Abstract
The nucleotide sequence of a 10,528-bp region comprising the chlorocatechol pathway gene cluster tetRtetCDEF of the 1,2,3,4-tetrachlorobenzene via the tetrachlorocatechol-mineralizing bacterium Pseudomonas chlororaphis RW71 (T. Potrawfke, K. N. Timmis, and R.-M. Wittich, Appl. Environ. Microbiol. 64:3798–3806, 1998) was analyzed. The chlorocatechol 1,2-dioxygenase gene tetC was cloned and overexpressed in Escherichia coli. The recombinant gene product was purified, and the α,α-homodimeric TetC was characterized. Electron paramagnetic resonance measurements confirmed the presence of a high-spin-state Fe(III) atom per monomer in the holoprotein. The productive transformation by purified TetC of chlorocatechols bearing chlorine atoms in positions 4 and 5 provided strong evidence for a significantly broadened substrate spectrum of this dioxygenase compared with other chlorocatechol dioxygenases. The conversion of 4,5-dichloro- or tetrachlorocatechol, in the presence of catechol, displayed strong competitive inhibition of catechol turnover. 3-Chlorocatechol, however, was simultaneously transformed, with a rate similar to that of the 4,5-halogenated catechols, indicating similar specificity constants. These novel characteristics of TetC thus differ significantly from results obtained from hitherto analyzed catechol 1,2-dioxygenases and chlorocatechol 1,2-dioxygenases.
Vast amounts of toxic chemicals have been released into the ecosphere in the last five decades (46). Substitution for hydrogen of normally readily biodegradable hydrocarbons with xenobiotic structural elements, such as halogens or nitro or sulfo groups, led to a significantly reduced or retarded microbial breakdown (1a) and, consequently, to the concomitant accumulation in the environment and the food chain. Therefore, halogenated pesticides and other halogenated aliphatics and aromatics are a major environmental concern.
Genetic analysis of bacterial catabolic loci encoding degradation of haloaromatic compounds, such as chlorinated benzenes, clearly underlined the role of incorporation and rearrangement of preexisting genetic material by transposition and other genetic events in the assembly of new catabolic capacities (29, 63, 64, 72). Recruitment of mobile genetic elements allows adaptation to the degradation of recalcitrant xenobiotic compounds and enhances the ability of bacteria to conquer new ecological niches for further evolution by a faster divergence of gene sequences.
The so-called modified ortho cleavage pathway is a central oxidative bacterial pathway that channels chlorocatechols, derived from the degradation of chlorinated benzoic acids, phenoxyacetic acids, phenols, benzenes, and other aromatics into the energy-generating tricarboxylic acid pathway (67). Gene clusters of the modified ortho cleavage pathway, clcRABD(E) (13, 21), tfdR(T)CDEF (47), and tcbRCDEF (65), have been shown to be evolutionarily related, as reflected by their conserved organization (56). The isolation of bacteria with catabolic transposable elements of almost identical cistronic organization indicates that global dissemination of genes encoding these chlorocatechol-degrading enzymes generally occurs.
These chlorocatechols represent key chemical structures, originating from the bacterial degradation of chlorobenzenes, chlorophenols, chlorobenzoates, and other chlorinated aromatics upon initial dioxygenation and dehydrogenation, and are further metabolized by either ortho or meta cleavage (24). Addition of molecular oxygen and subsequent cleavage between two adjacent hydroxyl groups has been shown to proceed through nonheme Fe(III)-dependent metalloenzymes, classified as intradiol dioxygenases (67). Protocatechuate 3,4-dioxygenase (25, 33), hydroxyquinol 1,2-dioxygenase (3, 13), catechol 1,2-dioxygenases (type I enzymes) (27, 38, 40), and chlorocatechol 1,2-dioxygenases (type II enzymes, of the so-called modified ortho pathway) (12, 21, 47, 68) have been extensively characterized. These investigations resulted in distinct information on kinetic features and respective substrate specificities as well as some structural information on catechol 1,2-dioxygenase (18, 60) and protocatechuate 3,4-dioxygenase (42, 43). The high-resolution crystal structure of protocatechuate 3,4-dioxygenase revealed two histidine and two tyrosine residues involved in the binding of the catalytic ferric iron (42, 62). These residues are conserved in all investigated intradiol dioxygenases. The iron atom in the pentacoordinated active center of this enzyme remains in the high-spin Fe(III) state during catalysis, as in enzyme-inhibitor complexes investigated so far. More detailed structural information deduced from analysis of the crystal structure of catechol 1,2-dioxygenase, and especially of chlorocatechol 1,2-dioxygenase, however, is scarce. X-ray absorption measurements have provided the first insights into the recognition of chemical structures by ortho-cleaving dioxygenases (7). The turnover of different substrates and recognition of key structures are assumed to be modulated through interactions of specific residues of the polypeptide with the substrate, not by the iron-coordinating ligands (7). The substrate specificities and affinities of hitherto characterized chlorocatechol 1,2-dioxygenases towards (highly) chlorinated catechols differ considerably, although most of these enzymes share relatively high sequence similarities (51).
Pseudomonas chlororaphis RW71 is the first microorganism shown to mineralize 1,2,3,4-tetrachlorobenzene via a 4,5-substituted chlorocatechol, (3,4,5,6-)tetrachlorocatechol. Biochemical analysis of the potential of this strain revealed unusual capacities, such as conversion not only of highly chlorinated catechols but also of 2,3,5-trichloromaleylacetate, a pathway product for which conversion has never been reported (49). In the present study, the tet operon from P. chlororaphis RW71 which specifies for enzymes that productively mineralize highly chlorinated catechols has been cloned and sequenced. The biochemical and spectroscopic characterization of recombinant TetC, the first catabolic enzyme of this operon, is also presented. Experimental data provide unequivocal evidence that 4,5-dichloro-, 3,4,5-trichloro-, and tetrachlorocatechol are productively converted by this enzyme. These chemicals were previously regarded as strong inhibitors of ortho-cleaving dioxygenases and found to have high recalcitrance towards aerobic bacterial catabolism (9, 16, 63).
MATERIALS AND METHODS
Strains, culture conditions, and general DNA techniques.
Bacterial strains used in this study were P. chlororaphis RW71 (49), Escherichia coli DH5α (Clontech), and E. coli BL21(DE3)(pLysS). Plasmids were pET9a (Novagen) and pBluescript II KS(+) and pCR-Script AMP SK(+) (Stratagene). P. chlororaphis RW71 was grown in mineral salts medium with 1,2,3,4-tetrachlorobenzene as the only carbon and energy source, as previously described (49). Liquid cultures were grown in baffled round-bottom flasks on a rotary shaker at 120 rpm at 28°C. Solid media contained 1% (wt/vol) purified agar (Oxoid). E. coli strains were grown at 37°C on Luria-Bertani (LB) medium containing the appropriate antibiotics. Standard techniques and DNA manipulations were carried out as described by Sambrook et al. (52). Oligonucleotide synthesis, DNA probe labeling, Southern blotting, colony lift hybridization, DNA elution from agarose gels, nucleotide sequencing, and sequence analysis were done essentially as described previously (3, 4). Plasmid DNA for sequencing was extracted with the Plasmid Max Kit (Qiagen) as recommended by the supplier. The accession number of the entire sequence is AJ271325 in the EMBL/DDBJ/GenBank database, and that of tetC is AJ132716.
Cloning of two overlapping DNA fragments encoding the tet operon.
For cloning of the chlorocatechol pathway genes, two probes were generated by PCR amplification using total DNA of RW71 extracted by means of a Qiagen Genomic DNA kit. A 745-bp fragment coding for a cycloisomerase was obtained using the degenerate primers TP2 (GTGCASMAGCAGAGCTA) and TP6 (TTGCAMAGCTTCAGCGA) as previously described for the detection of chlorocatechol degradation genes (48). A second probe coding for an 887-bp fragment within the genes of the dienelactone hydrolase and the maleylacetate reductase was amplified by using the oligonucleotides TP31 (CGCGGTGATCGGCTATTGCCTG) and TP32 (CCCTCAACCGCGTGCGCGATCG) priming the chlorocatechol pathway operon tcbRCDEF (65, 66). Both probes were sequenced in order to confirm the similarity to known homologues. Southern blots of either BamHI- or KpnI-digested total DNA of RW71 were hybridized under stringent conditions with the two PCR-amplified probes. Positive 9-kb BamHI and 8-kb KpnI fragments were revealed by both probes. For construction of a partial gene library, total DNA of RW71 was digested with the above-described restriction enzymes and electrophoretically separated on a 0.7% agarose gel. Slices containing DNA fragments of 8 to 11 kb for the BamHI digest and of 7 to 9 kb for the KpnI digest were excised from the gel. The DNA was electroeluted and cloned into pBluescript II KS(+), cleaved with either BamHI or KpnI, and dephosphorylated with calf intestine phosphatase. The two partial gene libraries obtained in E. coli DH5α cells were spread onto LB plates containing 1.0 mM isopropyl-β-d-thiogalactopyranoside (IPTG), 0.004% (wt/vol) 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, and 0.1 mg of ampicillin/ml. Colony hybridization with the two probes allowed selection of positive clones. The plasmids, pTP18 and pTP19, corresponding to the 9-kb BamHI and 8-kb KpnI fragments in pBluescript II KS(+), were mapped with various restriction enzymes and sequenced on both strands. The sequence analysis of the two cloned fragments from pTP18 and pTP19 revealed a 4.75-kb overlap between the two fragments.
Generation of the expression vector pTP20.
The gene tetC of strain RW71 was PCR amplified using the oligonucleotides TP74 (GCCCCATATGAACGAACGAGTGAAGC) and TP75 (GCGCAGATCTCATGCGTGCTCCCGGGG) with plasmid pTP19 as the template. These two primers contained a 5′ extension of 7 bp with NdeI and BglII restriction sites, respectively. PCR was performed according to the standard procedure, except that proofreading Pfu polymerase (Boehringer Mannheim) was used and 5% (vol/vol) dimethyl sulfoxide was added. Fragments of the expected size were excised after resolution of PCR products on 1.5% agarose gels, purified, and cloned into pCR-Script AMP SK(+), generating plasmid pTP15. The integrity of the sequence information of the 756-bp insert was checked, and the fragment was excised from pTP15 by restriction with NdeI and BglII. Purification on agarose gel and ligation with pET9a, previously digested with NdeI and BamHI, resulted in the expression plasmid pTP20.
Purification of recombinant TetC.
