Abstract
The light organ of the squid Euprymna scolopes is specifically colonized to a high density by the marine bacterium Vibrio fischeri. To date, only a few factors contributing to the specificity of this symbiosis have been identified. Using a genetic screen for random transposon mutants defective in initiating the symbiotic association or in colonizing the light organ to high density, we identified a mutant of V. fischeri that exhibited an apparent defect in symbiosis initiation. This mutant was not defective in motility, luminescence, or growth in minimal medium, suggesting that it lacks an essential, previously unidentified symbiotic function. By sequence analysis, we showed that the locus inactivated in this mutant encodes a predicted 927-amino-acid protein with a high degree of similarity to the sensor component of hybrid two-component regulatory systems. We have therefore designated this locus rscS, for regulator of symbiotic colonization—sensor. Sequence analysis revealed two hydrophobic regions which may result in the formation of a periplasmic loop involved in signal recognition; PhoA fusion data supported this proposed membrane topology. We have investigated the start site of rscS transcription by primer extension and identified a putative promoter region. We hypothesize that RscS recognizes a signal associated with the light organ environment and responds by stimulating a putative response regulator that controls protein function or gene expression to coordinate early colonization events. Further studies on RscS, its cognate response regulator, and the signaling conditions will provide important insight into the interaction between V. fischeri and E. scolopes.
The bioluminescent marine bacterium Vibrio fischeri forms an intimate symbiotic association with the Hawaiian squid Euprymna scolopes. Juvenile E. scolopes squid hatch from eggs uncolonized by V. fischeri cells but become rapidly and exclusively colonized by this bacterium despite potential competition from the many other bacterial species present in seawater (46, 48). Initiation of this mutualism typically requires only a short exposure (1 to 3 h) to V. fischeri, after which normal colonization will result even if the squid are removed to Vibrio-free seawater (40). Within 24 h, the V. fischeri cells reach a population of between 105 and 106 CFU in the symbiotic light organ, equivalent to approximately 108 CFU/cm3 (40).
Because of the exclusive nature of the relationship between V. fischeri and E. scolopes, it is likely that both bacterial and host genes function in actively establishing this specific, long-term association. Through the study of bacterial mutants, three major stages of symbiotic colonization have been identified. These stages include (i) initiation of the association (initiation), (ii) colonization to the high cell density typically achieved by the wild type (accommodation), and (iii) persistence at a high cell density (persistence) (reviewed in reference 39).
Mutants defective for each of the stages have been identified. Initiation mutants include those defective for motility, because neither nonflagellated nor flagellated but nonmotile mutants can colonize the host (21). It is thought that a thick mucus layer present in the ducts leading to the light organ crypts, where colonization occurs, may prevent entry of nonmotile bacteria (47). Accommodation mutants include bacteria defective in one of several amino acid-biosynthetic genes. The ability of these strains to colonize, albeit at lower levels, suggests that the light organ environment is nutrient rich and that the required amino acids are being supplied by the squid host. The discovery of peptides present at high concentration in the fluid of the light organ supports this hypothesis (22).
Persistence mutants include two distinct subclasses. One subclass mutant lacks the ability to produce and take up siderophores. This mutant exhibited a decreasing level of colonization over the course of 3 days (23), suggesting that sequestration of iron from the light organ environment is an important factor in maintaining the symbiotic association. The second subclass includes bioluminescence mutants, defective in either structural or regulatory genes. The bioluminescence mutants colonize to a level comparable to the wild-type strain during the first day and thereafter show a decreased level of colonization (45, 47). At present, the exact relationship between defects in light production and reduced colonization remains unclear, but it has been speculated that utilization of oxygen by the bioluminescence machinery may reduce host-imposed oxidative stress (45).
Although these symbiosis-impaired mutants have provided valuable clues to the environmental conditions in the light organ, no comprehensive screen for bacterial genetic determinants of symbiotic competence has been described. We anticipate the existence of bacterial genes that are essential to initiate the symbiosis, including surface markers or receptors, as well as additional genes required for accommodation. It is also likely that regulatory factors controlling one or more regulons involved in the symbiosis will be found, particularly in view of the developmental events taking place in both partners as their association progresses (33, 39).
In the present study, we have screened the ability of mutants generated by a random transposon insertion mutagenesis to colonize juvenile E. scolopes. We present here the characterization of one mutant with a severe symbiosis defect. This mutant colonizes poorly and thus may be defective in the ability to overcome a barrier(s) associated with early colonization events. We also present sequence data that predict that the defective gene interrupted by the transposon insertion in this mutant strain may encode a regulatory protein. We hypothesize that this putative regulator may play a key role in the expression of genes essential for, or even induced by, this bacterium-animal association.
MATERIALS AND METHODS
Strains and media.
V. fischeri strain ES114 (9) and its rifampin-resistant derivative ESR1 (21) were the parent strains used in this study. Escherichia coli strains DH5α and CC118λpir (27) were used as host strains for cloning, and S17-1λpir (43) was used as the donor strain in conjugations.
V. fischeri cells were grown in SWT (9), LBS (15), or HEPES-minimal medium (41) containing ribose as a carbon source. E. coli strains were grown in Luria-Bertani (LB) medium (12). Conditioned medium (CM) was used to determine whether the colonization mutant KV712 was defective for bioluminescence in culture and was made as follows. V. harveyi B392 (38) was grown in LM (35) to an optical density of approximately 0.8 at 600 nm. The cells were removed by centrifugation followed by passage of the supernatant through a 0.2-μm-pore-sized filter. Where appropriate, antibiotics were added to the following final concentrations: ampicillin, 100 μg/ml; chloramphenicol (CHL), 1 to 5 μg/ml for V. fischeri and 30 μg/ml for E. coli; erythromycin, 1 to 5 μg/ml for V. fischeri and 150 μg/ml for E. coli; rifampin, 100 μg/ml; and tetracycline (TET), 5 μg/ml for V. fischeri and 15 μg/ml for E. coli. Agar was added to a final concentration of 1.5% for solid medium or 0.25% for motility plates.
