Abstract
Protocatechuate degradation is accomplished in a multistep inducible catabolic pathway in Acinetobacter sp. strain ADP1. The induction is brought about by the transcriptional regulator PcaU in concert with the inducer protocatechuate. PcaU, a member of the new IclR family of transcriptional regulators, was shown to play a role in the activation of transcription at the promoter for the structural pca genes, leaving open the participation of additional activators. In this work we show that there is no PcaU-independent transcriptional activation at the pca gene promoter. The minimal inducer concentration leading to an induction response is 10−5 M protocatechuate. The extent of expression of the pca genes was observed to depend on the nature of the inducing carbon source, and this is assumed to be caused by different internal levels of protocatechuate in the cells. The basal level of expression was shown to be comparatively high and to vary depending on the noninducing carbon source independent of PcaU. In addition to the activating function, in vivo results suggest a repressing function for PcaU at the pca gene promoter in the absence of an elevated inducer concentration. Expression at the pcaU gene promoter is independent of the growth condition but is subject to strong negative autoregulation. We propose a model in which PcaU exerts a repressor function both at its own promoter and at the structural gene promoter and in addition functions as an activator of transcription at the structural gene promoter at elevated inducer concentration.
The soil bacterium Acinetobacter sp. strain ADP1 is able to utilize a wide range of aromatic compounds. After an initial conversion into the central metabolites protocatechuate and catechol, all aromatic compounds are funneled through the branched β-ketoadipate pathway (24, 38). The enzymes necessary for the protocatechuate branch of the pathway are synthesized only at elevated levels of protocatechuate (2, 3). Recently, the transcriptional activator protein PcaU has been described as the central actor in the induction of the protocatechuate-degrading enzymes (20). The protein could not be included in any of the groups of regulators described so far but showed similarity to a few regulators, like IclR from Escherichia coli (regulation of the glyoxylate cycle) (18), GylR from Streptomyces coelicolor (regulation of glycerol degradation) (25), and KdgR from Erwinia chrysanthemi (regulation of pectin degradation) (7). Higher similarities exist with a growing number of regulators of pathways connected with degradation of aromatic compounds: PcaR from Pseudomonas putida (regulation of protocatechuate degradation) (40), CatR and PcaR from Rhodococcus opacus (probably regulation of catechol and protocatechuate catabolism) (16, 17), and PobR from Acinetobacter sp. strain ADP1 (regulation of p-hydroxybenzoate hydroxylase) (9). This new group of regulatory proteins has been referred to as the IclR family or PobR subfamily and carries a helix-turn-helix motif at the N terminus. The understanding of the mechanisms that govern these proteins' regulatory function is only in the beginning stages. The PcaR protein has been purified, and its binding at two promoters has been analyzed (22). A mutational analysis of PobR lead to the model of a domain structure (31). We are using the PcaU protein to elucidate its effects on gene regulation as well as some of the factors determining function.
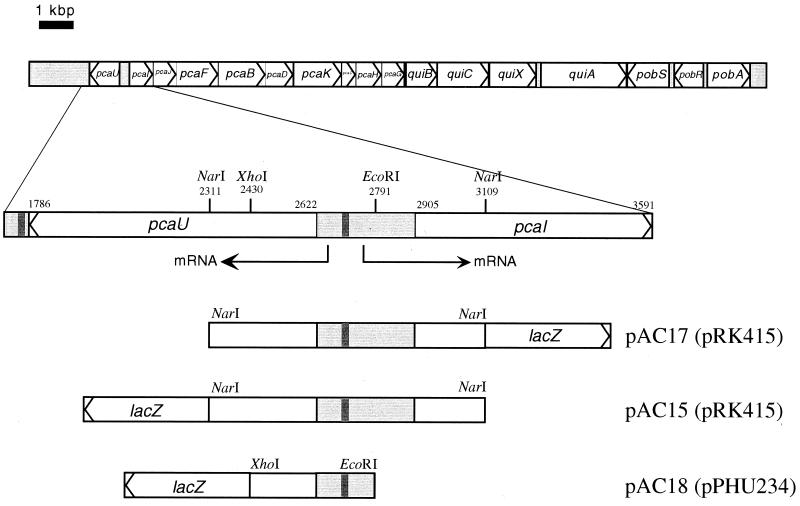
The genes for the protocatechuate-degrading enzymes (pca genes) are clustered on the chromosome of Acinetobacter sp. strain ADP1 (33, 39) (Fig. 1). The gene for the regulator lies upstream of the pca gene cluster and is transcribed in the opposite direction. The PcaU protein was shown to bind to DNA containing part of the 282-bp intergenic region in a protocatechuate-independent manner. The intergenic region has been characterized for the transcriptional start sites upstream of pcaU and pcaI (20). It contains a 19-bp palindromic sequence with high similarity to the PobR binding site as well as to the PcaR binding site (11, 22). A sequence differing from the intergenic palindrome in only two positions is located directly downstream of the pcaU gene. DNA containing this area also binds PcaU (20).
FIG. 1.
Overview of the clustered organization of genes encoding enzymes for protocatechuate degradation (pca genes), quinate oxidation (qui genes), and p-hydroxybenzoate hydroxylase (pob genes) in Acinetobacter sp. strain ADP1. The area containing the pcaU gene, the pcaI-pcaU intergenic region, and the beginning of pcaI is magnified underneath the gene clusters, including restriction sites used in the current study and the location of the potential PcaU binding sites. The numbers refer to the numbering of nucleotides as used in GenBank file L05770. The three bars at the bottom show DNA fragments which are parts of the magnified area and have been cloned in front of lacZ reporter gene cassettes as indicated in the current study (lacZ not drawn to scale). The names of the resulting lacZ fusion plasmids are given, with the respective vector in parentheses. The dark shaded areas indicate the locations of potential PcaU binding sites.
This investigation consists of a thorough description of the expression patterns at the promoter of the pca genes (pcalp) and the promoter of the regulator gene (pcaUp). Former investigations did not give a clear picture as to whether protocatechuate-dependent regulation of the pca genes can occur in the absence of a functional PcaU. Results presented in this study give clear evidence, that this is not the case. Furthermore, the data allow us for the first time to quantify the expression at the two promoters under a variety of different growth conditions in the presence or absence of PcaU. PcaU turns out to be a bifunctional regulator repressing and activating at pcaIp and in addition repressing expression of its own gene.
MATERIALS AND METHODS
Organisms, plasmids, and growth conditions.
