Abstract
We have examined the role of the active-site CXXC central dipeptides of DsbA and DsbC in disulfide bond formation and isomerization in the Escherichia coli periplasm. DsbA active-site mutants with a wide range of redox potentials were expressed either from the trc promoter on a multicopy plasmid or from the endogenous dsbA promoter by integration of the respective alleles into the bacterial chromosome. The dsbA alleles gave significant differences in the yield of active murine urokinase, a protein containing 12 disulfides, including some that significantly enhanced urokinase expression over that allowed by wild-type DsbA. No direct correlation between the in vitro redox potential of dsbA variants and the urokinase yield was observed. These results suggest that the active-site CXXC motif of DsbA can play an important role in determining the folding of multidisulfide proteins, in a way that is independent from DsbA's redox potential. However, under aerobic conditions, there was no significant difference among the DsbA mutants with respect to phenotypes depending on the oxidation of proteins with few disulfide bonds. The effect of active-site mutations in the CXXC motif of DsbC on disulfide isomerization in vivo was also examined. A library of DsbC expression plasmids with the active-site dipeptide randomized was screened for mutants that have increased disulfide isomerization activity. A number of DsbC mutants that showed enhanced expression of a variant of human tissue plasminogen activator as well as mouse urokinase were obtained. These DsbC mutants overwhelmingly contained an aromatic residue at the C-terminal position of the dipeptide, whereas the N-terminal residue was more diverse. Collectively, these data indicate that the active sites of the soluble thiol- disulfide oxidoreductases can be modulated to enhance disulfide isomerization and protein folding in the bacterial periplasmic space.
The formation of stable disulfide bonds in gram-negative bacteria is catalyzed by the Dsb thiol-disulfide oxidoreductase enzymes (30, 31). The oxidation of protein thiols in newly secreted proteins is catalyzed by the periplasmic enzyme DsbA. However, the formation of protein disulfide bonds by DsbA occurs very rapidly and with little regard for the pairing of protein cysteines in the native three-dimensional structure (41). Disulfide bonds between cysteines that are not linked in the native structure must be rearranged, a function that is catalyzed by two homologous, homodimeric enzymes, DsbC and DsbG (8, 43). Both enzymes are maintained in a reduced state in the periplasm through the action of the integral membrane protein, DsbD, which transfers electrons from thioredoxin in the cytoplasm to the active site thiols of DsbC and DsbG in the periplasmic space.
All the soluble Dsb proteins (DsbA, DsbC, and DsbG), as well as one subdomain of the integral membrane protein DsbD, have been shown or predicted to possess a common structural motif, the thioredoxin fold. In the thioredoxin superfamily the catalytic cysteines are located within a Cys-X-X-Cys motif. The two central amino acids in the Cys-X-X-Cys motif strongly influence the intrinsic redox potential of the active-site disulfide bond of thioredoxin family members (12). Holmgren and coworkers initially showed that substituting the dipeptide of Escherichia coli thioredoxin 1 with that found in eukaryotic protein disulfide isomerase (PDI) could alter the redox properties of thioredoxin in the direction of PDI (22, 23). The dipeptide was subsequently found to be important in determining the redox properties of many other members of the thioredoxin family (11, 15, 18, 25, 26).
The identity of the active-site dipeptide in thiol-disulfide oxidoreductases has also been shown to modulate oxidative protein folding in vivo in the eukaryotic endoplasmic reticulum and in the bacterial periplasm (11, 15, 21). Grauschopf et al. constructed a library of DsbA mutants in which the active-site CXXC dipeptide sequence had been randomized (15). This mutant library was screened for the degree of complementation of a dsbA null mutant, using a modification of the screen originally employed in the genetic isolation of the dsbA and dsbB genes (38). Biochemical characterization of these mutants showed that all had lower redox potentials than wild-type DsbA. The pKa of the N-terminal cysteine of the CXXC could be used to accurately predict the oxidizing power of each mutant. However, no clear correlation between the redox potential of the DsbA variants and the degree of complementation of a dsbA null mutant could be discerned. More recently, variants of thioredoxin 1 (TrxA) expressed in the periplasm and containing the active-site dipeptide normally found in DsbA, PDI, or the glutaredoxins (containing, respectively, the sequence -PH-, -GH-, or -PY-) were shown to partially alleviate two phenotypic defects in dsbA mutants (21). Complementation was dependent on the turnover of the more oxidizing TrxA variants by DsbB (13, 21). However, in the above-mentioned studies, the TrxA and DsbA variants were expressed from multicopy plasmids at levels significantly higher than the physiological levels of DsbA, thus making it difficult to discern effects resulting from protein concentration versus intrinsic catalytic properties.
In bacteria, disulfide isomerization represents the rate-limiting step in the expression of complex eukaryotic proteins having multiple disulfide bonds (27, 29). DsbC plays a major role in the folding of multidisulfide proteins in the E. coli periplasm. For example, the formation of active mouse urokinase, which contains 12 disulfide bonds in its native state, is severely impaired in dsbC strains (33). Furthermore, human tissue plasminogen activator having 17 disulfide bonds cannot be produced in active form in E. coli unless DsbC is overexpressed at moderately high levels (29). DsbA does not normally appear to catalyze disulfide bond isomerization in vivo. Rather, it is responsible for the formation of most of the aberrant disulfides in the first place. Recent in vitro studies by Jonda et al. showed that the catalytic proficiency of DsbA results in the formation of incorrect disulfide bonds in hirudin. In contrast, catalysis of disulfide bond formation by TrxA variants with less oxidizing redox potentials than DsbA's resulted in higher yields of correctly folded hirudin (21).
Given the significance of the active-site dipeptide on the redox potential of thioredoxin superfamily members, we sought to examine its effects on disulfide bond isomerization in vivo. Here we present a systematic study on the effect of varying the active-site dipeptide on the effectiveness of disulfide bond isomerization within model protein substrates in the periplasm. We hypothesized that altering the oxidizing power of DsbA and DsbC by mutagenesis might enhance the in vivo folding of eukaryotic proteins that contain multiple disulfides. We have succeeded in identifying several mutants of DsbA and DsbC that significantly enhance the yield of multidisulfide proteins in the periplasm. However, at least for the DsbA variants, the effect on the folding of multidisulfide protein substrates showed no simple correlation to the redox potential of the enzyme.
