Abstract
Neisserial lipooligosaccharide (LOS) contains three oligosaccharide chains, termed the α, β, and γ chains. We used Southern hybridization experiments on DNA isolated from various Neisseria spp. to determine if strains considered to be nonpathogenic possessed DNA sequences homologous with genes involved in the biosynthesis of these oligosaccharide chains. The presence or absence of specific genes was compared to the LOS profiles expressed by each strain, as characterized by their mobilities on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and their reactivities with various LOS-specific monoclonal antibodies. A great deal of heterogeneity was seen with respect to the presence of genes encoding glycosyltransferases in Neisseria. All pathogenic species were found to possess DNA sequences homologous with the lgt gene cluster, a group of genes needed for the synthesis of the α chain. Some of these genes were also found to be present in strains considered to be nonpathogenic, such as Neisseria lactamica, N. subflava, and N. sicca. Some nonpathogenic Neisseria spp. were able to express high-molecular-mass LOS structures, even though they lacked the DNA sequences homologous with rfaF, a gene whose product must act before gonococcal and meningococcal LOS can be elongated. Using a PCR amplification strategy, in combination with DNA sequencing, we demonstrated that N. subflava 44 possessed lgtA, lgtB, and lgtE genes. The predicted amino acid sequence encoded by each of these genes suggested that they encoded functional proteins; however, structural analysis of LOS isolated from this strain indicated that the bulk of its LOS was not modified by these gene products. This suggests the existence of an additional regulatory mechanism that is responsible for the limited expression of these genes in this strain.
Neisseria spp. are able to colonize a variety of human mucosal membranes, yet only a few species are considered to be pathogenic. The ability of Neisseria gonorrhoeae and N. meningitidis to cause disease depends on the expression of specific surface components, the absence of which correlates with a decreased or lack of ability to cause disease. In this context, commensal organisms probably fail to cause disease in healthy individuals because they lack one or more of the needed virulence determinants (6).
Lipooligosaccharide (LOS) is an important virulence determinant in N. gonorrhoeae and N. meningitidis. It is an outer membrane glycolipid involved in immune system evasion, attachment to epithelial tissue, host cell invasion, mediation of toxic damage in the fallopian tube, and stimulation of the production of bactericidal antibodies (2, 15, 20, 41, 44, 45). Biosynthesis of gonococcal and meningococcal LOS occurs via the same biosynthetic pathway producing a branched oligosaccharide attached to lipid A via two 3-deoxy-d-manno-2-octulosonic acid (KDO) molecules (12, 13, 21, 22, 48). The structure of LOS in the gonococcus and meningococcus shows a great deal of heterogeneity both within and among strains. Variation in the number of LOS components expressed and their relative concentrations and in the sugar compositions of the individual LOS molecules is observed (10, 12, 13, 21, 26, 27, 33, 47). The number of branches and the length of each oligosaccharide in the branch vary, and this variation is important in determining the pathogenic potential of the expressing strain (41, 47). Additionally, gonococci and meningococci can modify their LOS by adding a sialic acid residue onto a terminal galactose that is found in the lacto-N-neotetraose structure (32). This addition effectively masks the LOS molecule from host immune functions, making the organism resistant to complement-mediated killing, i.e., serum resistant (29). Certain LOS structural features can be deduced from their ability to react with monoclonal antibodies (MAbs) (9, 27, 28) and their mobility on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (19, 43).
In recent years, several LOS biosynthesis genes have been cloned and characterized. The rfaC gene product adds the first heptose (Hep I) to KDO (50). The rfaF gene product adds the second Hep onto Hep I (35, 40) and is required for α-chain elongation (12, 34). The first Glc of the α chain is added by gene lgtF (24). Genes lgtA, lgtB, lgtC, lgtD, and lgtE (called lgt locus or lgt gene cluster) are responsible for the synthesis of different α chains (14). Genes lgtA, lgtC, and lgtD contain poly(G) tracts (5, 7, 49). When the number of guanines found in these genes changes during DNA replication, alterations in the coding sequence may occur, making translation of the proteins encoded by these genes susceptible to premature termination. Loss of function of any of these genes effects changes in the structure of LOS. Similarly, lgtG, required for addition of the first Glc of the β chain, contains a poly(C) tract. When this gene encodes a functional protein, strains can extend the β chain (3). The β chain may be composed of a single glucose or lactose or lactose with additional sugars (13, 47, 48). Finally, the rfaK gene product is required for γ-chain elongation (23, 24).
While commensal Neisseria spp. are rarely associated with disease, some strains occasionally cause clinical illnesses (e.g., endocarditis caused by N. sicca (11) and arthritis caused by N. subflava (1). We have recently shown that the expression of the lacto-N-neotetraose LOS is able to promote gonococcal invasion into tissue culture cells (41), indicating that the expression of the lacto-N-neotetraose glycoform is important for the establishment of invasive infections. Since most commensal Neisseria spp. express LOS molecules that fail to bind LOS-specific MAbs that recognize this LOS structure, we hypothesized that commensal strains lack the genes needed to make this LOS moiety. Since LOS plays such a prominent role in gonococcal and meningococcal disease, we believe that commensal Neisseria spp. rarely cause disease because they lack the ability to make this essential virulence component. This study was designed to test the hypothesis that LOS-biosynthetic pathways utilized by pathogenic strains are conserved within the Neisseriaceae. In this study, we used several cloned gonococcal LOS-biosynthetic genes as probes to determine if nonpathogenic Neisseria spp. possess homologous genes. Because N. subflava 44 was found to possess genes with homology to several LOS-biosynthetic genes in this study yet failed to produce the expected LOS molecules (42), we studied the organization and expression of some of its LOS-biosynthetic genes in greater detail.
MATERIALS AND METHODS
Bacterial strains, plasmids, oligonucleotides, and culture conditions.
All bacterial strains, plasmids, and oligonucleotide primers used in this study are listed in Tables 1 and 2. All Neisseria strains were grown in phosphate-buffered gonococcal broth plus growth supplements (46)–0.042% sodium bicarbonate or on gonococcal agar base (Difco) plus growth supplements (46) in a 37°C CO2 incubator. Escherichia coli strains were grown on Luria-Bertani plates (39). Kanamycin was used at 30 μg/ml, and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was used at 35 μg/ml.
TABLE 1.
