Sigma products Tergitol NP-40 and Tyloxapol were used during the sterol uptake experiments.
Strains and culture conditions.
All strains used during the following experiments are listed in Table 1. Yeast cultures were grown at 30°C in either rich or synthetic complete selective media prepared as previously described (18).
Insertional inactivation of YLR228c.
The procedure used to insertionally inactivate the YLR228c ORF, which we call the Belgian technique (4, 6), involves creating a DNA fragment that contains upstream and downstream sequences of the ORF in question surrounding an auxotrophic cassette. This fragment is used to transform the appropriate yeast strain. Colonies are selected for the expression of the specific auxotrophic cassette. For these experiments PCR was performed with the following components added to each reaction: 2 μg of primers YLR-5 (5′-ACAAGACTGCAGATACTAGAGAAACATCAA-3′) and YLR-3 (5′-TGCTATAAGCTTCTGGCACATTAACGTATG-3′), 1× PCR buffer (1.5 mM MgCl2), 1 × Q-solution, a 200 μM deoxynucleoside triphosphate mix, and 1.25 U of Taq DNA polymerase. Sterile distilled water was added to a final volume of 50 μl. The following conditions were used in a Perkin-Elmer 2400 thermal cycler as follows: 5 min at 94°C followed by 35 cycles of 30 s at 94°C, 30 s at 50°C, and 2 min at 72°C. The reaction was completed with a 7-min hold at 72°C. The amplified region consisted of 610 bp upstream of the ORF, the YLR228c ORF (2,445 bp), and 577 bp downstream of YLR228c. This 3.6-kb PCR fragment was purified using the GeneClean III Kit from Bio 101. The YLR-5 and YLR-3 primers contain PstI and HindIII restriction sites, respectively, which were used to clone the PCR product into the pUC18 vector which had been cut with the same enzymes. This construct was used as a template for PCR to amplify the upstream and downstream portions of YLR228c. Primers were constructed that amplified away from the ORF, resulting in a linear fragment that contained an upstream fragment, the entire pUC18 plasmid, and the downstream fragment (in this order). The primers used for this reaction were YLRKO-5 (5′-TTCGATGGATCCGTGCTACAGAATGTGAAC-3′) and YLRKO-3 (5′-TTCCCAGGATCCGATGTCATGTTATGTTAG-3′). For PCR, the same conditions were used as before, except the annealing temperature was changed to 45°C. The PCR product was cut upstream at −19 with BglII and downstream at +185 with BclI. The resulting product contained 39 bp (19 upstream and 20 at the beginning of the ORF), the entire pUC18 plasmid, and 185 bp of the downstream sequence (starting 25 bp downstream from the stop codon). The BglII/BclI-cut product was ligated to the LEU2 cassette, which had been cut out of YDp-L using BamHI. To create a linear fragment for the transformation of yeast, the construct was excised, using the PstI and HindIII sites that were incorporated during the original amplification, to ligate the 3.6-kb product into pUC18. This fragment was transformed into a diploid wild-type yeast strain (463-1C × 463-1D). Leucine prototrophs were selected and allowed to sporulate. Tetrads were dissected and scored again for leucine prototrophy. Deletion of the YLR228c ORF was then confirmed by Southern blot.
Creation of the upc2-1 point mutation in the YLR228c ORF.
