Abstract
The KorB and TrbA proteins of broad-host-range plasmid RK2 are key regulators of the plasmid genes required for conjugative transfer. trbBp is the primary promoter responsible for expression of mating pair formation genes. We show that despite the targets for KorB and TrbA at trbBp being about 165 bp apart, 189 bp upstream of the transcription start point and overlapping the −10 region, respectively, these two proteins show up to 10-fold cooperativity for the repression of trbBp. Deletion analysis of TrbA showed that the C-terminal domain (CTD), which has a high degree of sequence conservation with the CTD of KorA, is required for this cooperativity with KorB. Western blotting demonstrated that the apparently mutual enhancement of repression is not due simply to elevation of repressor level by the presence of the second protein, suggesting that the basis for cooperativity is interaction between KorB and TrbA bound at their respective operators.
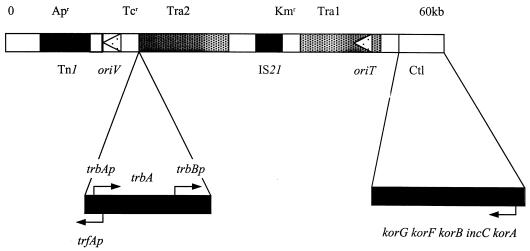
The self-transmissible, broad-host-range plasmid RK2 (indistinguishable from RP4 and RP1) has been studied in great detail (17). Its replication, partitioning, and transfer functions show many features of interest, but its most striking aspect is the complex regulatory circuitry which coordinates expression of the genes for all these basic functions. The regulatory proteins responsible for this coordination are KorA and KorB, encoded in the central control operon; KorC, encoded in the klc operon; and TrbA, encoded before the trb operon. The study described in this paper focused on the interactions between KorB and TrbA at trbBp (Fig. 1), since this is a key point in regulating the expression of genes for conjugative transfer (26): trbBp directs transcription of the whole of the trb operon, which encodes all but one of the genes needed for pilus synthesis and mating pair formation.
FIG. 1.
Linear map of the RK2 plasmid showing the main features of its 60-kb genome: transposable elements Tn1 and IS21 (black blocks), antibiotic resistance markers as indicated in Materials and Methods, origin of vegetative replication (oriV), and two blocks of transfer genes, Tra1 and Tra2 (dark grey blocks), including origin of transfer oriT. Arrowheads for oriV and oriT indicate the direction of replication or transfer. Enlarged are the trfAp-trbAp-trbBp region with the trbA gene and the central control operon, Ctl. Arrows by trfAp, trbAp, and trbBp indicate transcription start points and direction of transcription. The arrow above korA indicates the single promoter which is responsible for transcription of all five cistrons in the central control operon.
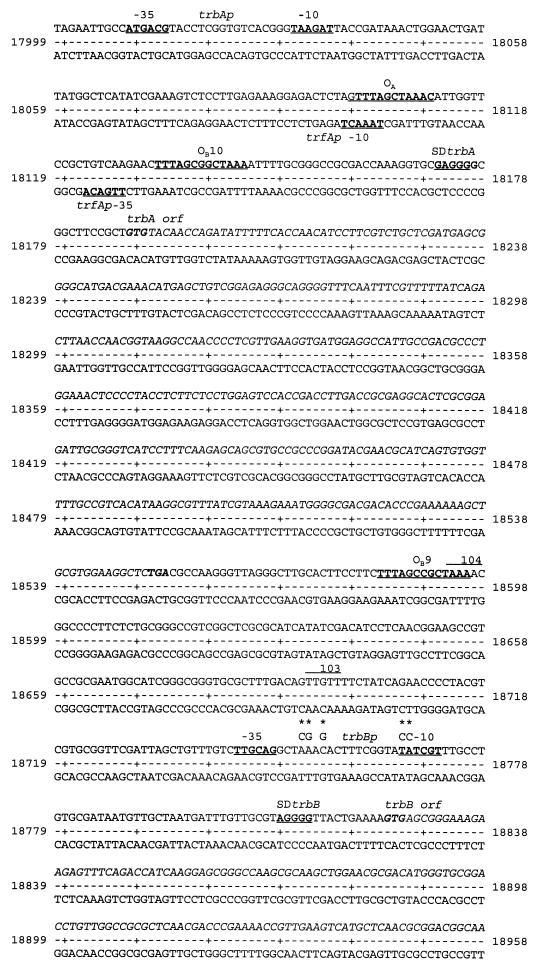
KorB binds specifically to twelve places on the plasmid genome (KorB operators OB1 to OB12) (1, 24), with a hierarchy of binding affinities (10). Five of these sites (OB4, OB5, OB7, OB8, and OB9) are within the regions encoding transfer genes, but none of them are very close to promoters, and only one of them (OB9) is known to mediate KorB repression of transcription. OB9 is 189 bp upstream of trbBp (Fig. 2) (24), and KorB is a strong repressor of trbBp (22). It has been proposed that KorB may cause looping between OB9 and a possible degenerate operator overlapping the trbBp −35 region (22), but this was excluded as a necessary element for KorB repression (8). This raises the possibility that KorB bound at a distance from the promoter makes direct contact with RNA polymerase through the creation of a DNA loop.
