Abstract
The anaerobic metabolism of 3-hydroxybenzoate was studied in the denitrifying bacterium Thauera aromatica. Cells grown with this substrate were adapted to grow with benzoate but not with 4-hydroxybenzoate. Vice versa, 4-hydroxybenzoate-grown cells did not utilize 3-hydroxybenzoate. The first step in 3-hydroxybenzoate metabolism is a coenzyme A (CoA) thioester formation, which is catalyzed by an inducible 3-hydroxybenzoate–CoA ligase. The enzyme was purified and characterized. Further metabolism of 3-hydroxybenzoyl-CoA by cell extract required MgATP and was coupled to the oxidation of 2 mol of reduced viologen dyes per mol of substrate added. Purification of the 3-hydroxybenzoyl-CoA reducing enzyme revealed that this activity was due to benzoyl-CoA reductase, which reduced the 3-hydroxy analogue almost as efficiently as benzoyl-CoA. The further metabolism of the alicyclic dienoyl-CoA product containing the hydroxyl substitution obviously required additional specific enzymes. Comparison of the protein pattern of 3-hydroxybenzoate-grown cells with benzoate-grown cells revealed several 3-hydroxybenzoate-induced proteins; the N-terminal amino acid sequences of four induced proteins were determined and the corresponding genes were identified and sequenced. A cluster of six adjacent genes contained the genes for substrate-induced proteins 1 to 3; this cluster may not yet be complete. Protein 1 is a short-chain alcohol dehydrogenase. Protein 2 is a member of enoyl-CoA hydratase enzymes. Protein 3 was identified as 3-hydroxybenzoate–CoA ligase. Protein 4 is another member of the enoyl-CoA hydratases. In addition, three genes coding for enzymes of β-oxidation were present. The anaerobic 3-hydroxybenzoate metabolism here obviously combines an enzyme (benzoyl-CoA reductase) and electron carrier (ferredoxin) of the general benzoyl-CoA pathway with enzymes specific for the 3-hydroxybenzoate pathway. This raises some questions concerning the regulation of both pathways.
Phenolic compounds comprise a large and diverse group of organic, water-soluble compounds that can serve as growth substrates for microorganisms. In recent years, it has been established that bacteria can make use of these compounds as carbon and energy source both aerobically and under anoxic conditions.
Aerobic metabolism of phenolic compounds requires molecular oxygen and oxygenases for the cleavage of the aromatic ring (for a recent review, see reference 19). Anaerobic metabolism differs in several aspects; most importantly, it is by definition an oxygen-independent process. Phenolic compounds such as phenol or o-cresol are converted to the corresponding hydroxybenzoic acids 4-hydroxybenzoate and 3-methyl-4-hydroxybenzoate by para carboxylation (reviewed in references 18, 21, and 36). Hydroxybenzoic acids are also formed from other aromatic compounds by bacteria; e.g., p-cresol is oxidized to 4-hydroxybenzoate, and m-cresol is oxidized to 3-hydroxybenzoate (7, 33).
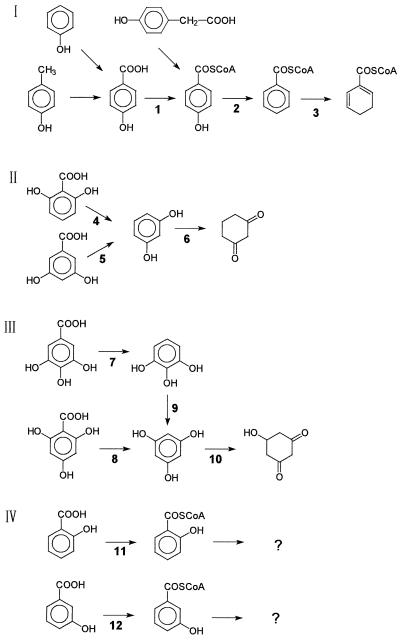
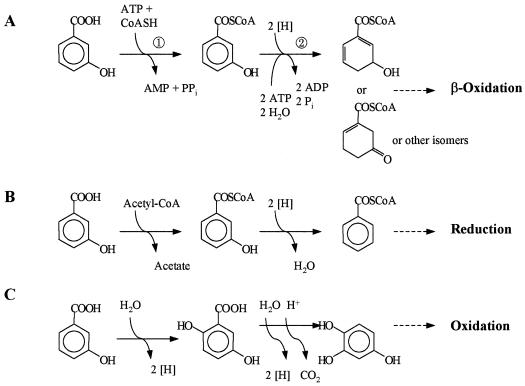
It appears that there are at least four ways to metabolize hydroxybenzoic acids further under anaerobic conditions (Fig. 1). Two directions require coenzyme A (CoA) thioester formation. One way is to reductively eliminate the hydroxyl group(s) yielding benzoyl-CoA. This process has been studied in some detail for 4-hydroxybenzoate metabolism in the bacterium Thauera aromatica; the reductive dehydroxylation of 4-hydroxybenzoyl-CoA is catalyzed by a new molybdenum enzyme (8, 13). 4-Hydroxybenzoyl-CoA reductase (dehydroxylating) is induced in cells grown with phenol, p-cresol, 4-hydroxybenzoate, or 4-hydroxyphenylacetate, which all are metabolized via 4-hydroxybenzoyl-CoA (20). The enzyme is specific for 4-hydroxybenzoyl-CoA and is inactive with 3-hydroxybenzoyl-CoA (13). Similarly, 4-hydroxybenzoate-CoA ligase (EC 6.2.1.27) is specific for 4-hydroxybenzoate and is inactive with 3-hydroxybenzoate (3). The second way is restricted to phenolic acids with hydroxyl groups in the meta position with respect to each other (36). 2,6-Dihydroxybenzoate and the 3,5-isomer are decarboxylated by specific enzymes, and the resulting 1,3-dihydroxybenzene (resorcinol) is directly reduced to cyclohexane-1,3-dion in fermenting bacteria (23, 39). Denitrifying bacteria oxidize resorcinol further to hydroxyquinone (32). The third way concerns trihydroxybenzoic acid isomers which, after decarboxylation, yield pyrogallol (1,2,3-trihydroxybenzene) or phloroglucinol (1,3,5-trihydroxybenzene) (36). These compounds are metabolized by fermenting bacteria via phloroglucinol, which is reduced by phloroglucinol reductase (14, 17). The fourth way, exemplified by 2-hydroxybenzoate and 3-hydroxybenzoate, is less clear and requires also CoA thioester formation (6, 7, 11).
FIG. 1.
Initial steps in the anaerobic metabolism of hydroxybenzoic acids in various bacteria. (I) 4-Hydroxybenzoate: 1, 4-Hydroxybenzoate–CoA ligase; 2, 4-hydroxybenzoyl-CoA reductase (dehydroxylating); 3, benzoyl-CoA reductase (dearomatizing). (II) Metabolism of dihydroxybenzoic acids with meta hydroxyl groups (resorcylic acids) via resorcinol in fermenting and denitrifying bacteria: 4 and 5, specific decarboxylases; 6, resorcinol reductase. (III) Metabolism of trihydroxybenzoic acids with meta hydroxyl groups via phloroglucinol: 7 and 8, specific decarboxylases; 9, transhydroxylase; 10, phloroglucinol reductase. (IV) Metabolism of 2- and 3-hydroxybenzoate: 11 and 12, specific CoA ligases. For explanations, see the text.
The present work is an exemplary study of anaerobic hydroxybenzoate metabolism using the denitrifying bacterium T. aromatica and 3-hydroxybenzoate as a model. A CoA ligase acting on 3-hydroxybenzoate was induced when grown on this substrate. Furthermore, a slow oxidation of reduced viologen dyes was observed when 3-hydroxybenzoyl-CoA was added to an in vitro assay. The product of this reaction was expected to be benzoyl-CoA, but this has not been identified. From a chemical point of view, the meta position of the phenolic hydroxyl group of 3-hydroxybenzoate makes this substrate different from the corresponding para- or ortho-substituted phenolic acids, suggesting possibly different enzymic solutions for its metabolism.
MATERIALS AND METHODS
Materials and bacterial strains.
Chemicals were obtained from Sigma-Aldrich (Deisenhofen, Germany), Merck (Darmstadt, Germany), Roth (Karlsruhe, Germany), or Pierce (Beijenland, The Netherlands); biochemicals were from Roche (Mannheim, Germany) or Gerbu (Craiberg, Germany). [phenyl-14C]benzoate was obtained from MSD Isotopes (Montreal, Quebec, Canada), and [phenyl-14C]3-hydroxybenzoate was from Biotrend (Cologne, Germany). Materials and equipment for solid-phase extraction and fast-performance liquid chromatography (FPLC) were obtained from ICT (Bad Homburg, Germany), Amersham Pharmacia Biotech (Freiburg, Germany), or Bio-Rad (Munich, Germany). High-pressure liquid chromatography (HPLC) equipment came from Waters (Eschborn, Germany), Merck, Perseptive (Freiburg, Germany), Grom (Herrenberg-Kayh, Germany), and Raytest (Straubenhardt, Germany). Enzymes used for cloning experiments were purchased from MBI Fermentas (St.-Leon-Rot, Germany), Pharmacia, and Roche. Amersham provided Hybond-N positively charged membranes used for λ-Zap gene library screening.
T. aromatica strain K172 (DSM 6984) (38) has been deposited in Deutsche Sammlung von Mikroorganismen (Braunschweig, Germany). Escherichia coli strains XL1-Blue and XL-OLR from Stratagene (Heidelberg, Germany) were used for phage screening.
Growth of bacterial cells and preparation of cell extracts.
T. aromatica was grown anoxically at 28°C in mineral salt medium. Benzoate or 3-hydroxybenzoate and nitrate served as sole sources of cell carbon and energy. The substrates were continuously fed in a molar ratio of 1:3.6 (benzoate-nitrate) or 1:3.5 (3-hydroxybenzoate–nitrate) from a concentrated stock solution at pH 7.4, e.g., containing 0.5 M benzoate and 1.8 M KNO3. Cultivation, cell harvesting, storage, and preparation of cell extracts were as described earlier (38).