To duplicate the point mutation from upc2-1 in the YLR228c ORF, the Quickchange Site-Directed Mutagenesis Kit from Stratagene was used. Primers YLR-A (5′-GATGAGTATAGTGGAGGTGGTGATATGCATATGATGCTGG-3′) and YLR-B (5′-CCAGCATCATATGCATATCACCACCTCCACTATACTCATC-3′) were used, and the manufacturer's directions for the kit were followed. The pUC18 vector, which contained the YLR228c ORF, along with upstream (610 bp) and downstream (577 bp) sequences, was used as the template for mutagenesis. This vector was used instead of the appropriate yeast shuttle vector (YEp351) because the efficiency of the mutagenesis protocol is dependent on size (Stratagene literature and catalog): 6.3 kb for pUC18 and 9.2 kb for YEp351. Once the point mutation was created, the 3.6-kb fragment was cut from pUC18 using PstI and HindIII. The fragment was then ligated to the YEp351 vector, which had been cut with the same enzymes. The YEp351 auxotrophic LEU2 marker was replaced with the HIS3 marker by cutting out a portion of LEU2 and ligating the HIS3 cassette into the disrupted marker. The vector was sequenced across the mutation by the Iowa State University sequencing facility to verify that the point mutation was present. The YLR228c::LEU2 knockout strain, H1B, was transformed with the YEp351-YLR228c (point mutation) construct. Leucine and histidine prototrophs were selected. This strain, H1B(+), was then crossed with other appropriate strains and used for sterol uptake experiments (Table 2). To construct a low-copy-number vector containing the YLR228c point mutation, PCR was performed using the following primers containing XhoI and NotI restriction sites, respectively: YLR-5 XhoI (5′-CCCGAAACTCGAGCAAGACGGAAAATACTAGAGAAACATCAAC-3′) and YLR-3 NotI (5′-CCCGCGGCCGCTGCTATATTCTCCTGGCACATTAACGTATGCC-3′). The PCR product was cloned into the pGEM-T vector by TA cloning, cut with XhoI and NotI, and ligated into pRS313 (low-copy vector) via the XhoI and NotI restriction sites. Sequencing was performed, as before, to verify the presence of the point mutation. This vector was then transformed into H1B and crossed with the appropriate strains for sterol uptake experiments.
TABLE 2.
Aerobic sterol uptake (high-copy vector)
| Straina | Genotype | Sterol uptakeb |
|---|---|---|
| 463-1C | Wild type | 1 |
| CJ2-A | upc2-1 | 12.40 |
| H1B | ylr228c::LEU2 | 1 |
| H1B (+) | ylr228c::LEU2 | 1.95 |
| KS93 (+) | ylr228c::LEU2 upc2-1 | 15.82 |
| KS94 (+) | ylr228c::LEU2 upc2::URA3 | 2.92 |
+, Strain was transformed with the YEp351 plasmid containing the YLR228c point mutation.
Sterol uptake is represented as the fold difference compared to the wild type from an experiment performed in triplicate.
[14C]cholesterol uptake.
Appropriate selective media were made with the addition of radiolabeled (hot) and nonlabeled (cold) cholesterol. The hot-cold mixture was made by adding 2 mg of cold cholesterol to 1 ml of toulene. Then, 2.5 μCi of [14C]cholesterol was added. The hot-cold mixture was completely dried with nitrogen gas. Once dried, 100 μl of 95% ethanol and 100 μl of Tyloxapol were added to the dried sample. The sample was resuspended by vigorous vortexing. The entire resuspended sample was transferred to 100 ml of media. Then, 5 ml of hot-cold media was transferred to 15-ml conical tubes, and 50 μl from the overnight cultures was inoculated into the media. The culture was grown aerobically with vigorous shaking at 30°C for 48 h. After 2 days of growth the culture was pelleted and washed with 5 ml of 0.5% Tergitol. The pellet was vortexed in the residual wash solution and lyophilized overnight or until the pellet was dry. The pellets were weighed and placed in 3 ml of scintillation fluid, and the accumulated [14C]cholesterol was determined using a Beckman LS6500 scintillation counter. Samples were compared by determining the amount of [14C]cholesterol taken up by the cells (in micrograms per milligram of dry weight of the cells). All samples were tested in triplicate, and an average was used for comparison.
Sterol analysis.
Sterols were extracted according to the method of Parks et al. (16), and the nonsaponifiable fraction was analyzed by using isothermal gas chromatography as described by Fenner and Parks (7).
RESULTS AND DISCUSSION
A GenBank database search, using the predicted amino acid sequence for Upc2p (YDR213w), resulted in the identification of an S. cerevisiae protein with 45% identity and 57% similarity across 945 amino acids (Fig. 1). This protein is encoded by the YLR228c ORF. All of the other matches were to small regions homologous to the Zn(II)2Cys6 DNA-binding domain near the amino terminus of Upc2p. It was not surprising to find proteins with homology to the DNA-binding domain, since this domain is unique to the fungi and well conserved and over 80 similar proteins have been described (21). The protein encoded by YLR228c has strong similarity to the DNA binding domain and a large region at the carboxy terminus of Upc2p (Fig. 1). In Gal4p and some other transcriptional activators the carboxy terminus acts as the major activation domain (9, 19).