E. coli cells containing pTP20 were grown at 37°C in LB medium containing 50 μM kanamycin until an absorbance at 600 nm of 1.0 was reached. Upon induction by 1 mM IPTG, the medium was supplemented with 0.1 mM FeCl3 to prevent excess accumulation of apodioxygenase. Cells were grown for three additional hours at 30°C and then harvested by centrifugation (10 min; 4°C; 10,000 × g). The cell pellet was washed with 33 mM Tris-HCl, pH 8.0, and stored at −70°C. For further processing, cells were thawed on ice, resuspended in 20 ml of 33 mM Tris-HCl, pH 8.0 (9), and broken by two passages through a chilled French pressure cell (Aminco, Silver Spring, Md.) at 10,000 lb/in2. All further purification steps were carried out at 4°C. A protocol for purification of TetC was designed as a three-step procedure avoiding high ammonium sulfate concentrations and including a fast-flow Pharmacia DEAE Sepharose column, a HiLoad 16/60 (Superdex 200) gel filtration column from Pharmacia, and a Ceramic Hyper DF column (BioSepra SA, Gergy Saint Christophe, France). The crude extract was centrifuged at 18,000 × g for 30 min, and the supernatant was applied on the DEAE column equilibrated with 50 mM Tris-HCl buffer, pH 8.0. Protein was eluted with a linear gradient of 0 to 700 mM NaCl in this buffer. The brownish fractions containing the desired activity were combined, concentrated, and desalted on an Amicon filtration unit with a 10,000-Da exclusion membrane. Gel filtration was performed with the above-described Tris-HCl buffer, containing 0.1 M NaCl, at a reduced flow rate of 0.5 ml/min. The Ceramic Hyper DF column was equilibrated with 50 mM Tris-HCl, pH 8.0, and the protein applied was eluted with a linear gradient of NaCl. Samples were pooled and desalted. Eventually, a Mono-P column (HR5/20) from Pharmacia which resolves pI differences of 0.02 pH unit was used for further purification. Equibration of the column was performed using 25 mM Tris-acetate buffer (pH 8.3). For resolution, a pH range of 8 to 5 was chosen by mixing Polybuffer 96 (30%) with Polybuffer 74 (70%) from Pharmacia and adjusting the pH at 5.0 with acetate. Highly purified protein from the Ceramic Hyper DF column was loaded and eluted with a linear flow of 0.5 ml/min for approximately 8 column volumes. The eluted protein was fractionated for further treatment, consisting of polybuffer removal by gel filtration, and concentrated.
Analytical methods.
The apparent subunit molecular mass of the polypeptide and the purity of fractions were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under denaturing conditions with 12% (wt/vol) polyacrylamide according to the method of Laemmli (30). SDS-PAGE was performed using an SE200 small vertical slab-gel unit (Hoefer Pharmacia). Gels were stained overnight with a commercial Coomassie blue solution (Roti-Blue; Roth) with a sensitivity comparable to that of silver staining, allowing detection of less than 30 ng of protein. The apparent molecular weight of the holoenzyme was determined by gel filtration on a calibrated Superdex 200 column (Pharmacia) eluted at a flow rate of 0.5 ml/min with 50 mM Tris-HCl, pH 8.0, containing 100 mM NaCl. Standards for calibration were from Bio-Rad.
The exact molecular masses of polypeptides were determined by electrospray ionization tandem mass spectrometry on a Finnigan MAT TSQ 700 triple quadrupole mass spectrometer equipped with an electrospray ion source (Finnigan MAT Corp., San José, Calif.). Desalted samples were dissolved in acetonitrile to approximately 10 pmol/ml and injected at a flow rate of 1 ml/min. A voltage of 5.5 kV was applied to the electrospray needle. The theoretical average mass of monomeric TetC including the initial methionine was calculated by means of the program PAWS (R. C. Beavis, New York University/Skirball Institute), taking into consideration the statistical isotope distribution.
The pI of the enzyme was determined under nondenaturing conditions, using the Pharmacia Phast system for isoelectric focusing, according to the protocol of the manufacturer. An isoelectric focusing kit was used as the standard marker. The same system was applied for electrophoretic titration curve analysis over a pH range of 3 to 9. All gels were stained with a commercial silver staining kit from Pharmacia.
For N-terminal sequencing, the protein was transferred to a microcentrifuge vial, dried in a Speed Vac, redissolved in 50% acetonitrile, applied to a Bioprene-treated, precycled glass fiber filter, and sequenced by automated Edman degradation with an Applied Biosystems sequencer (pulsed-liquid; model 473A) with on-line high-performance liquid chromatography (HPLC) detection of phenylthiohydantoin-derivatized amino acids.
EPR and UV-visible spectroscopy.
Electron paramagnetic resonance (EPR) spectra were recorded on a Varian E 109 spectrometer equipped with a helium flow cryostat (ESR 900; Oxford Instruments). Spin concentrations were determined by double integration of the signals and compared to a reference of multiple iron concentrations. UV-visible absorption spectra were recorded on an HP8452A diode array spectrophotometer (Hewlett-Packard), the cuvette holder of which was connected to an anaerobic glove box through optic fibers and adapters (Spectrofip 8452; Photonetics). The raw data were corrected with a cubic spline fit using the program Kaleidagraph (Synergy Software, Inc.).
Enzyme assays and kinetic measurements.
Protein concentrations were determined by the method of Bradford with bovine serum albumin as a standard (6). All enzyme assays were performed at 25°C with a spectrophotometer (UV 2100; Shimadzu Corp., Kyoto, Japan) as described previously (49). Specific activities are expressed as units per milligram of protein; 1 U is defined as 1 μmol of substrate transformed per min. The extinction coefficients for chloromuconates reported by Dorn and Knackmuss (16) and Sander et al. (54) were used for the determination of catechol 1,2-dioxygenase (C120; EC 1.13.11.1) activity and that of its derivatives. Slow reactions were monitored by HPLC. Kinetic data were analyzed with the ENZPACK computer software (P. A. Williams, ENZPACK manual, Elsevier-Biosoft Cambridge, United Kingdom). Considerations of interaction of substrates with proteins in regard to mechanisms of inhibition followed previously published outlines (20).
Transformation experiments with chlorocatechols of the 4,5-halogenation type.
For turnover experiments, 0.1 to 1 mM concentrations of 4,5-dichloro-, 3,4,5-trichloro-, or tetrachlorocatechol were incubated with purified protein (10.6 to 53 μg) in 1 ml of 33 mM Tris-HCl buffer (pH 8.0) for 12 h. Samples from these assays were analyzed by HPLC as described previously (49). The net elution volumes of metabolites by a 36% methanol–H2O solvent system were 3.4 min for 2,3,5-trichlorodienelactone and 9.1 min for 3,4-dichloro-cis,cis-muconic acid. Conversion of tetrachlorocatechol to the corresponding tetrachloromuconic acid was detectable as its cyclization product, 2,3,5-trichlorodienelactone, only after the sample was acidified to pH 1.5. This allows the prevention of on-column cycloisomerization caused by the acidic conditions used for separation by HPLC (49). The structural identity was confirmed by gas chromatography-mass spectrometry (GC-MS) analysis of the respective products after extraction and derivatization of samples from enzyme assays as described earlier (49). Quantification of 3,4,5-trichlorocatechol transformation was performed by substrate depletion analysis, due to instability of the corresponding chloromuconic acid under conditions of separation by HPLC.
Insertion of Fe(III) into apodioxygenase.
Experimental removal of Fe(III) from the catalytic center of TetC was carried out through chromatography of this polypeptide on a Mono-P column (Pharmacia) as described above and subsequent gel filtration, also as described above. Experiments for reconstitution of the activity of apo TetC were performed with protein concentrations of 10 to 27 μg/ml in 33 mM Tris-HCl buffer (pH 8.0) containing 0.1 or 0.2 mM Fe(III) in the presence of different concentrations of ammonium sulfate (0.5 to 2.0 M) and with 3-chlorocatechol as a substrate. Different incubation temperatures were tested (25 to 40°C), as were different incubation times (1 to 20 min) at appropriate intervals.
Chemicals.
Tetrachlorocatechol and 3,4,5-trichlorocatechol were purchased from Aldrich (Steinheim, Germany) and Promochem (Wesel, Germany), respectively. 3-Chloro-, 4-chloro-, 3,5-dichloro-, 3,6-dichloro-, 4,5-dichloro-, and 3,4,6-trichlorocatechol were kindly provided by H.-A. Arfmann (GBF). Stock solutions of chlorocatechols were prepared in dimethyl sulfoxide. All chemicals used in this study were of the highest commercially available grade.
RESULTS AND DISCUSSION
Genetic organization of the tet cluster.
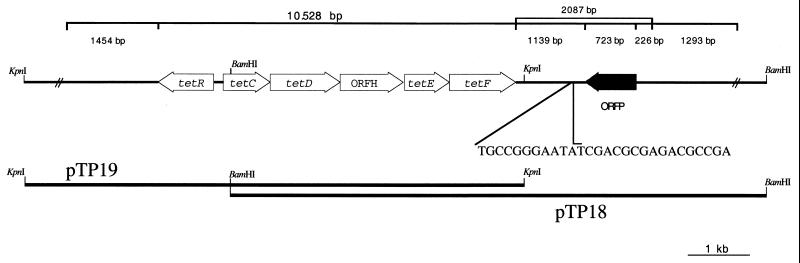
From total DNA extracted from P. chlororaphis RW71, a region encoding a cycloisomerase and other chlorocatechol degradation genes was cloned and sequenced (Fig. 1). Among the six open reading frames (ORFs) detected on the 8.1-kb KpnI fragment of plasmid pTP19, five ORFs were assigned by a homology search to the chlorocatechol operon genes and named tetRtetCDEF (Fig. 1). The gene tetR, encoding a regulatory protein (transcriptional activator), is divergently transcribed from the genes specifying the chlorocatechol 1,2-dioxygenase (tetC), the chloromuconate cycloisomerase (tetD), the ORFH, to which no function was assigned, the chlorodienelactone hydrolase (tetE), and the chloromaleylacetate reductase (tetF). The chlorocatechol operon tet identified in RW71 is therefore organized identically to the corresponding clusters of pP51 (68), pNH91 (41), pAC27 (21), pJP4 (14, 31) and, to a lesser extent, pEST4011 (28). An additional ORF was detected on the 8.7-kb BamHI fragment (pTP18) and termed ORFP. It was identified about 1.1 kb downstream of tetF and appeared to be divergently transcribed. A similar structural element was recently reported at the same position on pNH91, a chlorocatechol pathway catabolic plasmid from Alcaligenes eutrophus (41). Comparison of the amino acid sequence specified by ORFP with sequences available in databases indicated some similarities with cellular transporter systems, possibly responsible for the uptake of branched-chain amino acids (1, 26). The 1,519-bp region upstream of ORFP and the 1,454-bp region downstream of tetR did not show any significant homology to published sequences.
FIG. 1.
Genetic organization of the catabolic tet operon and its environment. The tetRCDEF genes and the open reading frame, ORFP, from plasmids pTP18 and pTP19 are schematically presented. The arrows show the location and direction of transcription. The upper line indicates the lengths of sequenced DNA fragments in front of and behind the degradative gene cluster.
Detailed analyses revealed that plasmids pTP18 and pNH91 share an almost identical 2,087-bp region downstream of tetF and cbnD, respectively, with the exception of a 28-bp insert specific to pTP18 between tetF and ORFP (Fig. 1). Downstream of this 2,087-bp region, the cbn locus ends with a sequence encoding the insertion element IS1600. No such structure was found on the remaining 1,293 bp sequenced from pTP18. The same nucleotide sequence was located 2 kb upstream of cbnR on pNH91, forming the chlorocatechol catabolic transposon Tn5707 with an overall size of 15 kb (41). Comparable structural elements were not reported from the flanking regions of the corresponding chlorocatechol operon structures of pP51, beyond tcbF. The extended analysis of the flanking regions of the chlorocatechol operon on pJP4 of Ralstonia eutropha JMP134 has also shown an insertion sequence, ISJP4, flanking the truncated regulatory genes tfdT(I) and tfdK(II) (32). A much larger conjugative element of 105 kb of the 3-chlorobenzoate-degrading strain Pseudomonas sp. B13, carrying the chlorocatechol pathway genes, has been reported by Ravatn et al. (50). Although they are not identified on pTP18 and pTP19, the existence of such mobilizing elements cannot be definitively excluded, since many catabolic sequences are located on transposon structures (61). The catabolic potential for 1,2,3,4-tetrachlorocatechol mineralization was found to be highly unstable in strain RW71 (49), suggesting localization of this genetic information on a plasmid: PCR analysis of total DNA extracted from strains of cured phenotype, using oligonucleotide pairs TP2-TP6 and TP31-TP32, did not detect the presence of any chlorocatechol operon genes. The loss of the initial chlorobenzene dioxygenase and cis-chlorobenzene dihydrodiol dehydrogenase system cannot be ruled out: such not very stable genes were already found on transposon Tn5280 of pP51 from Pseudomonas sp. strain P51 (64, 69) and may be present on unstable or transmissible transposon structures in RW71.