Genetic techniques.
Transposon mutagenesis was carried out using the Tn10-lacZ delivery plasmid pKV124 (see below). Conjugations were performed as follows. The donor strain (S17-1λpir carrying the plasmid to be delivered) and the recipient V. fischeri strain (typically either ES114 or ESR1) were grown to mid-log phase at 37°C in LB and at 28°C in LBS, respectively. The strains were mixed, concentrated by centrifugation, and allowed to mate on solid medium. After an incubation of 6 to 14 h at 28°C, cells were diluted in LBS and transferred to selective medium. In some cases, triparental conjugations were performed using the appropriate V. fischeri recipient strain and two E. coli strains, DH5α carrying the plasmid to be transferred and DH5α carrying pRK2013 (13, 17), to supply conjugation functions.
Molecular techniques.
All plasmid constructions were carried out by standard molecular biology techniques, using restriction and modifying enzymes obtained from New England Biolabs (Beverly, Mass.) or Promega (Madison, Wis.). The mini-Tn10-lacZ transposon in pKV124 was derived from pBSL181 (1) by the insertion of both a promoterless lacZ gene and oriR6K, the pir-dependent origin of replication from plasmid R6K (31). This plasmid is therefore a “suicide” plasmid in hosts lacking the pir gene, including V. fischeri. Cloning of DNA flanking the transposon insertion in V. fischeri strain KV712 was accomplished using the oriR6K and CHL resistance elements contained within the transposon. Chromosomal DNA was isolated and digested with a restriction enzyme that does not cleave within the insertion sequences. Fragments were then self-ligated using T4 DNA ligase and transformed into the permissive host CC118λpir or S17-1λpir. Clones carrying the transposon and flanking DNA were selected on CHL-containing LB plates. Two plasmids, pKV132 (containing about 3.8 kb of flanking DNA) and pKV133 (containing about 8.0 kb of flanking DNA), were obtained by digestion with NheI and BsrGI, respectively. DNA fragments from clones pKV132 and pKV133 were subcloned into pBluescript (Stratagene, La Jolla, Calif.), pBC (Stratagene), or pEVS79 (E. V. Stabb and E. G. Ruby, submitted for publication). One of these subclones, pLMS22, contained a 2-kb HindIII-PstI fragment; this plasmid was recombined into V. fischeri strain ESR1. Chromosomal DNA isolated from the resulting recombinant was used to clone the wild-type copy of the rscS gene and flanking DNA, resulting in pLMS25. Plasmid pLMS26 was derived from pLMS25 by the insertion of a 6-kb (rscS+) fragment into pKV69, a TETr CHLr mobilizable vector.
Sequencing was carried out using forward and reverse primers complementary to vector and insert sequences. Manual sequencing for primer extension was carried out using the Sequenase sequencing kit (Amersham-Pharmacia Biotech, Piscataway, N.J.). Similarity searches were performed using the NCBI Blast program (2). Oligonucleotides were obtained from either the Loyola Macromolecular Analysis Facility or Integrated DNA Technologies (Coralville, Iowa).
Southern blotting.
Chromosomal DNA isolated from V. fischeri strain KV712 was digested with EcoRV, which cuts once within the mini-Tn10-lacZ transposon, or BsrGI, which cuts outside the transposon. DNA fragments were separated using a 0.6% agarose gel, transferred onto a nylon membrane (Hybond XL; Amersham-Pharmacia Biotech), and UV cross-linked. Detection was carried out using the Boehringer Mannheim digoxigenin DNA labeling kit (Roche Molecular Biochemicals, Indianapolis, Ind.) as follows. Random priming was used to generate biotinylated DNA fragments complementary to the transposon sequences, and these fragments were hybridized to the chromosomal DNA in a buffer containing 50% formamide, 0.1% N-lauroylsarcosine, 0.02% sodium dodecyl sulfate, 750 mM NaCl, and 75 mM sodium citrate. Blocking reagent was added to 2% for the prehybridization step. Washing, blocking, treatment with primary and secondary antibodies, and colorimetric detection were carried out under conditions of high stringency according to the manufacturer's instructions.
Primer extension.
A 25-ml culture of V. fischeri strain ES114 was grown in LBS to an optical density of 1 at 600 nm. The cells were lysed in GITCN lysis buffer (44), and mRNA was isolated by centrifugation through a cesium chloride gradient as previously described (44). An oligonucleotide primer, 10R3 (5′-GATTGTGATAAGGCTATAACG-3′), complementary to the 5′ end of the rscS locus was radiolabeled by treatment with T4 polynucleotide kinase (Amersham-Pharmacia Biotech) in the presence of [γ-32P]ATP (Amersham-Pharmacia Biotech). The radiolabeled primer was hybridized to the mRNA and incubated with Moloney murine leukemia virus (MMLV) reverse transcriptase (Stratagene) and nucleotides per the manufacturer's instructions. Extension products were visualized by autoradiography after separation on an 8% polyacrylamide gel and compared to a sequencing ladder generated using the same oligonucleotide primer to sequence from plasmid pLMS35, containing the rscS locus and flanking DNA.
Colonization assays.
To determine whether mutant V. fischeri strains were able to form a symbiotic association with E. scolopes, juvenile squid were placed in artificial seawater (Instant Ocean; Aquarium Systems, Mentor, Ohio) containing an inoculum of 1,000 to 5,000 cells of the desired V. fischeri strain per ml of fluid as previously described (39). Luminescence was monitored over the course of 16 to 24 h using a Packard Tri-carb 2100TR scintillation counter (Packard Instruments, Meriden, Conn.) modified to detect bioluminescence. At specific intervals, juvenile E. scolopes were sacrificed by homogenization to directly quantify the number of bacteria present in the light organ. Homogenates were serially diluted, and aliquots were spread on SWT agar for viable-cell counts. The limit of detection is 14 V. fischeri cells per squid.
