SUMMARY
Facultative multicellular behaviors expand the metabolic capacity and physiological resilience of bacteria. Despite their ubiquity in nature, we lack an understanding of how these behaviors emerge from cellular-scale phenomena. Here, we show how the coupling between growth and resource gradient formation leads to the emergence of multicellular lifecycles in a marine bacterium. Under otherwise carbon-limited growth conditions, Vibrio splendidus 12B01 forms clonal multicellular groups to collectively harvest carbon from soluble polymers of the brown-algal polysaccharide alginate. As they grow, groups phenotypically differentiate into two spatially distinct subpopulations: a static ‘shell’ surrounding a motile, carbon storing ‘core’. Differentiation of these two subpopulations coincides with the formation of a gradient in nitrogen-source availability within clusters. Additionally, we find that populations of cells containing a high proportion of carbon storing individuals propagate and form new clusters more readily on alginate than do populations with few carbon-storing cells. Together, these results suggest that local metabolic activity and differential partitioning of resources leads to the emergence of reproductive cycles in a facultatively multicellular bacterium.
Keywords: morphogenesis, cooperation, division of labor, self-organization, marine microbes, alginate, Vibrio, nanoSIMS, PHA, motility, type IV adhesin
eTOC Blurb
Schwartzman et al. demonstrate how phenotypic heterogeneity helps a marine bacterium break down organic matter by forming multicellular clusters. The heterogeneity that emerges within clonal clusters of the marine bacterium Vibrio splendidus 12B01 spatially allocates resources and promotes ‘lifecycles’ of growth and dispersal.
INTRODUCTION
Clonal groups of microbes often form transient multicellular structures such as biofilms and fruiting bodies1,2. The emergent behaviors of these structures underly ecologically important functions such as antimicrobial resistance3, immune evasion4, and the mobilization of complex nutrients5,6. In addition to these benefits, life at high cell density is also an inevitable source of cellular conflict. Even when cells cooperate, competition for access to other growth-essential nutrients is inevitable. Thus, cells in transient multicellular states must balance cooperation and conflict for the population to derive an ecological benefit. One possible mechanism through which this is achieved within clonal populations is the emergence of phenotypic heterogeneity, which is a ubiquitous attribute of microbial populations7,8. However, we have little understanding of when heterogeneity provides an ecological benefit9. Here, we explore how the emergence of phenotypic heterogeneity in transient multicellular structures formed by a marine bacterium enhances resource partitioning and gives rise to complex reproductive cycles.
The focus of our study is the transient multicellularity that emerges during bacterial decomposition. Decomposers have an ecological advantage in resource-poor environments because they can access otherwise recalcitrant nutrients10. Because decomposition takes place outside of cells, the ability to pool enzymes accelerates the rate of decomposition, and in turn, enhances the recovery of breakdown products. It is common for this behavior to be positively dependent on cell density; strong positive-density dependence results in critical population thresholds below which cells die off5. In this context, the ecological benefit of locally increasing cell density can exert strong evolutionary selection for multicellular behavior11, suggesting that multicellular behaviors are likely widespread among bacterial degraders in nature12.
Here, we demonstrate how phenotypic heterogeneity within multicellular groups shapes resource allocation, growth, and reproduction in a marine polysaccharide decomposer. The Gammaproteobacterium Vibrio splendidus 12B01 grows on the soluble brown-algae derived polysaccharide alginate through the formation of cooperative multicellular clusters. In previous work, we and others have shown that growth of 12B01 on alginate polysaccharide is strongly dependent on local population density13,14. Here, we show, using a combination of transcriptomics, quantitative light microscopy, and single cell stable isotope probing, that cells in clusters exist in a mix of phenotypic states that form reproducible multicellular structures up to 40 μm in radius. We show that phenotypic differentiation promotes sharing of polysaccharide-derived carbon and mitigates competition for other growth-essential nutrients. Our results highlight the role of phenotypic differentiation in the context of polysaccharide decomposition, a common microbial group behavior.
RESULTS
Multicellular growth on alginate is initiated by clonal groups and involves formation of phenotypically distinct sub-populations.
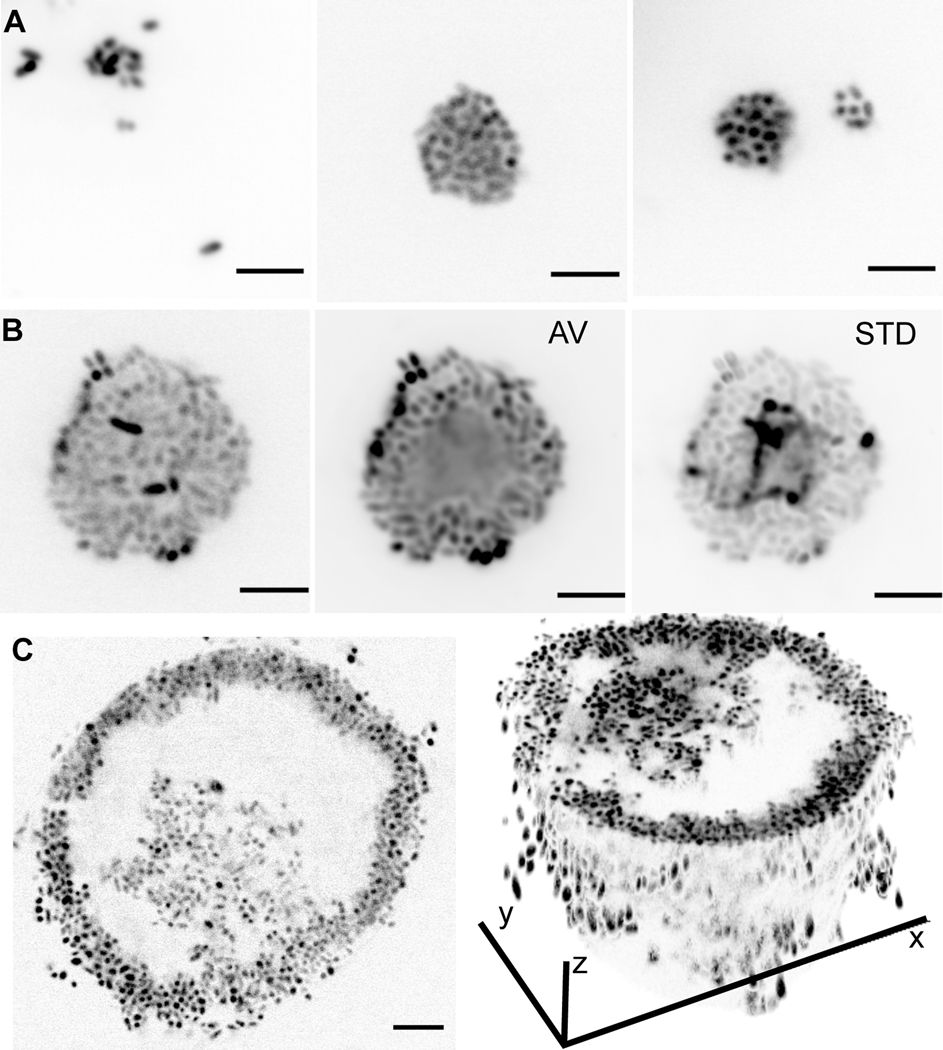
Alginate decomposition is a common trait among marine Vibrio13, and the molecular mechanisms by which 12B01 decomposes alginate are well characterized15–18. A striking feature of 12B01 growth on dissolved alginate is the emergence of motility within cell clusters14 (Video S1). To better understand the process leading to the emergence of motility, we monitored the formation of clusters during growth of 12B01 on alginate in shaking flasks. The population was initially composed of solitary cells, but when growth was detected, it was accompanied by the emergence of tightly packed clusters of cells. Clusters underwent three distinct stages as they grew larger: i) initial formation clusters containing only a few cells (Figure 1A), ii) self-organization of larger multicellular clusters into layered structures composed of an outer sub-population with clear cell-cell adhesion and an inner motile sub-population (Figure 1B, Video S1), and iii) rupture of layered structures accompanied by the release of motile internal contents (Figure 1C). This behavior is reminiscent of the hollowing and seeding dispersal observed in diverse biofilm-forming bacteria19.
Figure 1. 12B01 growth on alginate involves formation of multicellular clusters with phenotypically distinct sub-populations.
A-C) Stages of 12B01 growth on alginate. Scale bars are 5 μm. A) In stage i, single cells slowly form undifferentiated groups. Panels show three examples of stage i groups. B) In stage ii, groups grow and differentiate into layered structures, with a solid shell and a mobile core. Panels show mixing in the center of groups. AV; average intensity projection of time-lapse, showing populations of cells that are motile and not motile (Video S1); STD, standard deviation projection, showing differences in pixel intensity between frames. C) Rupture of clusters leads to hollowing in stage iii. Left: optical section through a cluster, showing absence of cells in center; right: 3D projection, showing structure of hollowed cluster. Additional imaging of 12B01 clusters is presented in Figure S1.
Both physical interactions with soluble strands of alginate polysaccharide and physiological changes resulting from the availability of alginate-derived carbon could contribute to the self-organization of 12B01. We tested these ideas by comparing the growth of cells on alginate polysaccharide, the oligosaccharides released from enzymatic digestion of alginate, or a mix of polysaccharide and oligosaccharides. Although the presence of alginate polysaccharide was sufficient to induce cellular aggregation, aggregates that formed in the presence of oligosaccharides differed greatly in their morphology compared to the compact clusters formed when only polysaccharide was available (Figure S1A). Staining with a cell-impermeant fluorescent dye specific for DNA revealed that these loose biofilm-like aggregates contained dead cells and extracellular DNA, in contrast to the tightly packed clusters that formed during growth on polysaccharides alone (Figure S1B). We infer from these results that while cellular interactions with alginate polysaccharide are likely to take place in many growth environments, alginate polysaccharide must be the only available carbon source for 12B01 to form into tightly packed structures with internal mixing and an external shell.
Formation of clusters coincides with a positive dependence of growth on cell density.
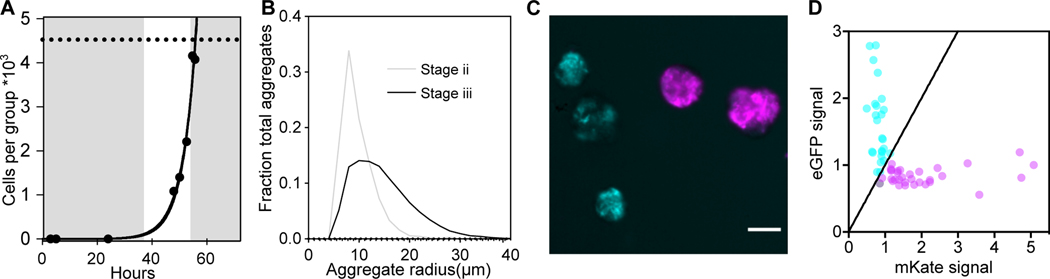
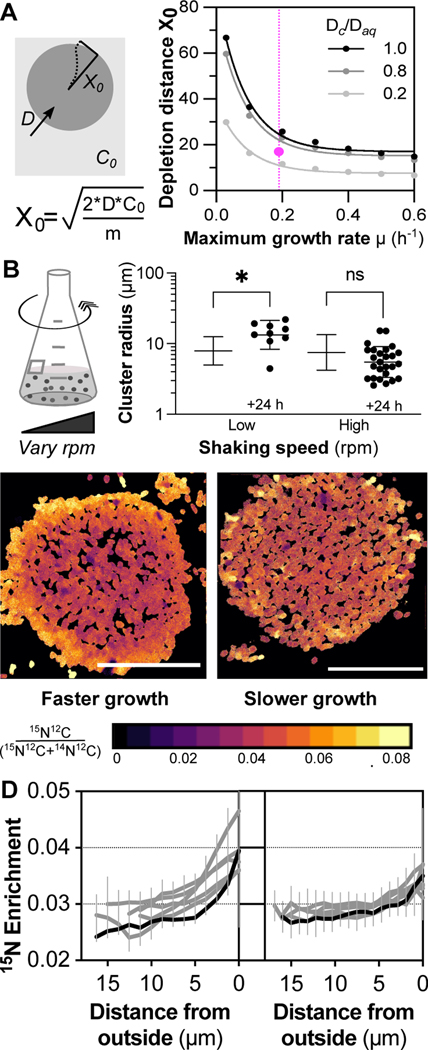
Cultures of 12B01 grown on alginate as the sole carbon source underwent a long lag phase before growth was detectable (Figure 2A). The maximum growth rate of individual clusters was 0.2 h−1 (Figure 2A) compared to the 0.6 h−1 maximum growth rate of cells on alginate oligosaccharides (Figure S2A). Quantification of cluster size distributions at the beginning and end of growth revealed an overall increase in mean size, and a broadening of the distribution of cluster sizes (Figure 2B). The increase in mean cluster size corresponded with a shift in cluster morphology, from a majority in stage ii to a majority in stage iii. Collectively, these results indicate that population-level growth is tied to the growth and morphogenesis of individual aggregates.