List of bacterial strains and plasmids used
| Strain or plasmid | Relevant genotype, phenotype, or description | Source or reference |
|---|---|---|
| Strains | ||
| N. gonorrhoeae F62 | Full LOS α chain, no β chain | P. Frederick Sparlinga |
| N. gonorrhoeae WR302 | Herman Schneiderb | |
| N. gonorrhoeae PID2 | Herman Schneider | |
| N. sicca 4318 | Herman Schneider | |
| N. sicca 342 | Herman Schneider | |
| N. subflava 4324 | Herman Schneider | |
| N. subflava 4325 | Herman Schneider | |
| N. subflava 4327 | Herman Schneider | |
| N. flavescens 4322 | Herman Schneider | |
| N. flavescens 4323 | Herman Schneider | |
| N. flavescens ATCC 13120 | H. Feldmanc | |
| N. subflava 44 | H. Feldman | |
| N. subflava 52 | H. Feldman | |
| N. cinerea 32824 | Virgina Clarkd | |
| N. cinerea 32165 | Virgina Clark | |
| N. lactamica 5841 | J. McLeod Griffisse | |
| N. meningitidis 89I | J. McLeod Griffiss | |
| N. gonorrhoeae FA19 | William Shaferf | |
| N. gonorrhoeae FA5100 | William Shafer | |
| E. coli DH5αMCR | Cloning host strain, F−mcrAΔ(mrr-hsdRMS-mcrBC) | BRLg |
| N. gonorrhoeae F62 ΔlgtA | 239-bp ApoI deletion in lgtA; truncated lactosyl α chain | 41 |
| N. gonorrhoeae F62 ΔlgtA ΔlgtE | 622-bp BspEI and AgeI deletion in lgtE of F62 ΔlgtA | 41 |
| N. gonorrhoeae F62TF#1 | pNS44lgtABE transformed into F62 ΔlgtA ΔlgtE; MAb 2-1-L8-reactive mutant | This work |
| Plasmids | ||
| pK18 | 37 | |
| pRFAK2-1 | F62 lgtF and rfaK fragment cloned into pK18-up (with uptake sequence) | This work |
| pNS44lgtABE | 44 lgtA, lgtB, and lgtE inserted into pK18 | This work |
| pNS44lgtABΔE | 44 lgtA, lgtB, and part of lgtE inserted into pK18 | This work |
| pF62TF#1lgtABE | N. gonorrhoeae F62TF#1 lgt fragment cloned into pK18 | This work |
| pNS44lgtG | 44 lgtG inserted into pK18 | This work |
University of North Carolina at Chapel Hill, Chapel Hill.
Walter Reed Army Institute for Research, Washington, D.C.
Albany Medical Center, Albany, N.Y.
University of Rochester, Rochester, N.Y.
University of California at San Francisco, San Francisco.
Emory Univery Medical School, Atlanta, Ga.
BRL, Life Technologies, Rockville, Md.
TABLE 2.
List of primers used in this study
| Primer | Sequence (5′–3′) | Descriptiona |
|---|---|---|
| JL14 | AACGCCGGCAAAGGCGCGAACAGTTAAT | 5′ end of F62 lgtC; to make probe for lgtC |
| JL15 | GCCGTCTGAAGGCTTCAGACGGCCTGCC | 3′ end of F62 lgtC; to make probe for lgtC |
| JL50 | CTGAATTCGGCCGACATCGCGCTTTTGGGCG | 5′ of start codon of F62 lgtA, with EcoRI site |
| JL51 | ATGGATCCGGGGCGATTTTACCTAGCAGATGAA | 200 bp downstream of stop codon of F62 lgtE, with BamHI site |
| Got5220R | GAATGACAGTGGATCCATTTCTGATTTTA | 3′ end of F62 lgtE, with BamHI site; to clone pNS44lgtABE |
| DA5 | GCCGTAAACTTTCTCAAGCTCCGCCT | Close to the 3′ end of F62 lgtE; to clone pNS44lgtABΔE |
| DA3 | AACTGTTCGCGCCTTTGCCGGCG | 3′ end of F62 lgtB |
| 196-1040R | CCCGAGCTCAAAGGATAAAGGCAAAATGCC | 5′ end of 15253 lgtG, with SacI site; to clone pNS44lgtG and make probe for lgtG |
| lgtG-1240R | AATGAATTCTGAAAACCCGTTCAGACGGCA | 3′ end of 15253 lgtG, with EcoRI site; to clone pNS44lgtG |
| 196-1530F | TTTGAGAATTCCCCGTTAGCTTTTTGCCG | 3′ end of 15253 lgtG, with EcoRI site; to make probe for lgtG |
| rfaK-147F | AAGCCCGGGCGTATGTTTGGGCTTTTTTGC | 5′ end of F62 rfaK operon, with SmaI site; to make probe for lgtF and rfaK |
| rfaK-3780R | GTGAAGCTTATATTGCATCCAATAATTTGTCG | 3′ end of F62 rfaK operon, with HindIII site; to make probe for lgtF and rfaK |
| Uptake-A | GATCAGAATTCAGACGGCT | Primer for inserting uptake sequence into BamHI site of plasmids |
| Uptake-B | GATCAGCCGTCTGAATTCT | Primer for inserting uptake sequence into BamHI site of plasmids |
Description includes the gene the primer was used to amplify, the location of the primer relative to the coding sequence, and any restriction enzyme sites that are incorporated at the 5′ end which would facilitate the cloning of the resultant amplicon.
Chemicals, reagents, and enzymes.
Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (Beverly, Mass.). All chemicals used for this study were reagent grade or better and were purchased from Sigma Chemical Co. (St. Louis, Mo.) unless otherwise specified. MAbs 2-1-L8, 25-1-LC1, and 17-1-L1 were generously provided by Wendell Zollinger, Walter Reed Army Institute of Research, Washington, D.C. MAb 1B2 was a gift from J. McLeod Griffiss, University of California, San Francisco. MAb 3G9 was graciously provided by Peter Rice, Boston University, Boston, Mass. MAb B5 was a gift from Margaret A. Gidney, Institute for Biological Sciences, National Research Council, Ottawa, Canada.
DNA isolation procedures.
Chromosomal DNA was isolated by a modification of the method of Rodriguez and Tait (38). This modification involved the use of proteinase K (Fc, 50 pg/ml) instead of pronase and subsequent incubation at 37°C overnight. Plasmid DNA was isolated by the alkaline lysis procedure of Birnboim and Doly (4).
Southern hybridization experiments.
