Abstract
Streptococcus mutans is a bacterium that has evolved to be dependent upon a biofilm “lifestyle” for survival and persistence in its natural ecosystem, dental plaque. We initiated this study to identify the genes involved in the development of genetic competence in S. mutans and to assay the natural genetic transformability of biofilm-grown cells. Using genomic analyses, we identified a quorum-sensing peptide pheromone signaling system similar to those previously found in other streptococci. The genetic locus of this system comprises three genes, comC, comD, and comE, that encode a precursor to the peptide competence factor, a histidine kinase, and a response regulator, respectively. We deduced the sequence of comC and its active pheromone product and chemically synthesized the corresponding 21-amino-acid competence-stimulating peptide (CSP). Addition of CSP to noncompetent cells facilitated increased transformation frequencies, with typically 1% of the total cell population transformed. To further confirm the roles of these genes in genetic competence, we inactivated them by insertion-duplication mutagenesis or allelic replacement followed by assays of transformation efficiency. We also demonstrated that biofilm-grown S. mutans cells were transformed at a rate 10- to 600-fold higher than planktonic S. mutans cells. Donor DNA included a suicide plasmid, S. mutans chromosomal DNA harboring a heterologous erythromycin resistance gene, and a replicative plasmid. The cells were optimally transformed during the formation of 8- to 16-h-old biofilms primarily consisting of microcolonies on solid surfaces. We also found that dead cells in the biofilms could act as donors of a chromosomally encoded antibiotic resistance determinant. This work demonstrated that a peptide pheromone system controls genetic competence in S. mutans and that the system functions optimally when the cells are living in actively growing biofilms.
Natural genetic transformation is a process by which bacteria are able to take up and integrate exogenous free DNA from their environment (30). This process enables the recipient organisms to acquire novel genes or heritable traits, thereby promoting the emergence of antibiotic resistance and genetic variation and the rapid evolution of virulence factors (10, 13, 15). Therefore, natural genetic transformation can be an important mechanism whereby bacteria adapt to changing environments. Natural transformation in Streptococcus mutans was first demonstrated in 1981, when Perry and Kuramitsu showed that three strains of S. mutans could be transformed to streptomycin resistance (45). They later found that a number of cariogenic properties, including the ability to synthesize water-insoluble glucan and the production of bacteriocins, were conferred by genetic transformation (46). These early works describing the natural transformation of S. mutans have allowed investigators to exploit this property to construct defined mutants and to analyze the functions of many genes in this organism.
Studies of the mitis group of the genus Streptococcus have demonstrated that these bacteria enter a physiologic state called genetic competence that allows them to transport and incorporate exogenous DNA. Induction of genetic competence in these streptococci is mediated by quorum sensing, which depends on a competence-stimulating peptide (CSP) signaling system (20, 21, 23, 32). Cell-cell signaling in these bacteria involves at least six gene products, encoded by comAB, comCDE, and comX (7, 25). Mutations in these genes result in a defect in competence development in Streptococcus pneumoniae (24). In S. pneumoniae, Streptococcus mitis, and Streptococcus oralis, the CSP signaling system is encoded by the comCDE genes, which correspond to the CSP precursor, a histidine kinase, and a response regulator, respectively.
Although attempts have been made to identify homologs of these genes in S. mutans, the location of the corresponding genetic locus has remained elusive in this species (20). In this report, we describe the locus involved in this process and the involvement of the genes in competence. Directed mutagenesis of the S. mutans comCDE genes followed by assays of transformation and competence stimulation by a chemically synthesized CSP clearly demonstrated the roles of these genes in genetic competence.
Competence development in S. mutans had been known to occur in a few easily transformable strains, although the transformation frequency was found to be much lower than for other streptococci (31, 41). Moreover, genetic competence in S. mutans has been assayed exclusively by growing the organism in fluid cultures. To date, there has been no attempt to examine the competence development of S. mutans growing in biofilms, its natural environment.
Although there has not been a systematic study of the relationship between genetic competence and biofilm formation, a recent study of Streptococcus gordonii showed that a mutation in comD (a sensor kinase gene) resulted in a defect in biofilm formation (29). These results suggested that quorum sensing via a signal peptide and a two-component system is important in genetic competence, biofilm formation, and likely other physiologic activities of surface-adherent streptococci.
Studying genetic competence in biofilms is a prerequisite for understanding the mechanism of horizontal gene transfer occurring in natural environments (9). Recent evidence suggests that the growth of bacteria in biofilms can facilitate horizontal gene transfer between bacterial species via either conjugation or transformation (8, 17, 50). To test this hypothesis, we have developed a system to measure and optimize the natural genetic transformation of biofilm-grown cells. Using this system, we have demonstrated that the formation of biofilms greatly enhances competence induction and the ability of S. mutans to transport and integrate foreign DNA.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Six wild-type or parent strains of S. mutans along with their relevant characteristics and sources are listed in Table 1. All the strains were subcultured from freeze-dried ampoules and maintained routinely on Todd-Hewitt agar plates supplemented with 0.3% yeast extract (THYE) (BBL; Becton Dickinson, Cockeysville, Md.). For selection of antibiotic-resistant colonies after genetic transformation, the medium was supplemented with either 10 μg of erythromycin or 500 μg of kanamycin (Sigma-Aldrich, St. Louis, Mo.)/ml. In the present study, two plasmids were employed as prepared donor DNA (Table 1). Plasmid pVA-GTFA was derived from the streptococcal integration vector pVA891 (35). It was constructed by cloning a 2.4-kb EcoRI fragment harboring a portion of the S. mutans gtfA gene from plasmid pSUCRI (5). Plasmid pDL289 (4) is an Escherichia coli-Streptococcus shuttle vector carrying a kanamycin resistance (Kmr) gene that is expressed in both species, conferring resistance to kanamycin in E. coli at 35 μg/ml and in S. mutans at 500 to 1,000 μg/ml.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| S. mutans | ||
| UA159 | Wt; Ems Kms | J. Ferretti, OU-ACGT |
| NG8 | Wt; Ems Kms | A. S. Bleiweis, University of Florida |
| JH1005 | Blis+ Ems Kms | 22 |
| BM71 | Wt; Ems Kms | G. H. Bowden, University of Manitoba |
| GB14 | Wt; Ems Kms | G. V. Kulkarni, University of Toronto |
| LT11 | Highly transformable mutant; Ems Kms | 49 |
| YD025 | UA159::pVA-GTFA; Emr | This study |
| SMCC1 | NG8::pComC-KO; ComC− Emr Kms | This study |
| SMCD1 | NG8::pComD-KO; ComD− Emr Kms | This study |
| SMCE1 | NG8::pComE-KO; ComE− Emr Kms | This study |
| E. coli XL1-Blue | Cloning host | Stratagene |
| Plasmids | ||
| pCR-Script | PCR cloning vector; Ampr | Stratagene |
| pTV32OK | PCR template for Tn917 Emr gene | 11 |
| pVA891 | Steptococcal integration plasmid; Emr | 35 |
| pVA8912 | Streptococcal integration plasmid derived from pVA891; Emr | Horst Malke, Jena University, Jena, Germany |
| pSUCRI | pMK3 harboring the S. mutans gtfA gene | 5 |
| pVA-GTFA | pVA891 harboring a 2.4-kb fragment containing the S. mutans gtfA gene | This study |
| pDL289 | E. coli-Streptococcus shuttle vector; Kmr | 4 |
| pComC-KO | pCR-Script harboring an inactivated comC gene; Ampr Emr | This study |
| pComD-KO | pVA8912 harboring a 292-bp internal comD fragment; Emr | This study |
| pComE-KO | pVA8912 harboring a 462-bp internal comE fragment; Emr | This study |
Wt, wild type.
Identification of the comCDE locus in S. mutans.