Phylogenetic relationship between (chloro-)catechol catabolic genes and enzymes.
The sequences of the genes specified by the tet operon share a high degree of similarity with those encoded by the operons tcb and cbn of plasmids pP51 and pNH9, respectively (Table 1). On the amino acid level, identities are superior to 99% for the regulator protein, the chloromuconate cycloisomerase, the chlorodienelactone hydrolase, and the maleylacetate reductase. The identity of the intracistronic ORF is also significantly high, and only the chlorocatechol 1,2-dioxygenase, TetC, exhibits slightly reduced identity (95%) with TcbC from Pseudomonas sp. strain P51. The sequences of the enzymes specified by the operons tet, tcb, and cbn show relatively low similarities with those of the type II enzymes (14, 57): from 52 to 57% for the transcriptional activators, from 58 to 68% for the chlorocatechol 1,2-dioxygenases, from 76 to 84% for the chloromuconate cycloisomerases, from 52 to 53% for the chlorodienelactone hydrolases, and from 55 to 59% for the chloromaleylacetate reductases. Similarities with genes encoding the catechol pathway (type I pathway) (8) of gram-negative and even of gram-positive bacteria (19) are in the range of only 30 to 40%. These three levels of similarity described above allow a clear discrimination of the different (chloro-)catechol catabolic enzymes into several groups: the tcb, cbn, and tet catabolic operons may be distinguished from the classical type I (cat) and type II (clc and tfd). Possibly we have to add to the above group of sequences those of the 1,2,4-trichloro- and 1,2,4,5-tetrachlorobenzene-degrading strains Burkholderia (Pseudomonas) sp. strain PS12 and Acidovorax (Pseudomonas) sp. strain PS14 (54): the sequence of the N-terminal amino acids of the chlorocatechol 1,2-dioxygenase of PS14 (28 amino acids) is identical to those of CbnA, TcbC, and TetC (63). Accordingly, similarities to ClcA and TfdC are below 55%. The 14 N-terminal amino acids of the chloromuconate cycloisomerase of PS14 (63) share a relatively high degree of similarity with TfdD of pJP4 (74%) and ClcB of pAC27 (83%), but this sequence is again much closer to those of both TetD of strain RW71 and CbnD of strain NH9 (>97%) and fully identical to that of TcbD of P51. Strains PS12 and PS14 share many of their catabolic features with strain RW71. This is possibly due to the fact that all three isolates were obtained from the same source, a chlorophenoxy herbicide and lindane production site where surplus waste isomers had been dumped (54).
TABLE 1.
Genes and corresponding gene products of the tet operon and of related homologous elements
| Gene | Position in sequence (nt) | Probable function or product | Protein with homologous sequence | Source | % Identitya | Reference for homologous proteinb |
|---|---|---|---|---|---|---|
| tetR | 1444–2328 | LysR-type regulatory protein transcriptional activator | TcbR | Pseudomonas sp. strain P51 | 99.6 | P27102 |
| CbnR | Ralstonia eutropha NH9 | 99.3 | BAA74529 | |||
| ClcR | Ralstonia sp. | 57.2 | CAA06967 | |||
| ClcR | Pseudomonas putida | 56.8 | Q05840 | |||
| ClcR | Pseudomonas aeruginosa JB2 | 56.5 | AAC69473 | |||
| ClcR | Pseudomonas putida | 56.5 | A40641 | |||
| TfdR | Alcaligenes eutrophus | 52.7 | A55212 | |||
| TfdR | Alcaligenes eutrophus | 52.7 | P10086 | |||
| TfdR | Variovorax paradoxus | 52.1 | BAA88068 | |||
| tetC | 2478–3233 | Chlorocatechol 1,2-dioxygenase | CbnA | Ralstonia eutropha NH9 | 99.2 | BAA74530 |
| TcbC | Pseudomonas sp. strain P51 | 94.8 | P27098 | |||
| ClcA | Pseudomonas putida | 67.8 | P11451 | |||
| ClcA | Pseudomonas aeruginosa | 67.4 | AAF00195 | |||
| ClcA | Pseudomonas putida | 67.4 | SO6449 | |||
| ClcA | Ralstonia sp. strain JS705 | 67.0 | CAA06968 | |||
| TfdCII | Ralstonia eutropha | 66.8 | AAC44730 | |||
| TfdC | Variovorax paradoxus | 64.0 | BAA88069 | |||
| TfdC | Alcaligenes eutrophus | 57.6 | P05403 | |||
| tetD | 3230–4342 | Chloromuconate cycloisomerase | TfdD | Comamonas acidovorans | 100 | AAD55083 |
| TfdD | Rhodoferax sp. strain P230 | 100 | AAD55081 | |||
| CbnD | Ralstonia eutropha NH9 | 100 | BAA74531 | |||
| TcbD | Pseudomonas sp. strain P51 | 96.7 | P27099 | |||
| TfdD | Variovorax paradoxus | 83.6 | BAA88065 | |||
| IjbD | Burkholderia cepacia | 83.0 | AAB86807 | |||
| ClcB | Pseudomonas putida | 76.4 | P11452 | |||
| ClcB | Pseudomonas aeruginosa | 76.4 | AAF00202 | |||
| ClcB | Pseudomonas aeruginosa | 76.4 | AAF00196 | |||
| ClcB | Pseudomonas aeruginosa | 76.4 | AAC69475 | |||
| ORFH | 4344–5354 | Unknownc | ORF | Ralstonia eutropha NH9 | 99.4 | BAA74532 |
| ORF | Pseudomonas sp. strain P51 | 97.0 | P27103 | |||
| Unnamed | Variovorax paradoxus | 75.5 | BAA88066 | |||
| ORF | Escherichia coli | 55.1 | AAA98283 | |||
| ORF | Pseudomonas aeruginosa 142 | 55.1 | AAF00197 | |||
| ORF | Pseudomonas aeruginosa JB2 | 55.0 | AAF00203 | |||
| ClcB-ClcD intergenic region | Pseudomonas putida | 54.2 | Q47100 | |||
| tetE | 5376–6092 | (Chloro-)dienelactone hydrolase | CbnC | Ralstonia eutropha NH9 | 100 | BAA74533 |
| TcbE | Pseudomonas sp. strain P51 | 99.5 | P27100 | |||
| TfdE | Alcaligenes eutrophus | 52.8 | P27136 | |||
| ClcD | Pseudomonas aeruginosa | 52.6 | AAC69477 | |||
| ClcD | Pseudomonas aeruginosa JB2 | 52.2 | AAF00204 | |||
| ClcD | Pseudomonas aeruginosa 142 | 52.2 | AAF00198 | |||
| Carboxymethylene butenolidase | Unnamed | Pseudomonas sp. | 52.2 | S02022 | ||
| Chlorodienelactone hydrolase | ClcD | Pseudomonas putida | 51.7 | P11453 | ||
| tetF | 6089–7147 | Maleylacetate reductase | CbnD | Ralstonia eutropha NH9 | 99.4 | BAA74534 |
| TcbF | Pseudomonas sp. strain P51 | 99.4 | P27101 | |||
| ClcE | Pseudomonas sp. strain B13 | 59.1 | O30847 | |||
| ClcE | Pseudomonas aeruginosa JB2 | 58.8 | AAC69478 | |||
| MacA | Ralstonia eutropha 335 | 56.9 | AAD55886 | |||
| TftE | Pseudomonas cepacia | 55.4 | Q45072 | |||
| Tff1 | Alcaligenes eutrophus | 54.8 | P27137 | |||
| Tff2 | Alcaligenes eutrophus | 49.2 | P94135 | |||
| MacA | Rhodococcus opacus | 43.2 | AAC38802 | |||
| ORFP | 4344–5354 | High-affinity branched-chain amino acid transporter | ORFL | Ralstonia eutropha NH9 | 100 | BAA11021 |
| LivF | Escherichia coli | 42.3 | P22731 | |||
| BraG | Pseudomonas aeruginosa | 38.8 | E36125 |
Percentage of amino acids that are identical when sequences are aligned with sequences listed in all nonredundant GenBank database and PDB/SwissProt/PIR/PRF formats.
GenBank database and PDB/SwissProt/PIR/PRF formats via the BLAST program of the National Center for Biotechnology Information (2).
No similarity to known proteins.
General biochemical characterization of recombinant TetC.
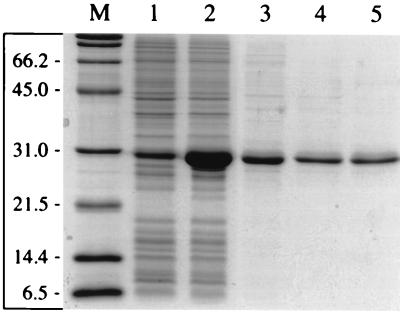
From plasmid pTP19 containing an 8.1-kb DNA fragment from P. chlororaphis RW71, the gene tetC was PCR amplified and inserted into the T7-based overexpression system pET9a. The resulting plasmid, pTP20, was introduced into E. coli strain BL21(DE3)(pLysS) cells. Supplementing the medium with 0.1 mM Fe(III) was found to significantly increase the production of holodioxygenase, since the specific activity of the crude extract, reproducibly, was then threefold higher. The purification resulted in an almost homogeneous preparation. The recombinant TetC migrated on this SDS–12% PAGE as a single band of 30 kDa (Fig. 2). About 10 to 15 mg of purified recombinant protein/liter of wet cells was obtained, equivalent to a 20% yield at a purification factor of 2.5.
FIG. 2.
SDS-PAGE analysis of purification of TetC. Crude extract from uninduced cells of E. coli BL21(DE3)(pLys S) harboring pTP20 are shown in lane 1; extracts from IPTG-induced cells are shown in lane 2. Lanes 3 to 5 show results from the purification: lane 3, fraction from DEAE chromatography; lane 4, fraction from gel filtration; lane 5, fraction from Ceramic Hyper DF column chromatography. Lane M, protein standards in kilodaltons. Approximately 1.5 μg of protein was loaded in each of lanes 3 to 5.
The N-terminal amino acid sequence of the purified protein was determined by Edman degradation and found to be fully identical to the sequence of the polypeptide specified by tetC and tcbC (65) and to the chlorocatechol 1,2-dioxygenase of 1,2,4,5-tetrachlorobenzene-degrading Acidovorax sp. strain PS14 (63). The average molecular mass of the TetC monomer obtained by electrospray ionization tandem mass spectrometry (27,474 Da) appears to be in full agreement with the theoretical value of the polypeptide, including the initial methionine. The apparent molecular mass of 67.5 ± 3 kDa determined by gel filtration indicated that the purified holoenzyme behaves in these experimental conditions as a homodimer.