MATERIALS AND METHODS
E. coli strains and plasmids.
The strains and plasmids used in the present study are listed in Table 1. Plasmids containing the dsbA mutant alleles are derivatives of pUG1 (15). The dsbA coding region was excised by BamHI restriction digestion and cloned into pS1080 at the unique BamHI site. The suicide vector pS1080 contains the conditional R6Kγ origin and ampicillin resistance selectable marker, as well as a counterselectable sacB gene, which confers sucrose sensitivity. Allele exchange was conducted using the protocol of Metcalf et al. (24) as modified by Bass et al. (4). Cointegrates were transferred into the SF100 dsbA::kan strain background and screened for loss of the kanr marker. Following sucrose counterselection, colonies that were ampicillin sensitive and sucrose resistant were screened by PCR using a forward primer complementary to the native dsbA active-site sequence. Those colonies that failed to amplify, presumably due to mismatches, were picked for further analysis. Allele exchange was finally confirmed by amplification of the entire dsbA reading frame followed by DNA sequencing. The degP41::kan allele was then transduced into the dsbA mutant strains using P1vir grown on SF110. The unmarked ΔdegP allele in strain PB402 was generated by PCR amplification of the genomic flanking regions of degP and ligation to create a deletion fragment in pS1080, which was used in allele exchange as described above. Likewise, a strain carrying a complete and unmarked deletion of dsbC (PB351) was constructed in an analogous fashion.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or features | Source or reference(s) |
|---|---|---|
| Strains | ||
| DHB4 | araD139 Δ(araA-leu)7679 Δ(codB-lac)X74 galE15 galK16 rpsL150 relA1 thi ΔphoA (PvuII) phoR ΔmalF3 F′[lac+(lacI) pro] | 9 |
| MC1000 | araD139 Δ(araA-leu)7679 Δ(codB-lac)X74 galE15 galK16 rpsL150 relA1 thi | 35 |
| JCB571 | MC1000 phoR zih12::Tn10 dsbA::kan | 3 |
| KS272 | F− ΔlacX74 galE galK rpsL ΔphoA (PvuII) | 37 |
| SF100 | KS272 Δ(ompT-entF) | 1 |
| SF110 | SF100 degP41 (ΔPstI Kanr) | 1 |
| PB350 | PB402 dsbA (APHA)a | This study |
| PB351 | PB402 ΔdsbC | This study |
| PB401 | SF100 dsbA::kan | This study |
| PB402 | SF100 ΔdegP (Kans) | This study |
| PB406 | SF100 dsbA (CPPC) | This study |
| PB410 | SF100 dsbA (CSFC) | This study |
| PB416 | SF100 dsbA (CPSC) | This study |
| PB419 | SF100 dsbA (CPLC) | This study |
| PB420 | SF110 dsbA (CPPC) | This study |
| PB421 | SF110 dsbA (CLQC) | This study |
| PB422 | SF110 dsbA (CTRC) | This study |
| PB423 | SF110 dsbA (CVLC) | This study |
| PB424 | SF110 dsbA (CSFC) | This study |
| PB425 | SF110 dsbA (CLLC) | This study |
| PB426 | SF110 dsbA (CQAC) | This study |
| PB427 | SF110 dsbA (CFLC) | This study |
| PB428 | SF110 dsbA (CALC) | This study |
| PB429 | SF110 dsbA (CQLC) | This study |
| PB430 | SF110 dsbA (CPSC) | This study |
| PB431 | SF110 dsbA (CSVC) | This study |
| PB432 | SF110 dsbA (CPRC) | This study |
| PB433 | SF110 dsbA (CPLC) | This study |
| Plasmids | ||
| pUG1 | DsbA mutant parent vector, pKK233-2 derivative, Ptrc, ColE1 ori, Ampr | 15 |
| pS1080 | Counter-selectable allele-exchange suicide vector, R6Kγ origin, sacB, Ampr | Steve Bass |
| pRDB8-A | Constitutive secreted expression of mouse urokinase-type plasminogen activator, ColE1 origin, Ampr | 5, 14 |
| puPA184 | Urokinase gene from pRDB8-A cloned into pACYC184 | 8 |
| pTrcdsbC | DsbC in pTrc99A | This study |
| pBADdsbC | DsbC in pBAD33 | 7 |
| pBADdsbC(G119P) | DsbC with active site CPYC in pBAD33 | This study |
| pBADdsbC(C118Stop) | DsbC in pBAD33 with stop codon in active site and PmlI site added | This study |
| pBADdsbC.CXXC | DsbC with randomized active site dipeptide codons in pBAD33 | This study |
| pTrcStIIvtPA | tPA(Δ6-175) with StII leader in pTrc99A | 7 |
| pBAD33 | araBAD promoter, Cmr, p15A origin | 16 |
| pTrc99A | trc promoter, Ampr, ColE1 origin | Amersham Pharmacia Biotech |
| pACYC184 | General cloning vector, p15A origin, Cmr | New England Biolabs, Beverly, Mass. |
Four-letter dipeptide sequence is given in parentheses.
DsbC mutant library construction and screening.
Plasmid pTrcdsbC was constructed as follows: the dsbC gene was amplified from the chromosome with PCR primers dsbC.f and dsbC.b (Table 2). The resulting product was digested with BspHI and HindIII and cloned into pTrc99A (Amersham Pharmacia Biotech) at sites NcoI and HindIII. Plasmid pBADdsbC(C118Stop) was constructed by first amplifying plasmid pTrcdsbC by PCR using primers dsbC(C118Stop).f and dsbC.b. The resulting 390-bp product was used as the reverse primer in a second reaction with the template pBADdsbC and forward primer pBAD.s. The 830-bp product of the second reaction was digested with XbaI and HindIII and cloned into pBADdsbC in place of the dsbC coding region to generate pBADdsbC(C118Stop). This plasmid contains DsbC with a mutation of cysteine codon 118 (TGT) to the opal stop codon (TGA), a mutation of cysteine codon 121 (TGC) to serine (AGC), and introduction of the unique PmlI restriction site (CACGTG) directly 5′ of the active site by silent mutagenesis. Codon numbering is from the ATG start codon of the open reading frame.
TABLE 2.
Oligonucleotide primers used in this study
| Oligonucleotide primer | Sequence |
|---|---|
| dsbC.f | CGTCTAGATCATGAAGAAAGGTTTTATGTTG |
| dsbC.b | ACGGGATCCAAGCTTGAATTATTTACCGCTGGTCA |
| dsbC(G119P).f | TACTGATATCACGTGTCCTTACTGCCACAAACTGCATGAG |
| dsbC(C118Stop).f | ACTGATATCACGTGAGGTTACAGCCACAAACTGCATGAG |
| dsbC(CXXC).f | TACTGATATCACGTGTNNSNNSTGTCACAAACTGCATGAG |
| pBAD.s | TCGCAACTCTCTACTGTTTC |
| rrnBT1T2.s | GGCTGAAAATCTTCTCTC |
For the library construction, part of dsbC was amplified from plasmid pTrcdsbC with the mutagenic primer dsbC(CXXC).f and reverse primer rrnBT1T2.s. The product was digested with PmlI and HindIII and ligated into the vector pBADdsbC(C118Stop) described above, resulting in the creation of pBADdsbC.CXXC.