Simultaneous adaptation experiments.
The simultaneous adaptation experiments were carried out under anaerobic conditions. Frozen cells (0.7 g, wet mass), grown anaerobically with benzoate or 3-hydroxybenzoate and nitrate, were suspended in mineral salts medium without growth substrate. After the cells were washed three times with mineral salts medium, the pellet was suspended in 25 ml of medium. The final cell density of the cell suspension corresponded to a ΔA578 (d = 1 cm) of 13. Aliquots (10 ml) of each suspension were dispensed anaerobically into Hungate tubes, which were closed with rubber stoppers. The addition of 10 mM nitrate and 1 mM benzoate or 3-hydroxybenzoate started the reaction after the temperature was adjusted to 30°C. Each cell suspension was tested for the degradation of both benzoic acid and 3-hydroxybenzoic acid. After different incubation periods, 0.5-ml samples were withdrawn, cooled on ice, and centrifuged at 10,000 × g (4°C). The supernatant was acidified to pH 2 by the addition of 40 μl of 1 M hydrochloric acid. The UV absorption spectrum of the supernatant was recorded between 220 and 350 nm. Detection of benzoic acid was done at 273 nm (ɛ = 0.97 mM−1 cm−1) and of 3-hydroxybenzoic acid at 294 nm (ɛ = 2.55 mM−1 cm−1).
Synthesis, purification, and HPLC analysis of CoA-thioesters. (i) Benzoyl-CoA.
Benzoyl-CoA was synthesized from CoA and benzoic acid anhydride under anaerobic conditions according to the general method of Schachter and Taggart (35). CoA (200 μmol) was dissolved in 20 ml of 0.1 M sodium bicarbonate (pH 8). After the addition of 500 μmol of benzoic acid anhydride, the reaction mixture was held for 4 h at room temperature. During this time, the pH value was controlled, and portions (100 to 500 μl) of 0.1 M sodium bicarbonate (pH 8) were added to maintain a pH of 8. The formation of the thioester was tested with the nitroprusside assay for CoA (37). After completion of the reaction, the CoA-thioester solution was acidified to pH 3.5 by the addition of ∼4 ml of 2 M HCOOH. Contaminant material was extracted three times with diethyl ether (50 ml each time). After freeze-drying, the reaction mixture was dissolved in 5 ml of 20 mM ammonium formate buffer (pH 3.5) containing 2% (by volume) methanol (solvent 1). For further purification, the sample was applied to a solid-phase extraction column (ICT; end-capped C18 material, 10 g; reservoir volume, 60 ml; flow rate, 0.5 ml min−1), which had been equilibrated with the same solvent (20°C). After the column was washed with 120 ml of solvent 1, benzoyl-CoA was eluted with 80% (by volume) aqueous methanol. After evaporation of methanol under vacuum at 30°C, the solution containing benzoyl-CoA was freeze-dried and stored at −20°C. The purity and amount were analyzed by comparison of the UV spectrum with published data (benzoyl-CoA, ɛ261 = 21,100 M−1 cm−1) (40). The yield of CoA-thioester synthesis and purification was 70 to 80%.
(ii) 3-Hydroxybenzoyl-CoA.
3-Hydroxybenzoyl-CoA was synthesized according to the method of Gross and Zenk (16) via the esterification of the corresponding free acid (5 mmol) with N-hydroxysuccinimide (5 mmol) in 25 ml of dried dioxane by adding dicyclohexylcarbodiimide (5 mmol, dissolved in 5 ml of dried dioxane). The insoluble reaction product was removed by filtration, and the filtrate containing the succinimidyl ester was evaporated at 35°C under reduced pressure. The transesterification of the succinimidyl ester to the CoA-thioester was performed under the same conditions as described above for unlabeled benzoyl-CoA using 200 μmol of CoA and 400 μmol of ester. The yield of CoA-thioester synthesis and purification was approximately 70%.
(iii) [phenyl-14C]benzoyl-CoA and [phenyl-14C]3-hydroxybenzoyl-CoA.
14C-labeled benzoyl-CoA and 3-hydroxybenzoyl-CoA were enzymatically synthesized from [phenyl-14C]benzoate (specific radioactivity, 4.5 GBq mmol−1) and [phenyl-14C]3-hydroxybenzoate (specific radioactivity, 4.5 GBq mmol−1), respectively, and CoA with enriched benzoate-CoA ligase (EC 6.2.1.25) from T. aromatica (1) or with extract of 3-hydroxybenzoate cells after two-steps precipitation by ammonium sulfate (35 and 65% saturation), respectively. The assay contained 700 μl of 100 mM potassium phosphate (pH 7.4), 5 mM MgCl2, 0.3 mM [phenyl-14C]benzoate or [phenyl-14C]3-hydroxybenzoate, 2 mM ATP, 1 mM CoA, 0.5 mM NADH, 2 mM phosphoenolpyruvate, 8 nkat of myokinase, 8 nkat of pyruvate kinase, and 20 nkat of lactate dehydrogenase. Addition of 1 nkat of enriched ligase started the reaction. The formation of the CoA-thioester was controlled by HPLC using an analytic RP-C18 column (Grom; Grom-Sil 120 ODS-4 HE, 5 μm; 120 by 4 mm). A linear 2 to 15% (by volume) acetonitrile gradient formed from acetonitrile and 50 mM potassium phosphate (pH 6.7) as solvent was used at a flow rate of 1 ml min-1. The effluent was monitored using a flowthrough scintillation counter with a solid scintillator cell. After 40 min of incubation at 37°C, the reaction was stopped by stirring the reaction mixture on ice.
Determination of enzyme activities. (i) Benzoate-CoA ligase activity.
The benzoate-, MgATP-, and CoA-dependent formation of AMP (which parallels benzoyl-CoA formation) was measured in a coupled spectrophotometric assay at 30°C as described earlier (1). The apparent Km value for benzoate was determined at saturating ATP (4 mM) and CoA (2 mM) concentrations, with aromatic substrate concentration varying from 15 to 100 μM.
(ii) 3-Hydroxybenzoate–CoA ligase (EC 6.2.1.-) activity.
The 3-hydroxybenzoate–CoA formation was measured in a coupled spectrophotometric assay at 30°C, in compliance with the benzoate-CoA ligase activity assay as described earlier (1). The apparent Km values for aromatic substrates were determined at saturating ATP (4 mM) and CoA (2 mM) concentrations, with aromatic substrate concentrations varying from 15 to 1,000 μM. The substrate specificity and Vmax was tested using 1 mM aromatic substrate.
(iii) Benzoyl-CoA reductase (EC 1.3.99.15) activity.
Benzoyl-CoA reductase activity was determined as described earlier (4). The continuous spectrophotometric assay followed the benzoyl-CoA- and MgATP-dependent oxidation of reduced methyl viologen. It was performed under strictly anaerobic conditions in stoppered glass cuvettes at 37°C.
(iv) 3-Hydroxybenzoyl-CoA reductase activity.
3-Hydroxybenzoyl-CoA reductase activity in enriched protein fractions was routinely measured as described above for benzoyl-CoA reductase activity. The 3-hydroxybenzoyl-CoA- and MgATP-dependent oxidation of reduced methyl viologen was determined in a continuous spectrophotometric assay at 730 nm (ɛ730 = 2,400 M-1 cm-1). It was performed under strictly anaerobic conditions in stoppered glass cuvettes at 37°C. Because of the high rate of methyl viologen oxidation in the absence of substrate, this test could only be used after passing the cell extract over a DEAE-Sepharose anion-exchange column (see below). The 0.5-ml standard assay mixture contained 150 mM morpholinopropanesulfonic acid (MOPS)-KOH (pH 7.3), 10 mM MgCl2, 5 mM ATP, 1 mM methyl viologen, 0.5 mM 3-hydroxybenzoyl-CoA, and 10 to 50 μl of enzyme solution. Before the addition of enzyme, methyl viologen was reduced with dithionite to an A730 value of approximately 1.4, corresponding to a concentration of reduced methyl viologen of about 0.6 mM. Adding either the substrate 3-hydroxybenzoyl-CoA or alternatively ATP started the reduction reaction.
The examination of 3-hydroxybenzoyl-CoA reductase activity in the cell extract was performed in a radioactive assay at 37°C under strictly anaerobic conditions. In brief, the consumption of [phenyl-14C]3-hydroxybenzoyl-CoA and the formation of a radioactive product were monitored. The radioactive 0.3-ml standard assay mixture contained 150 mM MOPS-KOH (pH 7.3), 10 mM MgCl2, 2.5 mM [phenyl-14C]3-hydroxybenzoyl-CoA (15 kBq), 8 mM titanium(III) citrate, 6 mM ATP, 8 mM phosphoenolpyruvate, 20 nkat of pyruvate kinase, and 40 μl of supernatant (100,000 × g) of cell extract. For HPLC analysis, samples of 80 μl were retrieved after 1, 2, 4, and 8 min of incubation at 37°C and adjusted to pH 4.0 by adding 15 μl of 2 M formic acid. After centrifugation, the supernatants were applied to an analytical HPLC column (Grom; Grom-Sil 120 ODS-4 HE, 5 μm; 120 by 4 mm) that was equilibrated with 2% (by volume) acetonitrile in 50 mM potassium phosphate (pH 6.7) (20°C). HPLC separation of the reaction mixture was performed with a linear 2 to 15% acetonitrile gradient formed from acetonitrile and 50 mM potassium phosphate (pH 6.7). phenyl-14C-labeled compounds were quantified from the values obtained with a flowthrough radioactivity monitor.
Purification of benzoyl-CoA reductase.