Southern analysis was performed as described by Sambrook et al. (39) using the Genius 7 nonradioactive nucleic acid labeling kit of Boehringer Mannheim (Indianapolis, Ind.). The probe comprising the sequence from the 3′ end of rfaF was made from a 577-bp StuI-EcoRI fragment from pRDM45 (8). The probes for lgtF and rfaK were made by PCR amplification of N. gonorrhoeae F62 chromosome DNA with primers rfak-147F and rfak-3780R. The PCR product was cloned into pK18up (pK18 [37] modified by the addition of a DNA sequence [using primers uptake-A and uptake-B, each containing a gonococcal transformation uptake sequence and an EcoRI site] to a pK18 BglII site), giving pRFAK2-1. This plasmid was digested with EcoRI, generating three fragments: 0.8 kb of lgtF, 1.6 kb of rfaK, and 2.1 kb of the vector. The fragments were separated on a 1.2% agarose gel by electrophoresis in Tris-acetate buffer (40 mM Tris, 20 mM acetic acid, 2 mM EDTA [pH 8.1]), and the appropriate fragment was purified from the gel and labeled for Southern hybridization. Other probes were made by labeling PCR products directly.
SDS-PAGE analysis.
Proteinase K-treated whole-cell lysates were prepared from 18- to 20-h cultures by the procedure of Hitchcock and Brown (19). Lysates containing approximately 200 ng of LOS were subjected to SDS-PAGE either on a 13% acrylamide gel in a Tris-glycine buffer (0.025 M Tris, 0.192 M glycine, 0.1% SDS, pH 8.3) at a constant current of 30 mA per gel for ∼4 h at 10°C or on a Tris-Tricine gel (16.5%; Bio-Rad) in Tris-Tricine buffer in accordance with the protocol suggested by the manufacturer. Gels were fixed overnight in 40% ethanol–5% acetic acid, and the LOS was visualized by silver staining (43).
PCR.
PCR was used to generate the DNA fragments employed in the Southern hybridization and gene cloning experiments. DNA amplifications were performed using the GeneAmp PCR kit from Perkin-Elmer Cetus (Norwalk, Conn.). PCR to amplify the lgtA-E gene cluster was performed using the Expand PCR kit from Boehringer Mannheim under recommended conditions. Primers were obtained from either the University of Maryland Protein and Nucleic Acids Facility or Bioserve, Inc. (Laurel, Md.).
Western blot and colony blot analyses.
After SDS-PAGE, LOSs were electrotransferred onto an Immobilon-P membrane (Millipore Corp., Bedford, Mass.) in a Tris-glycine-methanol buffer (0.025 M Tris, 0.192 M glycine, 20% methanol) at a constant voltage of 100 V for 1 h in accordance with the protocol provided by Bio-Rad Corp. After being air dried for 1 h, the membrane was processed by the same procedure as that used for colony blotting, which was described previously (40).
Transformation.
Recombinant DNA transformation into E. coli DH5αMCR was done according to the standard protocols (39). Recombinant DNA transformation into N. gonorrhoeae F62 was done by spot transformation (18). After overnight growth, colonies were transferred to a nitrocellulose membrane (Schleicher & Schuell, Keene, N.H.) and screened for reactivity to MAb 2-1-L8.
Recombinant DNA methods.
Primers JL50 and Got5220R were used to amplify N. subflava 44 chromosome DNA via PCR. The 3-kb PCR product was digested with EcoRI and BamHI and inserted into the corresponding sites in pK18. The recombinant plasmid was named as pNS44lgtABE. Another plasmid, pNS44lgtABΔE, was generated by amplifying a fragment of N. subflava 44 chromosome DNA by PCR using primers JL50 and DA5, digesting the amplicon with EcoRI and AgeI, and cloning the fragment into pK18. Primers 196-N-1040R and lgtG-1260R were used to amplify the lgtG gene of N. subflava 44. The 1.2-kb fragment was cloned into pK18 and named pNS44lgtG. All of these recombinant plasmids (except pNS44lgtABΔE) were sequenced by the DNA Sequence Facility/Center for Agricultural Biotechnology, University of Maryland, College Park.
Nucleotide sequence accession numbers.
The DNA sequences of lgtABE and lgtG from N. subflava 44 appear in GenBank under accession no. AF240672 and AF241526, respectively.
RESULTS AND DISCUSSION
SDS-PAGE profiles of various Neisseria strains.
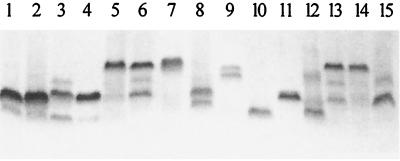
Neisserial LOS exhibits significant heterogeneity when analyzed by SDS-PAGE. The data presented in Fig. 1 show the SDS-PAGE profiles of LOSs expressed by several commensal species. These data demonstrate that all of the strains examined express LOS molecules that are larger than the LOS made by FA5100 (Fig. 1, lane 10). Since the structure of LOS made by FA5100 contains a single Hep added onto the lipid A-KDO core (12), we concluded that the LOSs isolated from all of the strains we examined must possess additional sugars.
FIG. 1.
SDS-PAGE profiles of LOS isolated from various Neisseria strains. Lanes: 1, N. cinerea 32864; 2, N. cinerea 32165; 3; N. lactamica 5841; 4, N. sicca 342; 5, N. sicca 4318; 6, N. flavescens 4322; 7, N. flavescens 4323; 8, N. flavescens ATCC 13120; 9, N. gonorrhoeae F62; 10, N. gonorrhoeae FA5100; 11, N. subflava 44; 12, N. subflava 52; 13, N. subflava 4324; 14, N. subflava 4325; 15, N. subflava 4327.
We performed Western blot and/or colony blot analysis of the LOSs made by these strains using MAb 1B2 (marker for a terminal Gal-GlcNAc on the lacto-N-neotetraose form of LOS [16]), MAb 2-1-L8 (marker for a terminal lactosyl group on the α chain [16]), MAb 3G9 (marker for a lactosyl group on the β chain [17]), MAb 17-1-L1 (marker for an alternate α-chain extension terminating in galactose [16]), MAb 25-1-LC1 (marker for the presence of β-chain glucose and α-chain lactose or sucrose [42]), and MAb B5 (marker for the presence of phosphoethanolamine on Hep II [36]). Of the nonpathogenic strains examined, N. lactamica 5841 contained LOS components that bound MAbs 1B2 and 2-1-L8 (Table 3). N. subflava 44 bound MAb 1B2 weakly on the colony blot, but we were able to visualize binding to a specific LOS band only when the SDS-PAGE gel was grossly overloaded (data not shown). These data suggest that LOSs isolated from most commensal organisms are significantly different structurally from LOSs isolated from pathogenic strains.
TABLE 3.