Approximately 3,000 chloramphenicol-resistant colonies were pooled, and DsbC plasmids were isolated and transformed to the strain DHB4, along with pTrcStIIvtPA expressing a truncated human tissue plasminogen activator, which contains nine disulfide bonds. The cells were inoculated into 200 μl of Luria-Bertani (LB) medium containing 50 μg of carbenicillin per ml and 25 μg of chloramphenicol per ml in the wells of standard 96-well microplates (Costar, Corning, N.Y.). The plates were incubated overnight at 30°C without shaking and with the low-evaporation lid in place. The following day, a 10-μl volume from each well was subcultured into 240 μl of fresh medium in a new plate. The plates were incubated for 5 h at 30°C, at which time the cells were induced by the addition of l-arabinose to a final concentration of 0.2% and of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. After an additional 3 h of incubation at 30°C, the plates were removed and the turbidity at 595 nm was measured. Cells were lysed by transferring 30 μl of the culture to the wells of a plate containing 20 μl per well of cell lysis reagent, BugBuster (Novagen), and shaking the plates for 30 min at room temperature before freezing them at −20°C. The plates were thawed at room temperature, and the human tissue plasminogen activator (tPA) activity of the lysate was measured by adding 200 μl of assay reagent (0.01 μg of human Glu-type plasminogen [American Diagnostica, Greenwich, Conn.] per μl, 0.1 mM Spectrozyme PL [American Diagnostica] in 50 mM Tris-HCl [pH 7.4, at 37°C], 0.01% Tween 80). After incubation at 37°C for 90 min, the absorbance at 405 nm was measured and normalized based on the optical density of the cells at 595 nm as measured prior to lysis. Clones that produced amounts of tPA similar to or greater than the amount produced by the positive control (overexpressed wild-type DsbC) were retrieved from the uninduced overnight plates and streaked onto solid media to isolate single colonies. Plasmid DNA was isolated, and the region encoding the dipeptide was sequenced using the primer pBAD.s. Subsequently, plasmids were retransformed to DH5α, and colonies were selected that were chloramphenicol resistant and carbenicillin sensitive, thus isolating the DsbC plasmid away from the truncated-tPA (vtPA) plasmid. Plasmids were prepared from the resulting colonies and used to retransform competent DHB4/pTrcStIIvtPA cells. The control plasmids expressing wild-type DsbC and truncated DsbC(C118Stop) were also transformed to the fresh background at this time.
The effect of the DsbC mutant alleles was evaluated by growing the cells in 125-ml shake flasks in 10 ml of LB medium with 50 μg of carbenicillin per ml and 25 μg of chloramphenicol per ml at 30°C. Cells were diluted 1:100 from an overnight culture, grown to an optical density at 600 nm (OD600) of 0.8 and arabinose was added to a final concentration of 0.2% to induce DsbC expression. Thirty minutes later, vtPA expression was initiated by the addition of IPTG to 1 mM. After three additional hours of growth, the cells were pelleted and frozen. Cell pellets were resuspended in cold phosphate-buffered saline and lysed with a French press. Following the removal of insoluble material by centrifugation (12,000 × g, 10 min at 4°C), the soluble protein concentration was determined by the Bradford assay (Bio-Rad) using bovine serum albumin (BSA) as standard. For tPA activity assays the lysates were diluted to 0.5 μg/μl in 50 mM Tris-HCl (pH 7.4 at 37°C)–0.01% Tween 80, and 10 μl of this diluted lysate was added to 250 μl of the same buffer containing 0.04 μg of human Glu-type plasminogen per μl and 0.4 mM Spectrozyme PL. To measure tPA activity, samples were incubated at room temperature, and the A405 was monitored as a function of time.
Motility assay.
Motility was assayed on M63 minimal salts soft agar (0.3%) supplemented with 18 amino acids (excluding cysteine and methionine), thiamine, and 0.2% glycerol. Overnight liquid cultures were diluted based on the optical density at 595 nm to normalize for cell number and were then inoculated into the center of a soft agar plate. After 24 h of incubation at 37°C, the diameter of the swarm was measured.
Enzymatic assays.
To test the effect of various dsbA or dsbC variants on the yield of active urokinase, cells were first transformed with plasmid pRDB8-A or puPA184, each of which constitutively expresses secreted murine urokinase-type plasminogen activator. Overnight cultures in LB medium with 50 μg of carbenicillin per ml were diluted 1:100 into 5 ml of the same medium and grown at 30°C in test-tube cultures. In the uninduced DsbA experiments, the cultures were incubated for approximately 5 h, and the cells were pelleted and frozen. For coexpression of DsbC variants, the DsbC expression was induced by arabinose (final concentration, 0.2% [wt/vol]) at an OD595 of 0.8, and the cultures were grown for 3 additional h before being pelleted and frozen. Cell pellets were resuspended in 50 mM Tris-HCl (pH 7.4 at 37°C)–0.01% Tween 80, and lysed with a French press. Following the removal of insoluble material by centrifugation (12,000 × g, 10 min. at 4°C), the soluble protein concentration was determined by the Bradford assay (Bio-Rad) using BSA as the standard. Samples were diluted to 0.25 μg/μl in the buffer described above, and a 50-μl volume was added to a well containing 100 μl of the same buffer with 0.05 μg of human glu-type plasminogen per μl, 0.2 mM Spectrozyme PL, and 3 mM 6-amino-n-hexanoic acid. The plate was incubated at ambient temperature, and the absorbance at 405 nm was monitored.
Alkaline phosphatase (AP) activity was measured essentially as previously described (10) except that 1 mM iodoacetamide was included in all buffers to prevent spontaneous oxidation of reduced AP. Fibrin plate clearance assays were performed as described previously (29), using 10 μg of soluble cell lysate.
RESULTS
Multicopy expression of DsbA active-site mutants.
Grauschopf and coworkers had constructed a library of plasmid-encoded DsbA variants in which the active-site dipeptide sequence was randomized (15). Mutants capable of supporting the oxidation of the cysteines in a MalF–β-galactosidase chimera, thus conferring a lacZ phenotype, were isolated. The presence of the MalF moiety in MalF–β-galactosidase targets the fusion protein to the membrane, and, as a result, a portion of the β-galactosidase portion is exported into the periplasm. In cells expressing inactive or weakly active DsbA, the transmembrane topology of MalF–β-galactosidase is unstable, and, therefore, β-galactosidase can retract into the cytoplasm where it is enzymatically active, giving rise to blue colonies. In contrast, cells expressing DsbA support the formation of disulfide bonds in the periplasmic portion of β-galactosidase which is thus misfolded and inactive, resulting in the formation of white colonies.