Purification was performed at 4°C under strictly anaerobic conditions in a glove box with N2-H2 (95:5, by volume) as the gas phase. The columns were cooled by an external water bath, and the fraction collector was placed in a small refrigerator. All buffers contained 0.25 mM dithionite and 1 mM dithioerythritol as reducing agents. Preparation of benzoyl-CoA reductase started with extracts from 200 g of cells of T. aromatica (wet mass) grown anaerobically with benzoate and nitrate. The purification procedure was performed in four steps, including anion-exchange chromatography on DEAE-Sepharose and Mono-Q, chromatography on hydroxyapatite, and gel filtration (4).
Purification of 3-hydroxybenzoyl-CoA reducing enzyme.
All of the following steps were performed at 4°C under anaerobic conditions. All buffers contained 0.25 mM dithionite plus 1 mM dithioerythritol. The extract from 15 g of cells (wet mass), which were grown anaerobically with 3-hydroxybenzoate and nitrate, was prepared as described earlier (4).
(i) DEAE-Sepharose chromatography.
The 100,000 × g supernatant of cell extract (32 ml) was applied to a DEAE-Sepharose column (Pharmacia, fast flow; diameter, 3.0 cm; volume, 85 ml) that had been equilibrated with 20 mM triethanolamine hydrochloride/KOH (pH 7.8)–10% (by volume) glycerol (referred to as buffer A) at a flow rate of 3 ml min−1. The column was washed with 2 bed volumes of buffer A and with 2 bed volumes of 80 mM KCl in buffer A. 3-Hydroxybenzoyl-CoA reductase activity was eluted with 120 mM KCl in buffer A in volume of 120 ml.
(ii) Hydroxyapatite chromatography.
The combined fractions from DEAE-Sepharose chromatography were applied to a FPLC column (diameter, 16 mm; volume, 20 ml) of Macro-Prep (Bio-Rad; ceramic hydroxyapatite with diameter of 40 μm) that had been equilibrated with buffer A at a flow rate of 2 ml min−1. The column was washed with 2 bed volumes of buffer A and 2 bed volumes of 1 M KCl in buffer A. After re-equilibration with buffer A (1 bed volume), 3-hydroxybenzoyl-CoA reductase activity was eluted with a linear 0 to 30 mM potassium phosphate gradient (5 bed volumes) formed from buffer A and 30 mM potassium phosphate (pH 7.8) containing 10% glycerol. The reductase eluted at 15 to 20 mM potassium phosphate in a 36-ml volume.
Purification of 3-hydroxybenzoate–CoA ligase.
Preparation of 3-hydroxybenzoate–CoA ligase started with extracts from 20 g of cells of T. aromatica (wet mass) grown anaerobically with 3-hydroxybenzoate and nitrate. The following purification steps were performed at 4°C.
(i) DEAE-Sepharose chromatography.
The 110,000 × g supernatant of cell extract (36 ml) was applied to a DEAE-Sepharose column (Pharmacia Fast Flow; diameter, 30 mm; volume, 85 ml) that had been equilibrated with 10 mM Tris-HCl (pH 7.8)–10% (by volume) glycerol–2 mM dithioerythritol (referred to as buffer B) at a flow rate of 3 ml min−1. The column was washed with 1 bed volume of buffer B and with 2 bed volumes of 80 mM KCl in buffer B. 3-Hydroxybenzoate–CoA ligase activity was eluted with 160 mM KCl in buffer B in a volume of 120 ml.
(ii) Q-Sepharose chromatography.
The combined fractions from DEAE-Sepharose chromatography were applied to a Q-Sepharose column (Pharmacia High Performance; diameter, 26 mm; volume, 58 ml) that had been equilibrated with buffer B at a flow rate of 3 ml min−1. The column was washed with 2 bed volumes of 160 mM KCl in buffer B. 3-Hydroxybenzoate–CoA ligase activity was eluted with 220 mM KCl in buffer B in a volume of 75 ml.
(iii) Hydroxyapatite chromatography.
The combined fractions from Q-Sepharose chromatography were applied in two runs to a FPLC column (diameter, 16 mm; volume, 10 ml) of Macro-Prep (Bio-Rad; ceramic hydroxyapatite with a diameter of 40 μm) that had been equilibrated with buffer B at a flow rate of 1 ml min−1. The column was washed with 2 bed volumes of buffer B. 3-Hydroxybenzoate–CoA ligase activity was eluted with a linear 0 to 30 mM potassium phosphate gradient (6 bed volumes) formed from buffer B and 30 mM potassium phosphate (pH 7.8) containing 10% glycerol and 2 mM dithioerythritol. The ligase eluted at 10 to 15 mM potassium phosphate in a 16-ml volume.
(iv) Reactive green chromatography.
The combined fractions from hydroxyapatite chromatography were applied in 2-ml portions to an affinity column (Sigma Reactive green 19; diameter, 5 mm; volume, 0.6 ml) that had been equilibrated with buffer B at a flow rate of 0.2 ml min−1. The column was washed with 1 bed volume of buffer B, 3 bed volumes of 150 mM potassium phosphate (pH 7.8) containing 10% glycerol and 2 mM dithioerythritol, 3 bed volumes of 180 mM KCl in buffer B (referred to as buffer C), 3 bed volumes of 1 mM NAD in buffer C, and 3 bed volumes of 2.5 mM benzoate and 2.5 mM 3-hydroxybenzoate in buffer C. After re-equilibration with 2 bed volumes of buffer C, 3-hydroxybenzoate–CoA ligase activity was eluted with 0.1 mM ATP in buffer C in a volume of 2.1 ml.
HPLC analysis and UV spectra of 3-hydroxybenzoyl-CoA and of the product of reduction.
A special 350-μl assay mixture containing 150 mM MOPS-KOH (pH 7.2), 10 mM MgCl2, 2.5 mM unlabeled or [phenyl-14C]3-hydroxybenzoyl-CoA, 10 mM titanium(III) citrate, 6 mM ATP, 8 mM phosphoenolpyruvate, and 10 nkat of pyruvate kinase was used for HPCL analysis. The reaction was started by the addition of 40 μl of cell extract or of 1 nkat of enriched reductase enzyme fraction obtained after hydroxyapatite chromatography of benzoyl-CoA reductase or of the 3-hydroxybenzoyl-CoA reducing enzyme. After incubation at 37°C, the reaction was stopped by adding 5 μl of 2 M formic acid to a 100-μl reaction mixture, and the denatured protein was centrifuged. After centrifugation, 40-μl samples of supernatant were applied to an analytical HPLC column (Grom; Grom-Sil 120 ODS-4 HE, 5 μm; 120 by 4 mm) which was equilibrated with 2% acetonitrile in 50 mM potassium phosphate (pH 6.7) (20°C). HPLC separation of the reaction mixture was performed with a linear 2 to 15% acetonitrile gradient formed from acetonitrile and 50 mM potassium phosphate (pH 6.7). Substrate and products were monitored using a radioactivity monitoring analyzer with a solid scintillator cell, as well as by a photo diode array detector. Retention times were as follows: product of 3-hydroxybenzoyl-CoA reduction, 20.3 min; 3-hydroxybenzoyl-CoA, 25.8 min.
Electrospray mass spectroscopy of 3-hydroxybenzoyl-CoA and of the product of reduction.
A 100-μl enzyme assay was used that contained 100 mM potassium phosphate (pH 7.4), 10 mM MgCl2, 3 mM [phenyl-14C]3-hydroxybenzoyl-CoA (or [phenyl-14C]benzoyl-CoA) (10 kBq each), 10 mM titanium(III) citrate, 6 mM ATP, 8 mM phosphoenolpyruvate, and 10 nkat of pyruvate kinase. The reaction was started by the addition of 0.1 nkat of enriched benzoyl-CoA reductase, either obtained from cells grown anaerobically on 3-hydroxybenzoate or from cells grown anaerobically on benzoate and nitrate, after hydroxyapatite chromatography. After 20 min of incubation at 37°C, the reaction was stopped by adding 5 μl of 2 M formic acid to the reaction mixture, and the denatured protein was centrifuged. For HPLC separation of substrates and products, 100 μl of the supernatant was applied to an analytical HPLC column (Grom; Grom-Sil 120 ODS-4 HE, 5 μm; 120 by 4 mm), which was equilibrated with 2% acetonitrile in 50 mM potassium phosphate (pH 6.7) (20°C). HPLC separation of the reaction mixture was performed with a linear 2 to 15% acetonitrile gradient formed from acetonitrile and 50 mM potassium phosphate (pH 6.7) at a flow rate of 1 ml min−1. The collected fractions were freeze-dried. The dried powder containing the CoA-thioester was dissolved in 20 μl of 5% acetonitrile (by volume) and 0.1% trifluoroacetic acid (by volume) at pH 2.0. For desalting, 5-μl portions of this sample were applied to an HPLC-RP-C18 column (Vydac; 5 μm; 150 by 8 mm) directly coupled to the mass spectrometer. Substrates and reduction products were eluted by a linear gradient of 0 to 55% acetonitrile in 0.1% trifluoroacetic acid (by volume) at a flow rate of 20 μl min−1, provided by a dual-piston pump 140B (Applied Biosystems). Electrospray mass spectroscopy analysis was performed on a Finnigan TSQ700 mass spectrometer with electrospray interface.
Cloning and screening a phage library.
Standard protocols were used for DNA cloning, transformation, amplification, and purification (2, 34). Preparative purification of plasmid DNA and PCR products was performed according to the instructions included with the Wizard Minicolumn (Promega, Mannheim, Germany). A λ-ZAP Express-gene library derived from the chromosomal DNA of T. aromatica was constructed according to the instructions of the manufacturer (Stratagene, Heidelberg, Germany). Genomic DNA, partially digested with restriction enzyme Sau3AI, was size fractionated via sucrose gradient centrifugation. The fraction containing the 5- to 10-kb fragments was subsequently used for ligation into the BamHI site of the λ-ZAP Express vector. The PCR product used as a probe for screening was labeled with digoxigenin-11-dUTP (Roche, Basel, Switzerland). Recombinant phagemid vectors, pBk-CMV [Ampr lacZ, fl(−), ColE1-ori], were directly sequenced and maintained in Escherichia coli XL-OLR or DH5α.