Strains and plasmids used in this study are presented in Table 1. Strains of Acinetobacter were grown at 30°C in nutrient broth or mineral medium (10 mM Na2HPO4, 8.8 mM KH2PO4, 9.3 mM NH4Cl, 0.8 mM MgSO4, 0.033 mM FeSO4, and 0.034 mM CaCl2) supplemented with 10 mM succinate, 10 mM glucose, 10 mM pyruvate, 5 mM quinate, 5 mM p-hydroxybenzoate, or 5 mM protocatechuate. E. coli strains were grown in Luria-Bertani medium at 37°C. E. coli K-12 was grown on M9 medium with 0.4% (wt/vol) lactose as the carbon source (37). Antibiotics were added as needed at the following final concentrations for growth of E. coli: ampicillin at 100 μg/ml, tetracycline at 12.5 μg/ml, and kanamycin at 25 μg/ml. If required, liquid Acinetobacter cultures contained tetracycline (3 μg/ml) and kanamycin (3 μg/ml). For solid medium, higher concentrations were used: tetracycline at 6 μg/ml and kanamycin at 12 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Acinetobacter strains | ||
| ADP1 | Wild type (strain BD413, ATCC 33305) | 29 |
| ADP331 | ΔpcaU2, a 552-bp deletion between NcoI and MluI restriction sites in pcaU | 20 |
| ADP197 | recA100::Tn5 | 21 |
| ADPU331 | ΔpcaU2 recA100::Tn5 | This study |
| E. coli strains | ||
| K-12 | Wild type | |
| DH5α | general cloning strain | 23 |
| S17-1 | Mobilizing strain | 50 |
| Plasmids | ||
| pRK415 | Tcr, lacp/o, shuttle vector | 30 |
| pUC19 | Apr, lacp/o | 55 |
| pKOK6 | Contains lacZ-Kmr cassette for construction of transcriptional fusions | 32 |
| pPHU234 | Tcr, broad-host-range translational lacZ fusion vector | 27 |
| pZR9 | pcaU′-pcaIJFBD′ from Acinetobacter sp. strain ADP1 | 20 |
| pAC3 | 798-bp NarI fragment from pZR9 inserted into the AccI site of pUC19, lacp downstream of pcaI′ | This study |
| pAC4 | 798-bp NarI fragment from pZR9 inserted into the AccI site of pUC19, lacp upstream of pcaI′ | This study |
| pAC5 | lacZ-Kmr cassette from pKOK6 inserted into PstI site of pRK415, lacp downstream of lacZ | This study |
| pAC12 | 740-bp XbaI fragment from pAC4 inserted into XbaI site of pRK415, lacp downstream of lacZ | This study |
| pAC15 | lacZ-Kmr cassette from pKOK6 inserted into PstI site of pAC12, transcriptional pcaU-lacZ fusion, lacp downstream of lacZ | This study |
| pAC16 | 740-bp XbaI-PstI fragment from pAC3 inserted between XbaI and PstI sites of pRK415 | This study |
| pAC17 | lacZ-Kmr cassette from pKOK6 inserted into PstI site of pAC16, transcriptional pcaI-lacZ fusion, lacp downstream of lacZ | This study |
| pAC18 | 361-bp EcoRI-XhoI fragment from pZR9 inserted between EcoRI and XhoI sites of pPHU234, translational pcaU-lacZ fusion | This study |
Gene transfer by natural transformation or by conjugation.
For transformation, Acinetobacter strains were grown in 5 ml of mineral medium with succinate overnight. After addition of 10 μl of 1 M succinate and additional growth for 30 min, 50 μl of the culture was transferred onto a polycarbonate filter (Costar Nucleopore, Cambridge, Mass.) placed on a nonselective plate together with 1 μg of linearized plasmid DNA or purified DNA fragment. After incubation for 5 to 6 h at 37°C, the cells were washed off the filter and spread on a selective plate. Plasmids were introduced into Acinetobacter by conjugation from E. coli S17-1, followed by selection for resistance to the appropriate antibiotics on a plate with mineral medium for Acinetobacter, which did not allow growth of E. coli. (49).
DNA manipulation and plasmid construction.
Recombinat DNA techniques were performed as described elsewhere (44). The boiling lysis method was used to isolate plasmid DNA from Acinetobacter (26). A transcriptional pcaI-lacZ fusion and a transcriptional pcaU-lacZ fusion were constructed on plasmids pAC17 and pAC15. For this purpose, the 798-bp NarI fragment from pZR9, containing the intergenic region and the beginnings of both the pcaU and the pcaI genes, was subcloned into pUC19. Plasmids with both orientations of the insert were needed for further applications plasmids (pAC4 and pAC3, respectively). For creation of the pcaU-lacZ fusion, the 750-bp DNA fragment between the XbaI site in pcaU and the XbaI site of the polylinker of pAC4 was ligated into the respective site of pRK415, creating plasmid pAC12. The promoterless lacZ-Kmr cassette of pKOK6 was inserted into the PstI site of pAC12, yielding plasmid pAC15. To form the pcaI-lacZ fusion, the 740-bp fragment between the XbaI site in pcaU and the PstI site of the polylinker of pAC3 was ligated between the respective sites of pRK415, leading to plasmid pAC16. The lacZ-Kmr cassette of pKOK6 was inserted into the PstI site of pAC16, creating pAC17. Plasmid pAC5 with the promoterless lacZ-Kmr cassette of pKOK6 inserted into pRK415 as a PstI fragment was constructed for determination of promoter activity without DNA inserted upstream of the lacZ-Kmr cassette. All three plasmids (pAC5, pAC15, and pAC17) carried the vector lacp downstream of lacZ to ensure that no transcriptional activity initiated at this promoter was detected in the assays. Translational pcaU-lacZ fusion plasmid pAC18 was made by cloning the 361-bp EcoRI-XhoI fragment from pZR9, consisting of part of the intergenic region and the beginning of the pcaU gene, into the respective sites of plasmid pPHU234. The latter plasmid without an insert was used as a control.
Determination of copy number of pRK415 derivatives in Acinetobacter.
Bacteria were grown in mineral medium with succinate as the carbon source to the mid-logarithmic growth phase. Total DNA was extracted as described elsewhere (19). DNA was digested with restriction endonuclease DdeI and analyzed by Southern blotting and hybridization using Hybond-N+ nylon membranes as recommended by the manufacturer (Amersham Pharmacia Biotech Europe, Freiburg, Germany). The DNA probe was labeled with [α-32P]dATP using a Random Primers DNA labeling system (Life Technologies, Munich, Germany). For detection of radioactivity bound to the membrane, a Bio Imager Fujix BAS 1000 (Fuji Photo Film Co., Ltd., Tokyo, Japan) was used. The software MacBAS (Fuji Photo Film Co., Ltd., Tokyo, Japan) was applied for quantitative exploitation. The DNA probe was the 798-bp NarI fragment from pZR9 contained in pAC15 and pAC17. DdeI restriction of either plasmid resulted in a 3-kbp hybridizing fragment, whereas the respective chromosomal fragment had a size of 2.4 kbp. The ratio of signal intensities of the plasmid band and the chromosomal band was the copy number.
Enzyme activity and protein determinations.
Previously described procedures were used for assay of protocatechuate 3,4-dioxygenase (13). Each extract was assayed 5 to 10 times, and the standard deviation between individual assays was no more then 4%. The standard deviation for protocatechuate 3,4-dioxygenase between different cultures grown under the same conditions was 10 to 15%. β-Galactosidase activity was measured as described by Miller using chloroform and sodium dodecyl sulfate to open the cells (37). Samples were taken in the course of growth or at the beginning of stationary growth phase. Each sample was assayed in duplicate or more, and the standard deviation was no more then 2%. Between different cultures of the same plasmid-harboring strain grown under the same conditions, there was a standard deviation of up to 15%.