Fourteen DsbA mutants that gave a lacZ phenotype were examined here for their ability to restore the phoA+ phenotype of the dsbA-null mutant strain JCB571. These DsbA mutants were encoded in plasmid pUG1 and transcribed from the inducible trc promoter. Cells were grown in minimal media with or without IPTG, harvested in mid-exponential phase, and the AP activity was determined. All 14 DsbA variants conferred AP activity equal to that conferred by DsbA containing the authentic dipeptide (data not shown). The same AP activity was obtained irrespective of whether the synthesis of DsbA was induced by IPTG. The in vivo redox state of the DsbA mutants was analyzed by reacting free thiols with 4-acetamido-4′-maleimidyl-stilbene-2,2′-disulfonic acid (AMS) and resolving the reduced and oxidized forms of the protein electrophoretically (19). When overexpressed, the wild-type DsbA accumulates predominantly in the oxidized state, but a small fraction of reduced protein can also be detected (19). An identical pattern was evidenced for all 14 DsbA active-site mutants examined here (data not shown). The mutants and wild-type DsbA expressed from the same vector accumulate to a comparable level in the periplasm. In all cases DsbC was maintained in the fully reduced state, confirming that the DsbA mutants did not affect the reduction of DsbC by DsbD.
The expression of enzymatically active mouse urokinase in the periplasm of E. coli is strongly dependent on DsbC. In a dsbC mutant strain, active urokinase accumulates at a level almost 100-fold lower than that in wild-type cells (32, 33, 36). The main in vitro function of DsbC is to catalyze the isomerization of nonnative disulfide bonds (43), and this process appears to represent the rate-limiting step in the formation of active urokinase. Therefore, the formation of active urokinase was used to probe the effect of DsbA active-site mutants on disulfide bond isomerization. The degP ompT strain SF110 was used for this study to reduce proteolysis of any unstable folding intermediates of murine urokinase. SF110 was cotransformed with the expression vectors encoding the 14 DsbA variants mentioned above and with puPA184, a pACYC184 derivative encoding constitutively expressed, secreted murine urokinase. Cultures grown in the presence of IPTG, which resulted in high levels of expression of the DsbA mutants, showed no statistically significant differences in urokinase activity, with the exception of the CPPC variant, which consistently gave much lower activity than the wild-type. On the other hand, certain differences in the yield of urokinase were detected in cultures grown without IPTG, where the DsbA mutants were expressed at a basal level due to the leakiness of the trc promoter. Most noticeably, DsbA variants with a CSVC or CPSC active site conferred a level of urokinase activity twofold higher than that conferred by the other mutants and the wild-type DsbA control (data not shown).
Chromosomal integration of DsbA mutants.
To distinguish between the influence of the active-site dipeptide sequence and the effect of the level of DsbA expression, we used allele replacement to transfer the set of 14 dsbA variants discussed above into the chromosome (Table 1). In addition, a mutant strain in which the two cysteines in the active site of the chromosomally encoded dsbA gene had been replaced with alanine (the APHA mutant) was also constructed. Allele replacements were confirmed by PCR amplification and DNA sequencing of the relevant region of the chromosome. In this manner, all the DsbA mutants were transcribed from the native promoter and translation was initiated from the wild-type ribosomal binding site.
In the study by Grauschopf et al., DsbA proteins having different active-site dipeptide sequences were ranked in terms of their oxidative capacity in vivo. The ranking was based on the ability of the DsbA variants to confer a lacZ phenotype on cells expressing MalF–β-galactosidase when grown in the presence of increasing concentrations of the reductant dithiothreitol (DTT). The following ranking of dipeptide sequences in terms of descending oxidative ability in vivo was obtained (the central dipeptides are indicated by the two-letter sequences): PH (wild type), PL, LQ > PR > PS, AL, QL, SV > LT > ST > PP, TR, VL, SF, LL, QA, RC, FL. According to the ranking, the least oxidizing mutants exhibited activities only slightly above background (dsbA) (15). This ranking employed multicopy expression of DsbA from an uninduced trc promoter (15). However, Jonda et al. recently showed that overexpression of DsbA under some conditions can lead to restored periplasmic protein oxidation and cell motility, even in a dsbB mutant where the catalytic cycle of DsbA cannot be completed (21). Therefore, it was of interest to reexamine whether the DsbA mutants, when expressed from the native promoter in the chromosome, could be ranked in a similar manner on the basis of physiologically more relevant phenotypes.
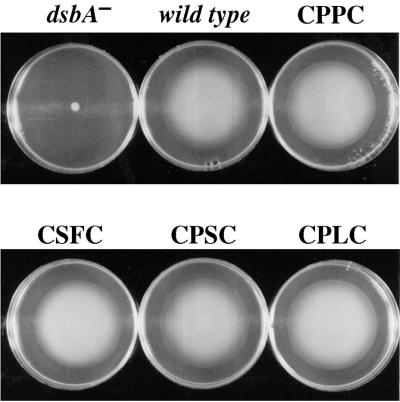
Strains lacking DsbA exhibit slow growth in minimal salts media, both in liquid and on agar plates. However, all the strains carrying active-site dsbA alleles (PB420 to PB433) exhibited growth essentially indistinguishable from that of the corresponding parental strain carrying the wild-type gene (not shown). The motility of the strains carrying the dsbA alleles was tested in minimal media. Representative data are shown in Fig. 1. All of the active-site mutants, regardless of their in vitro redox potentials, displayed motility indistinguishable from that of the strain having the wild-type dsbA allele, whether in a degP+ or degP background. On the other hand, a dsbA mutant and a dsbA(APHA) allele both displayed essentially no motility, as expected.
FIG. 1.
Motility assay for various chromosomal dsbA mutant alleles. The identity of the DsbA active-site dipeptide is indicated. Strains used were PB401 (dsbA-null mutant), SF100 (wild type), PB406 (CPPC dipeptide), PB410 (CSFC dipeptide), PB416 (CPSC dipeptide), and PB419 (CPLC dipeptide).