FIG. 2.
Sequence of the trfAp-trbAp-trbBp region. Coordinates refer to the complete sequence of RK2, available as GenBank accession no. LZ7758. −35 and −10 regions of relevant promoters are indicated. OA, KorA operator; OB, KorB operator; SD, Shine-Dalgarno sequence. Asterisks and alternative letters show mutations in either the putative degenerate KorB operator (first three, from left to right) or in the −10 region of the trbBp region (fourth and fifth). The primers used for amplification of promoter fragments and TrbA deletions by PCR (described in Materials and Methods) correspond to the following coordinates: 1, 18190 to 18212; 2, 18259 to 18278; 3, 18242 to 18263; 4, 18449 to 18431; 5, 18498 to 18477; 6, 18506 to 18489; 7, 18514 to 18493; 8, 18518 to 18501; 9, 18527 to 18508; 10, 18554 to 18534; 11, 18716 to 18735; 12, 18742 to 18759; 13, 18790 to 18776; 14, 18915 to 18896; and 15, 18397 to 18414. orf, open reading frame.
The binding site for TrbA is not known, but it definitely represses transcription from the promoters traGp, traJp, traKp (25), and trbBp (7). Evidence presented in this paper indicates that the target for TrbA is within the trbB promoter itself, overlapping the −10 region. In addition to defining the targets for KorB and TrbA alone more closely and thus making it clear that these sites are separated by at least 140 bp, we demonstrate that there is cooperativity between KorB and TrbA and that this cooperativity depends on a C-terminal domain (CTD) of TrbA which shows a high degree of conservation with the CTD of KorA. The results suggest that KorB and TrbA can interact at a distance and thus have wider significance, because this leads to hypotheses about the role of the orphan KorB binding sites in the Tra1 and Tra2 region and may help to provide clues to the logic of the regulatory organization of the RK2 genome.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The Escherichia coli K-12 strain used was C600K (thr-1 leu-6 thi-1 lacYL supE44 tonA2 galK3) (13). The plasmids used are listed in Table 1. Bacteria were grown in L broth (9) with shaking at 37°C. Solid medium was obtained by the addition of agar (1.5% [wt/vol]). Antibiotic resistance was selected by the addition of penicillin G (Pnr) (sodium salt at 100 μg/ml) in liquid medium and 300 μg/ml in solid medium), kanamycin (Kmr) (50 μg/ml), streptomycin (Smr) (30 μg/ml), and tetracycline (Tcr) (25 μg/ml).
TABLE 1.