The isoelectric point of the holoenzyme was determined experimentally to be 5.8. Titration curve analyses (IEF) of freshly purified TetC were performed on polyacrylamide gels run under nondenaturing conditions, in order to confirm the homogeneity of the preparation. Four dominant bands were observed, although the polypeptide content was homogeneous according to SDS-PAGE and mass spectrometric analysis. In light of these results, an additional chromatofocusing step for further purification of TetC, followed by removal of polybuffer by means of gel filtration, was carried out. During this step the protein lost its characteristic reddish color and became inactive, as only ca. 0.1% of the initial activity was remaining. Constituents of the amphoteric buffer system seemed to inhibit the enzyme by chelating and removing the Fe(III) ion from the active site. Whereas this fraction of inactive TetC lacking its ferric iron could not be crystallized, reddish-brown crystals of TetC were obtained in a glycine-NaOH buffer (pH 9.1) with sodium formate as the precipitant, using the purest active fraction, corresponding to lane 5 of Fig. 2. In the following isoelectric focusing titration of such a crystal dissolved in water, only a single major band was detected, and when the gel was highly overloaded in order to visualize impurities, only a single, weakly contaminating band was visible. The major band should represent the catalytically active holodioxygenase. Another single band at a different position in a focusing gel was obtained from colorless, catalytically inactive TetC from the above-described chromatofocusing procedure. The heterogeneous pattern of four major bands mentioned above is, therefore, assumed to be composed of monomeric and dimeric apo- and holodioxygenases.
Measurement of the iron content of recombinant TetC and attempts at reconstitution of the holoenzyme.
Earlier determinations of the iron content of several catechol 1,2-dioxygenases and chlorocatechol 1,2-dioxygenases did not clarify if one iron ion was present in each monomer or one single iron atom was bound at the interface or only in one monomer per holoenzyme. Ratios per homodimer of one iron atom (9, 36, 37, 55), two iron atoms (8, 39, 45), or from 0.6 to 0.89 iron atom (5, 60) were reported. Quantitation of the iron content of different TetC preparations was carried out by means of EPR spectroscopy, the isotropy at maximal rhombicity allowing for the detection of small quantities of bound ferric iron by comparison with a standard (22). Assuming from the isoelectric focusing analysis that less than one third of the protein sample was in a holoform in the first two samples, the iron content measured was close to 0.9 iron atom per monomer. For another sample exhibiting a 1.3-fold higher specific activity for 3-chlorocatechol, a signal consistent with 1.1 iron atoms per monomer was measured. Unspecific binding of additional iron on the protein surface may have led to this minute overestimation.
Previously reported reconstitution experiments on intradiol apodioxygenases had indicated that ferrous iron could be incorporated into the enzyme and then oxidized to the active ferric form (39), a mechanistic study not confirmed up to now. Zaborina et al. (73) have reported reconstitution of 40% of the activity of the intradiol-cleaving 6-chlorohydroxyquinol 1,2-dioxygenase from Streptomyces rochei 303 by the addition of Fe(II) in a time-dependent reaction after the initial activity had been completely inhibited by the chelator Tiron at a 0.1 mM concentration. Attempts at insertion of Fe(II) and Fe(III) into the active site of the apodioxygenase were carried out with the apoprotein obtained by means of MonoP chromatography. Different parameters were checked, such as Fe(II), Fe(III), and ammonium sulfate concentrations, the incubation temperature, and the incubation time. Reconstitution of holoTetC was monitored through measurement of 3-chlorocatechol dioxygenation activity, but even in the best conditions [incubation of apo-TetC with 0.1 mM Fe(III) and 1.3 M ammonium sulfate], no reactivation of more than 2% of the maximal activity of the holoenzyme preparation was obtained.
Kinetic properties of recombinant TetC.
Chlorocatechols of the 4,5-halogenation type have been classified as high-affinity inhibitors for diverse (chloro-)catechol 1,2-dioxygenases (9, 16, 23, 58, 63). Ki values for 4,5-dichlorocatechol and tetrachlorocatechol were 30 and 5 nM, respectively, for the dioxygenase of the chlorobenzoate-degrading strain Pseudomonas putida AC27 (9) and 4 and 108 nM for the dioxygenase of Acidovorax (formerly Pseudomonas) sp. strain PS14 (63). The latter enzyme was shown to be competitively inhibited by 4,5-dichloro- and 3,4,5-trichlorocatechol but uncompetitively inhibited by tetrachlorocatechol, with catechol as the substrate.
(i) Spectrum of substrates cleaved by TetC.
Specific activities and kinetic parameters of recombinant TetC were determined for the whole range of chlorinated catechols (Table 2). The ratio of specific activities for 3-chlorocatechol to those for catechol for TetC (3.16) is about 50% higher than that of the chlorocatechol 1,2-dioxygenase from Burkholderia (formerly Pseudomonas) sp. strain PS12 (2.17) (54) and much higher than those of the others so far described in the literature (reviewed in reference 51). From results shown in Table 2 it is evident that 3,5-dichlorocatechol is the preferred substrate, followed by 3-chlorocatechol, as indicated by high values for the ratio kcat/Km. For the 4,5-halogenated chlorocatechols, exact Michaelis-Menten constants during long-term HPLC-based depletion measurements could not be determined. They have to be assumed to be in the lower micromolar range. Consequently, no specificity constants were calculated, but they should be similar to that for 3-chlorocatechol, since there was no inhibition of 3-chlorocatechol turnover in their presence.
TABLE 2.
Kinetic parameters for P. chlororaphis chlorocatechol 1,2-dioxygenase
| Substrate | kcat (s−1)a | Vmax (U/mg)b | Km (μM) | kcat/Km (s × μM)−1 |
|---|---|---|---|---|
| Catechol | 2.01 | 2.2 (1.00) | 1.24 | 1.62 |
| 3-Chlorocatechol | 6.41 | 6.95 (3.16) | 0.37 | 17.33 |
| 4-Chlorocatechol | 3.3 | 3.60 (1.64) | 0.27 | 12.22 |
| 3,4-Dichlorocatechol | 1.12 | 1.23 (0.56) | 0.16 | 7.00 |
| 3,5-Dichlorocatechol | 3.42 | 3.74 (1.70) | 0.15 | 22.80 |
| 3,6-Dichlorocatechol | 1.76 | 1.94 (0.88) | 0.20 | 8.80 |
| 4,5-Dichlorocatecholc | 0.07 | 0.07 (0.03) | NDd | ND |
| 3,4,5-Trichlorocatecholc | 0.04 | 0.04 (0.02) | ND | ND |
| 3,4,6-Trichlorocatechol | 1.00 | 1.10 (0.50) | 0.16 | 6.25 |
| Tetrachlorocatecholc | 0.08 | 0.08 (0.04) | ND | ND |
The number of active centers of the dimeric enzyme was taken as two.
Relative to Vmax with catechol.
Activities determined by HPLC. Data represent means of at least two independently performed experiments.
ND, not determined.
The specificity constant for the TetC preparation with 3,5-dichlorocatechol is threefold higher than that reported for the so-called 3,5-dichlorocatechol 1,2-dioxygenase of Burkholderia (formerly Pseudomonas) cepacia strain CSV90 (5). It should be threefold higher for the pure TetC holoenzyme. The enzyme of Pseudomonas chlororaphis RW71 exhibits a significantly broader substrate tolerance compared to the substrate pattern of other enzymes (51), such as that of the already mentioned strain PS12 (54) or of Pseudomonas sp. strain P51 (65).
(ii) 4,5-substituted chlorocatechols are transformed by TetC.
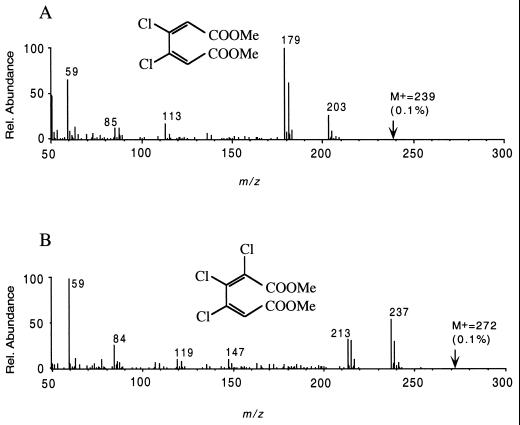
The major part of the above-cited literature reported that chlorocatechols of the 4,5-substitution pattern were not productively transformed by (chloro-)catechol dioxygenases. It was shown recently that strain RW71 degrades 1,2,3,4-tetrachlorobenzene via tetrachlorocatechol (49). This obvious contradiction, therefore, needs to be elucidated in more detail. Spectral changes during conversion of 4,5-dichloro-, 3,4,5-trichloro-, and tetrachlorocatechol, respectively, were monitored in the UV range (230 to 350 nm; plots not shown). Overlay spectra displaying prolonged transformation of the three compounds were recorded and revealed new maxima at 268 nm for 3,4-dichloromuconate, at 273 nm for 2,3,4-trichloromuconate, and at 275 nm for tetrachloromuconate, in accordance with the typical increase of absorption for (chloro-)muconic acids (15, 53). The products from these conversions were resolved by reverse-phase HPLC. Acidified samples were extracted with ethyl acetate and methylated for structure elucidation by GC-MS. Interpretation of results from fragmentation revealed dimethylated 3,4-dichloromuconic acid, dimethyl 2,3,4-trichloromuconate (Fig. 3), and dimethyl tetrachloromuconate, respectively. The molecular ion of dimethylated 3,4-dichloromuconate (m/z = 239) is of very low intensity (0.1%); loss of one carboxymethoxy group from the molecular ion forms the base peak at m/z = 179. The peak observed at m/z = 203 is formed due to the loss of a first chlorine (Fig. 3A). The mass spectrum of the dimethylester of 2,3,4-trichloromuconic acid (Fig. 3B) indicates the loss of chlorine from the molecular peak at m/z = 272 (0.3%), resulting in the signal at m/z = 237. The signal at m/z = 213 represents the loss of one carboxymethoxy group (m/z = 59) from the molecular ion and corresponds to the theoretical value for a trichlorinated substance; the one at m/z = 237 is that of a dichlorinated structure. The general cis,cis configuration is assumed for all muconates. The product from the conversion of tetrachlorocatechol has already been characterized (49). Traces of these chloromuconates had already been identified in transformation experiments with crude cell extracts of Burkholderia (formerly Pseudomonas) sp. strain PS12 (53). The novelty of TetC is a substrate pattern that includes the chlorocatechols with chlorine atoms at positions 4 and 5 and their productive transformation in the cascade of the chlorocatechol pathway, allowing RW71 to grow at the expense of 1,2,3,4-tetrachlorobenzene. The 4,5-halogenated chlorocatechols have previously been shown to be transformed by other organisms or their corresponding enzymes at extremely low rates at high concentrations of enzyme only (11, 46, 53, 55).
FIG. 3.
Identification of products from transformation of 4,5-substituted chlorocatechols. Characteristic fragmentation pattern from GC-MS analysis of methylated extracts from conversion of 4,5-dichlorocatechol and 3,4,5-trichlorocatechol by recombinant TetC are depicted. 70 eV mass spectra of 3,4-dichloromuconic acid dimethylester (A) and 2,3,4-trichloromuconic acid dimethylester (B) are shown. Rel., relative.