Luminescence assays.
KV712 and ESR1 were diluted 1:100 and grown in CM prepared as described above, in the presence or absence of a synthetic V. fischeri autoinducer (600 ng of 3-oxo-hexanoyl-l-homoserine lactone [Sigma, St. Louis, Mo.] per ml). At various times after inoculation, 1-ml samples were taken for luminescence and optical density measurements. A TD-20/20 luminometer (Turner Designs, Sunnyvale, Calif.) was used to determine the level of bioluminescence of KV712 and its parent.
Construction and analysis of phoA fusions.
A mini-Tn10 derivative carrying the phoA and lacZ genes fused in frame was obtained from M. Alexeyev. This transposon, carried by pMA651, was introduced by conjugation into DH5α carrying pBlueScript with a 3-kb rscS+ insert. After conjugation and selection for recipient strains carrying the transposon (carried out as described above for V. fischeri), the selected cells were pooled and subjected to a plasmid extraction procedure. The pooled plasmids were sequentially introduced into DH5α and CC118λpir which were then grown on LB medium containing CHL and ampicillin to select for the presence of plasmids containing a transposon insertion. CC118λpir recipient cells were plated on LB containing CHL and 5-bromo-4-chloro-3-indolylphosphate, an indicator of alkaline phosphatase activity. Colonies that turned blue on this medium (indicating an active fusion to PhoA) were identified; the plasmids from these colonies were analyzed by restriction digestion to ascertain the location of the transposon fusion. Fusions that mapped to the rscS gene were further localized by sequence analysis.
Nucleotide sequence accession number.
The GenBank accession number assigned to the rscS sequence is AF319618.
RESULTS
Identification of a novel symbiotic locus.
We have developed a genetic screen to identify bacterial genes important for the symbiotic interaction between V. fischeri and E. scolopes. Upon entering the light organ of a newly hatched juvenile squid, wild-type V. fischeri rapidly reaches a high cell density (40). A cell density-dependent (quorum-sensing) mechanism regulates V. fischeri bioluminescence; therefore, the amount of light produced by the bacteria in the light organ serves as an indirect measure of colonization. We constructed transposon insertion mutants of V. fischeri strain ESR1 (see Materials and Methods) and inoculated newly hatched E. scolopes juveniles with individual mutant strains. Automated monitoring of luminescence over a 16- to 24-h period was used to determine the ability of these mutants to enter and proliferate within the light organ. This screen should identify mutants of the initiation class, which would be unable to colonize the light organ. The screen should also identify mutants of the accommodation class, which would be unable to reach normal cell densities after initial colonization.
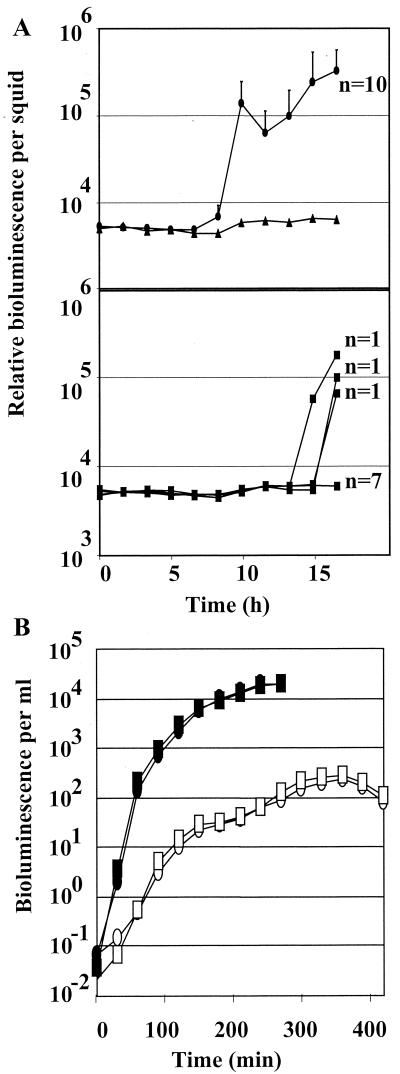
From a collection of approximately 2,000 transposon insertion mutants, we identified a strain, KV712, that failed to exhibit the wild-type pattern (Fig. 1A, top) of bioluminescence in the symbiotic association: during the first 16 h post inoculation, 7 of 10 juvenile squid exposed to this mutant failed to emit light, while 3 exhibited delayed onset of bioluminescence (Fig. 1A, bottom). However, KV712 showed no detectable defect relative to the parent strain ESR1 in producing bioluminescence in culture or in inducing luminescence in response to the addition of an autoinducer (3-oxo-hexanoyl l-homoserine lactone) (Fig. 1B). Thus, the symbiotic luminescence defect apparently did not result from a mutation in the lux operon, which encodes the structural genes for bioluminescence.
FIG. 1.
Luminescence levels of colonization mutant KV712 and its parent. (A) Newly hatched juvenile E. scolopes squid were incubated in artificial seawater containing no V. fischeri (solid triangles, top panel) or either the parent strain ESR1 (solid circles, top panel) or the colonization mutant KV712 (solid squares, bottom panel). Bioluminescence was measured over time. The light levels of 10 animals inoculated with ESR1 (indicated by n = 10) and of 7 animals inoculated with KV712 (indicated by n = 7), were averaged, while three squid colonized by KV712 are represented singly (indicated by n = 1). Error bars are shown, representing the standard deviation of luminescence readings for ESR1-colonized animals; the level of light observed for seven of the KV712-inoculated squid was not above the limit of detection for the machine, and thus error bars are not displayed. (B) ESR1 (circles) and KV712 (squares) were grown in CM in the absence (open symbols) or presence (solid symbols) of an autoinducer of bioluminescence (3-oxo-hexanoyl-l-homoserine lactone). The level of luminescence was measured using a Turner 20/20 luminometer. The two strains grew at essentially the same rate.