Figure 2. Population-level growth of 12B01 on alginate occurs through formation of clonal clusters.
A) Growth of 12B01 in groups, expressed as cells per group. Grey shaded regions delineate growth stages. Exponential growth rate of cells in stage ii (fit line): μmax=0.19 +/− 0.03 h−1. Dashed line indicates size at which growth of clusters ceased due to rupture. Maximum growth rate on alginate oligosaccharides is shown in Figure S2A. B) Histograms showing the change in the distribution of cluster radii at different timepoints of 12B01 growth on alginate. ‘Stage ii’ represents a timepoint when most clusters are growing and have a motile core, and ‘Stage iii’ represents a timepoint when most clusters are ruptured. Clusters measured per timepoint: Stage ii=1085, Stage iii=6105. C) Formation of 12B01 groups tracked using isogenic strains that express fluorescent proteins (Figure S2A). Representative images show distribution of fluorescent protein among eGFP (cyan) or mKate (magenta)-labeled groups at stage ii. Scale bar= 20 μm. D) Quantification of fluorescence intensity within 12B01 groups. Each point represents the total intensity per group, normalized by the mean intensity of groups in each channel. Black diagonal line=equal proportion of both strains. Figure S2B,C provide additional information about cluster formation by the eGFP and mKate-expressing strains.
Biofilms, swarms, and other bacterial social structures often form by the coalescence of many individual cells. Unlike clonal structures, conflict can emerge within aggregative social structures due to the presence of multiple independent genotypes20–23. This led us to investigate whether 12B01 clusters were aggregative or clonal. We mixed isogenic populations of 12B01 each expressing a fluorescent protein and monitored the proportion of clusters that formed with each fluorescent protein, or with a mix of both (Figure 2C, Figure S2B). Clusters were found to express only one fluorescent protein rather both (Figure 2D, Figure S2C), demonstrating that the emergent cooperation of 12B01 clusters on alginate polymer is achieved by division of a progenitor cell into a clonal collective. One consequence of this clonality is that cooperative resource sharing and phenotypic heterogeneity emerge from interactions among individuals with high genetic relatedness.
Cluster stages and sub-populations are transcriptionally distinct.
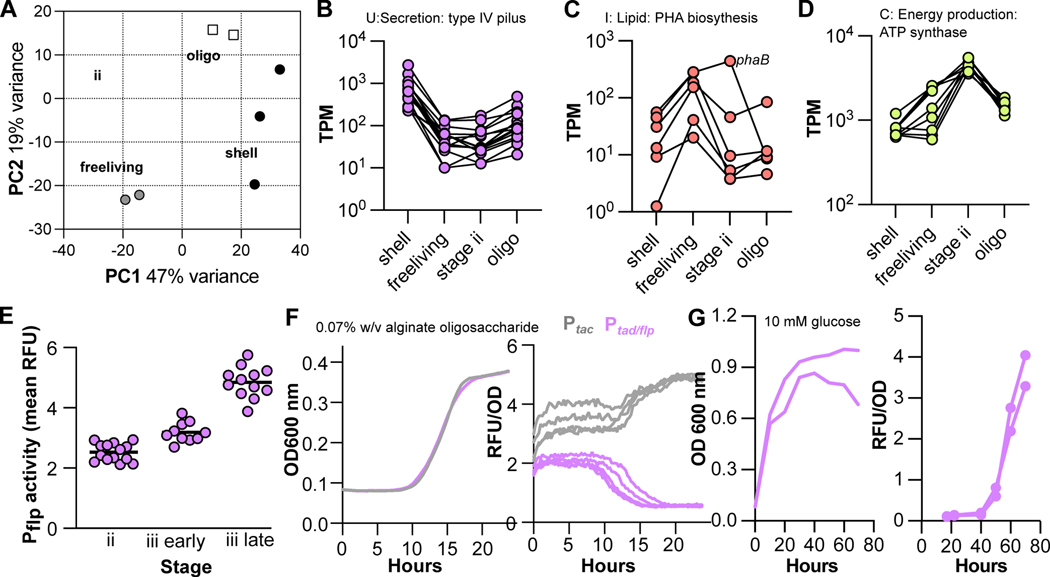
To define physiological differences between sub-populations, we profiled global transcription at different stages of 12B01 growth. The timing of cluster formation in our experimental setup enabled collection of RNA from populations of clusters such that the majority were in a similar stage of morphogenesis. We sampled early stage ii, cells remaining in the shells of late-stage iii clusters, and cells released from late-stage iii clusters (Figure 2B). These samples were chosen to maximize the homogeneity within the sampled population while capturing distinct stages of growth. Free-living stage iii samples were collected no more than 30 minutes after the first signs of rupture as assessed by microscopy and hence likely reflected some aspects of the transcriptional state of the ‘core’ subpopulation directly prior to rupture. We also profiled the transcription of cells grown on alginate oligosaccharides to exponential phase to provide a comparison with a well-mixed planktonic growth state. The full analysis of pair-wise differential gene expression for all conditions is provided in Table S1. Globally, the transcriptional signatures of 12B01 cells differed between samples representing different morphogenic stages and sub-populations (Figure 3A). Transcriptomes of stage ii and stage iii shell samples were most like samples of 12B01 during planktonic growth on alginate oligosaccharides, while the transcription of stage iii free-living cells was more divergent (Figure 3A).
Figure 3. Transcriptional differences define cluster sub-populations.
A) Principal component analysis of normalized gene expression among samples. B-D Transcription of gene clusters specifically transcribed by cluster sample types. TPM, transcripts per million. Lines link the same gene across conditions. See Figure S3A for an analysis of all differentially expressed genes between pairs of samples. B) Transcription of type IV pili genes across sample types (V12B01_22456- V12B01_22516). C) Transcription of polhydroxybutyrate biosynthesis genes (V12B01_15106-V12B01_15126 and V12B01_24289) across sample types. D) Transcription of ATP synthase genes (VF12B01_01867–01907) across sample types. E) Transcriptional activity of tad/flp pili within clusters at timepoints during growth of 12B01 on alginate. Each data point represents the mean activity measured within an individual cluster. Timepoints were taken in the transition from stage ii to stage iii, as noted on the X axis. F) Activity of the tad/flp promoter or synthetic tac promoter during growth on alginate oligosaccharides. Growth of the reporter-containing strains is shown on the left, and fluorescent signal from the transcriptional reporters normalized by cell density (OD 600 nm) is shown on the right. Three replicates are plotted. G) Activity of the tad/flp promoter during growth and stationary phase of 12B01 in glucose defined minimal medium. As in F, cell density (right) and reporter activity (left) are plotted. Two replicates are shown. Figure S3B provides detailed information about the transcription of other biofilm-associated genes, and Figure S3C shows additional controls necessary for the development of the transcriptional reporter construct. See also Tables S1–S3.
Each sample type was characterized by distinct profiles of gene expression. To understand the specific transcriptional differences between cluster stages and sub-populations, we defined sets of genes differentially expressed in a particular sample type relative to all other conditions sampled (Table S2). We further binned these ‘sample-type specific’ genes into categories defined by cellular process using the COG ontology (Figure S3A, Table S2), revealing functional categories that were highly represented in each sample type (Figure S3A). Within each functional category, highly represented functions often reflected the transcription of groups of genes co-localized on the 12B01 chromosome. Examples of these sample-type defining gene groups included a putative type IV tad/flp pilus, which was more highly expressed by stage iii shell samples than in any other sample type (Figure 3B). Stage iii free-living samples transcribed a group of genes related to the biosynthesis of lipid-rich storage compounds known as polyhydroxyalkanoates (PHAs) (Figure 3C). PHA-associated genes were grouped on the chromosome, with the exception of a phaB homolog, which was located near a previously characterized15 alginate catabolism gene cluster. Together, these genes encode machinery to synthesize PHA storage organelles called carbonosomes24,25. Stage ii samples highly expressed F1F0 ATP synthase genes (Figure 3D). In Escherichia coli, ATP synthase genes are subject to growth-rate dependent transcriptional regulation26, consistent with the observation that stage ii clusters are a state where rapid growth occurs. Together, these transcriptional signatures suggest that metabolic activity may change during cluster morphogenesis, and that sub-populations differentiate by expressing genes related to adhesion and carbon storage.
The formation of 12B01 clusters requires cellular adhesion, but it was unclear whether additional biofilm-associated genes participate in cluster formation was unclear. To better address this, we investigated the transcription of Vibrio biofilm formation gene homologs27–29, including genes encoding Syp-like biofilm matrix polysaccharide, an MSH-type pilus, a Lap-like adhesin, and two separate loci encoding type IV Tad/Flp pili (Table S3). The syp-like cluster and one tad/flp-like cluster were transcribed at a very low or undetectable level in our RNASeq profiles (Table S3). Transcription from the msh genes showed no consistent trend across experimental conditions, and the lap gene cluster was elevated in stage ii and stage iii samples (Figure S3A). Only genes belonging to the second tad/flp loci were significantly more expressed in stage iii shell samples relative to other sample types (Figure S3A). Collectively, these results suggest that formation of clusters during 12B01 growth on alginate is unlikely to involve excretion of extracellular polysaccharide matrix but could involve a Lap-like adhesin and a specific role for type IV Tad/Flp pili during the emergence of the shell subpopulation.
To validate the stage iii shell-specific transcription of tad/flp genes, we developed a fluorescent transcriptional reporter (Figure S3C). We found that tad/flp pilus promoter activity was elevated in clusters sampled in later stages of formation (Figure 3E). To understand the growth conditions under which elevated transcription of tad/flp occurs, we characterized transcriptional activity as a function of growth for planktonic cultures. The activity of the Pflp promoter decreased in exponential phase during growth on alginate oligosaccharides (Figure 3F). This decrease in reporter activity was not shared by a constitutively expressed synthetic promoter (Figure 3F), supporting the interpretation that the decrease in tad/flp reporter activity reflects lower transcription during exponential growth. We additionally observed that Pflp reporter activity continued to increase as cells were maintained in stationary phase (Figure 3G). Together, these data suggest that cellular growth state is a regulatory cue for transcription of the tad/flp genes. In the context of cluster formation, the increase in transcription of the tad/flp locus during formation of 12B01 clusters may be linked to slower growth in the stage iii shell sub-population.
Growth of clusters is accompanied by accumulation of carbon storage granules.
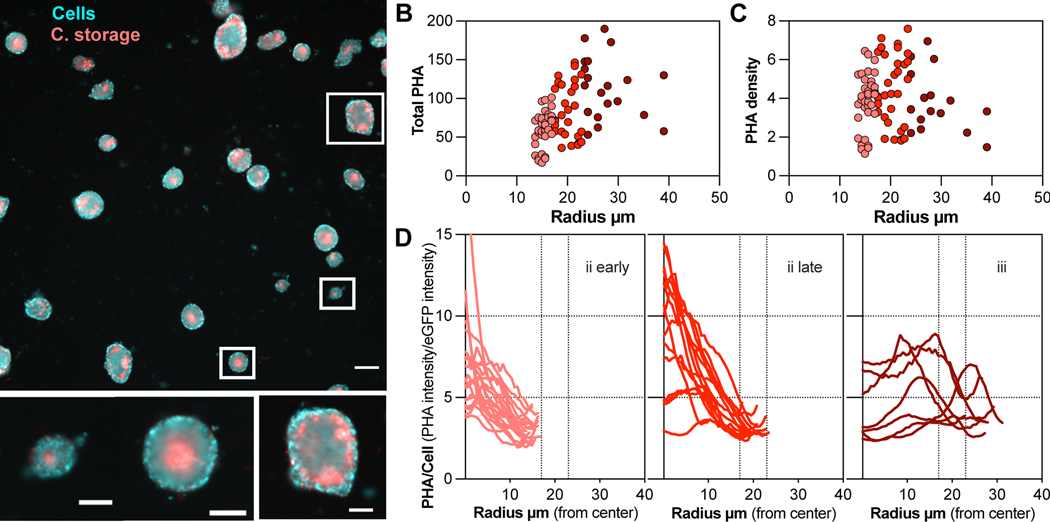
The elevated transcription of PHA biosynthesis genes in stage iii free-living samples led us to predict that cells in this subpopulation stored carbon. We experimentally validated this prediction by staining clusters with the lipophilic fluorescent dye Nile Red (Figure 4A). We found that cells stained brightly with Nile Red, a presumptive PHA-storing sub-population, were indeed localized within the cluster core. The total intensity of Nile Red staining was positively correlated with cluster size (Figure 4B), indicating that the amount of PHA stored by clusters increases as they grow. However, no increase in Nile Red intensity was noted when the signal intensity was normalized by cluster area (Figure 4C), suggesting that accumulation of PHA per cell does not increase greatly as clusters grow. An investigation of the radial intensity profiles of individual clusters supported this idea: although PHA intensity was elevated towards the center of larger stage ii clusters, the magnitude of the difference was modest (Figure 4D). Together, these observations suggest that the microenvironment in the core of stage ii clusters induces accumulation of PHA during growth in this multicellular state.