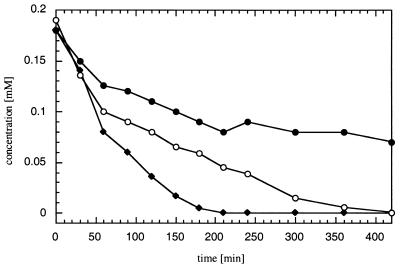
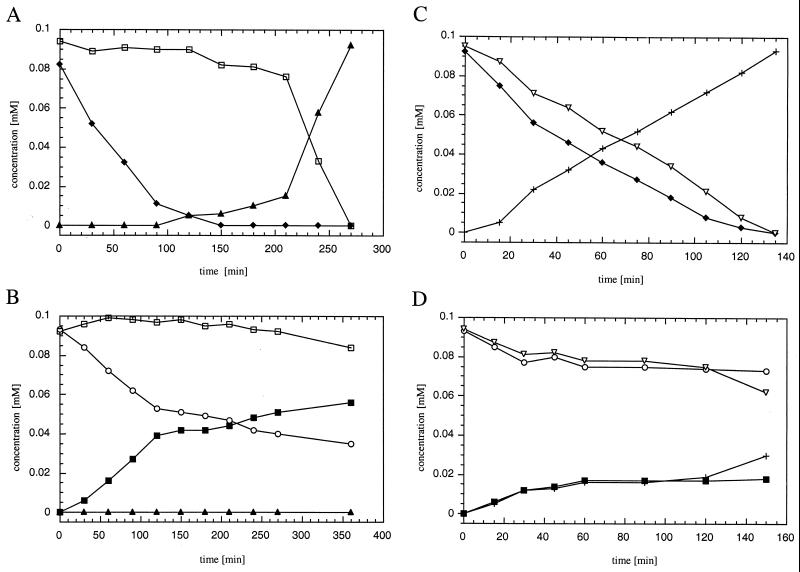
In order to determine the consistency of the catalytic activity of TetC over a prolonged period of time, conversions by recombinant TetC of 0.19 mM solutions of 4,5-dichloro-, 3,4,5-trichloro-, and tetrachlorocatechol were followed by HPLC analysis for 420 min (Fig. 4). Depletion of tetrachlorocatechol was completed after 200 min, and 4,5-dichlorocatechol was transformed within 420 min, whereas 39% of the initial concentration of 3,4,5-trichlorocatechol was still detected after this time span. Further slow conversion until complete depletion was noticed when measured upon overnight incubation. At a protein concentration in this assay of 21.2 μg/ml, which corresponds to 1 μM, the turnover efficiency of the enzyme with these three substrates was determined to be relatively low, corresponding to a rate of about 2 to 3% of the turnover rates of the other chlorocatechols. Initial specific transformation rates were relatively stable over the first 60 min and were 70, 42, and 78 mU per mg of protein for 4,5-dichlorocatechol, 3,4,5-trichlorocatechol, and tetrachlorocatechol, respectively. Interestingly, conversion rates showed a sudden decrease after 1 h, probably due to the formation of highly stable enzyme product complexes of small dissociation constants, formerly identified as the rate-limiting step of substrate turnover, by catechol 1,2-dioxygenase (71) and protocatechuate 3,4-dioxygenase (10, 11, 70). On the other hand, inactivation of the biocatalyst under the test conditions in the presence of an excess of the phenolic substrates cannot be ruled out.
FIG. 4.
Kinetics of transformation of 4,5-substituted chlorocatechols. The time course of the conversion by purified TetC of 0.2 mM 4,5-dichlorocatechol (○), 3,4,5-trichlorocatechol (●), and tetrachlorocatechol (⧫) at 21.2 μg of protein per ml is shown.
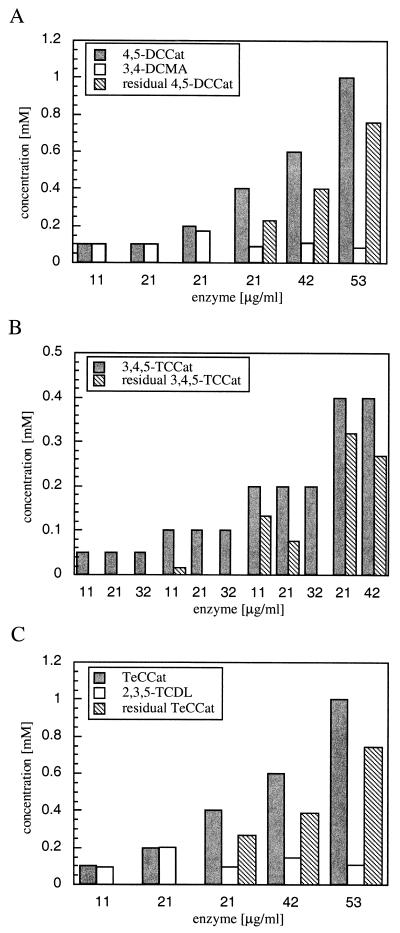
Therefore, more detailed transformation assays were performed over 12 h with up to 1 mM concentrations of 4,5-halogenated catechols. With a high concentration of substrate, such as 0.4 mM, the addition of 21 μg of protein per ml led to a 30% decrease in the substrate concentration. However, at concentrations of 0.2 mM or below, substrates were productively transformed (Fig. 5A to C). Further, we could clearly demonstrate with the substrate 3,4,5-trichlorocatechol that the ratio of substrate to enzyme is crucial (Fig. 5B), providing evidence for inactivation by the substrate. Control experiments revealed that none of these substrates have been absorbed onto glass surfaces or transformed in the absence of TetC. Similar observations for other 1,2-dioxygenases have not yet been reported, and therefore these results cannot be compared with those from other studies. In terms of amenability of this enzyme to applications, however, such parameters appear important.
FIG. 5.
Concentration-dependent effects on the efficiency of TetC. Depletion of halocatechols and formation of halomuconates at incubations of various concentrations of 4,5-dichlorocatechol (4,5-DCCat) (A), 3,4,5-trichlorocatechol (3,4,5-TCCat) (B), and tetrachlorocatechol (TeCCat) (C) by TetC are shown. Initial and residual concentrations were determined after 12 h by HPLC. Tetrachloromuconic acid formation was monitored by its cycloisomerization product, 2,3,5- trichlorodienelactone, which was formed after acidification of the assay. Transformation of 3,4,5-trichlorocatechol was followed by substrate depletion because of the high level of instability of its corresponding product, 2,3,4-trichloromuconic acid. Protein amounts displayed are rounded to whole numbers.
(iii) Inhibitory features.
In order to elucidate aspects of the inhibitory features of 4,5-dihalogenated catechols on the conversion of catechol and its lower chlorinated derivatives, transformation experiments with a mixture of catechol and 3-chlorocatechol, 4,5-dichlorocatechol, or tetrachlorocatechol were performed. These may provide deeper insight into the process of transformation than simple Ki determinations. Results shown in Fig. 6A clearly indicate that the conversion of catechol was completely inhibited until depletion of tetrachlorocatechol was exhaustive. This inhibition was clearly competitive. However, when tetrachlorocatechol and 3-chlorocatechol were simultaneously offered to TetC, the two compounds were converted at similar rates. Interestingly, the conversion of the latter compound was slightly slower than that of the former (Fig. 6C). Although there was some inactivation of TetC after 2 h, visible through a sharp decline of the rate of 4,5-dichlorocatechol turnover into 3,4-dichloromuconate, catechol was not significantly transformed into muconate (Fig. 6B). This, however, proceeded upon overnight incubation, after complete turnover of 4,5-dichlorocatechol (not shown). Similar to the results described for tetrachlorocatechol, the transformation of 3-chloro- and 4,5-dichlorocatechol proceeded almost in parallel, with simultaneous production of both corresponding chloromuconates (Fig. 6D). The formation of corresponding (chloro-)muconate was confirmed in all these experiments, except for tetrachloromuconate, which cycloisomerizes during analysis by HPLC. The inhibitory features described for TetC have never been reported for all hitherto known (chloro-)catechol dioxygenases.
FIG. 6.
Effects of 4,5-substituted halocatechols on conversion of catechol and 3-chlorocatechol. Transformation by TetC (21.2 μg of protein per ml) of 0.1 mM catechol is shown in the presence of equimolar concentrations of tetrachlorocatechol (A) and 4,5-dichlorocatechol (B). Muconate was detected as its corresponding dienelactone. Transformation of 0.1 mM 3-chlorocatechol was in the presence of equimolar concentrations of tetrachlorocatechol (C) and 4,5-dichlorocatechol (D). Concentrations of catechol (□), 3-chlorocatechol (▿), 4,5-dichlorocatechol (○), tetrachlorocatechol (⧫), muconic acid (▴), 2-chloromuconic acid (+), and 3,4-dichloromuconic acid (■) are shown.
(iiii) Stability of TetC over time.
The stability of the enzyme was monitored. A loss of 40% of the initial activity was detected when recombinant TetC was stored at 4°C. Storage at 20°C over the same period of time resulted in a loss of activity of 93%. Aliquots of the protein stored for 1 week at −70°C and for a long period, about 5 months, showed no measurable loss of specific activity after thawing and after subsequent storage on ice within 12 h.
Spectroscopic properties of recombinant TetC in the presence of its substrates.
The catalytic properties of TetC, featuring productive breakdown of 4,5-dihalogenated catechols, deserve a closer view at the active site. The absorption spectra of TetC were measured in the presence of these substrates, as were its EPR parameters. Such investigations had been performed earlier with the type II catechol 1,2-dioxygenase of P. putida AC27 but with catechol as the substrate only (9). Halogenated catechols and particularly those considered as strong inhibitors of this class of enzymes should be tested, therefore, with TetC.
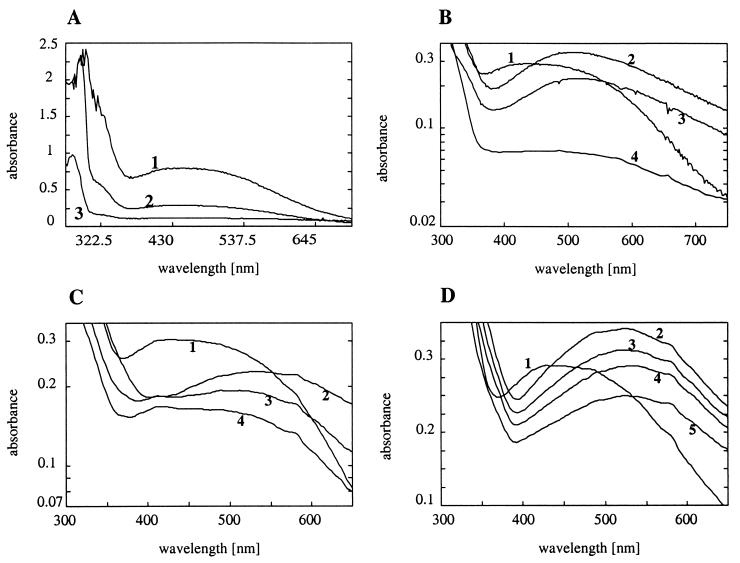
The electronic absorption spectra of the native enzyme and the enzyme-substrate complexes maintained under anaerobic conditions are shown in Fig. 7. The broad absorption spectrum of TetC with a maximum of 444 nm is typical for a ligand to Fe(III) charge-transfer transition due to tyrosinate coordination (Fig. 7A) (9). The addition of 3-chlorocatechol to the protein incubated in an anaerobic environment resulted in an increase of its absorbance and led to a red shift of its initial absorption maximum to 506 nm (Fig. 7B). Successive dilution of the anaerobic reaction mixture with air-saturated buffer resulted in an increasing absorption at 260 nm, the absorption maximum of the reaction product, 2-chloromuconic acid. Depletion of 3-chlorocatechol, however, did not result in the expected clear back-shift to its initial absorption spectrum. The addition of 0.2 mM tetrachlorocatechol to the relatively high concentration, 0.1 mM, of TetC (Fig. 7C) shifted the maximum absorption to 560 nm, but with significantly reduced intensity. Successive dilution with air-saturated buffer then backshifted the maximum absorption to the initial maximum of the protein, indicative of complete transformation of the substrate. The experimental complex of TetC with 4,5-dichlorocatechol showed a maximum absorption at 524 nm (Fig. 7D). No change of the absorption spectrum was recorded during addition of air-saturated buffer, although some slow transformation of this substrate had been detected as described above by spectrophotometrical tests and HPLC analysis. It seems probable that residual substrate was still bound to the active center of the enzyme, confirming the strong inhibitory character of 4,5-dichlorocatechol. Not only sterical but also electronic features with regard to inhibition of ortho-cleaving enzymes have to be taken into account, and 4,5-dichlorocatechol should better fit into the active site cavity of TetC than the more bulky substrate tetrachlorocatechol. An excellent and detailed study on inhibitory complexes with protocatechuate 3,4-dioxygenase has been published recently (44), with halogenated substitutes of protocatechuate being the most potent inhibitors.