RESULTS
Dynamics of the activities of protocatechuate 3,4-dioxygenase and E. coli β-galactosidase expressed in a reporter gene system in Acinetobacter sp. strain ADP1.
For quantification of expression of the pca genes, we measured the activity of protocatechuate 3,4-dioxygenase. To enable the accurate determination of low expression levels, we used E. coli β-galactosidase as a reporter for pcaIp-driven expression in addition to the activity of protocatechuate 3,4-dioxygenase. For this purpose, plasmid pAC17 was constructed (Fig. 1). Plasmid pAC15 contained a transcriptional pcaU-lacZ fusion for measurements of pcaUp-driven expression. A plasmid without any Acinetobacter DNA in front of the lacZ-Kmr cassette was made as a control (pAC5). For measurement of pcaUp-driven expression, we also constructed a translational pcaU-lacZ fusion using broad-host-range lacZ fusion vector pPHU234 (pAC18). These plasmids were introduced into derivatives of Acinetobacter sp. strain ADP1 without a functional RecA. Interruption of the recA gene reduced the frequency of recombinational events by 100-fold and thus enabled stable maintenance of plasmids carrying DNA that corresponds to chromosomal DNA of the host (21). The resulting strains carried the described reporter plasmids in addition to the chromosome. Thus, all the components contributing to the expression of the pca genes were there in the same amount as in the wild type; in addition, they contained the respective fragment of the pca intergenic region carried on the individual plasmid in a number corresponding to the copy number of the vector in Acinetobacter sp. strain ADP197. The copy number for pRK415-based plasmids (pAC15 and pAC17) in Acinetobacter sp. strain ADP197 was found to be 2 (data not shown). The copy number of pPHU234 and its derivative pAC18 can be assumed to be 2 because pRK415 and pPHU234 both are derived from the same plasmid (pRK290) (12). Determination of β-galactosidase activity of strains with the control plasmids pAC5 and pPHU234 showed that there was no detectable activity in either of these strains. Thus, any activity measured using strains carrying plasmids with inserted DNA in front of the lacZ gene was caused by promoter activity in these inserts.
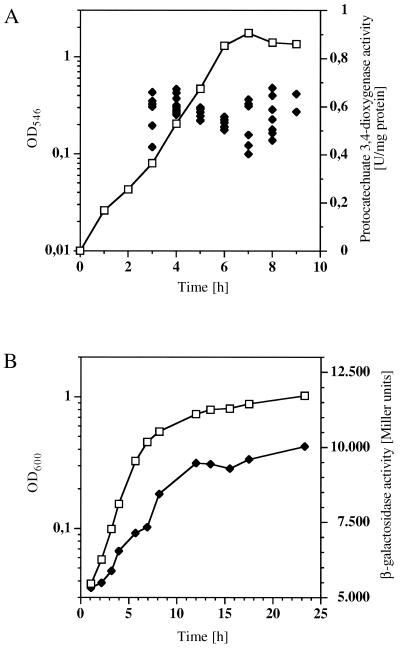
Preliminary determinations of the activity of both enzymes introduced above were performed to define the conditions of measurement with respect to expression in the course of growth. Acinetobacter sp. strain ADP1 was grown on mineral medium containing different carbon sources metabolized via protocatechuate (p-hydroxybenzoate, quinate, and protocatechuate), and the activity of protocatechuate 3,4-dioxygenase was determined throughout all stages of growth. As an example, the result for growth on mineral medium with quinate is shown in Fig. 2. Specific protocatechuate 3,4-dioxygenase activity stayed at the same level during all stages of growth (with an optical density of 0.1 as the earliest harvest time), and the same was true for the other growth conditions (data not shown). This allowed us to conduct the cell harvest at optical densities that were not exactly identical for different cultures.
FIG. 2.
Activities of the enzymes protocatechuate 3,4-dioxygenase (A) and β-galactosidase (B) used to measure pca gene expression in the course of growth of Acinetobacter sp. strain ADP1 (wild type) (A) or ADP197 (recA-negative derivative of the wild type) (B). For measurement of pcalp-driven β-galactosidase activity, plasmid pAC17 was introduced into strain ADP197. Cells were grown on mineral medium with quinate as the carbon source. Optical density at 546 nm (OD546) or 600 nm (open squares) and enzyme activity (solid diamonds) are shown. Each symbol stands for the enzyme activity of an individual batch of cells harvested at the time indicated. In the case of β-galactosidase, data are given for one representative culture.
Corresponding experiments were performed with strains containing the reporter gene fusions introduced above. Acinetobacter sp. strain ADP197 containing plasmid pAC15, pAC17, or pAC18 was grown on mineral medium with carbon sources metabolized through protocatechuate (p-hydroxybenzoate, quinate, and protocatechuate) as well as carbon sources not metabolized through the β-ketoadipate pathway (succinate, pyruvate, and nutrient broth). Cultures used as inocula were grown under the same conditions. In all cases we found an increase in β-galactosidase activity during the logarithmic growth phase of two- to fourfold, followed by constant β-galactosidase activities in the stationary phase (Fig. 2). The level of β-galactosidase activity in the stationary phase was constant between individual cultures grown under the same conditions. Therefore, we used the β-galactosidase activity reached in the stationary phase as a measure for expression from each plasmid. For comparison, we also followed the course of expression of β-galactosidase from the same plasmids in E. coli DH5α. Expression was constant during the logarithmic growth phase. Expression of the endogenous β-galactosidase in wild-type E. coli (strain K-12) stayed on the same level as well (data not shown). All measurements of pcaIp-driven gene expression presented in this study were performed using both protocatechuate 3,4-dioxygenase and β-galactosidase activity determinations and were consistent in all cases. When possible, we present the protocatechuate 3,4-dioxygenase activity determinations; for experiments with measurements below the detection limit of this enzyme, we present the β-galactosidase data.
PcaU is required for protocatechuate-dependent pca gene expression and needs an external inducer concentration of 10 μM for response.