The comD and comE genes were identified in the partially completed S. mutans genome database, available from the University of Oklahoma Advanced Center for Genome Technology (OU-ACGT; http://www.genome.ou.edu/smutans.html), by comparison with their homologs in S. pneumoniae (47) using the tblastn program (1). A putative comC sequence was identified 148 nucleotides 5′ proximal to the stop codon of the comD homolog on the opposite strand. A pair of PCR primers, ComC-F5 and ComC-B5 (Table 2), was designed to amplify a 651-bp fragment containing the entire comC gene from six strains of S. mutans. Chromosomal DNA, isolated from each strain by the method of Chen et al. (6), was used as a template. The amplified products were purified using the StrataPrep PCR Product Purification Kit (Stratagene, La Jolla, Calif.) following the manufacturer's instructions. The purified PCR products were subsequently sequenced from both ends with the original amplification primers and either a Pharmacia ALF or a Perkin-Elmer-ABI Prism 377, using either dye-primer or dye terminator chemistry (DNA Sequencing Facility at The Centre for Applied Genomics, The Hospital for Sick Children, Toronto, Canada). The gene sequence of comC was used to deduce the protein sequence of the putative CSP precursor. The mature CSP sequence was then determined as the cleavage product of its precursor protein, with the cleavage site located after a glycine-glycine consensus sequence commonly found in the leader sequence of gram-positive peptide signal molecules (19).
TABLE 2.
PCR primers used in sequence determination and mutant construction
| Primer | Nucleotide sequencea | Amplicon size (bp) |
|---|---|---|
| ComC-F5 | 5′-AGTTTTTTGTCTGGCTGCG-3′ | 651 |
| ComC-B5 | 5′-TCCACTAAAGGCTCCAATCG-3′ | |
| ComC′-F1 | 5′-GACTAGTCATTGGCGGAAGCGGAAGCCTATCAAC-3′ | 3,423 |
| ComC′-B1 | 5′-GCTCTAGAGCTCAGAACATCAAAAATGACCGTTTAGGAC-3′ | |
| Erm2-F3 | 5′-GACTAGTCCAAACAGGTAACGGTTATTGCAGG-3′ | 1,291 |
| Erm2-B1 | 5′-GCTCTAGAGCCCTCTTTAGCTCCTTGGAAGCTGT-3′ | |
| ComD-F1 | 5′-CGGGATCCCGCTAAGTTACCTCTTTTCTCAGTG-3′ | 292 |
| ComD-B1 | 5′-GGAATTCCGCTTCCTTTTGTGCCATTATC-3′ | |
| ComE-F1 | 5′-CGGGATCCCCTGAAAAGGGCAATCACCAG-3′ | 462 |
| ComE-B1 | 5′-GGAATTCCGCGATGGCACTGAAAAAGTCTC-3′ |
Engineered restriction sites are underlined; actual endonuclease recognition sequences are in boldface. Restriction endonuclease recognition sequences are as follows: BamHI, G/GATCC; EcoRI, G/AATTC; Spel, A/CTAGT; Xbal, T/CTAGA.
Construction of mutants.
A knockout mutant of comC was created by allelic exchange via insertion of an erythromycin resistance (Emr) determinant into the comC locus. The comC PCR fragment from strain NG8 previously amplified for nucleotide sequencing was cloned into plasmid PCR-Script Amp SK(+) (Stratagene) at the SrfI site. A pair of PCR primers, ComC′-F1–ComC′-B1 (Table 2), was designed to amplify the PCR-Script-comC plasmid outward from the flanking regions of the comC gene. This resulted in a linear plasmid harboring flanking S. mutans DNA but devoid of the comC open reading frame (ORF). Primer pair Erm2-F3–Erm2-B1 was used to amplify an erythromycin resistance cassette from plasmid pTV32-OK (11) and was inserted into the PCR-Script-comC vector via XbaI and SpeI restriction sites. The resultant construct, designated pComC-KO, was transformed into Epicurian coli XL1-Blue supercompetent cells (Stratagene) for propagation. Transformed colonies were screened on Luria-Bertani–erythromycin (300 μg/ml) agar plates to select those harboring pComC-KO plasmids, which were then isolated from the selected clones by precipitation with polyethylene glycol (Applied Biosystems, Foster City, Calif.). Purified pComC-KO plasmids were then linearized by ScaI digestion to disrupt the beta-lactamase gene, purified following agarose gel electrophoresis, and used to transform the S. mutans wild-type strain NG8. Transformed colonies were screened on THYE-erythromycin (10 μg/ml) agar plates to identify mutants harboring the integrated Emr determinant.
Knockout mutants of comD and comE were constructed by insertion-duplication mutagenesis. The primer pairs ComD-F1–ComD-B1 and ComE-F1–ComE-B1 (Table 2) were designed to amplify internal regions of comD and comE, respectively. Each amplicon was then ligated to the integration plasmid pVA8912 via BamHI and EcoRI sites. The recombinant constructs, designated pComD-KO and pComE-KO, were individually transformed into Epicurian coli XL1-Blue supercompetent cells for propagation. Colonies with pComD-KO or pComE-KO were selected on Luria-Bertani–erythromycin (300 μg/ml) agar plates and purified as described above. Purified pComD-KO or pComE-KO plasmids were used to transform the wild-type S. mutans strain NG8. Mutants harboring the integrated plasmids were selected on THYE-erythromycin (10 μg/ml) agar plates.
Confirmation of plasmid insertions causing gene disruption was performed either by Southern hybridization or by a rapid protocol involving PCR. Southern blotting was carried out with digoxygenin (DIG)-labeled PCR products corresponding to the targeted genes and the Emr determinant as probes, using the DIG Non-Radioactive Nucleic Acid Detection Kit (Roche Diagnostics, Laval, Canada). For PCR verification, primers previously designed for the targeted gene fragments were used in combination with those made for the Emr cassette (Table 2) to test whether gene segments could be amplified from wild-type and mutant S. mutans chromosomal DNA. The recombinant plasmids used for transformation were employed as PCR templates for positive controls. Correct gene disruption could be shown by mutants with a pattern of amplification identical to that seen in the positive control.
Synthesis and use of CSP.
The CSP precursor amino acid sequence (46 amino acids) was derived using the universal genetic code based on the consensus nucleotide sequence of the comC gene obtained from various S. mutans strains (Fig. 1). The Gly-Gly cleavage site was deduced, and the resultant mature CSP consisting of 21 amino acids was synthesized using automated 9-fluorenylmethoxy carbonyl chemistry and confirmed by reverse-phase high-performance liquid chromatography and mass spectrometry profiles (Biotechnology Service Centre, University of Toronto). The synthetic CSP (SCSP) was freeze-dried and stored at −20°C.
FIG. 1.
Deduced amino acid sequences of CSPs from various S. mutans strains. Boldface type indicates variant amino acids. ^, predicted cleavage site.
To evaluate enhancement of transformation efficiency by CSP, the synthetic peptide was freshly dissolved in sterile distilled deionized water at a concentration of 1 mg/ml. This peptide solution was then added to early- to mid-log-phase cultures at final concentrations ranging from 10 to 1,000 ng/ml. The cultures were preincubated with the peptide at 37°C for 30 min before DNA was added. The cultures were incubated for an additional 2 h and plated on antibiotic selective plates. The transformation efficiency was assessed after 2 days of incubation at 37°C in an atmosphere of 5% CO2.
Biofilm formation and quantification.
Biofilms were developed on polystyrene microtiter plates to provide a simple and rapid method for assaying genetic transformation. A 4×-diluted THYE medium supplemented with final concentrations of 0.01% hog gastric mucin (Sigma-Aldrich) and 5 mM glucose was used as the biofilm medium (BM) (27). The formation of biofilms was initiated by inoculating 20 μl of cell suspension (approximately 2.5 × 105 viable cells) into each well containing 2 ml of BM. Four wells were set up simultaneously: two for assaying transformation and two for quantification of biofilms by viable-cell count. After the cultures were incubated at 37°C with 5% CO2 for 20 h, fluid medium was removed for quantifying viable cells using a modification of the ultrasonic dispersion technique established for enumeration of oral streptococci (43). The wells were rinsed once with 10 mM KPO4 buffer (PB; pH 7.2), and biofilm cells were collected in 2 ml of PB after gentle sonication using the BioSonik IV (Bronwill, Rochester, N.Y.) with a low power output at a setting of 20 for 10 s. This procedure removed >99% of the attached cells as estimated by comparing plate counts to residual cell numbers revealed by scanning electron microscopy (SEM) and light microscopy. Planktonic-phase cells were also subjected to sonication. This procedure was used to disassociate chains before plating. The cells were examined by light microscopy following Gram staining to ensure adequate disruption of chains and aggregates and the integrity of the cells. Greater than 90% of the cells were present as individual cocci. Both biofilm and planktonic cells were immediately spread on THYE plates using the model DU2 spiral plater (Spiral Systems Inc., Cincinnati, Ohio) and incubated at 37°C in 5% CO2. Biofilms were quantified by viable-cell counts after the plates were incubated for 48 h. The results were expressed as the mean CFU ± standard deviation of four independent cultures.