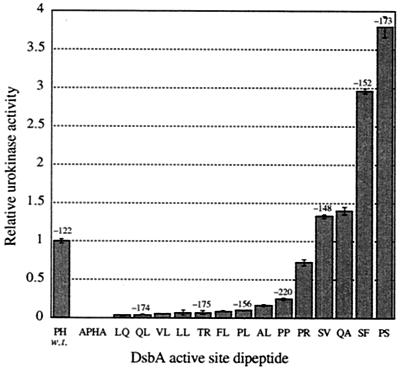
The above-mentioned results demonstrate that dsbA mutants exhibiting redox potentials as low as −220 mV (the PP mutant), compared to −122 mV for the wild-type enzyme, nonetheless appear completely normal with respect to two phenotypes dependent on the oxidation of periplasmic proteins. The steady-state level of a protein having one disulfide bond (β-lactamase) was indistinguishable among the dsbA alleles both in a degP+ background (strains PB420 to PB433) and in a degP background (strains PB401 to 419). In contrast, the formation of active murine urokinase was markedly affected by the sequence of the active-site dipeptide. This was especially evident with the chromosomal dsbA alleles. For the chromosomal dsbA alleles, urokinase activities ranged from negligible to significantly higher than those of the parental strain (Fig. 2). Two of the less-oxidizing mutants, SF (−152 mV) and PS (−173 mV) displayed levels of urokinase activity that were three- to four-fold higher than those displayed by the wild-type strain, PH. However, mutants with similar redox potentials resulted in widely different levels of active urokinase (Fig. 2). Most notably, the QL mutant that has an in vitro redox potential similar to that of PS gave very low activity. The least-oxidizing mutant PP (−220 mV) fell in the middle of the range in terms of the urokinase yield. Likewise, the urokinase activity did not correlate with the pKa of the reactive cysteine of the DsbA (15). Thus, it is clear that while the CXXC motif of DsbA strongly affects the ability of the cell to fold urokinase, it does so in a way that is not directly related to the effects of these mutations on the redox potential.
FIG. 2.
Yields of active mouse urokinase obtained in strains with altered chromosomal dsbA alleles. Strains SF110, PB350, and PB420 to PB433 were grown in rich media as described in Materials and Methods, and the activation of human plasminogen by cell lysates was determined using a coupled assay. Where known, the in vitro standard redox potential of the DsbA variant as calculated from Grauschopf et al. (15) is indicated (in millivolts). Error bars, standard deviations of four experiments.
Effect of active-site dipeptide of DsbC on disulfide isomerization in vivo.
As is the case with DsbA and all other proteins that have a thioredoxin fold, the active site of the bacterial disulfide isomerase, DsbC, contains a CXXC sequence. Presently there is no information on how the active-site dipeptide of DsbC affects the redox potential and the catalytic proficiency of the enzyme. To explore this issue, a library of DsbC mutants was constructed by randomizing the dipeptide sequence. This library was screened for clones that increase the yield of correctly folded heterologous proteins. No physiological substrates are known for DsbC, and null mutants do not exhibit any obvious phenotype. For this reason, the role of DsbC in catalyzing disulfide isomerization in vivo in E. coli was evaluated by its effect on the yield of heterologous proteins having multiple disulfides. These substrates include murine urokinase, bovine pancreatic trypsin inhibitor (BPTI) and human tissue plasminogen activator (29, 33, 44). Bessette et al. recently used a truncated form of tPA (vtPA) that consists of two (protease and kringle 2) of the five subdomains of the full-length protein and has nine disulfide bonds (7). When expressed in the bacterial periplasm, active vtPA accumulates to a very low level; however, cooverexpression of DsbC results in 100-fold higher yields of active protein. We decided to screen the DsbC library for mutants that upon overexpression result in a high yield of active vtPA for the following two reasons. (i) Given that a high intracellular amount of DsbC increases the yield of active vtPA, it is plausible that mutants with enhanced catalytic activity for disulfide bond formation and isomerization will result in even greater yields of active protein. This hypothesis was proven correct (see below). (ii) We have developed a convenient and sensitive assay for the yield of vtPA, thus enabling the screening of the DsbC library in a facile manner.
A library of DsbC mutants encoding a randomized active-site dipeptide sequence was constructed as described in Materials and Methods. Individual clones were grown in microtiter well plates to late exponential phase, the expression of DsbC and vtPA was induced for 3 h, and cells were lysed by a combination of chemical (nonionic detergent) and freeze-thaw lysis. Plasminogen activator activity was measured by a coupled chromogenic assay, and the results were normalized with respect to cell density. About 85% of the mutant clones were inactive in that they displayed no vtPA activity over background (i.e., cells without DsbC overexpression). Clones displaying a level of vtPA activity similar to (at least 90%) or greater than that displayed by the positive control were analyzed further.
Plasmids were isolated, sequenced, and retransformed into fresh cells together with the vtPA expression vector. The cells were grown in shake flask cultures, and the DsbC mutants were ranked qualitatively based on the fibrin plate clearance assay (Table 3). For selected clones the vtPA level was also determined quantitatively, and the ranking was found to be consistent with the fibrin plate assay results (data not shown).
TABLE 3.
DsbC active-site mutants isolated from the librarya
| Amino acid sequence of CXXC central dipeptide | DNA sequence | vtPA activity |
|---|---|---|
| GYb | GGT TAC | + |
| NY | AAC TAC | ++ |
| SF | TCG TTC | ++ |
| SF | TCC TTC | ND |
| TFc | ACC TTC | ++ |
| MF | ATG TTC | ++ |
| GF | GGG TTC | ++ |
| HHc | CAC CAC | ++ |
| VH | GTG CAC | ++ |
| SH | TCC CAC | ++ |
| RF | CGG TTC | +/− |
| RF | CGC TTC | ND |
| FA | TTC GCG | +/− |
| GA | GGC GCC | + |
| MA | ATG GCG | + |
| GI | GGG ATC | + |
| AV | GCG GTG | + |
DsbC mutants isolated from the library were grown in shake flask cultures and the vtPA activity was determined by the fibrin plate assay. ND, not determined. +, weak activity; ++, strong activity; +/−, indefinite activity.
Wild-type sequence.
Isolated twice.
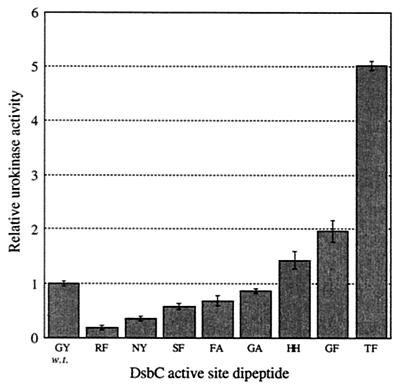
Western blot analysis revealed no differences in DsbC expression levels among various active-site dipeptide mutants tested (6). Finally, the ability of several active mutants to restore urokinase activity in a dsbC genetic background (PB351) was also determined. It should be noted that, unlike vtPA, with murine urokinase, the same yield of active protein was obtained regardless of whether DsbC was expressed at a low level from the chromosomal gene or was transcribed from a strong promoter in a multicopy plasmid. However, several DsbC active-site dipeptide mutants conferred yields of murine urokinase that were substantially higher than those conferred by co-expression of the wild-type enzyme (Fig. 3). The rank-ordering of the DsbC mutants with respect to their effect on the yield of murine urokinase and vtPA was similar for six out of eight mutants (Fig. 3 and Table 3). However, DsbC having an NY or SF active-site dipeptide supported a high yield of vtPA but not murine urokinase.