In addition to lux mutants, nonmotile mutants and some amino acid auxotrophs fail to establish normal levels of colonization (21, 23). When tested for motility in a soft agar plate, strain KV712 exhibited motility identical to that of the parent strain (data not shown). Growth in a minimal medium was also comparable to that of ESR1 (data not shown). These data suggested that KV712 carries a mutation in a previously unidentified locus that affects the symbiotic interaction.
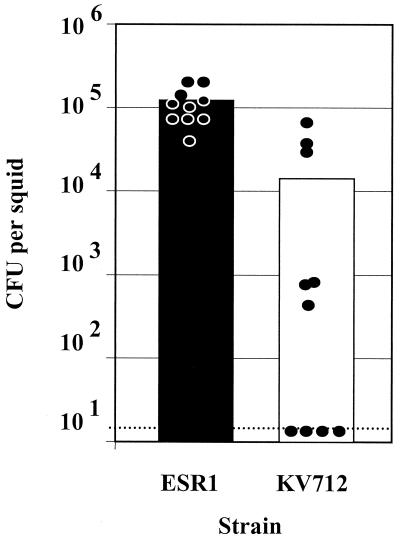
To confirm a potential colonization defect of KV712, we determined the number of CFU present in the light organ of the juvenile squid at 19 h postinoculation (Fig. 2). Of the 10 juveniles inoculated with KV712, 4 exhibited no detectable colonization (the limit of detection in this experiment was approximately 14 CFU), while an additional 3 were colonized at a level approximately 100-fold reduced from that of juveniles exposed to the wild-type strain. The three remaining juveniles, which had exhibited a delay in onset of bioluminescence (Fig. 1A), contained V. fischeri cells at a level comparable to that of the wild-type-colonized squid.
FIG. 2.
Symbiotic colonization by V. fischeri strain KV712 and its parent, ESR1. Newly hatched juvenile E. scolopes were exposed for 15 h to either KV712 or ESR1. The level of colonization achieved by these strains was determined by homogenization and plating 19 h after the organisms were placed together. Each circle represents the number of V. fischeri cells in an individual squid, and the bar indicates the average colonization level of the 10 squid in each inoculation condition (2,300 cells per ml). The level of colonization of four squid inoculated with KV712 was below the limit of detection, as indicated by the dashed line.
These data suggest that although KV712 is capable of bioluminescence, in squid assays it fails to emit the typical wild-type pattern of bioluminescence due to a symbiotic defect. We hypothesize that this mutant lacks the ability to properly initiate the symbiotic association. Occasionally, however, some mutant cells succeed in bypassing the “block” to initiation, after which colonization (i.e., multiplication) within the squid proceeds normally. In support of this idea, we observed that squid in which mutant cells apparently overcame the block more quickly (as judged by luminescence [Fig. 1A]) exhibited higher colonization levels at the assay time than those that experienced a greater delay. Furthermore, in preliminary experiments we have observed that the relative frequency of high-level colonization by KV712 was reduced with smaller inoculum and a shorter duration of inoculation (Skoufos and Visick, unpublished data). Thus, we believe that the observed variation in the data represents random events during the interaction of a homogeneous population of mutants with an animal host.
Molecular characterization of KV712.
Our data suggested that KV712 carries a mutation in a novel symbiosis locus. Because the mutation in this strain was generated by transposon mutagenesis, the number and location of transposon insertions could be easily determined by Southern blot analysis. DNA probes complementary to the transposon and its delivery plasmid were hybridized to chromosomal DNA isolated from KV712 and digested either with BsrGI (which does not cut within the inserted sequences) or EcoRV (which recognizes a single site within the transposon). Consistent with a single transposon insertion event in KV712, we observed a single band for BsrGI-digested DNA and two bands for EcoRV-digested DNA (approximately 5 and 7kb) (data not shown). When the same probe was hybridized to EcoRV-digested chromosomal DNA extracted from ESR1, no bands were observed (data not shown).
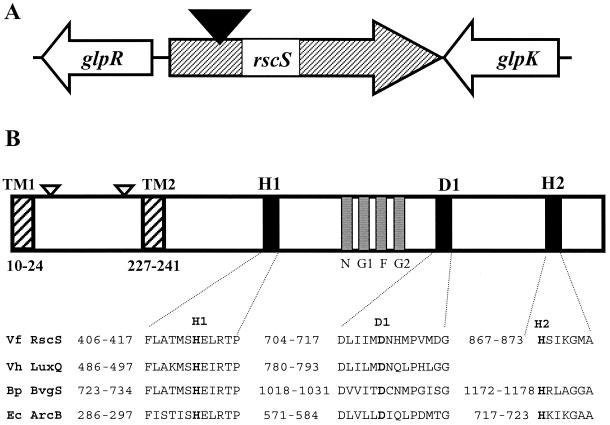
To further investigate the locus that was disrupted in KV712, we cloned the transposon and flanking chromosomal DNA. The presence of an origin of replication (oriR6K) and a CHL resistance marker within the transposon permitted rapid cloning by recircularization of chromosomal DNA fragments and transformation into a permissive E. coli host strain. Subcloning and sequencing revealed that the transposon had inserted into a large open reading frame (ORF), encoding a predicted 105-kDa protein of 927 amino acids.