Relevant properties of strains employed in this study
| Strain | Relative LOS size(s) (kDa) | MAb(s) binding | Presencea of:
|
|||||
|---|---|---|---|---|---|---|---|---|
| rfaE | rfaF | lgtA and lgtD | lgtC | lgtB | lgtE | |||
| N. gonorrhoeae F62 | 4.5, 4.8 | 1B2, 1-1-M, B5 | + | + | + | + | + | + |
| N. gonorrhoeae FA5100 | 2.2 | B5 | + | + | + | + | + | + |
| N. gonorrhoeae FA19 | 3.6, 4.3, 4.5 | 2-1-L8, 17-1-L1, 1B2, B5 | + | + | + | + | + | + |
| N. gonorrhoeae WR302 | 3.6, 4.3, 4.5 | 2-1-L8, 17-1-L1, 1B2, 25-1-LC1, B5 | + | + | No lgtDe | + | + | + |
| N. gonorrhoeae PID2 | 3.0, 3.6, 4.3, 4.5, 4.8, 5.1 | 1B2 | + | + | No lgtD | + | + | + |
| N. meningitidis 89I | 4.8, 4.9 | 1B2, B5 | + | + | + | + | + | + |
| N. subflava 44 | 3.0, 3.2 | 1B2,c 25-1-LC1 | + | + | No lgtD | − | + | + |
| N. lactamica 5841 | 3.6, 4.5 | 1B2, 2-1-L8, B5 | + | + | +b | + | Nd | N |
| N. sicca 342 | 3.6 | B5 | + | + | + | − | N | N |
| N. cinerea 32824 | 3.4 | 25-1-LC1, B5 | + | + | − | − | N | N |
| N. cinerea 32165 | 3.4 | None | + | + | − | − | N | N |
| N. sicca 4318 | 5.0 | None | − | − | − | − | N | N |
| N. subflava 52 | 4.5, 2.5 | None | − | − | − | − | N | N |
| N. subflava 4324 | 3.0, 3.8, 4.8 | None | − | − | − | − | N | N |
| N. subflava 4325 | 4.8 | None | − | − | − | − | N | N |
| N. subflava 4327 | 3.0, 3.8 | None | − | − | − | − | N | N |
| N. flavescens 4322 | 3.6, 4.2, 4.8 | None | − | − | − | − | N | N |
| N. flavescens 4323 | 5.1 | None | − | − | − | − | N | N |
| N. flavescens ATCC 13120 | 2.8, 3.6 | None | − | − | − | − | N | N |
Genes were found by Southern hybridization or gene sequencing. +, positive by Southern hybridization; −, negative by Southern hybridization.
Multiple bands were detected indicating multiple copies of the genes were present.
positive on colony blot only.
N, not determined.
No lgtD, lgtA only.
Determination of the presence of rfaF.
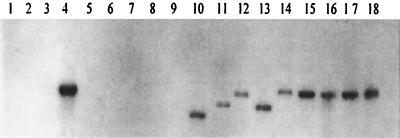
One of the key steps in the externalization of gonococcal LOS is the addition of Hep II onto Hep I: failure to make a diheptosyl structure results in the expression of LOS molecules that are terminated after a single Hep. In order for the gonococcus to express an LOS molecule that has sugar additions onto C-4 of Hep I, it must express rfaF (34, 40). If all Neisseria spp. express their LOSs using the same biosynthetic pathway, then homologs to rfaF should be found in all strains. Southern hybridizations were conducted on HincII-digested chromosomal DNA under conditions that should detect DNA sequences with limited homology to rfaF. The data presented in Fig. 2 indicate that DNA sequences with significant homology to the probe were found in about half of the strains examined. While all of the gonococcal strains, N. meningitidis 891, N. lactamica 5841, N. subflava 44, N. sicca 342, and the N. cinerea strains, possessed this gene, N. subflava 52, 4324, 4325, and 4327, N. sicca 4318, and the N. flavescens strains lacked this gene. If the rfaF gene in N. subflava 52, 4324, 4325, and 4327, N. sicca 4318, and the N. flavescens strains were to encode a functional protein and if the appropriate biosynthetic precursor were made, these strains should produce LOS that contains at least a diheptosyl structure linked to KDO. These strains should then make an LOS with an apparent mobility greater than that seen for strain FA5100. The data presented in Fig. 1 and 2 demonstrate that all strains that contained the DNA sequence that corresponded to rfaF expressed LOSs with higher molecular weights. Since all of the strains that lacked DNA sequences homologous to rfaF were also able to make LOSs that were significantly larger than the LOS made by strain FA5100, a strain whose LOS possesses a single Hep (12), this indicates that sugars can be added onto Hep I in the absence of the rfaF gene product or that a nonhomologous functional equivalent to rfaF exists in these strains.
FIG. 2.
Southern hybridization to test for the presence of rfaF DNA in various Neisseria strains. Lanes: 1, N. subflava 52; 2, N. subflava 4325; 3, N. subflava 4324; 4, N. subflava 44; 5, N. subflava 4327; 6, N. flavescens ATCC 13120; 7, N. flavescens 4323; 8, N. flavescens 4322; 9, N. sicca 4318; 10, N. sicca 342; 11, N. lactamica 5841; 12, N. meningitidis 89I; 13, N. cinerea 32165; 14, N. cinerea 32824; 15, N. gonorrhoeae FA19; 16, N. gonorrhoeae WR302; 17, N. gonorrhoeae PID2; 18, N. gonorrhoeae F62.
Determination of the presence of the lgt gene cluster.
Most of the genes required for the synthesis of the α-chain oligosaccharide in N. gonorrhoeae are found in a gene cluster consisting of lgtA-E (7, 14). We used a probe containing DNA sequences homologous to lgtA and D to test for their presence in the various Neisseria spp. We hypothesized that strains that possess these sequences would have the genetic potential to express the lacto-N-neotetraose and related structures and, as such, could make LOS components that react with the appropriate MAbs. A 954-bp fragment internal to lgtA was used to screen for the presence of lgtA and/or lgtD by Southern hybridization. The probe used contained all of lgtA except the first 49 bp and the last 45 bp. Under the stringency conditions employed, both lgtA and lgtD would bind the probe, due to the high degree of homology between the two genes. However, the binding to lgtA would be more efficient than the binding to lgtD due to the greater sequence identity between lgtA and the probe. Strains that failed to bind the probe were presumed to lack both lgtA and lgtD.
To determine if lgtA, lgtD, or both genes were present in the strains under study, chromosomal DNAs were digested with SspI. Since the DNA sequence of this region is known for N. gonorrhoeae F62 (14) and FA19 (D. C. Stein, unpublished data), we expected the probe to hybridize to a 2.3-kb SspI fragment containing lgtA and an 822-bp fragment internal to lgtD. The two other SspI fragments, containing the remaining lgtD sequences, would not be detected because of the small amount of homology that they have with the probe. DNA from F62 and FA19 exhibited the expected binding profiles, both in size and in relative intensity, indicating that they both contained a single copy of lgtA and lgtD (data not shown). The hybridization data also indicated that N. meningitidis 891, N. lactamica 5841, N. subflava 44, and N. sicca 342 possessed DNA sequences with significant homology to our probe (Table 3).