Plasmids used in this study
| Plasmid | Size (kb) | Replicon | Selective markera | Description | Reference |
|---|---|---|---|---|---|
| pGBT30 | 6.3 | pMB1 | Pnr | Expression vector | 6 |
| pGBT63 | 10.6 | pSC101 | Kmr | trbBp-xylE | 7 |
| pGBT66 | 10.6 | pSC101 | Kmr | trbB-xylE, transposon insertion 103 | 8 |
| pGBT100 | 6.3 | pMB1 | Pnr | Dual xylE-galK promoter probe | 4 |
| pGBT178 | 8.8 | pMB1 | Pnr | trbBp-xylE, transposon insertion 104 | This study |
| pGBT278 | 8.8 | pMB1 | Pnr | trbBp-xylE, transposon insertion 104 | This study |
| pGBTΔ66/78 | 8.7 | pMB1 | Pnr | trbBp-xylE, deletion between transposon insertions 103 and 104 | This study |
| pPTO1 | 10.1 | pSC101 | Kmr | xylE promoter probe | 23 |
| pDM1.2 | 14.0 | IncQ | Smr | Expression vector | 12 |
| pDM1.21 | 14.0 | IncQ | Smr | KorB overexpression | 12 |
| pMZT11 | 11.0 | pSC101 | Kmr | trfA trbA trbB xylE | 26 |
| pMZT14 | 10.6 | pSC101 | Kmr | trbBp-1-xylE | 26 |
| pMZT15 | 8.0 | M13 mp8RF | Substrate for site-directed mutagenesis | 26 | |
| pMZT24 | 6.6 | pMB1 | Pnr | TrbA (wt) overexpression | 26 |
| pMZT37 | 10.6 | pSC101 | Kmr | trbB-xylE, transposon insertion 104 | This study |
| pMZT38 | 10.5 | pSC101 | Kmr | trbB-xylE, internal deletion between insertions 103 and 104 | This study |
| pMZT39 | 10.6 | pSC101 | Kmr | trbB-xylE, mutation in putative degenerate KorB operator | This study |
| pMZT40 | 10.4 | pSC101 | Kmr | trbB-xylE (RK2 coordinates 18594 to 18915) | This study |
| pMZT41 | 10.3 | pSC101 | Kmr | trbB-xylE (RK2 coordinates 18715 to 18915) | This study |
| pMZT42 | 10.2 | pSC101 | Kmr | trbB-xylE (RK2 coordinates 18742 to 18790) | This study |
| pMZT43 | 6.6 | pMB1 | Pnr | TrbA-CΔ8 | This study |
| pMZT44 | 6.6 | pMB1 | Pnr | TrbA-CΔ11 | This study |
| pMZT45 | 6.6 | pMB1 | Pnr | TrbA-CΔ13 | This study |
| pMZT46 | 6.6 | pMB1 | Pnr | TrbA-CΔ15 | This study |
| pMZT47 | 6.6 | pMB1 | Pnr | TrbA-CΔ18 | This study |
| pMZT48 | 6.6 | pMB1 | Pnr | TrbA-CΔ33 | This study |
| pMZT49 | 6.6 | pMB1 | Pnr | TrbA-NΔ18 | This study |
| pMZT50 | 6.6 | pMB1 | Pnr | TrbA-NΔ23 | This study |
| pMZT51 | 11.0 | pSC101 | Kmr | trfA trbA trbB (18007 to 18915) | This study |
Pnr, resistance to penicillin G; Kmr, resistance to kanamycin; Smr, resistance to streptomycin.
Isolation of DNA, genetic manipulation, and sequencing.
Plasmid DNA was prepared on both small and large scales by the method of Birnboim and Doly (2) and, if necessary, by CsCl-ethidium bromide equilibrium density gradient centrifugation. DNA manipulations were carried out by standard techniques (19) using enzymes according to the manufacturer's instructions. Sequencing was performed automatically on an Applied Biosystems 373A DNA sequencer using a dye terminator kit supplied by the manufacturer.
Primers used for PCR amplification of relevant fragments.
The primers used for PCR amplification were as follows: 1, 5′ CGGAATTCATGTACAACCAGATATTTTTCACC 3′; 2, 5′ GGGAATTCATGTCGGAGAGGGCAGGGGTT3′; 3, 5′ GGGAATTCATGACGAAACATGAGCTGTCG 3′; 4, 5′ GGGTCGACTCACGGCACGCTGCTCTTGAAAGG 3′; 5, 5′ GGGTCGACTCACGCCTTATGTGACGGCAAAAC 3′; 6, 5′ GGGTCGACTCATACGATAAACGCCTTATG 3′; 7, 5′ GGGTCGACTCATTTCTTTACGATAAACGCCT 3′; 8, 5′ GGGTCGACTCAGCCCCATTTCTTTACGAT 3′; 9, 5′ GGGTCGACTCAGGTGTCGTCGCCCCATTTCT 3′; 10, 5′ GGGTCGACTCAGAGCCTTCCACGCAGCTT 3′; 11, 5′ GGGGGATCCGAATTCCGTCGTGCGGTTCGATTAGC 3′; 12, 5′ GGAGATCTAGATCTTGCAGGCTCGAGACT 3′; 13, 5′ GGGGATCCACATTATCGCACAGG 3′; 14, 5′ CGGGATCCTGAGCGCGGCCAACAGGTCC 3′; and 15, 5′ CGGGATCCTTGACCGCGAGGCACTCG 3′.