Sequencing of flanking DNA demonstrated that the genes adjacent to this ORF possess a high degree of similarity to E. coli genes involved in glycerol metabolism and regulation. Located 581 nucleotides upstream of the putative translational start of the large ORF, there is an apparent homolog of the glpR gene (10), while 68 nucleotides downstream there is a glpK analog (37). Both genes are transcribed divergently relative to the large ORF (Fig. 3A), precluding the possibility that the ORF is part of an operon and suggesting that the transposon insertion in strain KV712 affects expression only of that single, large ORF. KV712 and its parent, ESR1, both fail to grow on glycerol as a carbon source, but a mutation of this ORF in ES114 (N. D. Montgomery and K. L. Visick, unpublished data) did not alter that strain's ability to grown on glycerol (data not shown).
FIG. 3.
Schematic diagram of the rscS locus and flanking genes. (A) The region of the chromosome flanking the transposon insertion in KV712 is indicated. The transposon (solid triangle) inserted in a large ORF, rscS, which is flanked by genes with high sequence similiarity to the E. coli glpR and glpK genes. The direction of transcription is indicated. (B) Noteworthy regions of the 927-amino-acid RscS protein are diagrammed. RscS exhibits sequence similarity to the V. harveyi (Vh) LuxQ, Bordetella pertussis (Bp) BvgS, and E. coli (Ec) ArcB proteins, particularly, as indicated below the black bars, in the regions flanking the histidine and aspartic acid residues (H1, D1, and H2) thought to be involved in the phosphorelay cascade. RscS also exhibits sequence similarity to the nucleotide-binding sites depicted by the gray boxes and labeled N, G1, F, and G2. Two putative transmembrane (TM) regions are indicated by striped bars and are designated TM1 and TM2. The locations of active phoA transposon insertions are indicated by open triangles.
Comparison of the ORF sequence to known sequences revealed significant amino acid sequence identity with the sensory component of hybrid two-component regulatory systems (Fig. 3B). Close matches included LuxQ (39% identity, 62% similarity) (7), ArcB (28% identity, 48% similarity) (30), and BvgS (26% identity, 45% similarity) (4), as well as a number of hypothetical V. cholerae sensor kinases (26). Each of these proteins (LuxQ, ArcB, and BvgS) plays a role in sensing an environmental signal and transducing the signal through a phosphorelay cascade (3, 29, 36) to a response regulator protein (LuxO [8], ArcA [14], and BvgA [4], respectively). Each response regulator protein then up- or downregulates transcription of a set of genes.
The domains involved in nucleotide binding and in the phosphorelay cascade are highly conserved among these proteins. The strongest similarity between the ORF disrupted in KV712 and these sensor kinases occurs in these domains (Fig. 3B). From these sequence alignments, we believe that this ORF likely encodes a hybrid two-component sensor kinase. Thus, we tentatively assign this locus the name rscS, for regulator of symbiotic colonization—sensor.
Complementation of the symbiotic defect.
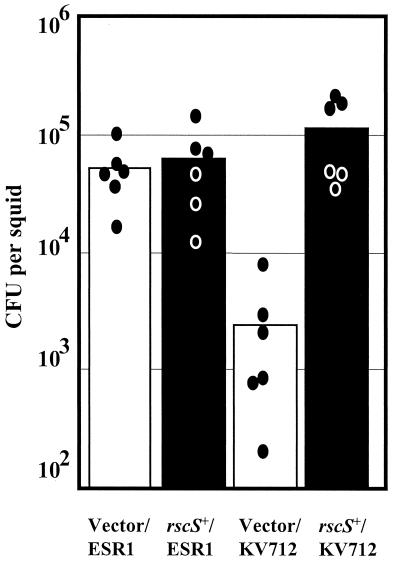
To determine whether the disruption of rscS caused the symbiotic defect of KV712, we cloned the wild-type copy of the locus. We performed complementation assays using a subclone, pLMS26, that contained rscS+ and approximately 3 kb of upstream DNA to ensure the presence of regulatory sequences. We infected juvenile E. scolopes with cells either wild-type (ESR1) or defective for rscS (KV712), carrying either the vector (pKV69) or the rscS+ complementing plasmid pLMS26. Plasmid pLMS26 but not pKV69 apparently restored symbiotic competence to KV712, as monitored indirectly by bioluminescence measurements over a 17-h period (data not shown). The presence of either plasmid did not affect symbiotic luminescence levels of the parent strain (data not shown).
After 17 h, we directly determined the level of colonization achieved by each strain. As predicted from the luminescence patterns, pLMS26 complemented the disrupted rscS gene in KV712: the complemented strain reached colonization levels comparable to those of the parent strain carrying either vector or pLMS26 (Fig. 4). We also observed wild-type levels of colonization when we inoculated juvenile squid with KV712 complemented with a construct containing only the 3-kb rscS+ locus (data not shown). In contrast, KV712 carrying the vector alone remained defective in properly initiating the symbiotic interaction. These results are consistent with the conclusion that the transposon insertion in rscS caused the symbiotic defect displayed by KV712.
FIG. 4.
Complementation of the colonization defect in KV712. Newly hatched juvenile E. scolopes were inoculated with one of four strains: KV712 or its parent ESR1, each carrying either vector pKV69 or rscS+ plasmid pLMS26. The squid were exposed to approximately 1,000 cells of V. fischeri per ml of seawater for 3 h. The level of colonization achieved by these strains 17 h after the organisms were placed together was determined by homogenization and plating as described in Materials and Methods. Each circle represents the number of V. fischeri cells in an individual squid, and the bar indicates the average colonization level of the six squid in each inoculation condition.
Analysis of the rscS promoter region.