Correlation of genetic data with SDS-PAGE profiles and MAb binding.
The presence of LOS components able to bind LOS-specific MAbs correlated with the presence or absence of genes located within the lgt gene cluster for all strains examined, with the exception of strains N. subflava 44 and N. sicca 342 (Table 3). While we were able to show that N. subflava 44 and N. sicca 342 contained DNA sequences with high homology to the lgt gene cluster, their LOS SDS-PAGE profiles (Fig. 1) and their lack of ability to bind LOS-specific MAbs suggested that the genes were nonfunctional.
Analysis of N. subflava 44 for the presence of other biosynthetic genes.
We chose to examine the genetic potential of N. subflava 44 to make LOS in greater detail because its LOS was able to bind MAb 1B2 on a colony blot yet failed to express a detectable amount of the lacto-N-neotetraose LOS component when its LOS was analyzed by SDS-PAGE. Furthermore, mass spectrometry combined with fluorophore-assisted carbohydrate electrophoresis monosaccharide composition analysis showed that N. subflava 44 expresses two major LOS components (LOSI and LOSII) (42). LOSI contains one glucose on both the α and β chains. LOSII is structurally related to LOSI and differs from it by the addition of a hexose (either glucose or galactose) on the α chain. Only a trace amount of LOS expressed in N. subflava 44 could further extend the α chain to form the lacto-N-neotetraose structure that could be detected on a colony blot. Based on our structural analysis of N. subflava 44 LOS (42), the structure of its core is identical to the gonococcal and meningococcal LOS core structures. In the gonococcus and the meningococcus, the proteins encoded by lgtF and rfaK are necessary for making the core LOS structure (24). Additional Southern hybridization experiments were performed on chromosomal DNA isolated from N. subflava 44. The data indicated that it contained DNA sequences that bound lgtF- and rfaK-specific probes with the same intensity as the other tested gonococcal strains, indicating that this strain possessed these genes (data not shown).
Analysis of the lgt gene cluster from N. subflava 44.
To study why N. subflava 44 only expressed a limited amount of lacto-N-neotetraose LOS, we characterized the lgt gene cluster. We used the DNA sequence of the N. gonorrhoeae F62 lgt gene cluster to design a pair of primers capable of amplifying this region from strain N. subflava 44 and cloned the amplicon into plasmid pK18 (named pNS44lgtABE). DNA sequence analysis of the PCR-amplified DNA showed that it contained three genes, corresponding to lgtA, lgtB, and lgtE. Each of these genes was >90% identical at the DNA level to its homologs found in N. gonorrhoeae and N. meningitidis. The predicted amino acid sequence of each of the three proteins possessed significant amino acid sequence identity to their homologs in N. meningitidis MC58, N. meningitidis 126E, and N. gonorrhoeae F62 (>98% identical) (data not shown).
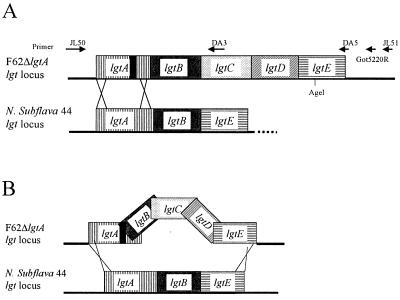
LgtE is responsible for the addition of galactose onto the α chain (49). In order to determine if lgtENS (the lgtE gene isolated from N. subflava 44) was functional, we used the spot transformation procedure to introduce lgtENS (contained on pNS44lgtABE) into N. gonorrhoeae F62 ΔlgtA ΔlgtE. We identified a transformant that acquired reactivity with MAb 2-1-L8 (named F62TF#1). The region surrounding these loci was amplified using PCR, and the DNA sequence of the amplicon was determined. The genetic organization of the recombinant, shown in Fig. 3, indicates that we not only replaced lgtEF62 (the lgtE gene isolated from N. gonorrhoeae F62) with lgtENS but also replaced lgtBF62 with lgtBNS. The SDS-PAGE profile of LOS expressed by the transformant (Fig. 4, lanes 4 and 5) indicates that, when lgtENS is introduced into N. gonorrhoeae F62 ΔlgtA ΔlgtE, LgtENS is functional.
FIG. 3.
Diagram of mechanisms of recombination between N. gonorrhoeae chromosome and plasmid DNA. (A) Description of the introduction of lgtANS into the chromosome of F62 ΔlgtA. (B) Description of the introduction of lgtBNS and lgtENS into the chromosome of F62 ΔlgtA ΔlgtE. Black box in lgtA, location of the deletion, if present.
FIG. 4.
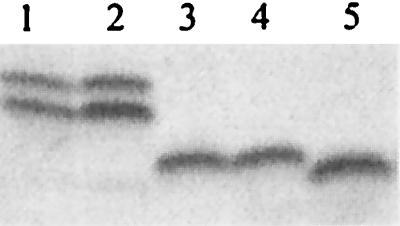
SDS-PAGE analysis of LOS isolated from various N. gonorrhoeae transformation mutants with pNS44lgtABE or pNS44lgtABΔE. The plasmid pNS44lgtABΔE gene was introduced into F62 ΔlgtA, and the plasmid pNS44lgtABE gene was introduced into F62 ΔlgtA ΔlgtE, by spot transformation. Transformants were isolated based on their changes in reactivity with MAb 2-1-L8, and LOS samples were isolated and analyzed on Tris-Tricine gel. Lanes: 1, N. gonorrhoeae F62; 2, N. gonorrhoeae F62 ΔlgtA transformed with pNS44lgtABΔE; 3, N. gonorrhoeae F62 ΔlgtA; 4, N. gonorrhoeae F62 ΔlgtA ΔlgtE transformed with pNS44lgtABE; 5, N. gonorrhoeae F62 ΔlgtA ΔlgtE.
Comparison of amino acid sequences of LgtE from N. gonorrhoeae F62, FA1090, FA19, 1291, and F62TF#1, N. meningitidis MC58 and 126E, and N. subflava 44 showed that there is no significant difference unique to strain N. subflava 44 (data not shown). Since the LgtE protein encoded by N. subflava 44 is able to restore complete function when expressed in a gonococcal background, this indicates that some unknown mechanism is limiting its function in N. subflava 44.