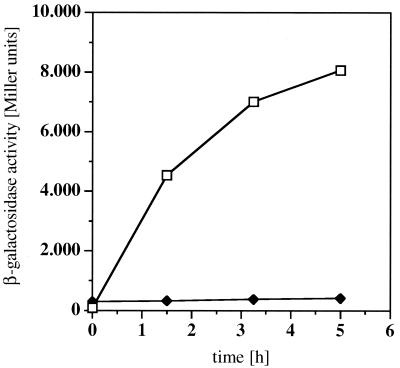
The initial description of PcaU had not ruled out a low level of inducible response to protocatechuate in strains of Acinetobacter lacking a functional PcaU (20). A PcaU-independent induction would have called for a second regulatory protein acting on the pca genes. One possible candidate was the PobR protein due to the high similarity of the two proteins as well as their suggested or determined DNA-binding sites. For clarification of this situation, induction experiments were performed. Cells were grown on mineral medium with carbon sources not inducing the pca genes (succinate, pyruvate, and glucose) and on a complex medium (nutrient broth). In the middle of the logarithmic growth phase, p-hydroxybenzoate was added (final concentration, 2 mM). p-Hydroxybenzoate is a direct precursor of protocatechuate (10) and was chosen because it is less toxic for the cells than protocatechuate. β-Galactosidase activity expressed from the plasmid-based transcriptional pcaI-lacZ fusion in the two recA-negative Acinetobacter sp. strains ADP197 (corresponding to the wild type) and ADPU331 (ΔpcaU derivative) was measured (Fig. 3). In the wild type, the addition of 2 mM p-hydroxybenzoate brought about an increase in β-galactosidase activity from 83 to 8,000 Miller units after 5 h; the ΔpcaU derivative showed a minimal increase from 290 to 410 Miller units within the same time period. We ascribe this to the increase observed generally during logarithmic growth with the reporter system as described above, since there was no change in protocatechuate 3,4-dioxygenase activity in the corresponding experiment (data not shown). Thus, the process of induction at pcaIp in response to elevated levels of protocatechuate as observed in the wild type is completely abolished in the strain without a functional PcaU. These experiments clearly show that there is no protocatechuate-dependent inducibility of the pca genes in the absence of the regulator PcaU. In addition, these data give another indication of regulation occurring at the transcriptional level, confirming data revealed by analysis of RNA (20).
FIG. 3.
Induction of pca gene expression determined by measurement of pcalp-driven β-galactosidase activity. Cells were grown on mineral medium with succinate as the carbon source. At mid-logarithmic growth phase (0 h), p-hydroxybenzoate was added (final concentration, 2 mM). Activity measurements are shown for the recA-negative derivative of the wild-type Acinetobacter sp. strain ADP197 (open squares) and for the ΔpcaU derivative ADPU331 (solid diamonds).
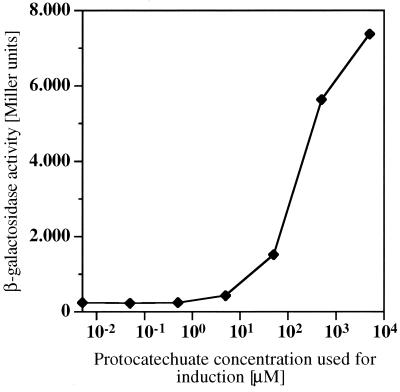
For determination of the minimal inducer concentration necessary to produce an inductive response at pcaIp, we used the recA-negative wild-type Acinetobacter sp. strain ADP197 containing plasmid pAC17. Cells were grown on mineral medium with pyruvate as the carbon source. In the middle of logarithmic growth, protocatechuate was added in different amounts. β-Galactosidase activity was determined after an additional 1 h. Induction was observed at a protocatechuate concentration in the medium of 10 μM or higher (Fig. 4).
FIG. 4.
Inductive response at pcalp dependent on the protocatechuate concentration in the medium. Cells of Acinetobacter sp. strain ADP197 with plasmid pAC17 were grown on mineral medium with pyruvate as the carbon source. Protocatechuate was added to the cultures in the middle of logarithmic growth, resulting in the concentrations indicated, and β-galactosidase activity was determined 60 min later.
Extent of induced or uninduced expression directed by the pca gene promoter varies depending on the growth substrate.
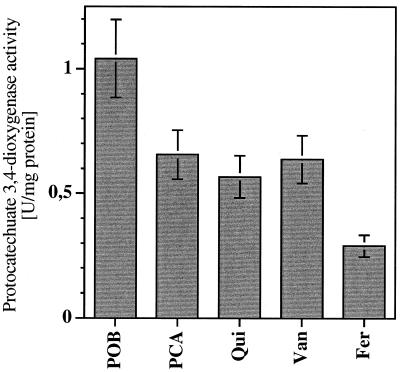
In the course of the initial experiments of this work, it became clear that the maximal level of expression of the pca genes was not constant on all substrates metabolized through the β-ketoadipate pathway but varied depending on the nature of the substrate. Comparing substrates that support growth of Acinetobacter sp. strain ADP1 after degradation via protocatechuate (p-hydroxybenzoate, protocatechuate, quinate, vanillate, and ferulate), we observed highest expression values in p-hydroxybenzoate-grown cells (1 U of protocatechuate 3,4-dioxygenase activity per mg, corresponding to 15,000 Miller units). Cells grown on protocatechuate, quinate, or vanillate had between 63 and 55% of the protocatechuate 3,4-dioxygenase activity of p-hydroxybenzoate-grown cells. Ferulate-grown cells had the lowest expression level (28% of the activity of p-hydroxybenzoate-grown cells; Fig. 5.)
FIG. 5.
Expression of the pca genes in Acinetobacter sp. strain ADP1 after growth on mineral medium with different inducing carbon sources (POB, p-hydroxybenzoate; PCA, protocatechuate; Qui, quinate; Van, vanillate; Fer, Ferulate). The level of protocatechuate 3,4-dioxygenase was used as the indicator of pca gene expression. Error bars indicate standard deviations between different cultures grown under the same conditions.
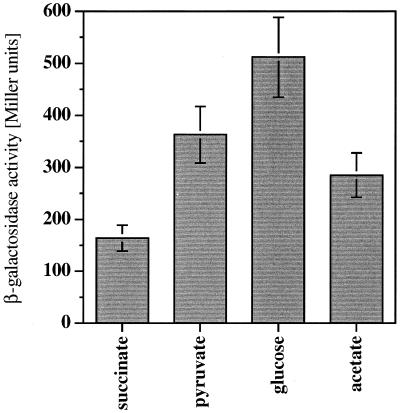
For definition of the level of expression at the pca gene promoter without the addition of inducing compounds, similar experiments were performed after growth of the cells on a variety of substrates (succinate, acetate, pyruvate, and glucose). pcaIp-driven β-galactosidase activity was highest after growth on glucose (512 Miller units); growth on succinate and acetate led to the lowest activities (164 and 285 Miller units, respectively) (Fig. 6). Taken together, these experiments show that there is not a uniform level of induced or uninduced expression of the pca genes, but there is variation depending on the individual growth substrate.
FIG. 6.
Expression of the pca genes in Acinetobacter sp. strain ADP197 after growth on mineral medium with different noninducing carbon sources. The level of β-galactosidase activity expressed by pcalp on plasmid pAC17 was used as the indicator of pca gene expression. Error bars indicate standard deviations between different cultures grown under the same conditions.
These observations are also critical for the definition of the degree of induction brought about by growth on substrates that are degraded via protocatechuate. For example, comparing glucose-grown cells and quinate-grown cells, pca gene expression increases by a factor of 18 when using the reporter enzyme measurements. Comparing growth on succinate and on p-hydroxybenzoate, the induction is 94-fold. This is the highest induction factor found in this work, since succinate-grown cells contain the lowest level and p-hydroxybenzoate-grown cells contain the highest level of pca gene expression.
Absence of PcaU and protocatechuate causes an enhanced basal level of expression at pcaIp.