FIG. 7.
Electronic absorption spectra of TetC. (A) The absorption of a preparation of purified enzyme in 33 mM Tris-HCl buffer–50 mM NaCl (pH 8.0) was recorded at room temperature. The dimer solution (curve numbers are in parentheses) was 0.3 mM (1), 0.1 mM (2), and 0.01 mM (3). (B) Electronic absorption spectra of 0.1 mM dimeric polypeptide TetC (1) and the enzyme-substrate complex with 0.1 mM 3-chlorocatechol in 33 mM Tris-HCl–50 mM NaCl (2). The addition of 100 μl of air-saturated buffer resulted in curve 3, and a fourfold dilution with buffer resulted in curve 4. (C) Absorption spectra of the enzyme (1) and the enzyme-substrate complex (2) with 0.2 mM tetrachlorocatechol. Spectra upon addition of 200 μl (3) and 400 μl (4) of air-saturated buffer are shown. (D) Native enzyme (1) and the enzyme-substrate complex (2) in the presence of 0.1 mM 4,5-dichlorocatechol. Spectra upon addition of 50 μl (3), 100 μl (4), and 200 μl (5) of air-saturated buffer are shown. All absorption spectra of enzyme-substrate complexes were anaerobically recorded at a concentration of 0.1 mM dimeric polypeptide.
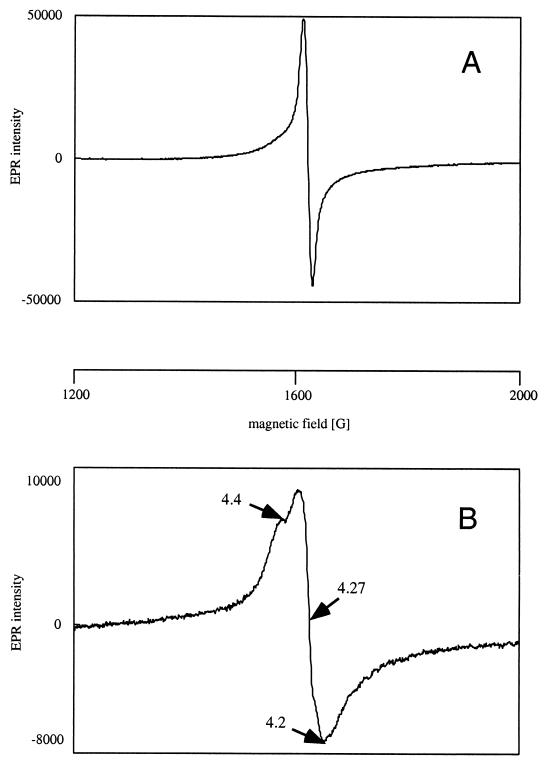
Further, EPR spectroscopic studies of the recombinant TetC dioxygenase were carried out in the presence of tetrachlorocatechol in order to partially characterize the environment of the Fe(III) ligands (Fig. 8). The sharp symmetrical electron spin resonance signal recorded for TetC without the substrate at 4.3 G (Fig. 8A) is in full agreement with high-spin Fe(III) in a rhombic environment as previously found for other intradiol dioxygenases (8, 9). Weak resonances centering at 9.8 G were observed outside the plotted range. The EPR analysis of the same sample maintained in an argon atmosphere immediately after addition of an excess of tetrachlorocatechol revealed different g values (Fig. 8B). The signal obtained for TetC is anisotropic with all possible g values from a particular Kramer's doublet: the system showed three different g values: gx = 4.27, gy = 4.2, and gz = 4.4. The low-field signal at 9.8 G broadened during the experiment and finally disappeared.
FIG. 8.
EPR spectra of native TetC. Spectra were recorded in the absence (A) and in the presence (B) of 1.0 mM tetrachlorocatechol. The concentration of the protein sample was made 0.3 mM by anaerobic dilution in 50 mM Tris-HCl, pH 8.0. The substrate solution was made anaerobic prior to addition and was added to the protein under argon flow. The protein was immediately frozen in liquid nitrogen and analyzed. Both spectra were recorded at 10 K, with a microwave frequency of 9,654 MHz, a microwave power of 0.1 mW, and a modulation amplitude of 10 G. The strength of the magnetic field is expressed in gauss (G).
Spectroscopic investigations on Fe(III) catecholate complexes indicated that the active-site iron should remain in the Fe(III) oxidation state during the whole catalytic cycle (17). The multidentate ligands influence the physical properties of the metal complex: electron-donating substituents of enzyme substrates foster an increase in catalytic activity, whereas electron-withdrawing groups, like the four chlorines of tetrachlorocatechol, decrease the dioxygen attack by forming a stable complex. Crystals of this compound with incorporated tetrachlorocatechol were shown to exhibit an asymmetrically chelated tetrachlorocatecholate ligand (17). Apparently, the iron content of the protein is not alone responsible for the differences in activity. Stabilizing effects through iron-catecholate complexes associated with conformational changes, or adverse effects (inactivation by denaturing agents was accelerated by catechol), were mentioned in earlier studies on intradiol dioxygenases (39). In order to elucidate the chelating attack of chlorocatechols of the 4,5-substitution type on the central catalytic iron atom leading to the inhibition of this enzyme, a possibility already discussed earlier (9), a preparation of TetC (ca. 1 μM) was incubated with an excess of tetrachlorocatechol (1 mM). A pinkish-violet color was formed, which had been observed earlier in some of the growth experiments performed with strain RW71 (49). This was possibly due to the inefficient induction of the expression of chlorocatechol pathway genes by the product of the TetC reaction, tetrachloromuconic acid, in the necessarily productive triggering by the transcriptional activator, TetR. In our experiments, the UV-visible spectrum of the tetrachlorocatechol-enzyme [iron(III)] complex proved to be identical with that obtained with ferric iron in the presence of 4,5-halogenated catechols, especially with tetrachlorocatechol. We found that the central ferric iron atom of recombinant TetC was removed from the active site of the holoenzyme in the presence of 1 mM tetrachlorocatechol. This observation was unambiguously proved by filtration of the solution through a 10,000-Da exclusion membrane and subsequent control of compartmentation of the polypeptide through protein determinations. Unfortunately, we were not able to apply this assay for the quantitative titration of the overall iron content of TetC.
Conclusions.
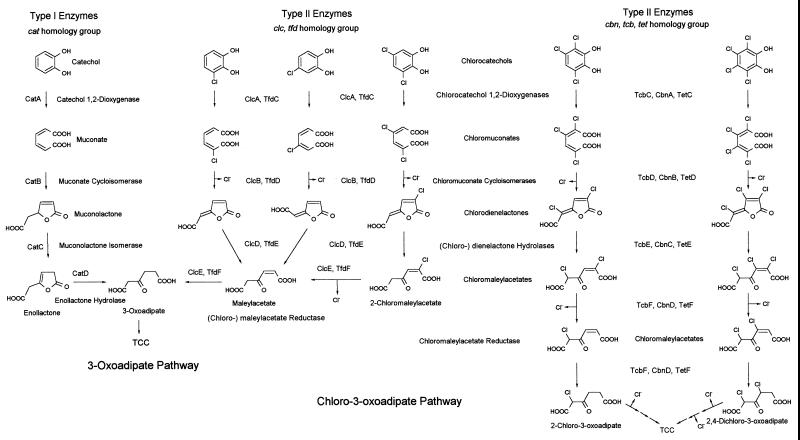
The catechol and chlorocatechol 1,2-dioxygenases are Fe(III)-containing dioxygenases. Based on their substrate ranges, they are historically distinguished as type I and type II enzymes, respectively. Type I represents enzymes that primarily convert catechol, whereas type II enzymes are induced during growth with (low-halogenated) chloromuconates. The type II enzymes have a wider substrate range and transform chlorinated catechols more rapidly than catechol. However, they are strongly inhibited by halocatechols simultaneously substituted in positions 4 and 5. Phylogenetic trees of ortho-cleaving dioxygenases have been published recently (3, 19). Based on sequence homologies, five subgroups may be distinguished within the chlorocatechol 1,2-dioxygenase family. This sequence distinction might be correlated with substrate specificities. Indeed, very few sequence changes, only 3 out of 260 amino acids, were shown to significantly contribute to alterations of the substrate preference of a chlorocatechol 1,2-dioxygenase (35). However, for such a functional comparison, the available experimental data for all members of this group are not sufficient, since the respective investigations did not comprise the testing of all relevant substrates. In the present study, TetC from P. chlororaphis RW71 was shown to exhibit a broader substrate range than classical type II dioxygenases and to be distinguished from these enzymes by its ability to productively convert 4,5-substituted chlorocatechols. For a functional comparison of this type of dioxygenase, a type III enzyme could be proposed, with TetC being the archetype, differentiating the chlorocatechol dioxygenases able to convert 4,5-substituted chlorocatechols. Additional experimental data on TcbC and CbnA are required and may confirm a high similarity of substrate range among these enzymes, possibly with those of strains PS12 and PS14 (63).
In order to better understand why catechol and chlorocatechol 1,2-dioxygenases exhibit such a diverse range of substrate preferences, three-dimensional structures of representatives of the different types described above should be compared. A more specific focus on the active site is required. For this purpose, the crystallization and three-dimensional structure resolution of recombinant TetC are currently under investigation. The specificity for the turnover of chlorocatechols of a high degree of halogenation and for substrates with halogen substituents at positions 4 and 5 of the catecholic ring system makes this group of enzymes at least significant among the already well-characterized catechol 1,2-dioxygenases and chlorocatechol 1,2-dioxygenases (51, 57). The subgroup partially characterized in this work thus may represent a novel alternative for the degradation of highly halogenated diphenolic compounds which are also mineralized through the chlorohydroxyhydroquinone pathway. The discussed reaction sequences are compared in Fig. 9 which, therefore, must be preliminary at present. It is not fully clear to which of the three branches those pathways for the mineralization of 3,4-dichloro- and 3, 6-dichlorocatechol (51, 59, 68) have to be assigned, because of the lack of experimental data. On the basis of specificity constants, the enzymes characterized by Broderick and O'Halloran (9) and Maltseva et al. (34) group with the classical type II pyrocatechase of Pseudomonas sp. strain B13 (16). All of those enzymes have 4-chlorocatechol as the preferred substrate and are evolutionarily distant from the proposed type III group. Consequently, some further work needs to be dedicated to the comparative experimental confirmation of the biochemical properties of all these enzymes, e.g., their expected high turnover rates and high affinities for polyhalogenated intermediates in the pathways for the degradation of polyhalocatechols. Apart from the halocatechols, these intermediary substrates are not commercially available and have to be produced biologically with the help of the corresponding enzymes. It is evident that the potentially different types of transcriptional activators especially may exhibit very high selectivity for the triggering compounds or show different and/or overlapping broad ranges of substrate tolerance. These polypeptides are assumed to discriminate between the individual structures of inducers, the monochloromuconic acids, and the more highly chlorinated homologues. It is quite probable that this is also true for the much more highly halogenated muconates in the breakdown of polychlorocatechols and, therefore, the specificity of transcriptional activation of these operons should be further investigated. Finally, our results suggest using strong caution in the interpretation of data derived from enzyme kinetics on wild-type and recombinant catechol 1,2-dioxygenases, especially with respect to the homogeneity of the preparation.