Biofilms were also grown in a chemostat-based biofilm fermentor to define and optimize the conditions for competence induction in biofilm-grown cells. The biofilm fermentor was modified in the Mechanical Engineering and Glass Blowing Shops, University of Toronto, based on a similar system described previously (27). The vessel was made of glass, with a working volume of 400 ml. The vessel lid was constructed of stainless steel and had 10 sampling ports, which allowed sterile insertion and retrieval of glass rods (approximately 4.0 cm2/per rod immersed in fluid medium), thereby providing abiotic surfaces for accumulation of biofilms. The temperature in the chemostat vessel was maintained at 37 ± 0.05°C by a temperature controller (model R-600F; Cole Parmer, Vernon Hill, Ill.). The culture pH was controlled by a pH control unit (Digital pH Meter/Controller [model 501-3400]; Barnant Corp., Barrington, Ill.) through the addition of 1 M KOH or 1 M HCl. The vessel was supported on a magnetic stirrer (Fisher Scientific, Nepean, Canada), and the culture was stirred by a polypropylene-coated magnetic stirrer bar (3 cm in length) at 200 rpm. Continuous cultures were established by pumping fresh BM into the vessel at the desired dilution rates. Daily maintenance of the chemostat included optical-density reading, viable-cell counts, and pH measurement in fluid cultures. When the cultures reached steady-state (at least 10 mean generation times), glass rods were aseptically inserted into them for the initiation of biofilm formation. Biofilms of different ages were then removed from the vessel to assay viable biofilm cell counts and genetic transformation. Sonication and microscopy were performed on the cells to ensure that chains and aggregates were dissociated as described above. Three rods from two independent cultures for each experimental condition were examined, and the results were expressed as the mean CFU ± standard deviation.
SEM of biofilms.
To verify the quantitative results, biofilms formed on the mucin-coated surfaces of polystyrene microtiter plates were examined by SEM. Biofilms that had accumulated for different times were washed once with 10 mM PB, fixed by adding 2 ml of 3.7% formaldehyde in 10 mM PB, and incubated at 20°C overnight. The samples were then dehydrated through a series of ethanol rinses (30, 50, 70, 95, and 100%), and critical-point dried with liquid CO2. The bottom surfaces of the wells were cut off, mounted, and sputter coated with gold. Samples were then examined at ×1,000 to 2,500 magnification using a scanning electron microscope (model S-2500; Hitachi Instruments, San Jose, Calif.). Biofilms accumulated on glass surfaces in the chemostat were not examined in this study, since the spatial distribution of S. mutans biofilms on glass rods over time under the same conditions had been reported previously (27).
Preparation of transforming DNA.
Both plasmid and chromosomal DNAs were used to assay natural genetic transformation. Plasmid DNAs were prepared from E. coli cultures by using a commercial plasmid preparation kit (Qiagen Inc., Mississauga, Canada). Homologous chromosomal DNA carrying an erythromycin resistance gene was isolated from the recombinant S. mutans strain YD025 (a derivative of strain UA159 harboring integrated pVA-GTFA) by the method of Chen et al. (6).
Transformation protocols.
In this study, two methods were used to assay and optimize the natural transformation of biofilm cells. Biofilms formed on polystyrene microtiter plates were prepared for transformation by carefully removing 2 ml of BM and replacing it with fresh THYE-HS. The cultures were incubated at 37°C for 2 h, and 1 μg of plasmid DNA/ml or 10 μg of chromosomal DNA/ml was added. The cultures were incubated for an additional 2 h before the fluid medium was removed and the planktonic-phase cells were collected. The wells were rinsed once with PB (pH 7.2), and the biofilm cells were collected in 2 ml of PB after gentle sonication. Suspensions of both the biofilm and planktonic cells were disrupted into single cells as described above and were centrifuged at 12,000 × g for 5 min and resuspended in 200 μl of fresh medium. An aliquot of each cell suspension was spread on THYE plates containing appropriate antibiotics, and the transformation frequency was determined after 48 h of incubation. The transformation frequency was expressed as the percentage of transformants over total viable recipient cells.
The genetic transformation of biofilm cells grown in the chemostat followed the same basic procedure described above, with the following exceptions. Glass rods with intact biofilm cells were removed and placed in 2 ml of prewarmed fresh THYE-HS medium and incubated at 37°C for 30 min before donor DNA was added. After a 2-h incubation, the biofilm cells were removed from the surface by gentle sonication. Two milliliters of planktonic cells was taken directly from the fluid phase of the chemostat for comparison of transformation frequencies. These cells were sonicated, examined by microscopy, centrifuged, and resuspended in 2 ml of fresh prewarmed THYE-HS.
Both biofilm and planktonic-cell suspensions were spread on THYE plates containing appropriate antibiotics for selection of transformants. Transformants were confirmed to harbor integrated DNA by colony hybridization using a DIG Colony Hybridization Kit (Roche Diagnostics) and a probe prepared using plasmid pVA891 (35), the parent plasmid of pVA-GTFA. To assay for a transformation-defective phenotype in comCDE mutants, overnight cultures of S. mutans grown in THYE were diluted 10-fold in THYE-HS and grown in 5% CO2 at 37°C for 1 h. Recipient cells were counted by spreading serial dilutions (10−5 to 10−7) on THYE agar plates containing erythromycin (10 μg/ml). A DNA solution containing saturating amounts of the replicative plasmid pDL289 was added (1 μg per ml of culture) before a further 2-h incubation. Aliquots were spread onto selective THYE agar plates (10 μg of erythromycin/ml; 500 μg of kanamycin/ml) and incubated in a 5% CO2 chamber for 48 h.
Transformation with heat-killed biofilm cells as a source of donor DNA.
To determine if dead cells in biofilms could act as a source of transforming DNA, plates coated with heat-killed biofilm cells were prepared. In this case, biofilms of the donor strain YD025 carrying an erythromycin resistance marker were developed on polystyrene microtiter plates. Before heat treatment, the fluid medium was removed and the biofilms were washed once with sterile water. The biofilms on plates were then incubated with 2 ml of preheated (80°C) sterile water for 20 min. This procedure was performed very carefully to reduce shearing of the biofilm cells from the surface. After heat treatment, the fluid was removed and the plates were dried and kept at −20°C until use. Samples from multiple wells were taken as described above for assessment of biofilm cell viability counts and plated on THYE agar to confirm cell death in heat-treated biofilms. There were no survivors after the treatment. The plates coated with heat-killed biofilms were then reused to grow biofilms and to assay genetic transformation. As a positive control, chromosomal DNA extracted from strain YD025 carrying an erythromycin resistance marker was used under saturating conditions (10 μg/ml). These conditions were used because they represented the maximal frequency of transformation that we could achieve with our in vitro biofilms.
Nucleotide sequence accession numbers.
The amino acid sequences of the CSP precursors in the seven strains of S. mutans under investigation have been submitted to the GenBank protein database (National Center for Biotechnology Information website at http://www.ncbi.nlm.nih.gov) under accession numbers AF277151 to AF277157.
RESULTS
Identification and confirmation of the comCDE locus in S. mutans.
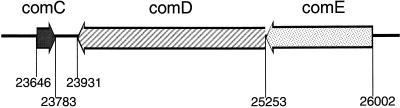
The comCDE locus in S. mutans was detected in contig 459 of the S. mutans genome database file dated 12, November 1999 (OU-ACGT website) by searching the S. mutans genome using tblastn with the comCDE region from S. pneumoniae (18) as the query sequence. A distinct organization of the three genes was notable from the ORF map (Fig. 2). comC was located 5′ proximal and in the opposite orientation to comD and comE. The precise locations of the comC, comD, and comE coding sequences were found to be at 23646 to 23783, 25253 to 23931, and 26002 to 25253, respectively, on the contig. Due to the shorter sequence of the comC gene, resulting in a lower tblastn score, and its unique reversed orientation with respect to comDE, an alignment of CSP precursors from several streptococcal species was performed to support its identity. This ORF appeared to encode a CSP precursor, since it had between 25 and 46% identity at the nucleotide level with CSP sequences from nine other species of oral streptococci (20) (Table 3).
FIG. 2.
Orientations and locations of the comC, comD, and comE genes in the partially completed S. mutans genome database, available from the OU-ACGT website. The numbers correspond to the base pair designations as they appear in contig 459, file date 12 November 1999.