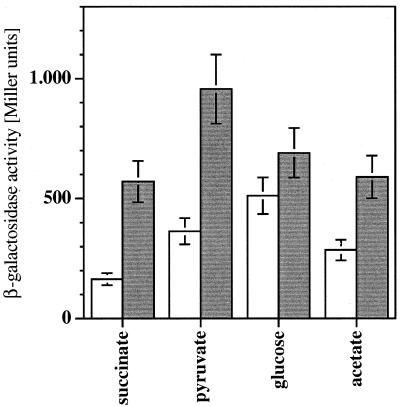
The PcaU protein is necessary to transduct the signal that there is an elevated protocatechuate concentration into the induced state of pca gene expression, as shown above; it therefore acts as an activator of pca gene transcription. On the other hand, we were interested to see whether PcaU has any effect on the pca gene promoter without an elevated level of protocatechuate. We set up a series of experiments in which we measured expression at the pca gene promoter after growth on carbon sources that are not degraded via protocatechuate in the wild type and in a strain missing two-thirds of the pcaU gene. After growth on succinate, pyruvate, and acetate, the ΔpcaU derivative expressed between 2- and 3.5-fold-higher enzyme levels than the wild type under the same conditions, reaching 6.2% of the activity of the maximally induced wild type (Fig. 7). After growth on glucose, the difference was smaller but still significant. We suggest that PcaU has a second function at the pca gene promoter: in the absence of an enhanced level of the inducer protocatechuate, it acts as a repressor. Thus, it appears to be a bifunctional protein, with a repressing and an activating effect on the expression at pcaIp depending on the internal level of protocatechuate.
FIG. 7.
Comparison of expression of the pca genes in Acinetobacter in the presence (open columns) or absence (shaded columns) of functional PcaU after growth on noninducing carbon sources. pAC17-containing Acinetobacter sp. strains ADP197 (PcaU+) ADPU331 (PcaU−) were used for measurement of pcalp-driven β-galactosidase activity. Error bars indicate standard deviations between different cultures grown under the same conditions.
pcaU is expressed constitutively at a low level and is repressed by its own gene product.
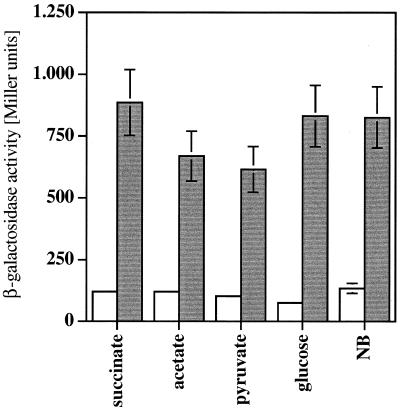
The pcaU gene had been shown to be expressed at elevated levels in response to protocatechuate addition to the medium and to be repressed by its own gene product by means of reporter gene assays (20). The regulated expression in response to different growth conditions has been supported by a primer extension experiment. The plasmid that was used for the reporter gene assays, pZR28, was problematic for two reasons. (i) The lac promoter of the vector pRK415 was located upstream of the reporter gene lacZ. The lac promoter from E. coli is functional in Acinetobacter sp. strain ADP1 (G. Trautwein and U. Gerischer, unpublished observation) and therefore is likely to have contributed to the β-galactosidase activity measured in cells containing this plasmid. (ii) The construct contained 4.7 kbp of Acinetobacter sp. strain ADP1 DNA upstream of the pcaU gene promoter, thus having an increased likelihood of carrying sequences which have promoter activity. For these reasons, the plasmid may have monitored unwanted promoter activities in addition to the pcaU gene promoter activity. We have prepared pcaU-lacZ transcriptional fusion plasmid pAC15, which contains 451 bp of Acinetobacter DNA upstream of the start point of the pcaU transcript (including the whole intergenic region between pcaU and pcaI and the first 205 bp of the open reading frame of the pcaI gene) (Fig. 1). The promoter of the vector is located downstream of the lacZ cassette, thus not interfering with the promoter under investigation. In addition, we used a translational pcaU-lacZ fusion based on plasmid pPHU234, which contained 138 bp of Acinetobacter DNA upstream of the start point of the pcaU transcript (pAC18; Fig. 1). Using strain ADP197 (the recA-negative derivative of the wild-type Acinetobacter sp. strain ADP1) with plasmid pAC15, we found constant β-galactosidase activity of about 100 Miller units on all carbon sources tested (p-hydroxybenzoate, protocatechuate, and quinate as compounds degraded via protocatechuate and succinate, acetate, pyruvate, glucose, and nutrient broth as carbon sources not requiring the β-ketoadipate pathway; data not shown). Comparing activities in ADP197 and ADPU331, we found activities between 6- and 11-fold higher in the strain without a functional PcaU after growth on neutral carbon sources (succinate, acetate, pyruvate, glucose, and nutrient broth; Fig. 8). Data obtained using translational pcaU-lacZ fusion plasmid pAC18 confirmed these results (data not shown). Taken together, these results are a clear indication that PcaU is a strong repressor of its own expression without any dependence on the presence of protocatechuate.
FIG. 8.
Comparison of pcaU gene expression in Acinetobacter sp. strain ADP197 (open bars) ADPU331 (shaded bars). Expression was determined by measurement of β-galactosidase activity expressed from transcriptional pcaU-lacZ fusion plasmid pAC15 in the two strains. Error bars indicate standard deviations between different cultures grown under the same conditions.
DISCUSSION
In this article, results are presented concerning the regulation of expression of the pca structural genes as well as the regulator gene pcaU of the soil bacterium Acinetobacter sp. strain ADP1. Two different methods have been used to quantify the expression of the pca structural genes: measurement of the enzymatic activity of protocatechuate 3,4-dioxygenase, the gene product of pcaHG, and plasmid-based reporter gene constructs using the activity of β-galactosidase transcriptionally fused to the pca gene promoter. The expression of the regulator gene has been studied using a transcriptional and a translational fusion between its promoter and the lacZ gene. The plasmids bearing the reporter gene constructs have a copy number of 2 in strains of Acinetobacter sp. strain ADP1. Preliminary experiments revealed that the two enzymes differed in their expression pattern upon culture development. Protocatechuate 3,4-dioxygenase was expressed at a constant level determined by the nature of the carbon source. Expression of E. coli β-galactosidase in Acinetobacter does not occur in a balanced state as it does in E. coli from the same plasmid. Similar observations have been made in other investigations (35, 46). The phenomenon may be based on different stabilities of the transcripts and proteins or on changes in the transcription or translation rates due to the newly created hybrid DNA molecules.
According to the initial description of PcaU, a chromosomal deletion of a major portion of the pcaU gene still allowed a small inducible response of pca gene expression (20). An explanation for such an observation would be a second regulatory factor causing elevated expression of the pca genes. One candidate was the closely related transcriptional activator PobR (9), which may bind not only to the binding site in front of the pobA gene but also to the binding site in front of the pca gene cluster and lead to the observed small activation of transcription. The two binding sites are very similar (16 identical positions out of 19), and the same is true for the effector molecules (protocatechuate and p-hydroxybenzoate). The experiments described in this study taking advantage of the high sensitivity of the β-galactosidase reporter system clearly show that there is no residual induction of the pca genes in the ΔpcaU derivative. As a conclusion from that, it can now be stated that there is no PcaU-independent inductive response at the pca gene promoter in Acinetobacter sp. strain ADP1. Only modifications of the proteins PobR and PcaU caused by mutations can bring about a change in their DNA-binding affinity or in their effect on initiation of transcription (31).