FIG. 3.
Yields of active mouse urokinase obtained in PB351/pRDB8-A with coexpression of DsbC active-site mutants. Error bars, standard deviations of four experiments.
DISCUSSION
The highly oxidizing state of the bacterial periplasm is maintained primarily through the action of DsbA, which is the strongest oxidant of all known thiol-disulfide oxidoreductases. The redox potentials of the other two periplasmic oxidoreductases, DsbC and DsbG, are quite comparable to that of DsbA (E0′ for DsbC = −129 mV and E0′ for DsbG = −124 mV [compare E0′ for DsbA = −122 mV]) (8, 39, 43). Even though DsbC and DsbG can be oxidized by DsbA in vitro, in the cell they are both maintained completely in a reduced state via the DsbD-TrxA-TrxB pathway and the consumption of NADPH (8, 31, 33).
The redox potential of enzymes belonging to the thioredoxin superfamily is critically dependent on the amino acid identity of the dipeptide within the Cys-X-X-Cys active site (12, 18, 22, 23, 25). A set of DsbA variants having different dipeptide sequences has been previously isolated (15). When expressed in multicopy, and in the presence of a reductant (DTT), these mutants support various levels of oxidation of the β-galactosidase moiety of a MalF–β-galactosidase fusion. Fourteen of these dsbA mutants were transferred into the E. coli chromosome by allele exchange. E. coli dsbA-null mutants are nonmotile, grow poorly in minimal media, and express reduced levels of periplasmic, disulfide-containing proteins such as β-lactamase. However, all the chromosomally encoded dsbA alleles fully complemented these defects to a level comparable to that afforded by the wild-type protein. Even DsbA variants poised at a very low redox potential (e.g., for the CPPC active-site mutant, E0′ = −220 mV) showed motility comparable to that of wild-type E. coli. These results suggest that the very high redox potential of DsbA is not essential for many of the physiological functions of this protein, at least under laboratory conditions. Most likely the catalytic proficiency of DsbA becomes important in environments rich in low-molecular-weight reductants. Along these lines, it should be noted that the DsbA active-site dipeptide mutants analyzed in the present study had been identified genetically on the basis of their ability to oxidize a MalF–β-galactosidase fusion in the presence of increasing concentrations of DTT (15). The more oxidizing DsbA variants could oxidize the β-galactosidase moiety of MalF–β-galactosidase in cells grown with up to 3 mM DTT. Under these conditions, less-oxidizing DsbA did not interfere with the folding of active β-galactosidase, resulting in the formation of blue colonies (15). Although, the β-galactosidase moiety of a MalF–β-galactosidase fusion is not a physiological substrate for DsbA, this result nonetheless demonstrates the importance of the wild-type enzyme in mediating the formation of disulfide bonds in cells grown in the presence of low-molecular-weight reductants.
Recently, Jonda et al. (21) reported that periplasmic targeting of TrxA variants with the CXXC sequence grafted from PDI, DsbA, or the glutaredoxins was able to partially restore the motility (and the lacZ) phenotype in a dsbA strain. These TrxA variants exhibit redox potentials comparable to or higher than that of the least-oxidizing DsbA variant examined in this study. The fact that the DsbA variants supported full complementation, while the TrxA variants did not, highlights the significance of kinetic considerations, in addition to the redox potential, with respect to disulfide bond formation in vivo.
While the DsbA mutants examined in this study had no obvious effect on the oxidation of native periplasmic proteins, they exerted a rather dramatic effect on the folding of a multidisulfide protein, murine urokinase. The formation of active urokinase in E. coli is critically dependent on the presence of both DsbA and DsbC in the periplasm; no urokinase activity is evident in dsbA mutants, and about 1% of the wild-type urokinase level is seen in dsbC mutants (2, 32). All of the DsbA variants having an altered active-site dipeptide increased the expression of functional urokinase above background (dsbA). However, the level of urokinase activity varied widely among the different mutants. The activity increase could not be correlated directly with the redox potential of DsbA. For example, the PS and QL mutants with essentially the same in vitro redox potential (−173 mV) gave urokinase activities differing by more than twenty-fold. Similarly, while the redox potentials of the SF and PL mutants have been experimentally determined as −152 mV and −156 mV, they gave, respectively, three-fold-higher and five-fold-lower active urokinase yields than the wild-type. These differences could be observed because the dsbA alleles were expressed from a chromosomal copy. However, multicopy expression obscured the true effect of the DsbA active-site dipeptide mutants. For example, in contrast to the results presented in Fig. 2, we found that upon multicopy expression, the yields of murine urokinase obtained with the CPLC mutant and wild-type DsbA were virtually indistinguishable.
Our data demonstrate that certain less-oxidizing DsbA mutants, while sufficient for supporting normal cellular processes, are actually more effective than the wild-type for the folding of proteins with several disulfides. The wild-type DsbA is an extremely efficient protein thiol oxidase and therefore causes the formation of aberrant disulfide bonds in proteins such as BPTI and hirudin (three disulfides) in vitro (40, 42). Jonda et al. have shown that the in vitro catalyzed oxidative folding of hirudin using TrxA variants with redox potentials between those of native TrxA (−270 mV) and native DsbA (−122 mV) is more efficient than that afforded by DsbA (21). Similarly, a circularly permuted DsbA variant with a redox potential of −179 mV also showed improved folding of hirudin in vitro (17). Interestingly, the refolding of hirudin with the TrxA variants was five times faster than oxidation by wild-type DsbA. Jonda et al. proposed that the TrxA variants promote the consecutive formation of disulfide bonds which is more efficient than the rapid, essentially random, thiol oxidation by DsbA. In the latter case, extensive disulfide isomerization must take place before the protein can reach the native state (21). However, the situation in vivo is more complicated. When thiol oxidation is slow, newly secreted polypeptide chains remain in a partially folded, less-stable conformation longer and, as a result, they are more vulnerable to degradation by the periplasmic proteolytic machinery (28). A kinetic balance must be reached to attain the maximum yield of multidisulfide proteins: oxidation needs to occur at a rate fast enough to support the formation of proteolytically stable intermediates but not so high that incorrect disulfide bonds prevent correct folding. Evidently, the CPSC and CSFC mutants allow protein oxidation to proceed at an optimal rate, resulting in greater protein yield, at least for the murine urokinase substrate.