FIG. 9.
Comparison of catabolic pathways. Degradative sequences are shown for the mineralization of catechol (type I enzymes), monochloro- and 3,5-dichlorocatechol(s) (type II enzymes), and the polychlorocatechols (possibly type III enzymes), possibly not being fully dehalogenated through the type II pathway sequence.
ACKNOWLEDGMENTS
Many thanks to Y. Jouanneau (CEA—Grenoble, Grenoble, France) for enabling a short stay in his laboratory, providing the possibility to record absorption spectra under strictly anaerobic conditions. We thank J. Gaillard (CEA—Grenoble) for recording the EPR spectra and are grateful for his enthusiastic discussions. We thank F. Halgand (CNRS-Institut de Chimie des Substances Naturelles, Gif/Yvette, France) for recording mass spectra of TetC and for the discussions on the iron-to-subunit ratio of TetC. We are indebted to H.-J. Hecht and S. Weissflog (GBF) for collaboration in initial crystallization attempts, to H.-A. Arfmann for synthesizing halocatechols, and to K. N. Timmis for providing bench space.
This work was mainly supported by the Deutsche Forschungsgemeinschaft (grants Wi 1226/2-1 and 1226/2-2).
REFERENCES
- 1.Adams M D, Wagner L M, Graddis T J, Landick R, Antonucci T K, Gibson A L, Oxender D L. Nucleotide sequence and genetic characterization reveal six essential genes for the LIV-1 and LS transport systems of Escherichia coli. J Biol Chem. 1990;265:11436–11443. [PubMed] [Google Scholar]
- 1a.Alexander M. Biodegradation of chemicals of environmental concern. Science. 1981;211:132–138. doi: 10.1126/science.7444456. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armengaud J, Timmis K N, Wittich R-M. A functional 4-hydroxysalicylate/hydroxyquinol degradative pathway gene cluster is linked to the initial dibenzo-p-dioxin pathway genes in Sphingomonassp. strain RW1. J Bacteriol. 1999;181:3452–3461. doi: 10.1128/jb.181.11.3452-3461.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beil S, Happe B, Timmis K N, Pieper D H. Genetic and biochemical characterization of the broad spectrum chlorobenzene dioxygenase from Burkholderiasp. strain PS12—dechlorination of 1,2,4,5-tetrachlorobenzene. Eur J Biochem. 1997;247:190–199. doi: 10.1111/j.1432-1033.1997.00190.x. [DOI] [PubMed] [Google Scholar]
- 5.Bhat M A, Ishida T, Horiike K, Vaidyanathan C S, Nozaki M. Purification of 3,5-dichlorocatechol 1,2-dioxygenase, a nonheme iron dioxygenase and a key enzyme in the biodegradation of a herbicide, 2,4-dichlorophenoxyacetic acid (2,4-D), from Pseudomonas cepaciaCSV90. Arch Biochem Biophys. 1993;300:738–746. doi: 10.1006/abbi.1993.1102. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Briganti F, Mangani S, Pedocci L, Scozzafava A, Golovleva L A, Jadan A P, Solyanikova L P. XAS characterization of the active sites of novel intradiol ring-cleaving dioxygenases: hydroxyquinol and chlorocatechol dioxygenases. FEBS Lett. 1998;433:58–62. doi: 10.1016/s0014-5793(98)00884-9. [DOI] [PubMed] [Google Scholar]
- 8.Briganti F, Pessione E, Giunta C, Scozzafava A. Purification, biochemical properties and substrate specificity of a catechol 1,2-dioxygenase from a phenol degrading Acinetobacter radioresistens. FEBS Lett. 1997;416:61–64. doi: 10.1016/s0014-5793(97)01167-8. [DOI] [PubMed] [Google Scholar]
- 9.Broderick J B, O'Halloran T. Overproduction, purification, and characterization of chlorocatechol dioxygenase, a non-heme iron dioxygenase with broad substrate tolerance. Biochemistry. 1991;30:7349–7358. doi: 10.1021/bi00243a040. [DOI] [PubMed] [Google Scholar]
- 10.Bull C, Ballou D. Purification and properties of protocatechuate 3,4-dioxygenase from Pseudomonas putida: a new iron to subunit stoichiometry. J Biol Chem. 1981;256:12673–12680. [PubMed] [Google Scholar]
- 11.Bull C, Ballou D P, Otsuka S. The reaction of oxygen with protocatechuate 3,4-dioxygenase from Pseudomonas putida: characterization of a new oxygenated intermediate. J Biol Chem. 1981;256:12681–12686. [PubMed] [Google Scholar]
- 12.Chatterjee D K, Kellogg S T, Hamada S, Chakrabarty A M. Plasmid specifying total degradation of 3-chlorobenzoate by a modified orthopathway. J Bacteriol. 1981;146:639–646. doi: 10.1128/jb.146.2.639-646.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daubaras D L, Hershberger C D, Kitano K, Chakrabarty A M. Sequence analysis of a gene cluster involved in metabolism of 2,4,5-trichlorophenoxyacetic acid by Burkholderia cepaciaAC1100. Appl Environ Microbiol. 1995;61:1279–1289. doi: 10.1128/aem.61.4.1279-1289.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Don R H, Weightman A J, Knackmuss H-J, Timmis K N. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophusJMP134(pJP4) J Bacteriol. 1985;161:85–90. doi: 10.1128/jb.161.1.85-90.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorn E, Knackmuss H-J. Chemical structure and biodegradability of halogenated aromatic compounds: two catechol 1,2-dioxygenases from a 3-chlorobenzoate-grown pseudomonad. Biochem J. 1978;174:73–84. doi: 10.1042/bj1740073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorn E, Knackmuss H-J. Chemical structure and biodegradability of halogenated aromatic compounds: substituent effects on 1,2-dioxygenation of catechol. Biochem J. 1978;174:85–94. doi: 10.1042/bj1740085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duda M, Pascaly M, Krebs B. A highly reactive functional model for catechol 1,2-dioxygenase: reactivity studies of iron (III) catecholate complexes of bis((2-pyridyl)methyl)((1-methylimidazol-2-yl)methyl) amine. Chem Commun. 1997;1997:835–836. [Google Scholar]
- 18.Earhart C A, Hall M D, Michaud-Soret I, Que L, Jr, Ohlendorf D H. Crystallization of catechol 1,2-dioxygenase from Pseudomonas arvillaC-1. J Mol Biol. 1994;236:377–378. doi: 10.1006/jmbi.1994.1144. [DOI] [PubMed] [Google Scholar]
- 19.Eulberg D, Kourbatova E M, Golovleva L A, Schlömann M. Evolutionary relationship between chlorocatechol catabolic enzymes from Rhodococcus opacus1CP and their counterparts in proteobacteria: sequence divergence and functional convergence. J Bacteriol. 1998;180:1082–1094. doi: 10.1128/jb.180.5.1082-1094.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fersht A. Enzyme structure and mechanism. 2nd ed. New York, N.Y: W. H. Freeman and Co.; 1985. [Google Scholar]
- 21.Frantz B, Chakrabarty A M. Organization and nucleotide sequence determination of a gene cluster involved in 3-chlorocatechol degradation. Proc Natl Acad Sci USA. 1987;84:4460–4464. doi: 10.1073/pnas.84.13.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagen W R. EPR spectroscopy of iron-sulfur proteins. Adv Inorg Chem. 1992;38:165–216. [Google Scholar]
- 23.Haigler B E, Nishino S F, Spain J C. Degradation of 1,2-dichlorobenzene by a Pseudomonassp. Appl Environ Microbiol. 1988;54:294–301. doi: 10.1128/aem.54.2.294-301.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harayama S, Rekik M. Bacterial aromatic ring-cleavage enzymes are classified into two different gene families. J Biol Chem. 1989;264:15328–15333. [PubMed] [Google Scholar]
- 25.Hartnett C, Neidle E L, Ngai K-L, Ornston L N. DNA sequences of genes encoding Acinetobacter calcoaceticusprotocatechuate 3,4-dioxygenase: evidence indicating shuffling of genes and of DNA sequences within genes during their evolutionary divergence. J Bacteriol. 1990;172:956–966. doi: 10.1128/jb.172.2.956-966.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoshino T, Kose K. Cloning, nucleotide sequences, and identification of products of the Pseudomonas aeruginosa PAO bragenes, which encode the high-affinity branched-chain amino acid transport system. J Bacteriol. 1990;172:5531–5539. doi: 10.1128/jb.172.10.5531-5539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kivisaar M, Kasak L, Nurk A. Sequence of the plasmid-encoded catechol 1,2-dioxygenase expressing gene, pheB, of phenol-degrading Pseudomonassp. strain EST1001. Gene. 1991;98:15–20. doi: 10.1016/0378-1119(91)90098-v. [DOI] [PubMed] [Google Scholar]
- 28.Koiv V, Marits R, Heinaru A. Sequence analysis of the 2,4-dichlorophenol hydroxylase gene tfdB and 3,5-dichlorocatechol 1,2-dioxygenase gene tfdCof 2,4-dichlorophenoxyacetic acid degrading plasmid pEST4011. Gene. 1996;174:293–297. doi: 10.1016/0378-1119(96)00043-1. [DOI] [PubMed] [Google Scholar]
- 29.Kukor J J, Olsen R H, Siak J-S. Recruitment of a chromosomally encoded maleylacetate reductase for degradation of 2,4-dichlorophenoxyacetic acid by plasmid pJP4. J Bacteriol. 1989;171:3385–3390. doi: 10.1128/jb.171.6.3385-3390.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Leveau J H J, van der Meer J R. The tfdR gene product can successfully take over the role of the insertion element-inactivated TfdT protein as a transcriptional activator of the tfdCDEF gene cluster, which encodes chlorocatechol degradation in Ralstonia eutrophaJMP134(pJP4) J Bacteriol. 1996;178:6824–6832. doi: 10.1128/jb.178.23.6824-6832.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leveau J H J, van der Meer J R. Genetic characterization of insertion sequence ISJP4 on plasmid pJP4 from Ralstonia eutrophaJMP134. Gene. 1997;202:103–114. doi: 10.1016/s0378-1119(97)00460-5. [DOI] [PubMed] [Google Scholar]
- 33.Lipscomb J D, Orville A M. Mechanistic aspects of dihydroxybenzoate dioxygenases. In: Sigel H, Sigel A, editors. Metal ions in biological systems. Vol. 28. New York, N.Y: Marcel Dekker, Inc.; 1992. pp. 243–298. [Google Scholar]
- 34.Maltseva O V, Solyanikova I P, Golovleva L A. Chlorocatechol 1,2-dioxygenase from Rhodococcus erythropolis1CP. Kinetic and immunochemical comparison with analogous enzymes from gram-negative strains. Eur J Biochem. 1994;226:1053–1061. doi: 10.1111/j.1432-1033.1994.01053.x. [DOI] [PubMed] [Google Scholar]