TABLE 3.
Identity and similarity of S. mutans comC nucleotide and protein sequences with those of nine other species of Streptococcus
| Species compared | Nucleotide identities (%) | Deduced amino acid sequence
|
||
|---|---|---|---|---|
| Identities (%) | Similarities (%) | Positives (%) | ||
| S. crista | 38 | 19 | 8 | 27 |
| S. gordonii | 35 | 20 | 11 | 31 |
| S. milleri | 38 | 24 | 12 | 36 |
| S. constellatus | 38 | 29 | 14 | 43 |
| S. anginosus | 40 | 29 | 18 | 47 |
| S. intermedius | 38 | 27 | 18 | 45 |
| S. oralis | 46 | 30 | 10 | 40 |
| S. sanguis | 25 | 6 | 10 | 16 |
| S. mitis | 40 | 23 | 8 | 31 |
The presence of the comCDE locus in all of the S. mutans strains under investigation was confirmed by PCR amplification. Primers designed to amplify internal regions of comD and comE based on the UA159 sequence were successful in generating PCR products of the predicted sizes (292 and 462 bp, respectively). As for the comparatively small comC, primers designed to amplify a region encompassing the entire gene and flanking regions were successful in yielding a predicted PCR product of 651 bp in the six different strains examined. The comC-containing PCR products were sequenced in both directions, and the conceptual translations of the comC ORFs were found to be essentially identical (Fig. 1).
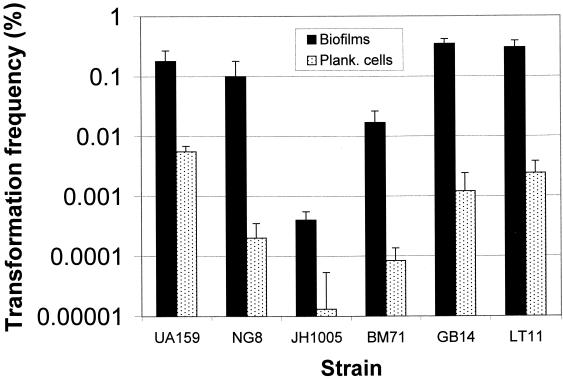
The consensus derived from these sequences (identical to the NG8 sequence) provided the basis for the design of an SCSP useful in the artificial induction of competence. The amino acid sequence of the CSP was deduced to be a 21-residue peptide at the carboxyl terminal of the CSP precursor (SGSLSTFFRLFNRSFTQALGK). The proteolytic cleavage site was predicted to arise immediately after a double-glycine consensus sequence, which is commonly observed at the end of leader peptides for nonlantibiotic peptide bacteriocins (19) and all CSPs produced by gram-positive bacteria that are known to date (20). The leader peptide in this case is the 25-amino-acid peptide at the amino terminus of the CSP precursor (MKKTLSLKNDFKEIKTDELEIIIGG). Interestingly, a premature stop codon in strain JH1005 was due to duplication of a stretch of 29 nucleotides (AGAATTTTACACAAGCTTTGGGAAAATAA) near the end of the comC coding sequence inserted after position 130. JH1005 was found to have a low transformation frequency compared with the other strains examined (Fig. 3).
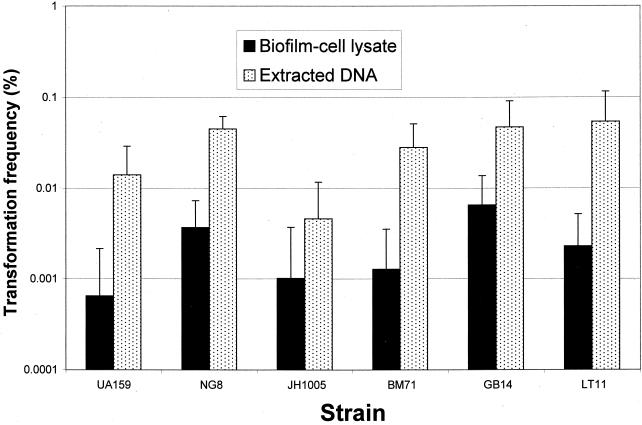
FIG. 3.
Natural genetic transformation of six S. mutans strains with saturating concentrations of integration plasmid pVA-GTFA as donor DNA (1 μg/ml). Biofilms accumulated on the surfaces of polystyrene microtiter plates for 20 h. Biofilm-grown cells of all strains were able to incorporate foreign DNA more efficiently (10- to 600-fold) than their planktonic (Plank.) counterparts. The transformation frequency is expressed as the percentage of viable cells transformed to erythromycin resistance. The results are expressed as the mean + standard deviation of four independent cultures.
The involvement of the comCDE genes in genetic competence was confirmed by assaying mutants for the ability to be transformed with plasmid DNA. The S. mutans strains SMCC1, SMCD1, and SMCE1 (comC, comD, and comE knockouts) were created from the wild-type strain NG8 (Table 4). Using the methods described above, a comC mutant clone was successfully isolated by double reciprocal crossover following transformation with linearized pComC-KO, while comD and comE mutants were derived from insertion-duplication mutagenesis with pComD-KO and pComE-KO, respectively. Disruption of the target genes in the three competence mutant strains was confirmed by positive amplification patterns using the rapid PCR screening protocol described above (data not shown). Using plasmid pDL289 for the transformation assay, all three mutants had dramatically decreased transformation efficiencies that were approximately 100-fold lower than that of the parent strain, NG8, in both planktonic (Table 4) and biofilm (data not shown) cultures.
TABLE 4.
Genetic transformation of comCDE mutant strains of S. mutans
| Straina | Transformant/recipient ratiob
|
|
|---|---|---|
| No SCSP | SCSP | |
| NG8 (wt) | 3.5 ± 5.2 × 10−5 | 3.9 ± 2.6 × 10−3 |
| SMCC1 | 3.6 ± 5.0 × 10−7 | 6.3 ± 5.2 × 10−4 |
| SMCD1 | 1.8 ± 3.1 × 10−7 | 2.5 ± 5.6 × 10−7 |
| SMCE1 | 2.3 ± 4.0 × 10−7 | 1.6 ± 5.8 × 10−7 |
| JH1005 | 5.2 ± 4.2 × 10−7 | 8.8 ± 6.0 × 10−4 |
| NG8(pVA-GTFA) | 4.2 ± 3.6 × 10−4 | 2.9 ± 2.5 × 10−2 |
wt, wild type. The donor DNA used for transformation was replicative plasmid pDL289 carrying a kanamycin resistance gene except with NG8, which was also transformed with the integrative plasmid pVA-GTFA.
All assays were performed with a saturating concentration of DNA (>1 μg/ml). The results are expressed as the mean value from three separate experiments ± standard deviation.
Effect of SCSP on transformation.
The addition of SCSP to S. mutans transformation reactions greatly increased the number of S. mutans transformants. For example, the transformation frequency of S. mutans NG8 (the wild-type strain) with the addition of SCSP was 100-fold higher than that of the cells transformed without SCSP (Table 4). Furthermore, the addition of SCSP to the transformation reactions restored the transformability of the comC mutant strain SMCC1 to levels near that of the wild-type strain with SCSP added. In another example of SCSP complementation, we were able to restore transformability to strain JH1005, which harbors a truncated comC, to levels approaching that of the wild-type strain with added SCSP (Table 4). When SCSP was added to biofilm cells, no increase in transformation efficiency was observed with strain NG8, but with the comC mutant strain SMCC1, transformation efficiency was restored to wild-type levels (data not shown).
Biofilm formation and quantification.