In addition to the effect of DsbA mutants, the formation of correctly folded multidisulfide proteins in E. coli is also greatly influenced by mutations in DsbC. A library of DsbC mutants with a randomized CXXC central dipeptide was screened for the ability to support the folding of a complex heterologous protein. The need to use a heterologous substrate was dictated by the fact that dsbC mutants exhibit no obvious phenotypes, and there are no specific physiological substrates that have been identified for this enzyme (32, 34). In contrast to the case of DsbA, where high-level expression can obscure the effect of mutations on disulfide bond isomerization, in the case of DsbC an elevated intracellular level of the enzyme (i.e., by high-level expression from a strong promoter), is essential for the formation of active vtPA in E. coli. For this reason, the library was screened for mutants that upon overexpression confer increased levels of vtPA activity. The majority of the clones analyzed (>85%) were inactive in the vtPA assay, indicating that only certain combinations of amino acids are tolerated within the active site. As can be seen in Table 3, a clear consensus for the sequence of the dipeptide emerged. The C-terminal amino acid in the active-site dipeptide (CXXC) exhibited a strong preference for hydrophobic amino acids and particularly for aromatic amino acids. Thirteen of 18 clones had phenylalanine, tyrosine, or histidine at the C-terminal position. Phenylalanine occurred at a much higher frequency (8 of 18) than the wild-type amino acid tyrosine (1 of 18). It is of interest to note that phenylalanine does not occur in that position in other known thioredoxin family members, except in the putative DsbC of certain Neisseria species, which contain the active site CPFC. The N-terminal position of the dipeptide (CXXC) was tolerant to a wider variety of amino acids, including basic, polar, and small hydrophobic residues. Acidic residues were not encountered in any of the clones that were scored in the screen.
Several mutants gave higher yields of murine urokinase and vtPA than the wild-type enzyme, indicating that they are at least equally, if not more, proficient in assisting the folding of multidisulfide proteins. The relative yields of vtPA and urokinase obtained with various DsbC variants was similar, indicating that the effect of the active-site mutations on disulfide bond isomerization is not limited to one polypeptide substrate. However, the precise molecular understanding of the effect of active-site mutations on the catalysis of disulfide bond isomerization will have to await detailed in vitro analyses of the equilibria and kinetics.
In summary, we have shown, for the first time, that mutations in the active sites of DsbA and DsbC can have a profound effect on the in vivo yield of multidisulfide proteins. The rate-limiting step in the folding of the proteins urokinase and vtPA in the E. coli periplasm appears to be the isomerization of incorrect disulfide bonds. The collection of dsbA alleles and the plasmids expressing DsbC mutants we have reported here are likely to be widely useful for aiding the periplasmic expression of multidisulfide proteins and for other biotechnology purposes.
ACKNOWLEDGMENTS
This work was supported by NSF GOALI 95-111 (G.G. and J.R.S.) and NIH 5RO1 GM55090-02. Paul Bessette was supported in part by an NIH Biotechnology Training Fellowship.
REFERENCES
- 1.Baneyx F, Georgiou G. In vivo degradation of secreted fusion proteins by the Escherichia coli outer membrane protease OmpT. J Bacteriol. 1990;172:491–494. doi: 10.1128/jb.172.1.491-494.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardwell J C, Lee J O, Jander G, Martin N, Belin D, Beckwith J. A pathway for disulfide bond formation in vivo. Proc Natl Acad Sci USA. 1993;90:1038–1042. doi: 10.1073/pnas.90.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell J C, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 4.Bass S, Gu Q, Christen A. Multicopy suppressors of Prc mutant Escherichia coli include two HtrA (DegP) protease homologs (HhoAB), DksA, and a truncated RlpA. J Bacteriol. 1996;178:1154–1161. doi: 10.1128/jb.178.4.1154-1161.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belin D, Vassalli J D, Combepine C, Godeau F, Nagamine Y, Reich E, Kocher H P, Duvoisin R M. Cloning, nucleotide sequencing and expression of cDNAs encoding mouse urokinase-type plasminogen activator. Eur J Biochem. 1985;148:225–232. doi: 10.1111/j.1432-1033.1985.tb08829.x. [DOI] [PubMed] [Google Scholar]
- 6.Bessette P H. Ph. D. dissertation. Austin: University of Texas at Austin; 2000. [Google Scholar]
- 7.Bessette P H, Aslund F, Beckwith J, Georgiou G. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc Natl Acad Sci USA. 1999;96:13703–13708. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bessette P H, Cotto J J, Gilbert H F, Georgiou G. In vivo and in vitro function of the Escherichia coli periplasmic cysteine oxidoreductase DsbG. J Biol Chem. 1999;274:7784–7792. doi: 10.1074/jbc.274.12.7784. [DOI] [PubMed] [Google Scholar]
- 9.Boyd D, Manoil C, Beckwith J. Determinants of membrane protein topology. Proc Natl Acad Sci USA. 1987;84:8525–8529. doi: 10.1073/pnas.84.23.8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brickman E, Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and phi80 transducing phages. J Mol Biol. 1975;96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 11.Chivers P T, Laboissiere M C, Raines R T. The CXXC motif: imperatives for the formation of native disulfide bonds in the cell. EMBO J. 1996;15:2659–2667. [PMC free article] [PubMed] [Google Scholar]
- 12.Chivers P T, Prehoda K E, Raines R T. The CXXC motif: a rheostat in the active site. Biochemistry. 1997;36:4061–4066. doi: 10.1021/bi9628580. [DOI] [PubMed] [Google Scholar]
- 13.Debarbieux L, Beckwith J. On the functional interchangeability, oxidant versus reductant, of members of the thioredoxin superfamily. J Bacteriol. 2000;182:723–727. doi: 10.1128/jb.182.3.723-727.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duvoisin R M, Belin D, Krisch H M. A plasmid expression vector that permits stabilization of both mRNAs and proteins encoded by the cloned genes. Gene. 1986;45:193–201. doi: 10.1016/0378-1119(86)90254-4. [DOI] [PubMed] [Google Scholar]
- 15.Grauschopf U, Winther J R, Korber P, Zander T, Dallinger P, Bardwell J C. Why is DsbA such an oxidizing disulfide catalyst? Cell. 1995;83:947–955. doi: 10.1016/0092-8674(95)90210-4. [DOI] [PubMed] [Google Scholar]