Cloning of promoter fragments.
For mapping of the TrbA binding site and the KorB secondary binding site, the following plasmids were prepared as described below. For pMZT37, the BamHI trbBp fragment from pGBT178 was subcloned into the BamHI site of pPTO1. pGBT178 carries the same fragment as pGBT78 (8), with transposon insertion 104 in position 18594 of RK2 (20, 21) cloned into pGBT100. For pMZT38, the BamHI trbBp fragment from pGBTΔ66/78 was subcloned into the BamHI site of pPTO1. Plasmid pGBTΔ66/78 was obtained by ligating together the BamHI/EcoRI fragments from pGBT78 (EcoRI site within transposon insertion 104) and the BamHI/EcoRI fragment from pGBT66 (EcoRI site within transposon insertion 103) (Fig. 2) and inserting the resulting BamHI fragment into pGBT100. For pMZT39, the fragment amplified by PCR with primers 15 and 14 (Fig. 2) on template pMZT51 was cloned as a BglII/BamHI fragment into pPTO1. pMZT51, which carries RK2 DNA from coordinates 18007 to 18915 (trfA-trbA-trbBp) cloned as a BamHI fragment into pPTO1, has mutations in the putative secondary KorB operator overlapping the trbBp −35 region. Mutation changes CTTGCAGGCTAAAC to CTTGCAGGCTCGAG (−35 region in bold) and introduces an XhoI site. The mutation was obtained by site-directed mutagenesis on a pMZT15 (26) template with primer 5′ GTCTTGCAGGCTCGAGACTTTCGGTATATCG 3′. For pMZT40, the trbBp BamHI fragment from a pGBT278 derivative in which the DNA upstream of the 104 insertion site (Fig. 2) had been deleted was subcloned into the BamHI site of pPTO1. For pMZT41, the fragment amplified by PCR with primers 11 and 14 (Fig. 2) was cloned into the BamHI site of pPTO1. For pMZT42, the fragment amplified by PCR with primers 12 and 13 (Fig. 2) was cloned as a BglII/BamHI fragment into the BamHI site of pPTO1. Primer 12 carries a mutation in the putative degenerate KorB operator identical to that in pMZT51. This mutation does not change the sensitivity to either KorB or TrbA and was used because it introduces an XhoI site, which is convenient for future purposes.
Construction of plasmids overexpressing truncated derivatives of TrbA.
Plasmids pMZT43 to pMZT50 were constructed using fragments amplified from the pMZT24 DNA template by PCR with primers 1 + 9, 1 + 8, 1 + 7, 1 + 6, 1 + 5, 1 + 4, 2 + 10, and 3 + 10, respectively (Fig. 2). The fragments were purified and cloned into the EcoRI/SalI sites on pGBT30 (6). TrbA was expressed from tacp induced with IPTG (isopropyl-β-d-thiogalactopyranoside).
PCRs.
Reactions were performed using standard procedures (16), with parameters as follows: a 5-min denaturation at 96°C followed by 25 rounds of temperature cycling (96°C for 15 s, 55°C for 30 s, 72°C for 90 s) and a final 5-min step at 72°C. Routinely, 10 pmol of each primer was used with 50 ng of template and 1 U of Taq polymerase in 50 μl of reaction buffer as recommended by the manufacturer (Northumbria Biologicals Ltd.). Amplified fragments were purified by electroelution and digested with the appropriate enzyme(s) before cloning. The identity of the inserted fragment was confirmed by DNA sequencing.
Measurement of XylE activity.
The level of xylE expression was determined by an enzymatic assay of catechol 2,3-oxygenase activity in overnight and logarithmically growing bacteria using the method of Zukowski et al. (27). One unit is defined as the amount of enzyme necessary to convert 1 mmol of substrate to product in 1 min under standard conditions. The XylE units presented in the tables were normalized for differences in reporter plasmid copy number. This was done by isolating plasmid DNA from each culture used in the assay, followed by digestion with a restriction enzyme, separation on an agarose gel, and determination of relative band intensity with Imagequant software on a phosphorimager. Protein concentration was determined by the Biuret method (3).
Measurement of promoter activity by Northern blotting.