The minimal inducer concentration in the medium necessary to cause activation of transcription at pcaIp was found to be 10 μM. Corresponding experiments have been performed for the closest homologue of PcaU, PobR (9). There, the threshold concentration of inducer for induction of the respective promoter was found to be 10−1 μM, 100-fold lower. This difference may be explained by the need for the cell to tolerate a certain protocatechuate level without induction of the genes encoding enzymes for protocatechuate degradation to prevent a disturbance in the biosynthesis of the aromatic amino acids (53).
As a prerequisite for further studies of regulation by PcaU, we analyzed pca gene promoter activity under a variety of conditions. The different inducing carbon sources used in the study all caused different expression levels, with p-hydroxybenzoate at the high end and ferulate at the low end. The reason for these differences may be different levels of the inducer protocatechuate in the cells, depending on the quality of the enzymatic steps leading to protocatechuate formation. Different internal levels of protocatechuate may be sensed by PcaU and cause different promoter activities. Highest expression levels should be observed in strains with a block at the protocatechuate 3,4-dioxygenase step. Indeed, all the enzymes encoded by the pca genes are expressed at 150% of the fully induced wild-type level after growth on succinate with or without added p-hydroxybenzoate in such strains (3). Data obtained with derivatives of Acinetobacter sp. strain ADP1 are comparable. A strain with a 128-bp deletion in pcaH (ΔpcaH19 [19]), leading to a dysfunctional protocatechuate 3,4-dioxygenase, and carrying pcaIp reporter plasmid pAC17 had a β-galactosidase activity of 19,000 Miller units after growth on succinate, or 126% of the fully induced wild-type level (unpublished observation). Only determination of the internal level of protocatechuate under defined growth conditions can verify the hypothesis discussed here. The situation is made more complex by the fact that protocatechuate is transported actively via the cell membrane by more than one transport protein, and furthermore, protocatechuate can also leave the cells under certain growth conditions (8). Another approach will be the determination of transcriptional initiation at pcaIp in vitro at different protocatechuate concentrations.
The expression level of the pca genes without addition of the inducer also differs significantly. Experiments with a strain of Acinetobacter sp. missing a functional PcaU showed an equivalent pattern of expression after growth on the same noninducing carbon sources, suggesting a PcaU-independent mechanism of transcriptional regulation as a background. Such a mechanism could be carbon catabolite repression. In the recent past, evidence has been brought forward that in the genus Pseudomonas, a mechanism of carbon catabolite repression may exist which is different from the mechanisms already examined in great detail in E. coli and in Bacillus (51). Acinetobacter, being closely related to Pseudomonas, may contain a similar mechanism. Succinate has been described in numerous publications as one of the preferred carbon sources causing repression of transcription of catabolic operons (4, 6, 56). As succinate is the carbon source with the lowest expression, it seems plausible to explain the expression differences observed here with a regulatory mechanism of carbon catabolite repression which is also active in the absence of the aromatic substrate. Experiments comparing pca gene expression as well as the expression of other genes after growth on an aromatic carbon source with or without succinate as a second carbon source indicate the presence of a mechanism of catabolite repression in Acinetobacter sp. strain ADP1 (S. Dal and U. Gerischer, unpublished observation). This theory can only be proven when the underlying mechanism becomes understood.
Besides its function as an activator of pca gene transcription, a repressing effect of PcaU on pca gene transcription in the absence of an elevated inducer concentration is suggested here. The pca gene promoter exhibits a very high basal activity, and this high basal expression is adjusted to a lower level by the repressing function of transcriptional regulator PcaU. The system most closely related to the PcaU-dependent regulation of the pca genes, the PobR-dependent regulation of pobA expression in the same organism, appears to differ from PcaU in this respect. Screening for mutant strains without an active protocatechuate 3,4-dioxygenase never resulted in strains with a dysfunctional PcaU (19). In contrast, screening for such pob mutants resulted in a significant percentage of mutant strains with a dysfunctional PobR in addition to strains with an impaired PobA function (9, 10). This presents indirect evidence for the absence of a relevant basal expression of the pobA gene in strains with a dysfunctional PobR. Despite the similarities of the primary sequences of PobR and PcaU as well as their potential activator-binding sites, the respective intergenic regions display significant differences, which most likely contain the reason for the observed differences in expression of their target genes. The promoter sequences of the pca gene cluster and the pobA gene are very different; the 5′ noncoding sequences of the two transcripts differ in length tremendously (the pobA transcript is 21 bp, and the pca transcript is 118 bp) (11, 20).
pca gene expression levels without added inducer are relatively high even in the wild type. In this respect it has to be kept in mind that PcaU may have to fulfill different requirements. (i) Protocatechuate has been shown to be an inducer of the qui genes; their gene products convert shikimate or quinate into protocatechuate (14, 15, 52). It must be assumed that a similar basal-level expression for qui gene expression (which may be protocatechuate/PcaU/regulated as well) is the necessary condition for induction of the qui genes, otherwise protocatechuate would not accumulate to a level high enough to induce gene expression. The same may be true for the expression of other genes encoding enzymes for the conversion of various aromatic substrates into the central metabolite protocatechuate. For example, vanAB from Acinetobacter sp. strain ADP1 have recently been described (47). They encode the enzyme converting vanillic acid into protocatechuate. For the respective genes from Pseudomonas putida, protocatechuate has been shown to be the strongest inducer (54). (ii) pcaK encodes a transport protein with specificity for p-hydroxybenzoate and protocatechuate. Strains without PcaK and VanK (proteins with overlapping specificities) are severely impaired for growth on protocatechuate (8). A basal level of pcaK expression may be a prerequisite for the cells to bring in enough aromatic substrate to enable induction.
PcaU-dependent regulation at the pcaI-pcaU intergenic region also affects the pcaU gene itself in the form of a strong carbon source-independent repression. A corresponding observation has been made for the closest homologues of PcaU, PobR and PcaR (11, 22). Despite the functional similarity in these three systems, the architecture of the regulatory regions is different in each case. PcaR binds DNA directly upstream of the transcriptional start site, completely covering the −10 area of its own promoter. PobR binds to sequences located between the transcriptional start and the translational start site. In the case of PcaU, the potential binding site is located between positions −40 and −80 (R. Popp and U. Gerischer, unpublished observation), suggesting different mechanisms causing the autorepression.