N. subflava 44 lgtA possesses the greatest degree of homology (∼98%) to the N. meningitidis lgtA gene. However, its lgtA gene lacked the polyguanine nucleotide sequence that has been shown to be involved in the variable expression of this gene. We wanted to know whether this alteration in the poly(G) tract might affect its function. Primers JL50 and DA5 (specific for the N. gonorrhoeae F62 lgt gene cluster) were used to amplify N. subflava 44 chromosome DNA via PCR. The amplicon was cloned into pK18, and this DNA (pNS44lgtABΔlgtE) was used to transform N. gonorrhoeae F62 ΔlgtA by the spot transformation procedure. Many transformants that had lost the ability to bind MAb 2-1-L8 and that had acquired reactivity with MAb 1B2 were identified, indicating that lgtANS encodes a functional protein. The complete replacement of lgtAF62 with lgtANS was verified by PCR amplification and DNA sequence analysis. Functional complementation of lgtAF62 with lgtANS was verified by examination of the LOS phenotype by SDS-PAGE (Fig. 4, lanes 2 and 3). These data indicate that the minor sequence differences between lgtANS and lgtAGC (the lgtA gene isolated from gonococci) do not have an effect on LgtA function. They also suggest that lgtANS should encode a functional LgtA protein in N. sublflava 44.
Determination of the presence of lgtG in N. subflava 44.
Structural analysis showed that the major LOSs expressed by N. sublflava 44 possess one glucose on the β chain (42). Banerjee et al. (3) recently described gene lgtG, whose product adds a glucose onto Hep II. We used a probe specific for this gene in Southern hybridization experiments. These experiments indicated that N. subflava 44 possessed DNA sequences homologous to lgtG (Table 3). The region corresponding to lgtG was amplified using PCR, the amplicon was cloned, and the sequence of the amplified DNA was determined. DNA sequence analysis indicated that this gene contained a polycytosine tract that would result in the expression of a functional protein. Comparison of the amino acid sequence of LgtG from strain 15253 to that of LgtG from N. subflava 44 indicated that these two proteins were identical, with the exception of some minor sequence differences in the first 20 amino acids. Additionally, from the predicted coding sequences, lgtGNS initiates expression from a TTG start codon, while lgtG15253 initiates expression from an ATG. Since structural studies have demonstrated the presence of a glucose in N. subflava 44 LOS (42), this indicates that lgtGNS is functional.
Correlation of Southern hybridization data with SDS-PAGE analysis.
Kim et al. (25) showed that N. lactamica LOS was immunologically related to gonococcal and meningococcal LOS. The data presented in Table 3 indicate that N. lactamica 5841 possesses DNA sequences homologous to the lgt gene cluster. This provides genetic evidence to support the immunological observations of Kim et al. (25). The failure of the two N. cinerea strains that we examined to bind the LOS-specific MAbs used as markers for α-chain extensions was expected since these strains lacked the DNA sequences homologous to the lgt gene cluster. Kim et al. (25) were able to identify strains of N. cinerea that reacted with MAb 3F11 (this antibody has the same specificity as MAb 1B2 used in this study). The ability of N. cinerea 32824 to bind MAb 25-1-LC1 suggests that this strain has a functional lgtG gene. The observations of Kim et al. (25), combined with our data, provide one genetic explanation for the structural variation in LOS seen in this species (i.e., the deletion or acquisition of the needed coding region).
We were able to demonstrate the variable presence of the lgt gene cluster in two other species of Neisseria. These genes were found in N. subflava 44 but not in four other N. subflava strains tested. Likewise, we were able to find these genes in N. sicca 342 but not N. sicca 4318. Since it appears that a small number of commensal organisms have acquired DNA sequences associated with gonococcal LOS biosynthesis, it is possible that the reverse has also occurred. Pathogens may have acquired additional biosynthetic potential from commensal strains. A small number of gonococci and meningococci that make LOS molecules that cannot be explained with our current knowledge of LOS genetics have been isolated. For example, the L5 LOS types of meningococci have alternate α-chain structures (30). Likewise, several gonococcal strains that express additional LOS structures have been identified (13).
Speciation of the Neisseriaceae has been based on phenotypic characteristics, including the ability to produce pigment, patterns of acid production from carbohydrates, production of a polysaccharide from sucrose, reduction of nitrate to nitrite, and the production of deoxyribonucleases (31). However, since many examples of localized interspecies recombination between the pathogenic and commensal species have been shown to have occurred (51) and since some commensal organisms are occasionally associated with disease, it seems possible that these apparent discrepancies in the presence of LOS-biosynthetic genes seen for some species can be attributed to the inherent genetic instability of the genus. The ability of some strains of N. subflava and N. sicca to express high-molecular-mass LOS in the absence of an rfaF homolog suggests that some of these species have acquired the ability to express alternate LOS structures.
Taking into account all the data, we have concluded that our current view of the biosynthetic potential and types of LOS structures that might be made by the Neisseriaceae is too limited. This has significant ramifications for researchers as they try to elucidate the role of LOS in the disease process, because they may inadvertently choose the wrong strain for their in vitro models and thereby conclude that a particular molecule is not important in the disease process, because their strain lacked the genetic capability to make the important variation of that structure. The nonpathogenic Neisseriaceae may act as a reservoir for LOS antigenic variants and thereby indirectly contribute to disease.
ACKNOWLEDGMENTS
We thank the members of the laboratory, especially James Levin, for all of their help.
This work was supported by a grant from the National Institutes of Health to D.C.S. (grant A124452). Dan Arking was supported by a grant from the Howard Hughes Medical Institute through the Undergraduate Biological Sciences Education Program.