Figure 4. Accumulation of polyhydroxyalkanoate (PHA) within 12B01 clusters.
A) Composite image showing localization of PHA (C. storage) in eGFP-labeled 12B01 clusters (cells). Three patterns of PHA storage corresponding to different stages of cluster morphogenesis are shown inset, below. Image obtained from a timepoint between those quantified in panel 2A. Scale bar main image= 50 μm, inset=20 μm. B-C) Measurement of total PHA per cluster, as a function of cluster size (B) and PHA density per cluster (total PHA per cluster divided by the cluster area (C). Colors indicate different cluster size bins. Total PHA is measured by the intensity of Nile Red staining, in arbitrary fluorescence units. D) Radial profiles of carbon storage intensity, normalized by cellular expression of GFP. Each trace represents a measurement from individual clusters. Measurements are made from the cluster center (zero) to the periphery. Vertical dashed line notes 17 μm radial distance. Traces are grouped by their size: numbers indicate the patterns represented by inset images in panel 4A. First panel, n=22, second panel n=13, third panel n=7. Colors are consistent with panels B and C.
Inner sub-populations within stage ii clusters become nitrogen limited.
The elevated transcription of genes related to amino acid transport, metabolism and biosynthesis in stage iii free-living samples (Table S2) led us to hypothesize, by analogy to studies of nitrogen limitation in E. coli30, that this sub-population may be nitrogen limited. We further reasoned that nitrogen limitation was most likely to arise while cells were inside clusters. To determine whether limitation in aggregates could be caused by cellular consumption, we made a numerical estimate of ammonium depletion in clusters. Importantly, our estimate assumed that cells in clusters were phenotypically undifferentiated and did not move, a simplification that allowed us to approximate the length scale over which cellular consumption could deplete ammonium supplied by diffusion from outside of a cell cluster (Figure 5A). For clusters growing at the maximum observed rate of 0.2 h−1 or faster, our estimate predicted that clusters could reach 15–20 μm in radius before cells in the center would be completely starved for ammonium (Figure 5A).
Figure 5. Carbon-storing inner sub-populations are nitrogen limited.
A) Estimates of gradients emerging within clusters due to cellular consumption of ammonium as it diffuses in from the bulk medium. Steady state approximation derived as in61. X0= distance from outer surface to point where no ammonium remains, Daq= diffusion coefficient of ammonium in water, Dc/Daq= ratiometric difference between diffusion coefficient in water and in the extracellular space of cell clusters estimated from62, C0=concentration of ammonium ion in medium, m=per cell consumption of ammonium, μ= cellular growth rate. Pink dot indicates cluster size where PHA storage is evident. Vertical pink dashed line indicates the growth rate of clusters with slow shaking (pink). See methods for explanation of cellular consumption rate. B) Growth of 12B01 clusters over a 24 h interval during culture with low or high shaking. Low shaking indicates the speed normally used to cultivate clusters. C) 15N-ammonium assimilation within stage ii clusters formed under low shaking (left- faster growth), or high shaking (right- slower growth) measured by nanoSIMS. Images show spatial patterns of 15N enrichment in an aggregate (fractional abundance 15N), where warmer colors represent higher levels of enrichment (see Figures S4 and S5 for all data). Cellular N assimilation=15N12C /(14N12C+15N12C). All Scale bars= 20 μm. D) Radial profiles of 15N-ammonium enrichment within 12B01 clusters grown under slow (left, n=6) or fast (right, n=7) shaking. Bars indicate standard deviation of pixel intensity. Black traces indicate profiles for images shown in Figure 5C.
We found that increasing the rotation speed with which flasks were shaken slowed down the growth of clusters (Figure 5B). This observation allowed us to test a prediction of our estimate: that a decrease in cellular growth rate would decrease cellular demand for nitrogen and flatten the formation of a nitrogen gradient due to cellular consumption (Figure 5A). We used Nanoscale secondary ion mass spectrometry (NanoSIMS) to measure the assimilation of 15N into biomass. 15N was introduced as ammonium, and clusters were incubated with the heavy isotope for 4 hours, which is equivalent to slightly more than one doubling of cluster biomass. We measured 15N assimilation into biomass in both slow-growing clusters (Figure S4) and fast-growing clusters (Figure S5). Profiles of 15N assimilation revealed a pronounced gradient in assimilation from the outer shell to inner core in fast-growing clusters (Figure 5C). In contrast, we observed a decrease in overall cellular assimilation of 15N as well as a flattening of the assimilation profile across the radius of slow-growing clusters (Figure 5C). Quantification of the nanoSIMS measurements revealed that the outer layers of cells assimilated more than twice the amount of ammonium than cells in the core of clusters (Figure 5D). Together, these measurements point towards the local density-dependent depletion of ammonia as a cue that can emerge in fast-growing clusters. This suggests that growth of clusters is mediated by cooperation for carbon-containing alginate and competition for nitrogen-containing ammonium.
Carbon storage enhances propagation of 12B01 on alginate polysaccharide.
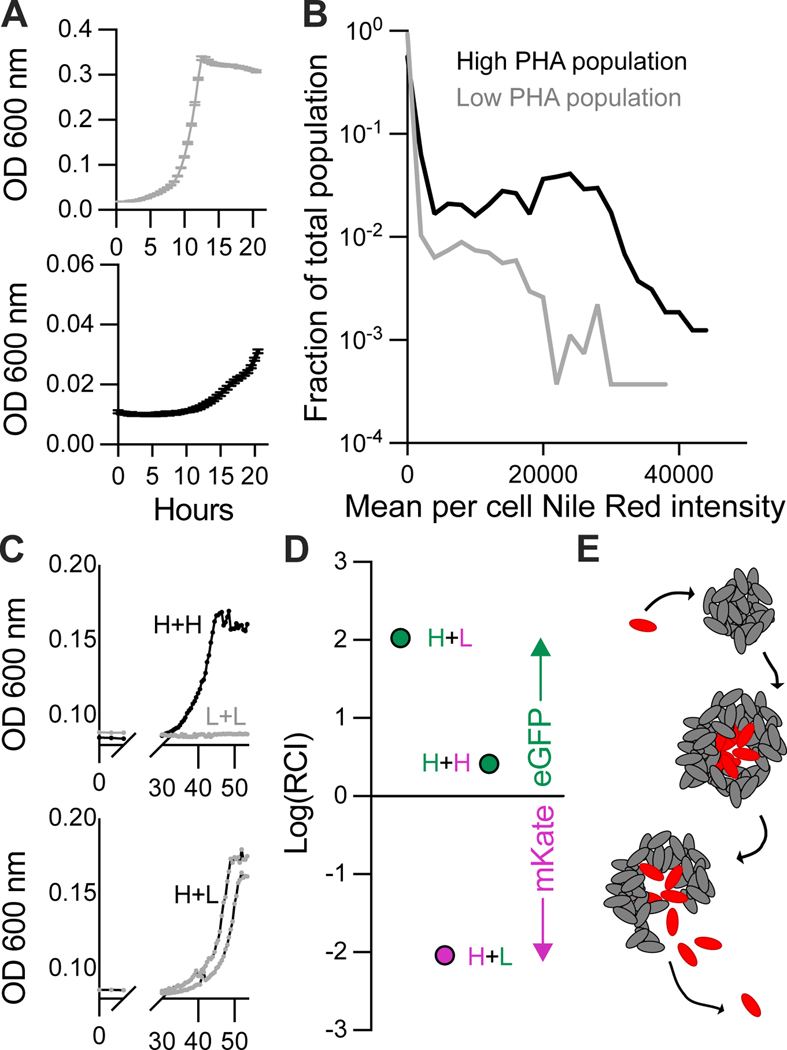
To determine conditions under which PHA accumulated in cells, we monitored PHA storage in populations of 12B01 during growth on alginate oligosaccharides (Figure 6A). We found that PHA storage was most favored under conditions where cells underwent more rounds of replication from their initial founding population: in medium containing alginate oligosaccharides as a carbon source and ammonium as a nitrogen source, a population in which cells underwent on average at least 12 doublings contained a higher proportion of cells that stained brightly with Nile Red (high-PHA) than a population that underwent only four doublings (Figure 6B). Importantly, cells sampled for RNASeq were inoculated into medium containing a concentration of alginate oligosaccharide that would support on average 6 divisions before sampling took place. While we do not yet understand the precise cues that induce carbon storage in 12B01, the relationship between the number of cellular divisions and PHA storage allowed us to reproducibly derive populations with a high or low fraction of cells storing PHA, as measured by Nile Red staining.
Figure 6. Carbon storage enhances propagation of 12B01 on alginate polysaccharide.
A) Growth curves of high-PHA and low-PHA 12B01 populations on alginate oligosaccharides. Black, high-PHA population; grey, low-PHA population. Bars indicate standard error, n=3. B) Fraction of total cell population from A, binned by the mean intensity of Nile Red staining per cell in arbitrary fluorescence units. Nile Red staining is taken as a measurement of PHA storage within cells. C) Growth of high-PHA and low-PHA populations on alginate polysaccharide. Populations were grown alone, or as admixtures. H+H, a 1:1 mixture of mKate and eGFP-expressing high-PHA populations; L+L a 1:1 mixture of low-PHA expressing populations; H+L mixtures of high-PHA and low-PHA populations. All cultures were inoculated with 1.3*106 cells/mL +/− 0.7*106 cells/mL. D) Composition of cultures containing eGFP and mKate admixtures. RCI, relative competitive index, or the initial ratio of eGFP/mKate cells divided by the final ratio or eGFP/mKate cells. H, high PHA; L, low PHA; pink, mKate; green, eGFP. E) Proposed ‘reproductive cycles’ supporting cooperative growth of 12B01 growth on alginate. Cells form clonal clusters. Local density promotes growth on alginate polysaccharide in a carbon-limited environment. As clusters grow, they phenotypically differentiate into ‘shell’ (grey) and ‘core’(red) sub-populations in response to resource gradients. Clusters rupture, releasing carbon-storing ‘core’ sub-population which can propagate to form new clonal clusters.
A question that arises from the observation of PHA-storing cells in the center of clusters is whether carbon storage increases the ability of individual cells to form new clusters. We addressed this question by comparing the growth of high-PHA and low-PHA populations on alginate polysaccharide (Figure 6C). We found that, in contrast to the high-PHA population the low-PHA population was unable to grow on alginate after 50 hours of incubation (Figure 6C). We also assessed the growth of mixed populations (Figure 6C), by deriving high-PHA and low-PHA populations expressing different fluorescent markers (Figure 6D). Our competition experiments revealed that high-PHA populations retained a competitive advantage even when mixed with low-PHA populations (Figure 6D). Together, these results suggest that carbon storage supports the reproductive lifecycles of 12B01 during multicellular growth on alginate polysaccharide.
DISCUSSION
Phenotypic heterogeneity is a property of most clonal populations of bacteria8,23,31. Where this heterogeneity allows cells to perform specific and synergistic tasks, it can give rise to a phenomenon called division of labor. However, it remains challenging to demonstrate that heterogenous cell sub-populations interact synergistically. Outside of experiments that have engineered such interactions in the lab32, biofilms9,33,34 and infection35, few natural examples of bacterial division of labor are known. We propose that the phenotypic differentiation of 12B01 cells during cooperative growth on alginate falls into the framework of a division of labor (Figure 6E). We demonstrate that the functions expressed by these two sub-populations cannot co-occur: Populations grown on alginate oligosaccharides do not highly transcribe type IV tad/flp pili and do not stick together: it is only when cells enter stationary phase during carbon-limited growth in batch culture that pili genes are highly transcribed. In our system, growth on alginate requires cellular cooperation as it is the combined enzymatic activity of cells and their proximity that supports growth on this polymeric carbon source. The differentiation of ‘shell’ and ‘core’ subpopulations provides the local density required for cooperative growth by preventing dispersal. This mechanism is unlike the oscillatory growth observed in Bacillus subtilis colony biofilms, where cellular demand for nitrogen fluctuates to support growth of different sub-populations34. While further work will be needed to capture the dynamics of cluster formation with higher temporal and spatial resolution, our data point to both the mechanical and physiological properties of clusters as critical aspects of division of labor in 12B01 clusters.