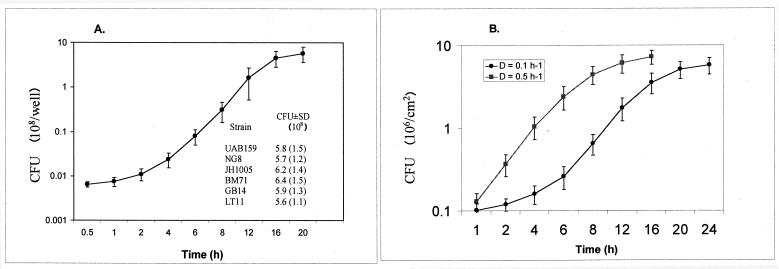
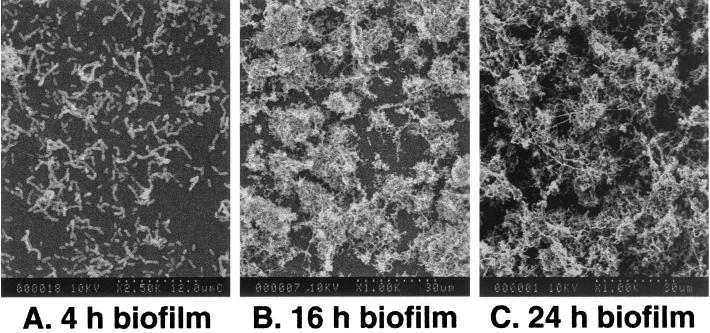
Previous studies showed that biofilm formation was usually favored when bacteria were grown in an oligotrophic environment or in a nutritionally limited or minimal medium (29, 44). We demonstrated that a highly diluted rich medium, such as commercially available Todd-Hewitt broth with proper supplements, also favored biofilm formation. All of the strains tested formed relatively stable and reproducible biofilms over time when grown in a 4×-diluted THYE medium (BM) supplemented with final concentrations of 0.01% hog gastric mucin and 5 mM glucose. It is noteworthy that hog gastric mucin supplemented in the medium was not used as a nutrient source but for conditioning the surface, since S. mutans alone is unable to degrade mucin (3, 27). Previous work demonstrated that the glucose concentration affects the biomass of the films, with increased glucose resulting in thicker films (27). The accumulation rate of strain UA159 in the biofilm fermentor and in microtiter wells is illustrated in Fig. 4. These growth curves are typical of those observed with the other S. mutans strains. After 20 h of accumulation, the mean cell numbers in biofilms on microtiter plates ranged from 5.6 to 6.4 × 108 CFU per well for all six strains tested (Fig. 4A and data not shown). SEM graphically illustrated the cell densities in relation to the viable biofilm cell counts (Fig. 5). Biofilms that had accumulated on the surface for 4 h generally showed a relatively even distribution, with single or a few layers of attached cells (Fig. 5A). As the biofilms accumulated, the cell numbers on the surface rapidly increased, with the formation of many microcolonies (Fig. 5B). Biofilms that had accumulated for 24 h consisted predominantly of dense layers of cells with visible extracellular matrix (Fig. 5C). The biofilms became more heterogeneous in appearance with time, with various chain lengths easily visible among the aggregates.
FIG. 4.
Kinetics of biofilm formation of S. mutans strain UA159 on polystyrene microtiter plates (A) and on glass rods suspended in the chemostat (B). The inset in panel A shows the mean viable biofilm cell counts of S. mutans strains during 20 h of accumulation. Biofilm formation by S. mutans grown in BM usually showed three accumulation phases: (i) the adherence phase (0 to 4 h), (ii) the active accumulation phase (4 to 20 h), and (iii) the slow or plateau accumulation phase (after 20 h). Results are expressed as mean CFU ± standard deviation (SD) of four independent cultures for polystyrene-grown biofilms and of three rods from each of two independent cultures for fermentor-grown biofilms.
FIG. 5.
Scanning electron micrographs of biofilms accumulated on polystyrene microtiter wells at various times.
The kinetics of biofilm accumulation of a representative strain, UA159, grown in the chemostat at different dilution rates is presented in Fig. 4B. Biofilm formation by this strain grown in BM at a dilution rate (D) of 0.1 h−1 showed three accumulation phases: (i) the adherence phase (0 to 4 h), (ii) the active accumulation phase (4 to 20 h), and (iii) the slow or plateau accumulation phase (after 20 h). These phases were similar to those described previously (27). The number of cells for 20-h biofilms was 5.8 ± 1.5 × 106 CFU/cm2, while the doubling time for biofilms during the active accumulation phase (6 to 12 h) was 2.3 to 4 h, about two- to threefold faster than that for planktonic cells (6.93 h) grown under the same conditions. The accumulation of biofilms grown at a D of 0.5 h−1 was much more rapid than that of biofilms grown at a D of 0.1 h−1. At a D of 0.5 h−1, the numbers of cells on the surfaces reached 8.6 ± 2.3 × 106 CFU/cm2 after only 8 h of accumulation, and the sequence of the accumulation phases was less apparent.
High efficiency of genetic transformation in S. mutans biofilms.
All the strains examined in this study were transformable with either plasmid or chromosomal DNA when they were grown in both biofilms and suspended fluid cultures. Strains UA159, NG8, GB14, and LT11 showed the highest transformation frequencies, whereas strain JH1005 gave the lowest frequency of transformation (Fig. 3 and 6). Remarkably, the transformation frequencies of biofilm-grown cells of all the strains were about 10- to 600-fold higher than those of the planktonic cells when DNA harboring S. mutans homologous sequence was used for the transformation. Similar increases in transformation frequency were observed in S. mutans biofilms when a replicative plasmid (pDL289) carrying no homologous DNA was used as the donor DNA (data not shown). However, the transformation efficiency with the replicative plasmid was about 1 to 2 log units lower than that with homologous donor DNA in both biofilm and planktonic cells. S. mutans cells growing in biofilms were evidently able to incorporate foreign DNA much more efficiently than their free-living counterparts in fluid cultures. To verify the incorporation of plasmid pVA-GTFA DNA in transformants, plates were randomly selected for colony hybridization and probed with pVA891, the parent plasmid of pVA-GTFA. The probe hybridized with lysed colonies on the transformant-selective plates and revealed a strong signal, but it did not hybridize to colonies on plates containing untransformed parent cells, confirming the integration of the vector in erythromycin-resistant colonies (data not shown).
FIG. 6.
Natural transformation with heat-killed biofilms of strain YD025 (strain UA159 harboring chromosomally integrated pVA-GTFA; Emr) as a source of donor DNA (lysate). Extracted chromosomal DNA (10 μg/ml) from the same strain was used as a control. Results are expressed as the mean + standard deviation of three independent experiments.
Similar frequencies of transformation of biofilms were also observed when they were incubated in THYE medium without horse serum, indicating that this component was not required when cells were grown in biofilms (data not shown).
Transformation with heat-killed biofilm cells as a source of donor DNA.
In natural ecosystems, exogenous transforming DNA for bacteria most likely originates from incompletely degraded DNA fragments released from dead cells in their immediate surroundings (30). We hypothesized that a high-cell-density biofilm community with bacterial species showing high genetic similarity would probably provide an environment conducive to acquiring exogenous DNA. To test this hypothesis, we established biofilms of strain YD025 harboring a chromosomally encoded erythromycin resistance marker, killed the organisms by heat, and initiated transformation by regrowing biofilms on the plates containing the dead biofilms. Transformation efficiency in both biofilm and planktonic cells exposed to the dead cells was assayed after 6 h of incubation (1:4 ratio of mid-log-phase cells to fresh slides containing the dead biofilms in THYE-HS). Biofilm cells of all strains used in this study were transformable on the dead-cell-coated plates (Fig. 6).
Effects of dilution rates, growth pH, and biofilm age on competence development.
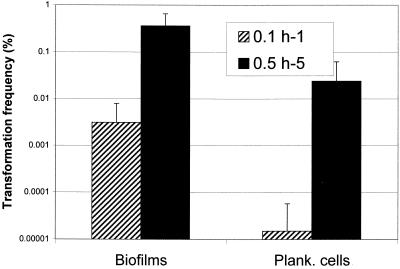
Competence development in streptococci is a transient physiologic event that is highly dependent on growth phase and cell density (21). The determination of optimal conditions for induction of competence in batch cultures is usually difficult. Consequently, a chemostat-based continuous-flow fermentor was used in this study to provide an advantage in defining and optimizing the conditions for competence development in S. mutans. The dilution rate, culture pH, and biofilm age all influenced the competence development of S. mutans growing in biofilms. Development of genetic competence was generally associated with actively growing cells or with cells during their active accumulation phase (4 to 20 h). For example, UA159 cells grown at a high dilution rate (D = 0.5 h−1) exhibited a much higher transformation efficiency than cells grown at a low dilution rate (D = 0.1 h−1), although biofilm cells grown under both conditions could become competent (Fig. 7). The highest frequency of natural transformation was observed with biofilms accumulated on surfaces for less than 20 h at a D of 0.5 h−1 at pH 7.0 to 8.0 (data not shown). It is notable that a high rate of transformation efficiency was also observed in planktonic-phase cells grown at the high dilution rate (D = 0.5 h−1) (Fig. 7). This probably resulted from the increased density of planktonic cells with the higher rate of medium turnover or possibly from the release of cells from the biofilms, which typically shed cells at a higher rate under these conditions. The transformation efficiency of biofilm cells grown at a constant pH of 6.0 was significantly decreased (data not shown). Moreover, no transformation was detected in the planktonic cells at pH 6.0. (data not shown). These results seemed to indicate that low pH impaired the competence induction of S. mutans.