Experimentally, the first step was to inactivate the YLR228c ORF. This was accomplished using the technique of Eberhardt and Hohmann (6) as modified by Crowley et al. (4). After the YLR228c knockout strain was created, the strain was tested for sterol uptake and cation sensitivity. As was shown for the UPC2 knockout (4), the YLR228c knockout strain resulted in a wild-type level of uptake and cation resistance. Next, the YLR228c knockout was crossed with the UPC2 knockout and the upc2-1 strain in preparation for sterol uptake experiments. Analysis of over 40 tetrads from three different matings did not result in a single ascospore that grew on synthetic complete plates lacking leucine and uracil. This indicates that the combination of YLR228c::LEU2 with UPC2::URA3 results in lethality. Independently, the auxotrophic markers used to knock out the genes segregated in a 2:2 manner. The lethality of the double knockout suggested that the two proteins have some essential overlapping function. The amino acid sequence homology between the carboxy termini (possible activation domain) further implied an overlap in function.
We had shown in a preliminary experiment that sterol uptake for a haploid strain containing the upc2-1 point mutation and the YLR228c::LEU2 knockout, strain KS93(−), resulted in a higher level of sterol uptake in comparison to upc2-1 alone. This being known, we wanted to determine the sterol uptake levels of strains containing defects in UPC2 and YLR228c (Table 2). The double point mutation where YLR228c was expressed from a high-copy vector resulted in a 15-fold increase in sterol uptake over the wild type and a slight but significant increase over a strain with only the upc2-1 mutation. The uptake from a strain containing only the YLR228c knockout was equivalent to the wild type. After we transformed the knockout strain with the YEp351-YLR228c (point mutation) vector the uptake doubled. The increased uptake shows that the duplicated upc2-1 mutation within the YLR228c amino terminus results in the aerobic sterol uptake phenotype similar to upc2-1, albeit at a much lower level. A strain containing the double knockout is nonviable, but when the YEp351-YLR228c (point mutation) vector is present, the strain is viable and uptake is threefold greater than with the wild type. The sterol uptake experiment shown in Table 2 was repeated twice, and the fold sterol uptake was similar in each experiment.
Sterol uptake experiments where the YLR228c point mutation was expressed from a low-copy centromeric vector (pRS313) did not result in increased uptake to our surprise (Table 3). The uptake experiment was repeated four times to verify the unexpected results. One possible explanation for the low-copy results is that the mechanism of sterol uptake may involve other minor contributory factors that are obvious only when the mutated YLR228c is overexpressed. Another scenario may be that YLR228c is actively repressed under specific environmental conditions. When YLR228c is overexpressed this repression would be lost and aerobic sterol uptake could occur.
TABLE 3.
Aerobic sterol uptake (low-copy vector)
| Straina | Genotype | Sterol uptakeb |
|---|---|---|
| 463-1C | Wild type | 1 |
| CJ2-A | upc2-1 | 11.4 |
| H1B | ylr228c::LEU2 | 1.5 |
| H1B (++) | ylr228c::LEU2 | 1.2 |
| KS122 (++) | ylr228c::LEU2 upc2-1 | 9.1 |
| KS119 (++) | ylr228c::LEU2 upc2::URA3 | 1.5 |
++, Strain was transformed with the pRS313 plasmid containing the YLR228c point mutation.
Sterol uptake is represented as the fold difference compared to the wild type from an experiment performed in triplicate.
The possibility remained that increased sterol uptake from the high-copy vector could be the result of the production of aberrant sterols instead of ergosterol. To verify that ergosterol was the consensus sterol being produced by each strain, gas chromatography was performed on the nonsaponifiable fraction of the various strains. These analyses showed that the sterols from all of the strains were similar to the wild-type profile (data not shown), where ergosterol was the consensus sterol produced.
The results presented here show that a haploid strain with combined point mutations in UPC2 and the YLR228c ORF expressed from a high-copy vector resulted in a modestly increased level of aerobic sterol uptake compared to a haploid strain with a single mutation in UPC2. Also, the combination of the UPC2 and YLR228c ORF knockouts was nonviable. However, this strain was rescued with the addition of one of the knocked-out proteins supplied in trans.