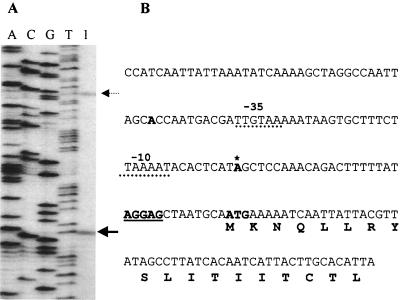
To confirm transcription of the rscS gene and to identify a putative promoter region, we mapped the transcriptional start of rscS using primer extension. A radiolabeled primer complementary to the 5′ region of the putative rscS ORF (spanning nucleotides 19 to 39 downstream from the putative translational start) was hybridized to mRNA isolated from the wild-type V. fischeri strain ES114. This served as the template for reverse transcription by MMLV reverse transcriptase. The resulting product was compared to a sequencing ladder generated from the same primer. We observed a major band (Fig. 5A), which places the transcriptional start site (+1) at the A residue located 34 nucleotides upstream of the predicted ATG translational start codon (Fig. 5B), and a minor band, which maps to an A residue 46 bases further upstream. The significance of the minor transcriptional start is unclear.
FIG. 5.
Promoter mapping by primer extension. (A) The primer extension products obtained using a primer complementary to the rscS coding sequence (see Materials and Methods) appear in lane 1 and are indicated by arrows. Contained within the adjacent lanes, labeled A, C, G, and T, are the bands obtained by DNA sequencing using the same primer. (B) The DNA sequence at the beginning of the rscS ORF and upstream is shown. The putative ATG translational start is indicated by the bold ATG. Centered 11 bp upstream of the possible translational start is a potential Shine-Dalgarno site, boldface and underlined. The possible transcriptional starts identified by primer extension are Mboldface; the more significant product is also indicated by an asterisk. A possible promoter region is indicated by a dashed underline and labeled above with −10 and −35.
We have identified possible regulatory elements upstream of the putative translational start, based on the location of the major transcriptional start site and consensus sequences found in E. coli. These assignments are tentative because there has been little research identifying such elements in V. fischeri. A possible Shine-Dalgarno sequence, AGGAGC (shown in boldface and underlined), is located 8 bases upstream of the proposed translational start and matches five of six bases of the consensus E. coli sequence AGGAGG (42). A putative −10 promoter sequence (dotted underline), TAAAAT, is centered at base −11 with respect to the transcriptional start. This sequence is similar to that identified by primer extension mapping of the V. fischeri lux operon (16) and is identical at five of six bases to the canonical E. coli −10 sequence, TATAAT (25). Fifteen nucleotides upstream of this sequence (dotted underline) is a potential −35 sequence, TTGTAA, that matches the E. coli −35 consensus (TTGACA) (25) at four of six bases. In addition to predicting a possible promoter, these primer extension results provide evidence that the rscS locus is transcriptionally active in cells grown in laboratory culture.
PhoA fusions predict a periplasmic loop.
LuxQ and BvgS hypothetically recognize their respective environmental signals by means of an amino-terminal periplasmic loop (4, 7). These periplasmic loops exhibit no sequence similarity to each other despite the strong conservation of other domains in the proteins, suggesting that this region gives each protein its unique function. Similarly, the amino terminus of the putative RscS shows no significant similarity to any other known protein.
Initial support for the hypothesis that RscS may possess a periplasmic loop resulted from a hydrophobicity analysis (dense alignment surface method) (11). Two highly hydrophobic regions were found in the N-terminal third of the protein (amino acids 10 to 24 and 227 to 241). A second program (SOUSI) (28) predicted two transmembrane (TM) regions, from 6 to 28 and 222 to 244, as well as a third TM from 312 to 334.
To investigate whether these hydrophobic regions result in a periplasmic loop, we mutagenized an E. coli strain (Materials and Methods) carrying a plasmid-borne copy of rscS with a mini-TnphoA transposon. Insertion of this transposon in frame into the rscS gene would result in synthesis of an RscS-PhoA fusion protein, but the PhoA portion would be active only if localized to the periplasm (32). An alkaline phosphatase indicator medium was used to identify fusions with alkaline phosphatase activity; four independent insertions which had this activity were identified. Two of the four active insertions were located after amino acid 40 of RscS, and the other two were both inserted after amino acid 200. Both of these sites are within the region flanked by the two putative membrane-spanning segments, providing strong evidence that this region lies in the periplasm. These data also demonstrate that rscS is not only transcribed but also translated in vivo.
DISCUSSION
This paper reports the first major result of a genetic screen for V. fischeri mutants defective in symbiotic initiation and accommodation: a novel symbiotic locus, rscS, that appears to play an important role in symbiotic initiation. A single transposon insertion in rscS severely impaired the ability of V. fischeri to initiate a symbiotic association with juvenile E. scolopes (Fig. 2), a defect that could be complemented by a wild-type copy of the rscS gene. We identified putative promoter and ribosome-binding sequences on the basis of the transcriptional start site and E. coli consensus sequences. Given a paucity of data for regulatory elements in V. fischeri genes, however, positive confirmation of these sequences will require mutational and other analyses.
Molecular characterization of rscS revealed a predicted 927-amino-acid protein, RscS, that exhibited strong sequence similarity (Fig. 3B) to hybrid two-component sensor kinases. Each of these kinases participates in a series of four phosphorelay reactions that occur in response to particular environmental conditions (such as a specific level of osmolarity or the presence of a particular ion [3, 36]). Initially, autophosphorylation occurs on a histidine residue (H1). This phosphate sequentially transfers to an aspartic acid (D1), another histidine (H2), and finally to another aspartic acid residue (D2).
Although as many as four distinct proteins may be utilized in this phosphorelay, there are typically two key proteins: a sensor that recognizes the environmental cue and begins the phosphorelay, and a response regulator that contains the conserved final aspartic acid residue and effects changes in gene expression or protein function (3, 29, 36). The module arrangement of the RscS protein most resembles that of BvgS and ArcB, both of which contain three of the four conserved phosphorylation domains (H1, D1, and H2). RscS therefore appears to be a sensor component belonging to the subclass of hybrid two-component regulators. The amino acid sequence in the periplasmic loop domain, however, diverges from other known sequences, suggesting that it may respond to a distinct environmental condition.