DNA sequencing and computer searches.
DNA sequences were determined using an Alfexpress sequencer (Amersham Pharmacia Biotech, Freiburg, Germany). Sequenced regions were extended by primer walking. Similar sequences were identified using the BLAST network service at the National Center for Biotechnology Information (Bethesda, Md.) The sequences were deposited in the EMBL Nucleotide Sequence Database under accession no. AJ278289.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 10 to 12% polyacrylamide) was performed as described by Laemmli (24). Protein separation by two-dimensional gel electrophoresis was performed according to the method of O'Farell (28). Proteins were visualized using the Coomassie blue staining technique (41).
Other methods.
Protein was routinely determined by using the BCA protein assay reagent (Pierce). In addition, the method of Bradford (10) using bovine serum albumin as a standard was used. N-terminal amino acid sequences were obtained by gas- and liquid-phase sequencing with an Applied Biosystems 473A sequencer, as described earlier (22).
RESULTS
Growth on 3-hydroxybenzoate.
T. aromatica grows not only with 3-hydroxybenzoate and oxygen but also with nitrate in the complete absence of molecular oxygen, producing cell mass, CO2, intermediary nitrite, N2O, and finally N2. The molar growth yield for anaerobic growth with 3-hydroxybenzoate and nitrate was approximately 60 g of cell (dry mass) formed per mol of 3-hydroxybenzoate consumed; the generation time was 6 to 12 h. The variation in generation time was due to fast growth during the nitrate consumption phase (leading to nitrite accumulation) and slower growth during the nitrite reduction phase. These values resulted in a specific substrate consumption rate of 32 to 64 nmol min−1 mg of cell protein−1.
The simultaneous adaptation of cells grown anaerobically on benzoate and nitrate for the metabolism of 3-hydroxybenzoate was investigated with dense suspensions of whole cells. Similarly, the utilization of benzoate by cells grown on 3-hydroxybenzoate and nitrate was studied. All cells were precultivated several times on either benzoate or 3-hydroxybenzoate and nitrate. The conversion rate of these aromatic acids by preadapted, suspended cells corresponded to the substrate consumption rate of growing cultures and amounted to 1 mM substrate consumed per 10 to 20 min by a cell suspension with a ΔA578 of 13.
Cells grown on 3-hydroxybenzoate immediately utilized benzoate and 3-hydroxybenzoate (1 mM each) completely within 20 min, with similar initial rates without a lag phase, and were therefore simultaneously adapted to metabolize benzoate. In contrast, cells grown on benzoate degraded benzoate within 10 min but consumed little 3-hydroxybenzoate within 60 min of incubation (<10%) (not shown). Cells grown with benzoate, therefore, seem to have only a poor capability, if any, to metabolize 3-hydroxybenzoate, and the full induction appears to take more than 60 min.
CoA ligase activities in extracts of cells grown with different aromatic substrates.
CoA ligase activities acting on benzoate, 3-hydroxybenzoate, and 4-hydroxybenzoate were comparatively investigated in cells grown with benzoate, 3-hydroxybenzoate, and 4-hydroxybenzoate (Table 1). 3-Hydroxybenzoate–CoA ligase activity was present only in cells grown with 3-hydroxybenzoate, whereas 4-hydroxybenzoate–CoA ligase activity was present in 3-hydroxybenzoate- and 4-hydroxybenzoate-grown cells. Both activities were missing in benzoate-grown cells. The product of ATP cleavage was AMP, and the reaction catalyzed was therefore as follows: 3-hydroxybenzoate + MgATP + CoASH → 3-hydroxybenzoyl-SCoA + MgAMP + PPi.
TABLE 1.
Activities of CoA ligases (AMP-forming) acting on various aromatic acids in cell extracts of T. aromatica after anaerobic growth with different substratesa
| Growth substrate | CoA ligase activity (nmol min−1 mg of protein−1) on:
|
||
|---|---|---|---|
| Benzoate | 3-Hydroxybenzoate | 4-Hydroxybenzoate | |
| Benzoate | 115 | <5 | <1 |
| 3-Hydroxybenzoate | 60 | 62 | 56 |
| 4-Hydroxybenzoate | 70 | 6 | 51 |
All extracts were precipitated by ammonium sulfate (65% saturation) to decrease nonspecific background activity. The ligase activity was determined in a coupled spectrophotometric assay at 30°C at pH 7.8 as described previously (1).
Since 4-hydroxybenzoate-CoA ligase is inactive with 3-hydroxybenzoate as substrate (3), the presence of 3-hydroxybenzoate–CoA ligase in 3-hydroxybenzoate-grown cells suggests the induction of an additional ligase acting on 3-hydroxybenzoate. The presence of 4-hydroxybenzoate–CoA ligase activity in 3-hydroxybenzoate-grown cells suggests that either the 3-hydroxybenzoate activating enzyme is unspecific and acts also on 4-hydroxybenzoate or 4-hydroxybenzoate–CoA ligase is unspecifically regulated and induced by 3-hydroxybenzoate as well. All cells contained benzoate-CoA ligase activity. Due to unknown inhibitory substances and high endogenous NADH oxidation these activities could only be measured in a coupled spectrophotometric assay, e.g., when the cell extract was precipitated by ammonium sulfate.
Purification, characterization, and N-terminal amino acid sequence of 3-hydroxybenzoate–CoA ligase.
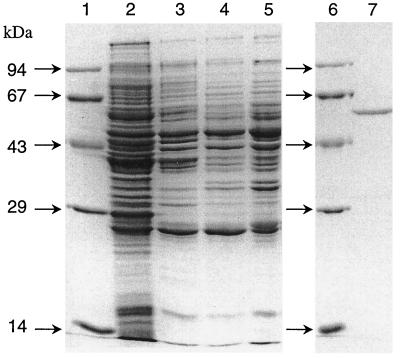
3-Hydroxybenzoate–CoA ligase was purified from 20 g of fresh cell mass with a 24% yield. The purification protocol is given in Table 2. The colorless enzyme had a specific activity of 4.1 μmol min−1 mg−1. A single band corresponding to a molecular mass of 60 kDa was observed in SDS-PAGE (Fig. 2), and a single symmetrical protein peak corresponding to the same mass was noted during gel filtration, suggesting a monomeric composition of the native enzyme (not shown).
TABLE 2.
Purification of 3-hydroxybenzoate–CoA ligase from extracts of T. aromatica grown anaerobically with 3-hydroxybenzoate and nitratea
| Purification step | Protein amt (mg) | Activity (μmol min−1) | Sp act (nmol min−1 mg of protein−1) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Cell extractb | 1,980 | 68 | 34 | 1.0 | 100 |
| DEAE-Sepharose | 523 | 65 | 124 | 3.6 | 96 |
| Q-Sepharose | 265 | 47 | 175 | 5.1 | 68 |
| Hydroxyapatite | 193 | 43 | 222 | 6.5 | 63 |
| Reactive green | 4 | 16.5 | 4,120 | 120 | 24 |
The enzyme activity was determined in a coupled spectrophotometric assay at 30°C at pH 7.8 as described in Materials and Methods.
110,000 × g supernatant.
FIG. 2.
SDS-PAGE analysis of different 3-hydroxybenzoate–CoA ligase fractions obtained after different purification steps. Lanes: 1, molecular mass standard proteins; 2, cell extract; 3, enzyme fraction after DEAE-Sepharose chromatography; 4, enzyme fraction after Q-Sepharose chromatography; 5, enzyme fraction after chromatography on hydroxyapatite; 6, molecular mass standard proteins; 7, enzyme fraction after affinity chromatography. Each lane contained 2 to 10 μg of protein except for lane 2 (20 to 25 μg of protein). For details regarding the purification and assay, see Materials and Methods.
Substrate specificity was tested with saturating ATP (4 mM), CoA (2 mM), and aromatic substrate (1 mM) concentrations. The enzyme acted on 3-hydroxybenzoate (100%), 4-hydroxybenzoate (83%), benzoate (8%), 4-aminobenzoate (6%), 3-aminobenzoate (4%), 3-fluorobenzoate (4%), 4-fluorobenzoate (4%), 3-chlorobenzoate (4%), and 4-chlorobenzoate (3%). Reactions with 3,4-dihydroxybenzoate, 2,3-dihydroxybenzoate, and 2-hydroxybenzoate were below the detection limit (<1%). 3-Hydroxybenzoate- and 4-hydroxybenzoate–CoA ligase activities in an extract of 3-hydroxybenzoate-grown cells (see above) therefore may well be explained by the presence of 3-hydroxybenzoate–CoA ligase alone, which nonspecifically acts also on 4-hydroxybenzoate.
The enzyme activity steadily increased with an increasing pH value of from 7 to 9; at a pH of >9 the coupled spectrophotometric assay became limited due to instability of the substrates. Activities at pH values of 7.0 and 8.0 were 20 and 65%, respectively, of the activity observed at pH 9.0. This indicates that the pH optimum of the enzymatic reaction is 9 or higher. The apparent Km value for the substrate 3-hydroxybenzoate was 60 ± 5 μM and for 4-hydroxybenzoate 65 ± 5 μM. The apparent Vmax values were 4.1 μmol min−1 mg−1 for 3-hydroxybenzoate and 3.4 μmol min−1 mg−1 for 4-hydroxybenzoate. The N-terminal amino acid sequence was SEQLQP; the expected N-terminal methionine was missing (see below, 3-hydroxybenzoate-induced protein 3).
Reduction of 3-hydroxybenzoyl-CoA.