Total RNA was isolated from 12-ml cultures of logarithmically growing bacteria. Pellets were quick-frozen in liquid nitrogen prior to extraction. Bacteria were resuspended in 200 μl of lysis buffer (0.02 M sodium acetate [pH 5.5], 0.5% sodium dodecyl sulfate [SDS], 1 mM EDTA), and then 200 μl of acid phenol was added. Samples were vortexed and then incubated at 60°C for 5 min. Phenol and aqueous phases were separated by centrifugation, and the aqueous phases were reextracted with hot phenol a further two times. Sodium chloride was added to 0.5 M, and the RNA was allowed to precipitate at −20°C for 1 h after the addition of 3 volumes of 100% ethanol. After centrifugation the RNA pellet was washed twice with 70% ethanol. Twenty micrograms of each RNA sample was fractionated on a formaldehyde denaturing gel (1.2% agarose) (19). Fractionated RNA was blotted onto a nylon membrane in a 20× SSC transfer buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and UV cross-linked to the membrane. A 32P-labeled xylE DNA probe was hybridized to the membrane overnight at 68°C in 7% SDS–0.25 M phosphate buffer (pH 7.2)–10 mM EDTA, followed by two 15-min washes in 2× SSC–0.1% SDS at 68°C. Hybridization signal intensity for each lane was determined using a phosphorimager and Quantity One software (Bio-Rad Laboratories).
Antibody preparation.
Rabbit polyclonal antibodies against KorB and TrbA were prepared as described previously for antibodies against KorA (5).
Western blot analysis.
The protein extracts were prepared as described for the XylE assay, and proteins were separated by SDS–20% polyacrylamide gel electrophoresis according to the method of Sambrook et al. (19). The proteins were electrotransferred onto nitrocellulose filters at 50 mA overnight according to the method of Sambrook et al. (19). The filters were used directly for Western blotting with a Bio-Rad detection kit.
RESULTS
KorB cooperates with TrbA in repression of trbBp.
It was previously observed that both KorB and TrbA are necessary to fully repress trbBp (26). To test whether there is cooperativity between these two repressors when both are present we constructed strains carrying three compatible plasmids: a reporter plasmid carrying trbBp linked to xylE (pGBT63), a plasmid overproducing TrbA (pMZT24) or the relevant control (pGBT30), and a plasmid overproducing KorB (pDM1.21) or the relevant control (pDM1.2) (Table 2). Both the trbA (pMZT24) and KorB (pDM1.2.1) open reading frames were cloned under the control of the tac promoter so that their expression could be simultaneously induced with IPTG. The activity of the reporter xylE was measured in the presence of either TrbA and KorB together or TrbA or KorB alone (Table 2). The repression (R) in the presence of both TrbA (A) and KorB (B) was much greater [R(AB) = 203.5-fold] than in the presence of TrbA only [R(A) = 2.3-fold] or KorB only [R(B) = 8.3-fold]. It was also much greater than the product of the repression indices of both proteins [R(A) × R(B) = 19.1-fold], which would be expected for the situation where the two repressors act independently of each other.
TABLE 2.
Cooperative interaction between KorB and TrbA proteins on trbBp-xylE fusion present in plasmid pGBT63a
| Repressorc | XylE assays of log cultures (0.05 mM IPTG)
|
XylE assyas of overnight cultures (0.03 mM IPTG)b
|
XylE assays of log cultures (0.03 mM IPTG)b
|
mRNA analysis of log cultures (0.03 mM IPTG)b
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| XylE units | RA+Bd | RA × RBe | CIf | XylE units | RA+Bd | RA × RBe | CIf | XylE units | RA+Bd | RA × RBe | CIf | mRNA signalg | RA+Bd | RA × RBe | CIf | |
| TrbA + KorB | 0.0057 ± 0.0007 | 203.5 | 19.1 | 10.7 | 0.0047 ± 0.001 | 1593.6 | 257 | 6.19 | 0.029 ± 0.011 | 47.2 | 2.9 | 16.3 | 27,861 ± 5,094 | 61.8 | 19.8 | 3.1 |
| KorB | 0.14 ± 0.032 | 8.3 | 0.32 ± 0.09 | 23.4 | 0.61 ± 0.10 | 2.2 | 174,792 ± 21,006 | 9.9 | ||||||||
| TrbA | 0.51 ± 0.13 | 2.3 | 0.68 ± 0.012 | 11 | 1.03 ± 0.05 | 1.3 | 879,996 ± 18,714 | 2.0 | ||||||||
| None | 1.06 ± 0.28 | 7.49 ± 1 | 1.37 ± 0.08 | 1,722,559 ± 173,010 | ||||||||||||
Each result comes from at least 3 repetitions.