The ability to form large, self-organized clusters and undergo reproductive cycles with coordinated bursts of dispersal may shape the ecological dynamics of 12B01. We do not know whether this strain can form similar structures in coastal seawater, however, the concentrations of alginate and ammonium chloride used in our experimental model fall within what can accumulate during algal blooms36. Blooms are also characterized by waves of bacteriophage infection. Biofilms and other bacterial structures have been proposed as barriers to phage infection37–39. Thus, the highly structured clusters formed by 12B01 may provide a barrier to bacteriophage infection, particularly of the cells in the interior of clusters. Blooms are also accompanied by increases in bacterivores such as protists. The grazing efficiency of protists has been shown to depend on the size and structural integrity of their prey, thus, the formation of dense 40 μm groups may enhance resistance to predation40. Future work will be needed to determine whether, in addition to accelerating cycles of reproduction, formation of phenotypically differentiated multicellular groups also enhances resistance to bacteriophage or protist predators.
The formation and rupture of the multicellular clusters described in this work are reminiscent of the formation and dispersal of surface-attached biofilms37,41–46. It is well known that bacteria integrate environmental cues and gradients into the regulation of biofilm formation47–49. Indeed, our work suggests that cellular adhesins associated with biofilm formation are also important for the formation of 12B01 clusters. Importantly, our data suggest that type IV pili are a structural component of 12B01 clusters, especially in the shell subpopulation. Type IV pili are known contribute to biofilm formation and DNA uptake in Vibrio cholerae50, and are often capable of dynamic extension and retraction in many bacteria51. The specific type IV pilus described in this work, Tad/Flp, has a previously characterized role in adhesion to cells and surfaces by diverse bacteria52–56, but has no known role in DNA exchange. Further work will be needed to determine the molecular mechanisms through which Tad/Flp and other cellular adhesins mediate cluster morphogenesis in 12B01.
The emergence of a motile core in 12B01 clonal groups recalls a well-known dispersal phenotype of proteobacterial biofilms sometimes called hollowing37. We show that the emergence of the hollowing-like phenotype in stage ii 12B01 clusters coincides with the formation of carbon-storage granules by this sub-population. Stored carbon can enhance the quality of each propagule by fueling biosynthesis or even transiently increasing the size of the propagule population, as has been observed in PHA-storing rhizobia isolates57,58. PHA-fueled growth may kick start the degradation of difficult to access carbon sources or allow for increased propagule survival or prolong the search-time for a population to find a suitable habitat patch to colonize. Our findings and other recent work58 suggest phenotypic differentiation and asymmetric resource allocation among cells is a strategy employed in natural bacterial populations. More work is needed to establish how well conserved this strategy is among bacteria in nature, and to understand how the integration of motility and carbon storage phenotypes are regulated.
The division of labor, and emergence of complex self-organized structures that characterize 12B01 growth on alginate are attributes shared by simple multicellular organisms59. In this context, the ability of 12B01 to grow in resource-limited environments as clonal structures is particularly intriguing: simulations have suggested that microbial decomposition is constrained both by resource availability and by the evolution of cheats60. We speculate that 12B01’s ability to form clonal, rather than aggregative, structures that phenotypically specialize in response to resource limitation stabilizes this strain’s strong cooperation, much like clonality stabilizes the evolution of multicellular complexity by minimizing cheating and evolutionary conflict21,22. Thus, the ‘lifecycles’ of 12B01 on alginate polysaccharide are likely to prove a useful model through which to understand the ecological and evolutionary consequences of phenotypic heterogeneity during growth in transient multicellular states.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Direct inquiries for further information, resources, and reagents to lead contact Otto X. Cordero (ottox@mit.edu), who will fulfill these requests.
Materials availability
Plasmids and strains generated in this study are available upon request from the lead contact.
Data and code availability
Transcriptomic data (raw trimmed reads, count tables) have been deposited in the NCBI GEO repository GSE190325 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Raw data reported in this paper will be shared by the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial strains | ||
| Vibrio splendidus | 76 | 12B01 |
| Escherichia coli PIR1 | Invitrogen, Waltham, MA, USA | Cat#C101010 |
| V. splendidus carrying a mKate-expressing, chloramphenicol resistant replicative plasmid pLL103 | This paper | 12B01 + pLL103 |
| Escherichia coli PIR1 carrying pLL103 | This paper | PIR1 + pLL103 |
| V. splendidus carrying an eGFP-expressing, chloramphenicol resistant replicative plasmid pLL104 | This paper | 12B01 + pLL104 |
| Escherichia coli PIR1 carrying pLL104 | This paper | PIR1 + pLL104 |
| V. splendidus carrying transcriptional reporter plasmid pJAS2020.1 | This paper | 12B01 + pJAS2020.1 |
| Escherichia coli PIR1 carrying pJAS2020.1 | This paper | PIR1 + pJAS2020.1 |
| Escherichia coli DH5-alpha lambda pir carrying conjugative plasmid pEVS104 | 68 | DH5-alpha lambda pir + pEVS104 |
| Chemicals, peptides, and recombinant proteins | ||
| KpnI-HF | NEB, Ipswich, MA, USA | Cat#R3142 |
| SphI | NEB, Ipswich, MA, USA | Cat#R3182 |
| Chloramphenicol | Sigma-Aldrich St. Louis, MO, USA | Cat#C0378 |
| Marine Broth | BD, Franklin Lakes, NJ, USA | Difco Cat# 2216 |
| Low viscosity alginate | Sigma-Aldrich St. Louis, MO, USA | Cat#A1112 |
| Alginate lyase | Sigma-Aldrich St. Louis, MO, USA | Cat#A1603 |
| Ammonium chloride | Sigma-Aldrich St. Louis, MO, USA | Cat#213330 |
| Nile Red | Sigma-Aldrich St. Louis, MO, USA | Cat#72485 |
| Propidium Iodide | Invitrogen Waltham, MA, USA | Cat#P1304MP |
| Toto-3 | Invitrogen Waltham, MA, USA | Cat#T3604 |
| Lysozyme | Sigma-Aldrich St. Louis, MO, USA | Cat#12650-88-3 |
| 15N-labeled ammonium, 99% | Cambridge Isotope Laboratories, Tewksbury, MA, USA | Cat#NLM-467-PK |
| 16% Paraformaldehyde, aqueous, for electron microscopy | Electron Microscopy Sciences, Hatfield, MA, USA | Cat#50-980-487 |
| Technovit 8100 resin | Heraeus Kulzer GmbH | Cat#64709012 |
| Critical commercial assays | ||
| RNeasy kit | Qiagen, Hilden, Germany | Cat#74004 |
| RNA Protect Bacterial Reagent | Qiagen, Hilden, Germany | Cat#76104 |
| Lysing Matrix B | MPBio, Santa Ana, CA, USA | Cat#116911050-CF |
| Turbo RNA free DNAse kit | Ambion, Austin, TX, USA | Cat#AM1907 |
| HS RNA screen tape | Agilent, Santa Clara, CA, USA | Cat#5067- 5579 |
| HS RNA screen tape buffer | Agilent, Santa Clara, CA, USA | Cat#5067- 5580 |
| HS RNA screen tape ladder | Agilent, Santa Clara, CA, USA | Cat#5067- 5581 |
| Ribominus Yeast and Bacteria transcriptome isolation kit | Invitrogen, Waltham, MA, USA | Cat#A47335 |
| TruSeq stranded mRNA-Seq kit | Illumina, San Diego, CA, USA | Cat#20020594 |
| TruSeq indexes for paired end reads | Illumina, San Diego, CA, USA | Cat#PE-121-1003 |
| Quant-iT RNA Assay kit | Invitrogen, Waltham, MA, USA | Cat#Q32884 |
| Deposited data | ||
| 12B01 genome assembly | 77 | NCBI Genome: ASM15276v1 |
| RNASeq data | This paper | GEO: GSE190325 |
| Github repostitory for image analysis code | This paper78 | N/A |
| Oligonucleotides | ||
| Pflp_SphI_F: CTTGCATGCTTACGATGCATAATAATTAGATTAGAAGTACCACAAAAAAG | This paper | N/A |
| Pflp_KpnI_R: TAGGTACCTTGCGTATCAGGCTTTGTTTACCTG | This paper | N/A |
| mKate_R_Seq: CTCTGAGGTGCACTTGA | This paper | N/A |
| 8F: AGAGTTTGATCCTGGCTCAG | 79 | N/A |
| 1492R: GGTTACCTTGTTACGACTT | 79 | N/A |
| pLL104_confirm_F:GGTGACCGGATCCTTACT | This paper | N/A |
| pLL103_confirm_F:TGACCGGATCCTCAACGG | This paper | N/A |
| pLL10X_confirm_R:ACTACGTTTCATAATGGTTATGAACTTT | This paper | N/A |
| Recombinant DNA | ||
| pLL103 | 65 | N/A |
| pLL104 | 65 | N/A |
| pJS2020.1 | This paper | N/A |
| pEVS104 | 68 | N/A |
| Software and algorithms | ||
| Metamorph | Molecular Devices, San Jose, CA, USA | Version 6.2.3.733 |
| ImageJ | 80 | Version 2.1.0/1.53h |
| Trimmomatic | 81 | Version 0.36 |
| FastQC | 82 | Version 0.11.9 |
| bbduk | 73 | Version 38.16 |
| Bowtie2 | 83 | Version 2.3.4.1 |
| SAMTools | 84 | Version 1.9 |
| HTSeq | 85 | Version 0.12.4 |
| DeSeq2 | 86 | Version 1.34.0 |
| R Studio | 87 | Version 1.2.5042 |
| TPM | 74 | N/A |
| Eggnogg mapper | 88,89 | Version 2.1 |
| Look@nanosims | 90 | N/A |
| MATLAB | Mathworks, Natick, MA, USA | Release R2020b |
| Prism | Graphpad, San Diego, CA, USA | Version 9.3.1 |
| Grid/collection ImageJ plugin | 91 | Version 1.2 |
All original code has been deposited and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL
Vibrio splendidus strain 12B01 was grown at 25 °C. Liquid cultures were established in Marine Broth from colonies grown on MB medium solidified with 1.5% agarose. For experimental measurements, cultures were grown following a previously established protocol63. Briefly, precultures were established in a defined minimal medium64, from MB liquid cultures that had been growing for 4–6 h from a single colony inoculum. The preculture medium contained 10 mM ammonium as sole source of nitrogen and 10 mM glucose unless otherwise noted. Cultures were grown to an optical density of 0.4–0.6, as measured at 600 nm with a 1 cm path cuvette on a Genesys 20 spectrophotometer (Thermo Fisher, Waltham, MA, USA), rinsed by pelleting cells at 5000 rcf for 1 min in a tabletop microcentrifuge (Eppendorf, Hamburg, Germany), and resuspended in carbon-free defined minimal medium such that the final OD was 1.0. To make alginate minimal medium, low viscosity alginate was added to defined minimal medium at a concentration of 700 mg/L (0.07% w/v). The medium was stirred in a sterile glass flask to dissolve the polysaccharide strands, then passed through a 5 μm syringe filter (Pall Corporation, Port Washington, NY, USA) to remove any alginate that was not dissolved. Experimental cultures were initiated by inoculating cells to an initial OD of 0.01 in 250 mL borosilicate glass Erelenmeyer flasks containing 70 mL of alginate minimal medium. ‘Fast’ shaking was established by rotation of flasks at setting 4 on an orbital shaker (0.75” rotational orbit, Model 3500, VWR, Radnor, PA, USA), which corresponds to ~200 rpm, ‘slow’ shaking was established at a setting of 2, corresponding to ~100 rpm. To pre-digest alginate polysaccharide and produce alginate oligosaccharide defined minimal medium, 0.01 μg/mL commercial alginate lyase was added to alginate defined minimal medium and incubated overnight with stirring at 37°C. Alginate lyase was removed prior to assay by filtering the medium through 3 kDa Amicon Ultra molecular weight cutoff filters (MilliPore Sigma, Burlington, MA, USA). To maintain plasmids in 12B01, chloramphenicol was added to growth media at final concentration of 12.5 μg/mL.