Sterol uptake from the growth media is precluded aerobically but is essential for yeast growth under anaerobic conditions. The SUT1 gene (YGL162w) is hypoxically controlled (3, 20) with an anaerobic/aerobic ratio of expression of 9.6. Modest but significant differences in expression of UPC2 and YLR228c have also been observed, when anaerobic versus aerobic growth conditions are compared, with ratios of 2.2 and 1.8, respectively. These data are available online (http://wwwimp.leidenuniv.nl/∼yeast/search.htm) for aerobic and anaerobic glucose-limited steady-state growth conditions. How the medium and other alterations of growth conditions would affect these results remain to be determined. These results suggest that UPC2 and YLR228c may be activated anaerobically, whereas it is clear that SUT1 is maximally expressed under anaerobic (or hypoxic) conditions.
The hypoxic regulation of SUT1 is controlled via the binding of the Rox1p to the putative consensus sequence within the SUT1 promoter under aerobic conditions (3). This binding results in decreased aerobic transcription. The Rox1p consensus sequence has not been identified in the promoter region for UPC2 or YLR228c, and it has not been shown that the two genes are responsive to Rox1p aerobic transcriptional repression. Therefore, it is evident that UPC2 and the YLR228c ORF are unique genes involved in aerobic sterol uptake with an entirely different mechanism of control than that of SUT1.
Currently, the genes that are regulated by Upc2p and the YLR228c gene product have not been determined. Therefore, it is difficult to assign a specific function to these putative transcriptional activators based merely on the fact that they are involved in a few phenotypic characteristics (sterol uptake, cation sensitivity, etc.). It has been shown by Northern analysis that the upc2-1 mutant is responsible for an increase in the production of the late ergosterol biosynthetic enzymes (10). However, it is not known whether Upc2p is directly or indirectly controlling the level of expression of these enzymes. Further experiments to determine the specific sequence with which the DNA-binding domain interacts and which genes contain this sequence need to be performed for a better understanding of the role of Upc2p and the YLR228c product. Recently, D'Alessio and Brandriss (5) showed that Gal4p was able to bind to the Put3p DNA consensus sequence and control activation in a PUT3-null mutant. Put3p is a member of the Zn(II)2Cys6 transactivator family which controls genes responsible for the utilization of proline as the sole nitrogen source in Saccharomyces. This is the first evidence of cross talk occurring within this unique family of transcriptional activators, which leaves the possibility that this could occur between Upc2p and the YLR228c gene product at their respective DNA consensus sequences in the absence of one of the proteins.
ACKNOWLEDGMENTS
This research was supported by the North Carolina Agricultural Research Service, by a grant from the U.S. Army Research Office (DAAAH04-9-003), and by an AASERTS award (DAAAH094-G0197).
REFERENCES
- 1.Andreason A A, Stier T J B. Anaerobic nutrition of Saccharomyces cerevisiae. I. Ergosterol requirements for growth in a defined medium. J Cell Comp Physiol. 1953;41:23–36. doi: 10.1002/jcp.1030410103. [DOI] [PubMed] [Google Scholar]
- 2.Baloch R I, Mercer E I, Wiggins T E, Baldwin B C. Inhibition of ergosterol biosynthesis in Saccharomyces cerevisiae and Ustilago maydis by tridemorph, fenpropimorph, and fenpropidin. Phytochemistry. 1984;23:2219–2226. [Google Scholar]
- 3.Bourot S, Karst F. Isolation and characterization of the Saccharomyces cerevisiae SUT1 gene involved in sterol uptake. Gene. 1995;165:97–102. doi: 10.1016/0378-1119(95)00478-o. [DOI] [PubMed] [Google Scholar]