The predicted RscS protein most resembles LuxQ of V. harveyi (39% identity). LuxQ functions as one of two “redundant” quorum-sensing proteins that sense cell density and regulate luminescence through the recognition of an autoinducer signaling molecule (6, 7). Individually, LuxQ and a second sensor, LuxN, funnel their signals through a common histidine phosphotransferase protein, LuxU, which then modulates phosphorylation of LuxO, a DNA-binding protein that represses transcription of the V. harveyi lux operon (8, 18, 19). In recent years, proteins with sequence similarity to these and other V. harveyi quorum-sensing components have been identified in V. fischeri. The V. fischeri AinR protein (20) shows some sequence similarity with a portion of the V. harveyi LuxN sensor kinase. Furthermore, proteins with high sequence identity to the V. harveyi LuxO (65%) and LuxU proteins have recently been identified in V. fischeri (34). The V. fischeri LuxO plays a role similar to that of the V. harveyi protein in repressing luminescence (34).
The identification of a V. fischeri protein, RscS, with sequence similarity to LuxQ makes it tempting to speculate that RscS plays a role in recognizing an autoinducer and participating in lux gene regulation. Unlike LuxQ, which carries only two of the modules predicted to be involved in a phosphorelay cascade (H1 and D1), RscS contains three of the four modules (H1, D1, and H2). Thus, a LuxU intermediate would likely be unnecessary. The two proteins, RscS and LuxQ, show no apparent sequence similarity in the proposed periplasmic loops, suggesting that they recognize different signals. This prediction is supported by a lack of cross-stimulation of V. harveyi bioluminescence by V. fischeri culture supernatants (5, 24). Furthermore, our data to date indicate no apparent defect in luminescence gene regulation (Fig. 1B). It is formally possible that, similar to the V. harveyi system, RscS and a second sensor kinase, such as AinR, may function redundantly to control lux expression. Thus, we would be unable to detect a defect by removing only one of these two proteins. Further work needs to be performed to test this hypothesis by constructing ainR and possibly luxO mutations singly and in combination with an rscS mutation in our squid-symbiont strain of V. fischeri. These mutants can then be tested for both luminescence in culture and colonization in the symbiosis.
Regardless of a hypothetical additional role for RscS in controlling luminescence, normal symbiotic colonization by V. fischeri requires this protein. Given the sequence similarity of RscS to two-component sensors and the colonization defect observed in the rscS mutant strain, we believe that RscS may be a key regulatory factor in the Vibrio-Euprymna symbiosis. We propose that RscS responds to some factor unique to the light organ environment and subsequently communicates to a hypothetical “RscR” the information that the cell now occupies a special niche. The phosphorelay hypothetically initiated by RscS could then alter gene expression and/or protein activity, switching the bacteria into symbiotic mode and perhaps activating the developmental programs that permit V. fischeri to permanently colonize its host.
We expect that further characterization of the role of RscS will contribute to a better understanding of the mechanism(s) by which V. fischeri senses the symbiotic environment and of the nature of the response that permits these bacteria to colonize their host. We anticipate the discovery of a cognate response regulator that acts in conjunction with RscS to control a symbiotic gene function(s) and the identification of these downstream targets. In so doing, we may learn the environmental cues that permit V. fischeri to sense that it has entered the light organ and modulate specific functions required for the interaction between the bacteria and their animal host.
ACKNOWLEDGMENTS
We thank C. Beatty for her assistance with the primer extension experiments, P. Smith for his assistance with the bioluminescence assays, and the following individuals for experimental ideas and comments on the manuscript: A. Driks, F. Catalano, J. Graf, N. Montgomery, E. Ruby, J. Visick, and A. Wolfe.
This work was supported by an internal award to K.L.V. from the Potts Foundation and by NIH grant 1 RO1 GM59690-01A1 to K.L.V.