- 16.Guzman L M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hennecke J, Sebbel P, Glockshuber R. Random circular permutation of DsbA reveals segments that are essential for protein folding and stability. J Mol Biol. 1999;286:1197–1215. doi: 10.1006/jmbi.1998.2531. [DOI] [PubMed] [Google Scholar]
- 18.Huber-Wunderlich M, Glockshuber R. A single dipeptide sequence modulates the redox properties of a whole enzyme family. Fold Des. 1998;3(3):161–171. doi: 10.1016/S1359-0278(98)00024-8. [DOI] [PubMed] [Google Scholar]
- 19.Joly J C, Swartz J R. In vitro and in vivo redox states of the Escherichia coli periplasmic oxidoreductases DsbA and DsbC. Biochemistry. 1997;36:10067–10072. doi: 10.1021/bi9707739. [DOI] [PubMed] [Google Scholar]
- 20.Joly J C, Swartz J R. Protein folding activities of Escherichia coli protein disulfide isomerase. Biochemistry. 1994;33:4231–4236. doi: 10.1021/bi00180a017. [DOI] [PubMed] [Google Scholar]
- 21.Jonda S, Huber-Wunderlich M, Glockshuber R, Mössner E. Complementation of DsbA deficiency with secreted thioredoxin variants reveals the crucial role of an efficient dithiol oxidant for catalyzed protein folding in the bacterial periplasm. EMBO J. 1999;18:3271–3281. doi: 10.1093/emboj/18.12.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krause G, Lundstrom J, Barea J L, Pueyo de la Cuesta C, Holmgren A. Mimicking the active site of protein disulfide-isomerase by substitution of proline 34 in Escherichia coli thioredoxin. J Biol Chem. 1991;266:9494–9500. [PubMed] [Google Scholar]
- 23.Lundstrom J, Krause G, Holmgren A. A Pro to His mutation in active site of thioredoxin increases its disulfide-isomerase activity 10-fold. New refolding systems for reduced or randomly oxidized ribonuclease. J Biol Chem. 1992;267:9047–9052. [PubMed] [Google Scholar]
- 24.Metcalf W W, Jiang W, Wanner B L. Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6K gamma origin plasmids at different copy numbers. Gene. 1994;138:1–7. doi: 10.1016/0378-1119(94)90776-5. [DOI] [PubMed] [Google Scholar]
- 25.Mössner E, Huber-Wunderlich M, Glockshuber R. Characterization of Escherichia coli thioredoxin variants mimicking the active-sites of other thiol/disulfide oxidoreductases. Protein Sci. 1998;7:1233–1244. doi: 10.1002/pro.5560070519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mössner E, Huber-Wunderlich M, Rietsch A, Beckwith J, Glockshuber R, Aslund F. Importance of redox potential for the in vivo function of the cytoplasmic disulfide reductant thioredoxin from Escherichia coli. J Biol Chem. 1999;274:25254–25259. doi: 10.1074/jbc.274.36.25254. [DOI] [PubMed] [Google Scholar]
- 27.Ostermeier M, De Sutter K, Georgiou G. Eukaryotic protein disulfide isomerase complements Escherichia coli dsbA mutants and increases the yield of a heterologous secreted protein with disulfide bonds. J Biol Chem. 1996;271:10616–10622. doi: 10.1074/jbc.271.18.10616. [DOI] [PubMed] [Google Scholar]
- 28.Ostermeier M, Georgiou G. The folding of bovine pancreatic trypsin inhibitor in the Escherichia coli periplasm. J Biol Chem. 1994;269:21072–21077. [PubMed] [Google Scholar]
- 29.Qiu J, Swartz J R, Georgiou G. Expression of active human tissue-type plasminogen activator in Escherichia coli. Appl Environ Microbiol. 1998;64:4891–4896. doi: 10.1128/aem.64.12.4891-4896.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raina S, Missiakas D. Making and breaking disulfide bonds. Annu Rev Microbiol. 1997;51:179–202. doi: 10.1146/annurev.micro.51.1.179. [DOI] [PubMed] [Google Scholar]
- 31.Rietsch A, Beckwith J. The genetics of disulfide bond metabolism. Annu Rev Genet. 1998;32:163–184. doi: 10.1146/annurev.genet.32.1.163. [DOI] [PubMed] [Google Scholar]
- 32.Rietsch A, Belin D, Martin N, Beckwith J. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:13048–13053. doi: 10.1073/pnas.93.23.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rietsch A, Bessette P, Georgiou G, Beckwith J. Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J Bacteriol. 1997;179:6602–6608. doi: 10.1128/jb.179.21.6602-6608.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shevchik V E, Condemine G, Robert-Baudouy J. Characterization of DsbC, a periplasmic protein of Erwinia chrysanthemi and Escherichia coli with disulfide isomerase activity. EMBO J. 1994;13:2007–2012. doi: 10.1002/j.1460-2075.1994.tb06470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 36.Stafford S J, Lund P A. Mutagenic studies on human protein disulfide isomerase by complementation of Escherichia coli dsbA and dsbC mutants. FEBS Lett. 2000;466:317–322. doi: 10.1016/s0014-5793(99)01728-7. [DOI] [PubMed] [Google Scholar]
- 37.Strauch K L, Beckwith J. An Escherichia coli mutation preventing degradation of abnormal periplasmic proteins. Proc Natl Acad Sci USA. 1988;85:1576–1580. doi: 10.1073/pnas.85.5.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian H, Boyd D, Beckwith J. A mutant hunt for defects in membrane protein assembly yields mutations affecting the bacterial signal recognition particle and sec machinery. Proc Natl Acad Sci USA. 2000;97:4730–4735. doi: 10.1073/pnas.090087297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wunderlich M, Glockshuber R. Redox properties of protein disulfide isomerase (DsbA) from Escherichia coli. Protein Sci. 1993;2:717–726. doi: 10.1002/pro.5560020503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wunderlich M, Otto A, Seckler R, Glockshuber R. Bacterial protein disulfide isomerase: efficient catalysis of oxidative protein folding at acidic pH. Biochemistry. 1993;32:12251–12256. doi: 10.1021/bi00096a039. [DOI] [PubMed] [Google Scholar]
- 41.Zapun A, Bardwell J C, Creighton T E. The reactive and destabilizing disulfide bond of DsbA, a protein required for protein disulfide bond formation in vivo. Biochemistry. 1993;32:5083–5092. doi: 10.1021/bi00070a016. [DOI] [PubMed] [Google Scholar]
- 42.Zapun A, Creighton T E. Effects of DsbA on the disulfide folding of bovine pancreatic trypsin inhibitor and alpha-lactalbumin. Biochemistry. 1994;33:5202–5211. doi: 10.1021/bi00183a025. [DOI] [PubMed] [Google Scholar]
- 43.Zapun A, Missiakas D, Raina S, Creighton T E. Structural and functional characterization of DsbC, a protein involved in disulfide bond formation in Escherichia coli. Biochemistry. 1995;34:5075–5089. doi: 10.1021/bi00015a019. [DOI] [PubMed] [Google Scholar]
- 44.Zhan X, Schwaller M, Gilbert H F, Georgiou G. Facilitating the formation of disulfide bonds in the Escherichia coli periplasm via coexpression of yeast protein disulfide isomerase. Biotechnol Prog. 1999;15:1033–1038. doi: 10.1021/bp990083r. [DOI] [PubMed] [Google Scholar]