Combining a repressing and an activating function in one protein is not uncommon among transcriptional regulatory proteins (5, 45, 48), but the different functions are usually observed at two different promoters. Models are discussed which go beyond the simple idea of steric hindrance of binding of the RNA polymerase in the case of a repressing function but rather suggest a repressor action through contact with promoter-bound RNA polymerase (41). Amino acid residues responsible for the activating or repressing function of the regulatory proteins cyclic AMP receptor protein and GcvA have been identified (28, 36); the orientation of a regulator-binding site with respect to the promoter has been varied, resulting in a switch between an activating and a repressing function of the regulator GalR (42). In the case of PcaU, in addition to the different functions at different promoters, a dual functionality of repression and activation is found at the same promoter. Such a situation has been found in one other case, MerR from E. coli (34). This regulator represses transcription from a structural gene promoter in the absence of the inducer. In its presence, the DNA-MerR-RNA polymerase ternary complex is converted from a state of repression to a state of activation, and evidence for the involvement of changes in the DNA conformation has been presented (1). Changes in the structure of the regulator caused by binding of the inducer can thus cause the conversion from a repressor into an activator. To what extent this model also applies to the way PcaU performs or whether there will be a different mechanism will only become understood after further molecular analysis.
ACKNOWLEDGMENTS
We are highly indebted to Iris Steiner for brilliant technical assistance. Süreyya Dal contributed several enzyme activity determinations. We thank Peter Dürre for critical reading of the manuscript.
This research was supported by grants from the Deutsche Forschungsgemeinschaft and from the University of Ulm.
REFERENCES
- 1.Ansari A Z, Bradner J E, O'Halloran T V. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature. 1995;374:371–375. doi: 10.1038/374370a0. [DOI] [PubMed] [Google Scholar]
- 2.Cánovas J L, Stanier R Y. Regulation of the enzymes of the β-ketoadipate pathway. 1. General aspects. Eur J Biochem. 1967;1:289–300. doi: 10.1007/978-3-662-25813-2_40. [DOI] [PubMed] [Google Scholar]
- 3.Cánovas J L, Wheelis M L, Stanier R Y. Regulation of the enzymes of the β-ketoadipate pathway in Moraxella calcoacetica. 2. The role of protocatechuate as inducer. Eur J Biochem. 1968;3:293–304. doi: 10.1111/j.1432-1033.1968.tb19529.x. [DOI] [PubMed] [Google Scholar]
- 4.Cases I, de Lorenzo V. Expression systems and physiological control of promoter activity in bacteria. Curr Opin Microbiol. 1998;1:303–310. doi: 10.1016/s1369-5274(98)80034-9. [DOI] [PubMed] [Google Scholar]
- 5.Celis R T. Repression and activation of arginine transport genes in Escherichia coli K-12 by the ArgP protein. J Mol Biol. 1999;294:1087–1095. doi: 10.1006/jmbi.1999.3308. [DOI] [PubMed] [Google Scholar]
- 6.Collier D N, Hager P W, Phibbs P V., Jr Catabolite repression control in the Pseudomonads. Res Microbiol. 1996;147:551–561. doi: 10.1016/0923-2508(96)84011-3. [DOI] [PubMed] [Google Scholar]
- 7.Condemine G, Robert-Baudouy J. Tn5 insertion in kdgR, a regulatory gene of the polygalacturonate pathway in Erwinia chrysanthemi. FEMS Microbiol Lett. 1987;42:39–46. [Google Scholar]
- 8.D'Argenio D A, Segura A, Coco W M, Bünz P V, Ornston L N. The physiological contribution of Acinetobacter PcaK, a transport system that acts upon protocatechuate, can be masked by the overlapping specificity of VanK. J Bacteriol. 1999;181:3505–3515. doi: 10.1128/jb.181.11.3505-3515.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiMarco A A, Averhoff B, Ornston L N. Identification of the transcriptional activator pobR and characterization of its role in the expression of pobA, the structural gene for p-hydroxybenzoate hydroxylase in Acinetobacter calcoaceticus. J Bacteriol. 1993;175:4499–4506. doi: 10.1128/jb.175.14.4499-4506.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiMarco A A, Averhoff B A, Kim E E, Ornston L N. Evolutionary divergence of pobA, the structural gene encoding p-hydroxybenzoate hydroxylase in an Acinetobacter calcoaceticus strain well-suited for genetic analysis. Gene. 1993;125:25–33. doi: 10.1016/0378-1119(93)90741-k. [DOI] [PubMed] [Google Scholar]
- 11.DiMarco A A, Ornston L N. Regulation of p-hydroxybenzoate hydroxylase synthesis by PobR bound to an operator in Acinetobacter calcoaceticus. J Bacteriol. 1994;176:4277–4284. doi: 10.1128/jb.176.14.4277-4284.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doten R C, Ngai K-L, Mitchell D J, Ornston L N. Cloning and genetic organization of the pca gene cluster from Acinetobacter calcoaceticus. J Bacteriol. 1987;169:3168–3174. doi: 10.1128/jb.169.7.3168-3174.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elsemore D A, Ornston L N. The pca-pob supraoperonic cluster of Acinetobacter calcoaceticus contains quiA, the structural gene for quinate-shikimate dehydrogenase. J Bacteriol. 1994;176:7659–7666. doi: 10.1128/jb.176.24.7659-7666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elsemore D A, Ornston L N. Unusual ancestry of dehydratases associated with quinate catabolism in Acinetobacter calcoaceticus. J Bacteriol. 1995;177:5971–5978. doi: 10.1128/jb.177.20.5971-5978.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eulberg D, Lakner S, Golovleva L A, Schlömann M. Characterization of a protocatechuate catabolic gene cluster from Rhodococcus opacus 1CP: evidence for a merged enzyme with 4-carboxymuconolactone-decarboxylating and 3-oxoadipate enol-lactone-hydrolyzing activity. J Bacteriol. 1998;180:1072–1081. doi: 10.1128/jb.180.5.1072-1081.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eulberg D, Schlömann M. The putative regulator of catechol catabolism in Rhodococcus opacus 1CP—an IcIR-type, not a LysR-type transcriptional regulator. Antonie van Leeuwenhoek. 1998;74:71–82. doi: 10.1023/a:1001755928637. [DOI] [PubMed] [Google Scholar]
- 18.Galinier A, Nègre D, Cortay J C, Marcandier S, Maloy S R, Cozzone A J. Sequence analysis of the iclR gene encoding the repressor of the acetate operon in Salmonella typhimurium. Nucleic Acids Res. 1990;18:3656. doi: 10.1093/nar/18.12.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerischer U, Ornston L N. Spontaneous mutations in pcaH and -G, structural genes for protocatechuate 3,4-dioxygenase in Acinetobacter calcoaceticus. J Bacteriol. 1995;177:1336–1347. doi: 10.1128/jb.177.5.1336-1347.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerischer U, Segura A, Ornston L N. PcaU, a transcriptional activator of genes for protocatechuate utilization in Acinetobacter. J Bacteriol. 1998;180:1512–1524. doi: 10.1128/jb.180.6.1512-1524.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregg-Jolly L A, Ornston L N. Properties of Acinetobacter calcoaceticus recA and its contribution to intracellular gene conversion. Mol Microbiol. 1994;12:985–992. doi: 10.1111/j.1365-2958.1994.tb01086.x. [DOI] [PubMed] [Google Scholar]