REFERENCES
- 1.Amsel B J, Moulijn A C. Nonfebrile mitral valve endocarditis due to Neisseria subflava. Chest. 1996;109:280–282. doi: 10.1378/chest.109.1.280. [DOI] [PubMed] [Google Scholar]
- 2.Apicella M A, Westerink M A J, Morse S A, Schneider H, Rice P A, Griffiss J M. Bactericidal antibody response of normal human serum to the lipooligosaccharides of Neisseria gonorrhoeae. J Infect Dis. 1986;153:520–526. doi: 10.1093/infdis/153.3.520. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee A, Wang R, Uljohn S, Rice P A, Gotschlich E C, Stein D C. Identification of the gene (lgtG) encoding the lipooligosaccharide β chain synthesizing glucosyl transferase from Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1998;95:10872–10877. doi: 10.1073/pnas.95.18.10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burch C L, Danaher R J, Stein D C. Antigenic variation in Neisseria gonorrhoeae: production of multiple lipooligosaccharides. J Bacteriol. 1997;179:982–986. doi: 10.1128/jb.179.3.982-986.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannon J G, Black W J, Nachamkin I, Stewart P W. Monoclonal antibody that recognizes an outer membrane antigen common to the pathogenic Neisseria species but not to most nonpathogenic Neisseria species. Infect Immun. 1984;43:994–999. doi: 10.1128/iai.43.3.994-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danaher R J, Levin J C, Arking D, Burch C L, Sandlin R, Stein D C. Genetic basis of Neisseria gonorrhoeae lipooligosaccharide antigenic variation. J Bacteriol. 1995;177:7275–7279. doi: 10.1128/jb.177.24.7275-7279.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danaher R J, Petricoin III E F, Stein D C. Use of xylE fusions to demonstrate that lsi-1, a Neisseria gonorrhoeae lipooligosaccharide biosynthetic gene, and lsi-3 are not transcriptionally linked. J Bacteriol. 1994;176:3428–3432. doi: 10.1128/jb.176.11.3428-3432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dudas K C, Apicella M A. Selection and immunochemical analysis of lipooligosaccharide mutants of Neisseria gonorrhoeae. Infect Immun. 1988;56:499–504. doi: 10.1128/iai.56.2.499-504.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gamian A, Beurret M, Michon F, Brisson J-R, Jennings H J. Structure of the L2 lipopolysaccharide core oligosaccharide of Neisseria meningitidis. J Biol Chem. 1992;267:922–925. [PubMed] [Google Scholar]
- 11.Geisler W M, Markovitz D M. Septic arthritis caused by Neisseria sicca. J Rheumatol. 1998;25:826–828. [PubMed] [Google Scholar]
- 12.Gibson B W, Melaugh W, Phillips N J, Apicella M A, Campagnari A A, Griffiss J M. Investigation of the structural heterogeneity of lipooligosaccharides from pathogenic Haemophilus and Neisseria species and of R-type lipopolysaccharides from Salmonella typhimurium by electrospray mass spectrometry. J Bacteriol. 1993;175:2702–2712. doi: 10.1128/jb.175.9.2702-2712.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson B W, Webb J W, Yamasaki R, Fisher S J, Burlingame A L, Mandrell R E, Schneider H, Griffiss J M. Structure and heterogeneity of the oligosaccharides from the lipopolysaccharides of a pyocin-resistant Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1989;86:17–21. doi: 10.1073/pnas.86.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gotschlich E C. Genetic locus for the biosynthesis of the variable portion of Neisseria gonorrhoeae lipooligosaccharide. J Exp Med. 1994;180:2181–2190. doi: 10.1084/jem.180.6.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregg C R, Melly M A, Hellerquist C G, Coniglio J G, McGee Z A. Toxic activity of purified lipopolysaccharide of Neisseria gonorrhoeae for human fallopian tube mucosa. J Infect Dis. 1981;143:432–439. doi: 10.1093/infdis/143.3.432. [DOI] [PubMed] [Google Scholar]
- 16.Griffiss, J. M. The role of bacterial lipooligosaccharides in the pathogenesis of human disease. Trends Glycosci. Glycotechnol. 7:461–478.
- 17.Gulati S, McQuillen D P, Mandrell R E, Jani D B, Rice P A. Immunogenicity of Neisseria gonorrhoeae lipooligosaccharide epitope 2C7, widely expressed in vivo with no immunochemical similarity to human glycosphingolipids J. Infect Dis. 1996;174:1223–1237. doi: 10.1093/infdis/174.6.1223. . (Erratum, 175:1027, 1997.) [DOI] [PubMed] [Google Scholar]
- 18.Gunn J S, Stein D C. Use of a non-selectable transformation technique to construct a multiple restriction modification deficient mutant of Neisseria gonorrhoeae. Mol Gen Genet. 1996;251:509–517. doi: 10.1007/BF02173639. [DOI] [PubMed] [Google Scholar]
- 19.Hitchcock P J, Brown T M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jennings M P, Hood D W, Peak I R, Virji M, Moxon E R. Molecular analysis of a locus for the biosynthesis and phase-variable expression of the lacto-N-neotetraose terminal lipopolysaccharide structure in Neisseria meningitidis. Mol Microbiol. 1995;18:729–740. doi: 10.1111/j.1365-2958.1995.mmi_18040729.x. [DOI] [PubMed] [Google Scholar]
- 21.John C M, Griffiss J M, Apicella M A, Mandrell R E, Gibson B W. The structural basis for pyocin resistance in Neisseria gonorrhoeae lipooligosaccharides. J Biol Chem. 1991;266:19303–19311. [PubMed] [Google Scholar]
- 22.Johnson K G, Perry M B, McDonald I J. Studies on the cellular and free lipopolysaccharides from Neisseria canis and Neisseria subflava. Can J Microbiol. 1976;22:189. doi: 10.1139/m76-026. [DOI] [PubMed] [Google Scholar]
- 23.Kahler C M, Carlson R W, Rahman M M, Martin L E, Stephens D S. Inner core biosynthesis of lipooligosaccharide (LOS) in Neisseria meningitidis serogroup B: identification and role in LOS assembly of the α1,2 N-acetylglucosamine transferase (RfaK) J Bacteriol. 1996;178:1265–1273. doi: 10.1128/jb.178.5.1265-1273.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kahler C M, Carlson R W, Rahman M M, Martin L E, Stephens D S. Two glycosyltransferase genes, lgtF and rfaK, constitute the lipooligosaccharide ice (inner core extension) biosynthesis operon of Neisseria meningitidis. J Bacteriol. 1996;178:6677–6684. doi: 10.1128/jb.178.23.6677-6684.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J J, Mandrell R E, Griffiss J M. Neisseria lactamica and Neisseria menigitidis share lipooligosaccharide epitopes but lack common capsular and class 1, 2, and 3 protein epitopes. Infect Immun. 1989;57:602–608. doi: 10.1128/iai.57.2.602-608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee F K, Stephens D S, Gibson B W, Engstrom J J, Zhou D, Apicella M A. Microheterogeneity of Neisseria lipooligosaccharide: analysis of a UDP-glucose 4-epimerase mutant of Neisseria meningitidis NMB. Infect Immun. 