METHOD DETAILS
Constructs and validation of replicative plasmids
Multicopy plasmids expressing eGFP or mKate2 from the synthetic tac promoter were constructed in previous work65. These constructs encoded Ptac-eGFP/mKate, the chloramphenicol resistance gene cat and a segment of plasmid pVSV208 released by restriction digest by SpeI and SphI. This segment of pVSV208 contained an origin of transfer for RP4 conjugation66, the R6K origin of replication for maintenance of the plasmid in certain E. coli strains, as well as an origin of replication from a Vibrio fischeri plasmid that is stably maintained in multiple species of Vibrio67. The resultant plasmids, pLL103 and pLL104, were transferred into V. splendidus 12B01 by conjugation using a previously described protocol68. Briefly, ‘donor’ cultures of Escherichia coli PIR1 carrying pLL103 or pLL104 were grown overnight in LB with 25 μg/mL chloramphenicol. A conjugative helper strain, E. coli DH5-alpha lambda pir carrying plasmid pEVS104, was cultivated overnight in LB with 50 μg/mL kanamycin sulfate. 12B01 ‘recipient’ cultures were grown overnight on MB. Equal parts donor, helper, and recipient cultures were collected, pelleted, and resuspended in MB medium to remove any residual antibiotic. The washed cells were combined and centrifuged once more to concentrate the cells. The pellet was resuspended in ~30 μl MB and plated on an MB agar plate to create a dense colony and promote conjugation. After 24 h of incubation at room temperature (21–25 °C), about half of the conjugation mix was resuspended in 250 μl fresh MB, and 100 μl aliquots of this resuspended cell mixture were plated onto MB agar plates containing 12.5 μg/mL chloramphenicol to select for plasmid-carrying transconjugates. Plates were incubated at room temperature for up to 72 h, or until colonies became visible. Colonies were screened for the expression of eGFP or mKate using a blue LED light box with an orange plexiglass filter. The putative transconjugates were struck onto fresh MB chloramphenicol agar plates to derive isolated pure cultures, and stocks were made of single colonies. The presence of pLL103 and pLL104 was confirmed by amplification of a plasmid specific locus using primer pairs pLL103_confirm_F/pLL10X_confirm_R or pLL104_confirm_F/pLL10X_confirm_R. The taxonomic identity of the transconjugates was confirmed by Sanger sequencing of the 16S rRNA gene using primers 8F/1492R. Constructs were maintained in Escherichia coli PIR1 due to their R6K origin of replication.
To build a transcriptional reporter for the tad/flp locus, we mapped mRNA-Seq reads onto the region of the 12B01 chromosome encoding this locus to define the transcriptional start site upstream of the first gene in the locus. We further mapped the ribosome binding site associated with this gene, the putative pilin ORF V12B01_22511, and used a 142 bp segment of DNA falling upstream of this site to promote transcription in our reporter construct. Primers Pflp_SphI_F and Pflp_KpnI_R were designed to amplify this segment of DNA from 12B01 genomic DNA, and to introduce restriction sites. To construct the transcriptional reporter, we digested plasmid pLL103 and the PCR product of Pflp_SphIF/Pflp_KpnI_R with KpnI and SphI restriction enzymes. We ligated the cut insert and vector together with T4 DNA ligase and transformed ligation mixtures into chemically competent PIR1 E. coli. Insertion of the tad/flp promoter region upstream of the mKate gene was confirmed by Sanger sequencing across the junction of mKate into the tad/flp promoter using primer mKate_R_seq. The reporter plasmid was conjugated into V. splendidus 12B01 as described above. The resulting construct was named pJS2020.1 and was maintained in E. coli PIR1 with 25 μg/mL chloramphenicol.
We confirmed the stability of pLL103, pLL104, and pJAS2020.1 by monitoring growth and fluorescence using a Spark plate reader with Te-cool element (Tecan, Männedorf, Switzerland). Specifically, we performed titration experiments to ensure that the amount of chloramphenicol used to maintain positive selection (12.5 μg/mL) was sufficient to prevent plasmid loss and did not alter growth kinetics relative to a no-antibiotic control (Figure S3B). For these experiments, strains were grown in defined minimal medium with 10 mM glucose and continuous 2 mm amplitude 150 rpm double orbital shaking at 25°C. 150 μl of each culture was cultivated in black walled 96-well plates with flat transparent bottoms (Grenier, Kremsmuenster, Austria). Readings were acquired every 15 minutes for optical density (absorbance at 600 nm), eGFP fluorescence from pLL104, and mKate fluorescence from pLL103 or pJAS2020.1. Settings to detect eGFP fluorescence: excitation, 485/20 nm; emission 535/20 nm; dichroic 510 nm. Settings to detect mKate fluorescence: excitation, 588/20 nm; emission, 635/20 nm; dichroic 625 nm. Gain and Z position were set by measuring a sample of each strain a density of OD 1.0.
Light microscopy and staining conditions
Cluster formation by 12B01 during growth on alginate polysaccharide was visualized using a ImageXpress Confocal high content microscope (Molecular Devices, San Jose, CA, USA) using with Metamorph Software, operating in widefield mode. Images were acquired with a 40x Ph2 ELWD objective (0.6 NA, Nikon), and filter sets: Ex 482/35 nm, Em: 536/40 nm, dichroic 506 nm to detect eGFP; Ex: 562/40 nm, Em:624/40 nm, dichroic 593 nm to detect mKate, Nile Red, and Priopidium Iodide; Ex:377/50 nm Em: 447/60 nm dichroic 409 nm to detect TOTO-3. The excitation light source was LED lines from a Spectra X light engine (Lumencore, Beaverton, OR, USA) set to 100% maximum intensity. Images were acquired with a 2048×2048 pixel Andor Xyla 4.2 sCMOS detector. Unless otherwise noted, images were acquired with exposure times of 100 ms.
Visualization of clusters using spinning disk confocal microscopy was performed at the Keck Imaging Facility at MIT’s Whitehead Institute on a AxioVert 200M inverted microscope (Carl Zeiss Instruments, Oberkochen, Germany), with a CSU-22 spinning disk (Yokogawa, Musashino, Tokyo, Japan). Images were acquired using Metamorph software, with 63x or 100× 1.4 NA Plan Apochromat oil objectives and illumination from a 488 nm 150 mM OPSL excitation laser with a 525/50 nm bandpass emission filter and 488 nm dichroic. Unless otherwise noted, images were collected with exposure times of 200 ms, and processed with ImageJ.
To visualize DNA in live cells, culture samples were stained with 5 μM of the DNA-binding dye SYTO9, which emits green fluorescence when bound to DNA. Extracellular DNA was stained as described previously69, except that 1 μM TOTO-3 iodide was used in place of TOTO-1 iodide (fluorescent molecules are both cell-impermeant DNA stains, but with different excitation/emission properties). Dead cells were detected by staining with 20 μM Propidium iodide. PHA accumulation within cells was stained with 0.5 μg/mL lipophilic fluorescent dye Nile Red70. Nile Red dye was resuspended in DMSO at 1 mg/mL, and a fresh 10 μg/mL working solution was created for each experiment to enhance solubility in experimental medium.
RNA isolation and RNA-Seq
Cultures were grown to an indicated stage of morphogenesis, as defined by microscopy. To separate stage iii samples (ruptured clusters) into ‘shell’ and ‘free-living’ fractions, samples were allowed to settle. The top fraction (free-living) and bottom fraction (shell) were collected using a transfer pipette and separated. Note that samples of core and shell in this study were not paired. To collect samples of 12B01 grown on alginate oligosaccharides, glucose-grown precultures were inoculated into 5 mL alginate oligosaccharide defined minimal medium in a 14 mm plastic test tube with a vented cap at an initial optical density of 0.01 and grown with 250 rpm shaking at 25 °C to an optical density of 0.37 prior to harvesting. 30 mL RNA Protect Bacterial Reagent was added to 15 mL cultures of clusters, or 10 mL RNA protect was added to 5 mL or alginate oligosaccharide cultures. Samples were incubated at 4 °C overnight. Cells were collected by centrifugation at 5000 rcf and 4°C in a Sorvall Legend XIR centrifuge, equipped with a Fiberlite F15–6×100g rotor for 30 min. Supernatant was decanted, leaving pelleted cell material which was frozen at −20°C.
To isolate total RNA we used a Qiagen RNeasy kit, following manufacturer’s protocol except for the following modifications related to lysis: Cells were resuspended in 15 mg/mL lysozyme in TE buffer and incubated for room temperature for 30 min prior to addition of buffer RLT. Samples were lysed by mechanical disruption using lysing matrix B. Samples in lysing matrix were shaken in a Fastprep-24 Classic homogenizer (MP Bio, Santa Ana, CA, USA) for 10×30 second intervals, taking care to keep samples from heating up. Samples were then treated with DNase digestion using a Turbo RNA free DNAse kit, following manufacturer’s instructions. The integrity and purity of total RNA was assessed running a sample on an Agilent 4200 Tapestation (Agilent, Santa Clara, CA, USA) in a HS RNA screen tape.
To derive mRNA from total RNA, we performed rRNA depletion with the Ribominus Yeast and Bacteria transcriptome isolation kit. We included an optional step in the manufacturer’s protocol to concentration the remaining mRNA by ethanol precipitation with glycogen. Ethanol precipitated samples were resuspended in FPF sample buffer from the TruSeq stranded mRNA-Seq kit reagents. mRNA abundance was quantified using a Quant-iT RNA Assay kit on the Spark plate reader, following manufacturer’s instructions. The quality of the mRNA was further assessed using an Agilent HS tape on a Bioanalyzer to confirm rRNA depletion and quality: samples were screened for quality by requiring an RNA integrity number (RIN) or more than 8. mRNA remaining from rRNA depletion of 2–10 μg total RNA was used as template to create sequencing libraries. Sequencing libraries were prepared using an Illumina TruSeq Stranded mRNA library preparation kit following the manufacturer’s protocol. Libraries were assessed for quality using Agilent HS screen tapes and fluorescence-based plate Qbit assay prior to pooling. Libraries were sequenced at the Whitehead sequencing core (Whitehead Institute, MIT) on an Illumina HiSeq 2500 in 60×60 in paired-end mode.
Stable isotope amendment experiments and nanoSIMS analysis
To trace the assimilation of ammonium into cellular biomass, 15N-labeled ammonium was added at a final concentration of 10 mM to cultures of 12B01 growing on 0.07% alginate. 15N labeling experiments were performed on 12B01 alginate cultures grown with varying shaking speeds-because the growth of these cultures occurs at different rates (Figure 5B), the addition of the heavy labeled isotope was timed such that clusters were of similar size. The cultures initially contained 10 mM of unlabeled ammonium as the sole nitrogen source, and at the time of 15N addition, some of this unlabeled ammonium likely remained in the culture medium. After a 4 h incubation, including a 1 h step in which aggregates settle out, samples were collected and processed following previously established protocols71 with minor modifications as follows: After removal of supernatant, an equal volume of 4% paraformaldehyde marine PBS (mPBS, 50 mM sodium phosphate pH 7.4, 0.4 M NaCl) was added to the concentrated aggregate mixture for 45 min. 30 min of this time was static, to allow aggregates to settle out. Three 1-hour rinses with 1xmPBS were performed, in which aggregates were incubated with rotation, then allowed to settle for 30 min. Following the final wash step, concentrated aggregates were combined with 1 part molten 9% noble agar in phosphate buffered saline. The agar-aggregate mixture was allowed to solidify in a 1.5 mL centrifuge tube. Samples were stored at 4°C prior to processing. Immobilized samples were dehydrated in ethanol and stored at −20 °C prior to embedding in Technovit 8100 resin and continuing with the manufacturer’s standard protocol. Embedded samples were sectioned into 1 μm thick slices with a glass knife using a microtome (Leica, Wetzlar, Germany), mounted onto poly-l-lysine-coated glass slides, and sputter coated with 40nm of gold using a Cressington sputter coater. Samples were analyzed on a CAMECA nanoSIMS 50L in the Center for Microanalysis at Caltech. Data acquisition began with a pre-sputtering step using a 90-pA primary Cs+ ion beam with aperture diaphragm setting of D1 = 1 until the 14N12C− ion counts stabilized. 14N12C− and 15N12C− ions were imaged along with secondary electron images. Data were collected using a 0.5-pA primary Cs+ beam with aperture diaphragm setting of D1 = 3 and the entrance slit set at (ES)=2. Dwell times ranged from 1–10ms/pixel with acquisition area between 10 and 40 microns.
Oligoalginate growth curves and determination of growth yield.
To measure the maximum rate at which 12B01 grows on alginate oligosaccharides, we inoculated alginate oligosaccharide defined minimal medium with 12B01 cells precultured on the same medium. Cultures were inoculated with an initial optical density of 0.01. Cultures were established in the same growth environment used to study 12B01 cluster formation (described above): 70 mL of medium in Erelenmeyer flasks with slow shaking. Growth was monitored by measuring the 600 nm absorbance of 0.6 mL samples in semi-micro cuvettes on a Genesys 20 spectrophotometer. A range of alginate oligosaccharide concentrations were used to ensure that the measured growth rate reflected growth under conditions where carbon limited growth yield but not growth rate (Figure S2A).
Our calculations of cellular demand required measurement of 12B01 yield with respect to ammonium. To measure growth yield with respect to ammonium, growth yield was measured by cultivating 12B01 on 0.07% w/vol alginate oligosaccharides with varying concentrations of ammonium chloride. As 0.07% alginate oligosaccharides and 10 mM ammonium results in carbon-limited growth (Figure S2A), we titrated ammonium concentrations down from this point. We also used these experiments to determine the number of cells per mL at OD 1.0 by diluting cultures and plating on MB agar. Direct colony counts were converted to cells/OD my multiplying by dilution factor and dividing by the measured OD of the cultures. This number was measured to be 5*10^8 cells/OD.
Experimental manipulation of PHA expression in 12B01 populations.