FIG. 7.
Effect of dilution rate on competence development of S. mutans UA159 grown in continuous cultures. Twenty-hour biofilms were assayed for transformation with pVA-GTFA as the donor DNA (1 μg/ml). Plank., planktonic.
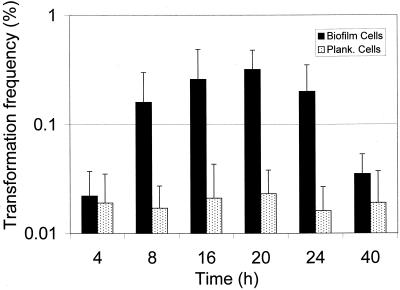
To determine if competence development was a common event in biofilms, we initiated a time course experiment to assay the natural transformation of S. mutans biofilm cells grown in a continuous culture. The data showed that higher frequencies of genetic transformation were usually observed in biofilms during their active accumulation phase (8 to 24 h) (Fig. 8). When biofilms that had accumulated on surfaces for 40 h or longer were assayed for transformability, they had decreased transformation efficiency, even when the cells were grown at a D of 0.5 h−1 (Fig. 8 and data not shown). Moreover, no transformation at all was detected in 5-day biofilms (data not shown).
FIG. 8.
Time course experiment of natural transformation of S. mutans UA159 grown at a D of 0.5 h−1, pVA-GTFA was used as the donor DNA (1 μg/ml). Plank., planktonic.
DISCUSSION
The implications of horizontal gene transfer for bacterial evolution and adaptation are far-reaching. The rapid emergence and spread of antibiotic resistance is probably the most commonly recognized manifestation of this process. The mechanisms operating in bacteria that permit the uptake and incorporation of foreign DNA include transformation, conjugation, and transduction. Although a great deal of work has provided insight into the molecular mechanisms that are involved in these processes, little is known about the functions of these systems in natural environments.
Since biofilms are more representative of bacterial growth in natural environments and S. mutans is an organism that relies on adherence to hard, nonshedding surfaces (teeth) to colonize, we set forth to investigate the ability of this bacterium to transport and integrate exogenous DNA when living in its biofilm state. We were fortunate to have the partially completed S. mutans database to search for and identify competence genes homologous to those recently described in other streptococci, with the best example coming from S. pneumoniae (18). The genetic competence mechanisms in S. pneumoniae are among the best-described cell-cell signaling systems in gram-positive bacteria. In streptococci, competence develops in the early to mid-exponential growth phase, with a wide variation in the optimal conditions for different species and strains. Details of genetic competence have been fairly well characterized for S. pneumoniae (20, 21) and S. gordonii (formerly some strains of Streptococcus sanguis) (32, 33, 34). Competence in these and several other streptococci involves the production, export, and subsequent uptake by neighboring cells of a small peptide signal molecule known as CSP in S. pneumoniae and competence factor (CF) in S. gordonii (26). S. pneumoniae CSP was found to be a 17-residue peptide derived from a 41-residue peptide precursor that is cleaved during export via the ComA transporter (18). In S. gordonii, the CSP-related CF had recently been characterized as a 19-amino-acid peptide, which is processed from a 50-residue prepeptide containing a 31-amino-acid double-glycine-type leader sequence encoded by comX (33). When taken up by the cell, CF or CSP initiates transcription of a cascade of competence-specific genes encoding at least 14 different proteins (40). The genes are generally grouped into early competence genes that encode proteins involved in cell-cell signaling and middle to late competence genes that encode proteins involved in DNA uptake and processing (21).
After examination of the S. mutans locus encompassing the comCDE genes, we found that the orientation of the genes in this region was clearly different from those described in other streptococci (20). In S. pneumoniae, S. mitis, S. oralis, S. gordonii, S. sanguis, and Streptococcus crista, as well as four organisms from the anginosus group, Streptococcus anginosus, Streptococcus intermedius, Streptoccus constellatus, and Streptococcus milleri, the comCDE genes are arranged in an operon flanked by the genes encoding the tRNA for arginine (targ) and glutamate (tglu). The S. mutans comCDE region was not detected when PCR primers based on the tRNA sequences were used to screen for the com locus (20), since they are not flanked by these sequences. In S. mutans, the comC gene is also encoded divergently on the strand complementary to the comDE genes, and there were also promoterlike sequences observed 5′ proximal to both the comC and comE genes, again a dissimilarity to the operonlike structures previously described. The nearest ORF located 5′ proximal to the comC gene was 497 bp away and had homology to the blpO gene encoding a hypothetical bacteriocin-like protein from S. pneumoniae (14). An ORF having homology to the dedA gene from E. coli (unknown function) was identified 497 bp 5′ proximal to the S. mutans comE gene (42).
We were able to demonstrate that the S. mutans comCDE homologs functioned in competence, since inactivation of the individual genes resulted in a competence-deficient phenotype and addition of the SCSP was able to restore transformability to the comC mutant SMCC1. These data provided strong evidence that the locus functions in the process of genetic competence. The addition of SCSP enhanced the transformation frequency by several orders of magnitude, even in wild-type cells, allowing us to exploit this property in the construction of mutants. Interestingly the comE mutant of S. mutans had a fairly high residual level of transformation (Table 4) compared with the comE mutants of S. pneumoniae, which had no detectable transformation (7, 47).
The sequence of the ComC peptide precursor revealed the characteristic Gly-Gly in the leader sequence typical of CSPs (18). We were able to deduce the sequence of the active peptide and to demonstrate its function in wild-type, chemically mutagenized (JH1005) and defined (SMCC1) comC mutant strains. Comparison of the sequences of six different S. mutans comC genes revealed little significant difference, with the exception of strain JH1005, which had a nonsense mutation resulting in a predicted peptide devoid of the three carboxyl-terminal residues. This strain was chemically mutagenized to increase bacteriocin production (22). This mutation probably explains the low frequency of transformation of strain JH1005 compared to the other strains examined (Fig. 2). Interestingly, strain JH1005 was transformed at rates similar to those of other wild-type cells when grown on dead cells as a source of transforming DNA (Fig. 6). One explanation for this observation is that JH1005 cells may have been activated by the intact peptide that was possibly associated with the dead biofilm.
S. pneumoniae is known to have many different pherotypes, or strains that produce a strain-specific peptide to facilitate communication linearly along clonal boundaries (20, 48). The conserved peptide sequence observed in S. mutans suggests that S. mutans species may not have different pherotypes, or strains that produce and recognize a CSP distinct from those of strains outside their pherotype. Although the CSP seems to be largely invariant in S. mutans, an examination of more strains will be necessary to confirm that only one active form of CSP exists within the species.
Since biofilms formed by gram-negative bacteria were previously observed to provide an optimal environment for quorum-sensing mechanisms to function in (12, 38), it seemed likely that a similar situation would occur with gram-positive biofilm-forming bacteria. S. mutans proved to be an excellent model organism to demonstrate the enhanced activity of a quorum-sensing system in a gram-positive bacterium. A novel, major finding of this study is that S. mutans cells growing in biofilms are able to incorporate foreign DNA much more efficiently than their free-living counterparts. Under the defined growth conditions, natural transformation of S. mutans could be readily assayed in biofilm populations, with transformation frequencies of biofilm-grown cells 10- to 600-fold higher than those of planktonic cells. To our knowledge, this report is the first to provide direct evidence that biofilm-grown bacteria of any species can be efficiently induced to become genetically competent for transformation. The evidence from this study suggests that the biofilm environments provide optimal conditions for quorum-sensing systems, as found with the induction of genetic competence, for bacteria that preferentially maintain a biofilm lifestyle in their natural ecosystem. Although we made no attempt to estimate the natural transformability of S. mutans strains in dental biofilms in vivo, our data strongly suggest that the transformability of S. mutans isolates previously determined in liquid culture might be underestimated (43, 45, 46). It seems that many “difficult or nontransformable” S. mutans strains may in fact be capable of natural transformation when grown as biofilms.