Many experiments were carried out to bring about an oxidative transformation of phenyl-14C-labeled 3-hydroxybenzoyl-CoA by using various oxidized coenzymes (NAD, NADP, and duroquinone) and artificial redox dyes. Two kinds of assays were used. First, the transformation of [14C]3-hydroxybenzoyl-CoA to labeled products was analyzed by HPLC and radiodetection. Second, the 3-hydroxybenzoyl-CoA-dependent reduction of coenzymes or various redox dyes was monitored spectrophotometrically. No 3-hydroxybenzoyl-CoA-dependent reduction of such dyes was observed, nor were labeled products detected by HPLC. The only product observed was 3-hydroxybenzoate, which was due to thioesterase activity in cell extracts. Since we could not obtain any evidence for oxidation of 3-hydroxybenzoyl-CoA in vitro (negative results not shown), the reductive conversion of unlabeled and phenyl-14C-labeled 3-hydroxybenzoyl-CoA was studied using different assays under anaerobic conditions with titanium(III) citrate as an electron donor. The formation of products was monitored by analytical HPLC using a radioactivity monitoring analyzer. The reduction of [phenyl-14C]benzoyl-CoA was studied in parallel.
3-Hydroxybenzoyl-CoA was converted by cell extracts under anaerobic conditions, provided that MgATP was added and oxygen was excluded; half of the enzyme activity was lost within minutes in air. The obtained product differed from benzoyl-CoA and from the product of benzoyl-CoA reduction, cyclohexa-1,5-diene-1-carbonyl-CoA, as deduced from the different retention times in HPLC. Benzoyl-CoA was converted by cell extract under the same conditions to the known intermediates of the anaerobic benzoyl-CoA pathway (25, 26). In addition, reaction mixtures contained a large fraction of labeled free acids. This may be due to a high thioesterase activity in 3-hydroxybenzoate-grown cells, which hydrolyzes the CoA thioester substrate and products. Interestingly, the reductive transformation of 3-hydroxybenzoyl-CoA to the unknown product was not only catalyzed by extracts of 3-hydroxybenzoate-grown cells but at least equally well also with benzoate-grown cells. Benzoate-grown cells contained much lower thioesterase activity which, in case of 3-hydroxybenzoate-grown cells, rapidly destroyed the substrate.
The electron stoichiometry of the reduction reaction was determined experimentally in spectrophotometric assays with enriched fractions containing the 3-hydroxybenzoyl-CoA reducing enzyme (the fraction after chromatography on hydroxyapatite) using reduced methyl viologen as electron donor; 2.4 mol of electrons were transferred per mol of 3-hydroxybenzoyl-CoA added.
Purification of 3-hydroxybenzoyl-CoA reducing enzyme and identification as benzoyl-CoA reductase.
The 3-hydroxybenzoyl-CoA reducing enzyme was purified under strictly anaerobic reducing condition from 3-hydroxybenzoate-grown cells. All enzyme activity was contained in the 110,000 × g supernatant. The two subsequent anaerobic purification steps comprised chromatography on DEAE and chromatography on hydroxyapatite. Consistently, 3-hydroxybenzoyl-CoA reducing activity coeluted with benzoyl-CoA reductase activity in a brownish-greenish protein band throughout the purification. The purified enzyme reduced 3-hydroxybenzoyl-CoA with 90 to 95% of the activity observed with benzoyl-CoA (100%). Both activities were strictly MgATP-dependent and oxygen sensitive, with a half-life of a few minutes. Since the spectrophotometric assay (using reduced methyl viologen as electron donor) could not be applied to cell extract due to unspecific oxidation reactions, a five- to sixfold enrichment was assumed for the first DEAE chromatography step, implying no loss of activity in this step. The purification protocol is given in Table 3. A 16-fold enrichment was achieved, and the yield was 66%. The specific activity was 71 nmol of 3-hydroxybenzoyl-CoA reduced min−1 mg of protein−1. The apparent Km value determined at a 5 mM concentration of ATP was 20 to 30 μM for 3-hydroxybenzoyl-CoA and 15 to 20 μM for benzoyl-CoA. Benzoyl-CoA reductase prepared from benzoate-grown cells was reinvestigated; the catalytic and molecular properties of the enzyme were undistinguishable from those of the 3-hydroxybenzoyl-CoA reducing enzyme, including the effective reduction of 3-hydroxybenzoyl-CoA, similar oxygen sensitivity, subunit composition, and molecular mass (see below). In former experiments, the 3-hydroxybenzoyl-CoA used in the assay of benzoyl-CoA reductase may possibly have been partly hydrolyzed resulting in low 3-hydroxybenzoyl-CoA reduction (20).
TABLE 3.
Purification of 3-hydroxybenzoyl-CoA reducing enzyme from extracts of T. aromatica grown anaerobically with 3-hydroxybenzoate and nitratea
| Purification step | Protein (mg) | Activity (μmol min−1) | Sp act (nmol min−1 mg of protein−1) | Purification (fold) | Yieldc (%) |
|---|---|---|---|---|---|
| Cell extractb | 1,760 | ||||
| DEAE-Sepharose | 350 | 6.1 | 17 | 5.5c | 100 |
| Hydroxyapatite | 110 | 4.0 | 71 | 16 | 66 |
The reducing enzyme activity was measured in a spectrophotometric assay at 37°C as described in Materials and Methods.
110,000 × g supernatant.
Assuming a 100% yield during DEAE chromatography.
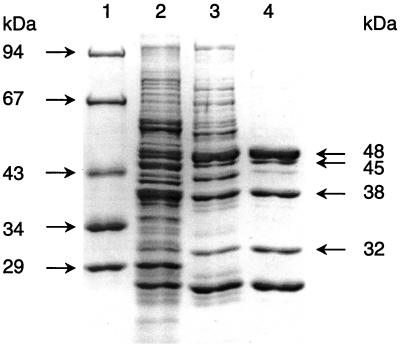
SDS-PAGE of the purified enzyme (after Coomassie blue staining) is shown in Fig. 3. It showed five protein bands a, b, c, d, and e of 48, 45, 38, 32, and 25 kDa, respectively. The molecular masses of proteins a, b, c, and d correlated with the four subunits α, β, γ, and δ of benzoyl-CoA reductase that was purified and characterized previously (4), indicating that this enzyme metabolized 3-hydroxybenzoyl-CoA. This conclusion was corroborated by showing that the N-terminal amino acid sequence of protein c (10 amino acids sequenced) was identical to the N-terminal amino acid sequence of benzoyl-CoA reductase subunit γ. The N-terminal amino acid sequence of the 25-kDa protein e was identical with one of the four 3-hydroxybenzoate-induced proteins (protein 1), which showed high similarity to the N-terminal amino acid sequences of different alcohol dehydrogenases (see below). This protein band e was missing when the enzyme was purified from benzoate-grown cells. The results indicated that the 3-hydroxybenzoyl-CoA reducing enzyme was identical with benzoyl-CoA reductase and that protein e copurified with benzoyl-CoA reductase in the first chromatographic steps. Obviously, 3-hydroxybenzoyl-CoA was nonspecifically reduced by benzoyl-CoA reductase in a two-electron step under anaerobic conditions. Further features arguing against two closely related isoenzymes, one acting on benzoyl-CoA and the other acting on 3-hydroxybenzoyl-CoA, are discussed below.
FIG. 3.
SDS-PAGE analysis of different enzyme fractions with 3-hydroxybenzoyl-CoA reducing activity obtained after different purification steps. Lanes: 1, molecular mass standard proteins; 2, cell extract; 3, enzyme fraction after DEAE-Sepharose chromatography; 4, enzyme fraction after chromatography on hydroxyapatite. Each lane contained 2 to 10 μg of protein except for lane 2 (20 to 25 μg of protein). For details regarding the purification and assay, see Materials and Methods.
Identification of products formed from 3-hydroxybenzoyl-CoA by purified benzoyl-CoA reductase and by the 3-hydroxybenzoyl-CoA reducing enzyme fraction containing the additional protein e.
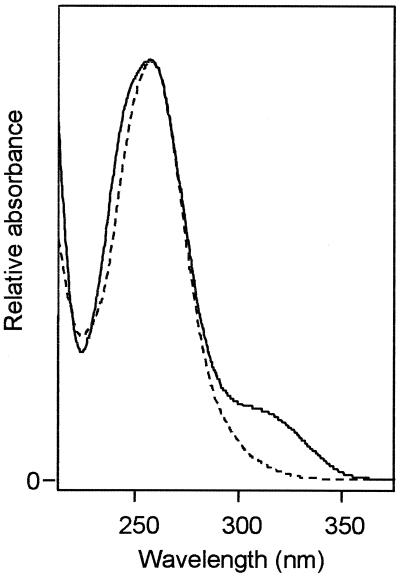
The enzymatic reduction of 3-hydroxybenzoyl-CoA was performed in different assays by purified benzoyl-CoA reductase and by the 3-hydroxybenzoyl-CoA reducing enzyme fraction containing the additional protein e. It should be noted that the benzoyl-CoA reductase preparation contained as an impurity also cyclohex-1,5-diene-1-carbonyl-CoA hydratase, which would convert approximately half of the dienoyl-CoA formed from benzoyl-CoA into 6-hydroxycyclohex-1-ene-1-carbonyl-CoA. With both enzyme preparations, only one product was formed which, according to the spectral properties, contained the CoA moiety. The substrate 3-hydroxybenzoyl-CoA and the obtained reduction product were purified by HPLC and analyzed by electrospray mass spectroscopy. 3-Hydroxybenzoyl-CoA showed a mass peak (mass plus H+) of 888.0 (theoretical value, 888.2); the two-electron reduction product showed a mass peak of 890.2 (theoretical value, 890.2). This latter value is identical to the expected molecular mass of 3-hydroxycyclohex-1,5-diene-1-carbonyl-CoA (which would be the 3-hydroxy analogue of the product of ring reduction of benzoyl-CoA) or an isomeric form of it. The reaction stoichiometry also is in support of a nonaromatic product that differs from the substrate by two additional H atoms. The UV spectra of the substrate 3-hydroxybenzoyl-CoA and the reduction product are shown in Fig. 4.