To prepare high-PHA or low-PHA populations of cells for inoculation into alginate medium, seed cultures of 12B01 carrying plasmids pLL103 or pLL104 were established in MB chloramphenicol medium from single colonies on MB chloramphenicol agar plates. Seed-cultures were diluted 1:100 into precultures containing 10 mM glucose defined minimal medium with chloramphenicol. Cultures between OD 0.2–0.6 were used to inoculate cultures of 12B01 on 0.07% alginate oligosaccharide defined minimal medium with chloramphenicol. Three serial 1:100 dilutions were established from a starting cell density of 0.01, so that the starting cell densities were 5*106, 5*104, and 5*102 cells/mL. Growth of the dilutions was monitored by plate reader, using a protocol described above for validation of pLL103 and pLL104 replicative plasmids with the following modifications: readings were taken at 30 min intervals, and the settings to detect eGFP and mKate were adjusted slightly to minimize autofluorescence from the defined minimal medium. Settings to detect eGFP were: excitation 486/15 nm, emission 525/15 nm, dichroic 510 nm. Settings to detect mKate fluorescence were: excitation 588/20 nm, emission 640/20 nm, dichroic 625 nm. Several technical replicates of each strain and dilution were established to allow for sufficient culture volume for measurement of carbonosomes and inoculation of cultures. The first serial dilution was harvested for low PHA populations, and the third serial dilution was harvested for high PHA populations.
To prepare cells for inoculation, the fraction of cell populations expressing PHA was measured for each dilution. PHA storage was visualized by Nile Red staining of eGFP-labeled cells using the epifluorescent light path of the ImageXpress Micro confocal microscope with the 40X ELWD objective, as described in more detail above. At least five fields of view were obtained for each dilution, and 16-bit images of eGFP intensity and Nile Red intensity were acquired. The intensity of Nile Red staining for each cell was quantified using custom analysis scripts described below. Cultures not stained with Nile red were collected from microtiter plate wells and centrifuged for 1 min at 5000 rcf in a tabletop microcentrifuge to pellet cells. The growth media was removed, and cells were resuspended at an optical density of 0.01 in alginate defined minimal medium. These suspensions were used to inoculate competition experiments.
To measure the contribution of PHA storage to the growth of 12B01 populations on alginate polysaccharide, we monitored growth and fluorescence of high-PHA and low-PHA populations through an additional plate reader experiment. We established growth on 0.07% alginate defined minimal medium with chloramphenicol in 150 μl volume of a 96-well plate. We measured fluorescence and absorbance at 600 nm in the Spark plate reader, using the same settings as for the transcriptional reporter assays. We inoculated individual strains into alginate minimal medium, as well as the following mixes: eGFP-expressing high-PHA and mKate expressing low PHA, eGFP-expressing low PHA and mKate-expressing high-PHA, eGFP and mKate expressing low-PHA populations, or eGFP and mKate expressing high-PHA populations. The cell density and initial ratio of eGFP and mKate-expressing cells in each condition was obtained from direct plate counts. Fluorescence intensity measurements were converted into approximations of cell density using the ratio of fluorescence/OD of the individual cultures. The ratios of mKate and eGFP-labeled cells in mixed cultures were calculated using these estimated cell densities. Relative competitive fitness was calculated by dividing the final ratio of initial eGFP/mKate expressing cells by the initial ratio of eGFP/mKate expressing cells in the inoculum.
QUANTIFICATION AND STATISTICAL ANALYSIS
Plotting data and statistical analysis
Unless otherwise noted, graphs and statistical analysis were generated in Prism.
Estimation of cellular ammonium consumption and depletion length-scale
To understand the conditions under which cells growing in close proximity start to compete for essential nutrients, we made an estimate based on a simplified scenario: a spherical clump of cells made up of individuals in homogenous physiological states. We estimated the length scale at which cells growing on alginate and ammonia would start to deplete their local environment of ammonia. In this scenario, ammonium diffuses into cell clusters from the bulk medium, and the concentration outside of the clusters is held constant at 10 mM. We derived a steady state approximation as in61. In this approximation, the key variables are the diffusion coefficient of ammonia, D, the concentration of ammonia outside the cluster (C0), and the cellular consumption rate, m. C0 is a constant, and 10 mM=1.8*10–3 fg/μm3. The length scale at which no ammonia reaches the center of the cell cluster, X0, is then equal to the square root of 2*D*C0/m. We considered the possibility that the diffusion coefficient of ammonium might vary in clusters and accounted for this by adjusting D by a linear factor such that it was 0.8 or 0.2 the diffusion coefficient in water. This coefficient Daq=1860 μm2/s. The key parameter of this estimate is cellular consumption, m, which is in units of femtograms ammonia consumed per cell volume per second. We calculated m by multiplying growth rate μ by approximations of cellular volume (0.2 μm3). Exponential growth rates are measured on alginate oligosaccharides (0.6 h-1, Figure S2A), and alginate (0.2 h-1, Figure 2A). We estimated mass for 12B01 as 500 fg from previously published range for stationary phase and exponentially growing 100–1000 fg V. splendidus 13B0172, a strain very closely related to 12B01. The yield of 12B01 with respect to ammonium was found to be 2.5 mM ammonium per OD during growth on alginate oligosaccharides (Figure S6). To build a 500 fg cell with this yield would require the cell to consume, 1/14 of its biomass, or 36 fg of ammonia.
Analysis of light microscopy images
Analysis scripts were written in Matlab, installed with the Image Analysis package.
Cluster growth was measured by segmenting images of aggregates obtained by brightfield microscopy. The images were obtained by focusing on the middle of the clusters so that the 2D area corresponded to a cross-section through mid-plane. Pixel size of the 16-bit images was 0.17 μm. A threshold was used to define the 2-dimensional area of aggregates, and this area was quantified. This threshold was established by segmenting the image by first smoothing the image using a gaussian filter, then segmenting based on pixel intensity. The resulting image was binarized. The remaining objects were filtered by their size (removing objects with a minor axis length smaller than 50 and a circularity less than 0.1). Objects close to the margins of the images were filtered out to exclude clusters not fully in the field of view. After these steps, the remaining objects represented clusters, and we derived a radius for each object from the 2D area, converted this length from pixels to μm, calculated a spherical volume, and converted this volume into a number of cells, using 0.2 μm3, as a conversion factor for volume per cell.
The composition of fluorescently labeled cells in aggregates was measured by segmenting images of eGFP and mKate expressing cells. 16-bit grayscale images of eGFP and mKate signal intensity were thresholded by first taking a gradient, then thresholding the resultant image by taking only pixels that were brighter than 4 times the mean intensity. Any pixels missing within objects were filled in to make a mask, and objects with an area smaller than 10 pixels2 were removed. At this point, masks for eGFP and mKate images were combined to create a mask for all clusters. The eGFP and mKate signal intensity was quantified within each cluster area, and the ratio of eGFP/mKate signal was also calculated, generating the data analyzed in this paper.
The mean radial intensity profile of PHA accumulation was quantified from separate 16-bit grayscale images of Nile red signal intensity (10 ms exposure) and eGFP signal intensity (300 ms exposure) taken with 40x magnification. The eGFP channel image was used to create a mask for clusters. Small objects were removed from the image by eroding with a sphere-shaped structural element of size 5. The ‘imfindcircles’ function was used to find the centers and radii of clusters. These coordinates were used to calculate the radial intensity profiles of clusters by walking out in concentric rings from the middle of a cluster, and for each ring, measuring the mean intensity of pixels. These mean radial profiles were calculated for eGFP and Nile red intensity to generate data used for downstream analysis.
The Nile red staining intensity of individual cells was acquired from images of eGFP-labeled 12B01 (200 ms) also stained with Nile Red. 16-bit grayscale images were acquired at 40x magnification. The image of eGFP signal intensity was used to create a mask defining cells. This mask was created by taking the gradient of the image, and then thresholding the gradient to retain pixels greater than 2.5 times the mean intensity. Any empty pixels within segmented cells were filled, and objects smaller than a cell were removed. The resulting mask was used to quantify eGFP and Nile Red signal intensity for each cell. The ratio of Nile Red to eGFP fluorescence was calculated as a relative measure of PHA per cell.
RNASeq data analysis
Paired-end Illumina reads were trimmed using Trimmomatic to remove sequencing adapters and low-quality reads (Phred >30) using parameters leading:30, trailing:30, sliding window 10:20, and minlen 36. PhiX reads were additionally removed using bbduk. The remaining paired reads were checked for quality using FastQC and mapped to the predicted coding regions of the V. splendidus 12B01 genome using Bowtie2, outputting SAM files. The overall alignment rate for reads was 93.06%. SAM files were sorted by position using SAMTools, and mapping was parsed using HTSeq to obtain count tables. HTSeq parameters were: -r name -a 0 -s yes -t CDS -i gene_id -f sam. Differential gene expression was assessed from count tables DeSeq2 (run in RStudio). Briefly, tables were processed using a variance stabilizing transformation to assess clustering of replicates, after which the statistical significance of log fold change between pairs of samples was assessed using a Wald test. P values were adjusted using the Benjamin-Hochberg method, to account for multiple tests. Normalized transcript abundance was also calculated from count tables with custom analysis scripts to implement the transcripts per kilobase million (TPM) method as described previously 74. Predicted 12B01 coding regions were further annotated using Eggnogg mapper to make functional inferences, including annotation of gene products using the COG ontology75.
NanoSIMS data analysis
Raw images were processed with Look@nanosims analysis scripts, to convert secondary ion count data output from the CAMECA into a format readable by MATLAB. Converted 14N12C and 15N12C secondary ion count data were further processed with custom analysis scripts to calculate the 15N fractional abundance and to calculate the average radial 15N fractional abundance. Where aggregates were imaged with multiple fields of view, images were stitched together in ImageJ using the Grid collection/stitching plugin.
The radial intensity profiles of stitched images were measured for the stitched 15N fractional abundance images. The intensity of the stitched image was first normalized by dividing pixel intensities by the maximum intensity in the image. The normalized image was used to create a mask, by thresholding the pixels to retain only those brighter than 0.5 times the mean intensity. Individual cells were removed from the image by a size threshold. Because the clusters were not circular at this magnification, the radial intensity profiles of the resulting segmented clusters were measured by defining bands through eroding the edges of the mask with a sphere structure factor of size 8. The eroded image defined an ‘inner’ portion of the cluster, which could be subtracted from the mask to allow for quantification of a band of pixels. By sequentially eroding the mask, we were able to measure the mean intensity in concentric rings. These positional data were used for downstream analysis of radial intensity profiles.
Supplementary Material
Bold entries indicate genes significantly differentially expressed in a pair. Condition 1,2 conditions compared; mean TPM 1,2, mean normalized gene expression within sample; stdev 1,2, standard deviation; log2FoldChange, output from DeSeq2, condition 1 vs. condition 2; lfcSE, log-fold change standard error; padj, adjusted P-value of DeSeq fold-change expression; COG_category, classification of gene function into COG ontology; Protein ID, NCBI identifier for protein; Annotation, predicted gene function from EggNOG; Description, more information about gene annotation from EggNOG; Preferred name, proposed gene name based on named homologs; PFAMs, protein families assigned to 12B01 predicted protein.
Column labels are the same as in Table S1. Columns H through Q are log2 fold change expression values from DeSeq. Shell, stage iii shell; ii stage ii clusters; free, stage iii free-living cells; oligo, cells grown planktonically on alginate oligosaccharides; Columns N-Q indicate the set of genes differentially expressed in free-living sample relative to all other samples: YES indicates that gene belongs to this set ‘NO’ indicates it does not.
Column labels are the same as in Table S1.
Cells constitutively express eGFP from plasmid pLL104. Scale bar= 10μm. A timestamp is shown in the video.
Highlights.
V. splendidus 12B01 forms clonal multicellular clusters to break down alginate. Phenotypic differentiation within clusters partitions limiting resources. Cluster sub-populations form a static shell and motile carbon-storing core. ‘Lifecycles’ of growth and rupture emerge during growth on alginate.
ACKNOWLEDGEMENTS
We thank the Polz lab for kindly providing Vibrio splendidus 12B01; Glen D’Souza, Jan Hendrick Hehemann and Andreas Sichert for advice about working with alginate; Steven Biller, Allison Coe, and members of the Chisholm lab for RNASeq advice and Yunbin Guan for assistance with nanoSIMS operations. We thank Terrence Hwa, Kapil Amernath, Ghita Ghessous and members of the Hwa lab, as well as Martin Ackermann for helpful discussions. A.E. acknowledges funding from Swiss National Science Foundation: Grants P2EZP2 175128 and P400PB_186751. Y.S. was funded through the Japan Society for the Promotion of Science KAKENHI (Grant Number 20H02291). O.X.C and J.S acknowledge support from the Kavli Institute of Theoretical Physics National Science Foundation Grant No. NSF PHY-1748958, NIH Grant No. R25GM067110, the Gordon and Betty Moore Foundation Grant No. 2919.02, National Science Foundation under Grant No. NSF PHY-1748958. This work was supported by Simons Foundation: Principles of Microbial Ecosystems (PriME) award numbers 542395 and 542393.