Based on our observation that a higher frequency of natural transformation occurred in actively growing biofilms than in planktonic cells, we conclude that the cell-cell signaling system controlling competence development in S. mutans is mediated by a cell density-dependent quorum-sensing mechanism that functions at optimal levels during the active accumulation phase of biofilms. The scanning electron micrographs corresponding to the times of optimal transformation clearly showed that the cells were living in microcolonies. In the 24-h-old mature biofilms, thick cell aggregates obviously provided an environment conducive to the secretion and detection of the natural signal peptide molecule capable of initiating the cascade needed for competence development. In the early growth phase preceding development of these microcolony structures (<6 h), the bacteria appeared as individual cells and chains from which a secreted molecule could easily diffuse into the external environment without contacting neighboring cells. In the older biofilms (40 h), the lower transformation frequency was possibly the result of a diffusion barrier caused by extracellular matrix (obviously visible in the electron micrographs) or by cells that were less metabolically active or dead. Since S. mutans is known to form extracellular polysaccharides from sucrose and not glucose, we suspect that the polymer may result from traces of sucrose available in the rich medium or from polysaccharide, possibly glycogen, released from cells by a yet-uncharacterized mechanism.
In addition to the genetic evidence presented, we conducted a careful quantitative and microscopic evaluation of S. mutans biofilm formation and transformability under steady-state culture conditions. Biofilm growth was characterized by an active accumulation phase following initial adherence to surfaces (Fig. 4). During the active accumulation phase, the number of biofilm cells increased logarithmically at either a low or high dilution rate. The logarithmic increase in cell numbers on the surfaces during this phase could be attributed to the growth of biofilm cells, since adherence alone did not result in a logarithmic increase in cell numbers on surfaces (27). A similar cell density-dependent multiplication during biofilm formation in S. gordonii was previously demonstrated by Liljemark et al. (28), who observed DNA synthesis using a [methyl-3 H]thymidine incorporation method. These authors found that the density-dependent cell division phase of biofilm formation in vivo contributed 90% of the biomass in the first 24 h of dental plaque biofilm growth.
The utility of polystyrene plates as a substratum for biofilm growth provided investigators a rapid and convenient method to grow and quantify biofilms and to screen for mutant strains defective in biofilm formation genes (29, 44). However, the use of microtiter plates is limited when applied to define the conditions required for the induction of genetic competence, a transient physiologic event usually occurring in the early to mid-log growth phase. The induction of genetic competence requires a wide variety of conditions for different species and strains (20, 21). In addition, this method seems to be biased towards the initial events of biofilm formation, which may not allow the identification of defects in late biofilm formation or of transient gene expression as part of the dynamic process of biofilm development (29). In contrast, a chemostat-based continuous-flow system allows researchers to observe physiologic activities of a bacterial population under constant growth conditions (16). By adjusting a single environmental or growth parameter, it is possible to define optimal conditions required for a certain function. Using these systems, we have determined that growth rate, culture pH, and biofilm age are important factors that influence the competence development of S. mutans growing in biofilms. In addition, the continuous-flow biofilm fermentor allowed us to generate reproducible quantitative data to compare differences between the physiologic properties of planktonic cells and biofilms of various ages.
Horizontal gene transfer by genetic transformation among bacteria has been confirmed in many natural ecosystems (30). Dental plaque is a complex biofilm community that harbors the most diverse resident microflora associated with humans (37). Bacteria in dental biofilms, including S. mutans, are frequently exposed to various stresses, such as extreme nutrient shortage or excess, low pH, high osmolarity, oxidation, and frequent consumption of antimicrobial agents by the host (2, 36). Adaptation to an environmental stress by genetic transformation was believed to be a very infrequent event. However, even a very infrequent event can be highly significant if the transforming DNA, such as an antibiotic resistance gene or a virulence factor, provides a selective advantage to the recipient cells. In the oral cavity, free DNA may be constantly available either from dead cells of the resident organisms, from incoming bacteria, or from foods or any other objects introduced into the mouth. Recent evidence also demonstrates that free DNA can survive for a significant length of time in the presence of human saliva (39). Since we demonstrated that dead cells can serve as a good source of transforming DNA in vitro, it is possible that S. mutans and other transformable streptococci living in dental plaque can acquire foreign DNA and hence new phenotypes from neighboring dead cells in the biofilm.
This study demonstrates that S. mutans uses a peptide pheromone quorum-sensing signal transduction system to stimulate the uptake and incorporation of foreign DNA. The signal peptide can be chemically synthesized and added to cultures to stimulate transformation. We have demonstrated that S. mutans is hypertransformable when grown in biofilms in vitro, suggesting that the plaque environment may provide optimal conditions for the function of this quorum-sensing system. We also demonstrated that living cells are able to acquire chromosomal DNA from dead cells of the same species. The concept that dental plaque may provide streptococci with a vast reservoir of genetic information which can be readily incorporated outside of their species boundaries has serious implications when considering the potential for the transfer of antibiotic resistance to pathogens that may transiently reside in dental plaque. Future work will focus on the ability of these commensal oral streptococci to act as donors of DNA to pathogenic streptococci.
ACKNOWLEDGMENTS
We thank J. Hillman for providing S. mutans strain JH1005, A. S. Bleiweis for NG8, G. V. Kulkarni for GB14, J. Ferretti for UA159, G. Bowden for BM71, L. Tao for LT11, D. LeBlanc for plasmid pDL289, H. Malke for pVA8912, and R. Burne for pSUCR1. We greatly appreciate public release of the Streptococcus mutans Genome Sequencing Project, funded by a USPHS-NIH grant from the National Institute of Dental and Craniofacial Research to B. A. Roe, R. Y. Tian, H. G. Jia, Y. D. Qian, S. P. Linn, L. Song, R. E. McLaughlin, M. McShan, and J. Ferretti from The University of Oklahoma.
Our work was supported by PHS grant DE 013230-01 from the National Institute of Dental and Craniofacial Research and grant MT-15431 from the Medical Research Council of Canada. J. H. Lee is the recipient of a University of Toronto Open Fellowship.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowden G H, Hamilton I R. Survival of oral bacteria. Crit Rev Oral Biol Med. 1998;9:54–85. doi: 10.1177/10454411980090010401. [DOI] [PubMed] [Google Scholar]
- 3.Bradshaw D J, Homer K A, Marsh P D, Beighton D. Metabolic cooperation in oral microbial communities during growth on mucin. Microbiology. 1994;140:3407–3412. doi: 10.1099/13500872-140-12-3407. [DOI] [PubMed] [Google Scholar]
- 4.Buckley N D, Lee L N, LeBlanc D J. Use of a novel mobilizable vector to inactivate the scrA gene of Streptococcus sobrinus by allelic replacement. J Bacteriol. 1995;177:5028–5034. doi: 10.1128/jb.177.17.5028-5034.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burne R A, Rubinfeld B, Bowen W H, Yasbin R E. Tight genetic linkage of a glucosyltransferase and dextranase of Streptococcus mutans GS-5. J Dent Res. 1986;65:1392–1401. doi: 10.1177/00220345860650120301. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y Y, Clancy K A, Burne R A. Streptococcus salivarius urease: genetic and biochemical characterization and expression in a dental plaque streptococcus. Infect Immun. 1996;64:585–592. doi: 10.1128/iai.64.2.585-592.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng Q, Campbell E A, Naughton A M, Johnson S, Masure H R. The com locus controls genetic transformation in Streptococcus pneumoniae. Mol Microbiol. 1997;23:683–692. doi: 10.1046/j.1365-2958.1997.2481617.x. [DOI] [PubMed] [Google Scholar]