FIG. 4.
UV spectra of 3-hydroxybenzoyl-CoA (—) and of the product of enzymatic two-electron reduction of this substrate (…).
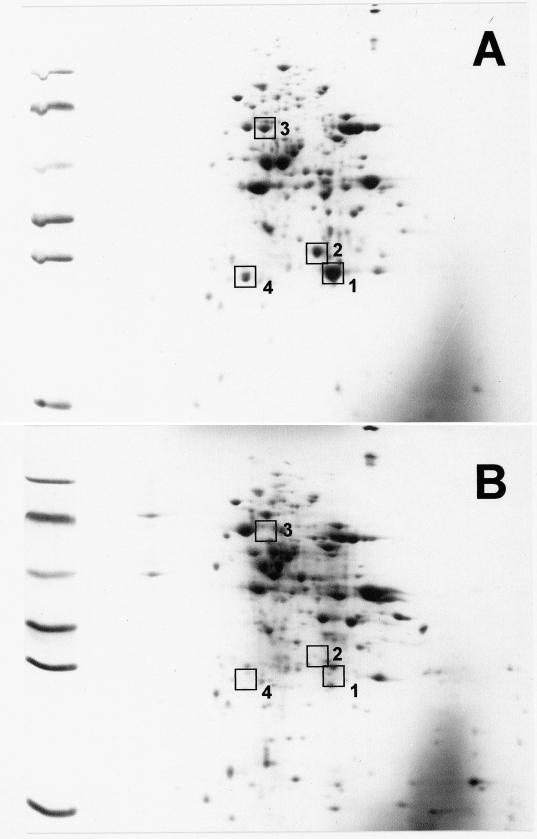
3-Hydroxybenzoate-induced proteins.
The results indicated that 3-hydroxybenzoyl-CoA was unspecifically reduced by benzoyl-CoA reductase. The induction of 3-hydroxybenzoate–CoA ligase during anaerobic growth with 3-hydroxybenzoate and the simultaneous adaptation experiments indicated that the 3-hydroxybenzoate metabolic pathway required additional specific gene products. These are not required for benzoate metabolism, and their synthesis is only induced during growth with 3-hydroxybenzoate. Therefore, we expected that the protein pattern of 3-hydroxybenzoate-grown cells differed from benzoate-grown cells. Comparison of two-dimensional gels of cell extracts revealed at least four proteins that were induced after growth with 3-hydroxybenzoate (Fig. 5); these proteins are considered to function in anaerobic 3-hydroxybenzoate metabolism. Vice versa, there were hardly any benzoate-induced additional proteins detectable. This suggests that the enzymes of anaerobic benzoate metabolism were induced also in 3-hydroxybenzoate-grown cells. This conclusion would be in line with the capacity of 3-hydroxybenzoate-grown cells to simultaneously metabolize benzoate. The N-terminal amino acid sequences of the most prominent 3-hydroxybenzoate-induced proteins were determined as follows: protein 1 (approximate molecular mass, 25 kDa), MRLEG KTAVV TGGAS GIGRA TAETL AAAGA HVVIG DLDQE (this protein was contaminant e of the benzoyl-CoA reductase preparation [see above]); protein 2 (approximate molecular mass, 30 kDa), MYKLK AADwH PEHFK LEVAN RVATI tlnrr drknpl; protein 3 (approximate molecular mass, 60 kDa), xEQLQ PQqxx; and protein 4 (approximate molecular mass, 25 kDa), qIxLN IDGAV AkaxL ERPxV. Lowercase characters denote residues of uncertain identity, “x” indicates unknown amino acids. A comparison with sequences in data banks indicated that protein 1 is likely to be a short-chain alcohol dehydrogenase and that protein 2 is a member of the enoyl-CoA hydratase enzymes.
FIG. 5.
Two-dimensional gel electrophoresis of extracts (150 μg of protein each) of cells grown with nitrate plus 3-hydroxybenzoate (A) and benzoate (B). The pI values of 3-hydroxybenzoate-induced proteins were determined from a two-dimensional gel with an immobilized pH gradient in the first dimension extended over a pH range from 3 (left) to 10 (right). The proteins were visualized by Coomassie blue staining. Proteins that were induced after growth with 3-hydroxybenzoate are marked by squares, and the numbers correspond to the numbers of the sequenced proteins. Molecular mass markers for the second dimension (11.5% polyacrylamide gel) are on the left margins (from top): phosphorylase b (94 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), lactate dehydrogenase (34 kDa), carboanhydrase (29 kDa), and lysozyme (14 kDa).
Cloning and sequencing of genes coding for 3-hydroxybenzoate-induced proteins.
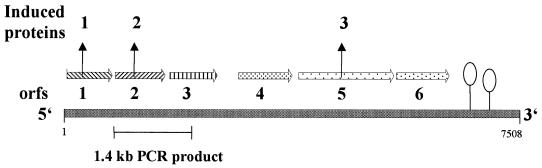
Degenerate oligonucleotides were derived from the N-terminal amino acid sequences of 3-hydroxybenzoate-induced proteins identified by two-dimensional gel electrophoresis. A PCR reaction (30 s at 95°C, 30 s at 50°C, and 30 s at 72°C; 30 cycles) with oligonucleotides derived from the deduced N-terminal amino acid sequences of protein 1 (sequence of the forward primer 30H5, 5′-CAC CCG GAR CAC TTC AAG CT-3′) and protein 2 (sequence of the reverse primer 3OH2, 5′-AGR TGC CCG ATS ACS ACR TG-3′) resulted in amplification of a 1.4-kbp DNA fragment. The sequence of the amplified PCR product confirmed the correctness of the probe as part of a gene coding for a short-chain alcohol dehydrogenase similar to 2-hydroxycyclohexane-1-carbonyl-CoA dehydrogenase from Rhodopseudomonas palustris (19, 31). The fragment contained almost the complete gene for protein 2 and part of an open reading frame (ORF) which codes for a protein whose N-terminal amino acid sequence was similar but not identical with that of protein 1. The-1.4 kbp DNA PCR product was used as a probe for screening a λ-ZAP Express-gene library.
Three positive clones were isolated that contained parts of a putative operon larger than 8 kb coding for proteins involved in the 3-hydroxybenzoate metabolism. The 8-kbp DNA which was double-strand sequenced contained a cluster of six genes lying in the same orientation (Fig. 6). The order of the genes was as follows: The first gene of 0.45 kb codes for a protein which showed high similarity to short-chain alcohol dehydrogenases, with the closest similarity to 2-hydroxycyclohexane-1-carbonyl-CoA dehydrogenase from R. palustris (19, 31). The deduced N-terminal amino acid sequence was identical with protein 1 (∼25 kDa). The second gene of 0.8 kb codes for a protein likely to be an enoyl-CoA hydratase, similar to PhaB from Pseudomonas putida (29). The N-terminal amino acid sequence was identical with protein 2 (∼30 kDa). The third gene of 0.8 kb codes for a protein which showed high sequence similarity with another 3-hydroxyacyl-CoA dehydrogenase (predicted molecular mass, 27.0 kDa). This gene is followed by a gene of 1.1 kb coding for an acyl-CoA dehydrogenase (predicted molecular mass, 42.4 kDa). The fifth gene of 1.6 kb codes for the 3-hydroxybenzoate–CoA ligase. The deduced N-terminal amino acid sequence was identical to the N-terminal sequence of the 3-hydroxybenzoate-induced protein 3 (∼60 kDa) and to that of the purified 3-hydroxybenzoate–CoA ligase. Finally, the cluster contained the gene coding for a putative ring hydrolase (26, 30), followed by stem-loop structures indicating the end of a possible operon. The 5′ end of this putative operon, as well as the gene for protein 4, is not located on the available clones.
FIG. 6.
Organization of genes likely to be involved in anaerobic 3-hydroxybenzoate metabolism in T. aromatica. ORFs 1, 2, and 5 code for 3-hydroxybenzoate-induced proteins 1, 2, and 3. Protein 3 was proven to be 3-hydroxybenzoate–CoA ligase. Downstream of ORF six potential stem-loop structures and no other ORF was found. For explanations, see the text.
DISCUSSION
In the present work the initial steps and the genes probably involved in anaerobic metabolism of 3-hydroxybenzoate were identified in the denitrifying bacterium T. aromatica. The capacity to metabolize 3-hydroxybenzoate is induced only when cells are grown on this substrate. Evidence for an interesting mixed pathway was provided (Fig. 7A). A relatively specific and substrate-induced CoA ligase catalyzes the first committed step, 3-hydroxybenzoyl-CoA formation. This intermediate is reduced by benzoyl-CoA reductase in a two-electron step (4, 5) to a cyclic dienoyl-CoA, a reaction which is coupled to the hydrolysis of 2 ATP. It remains to be shown which isomeric form of the dienoyl-CoA is formed. The genes coding for benzoyl-CoA reductase, for the electron donor ferredoxin, and for three additional enzymes of the benzoyl-CoA pathway form a separate cluster of eight genes (12); the transcriptional control of this putative operon in T. aromatica is not known. The metabolism of 3-hydroxybenzoate requires another gene cluster which is 3-hydroxybenzoate specific. Gene products of both clusters are required to form a functional pathway for 3-hydroxybenzoate.
FIG. 7.
Initial steps in the anaerobic bacterial metabolism of 3-hydroxybenzoate. (A) Denitrifying T. aromatica (this work). Step 1, 3-Hydroxybenzoate–CoA ligase; step 2, benzoyl-CoA reductase (dearomatizing). (B) Fermenting S. hydroxybenzoicum (11, 27). (C) Denitrifying strain BoNHB (33).
3-Hydroxybenzoyl-CoA reducing enzyme.