0.03 mM IPTG was used since at 0.05 mM the activity of trbBp was repressed so much as to be unmeasurable.
TrbA was expressed by pMZT24 (tacp-trbA), while KorB was expressed by pDM1.21 (tacp-korB). The equivalent expression vector without repressor gene for each plasmid was pGBT30 and pDM1.2.
Experimentally established repression in the presence of both TrbA and KorB.
Product of indices of repression by TrbA and KorB illustrates the expected repression after introduction of the two repressors together to the same cell if there is no interaction between them.
The ratio of RA+B/RA × RB illustrates the fold enhancement of the repression that the two repressors can mediate when they act together.
Hybridization signal intensity (± standard deviation) of xylE probe hybridized to total RNA.
The in vivo cooperativity index (CI) was calculated as described by Kostelidou et al. (11) as a ratio of experimentally obtained repression in the presence of both proteins [R(AB)] to the product of the repression indices of individual proteins [R(A) × R(B)], so CI = R(AB)/[R(A) × R(B)]. The CI reveals the degree of enhancement of repression activity that the two repressors can achieve when they act together. Table 2 shows that for assays performed on logarithmically growing cultures, CI = 10.7, which means that the repression by TrbA and KorB is 10.7-fold stronger than the product of their individual repression indices. The simplest explanation is that there is cooperative interaction between them.
The results presented in Table 2 for logarithmically growing bacteria were obtained in the presence of 0.05 mM IPTG. At this low concentration the repression by KorB and TrbA individually is 8.3- and 2.3-fold, respectively. At higher IPTG concentrations a greater repression was observed for both TrbA and KorB alone, but since the residual promoter activity was very low we could not calculate accurately the degree of enhancement that the repressors can achieve together. Similar experiments were done on stationary-phase cultures using 0.03 mM IPTG (Table 2). A very strong repression was again observed in the presence of both proteins, and the CI was calculated to be 6.19.
Since it is possible that the XylE activity measured does not give an accurate picture of transcription from the promoter (for example, if the relationship between enzyme activity and mRNA level is significantly nonlinear), we checked these results by Northern blotting followed by phosphorimager quantitation as described in Materials and Methods. An E. coli (strain C600) control was included on the Northern blot to allow for subtraction of nonspecific background hybridization from the values obtained. The mRNA hybridization values reported in Table 2 were normalized for differences in reporter plasmid copy number as described for measurements of XylE activity. We also used the same cultures in parallel for XylE assays. The results showed that while the apparent level of cooperativity was different between the two methods, a significant level of cooperativity was confirmed by the Northern blotting. This figure may be on the conservative side, because when a strain carrying the reporter plasmid without an inserted promoter was used as a negative control it actually gave a signal higher than the repressed level of mRNA from trbBp. This led to a calculated mRNA level of zero or less, which gives a level of cooperativity of >20.
KorB represses trbBp at a distance.
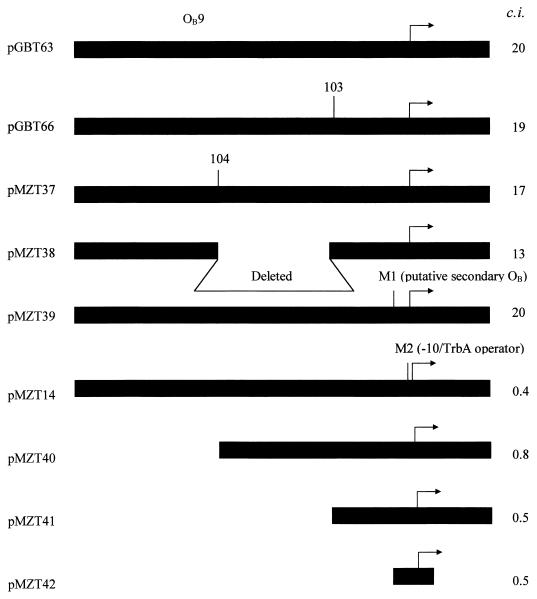