- 8.Christensen B B, Sternberg C, Andersen J B, Eberl L, Moller S, Givskov M, Molin S. Establishment of new genetic traits in a microbial biofilm community. Appl Environ Microbiol. 1998;64:2247–2255. doi: 10.1128/aem.64.6.2247-2255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costerton J W, Lewandowski Z, DeBeer D, Caldwell D, Korber D, James G. Biofilms, the customized microniche. J Bacteriol. 1994;176:2137–2142. doi: 10.1128/jb.176.8.2137-2142.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cvitkovitch, D. G. Genetic competence and transformation in oral streptococci. Crit. Rev. Oral Biol. Med., in press. [DOI] [PubMed]
- 11.Cvitkovitch D G, Gutierrez J A, Behari J, Youngman P J, Wetz J E, Crowley P J, Hillman J D, Brady L J, Bleiweis A S. Tn917-lac mutagenesis of Streptococcus mutans to identify environmentally regulated genes. FEMS Microbiol Lett. 2000;182:149–154. doi: 10.1111/j.1574-6968.2000.tb08889.x. [DOI] [PubMed] [Google Scholar]
- 12.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 13.Davison J. Genetic exchange between bacteria in the environment. Plasmid. 1999;42:73–91. doi: 10.1006/plas.1999.1421. [DOI] [PubMed] [Google Scholar]
- 14.De Saizieu A, Gardes C, Flint N, Wagner C, Kamber K, Mitchell T J, Keck W, Amrein K E, Lange R. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J Bacteriol. 2000;182:4696–4703. doi: 10.1128/jb.182.17.4696-4703.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dowson C G, Barcus, King S, Pickerill P, Whatmore A, Yeo M. Horizontal gene transfer and the evolution of resistance and virulence determinants in Streptococcus. Soc Appl Bacteriol Symp Ser. 1997;26:42S–51S. [PubMed] [Google Scholar]
- 16.Gottschal J C. Continuous culture. Vol. 1. New York, N.Y: Academic Press Inc.; 1992. [Google Scholar]
- 17.Hausner M, Wuertz S. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl Environ Microbiol. 1999;65:3710–3713. doi: 10.1128/aem.65.8.3710-3713.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Håvrstein L S, Coomaraswamy G, Morrison D A. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Håvrstein L S, Diep D B, Nes I F. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol. 1995;16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 20.Håvrstein L S, Hakenbeck R, Gaustad P. Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J Bacteriol. 1997;179:6589–6594. doi: 10.1128/jb.179.21.6589-6594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Håvarstein L S, Morrison D A. Quorum sensing and peptide pheromones in streptococcal competence for genetic transformation. In: Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C.: ASM Press; 1999. pp. 9–192. [Google Scholar]
- 22.Hillman J D, Dzuback A L, Andrews S W. Colonization of the human oral cavity by a Streptococcus mutans mutant producing increased bacteriocin. J Dent Res. 1987;66:1092–1094. doi: 10.1177/00220345870660060101. [DOI] [PubMed] [Google Scholar]
- 23.Kleerebezem M, Quadri L E, Kuipers O P, de Vos W M. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 24.Lee M S, Dougherty B A, Madeo A C, Morrison D A. Construction and analysis of a library for random insertional mutagenesis in Streptococcus pneumoniae: use for recovery of mutants defective in genetic transformation and for identification of essential genes. Appl Environ Microbiol. 1999;65:1883–1890. doi: 10.1128/aem.65.5.1883-1890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee M S, Morrison D A. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J Bacteriol. 1999;181:5004–5016. doi: 10.1128/jb.181.16.5004-5016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leonard C G. Early events in development of streptococcal competence. J Bacteriol. 1973;114:1198–1205. doi: 10.1128/jb.114.3.1198-1205.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y H, Bowden G H. Characteristics of accumulation of oral gram-positive bacteria on mucin-conditioned glass surfaces in a model system. Oral Microbiol Immunol. 1994;9:1–11. doi: 10.1111/j.1399-302x.1994.tb00207.x. [DOI] [PubMed] [Google Scholar]
- 28.Liljemark W F, Bloomquist C G, Reilly B E, Bernards C J, Townsend D W, Pennock A T, LeMoine J L. Growth dynamics in a natural biofilm and its impact on oral disease management. Adv Dent Res. 1997;11:14–23. doi: 10.1177/08959374970110010501. [DOI] [PubMed] [Google Scholar]
- 29.Loo C Y, Corliss D A, Ganeshkumar N. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J Bacteriol. 2000;182:1374–1382. doi: 10.1128/jb.182.5.1374-1382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorenz M G, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lunsford R D. Streptococcal transformation: essential features and applications of a natural gene exchange system. Plasmid. 1998;39:10–20. doi: 10.1006/plas.1997.1323. [DOI] [PubMed] [Google Scholar]
- 32.Lunsford R D, London J. Natural genetic transformation in Streptococcus gordonii: comX imparts spontaneous competence on strain wicky. J Bacteriol. 1996;178:5831–5835. doi: 10.1128/jb.178.19.5831-5835.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lunsford R D, Nguyen N, London J. DNA-binding activities in Streptococcus gordonii: identification of a receptor-nickase and a histonelike protein. Curr Microbiol. 1996;32:95–100. doi: 10.1007/s002849900017. [DOI] [PubMed] [Google Scholar]
- 34.Lunsford R D, Roble A G. comYA, a gene similar to comGA of Bacillus subtilis, is essential for competence-factor-dependent DNA transformation in Streptococcus gordonii. J Bacteriol. 1997;179:3122–3126. doi: 10.1128/jb.179.10.3122-3126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macrina F L, Evans R P, Tobian J A, Hartley D L, Clewell D B, Jones K R. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene. 1983;25:145–150. doi: 10.1016/0378-1119(83)90176-2. [DOI] [PubMed] [Google Scholar]
- 36.Marsh P D. Oral ecology and its impact on oral microbial diversity. In: Kuramitsu H K, Ellen R P, editors. Oral bacterial ecology: the molecular basis. Wymondham, Norfolk, United Kingdom: Horizon Scientific Press; 2000. pp. 11–65. [Google Scholar]
- 37.Marsh P D. The role of microbiology in models of dental caries. Adv Dent Res. 1995;9:244–254. doi: 10.1177/08959374950090030901. [DOI] [PubMed] [Google Scholar]
- 38.McLean R J, Whiteley M, Stickler D J, Fuqua W C. Evidence of autoinducer activity in naturally occurring biofilms. FEMS Microbiol Lett. 1997;154:259–263. doi: 10.1111/j.1574-6968.1997.tb12653.x. [DOI] [PubMed] [Google Scholar]
- 39.Mercer D K, Scott K P, Bruce-Johnson W A, Glover L A, Flint H J. Fate of free DNA and transformation of the oral bacterium Streptococcus gordonii DL1 by plasmid DNA in human saliva. Appl Environ Microbiol. 1999;65:6–10. doi: 10.1128/aem.65.1.6-10.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morrison D A, Baker M F. Competence for genetic transformation in pneumococcus depends on synthesis of a small set of proteins. Nature. 1979;282:215–217. doi: 10.1038/282215a0. [DOI] [PubMed] [Google Scholar]
- 41.Murchison H H, Barrett J F, Cardineau G A, Curtiss R D., III Transformation of Streptococcus mutans with chromosomal and shuttle plasmid (pYA629) DNAs. Infect Immun. 1986;54:273–282. doi: 10.1128/iai.54.2.273-282.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nonet M L, Marvel C C, Tolan D R. The hisT-purF region of the Escherichia coli K-12 chromosome. Identification of additional genes of the hisT and purF operons. J Biol Chem. 1987;25:12209–12217. [PubMed] [Google Scholar]
- 43.Olsen I, Socransky S S. Ultrasonic dispersion of pure cultures of plaque bacteria and plaque. Scand J Dent Res. 1981;89:307–312. doi: 10.1111/j.1600-0722.1981.tb01687.x. [DOI] [PubMed] [Google Scholar]
- 44.O'Toole G A, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 45.Perry D, Kuramitsu H K. Genetic transformation of Streptococcus mutans. Infect Immun. 1981;32:1295–1297. doi: 10.1128/iai.32.3.1295-1297.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perry D, Wondrack L M, Kuramitsu H K. Genetic transformation of putative cariogenic properties in Streptococcus mutans. Infect Immun. 1983;41:722–727. doi: 10.1128/iai.41.2.722-727.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pestova E V, Håvrstein L S, Morrison D A. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol. 1996;21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- 48.Ramirez M, Morrison D A, Tomasz A. Ubiquitous distribution of the competence related genes comA and comC among isolates of Streptococcus pneumoniae. Microb Drug Resist. 1997;3:39–52. doi: 10.1089/mdr.1997.3.39. [DOI] [PubMed] [Google Scholar]
- 49.Tao L, Tanzer J M. Transformation efficiency of EMS-induced mutants of Streptococcus mutans of altered cell shape. J Dent Res. 1993;72:1032–1039. doi: 10.1177/00220345930720060701. [DOI] [PubMed] [Google Scholar]
- 50.Williams H G, Day M J, Fry J C, Stewart G J. Natural transformation in river epilithon. Appl Environ Microbiol. 1996;62:2994–2998. doi: 10.1128/aem.62.8.2994-2998.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]