In a previous study of 4-hydroxybenzoate metabolism (8, 9), an enzyme activity was reported in 3-hydroxybenzoate-grown cells that catalyzed the oxidation of reduced viologen dyes upon addition of 3-hydroxybenzoyl-CoA in the absence of ATP. This finding could not be reproduced. It is likely that the former preparation of 3-hydroxybenzoyl-CoA contained CoA-disulfide which became reduced. In this study we observed an ATP-dependent oxidation of reduced viologen dyes upon addition of 3-hydroxybenzoyl-CoA. Since not even traces of benzoyl-CoA were observed during the reductive transformation of 3-hydroxybenzoyl-CoA, this substrate obviously was not dehydroxylated, as was shown for 4-hydroxybenzoyl-CoA that is reductively converted to benzoyl-CoA. All experimental evidence indicated that benzoyl-CoA reductase is the 3-hydroxybenzoyl-CoA reducing enzyme. (i) The transformation was catalyzed also by extract of benzoate-grown cells. Whole benzoate-grown cells did not metabolize 3-hydroxybenzoate, one reason being the lack of 3-hydroxybenzoate–CoA ligase (see below). (ii) Purified enzyme from benzoate-grown cells also acted on 3-hydroxybenzoyl-CoA forming the same product. In a previous study, the activity of the enzyme with 3-hydroxybenzoyl-CoA was hardly detectable, probably due to a partially hydrolyzed CoA-thioester substrate. (iii) Purification of the 3-hydroxybenzoyl-CoA reducing activity from 3-hydroxybenzoate-grown cells yielded benzoyl-CoA reductase (subunits a to d) (4). The preparation contained an additional, 3-hydroxybenzoate-induced protein e which appears to be an alcohol dehydrogenase. Despite the presence of this impurity, the product formed from 3-hydroxybenzoyl-CoA by the 3-hydroxybenzoyl-CoA reducing enzyme preparation was identical to the one observed with pure benzoyl-CoA reductase. (iv) The subunit size, catalytic properties, oxygen sensitivity, and N-terminal amino acid sequence of subunit c of benzoyl-CoA reductase and 3-hydroxybenzoyl-CoA reductase were indistinguishable.
The presence of closely related isoenzymes for benzoyl-CoA and 3-hydroxybenzoyl-CoA reduction cannot completely be ruled out. However, Southern hybridization with probes derived from benzoyl-CoA reductase genes in previous work revealed hybridizing DNA fragments that correlated to only one operon. Furthermore, various PCR experiments on the benzoyl-CoA reductase operon in previous work yielded only defined amplificates. Mutagenesis of benzoyl-CoA reductase genes and tests for growth of the mutants with both substrates are desirable.
The following reactions transforming the dienoyl-CoA product appear to require additional specific enzymes coded by the cluster of at least six additional genes, including the gene for 3-hydroxybenzoate–CoA ligase. Hence, the overall 3-hydroxybenzoate pathway combines enzyme (benzoyl-CoA reductase) and electron donor (ferredoxin) of the general benzoyl-CoA pathway with enzymes that are specific for the 3-hydroxybenzoate pathway. We hypothesize that glutaryl-CoA is an intermediate, as it is in benzoyl-CoA metabolism; this conclusion derives from the fact that both benzoate-grown and 3-hydroxybenzoate-grown cells contained active glutaryl-CoA dehydrogenase (20). However, all intermediates following 3-hydroxybenzoyl-CoA remain to be identified.
3-Hydroxybenzoate–CoA ligase.
The ligase was strictly 3-hydroxybenzoate induced, although it acted on 3-hydroxybenzoate and 4-hydroxybenzoate almost equally well; this holds true for the apparent Km values and the apparent Vmax values as well. This explains why cells grown with 3-hydroxybenzoate contained both ligase activities in a similar ratio. Obviously, neither 4-hydroxybenzoate–CoA ligase nor 4-hydroxybenzoyl-CoA reductase are induced during growth with 3-hydroxybenzoate (20). Our data show that the ligase belongs to the substrate-induced proteins (protein 3). The enzyme is a new member of the growing family of aromatic acid-CoA ligases which hydrolyze ATP to AMP and PPi, with a postulated intermediary acyl-AMP formation. The alkaline pH optimum is common in this class of enzymes and reflects the requirement of a cysteine thiolate anion in catalysis. 4-Hydroxybenzoate–CoA ligase does not act on 3-hydroxybenzoate, and benzoate-CoA ligase does not act on either of the two hydroxy analogues. This is one reason why cells grown with benzoate or 4-hydroxybenzoate are not adapted to grow on 3-hydroxybenzoate simply because the corresponding ligase is missing. Sequence comparison revealed that the 3-hydroxybenzoate–CoA ligase shows highest similarity with 4-hydroxybenzoate–CoA ligase from R. palustris (15).
3-Hydroxybenzoate-specific enzymes.
The gene cluster coding for 3-hydroxybenzoate-induced proteins contained not only the gene for the CoA ligase but at least five additional ORFs coding for putative proteins. This indicates that the enzymes of the benzoyl-CoA pathway are not sufficient to metabolize further the product of ring reduction. This is another reason why cells grown on benzoate are not adapted to grow with 3-hydroxybenzoate. Obviously, cyclohexa-1,5-diene-1-carbonyl-CoA hydratase, the next enzyme in the benzoyl-CoA pathway in T. aromatica, does not react with the ring reduction product of 3-hydroxybenzoyl-CoA; otherwise, our preparation of benzoyl-CoA reductase containing this hydratase as an impurity should have formed a mixture of the ring reduction product and the product of the following hydration step. However, HPLC analysis revealed only one labeled product derived from [14C]3-hydroxybenzoyl-CoA. This is in contrast to [14C]benzoyl-CoA reduction, where two labeled products were consistently observed: the dienoyl-CoA and the product obtained by water addition by dienoyl-CoA hydratase.
At present one can only speculate on the fate of the ring reduction product which may require the gene products of the additional ORFs. They code for putative hydroxyacyl-CoA dehydrogenase, enoyl-CoA hydratase, another alcohol dehydrogenase, acyl-CoA dehydrogenase, and another member of the enoyl-CoA hydratase family possible catalyzing a hydrolytic ring opening (26, 30). Three of these gene products were indeed identified as 3-hydroxybenzoate-induced proteins. The gene for the fourth 3-hydroxybenzoate-induced protein was not contained on the available clones, indicating that the gene cluster is still incomplete. The set of enzymes may catalyze a β-oxidation-like reaction sequence, finally forming glutaryl-CoA or a more central metabolic intermediate. It remains to be shown that the gene cluster forms an operon and how transcription is controlled. It seems that 3-hydroxybenzoate, 3-hydroxybenzoyl-CoA, or a product derived thereof acts also as inducer of the benzoyl-CoA pathway, in addition to the postulated inducer benzoyl-CoA. The deduced secondary structure following the last ORF could well form a transcription terminating loop structure.
Metabolic diversity.
It has been proposed that the hydroxyl group of 3-hydroxybenzoyl-CoA could be reductively eliminated, yielding benzoyl-CoA, as reported for 4-hydroxybenzoyl-CoA. This would allow further metabolism to acetyl-CoA through the benzoyl-CoA pathway (8). The reductive elimination of the m-hydroxyl group has only been demonstrated so far in a fermenting bacterium, Sporotomaculum hydroxybenzoicum (11, 27) (Fig. 7B). The previous postulate of such a reaction in the denitrifying T. aromatica was based on indirect evidence (9) and could not be confirmed.
A third pathway has been reported in the denitrifying bacterium strain BoNHB (36) (Fig. 7C). This bacterium seems to hydroxylate the substrate para to the phenolic hydroxyl group, yielding 2,5-dihydroxybenzoate (gentisate). Gentisate may be further hydroxylated and decarboxylated, forming hydroxyhydroquinone (1,2,4-trihydroxybenzene). The further fate of hydroxyhydroquinone is not exactly known. It is likely to be oxidized before the ring is opened hydrolytically (36).
These examples show that the anaerobic metabolism of 3-hydroxybenzoate may follow quite different strategies, and this may hold true for many other aromatic substrates which have not been studied in different organisms. The advantage of the T. aromatica variant pathway is obscure; it may simply be difficult to eliminate the m-hydroxyl group, in contrast to the p-hydroxyl group. In case of the fermenting bacterium the reductive elimination seems to be feasible, and the reduction of benzoyl-CoA may be ATP independent (as discussed in references (4, 18, and 36). The advantage of the hydroxyhydroquinone path is not obvious; this pathway requires a positive electron acceptor such as nitrate to afford the proposed anaerobic hydroxylation and oxidation reactions at the aromatic ring.
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
We thank Juliane Alt-Mörbe, Labor für DNA-Analytik, Freiburg, Germany, for DNA sequencing; Patrick Hörth and Wolfgang Haehnel, Freiburg, Germany, for mass spectrometry; and Petra Häussermann, Freiburg, Germany, and Jan Wesche, Greifswald, Germany, for help in the purification of 3-hydroxybenzoate–CoA ligase. We are especially grateful to Johann Heider for helpful suggestions and discussions.