REFERENCES
- 1.Alexeyev M F, Shokolenko I N. Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of Gram-negative bacteria. Gene. 1995;160:59–62. doi: 10.1016/0378-1119(95)00141-r. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appleby J L, Parkinson J S, Bourret R B. Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell. 1996;86:845–848. doi: 10.1016/s0092-8674(00)80158-0. [DOI] [PubMed] [Google Scholar]
- 4.Arico B, Scarlato V, Monack D M, Falkow S, Rappuoli R. Structural and genetic analysis of the bvg locus in Bordetella species. Mol Microbiol. 1991;5:2481–2491. doi: 10.1111/j.1365-2958.1991.tb02093.x. [DOI] [PubMed] [Google Scholar]
- 5.Bassler B L, Greenberg E P, Stevens A M. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol. 1997;179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassler B L, Wright M, Showalter R E, Silverman M R. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9:773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- 7.Bassler B L, Wright M, Silverman M R. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 8.Bassler B L, Wright M, Silverman M R. Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi. Mol Microbiol. 1994;12:403–412. doi: 10.1111/j.1365-2958.1994.tb01029.x. [DOI] [PubMed] [Google Scholar]
- 9.Boettcher K J, Ruby E G. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol. 1990;172:3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi Y L, Kawase S, Nishida T, Sakai H, Komano T, Kawamukai M, Utsumi R, Kohara Y, Akiyama K. Nucleotide sequence of the glpR gene encoding the repressor for the glycerol-3-phosphate regulon of Escherichia coli K12. Nucleic Acids Res. 1988;16:7732. doi: 10.1093/nar/16.15.7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in procaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 12.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 13.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drury L S, Buxton R S. DNA sequence analysis of the dye gene of Escherichia coli reveals amino acid homology between the Dye and OmpR proteins. J Biol Chem. 1985;260:4236–4242. [PubMed] [Google Scholar]
- 15.Dunlap P V. Regulation of luminescence by cyclic AMP in cya-like and crp-like mutants of Vibrio fischeri. J Bacteriol. 1989;171:1199–1202. doi: 10.1128/jb.171.2.1199-1202.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egland K A, Greenberg E G. Quorum sensing in Vibrio fischeri: elements of the luxI promoter. Mol Microbiol. 1999;31:1197–1204. doi: 10.1046/j.1365-2958.1999.01261.x. [DOI] [PubMed] [Google Scholar]
- 17.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freeman J A, Bassler B L. Sequence and function of LuxU, a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J Bacteriol. 1999;181:899–906. doi: 10.1128/jb.181.3.899-906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freeman J A, Lilley B N, Bassler B L. A genetic analysis of the functions of LuxN: a two-component hybrid sensor kinase that regulates quorum sensing in Vibrio harveyi. Mol Microbiol. 2000;35:139–149. doi: 10.1046/j.1365-2958.2000.01684.x. [DOI] [PubMed] [Google Scholar]
- 20.Gilson L, Kuo A, Dunlap P V. AinS and a new family of autoinducer synthesis proteins. J Bacteriol. 1995;177:6946–6951. doi: 10.1128/jb.177.23.6946-6951.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graf J, Dunlap P V, Ruby E G. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol. 1994;176:6986–6991. doi: 10.1128/jb.176.22.6986-6991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graf J, Ruby E G. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc Natl Acad Sci USA. 1998;95:1818–1822. doi: 10.1073/pnas.95.4.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graf J, Ruby E G. Novel effects of a transposon insertion in the Vibrio fischeri glnD gene: defects in iron uptake and symbiotic persistence in addition to nitrogen utilization. Mol Microbiol. 2000;37:168–179. doi: 10.1046/j.1365-2958.2000.01984.x. [DOI] [PubMed] [Google Scholar]
- 24.Greenberg E P, Hastings J W, Ulitzur S. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch Microbiol. 1979;120:87–91. [Google Scholar]
- 25.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L A, Gill S R, Nelson K E, Read T D, Tettelin H, Richardson D, Ermolaeva M D, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann R D, Nierman W C, White O, Salzberg S L, Smith H O, Colwell R R, Mekalanos J J, Venter J C, Fraser C M. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 29.Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 30.Iuchi S, Matsuda Z, Fujiwara T, Lin E C C. The arcB gene of Escherichia coli encodes a sensor-regulator protein for anaerobic repression of the arc modulon. Mol Microbiol. 1990;4:715–727. doi: 10.1111/j.1365-2958.1990.tb00642.x. [DOI] [PubMed] [Google Scholar]
- 31.Kolter R, Inuzuka M, Helinski D R. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell. 1978;15:1199–1208. doi: 10.1016/0092-8674(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 32.Manoil C, Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986;233:1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- 33.McFall-Ngai M J. Consequences of evolving with bacterial symbionts: insights from the squid-vibrio associations. Annu Rev Ecol Syst. 1999;30:235–256. [Google Scholar]
- 34.Miyamato C M, Lin Y H, Meighen E A. Control of bioluminescence in Vibrio fischeri by the LuxO signal response regulator. Mol Microbiol. 2000;36:594–607. doi: 10.1046/j.1365-2958.2000.01875.x. [DOI] [PubMed] [Google Scholar]
- 35.Nealson K H. Isolation, identification, and manipulation of luminous bacteria. Methods Enzymol. 1978;57:153–166. [Google Scholar]
- 36.Parkinson J S. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 37.Pettigrew D W, Ma D P, Conrad C A, Johnson J R. Escherichia coli glycerol kinase: cloning and sequencing of the glpK gene and the primary structure of the enzyme. J Biol Chem. 1988;263:135–139. [PubMed] [Google Scholar]
- 38.Reichelt J L, Baumann P. Taxonomy of the marine, luminous bacteria. Arch Mikrobiol. 1973;94:283–330. doi: 10.1007/BF00424970. [DOI] [PubMed] [Google Scholar]
- 39.Ruby E G. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu Rev Microbiol. 1996;50:591–624. doi: 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- 40.Ruby E G, Asato L M. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol. 1993;159:160–167. doi: 10.1007/BF00250277. [DOI] [PubMed] [Google Scholar]
- 41.Ruby E G, Nealson K H. Pyruvate production and excretion by the luminous marine bacteria. Appl Environ Microbiol. 1977;34:164–169. doi: 10.1128/aem.34.2.164-169.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 44.Totten P A, Lory S. Characterization of the type a flagellin gene from Pseudomonas aeruginosa PAK. J Bacteriol. 1990;172:7188–7199. doi: 10.1128/jb.172.12.7188-7199.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Visick K L, Foster J, Doino J, McFall-Ngai M, Ruby E G. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J Bacteriol. 2000;182:4578–4586. doi: 10.1128/jb.182.16.4578-4586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Visick K L, McFall-Ngai M J. An exclusive contract: specificity in the Vibrio fischeri-Euprymna scolopes partnership. J Bacteriol. 2000;182:1779–1787. doi: 10.1128/jb.182.7.1779-1787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Visick K L, Ruby E G. Construction and symbiotic competence of a luxA-deletion mutant of Vibrio fischeri. Gene. 1996;175:89–94. doi: 10.1016/0378-1119(96)00129-1. [DOI] [PubMed] [Google Scholar]
- 48.Wei S L, Young R E. Development of symbiotic bacterial bioluminescence in a nearshore cephalopod, Euprymna scolopes. Mar Biol. 1989;103:541–546. [Google Scholar]