- 35.Miguez C B, Greer C W, Ingram J M. Purification and properties of chlorocatechol 1,2-dioxygenase from Alcaligenes denitrificansBRI6011. Can J Microbiol. 1993;39:1–5. [Google Scholar]
- 36.Nakai C, Horiike K, Kuramitsu S, Kagamiyama H, Nozaki M. Three isoenzymes of catechol 1,2-dioxygenase (pyrocatechase), αα, αβ, and ββ, from Pseudomonas arvillaC-1. J Biol Chem. 1990;265:660–665. [PubMed] [Google Scholar]
- 37.Nakai C, Nakazawa T, Nozaki M. Purification and properties of catechol 1,2-dioxygenase (pyrocatechase) from Pseudomonas putida mt-2 in comparison with that from Pseudomonas arvillaC-1. Arch Biochem Biophys. 1988;267:701–713. doi: 10.1016/0003-9861(88)90079-3. [DOI] [PubMed] [Google Scholar]
- 38.Nakai C, Uyeyma H, Kagamiyama H, Nakazawa T, Inouye S, Kishi F, Nakazawa A, Nozaki M. Cloning, DNA sequencing, and amino acid sequencing of catechol 1,2-dioxygenases (pyrocatechase) from Pseudomonas putida mt-2 and Pseudomonas arvillaC-1. Arch Biochem Biophys. 1995;321:353–362. doi: 10.1006/abbi.1995.1405. [DOI] [PubMed] [Google Scholar]
- 39.Nakazawa T, Nozaki M, Hayaishi O. Studies on pyrocatechase. J Biol Chem. 1969;244:119–125. [PubMed] [Google Scholar]
- 40.Neidle E L, Hartnett C, Bonitz S, Ornston L N. DNA sequence of the Acinetobacter calcoaceticus catechol 1,2-dioxygenase I structural gene catA: evidence for evolutionary divergence of intradiol dioxygenases of DNA sequence repetitions. J Bacteriol. 1988;170:4874–4880. doi: 10.1128/jb.170.10.4874-4880.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogawa N, Miyashita K. The chlorocatechol-catabolic transposon Tn5707 of Alcaligenes eutrophus NH9, carrying a gene cluster highly homologous to that in the 1,2,4-trichlorobenzene-degrading bacterium Pseudomonassp. strain P51, confers the ability to grow on 3-chlorobenzoate. Appl Environ Microbiol. 1999;65:724–731. doi: 10.1128/aem.65.2.724-731.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohlendorf D H, Lipscomb J D, Weber P C. Structure and assembly of protocatechuate 3,4-dioxygenase. Nature. 1988;336:403–405. doi: 10.1038/336403a0. [DOI] [PubMed] [Google Scholar]
- 43.Ohlendorf D H, Orville A M, Lipscomb J D. Structure of protocatechuate 3,4-dioxygenase from Pseudomonas aeruginosaat 2.15 Å resolution. J Mol Biol. 1994;244:586–608. doi: 10.1006/jmbi.1994.1754. [DOI] [PubMed] [Google Scholar]
- 44.Orville A M, Elango N, Lipscomb J D, Ohlendorf D H. Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site. Biochemistry. 1997;36:10039–10051. doi: 10.1021/bi970468n. [DOI] [PubMed] [Google Scholar]
- 45.Patel R N, Hou C T, Felix A, Lillard M O. Catechol 1,2-dioxygenase from Acinetobacter calcoaceticus: purification and properties. J Bacteriol. 1976;127:536–544. doi: 10.1128/jb.127.1.536-544.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pearson C R. Halogenated aromatics. In: Hutzinger O, editor. The handbook of environmental chemistry—anthropogenic compounds. New York, N.Y: Springer Verlag; 1982. pp. 86–116. [Google Scholar]
- 47.Perkins E J, Gordon M P, Caceres O, Lurquin P F. Organization and sequence analysis of the 2,4-dichlorophenol hydroxylase and dichlorocatechol oxidative operons of plasmid pJP4. J Bacteriol. 1990;172:2351–2359. doi: 10.1128/jb.172.5.2351-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Potrawfke T, Löhnert T-H, Timmis K N, Wittich R-M. Mineralization of low-chlorinated biphenyls by Burkholderiasp. strain LB400 and by a two-membered consortium upon directed interspecies transfer of chlorocatechol pathway genes. Appl Microbiol Biotechnol. 1998;50:440–446. [Google Scholar]
- 49.Potrawfke T, Timmis K N, Wittich R-M. Degradation of 1,2,3,4-tetrachlorobenzene by Pseudomonas chlororaphisRW71. Appl Environ Microbiol. 1998;64:3798–3806. doi: 10.1128/aem.64.10.3798-3806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ravatn R, Studer S, Springael D, Zehnder A J B, van der Meer J R. Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonassp. strain B13. J Bacteriol. 1998;180:4360–4369. doi: 10.1128/jb.180.17.4360-4369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reineke W. Development of hybrid strains for the mineralization of chloroaromatics by patchwork assembly. Annu Rev Microbiol. 1998;52:287–331. doi: 10.1146/annurev.micro.52.1.287. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 53.Sander P. Bakterieller Abbau halogenierter Benzole. Ph.D. thesis. Hamburg, Germany: University of Hamburg; 1991. [Google Scholar]
- 54.Sander P, Wittich R-M, Fortnagel P, Wilkes H, Francke W. Degradation of 1,2,4-trichloro- and 1,2,4,5-tetrachlorobenzene by Pseudomonasstrains. Appl Environ Microbiol. 1991;57:1430–1440. doi: 10.1128/aem.57.5.1430-1440.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sauret-Ignaci G, Gagnon J, Béguin C, Barelle M, Markowicz Y, Pelmont J, Toussaint A. Characterization of a chromosomally encoded catechol 1,2-dioxygenase (E. C. 1.13.11.1) from Alcaligenes eutrophusCH34. Arch Microbiol. 1996;166:42–50. doi: 10.1007/s002030050353. [DOI] [PubMed] [Google Scholar]
- 56.Schlömann M. Evolution of chlorocatechol catabolic pathways. Biodegradation. 1994;5:301–320. doi: 10.1007/BF00696467. [DOI] [PubMed] [Google Scholar]
- 57.Schlömann M, Pieper D H, Knackmuss H-J. Enzymes of haloaromatics degradation: variations of Alcaligenes on a theme by Pseudomonas. In: Silver S, Chakrabarty A M, Iglewski B, Kaplan S, editors. Pseudomonas: biotransformations, pathogenesis, and evolving biotechnology. Washington, D.C.: American Society for Microbiology; 1990. pp. 185–196. [Google Scholar]
- 58.Sommer C, Görisch H. Enzymology of the degradation of (di) chlorobenzenes by Xanthobacter flavus14pl. Arch Microbiol. 1997;167:384–391. doi: 10.1007/s002030050459. [DOI] [PubMed] [Google Scholar]
- 59.Spain J C. Metabolic pathways for biodegradation of chlorobenzenes. In: Silver S, Chakrabarty A M, Iglewski B, Kaplan S, editors. Pseudomonas: biotransformations, pathogenesis, and evolving biotechnology. Washington, D.C.: American Society for Microbiology; 1990. pp. 197–206. [Google Scholar]
- 60.Strachan P D, Freer A A, Fewson C A. Purification and characterization of catechol 1,2-dioxygenase from Rhodococcus rhodochrous NCIMB 13259 and cloning and sequencing of its catAgene. Biochem J. 1998;333:741–747. doi: 10.1042/bj3330741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan H-M. Bacterial catabolic transposons. Appl Microbiol Biotechnol. 1999;51:1–12. doi: 10.1007/s002530051356. [DOI] [PubMed] [Google Scholar]
- 62.True A E, Orville A M, Pearce L L, Lipscomb J D. An EXAFS study of the interaction of substrate with the ferric active site of protocatechuate 3,4-dioxygenase. Biochemistry. 1990;29:10847–10854. doi: 10.1021/bi00500a019. [DOI] [PubMed] [Google Scholar]
- 63.Uhde H. Aufreinigung und Charakterisierung der Chlorcatechol 1,2-dioxygenase und der Chlormuconatcycloisomerase aus Acidovorax sp. PS14. Ph.D. dissertation. Hamburg, Germany: University of Hamburg; 1998. [Google Scholar]
- 64.van der Meer J R. Evolution of novel metabolic pathways for the degradation of chloroaromatic compounds. Antonie van Leeuwenhoek. 1997;71:159–178. doi: 10.1023/a:1000166400935. [DOI] [PubMed] [Google Scholar]
- 65.van der Meer J R, Eggen R I L, Zehnder A J B, de Vos W M. Sequence analysis of the Pseudomonas sp. strain P51 tcbgene cluster, which encodes metabolism of chlorinated catechols: evidence for specialization of catechol 1,2-dioxygenases for chlorinated substrates. J Bacteriol. 1991;173:2425–2434. doi: 10.1128/jb.173.8.2425-2434.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van der Meer J R, Frijters A C J, Leveau J H J, Eggen R I L, Zehnder A J B, de Vos W M. Characterization of the Pseudomonas sp. strain P51 gene tcbR, a LysR-type transcriptional activator of the tcbCDEFchlorocatechol oxidative operon, and analysis of the regulatory region. J Bacteriol. 1991;173:3700–3708. doi: 10.1128/jb.173.12.3700-3708.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van der Meer J R, Harayama S, Zehnder A J B, de Vos W M. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev. 1992;56:677–694. doi: 10.1128/mr.56.4.677-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van der Meer J R, van Nerven A R W, de Vries E J, de Vos W M, Zehnder A J B. Cloning and characterization of plasmid-encoded genes for the degradation of 1,2-dichloro-, 1,4-dichloro-, and 1,2,4-trichlorobenzene of Pseudomonassp. strain P51. J Bacteriol. 1991;173:6–15. doi: 10.1128/jb.173.1.6-15.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van der Meer J R, Zehnder A J B, de Vos W M. Identification of a novel composite transposable element, Tn5280, carrying chlorobenzene dioxygenase genes of Pseudomonassp. strain P51. J Bacteriol. 1991;173:7077–7083. doi: 10.1128/jb.173.22.7077-7083.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walsh T A, Ballou D P. Halogenated protocatechuates as substrates for protocatechuate dioxygenase from Pseudomonas cepacia. J Biol Chem. 1983;258:14413–14421. [PubMed] [Google Scholar]
- 71.Walsh T A, Ballou D P, Mayer R, Que L., Jr Rapid reaction studies on the oxygenation reactions of catechol dioxygenase. J Biol Chem. 1983;258:14422–14427. [PubMed] [Google Scholar]
- 72.Wyndham R C, Cashore A E, Nakatsu C H, Peel M C. Catabolic transposons. Biodegradation. 1994;5:323–334. doi: 10.1007/BF00696468. [DOI] [PubMed] [Google Scholar]
- 73.Zaborina O, Latus M, Eberspächer J, Golovleva L A, Lingens F. Purification and characterization of 6-chlorohydroxyquinol 1,2-dioxygenase from Streptomyces rochei 303: comparison with an analogous enzyme from Azotobactersp. strain GP1. J Bacteriol. 1995;177:229–234. doi: 10.1128/jb.177.1.229-234.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]