- 4.Crowley J H, Leak F W, Jr, Shianna K V, Tove S, Parks L W. A mutation in a purported regulatory gene affects control of sterol uptake in Saccharomyces cerevisiae. J Bacteriol. 1998;180:4177–4183. doi: 10.1128/jb.180.16.4177-4183.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Alessio M, Brandriss M C. Cross-pathway regulation in Saccharomyces cerevisiae: activation of the proline utilization pathway by Gal4p in vivo. J Bacteriol. 2000;182:3748–3753. doi: 10.1128/jb.182.13.3748-3753.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eberhardt I, Hohmann S. Strategy for deletion of complete open reading frames in Saccharomyces cerevisiae. Curr Genet. 1995;27:306–308. doi: 10.1007/BF00352097. [DOI] [PubMed] [Google Scholar]
- 7.Fenner G P, Parks L W. Gas chromatographic analysis of intact steryl esters in wild-type Saccharomyces cerevisiae and in an ester accumulating mutant. Lipids. 1989;24:625–629. doi: 10.1007/BF02535079. [DOI] [PubMed] [Google Scholar]
- 8.Gollub E G, Liu K P, Dayan J, Adlersberg M, Sprinson D B. Yeast mutants deficient in heme biosynthesis and a heme mutant additionally blocked in cyclization of 2,3-oxidosqualene. J Biol Chem. 1977;252:2846–2854. [PubMed] [Google Scholar]
- 9.Johnston M, Carlson M. Regulation of carbon and phosphate utilization. In: Jones E, Pringle J, Broach J, editors. The molecular and cellular biology of the yeast Saccharomyces: gene expression. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 193–281. [Google Scholar]
- 10.Leak Frank W., Jr . M.s. thesis. Raleigh: North Carolina State University; 1997. [Google Scholar]
- 11.Lewis T L, Keesler G A, Fenner G P, Parks L W. Pleiotropic mutations in Saccharomyces cerevisiae affecting sterol uptake and metabolism. Yeast. 1988;4:93–106. doi: 10.1002/yea.320040203. [DOI] [PubMed] [Google Scholar]
- 12.Lorenz R T, Parks L W. Involvement of heme components in sterol metabolism of Saccharomyces cerevisiae. Lipids. 1991;26:598–603. doi: 10.1007/BF02536423. [DOI] [PubMed] [Google Scholar]
- 13.Mercer I, Baloch R. Inhibition of sterol δ8-7 isomerase and δ14 reductase by fenpropimorph, tridemorph, and fenpropidin in cell-free enzyme systems from Saccharomyces cerevisiae. Phytochemistry. 1987;23:663–668. [Google Scholar]
- 14.Parks L W. Metabolism of sterols in yeast. Crit Rev Microbiol. 1978;6:301–41. doi: 10.3109/10408417809090625. [DOI] [PubMed] [Google Scholar]
- 15.Parks L W, Casey W M. Physiological implications of sterol biosynthesis in yeast. Annu Rev Microbiol. 1995;49:95–116. doi: 10.1146/annurev.mi.49.100195.000523. [DOI] [PubMed] [Google Scholar]
- 16.Parks L W, Bottema C D, Rodriguez R J, Lewis T A. Yeast sterols: yeast mutants as tools for the study of sterol metabolism. Methods Enzymol. 1985;111:333–346. doi: 10.1016/s0076-6879(85)11020-7. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez R J, Low C, Bottema C D, Parks L W. Multiple functions for sterols in Saccharomyces cerevisiae. Biochim Biophys Acta. 1985;837:336–343. doi: 10.1016/0005-2760(85)90057-8. [DOI] [PubMed] [Google Scholar]
- 18.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 19.Schjerling P, Holmberg S. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 1996;24:4599–4607. doi: 10.1093/nar/24.23.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.ter Linde J J, Liang H, Davis R W, Steensma H Y, van Dijken J P, Pronk J T. Genome-wide transcriptional analysis of aerobic and anaerobic chemostat cultures of Saccharomyces cerevisiae. J Bacteriol. 1999;181:7409–7413. doi: 10.1128/jb.181.24.7409-7413.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Todd R B, Andrianopoulos A. Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet Biol. 1997;21:388–405. doi: 10.1006/fgbi.1997.0993. [DOI] [PubMed] [Google Scholar]
- 22.Trocha P J, Sprinson D B. Location and regulation of early enzymes of sterol biosynthesis in yeast. Arch Biochem Biophys. 1976;174:45–51. doi: 10.1016/0003-9861(76)90322-2. [DOI] [PubMed] [Google Scholar]