- 22.Guo Z, Houghton J E. PcaR-mediated activation and repression of pca genes from Pseudomonas putida are propagated by its binding to both the −35 and the −10 promoter elements. Mol Microbiol. 1999;32:253–263. doi: 10.1046/j.1365-2958.1999.01342.x. [DOI] [PubMed] [Google Scholar]
- 23.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 24.Harwood C S, Parales R E. The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 25.Hindle Z, Smith C P. Substrate induction and catabolite repression of the Streptomyces coelicolor glycerol operon are mediated through the GylR protein. Mol Microbiol. 1994;12:737–745. doi: 10.1111/j.1365-2958.1994.tb01061.x. [DOI] [PubMed] [Google Scholar]
- 26.Holmes D S, Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 27.Hübner P, Willison J C, Vignais P M, Bickle T A. Expression of regulatory nif genes in Rhodobacter capsulatus. J Bacteriol. 1991;173:2993–2999. doi: 10.1128/jb.173.9.2993-2999.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jourdan A D, Stauffer G V. Mutational analysis of the transcriptional regulator GcvA: amino acids important for activation, repression, and DNA binding. J Bacteriol. 1998;180:4865–4871. doi: 10.1128/jb.180.18.4865-4871.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juni E, Janik A. Transformation of Acinetobacter calco-aceticus (Bacterium anitratum) J Bacteriol. 1969;98:281–288. doi: 10.1128/jb.98.1.281-288.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 31.Kok R G, D'Argenio D A, Ornston L N. Mutation analysis of PobR and PcaU, closely related transcriptional activators in Acinetobacter. J Bacteriol. 1998;180:5058–5069. doi: 10.1128/jb.180.19.5058-5069.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kokotek W, Lotz W. Construction of lacZ-kanamycin-resistance cassette, useful for site-directed mutagenesis and as a promoter probe. Gene. 1989;84:467–471. doi: 10.1016/0378-1119(89)90522-2. [DOI] [PubMed] [Google Scholar]
- 33.Kowalchuk G A, Hartnett G B, Benson A, Houghton J E, Ngai K-L, Ornston L N. Contrasting patterns of evolutionary divergence within the Acinetobacter calcoaceticus pca operon. Gene. 1994;146:23–30. doi: 10.1016/0378-1119(94)90829-x. [DOI] [PubMed] [Google Scholar]
- 34.Kulkarni R D, Summers A O. MerR cross-links to the alpha, beta, and sigma 70 subunits of RNA polymerase in the preinitiation complex at the merTPCAD promoter. Biochemistry. 1999;38:3362–3368. doi: 10.1021/bi982814m. [DOI] [PubMed] [Google Scholar]
- 35.Liang S T, Dennis P P, Bremer H. Expression of lacZ from the promoter of the Escherichia coli spc operon cloned into vectors carrying the W205 trp-lac fusion. J Bacteriol. 1998;180:6090–6100. doi: 10.1128/jb.180.23.6090-6100.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meibom K L, Sogaard-Andersen L, Mironov A S, Valentin-Hansen P. Dissection of a surface-exposed portion of the cAMP-CRP complex that mediates transcription activation and repression. Mol Microbiol. 1999;32:497–504. doi: 10.1046/j.1365-2958.1999.01362.x. [DOI] [PubMed] [Google Scholar]
- 37.Miller J H. A short course in bacterial genetics. Cold Spring Havbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 38.Ornston L N, Neidle E L. Evolution of genes for the β-ketoadipate pathway in Acinetobacter calcoaceticus. In: Towner K J, editor. The biology of Acinetobacter. New York, N.Y: Plenum Press; 1991. pp. 201–237. [Google Scholar]
- 39.Parke D, D'Argenio D A, Ornston L N. Bacteria are not what they eat: that is why they are so diverse. J Bacteriol. 2000;182:257–263. doi: 10.1128/jb.182.2.257-263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romero-Steiner S, Parales R E, Harwood C S, Houghton J E. Characterization of the pcaR regulatory gene from Pseudomonas putida, which is required for the complete degradation of p-hydroxybenzoate. J Bacteriol. 1994;176:5771–5779. doi: 10.1128/jb.176.18.5771-5779.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roy S, Garges S, Adhya S. Activation and repression of transcription by differential contact: two sides of a coin. J Biol Chem. 1998;273:14059–14062. doi: 10.1074/jbc.273.23.14059. [DOI] [PubMed] [Google Scholar]
- 42.Ryu S, Fujita N, Ishihama A, Adhya S. GalR-mediated repression and activation of hybrid lacUV5 promoter: differential contacts with RNA polymerase. Gene. 1998;223:235–245. doi: 10.1016/s0378-1119(98)00237-6. [DOI] [PubMed] [Google Scholar]
- 43.Saier M H., Jr A functional-phylogenetic system for the classification of transport proteins. J Cell Biochem Suppl. 1999;32–33:84–94. doi: 10.1002/(sici)1097-4644(1999)75:32+<84::aid-jcb11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 45.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 46.Schwartz E, Gerischer U, Friedrich B. Transcriptional regulation of Alcaligenes eutrophus hydrogenase genes. J Bacteriol. 1998;180:3197–3204. doi: 10.1128/jb.180.12.3197-3204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Segura A, Bünz P V, D'Argenio D A, Ornston L N. Genetic analysis of a chromosomal region containing vanA and vanB, genes required for conversion of either ferulate or vanillate to protocatechuate in Acinetobacter. J Bacteriol. 1999;181:3494–3504. doi: 10.1128/jb.181.11.3494-3504.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharrocks A D, Green J, Guest J R. FNR activates and represses transcription in vitro. Proc R Soc Lond B Biol Sci. 1991;245:219–226. doi: 10.1098/rspb.1991.0113. [DOI] [PubMed] [Google Scholar]
- 49.Simon R, O'Connell M, Labes M, Pühler A. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol. 1986;118:640–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- 50.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 51.Stülke J, Hillen W. Carbon catabolite repression in bacteria. Curr Opin Microbiol. 1999;2:195–201. doi: 10.1016/S1369-5274(99)80034-4. [DOI] [PubMed] [Google Scholar]
- 52.Tresguerres M E F, De Torrontegui G, Ingledew W M, Cánovas J L. Regulation of the enzymes of the β-ketoadipate pathway in Moraxella: control of quinate oxidation by protocatechuate. Eur J Biochem. 1970;14:445–450. doi: 10.1111/j.1432-1033.1970.tb00309.x. [DOI] [PubMed] [Google Scholar]
- 53.Tresguerres M E F, Ingledew W M, Cánovas J L. Potential competition for 5-dehydroshikimate between the aromatic biosynthetic route and the catabolic hydroaromatic pathway. Arch Mikrobiol. 1972;82:111–119. [Google Scholar]
- 54.Venturi V, Zennaro F, Degrassi G, Okeke B C, Bruschi C V. Genetics of ferulic acid bioconversion to protocatechuic acid in plant-growth-promoting Pseudomonas putida WCS358. Microbiology. 1998;144:965–973. doi: 10.1099/00221287-144-4-965. [DOI] [PubMed] [Google Scholar]
- 55.Yanisch-Perron C, Vieíra J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 56.Yuste L, Canosa I, Rojo F. Carbon source-dependent expression of the PalkB promoter from the Pseudomonas oleovorans alkane degradation pathway. J Bacteriol. 1998;180:5218–5226. doi: 10.1128/jb.180.19.5218-5226.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]