Footnotes
INCLUSION AND DIVERSITY STATEMENT
One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community.
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Claessen D., Rozen DE., Kuipers OP., Søgaard-Andersen L., and van Wezel GP. (2014). Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat. Rev. Microbiol 12, 115–124. [DOI] [PubMed] [Google Scholar]
- 2.Shapiro JA (1998). Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol 52, 81–104. [DOI] [PubMed] [Google Scholar]
- 3.Høiby N, Bjarnsholt T, Givskov M, Molin S, and Ciofu O. (2010). Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35, 322–332. [DOI] [PubMed] [Google Scholar]
- 4.Arciola CR, Campoccia D, and Montanaro L. (2018). Implant infections: adhesion, biofilm formation and immune evasion. Nat. Rev. Microbiol 16, 397–409. [DOI] [PubMed] [Google Scholar]
- 5.Ratzke C, and Gore J. (2016). Self-organized patchiness facilitates survival in a cooperatively growing Bacillus subtilis population. Nat Microbiol 1, 16022. [DOI] [PubMed] [Google Scholar]
- 6.Koschwanez JH, Foster KR, and Murray AW (2013). Improved use of a public good selects for the evolution of undifferentiated multicellularity. Elife 2, e00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vlamakis H, Chai Y, Beauregard P, Losick R, and Kolter R. (2013). Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol 11, 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dar D, Dar N, Cai L, and Newman DK (2021). Spatial transcriptomics of planktonic and sessile bacterial populations at single-cell resolution. Science 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Gestel J, Vlamakis H, and Kolter R. (2015). Division of Labor in Biofilms: the Ecology of Cell Differentiation. Microbiol Spectr 3, MB-0002–2014. [DOI] [PubMed] [Google Scholar]
- 10.Sichert A, and Cordero OX (2021). Polysaccharide-Bacteria Interactions From the Lens of Evolutionary Ecology. Front. Microbiol 12, 2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koschwanez JH, Foster KR, and Murray AW (2011). Sucrose utilization in budding yeast as a model for the origin of undifferentiated multicellularity. PLoS Biol. 9, e1001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Souza GG, Povolo VR, Keegstra JM, Stocker R, and Ackermann M. (2021). Nutrient complexity triggers transitions between solitary and colonial growth in bacterial populations. ISME J. 15, 2614–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hehemann J-H, Arevalo P, Datta MS, Yu X, Corzett CH, Henschel A, Preheim SP, Timberlake S, Alm EJ, and Polz MF (2016). Adaptive radiation by waves of gene transfer leads to fine-scale resource partitioning in marine microbes. Nat. Commun 7, 12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebrahimi A, Schwartzman J, and Cordero OX (2019). Multicellular behaviour enables cooperation in microbial cell aggregates. Philos. Trans. R. Soc. Lond. B Biol. Sci 374, 20190077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wargacki AJ, Leonard E, Win MN, Regitsky DD, Santos CNS, Kim PB, Cooper SR, Raisner RM, Herman A, Sivitz AB, et al. (2012). An engineered microbial platform for direct biofuel production from brown macroalgae. Science 335, 308–313. [DOI] [PubMed] [Google Scholar]
- 16.Jagtap SS, Hehemann J-H, Polz MF, Lee J-K, and Zhao H. (2014). Comparative biochemical characterization of three exolytic oligoalginate lyases from Vibrio splendidus reveals complementary substrate scope, temperature, and pH adaptations. Appl. Environ. Microbiol 80, 4207–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Badur AH., Plutz MJ., Yalamanchili G., Jagtap SS., Schweder T., Unfried F., Markert S., Polz MF., Hehemann J-H., and Rao CV. (2017). Exploiting fine-scale genetic and physiological variation of closely related microbes to reveal unknown enzyme functions. J. Biol. Chem 292, 13056–13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Badur AH, Jagtap SS, Yalamanchili G, Lee J-K, Zhao H, and Rao CV (2015). Alginate lyases from alginate-degrading Vibrio splendidus 12B01 are endolytic. Appl. Environ. Microbiol 81, 1865–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan JB (2010). Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res 89, 205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grosberg RK, and Strathmann RR (2007). The Evolution of Multicellularity: A Minor Major Transition? Annu. Rev. Ecol. Evol. Syst 38, 621–654. [Google Scholar]
- 21.Márquez-Zacarías P, Conlin PL, Tong K, Pentz JT, and Ratcliff WC (2021). Why have aggregative multicellular organisms stayed simple? Curr. Genet 67, 871–876. [DOI] [PubMed] [Google Scholar]
- 22.Pentz JT, Márquez-Zacarías P, Bozdag GO, Burnetti A, Yunker PJ, Libby E, and Ratcliff WC (2020). Ecological Advantages and Evolutionary Limitations of Aggregative Multicellular Development. Curr. Biol 30, 4155–4164.e6. [DOI] [PubMed] [Google Scholar]
- 23.Nadell CD, Drescher K, and Foster KR (2016). Spatial structure, cooperation and competition in biofilms. Nat. Rev. Microbiol 14, 589–600. [DOI] [PubMed] [Google Scholar]
- 24.Jendrossek D. (2009). Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J. Bacteriol 191, 3195–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greening C, and Lithgow T. (2020). Formation and function of bacterial organelles. Nat. Rev. Microbiol 18, 677–689. [DOI] [PubMed] [Google Scholar]
- 26.Kasimoglu E, Park SJ, Malek J, Tseng CP, and Gunsalus RP (1996). Transcriptional regulation of the proton-translocating ATPase (atpIBEFHAGDC) operon of Escherichia coli: control by cell growth rate. J. Bacteriol 178, 5563–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christensen DG, Marsden AE, Hodge-Hanson K, Essock-Burns T, and Visick KL (2020). LapG mediates biofilm dispersal in Vibrio fischeri by controlling maintenance of the VCBS-containing adhesin LapV. Mol. Microbiol 114, 742–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teschler JK, Zamorano-Sánchez D, Utada AS, Warner CJA, Wong GCL, Linington RG, and Yildiz FH (2015). Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat. Rev. Microbiol 13, 255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yildiz FH, and Visick KL (2009). Vibrio biofilms: so much the same yet so different. Trends Microbiol. 17, 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pahel G, Rothstein DM, and Magasanik B. (1982). Complex glnA-glnL-glnG operon of Escherichia coli. J. Bacteriol 150, 202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ackermann M. (2015). A functional perspective on phenotypic heterogeneity in microorganisms. Nat. Rev. Microbiol 13, 497–508. [DOI] [PubMed] [Google Scholar]
- 32.Dragoš A, Kiesewalter H, Martin M, Hsu C-Y, Hartmann R, Wechsler T, Eriksen C, Brix S, Drescher K, Stanley-Wall N, et al. (2018). Division of Labor during Biofilm Matrix Production. Curr. Biol 28, 1903–1913.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geerlings NMJ., Karman C., Trashin S., As KS., Kienhuis MVM., Hidalgo-Martinez S., Vasquez-Cardenas D., Boschker HTS., De Wael K., Middelburg JJ., et al. (2020). Division of labor and growth during electrical cooperation in multicellular cable bacteria. Proc. Natl. Acad. Sci. U. S. A 117, 5478–5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Prindle A, Humphries J, Gabalda-Sagarra M, Asally M, Lee D-YD, Ly S, Garcia-Ojalvo J, and Süel GM (2015). Metabolic co-dependence gives rise to collective oscillations within biofilms. Nature 523, 550–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ackermann M, Stecher B, Freed NE, Songhet P, Hardt W-D, and Doebeli M. (2008). Self-destructive cooperation mediated by phenotypic noise. Nature 454, 987–990. [DOI] [PubMed] [Google Scholar]
- 36.Martin-Platero AM, Cleary B, Kauffman K, Preheim SP, McGillicuddy DJ, Alm EJ, and Polz MF (2018). High resolution time series reveals cohesive but short-lived communities in coastal plankton. Nat. Commun 9, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rumbaugh KP, and Sauer K. (2020). Biofilm dispersion. Nat. Rev. Microbiol 18, 571–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vidakovic L, Singh PK, Hartmann R, Nadell CD, and Drescher K. (2018). Dynamic biofilm architecture confers individual and collective mechanisms of viral protection. Nat Microbiol 3, 26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDougald D, Rice SA, Barraud N, Steinberg PD, and Kjelleberg S. (2011). Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol 10, 39–50. [DOI] [PubMed] [Google Scholar]
- 40.Pernthaler J. (2005). Predation on prokaryotes in the water column and its ecological implications. Nat. Rev. Microbiol 3, 537–546. [DOI] [PubMed] [Google Scholar]
- 41.Mai-Prochnow A, Evans F, Dalisay-Saludes D, Stelzer S, Egan S, James S, Webb JS, and Kjelleberg S. (2004). Biofilm development and cell death in the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol 70, 3232–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunt SM, Werner EM, Huang B, Hamilton MA, and Stewart PS (2004). Hypothesis for the role of nutrient starvation in biofilm detachment. Appl. Environ. Microbiol 70, 7418–7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaplan JB, Meyenhofer MF, and Fine DH (2003). Biofilm growth and detachment of Actinobacillus actinomycetemcomitans. J. Bacteriol 185, 1399–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart PS, Rani SA, Gjersing E, Codd SL, Zheng Z, and Pitts B. (2007). Observations of cell cluster hollowing in Staphylococcus epidermidis biofilms. Lett. Appl. Microbiol 44, 454–457. [DOI] [PubMed] [Google Scholar]
- 45.Purevdorj-Gage B, Costerton WJ, and Stoodley P. (2005). Phenotypic differentiation and seeding dispersal in non-mucoid and mucoid Pseudomonas aeruginosa biofilms. Microbiology 151, 1569–1576. [DOI] [PubMed] [Google Scholar]
- 46.Sauer K, Camper AK, Ehrlich GD, Costerton JW, and Davies DG (2002). Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol 184, 1140–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerstel U, and Römling U. (2001). Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella typhimurium. Environ. Microbiol 3, 638–648. [DOI] [PubMed] [Google Scholar]
- 48.Glick R., Gilmour C., Tremblay J., Satanower S., Avidan O., Déziel E., Greenberg EP., Poole K., and Banin E. (2010). Increase in rhamnolipid synthesis under iron-limiting conditions influences surface motility and biofilm formation in Pseudomonas aeruginosa. J. Bacteriol 192, 2973–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zacharia VM, Ra Y, Sue C, Alcala E, Reaso JN, Ruzin SE, and Traxler MF (2021). Genetic Network Architecture and Environmental Cues Drive Spatial Organization of Phenotypic Division of Labor in Streptomyces coelicolor. MBio 12, e00794–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams DW, Stutzmann S, Stoudmann C, and Blokesch M. (2019). DNA-uptake pili of Vibrio cholerae are required for chitin colonization and capable of kin recognition via sequence-specific self-interaction. Nat Microbiol 4, 1545–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCallum M, Burrows LL, and Howell PL (2019). The Dynamic Structures of the Type IV Pilus. Microbiol Spectr 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nika JR, Latimer JL, Ward CK, Blick RJ, Wagner NJ, Cope LD, Mahairas GG, Munson RS Jr, , and Hansen EJ (2002). Haemophilus ducreyi requires the flp gene cluster for microcolony formation in vitro. Infect. Immun 70, 2965–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pönisch W, Eckenrode KB, Alzurqa K, Nasrollahi H, Weber C, Zaburdaev V, and Biais N. (2018). Pili mediated intercellular forces shape heterogeneous bacterial microcolonies prior to multicellular differentiation. Sci. Rep 8, 16567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pu M, and Rowe-Magnus DA (2018). A Tad pilus promotes the establishment and resistance of Vibrio vulnificus biofilms to mechanical clearance. NPJ Biofilms Microbiomes 4, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duong-Nu T-M, Jeong K, Hong SH, Puth S, Kim SY, Tan W, Lee KH, Lee SE, and Rhee JH (2019). A stealth adhesion factor contributes to Vibrio vulnificus pathogenicity: Flp pili play roles in host invasion, survival in the blood stream and resistance to complement activation. PLoS Pathog. 15, e1007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sangermani M, Hug I, Sauter N, Pfohl T, and Jenal U. (2019). Tad Pili Play a Dynamic Role in Caulobacter crescentus Surface Colonization. MBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ratcliff WC, Kadam SV, and Denison RF (2008). Poly-3-hydroxybutyrate (PHB) supports survival and reproduction in starving rhizobia. FEMS Microbiol. Ecol 65, 391–399. [DOI] [PubMed] [Google Scholar]