1995;63:2508–2515. doi: 10.1128/iai.63.7.2508-2515.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandrell R, Schneider H, Apicella M A, Zollinger W, Rice P A, Griffiss J M. Antigenic and physical diversity of Neisseria gonorrhoeae lipooligosaccharides. Infect Immun. 1986;54:63–69. doi: 10.1128/iai.54.1.63-69.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandrell R E. Further antigenic similarities of Neisseria gonorrhoeae lipooligosaccharides and human glycosphingolipids. Infect Immun. 1992;60:3017–3020. doi: 10.1128/iai.60.7.3017-3020.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandrell R E, Lesse A J, Sugai J V, Shero M, Griffiss J M, Cole J A, Parsons N J, Smith H, Morse S A, Apicella M A. In vitro and in vivo modification of Neisseria gonorrhoeae lipooligosaccharide epitope structure by sialylation. J Exp Med. 1990;171:1649–1664. doi: 10.1084/jem.171.5.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michon F, Beurret M, Gamian A, Brisson J R, Jennings H J. Structure of the L5 lipopolysaccharide core oligosaccharides of Neisseria meningitidis. J Biol Chem. 1990;265:7243–7247. [PubMed] [Google Scholar]
- 31.Morse S A, Knapp J S. The Genus Neisseria. In: Balews A, Trüper H G, Dworkin M, Harder W, Schliefer K H, editors. The prokaryotes: a handbook on the biology of bacteria. Berlin, Germany: Springer-Verlag; 1991. pp. 2495–2529. [Google Scholar]
- 32.Parsons N J, Andrade J R C, Patel P V, Cole J A, Smith H. Sialylation of lipopolysaccharide and loss of absorption of bactericidal antibody during conversion of gonococci to serum resistance by cytidine 5′-monophosphate-N-acetyl neuraminic acid. Microb Pathog. 1989;7:63–72. doi: 10.1016/0882-4010(89)90112-5. [DOI] [PubMed] [Google Scholar]
- 33.Pavliak V, Brisson J R, Michon F, Uhrin D, Jennings H J. Structure of the sialylated L3 lipopolysaccharide of Neisseria meningitidis. J Biol Chem. 1993;268:14146–14152. [PubMed] [Google Scholar]
- 34.Petricoin E F, III, Danaher R J, Stein D C. Analysis of the lsi region involved in lipooligosaccharide biosynthesis in Neisseria gonorrhoeae. J Bacteriol. 1991;173:7896–7902. doi: 10.1128/jb.173.24.7896-7902.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petricoin E F, III, Stein D C. Molecular analysis of lipooligosaccharide biosynthesis in Neisseria gonorrhoeae. Infect Immun. 1989;57:2847–2852. doi: 10.1128/iai.57.9.2847-2852.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plested J S, Makepeace K, Jennings M P, Gidney M A J, Lacelle S, Brisson J-R, Cox A D, Martin A, Bird A G, Tang C M, Mackinnon F M, Richards J C, Moxon E R. Conservation and accessibility of an inner core lipopolysaccharide epitope of Neisseria meningitidis. Infect Immun. 1999;67:5417–5426. doi: 10.1128/iai.67.10.5417-5426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pridmore R D. New and versatile vectors with kanamycin-resistance marker. Gene. 1987;56:309–312. doi: 10.1016/0378-1119(87)90149-1. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez R L, Tait R C. Isolation of chromosomal DNA, recombinant DNA techniques: an introduction. Reading, Mass: Addison Wesley Publishing Company; 1983. [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sandlin R C, Danaher R C, Stein D C. Genetic basis of pyocin resistance in Neisseria gonorrhoeae. J Bacteriol. 1994;176:6869–6876. doi: 10.1128/jb.176.22.6869-6876.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song W, Ma L, Chen R, Stein D C. Role of lipooligosaccharide in Opa-independent invasion of Neisseria gonorrhoeae into human epithelial cells. J Exp Med. 2000;191:949–959. doi: 10.1084/jem.191.6.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tong Y, Reinhold V, Reinhold B, Brandt B, Stein D C. Structural and immunochemical characterization of the lipooligosaccharides expressed by Neisseria subflava 44. J Bacteriol. 2001;183:942–950. doi: 10.1128/JB.183.3.942-950.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai C M, Frasch C E. A sensitive silver stain for detecting lipooligosaccharide in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 44.Ward M W, Watt P J, Robertson J N. The human fallopian tube: a laboratory model for gonococcal infection. J Infect Dis. 1974;129:650–659. doi: 10.1093/infdis/129.6.650. [DOI] [PubMed] [Google Scholar]
- 45.Watt P J, Ward M E, Heckels J E, Trust T J. Surface properties of Neisseria gonorrhoeae: attachment to and invasion of mucosal surfaces. In: Brooks G F, Gotschlich E C, Holmes K K, Sawyer W D, Young F E, editors. Immunobiology of Neisseria gonorrhoeae. Washington, D.C.: American Society for Microbiology; 1978. pp. 253–257. [Google Scholar]
- 46.White L A, Kellogg D S., Jr Neisseria gonorrhoeae identification in direct smears by fluorescent antibody counterstain method. Appl Microbiol. 1965;13:171–174. doi: 10.1128/am.13.2.171-174.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamasaki R, Kerwood D E, Schneider H, Quinn K P, Griffiss J M, Mandrell R E. The structure of lipooligosaccharide produced by Neisseria gonorrhoeae, strain 15253, isolated from a patient with disseminated infection: evidence for a new glycosylation pathway of the gonococcal lipooligosaccharide. J Biol Chem. 1994;269:30345–30351. [PubMed] [Google Scholar]
- 48.Yamasaki R, Koshino H, Kurono S, Nishinaka Y, McQuillen D P, Kume A, Gulati S, Rice P A. Structural and immunological characterization of Neisseria gonorrhoeae epitope defined by a monoclonal antibody 2C7; the antibody recognizes a conserved epitope on specific lipooligosaccharides in spite of the presence of human carbohydrate epitopes. J Biol Chem. 1999;274:36550–36558. doi: 10.1074/jbc.274.51.36550. [DOI] [PubMed] [Google Scholar]
- 49.Yang Q L, Gotschlich E C. Variation of gonococcal lipooligosaccharide structures is due to alterations in poly-G tracts in lgt genes encoding glycosyl transferases. J Exp Med. 1996;183:323–327. doi: 10.1084/jem.183.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou D, Lee N G, Apicella M A. Lipooligosaccharide biosynthesis in Neisseria gonorrhoeae: cloning, identification and characterization of the alpha 1,5 heptosyltransferase I gene (rfaC) Mol Microbiol. 1994;14:609–618. doi: 10.1111/j.1365-2958.1994.tb01300.x. [DOI] [PubMed] [Google Scholar]
- 51.Zhou J, Bowler L D, Spratt B G. Interspecies recombination, and phylogenetic distortions, within the glutamine synthetase and shikimate dehydrogenase genes of Neisseria meningitidis and commensal Neisseria species. Mol Microbiol. 1997;23:799–812. doi: 10.1046/j.1365-2958.1997.2681633.x. [DOI] [PubMed] [Google Scholar]