REFERENCES
- 1.Altenschmidt U, Oswald B, Fuchs G. Purification and characterization of benzoate-coenzyme A ligase and 2-aminobenzoate-coenzyme A ligase from a denitrifying Pseudomonas sp. J Bacteriol. 1991;137:5494–5501. doi: 10.1128/jb.173.17.5494-5501.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 3.Biegert T, Altenschmidt U, Eckerskorn C, Fuchs G. Enzymes of anaerobic metabolism of phenolic compounds. 4-Hydroxybenzoate-CoA ligase from a denitrifying Pseudomonas species. Eur J Biochem. 1993;213:555–561. doi: 10.1111/j.1432-1033.1993.tb17794.x. [DOI] [PubMed] [Google Scholar]
- 4.Boll M, Fuchs G. Benzoyl-coenzyme A reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. ATP dependence of the reaction, purification and some properties of the enzyme from Thauera aromatica strain K-172. Eur J Biochem. 1995;234:921–933. doi: 10.1111/j.1432-1033.1995.921_a.x. [DOI] [PubMed] [Google Scholar]
- 5.Boll M, Fuchs G. Benzoyl-coenzyme A reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. Identification and characterization of the natural electron donor ferredoxin and of FAD as a possible prosthetic group. Eur J Biochem. 1998;251:946–958. doi: 10.1046/j.1432-1327.1998.2510946.x. [DOI] [PubMed] [Google Scholar]
- 6.Bonting C F C, Fuchs G. Anaerobic metabolism of 2-hydroxybenzoic acid (salicylic acid) by a denitrifying bacterium. Arch Microbiol. 1996;165:402–408. doi: 10.1007/s002030050344. [DOI] [PubMed] [Google Scholar]
- 7.Bonting C F C, Schneider S, Schmidtberg G, Fuchs G. Anaerobic degradation of m-cresol via methyl oxidation to 3-hydroxybenzoate by a denitrifying bacterium. Arch Microbiol. 1995;164:63–69. [Google Scholar]
- 8.Brackmann R A C, Fuchs G. Enzymes of anaerobic metabolism of phenolic compounds: 4-hydroxybenzoyl-CoA reductase (dehydroxylating) from a denitrifying Pseudomonas species. Eur J Biochem. 1993;213:563–571. doi: 10.1111/j.1432-1033.1993.tb17795.x. [DOI] [PubMed] [Google Scholar]
- 9.Brackmann R A C. 4-Hydroxybenzoyl-CoA Reduktase: Ein Aromaten dehydroxylierendes Enzym und seine Rolle im anaeroben Stoffwechsel phenolischer Verbindungen. Ph. D. dissertation. Ulm, Germany: Universität Ulm; 1994. [Google Scholar]
- 10.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 11.Braumann A, Müller J A, Garcia J-L, Brune A, Schink B. Fermentative degradation of 3-hydroxybenzoate in pure culture by a novel strictly anaerobic bacterium, Sporotomaculum hydroxybenzoicum gen. nov. sp. nov. Int J Syst Bacteriol. 1998;48:215–221. doi: 10.1099/00207713-48-1-215. [DOI] [PubMed] [Google Scholar]
- 12.Breese K, Boll M, Alt-Mörbe J, Schägger H, Fuchs G. Genes coding for the benzoyl-CoA pathway of anaerobic aromatic metabolism in the bacterium Thauera aromatica. Eur J Biochem. 1998;256:148–154. doi: 10.1046/j.1432-1327.1998.2560148.x. [DOI] [PubMed] [Google Scholar]
- 13.Breese K, Fuchs G. 4-Hydroxybenzoyl-CoA reductase (dehydroxylating) from the denitrifying bacterium Thauera aromatica: prosthetic groups, electron donor, and genes of a member of the molybdenum-flavin-iron-sulfur proteins. Eur J Biochem. 1998;251:916–923. doi: 10.1046/j.1432-1327.1998.2510916.x. [DOI] [PubMed] [Google Scholar]
- 14.Brune A, Schink B. Phloroglucinol pathway in the strictly anaerobic Pelobacter acidigallici: fermentations of trihydroxybenzenes to acetate via triacetic acid. Arch Microbiol. 1992;157:417–424. [Google Scholar]
- 15.Gibson J, Dispensa M, Fogg G C, Evans D T, Harwood C S. 4-Hydroxybenzoate-coenzyme A ligase from Rhodopseudomonas palustris: purification, gene sequence, and role in anaerobic degradation. J Bacteriol. 1994;176:634–641. doi: 10.1128/jb.176.3.634-641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gross G G, Zenk M H. Darstellung and Eigenschaften von Coenzym A-Thioestern substituierter Zimtsäuren. Z Naturforsch. 1966;21b:683–690. [Google Scholar]
- 17.Haddock J D, Ferry J G. Purification and properties of phloroglucinol reductase from Eubacterium oxidoreducens G-41. J Biol Chem. 1989;264:4423–4427. [PubMed] [Google Scholar]
- 18.Harwood C S, Burchhardt G, Herrmann H, Fuchs G. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol Rev. 1998;22:439–458. [Google Scholar]
- 19.Harwood C S, Parales R E. The beta-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol. 1996;50:533–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 20.Heider J, Boll M, Breese K, Breinig S, Ebenau-Jehle C, Feil U, Gad'on N, Laempe D, Leuthner B, Mohamed M E-S, Schneider S, Burchhardt G, Fuchs G. Differential induction of enzymes involved in anaerobic metabolism of aromatic compounds in the denitrifying bacterium Thauera aromatica. Arch Microbiol. 1998;170:120–131. doi: 10.1007/s002030050623. [DOI] [PubMed] [Google Scholar]
- 21.Heider J, Fuchs G. Anaerobic metabolism of aromatic compounds. Eur J Biochem. 1997;243:577–596. doi: 10.1111/j.1432-1033.1997.00577.x. [DOI] [PubMed] [Google Scholar]
- 22.Hirsch W, Schägger H, Fuchs G. Phenylglyoxylate: NAD+ oxidoreductase (CoA benzoylating), a new enzyme of anaerobic phenylalanine metabolism. Eur J Biochem. 1998;251:907–915. doi: 10.1046/j.1432-1327.1998.2510907.x. [DOI] [PubMed] [Google Scholar]
- 23.Kluge C, Tschech A, Fuchs G. Anaerobic metabolism of resorcylic acids (m-dihydroxybenzoic acids) and resorcinol (1,3-benzenediol) in a fermenting and in a denitrifying bacterium. Arch Microbiol. 1990;155:68–74. [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Laempe D, Eisenreich W, Bacher A, Fuchs G. Cyclohexa-1,5-diene-1-carboxyl-CoA hydratase, a new enzyme involved in anaerobic metabolism of benzoyl-CoA in the denitrifying bacterium Thauera aromatica. Eur J Biochem. 1998;255:618–627. doi: 10.1046/j.1432-1327.1998.2550618.x. [DOI] [PubMed] [Google Scholar]
- 26.Laempe D, Jahn M, Fuchs G. 6-Hydroxycyclohex-1-ene-1-carbonyl-CoA dehydrogenase and 6-oxocyclohex-1-ene-1-carbonyl-CoA hydrolase, enzymes of the benzoyl-CoA pathway of anaerobic aromatic metabolism in the denitrifying bacterium Thauera aromatica. Eur J Biochem. 1999;263:420–429. doi: 10.1046/j.1432-1327.1999.00504.x. [DOI] [PubMed] [Google Scholar]
- 27.Müller J A, Schink B. Initial steps in the fermentation of 3-hydroxybenzoate by Sporotomaculum hydroxybenzoicum. Arch Microbiol. 2000;173:288–295. doi: 10.1007/s002030000148. [DOI] [PubMed] [Google Scholar]
- 28.O'Farell P H. High-resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1995;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 29.Olivera E R, Minambres B, Garcia B, Muniz C, Moreno M A, Ferrandez A, Diaz E, Garcia J L, Luengo J M. Molecular characterization of the phenylacetic catabolic pathway in Pseudomonas putida: the phenylacetyl-CoA catabolon. Proc Natl Acad Sci USA. 1998;95:6419–6424. doi: 10.1073/pnas.95.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pelletier D A, Harwood C S. 2-Ketocyclohexanecarboxyl coenzyme A hydrolase, the ring cleavage enzyme required for anaerobic benzoate degradation by Rhodopseudomonas palustris. J Bacteriol. 1998;180:2330–2336. doi: 10.1128/jb.180.9.2330-2336.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pelletier D A, Harwood C S. 2-Hydroxycyclohexanecarboxyl coenzyme A dehydrogenase, an enzyme characteristic of the anaerobic benzoate degradation pathway used by Rhodopseudomonas palustris. J Bacteriol. 2000;182:2753–2760. doi: 10.1128/jb.182.10.2753-2760.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Philipp B, Schink B. Evidence of two oxidative reaction steps initiating anaerobic degradation of resorcinol (1,3-dihydroxybenzene) by the denitrifying bacterium Azoarcus anaerobius. J Bacteriol. 1998;180:3644–3649. doi: 10.1128/jb.180.14.3644-3649.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rudolphi A, Tschech A, Fuchs G. Anaerobic degradation of cresols by denitrifying bacteria. Arch Microbiol. 1991;155:238–248. doi: 10.1007/BF00252207. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook T, Fritsch E F, Maniatis J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 35.Schachter D, Taggart J V. Benzoyl coenzyme A and hippurate synthesis. J Biol Chem. 1976;203:925–933. [PubMed] [Google Scholar]
- 36.Schink B, Philipp B, Müller J A. Anaerobic degradation of phenolic compounds. Naturwissenschaften. 2000;87:12–23. doi: 10.1007/s001140050002. [DOI] [PubMed] [Google Scholar]
- 37.Stadtman E R. Preparation and assay of acyl coenzyme A and other thiol esters: use of hydroxylamine. Methods Enzymol. 1957;3:931–941. [Google Scholar]
- 38.Tschech A, Fuchs G. Anaerobic degradation of phenol by pure cultures of newly isolated denitrifying pseudomonas. Arch Microbiol. 1987;148:213–217. doi: 10.1007/BF00414814. [DOI] [PubMed] [Google Scholar]
- 39.Tschech A, Schink B. Fermentative degradation of resorcinol and resorcylic acids. Arch Microbiol. 1986;143:52–59. [Google Scholar]
- 40.Webster L T, Mieyal J J, Siddiqui U A. Benzoyl and hydroxybenzoyl esters of coenzyme A. Ultraviolet characterization and reactions mechanisms. J Biol Chem. 1974;249:2641–2645. [PubMed] [Google Scholar]
- 41.Zehr B D, Savin T J, Hall R E. A one-step, low-background Coomassie staining procedure for polyacrylamide gels. Anal Biochem. 1989;182:157–159. doi: 10.1016/0003-2697(89)90734-3. [DOI] [PubMed] [Google Scholar]