- 58.Muller KE, and Denison RF (2018). Resource acquisition and allocation traits in symbiotic rhizobia with implications for life-history outside of legume hosts. R Soc Open Sci 5, 181124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szathmáry E, and Smith JM (1995). The major evolutionary transitions. Nature 374, 227–232. [DOI] [PubMed] [Google Scholar]
- 60.Allison SD (2005). Cheaters, diffusion and nutrients constrain decomposition by microbial enzymes in spatially structured environments. Ecol. Lett 8, 626–635. [Google Scholar]
- 61.Crank J. (1975). The Mathematics of Diffusion (Oxford University Press; ). [Google Scholar]
- 62.Stewart PS (2003). Diffusion in biofilms. J. Bacteriol 185, 1485–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amarnath K., Narla AV., Pontrelli S., Dong J., Caglar T., Taylor BR., Schwartzman J., Sauer U., Cordero OX., and Hwa T. (2021). Stress-induced cross-feeding of internal metabolites provides a dynamic mechanism of microbial cooperation. bioRxiv, 2021.06.24.449802. [Google Scholar]
- 64.Ebrahimi A, Schwartzman J, and Cordero OX (2019). Cooperation and spatial self-organization determine rate and efficiency of particulate organic matter degradation in marine bacteria. Proc. Natl. Acad. Sci. U. S. A 116, 23309–23316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pollak S, Gralka M, Sato Y, Schwartzman J, Lu L, and Cordero OX (2020). Public good exploitation in natural bacterioplankton communities. bioRxiv, 2020.12.13.422583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dunn AK, Millikan DS, Adin DM, Bose JL, and Stabb EV (2006). New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol 72, 802–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Le Roux F, Davis BM, and Waldor MK (2011). Conserved small RNAs govern replication and incompatibility of a diverse new plasmid family from marine bacteria. Nucleic Acids Res. 39, 1004–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stabb EV, and Ruby EG (2002). RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358, 413–426. [DOI] [PubMed] [Google Scholar]
- 69.Jemielita M, Wingreen NS, and Bassler BL (2018). Quorum sensing controls Vibrio cholerae multicellular aggregate formation. Elife 7, e42057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spiekermann P, Rehm BH, Kalscheuer R, Baumeister D, and Steinbüchel A. (1999). A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch. Microbiol 171, 73–80. [DOI] [PubMed] [Google Scholar]
- 71.McGlynn SE, Chadwick GL, O’Neill A, Mackey M, Thor A, Deerinck TJ, Ellisman MH, and Orphan VJ (2018). Subgroup Characteristics of Marine Methane-Oxidizing ANME-2 Archaea and Their Syntrophic Partners as Revealed by Integrated Multimodal Analytical Microscopy. Appl. Environ. Microbiol 84, e00399–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cermak N, Becker JW, Knudsen SM, Chisholm SW, Manalis SR, and Polz MF (2017). Direct single-cell biomass estimates for marine bacteria via Archimedes’ principle. ISME J. 11, 825–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bushnell B. (2014). BBMap: a fast, accurate, splice-aware aligner (Lawrence Berkeley National Lab.(LBNL), Berkeley, CA (United States)). [Google Scholar]
- 74.Sato Y, Hori T, Koike H, Navarro RR, Ogata A, and Habe H. (2019). Transcriptome analysis of activated sludge microbiomes reveals an unexpected role of minority nitrifiers in carbon metabolism. Commun Biol 2, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tatusov RL, Galperin MY, Natale DA, and Koonin EV (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Le Roux F., Zouine M., Chakroun N., Binesse J., Saulnier D., Bouchier C., Zidane N., Ma L., Rusniok C., Lajus A., et al. (2009). Genome sequence of Vibrio splendidus: an abundant planctonic marine species with a large genotypic diversity. Environ. Microbiol 11, 1959–1970. [DOI] [PubMed] [Google Scholar]
- 77.Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, and Polz MF (2008). Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320, 1081–1085. [DOI] [PubMed] [Google Scholar]
- 78.Schwartzman J. Github repository for 12B01 image analysis code with example images. (2022) https://github.com/jaschwartzman/12B01/ DOI: 10.5281/zenodo.6490838 [DOI]
- 79.Turner S, Pryer KM, Miao VP, and Palmer JD (1999). Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol 46, 327–338. [DOI] [PubMed] [Google Scholar]
- 80.Rasband WS, Image J, U. S. National Institutes of Health, Bethesda, Maryland, USA: (1997-2018). https://imagej.nih.gov/ij/. [Google Scholar]
- 81.Bolger AM, Lohse M, and Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brown J, Pirrung M, and McCue LA (2017). FQC Dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 33, 3137–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. (2021). Twelve years of SAMtools and BCFtools. Gigascience 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anders S, Pyl PT, and Huber W. (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Love MI, Huber W, and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.RStudio Team (2020). RStudio. RStudio: Integrated Development for R RStudio, PBC, Boston, MA. https://www.rstudio.com/. [Google Scholar]
- 88.Huerta-Cepas J., Szklarczyk D., Heller D., Hernández-Plaza A., Forslund SK., Cook H., Mende DR., Letunic I., Rattei T., Jensen LJ., et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, and Huerta-Cepas J. (2021). eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol 38, 5825–5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Polerecky L, Adam B, Milucka J, Musat N, Vagner T, and Kuypers MMM (2012). Look@NanoSIMS--a tool for the analysis of nanoSIMS data in environmental microbiology. Environ. Microbiol 14, 1009–1023. [DOI] [PubMed] [Google Scholar]
- 91.Preibisch S, Saalfeld S, and Tomancak P. (2009). Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25, 1463–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bold entries indicate genes significantly differentially expressed in a pair. Condition 1,2 conditions compared; mean TPM 1,2, mean normalized gene expression within sample; stdev 1,2, standard deviation; log2FoldChange, output from DeSeq2, condition 1 vs. condition 2; lfcSE, log-fold change standard error; padj, adjusted P-value of DeSeq fold-change expression; COG_category, classification of gene function into COG ontology; Protein ID, NCBI identifier for protein; Annotation, predicted gene function from EggNOG; Description, more information about gene annotation from EggNOG; Preferred name, proposed gene name based on named homologs; PFAMs, protein families assigned to 12B01 predicted protein.
Column labels are the same as in Table S1. Columns H through Q are log2 fold change expression values from DeSeq. Shell, stage iii shell; ii stage ii clusters; free, stage iii free-living cells; oligo, cells grown planktonically on alginate oligosaccharides; Columns N-Q indicate the set of genes differentially expressed in free-living sample relative to all other samples: YES indicates that gene belongs to this set ‘NO’ indicates it does not.
Column labels are the same as in Table S1.
Cells constitutively express eGFP from plasmid pLL104. Scale bar= 10μm. A timestamp is shown in the video.
Data Availability Statement
Transcriptomic data (raw trimmed reads, count tables) have been deposited in the NCBI GEO repository GSE190325 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Raw data reported in this paper will be shared by the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial strains | ||
| Vibrio splendidus | 76 | 12B01 |
| Escherichia coli PIR1 | Invitrogen, Waltham, MA, USA | Cat#C101010 |
| V. splendidus carrying a mKate-expressing, chloramphenicol resistant replicative plasmid pLL103 | This paper | 12B01 + pLL103 |
| Escherichia coli PIR1 carrying pLL103 | This paper | PIR1 + pLL103 |
| V. splendidus carrying an eGFP-expressing, chloramphenicol resistant replicative plasmid pLL104 | This paper | 12B01 + pLL104 |
| Escherichia coli PIR1 carrying pLL104 | This paper | PIR1 + pLL104 |
| V. splendidus carrying transcriptional reporter plasmid pJAS2020.1 | This paper | 12B01 + pJAS2020.1 |
| Escherichia coli PIR1 carrying pJAS2020.1 | This paper | PIR1 + pJAS2020.1 |
| Escherichia coli DH5-alpha lambda pir carrying conjugative plasmid pEVS104 | 68 | DH5-alpha lambda pir + pEVS104 |
| Chemicals, peptides, and recombinant proteins | ||
| KpnI-HF | NEB, Ipswich, MA, USA | Cat#R3142 |
| SphI | NEB, Ipswich, MA, USA | Cat#R3182 |
| Chloramphenicol | Sigma-Aldrich St. Louis, MO, USA | Cat#C0378 |
| Marine Broth | BD, Franklin Lakes, NJ, USA | Difco Cat# 2216 |
| Low viscosity alginate | Sigma-Aldrich St. Louis, MO, USA | Cat#A1112 |
| Alginate lyase | Sigma-Aldrich St. Louis, MO, USA | Cat#A1603 |
| Ammonium chloride | Sigma-Aldrich St. Louis, MO, USA | Cat#213330 |
| Nile Red | Sigma-Aldrich St. Louis, MO, USA | Cat#72485 |
| Propidium Iodide | Invitrogen Waltham, MA, USA | Cat#P1304MP |
| Toto-3 | Invitrogen Waltham, MA, USA | Cat#T3604 |
| Lysozyme | Sigma-Aldrich St. Louis, MO, USA | Cat#12650-88-3 |
| 15N-labeled ammonium, 99% | Cambridge Isotope Laboratories, Tewksbury, MA, USA | Cat#NLM-467-PK |
| 16% Paraformaldehyde, aqueous, for electron microscopy | Electron Microscopy Sciences, Hatfield, MA, USA | Cat#50-980-487 |
| Technovit 8100 resin | Heraeus Kulzer GmbH | Cat#64709012 |
| Critical commercial assays | ||
| RNeasy kit | Qiagen, Hilden, Germany | Cat#74004 |
| RNA Protect Bacterial Reagent | Qiagen, Hilden, Germany | Cat#76104 |
| Lysing Matrix B | MPBio, Santa Ana, CA, USA | Cat#116911050-CF |
| Turbo RNA free DNAse kit | Ambion, Austin, TX, USA | Cat#AM1907 |
| HS RNA screen tape | Agilent, Santa Clara, CA, USA | Cat#5067- 5579 |
| HS RNA screen tape buffer | Agilent, Santa Clara, CA, USA | Cat#5067- 5580 |
| HS RNA screen tape ladder | Agilent, Santa Clara, CA, USA | Cat#5067- 5581 |
| Ribominus Yeast and Bacteria transcriptome isolation kit | Invitrogen, Waltham, MA, USA | Cat#A47335 |
| TruSeq stranded mRNA-Seq kit | Illumina, San Diego, CA, USA | Cat#20020594 |
| TruSeq indexes for paired end reads | Illumina, San Diego, CA, USA | Cat#PE-121-1003 |
| Quant-iT RNA Assay kit | Invitrogen, Waltham, MA, USA | Cat#Q32884 |
| Deposited data | ||
| 12B01 genome assembly | 77 | NCBI Genome: ASM15276v1 |
| RNASeq data | This paper | GEO: GSE190325 |
| Github repostitory for image analysis code | This paper78 | N/A |
| Oligonucleotides | ||
| Pflp_SphI_F: CTTGCATGCTTACGATGCATAATAATTAGATTAGAAGTACCACAAAAAAG | This paper | N/A |
| Pflp_KpnI_R: TAGGTACCTTGCGTATCAGGCTTTGTTTACCTG | This paper | N/A |
| mKate_R_Seq: CTCTGAGGTGCACTTGA | This paper | N/A |
| 8F: AGAGTTTGATCCTGGCTCAG | 79 | N/A |
| 1492R: GGTTACCTTGTTACGACTT | 79 | N/A |
| pLL104_confirm_F:GGTGACCGGATCCTTACT | This paper | N/A |
| pLL103_confirm_F:TGACCGGATCCTCAACGG | This paper | N/A |
| pLL10X_confirm_R:ACTACGTTTCATAATGGTTATGAACTTT | This paper | N/A |
| Recombinant DNA | ||
| pLL103 | 65 | N/A |
| pLL104 | 65 | N/A |
| pJS2020.1 | This paper | N/A |
| pEVS104 | 68 | N/A |
| Software and algorithms | ||
| Metamorph | Molecular Devices, San Jose, CA, USA | Version 6.2.3.733 |
| ImageJ | 80 | Version 2.1.0/1.53h |
| Trimmomatic | 81 | Version 0.36 |
| FastQC | 82 | Version 0.11.9 |
| bbduk | 73 | Version 38.16 |
| Bowtie2 | 83 | Version 2.3.4.1 |
| SAMTools | 84 | Version 1.9 |
| HTSeq | 85 | Version 0.12.4 |
| DeSeq2 | 86 | Version 1.34.0 |
| R Studio | 87 | Version 1.2.5042 |
| TPM | 74 | N/A |
| Eggnogg mapper | 88,89 | Version 2.1 |
| Look@nanosims | 90 | N/A |
| MATLAB | Mathworks, Natick, MA, USA | Release R2020b |
| Prism | Graphpad, San Diego, CA, USA | Version 9.3.1 |
| Grid/collection ImageJ plugin | 91 | Version 1.2 |
All original code has been deposited and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.