The identified KorB binding site in the trbBp region, OB9, lies 189 bp upstream of the trbBp transcription start point, and binding of purified KorB at this site has been explored in vitro (8). In thinking about KorB-TrbA cooperativity it was important to explore the constraints on the KorB action on trbBp in vivo. To test the role of sequences that flank OB9 we used plasmids pGBT66 (8) and pMZT37 carrying transposon insertions 103 and 104, which involve duplications of the DNA between coordinates 18694 to 18698 and 18594 to 18598, respectively, and introduce convenient EcoRI sites which are situated 15 bp into the transposon DNA (15, 20, 21). After excision of the transposon, the 5-bp duplication of target sequence and 15 bp from each end of the transposon to the EcoRI site remained. Transposon insertions 103 and 104 destroyed parts of the region which was bound with increased affinity by KorB and was implicated in formation of higher order complexes (8). We also prepared the internal deletion which removed DNA between the transposon insertions 103 and 104 (between RK2 coordinates 18694 and 18598). In this construct, pMZT38, the region downstream of OB9, which was involved in increased affinity and formation of higher order complexes, is completely removed. We found that neither of the insertions nor the internal deletion affects the ability of KorB to repress trbBp in vivo. In fact, the promoter fragment with the internal deletion is repressed better than the wild-type fragment (Table 3). We have also prepared a set of deletions from the 5′ end of trbBp. In pMZT40, OB9 is removed, but the region downstream remains intact. In pMZT41, the whole region upstream of the promoter is removed, and the fragment cloned corresponds to RK2 coordinates 18715 to 18915 (Fig. 2), while pMZT42 contains an approximately 50-bp fragment spanning trbBp (coordinates 18743 to 18790). In all cases when OB9 was removed, we observed no repression by KorB in overnight cultures, but there was still weak repression (up to 2.6-fold) in logarithmic cultures. This included pMZT42, which gave twofold repression.
TABLE 3.
Mapping of sites needed for repression of trbBp by KorB and TrbAa
| Reporter plasmid | KorBb
|
TrbAd
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Presence (+) or absence (−) of repressor | Overnight culture (+ 0.05 mM IPTG)
|
Logarithmic culture (+ 0.5 mM IPTG)
|
Presence (+) or absence (−) of repressor | Overnight culture (+ 0.05 mM IPTG)
|
Logarithmic culture (+ 0.5 mM IPTG)
|
|||||
| XylE unitsa | Rc | XylE unitse | Rc | XylE unitse | Rc | XylE unitse | Rc | |||
| pGBT63 | + | 0.53 | 24.3 | 0.053 | 48.9 | + | 0.84 | 18 | 0.39 | 3.54 |
| − | 12.9 | 2.59 | − | 15.1 | 1.38 | |||||
| pGBT66 | + | 0.21 | 31.4 | 0.032 | 59.6 | + | 0.81 | 14.7 | 0.34 | 4.3 |
| − | 6.6 | 1.91 | − | 11.9 | 1.46 | |||||
| pMZT37 | + | 0.23 | 22.6 | 0.025 | 78.8 | + | 0.93 | 12.7 | 0.25 | 3 |
| − | 5.2 | 1.97 | − | 11.8 | 0.74 | |||||
| pMZT38 | + | 0.22 | 31.3 | 0.02 | 112 | + | 0.8 | 14.4 | 0.23 | 5.04 |
| − | 6.9 | 2.24 | − | 11.5 | 1.16 | |||||
| pMZT39 | + | 0.29 | 19.3 | 0.025 | 96 | + | 0.99 | 13.9 | 0.43 | 3.6 |
| − | 5.6 | 2.4 | − | 13.8 | 1.56 | |||||
| pMZT14 | + | 0.009 | 38.9 | 0.0045 | 37.8 | + | 0.45 | 1.0 | 0.06 | 1.5 |
| − | 0.35 | 0.17 | − | 0.45 | 0.09 | |||||
| pMZT40 | + | 4.22 | 1.2 | 0.94 | 2.6 | + | 0.77 | 15.1 | 0.27 | 3.9 |
| − | 5.0 | 2.45 | − | 11.6 | 1.05 | |||||
| pMZT41 | + | 4.31 | 1.2 | 1.1 | 1.8 | + | 0.45 | 17 | 0.24 | 5.6 |
| − | 5.1 | 2.01 | − | 7.64 | 1.34 | |||||
| pMZT42 | + | 3.27 | 1.4 | 0.82 | 2.1 | + | 0.93 | 17.5 | 0.12 | 7.3 |
| − | 4.6 | 1.74 | − | 16.3 | 0.84 | |||||
Results come from at least three experiments. The RK2 DNA present in the reporter plasmids is shown in Fig. 3.
KorB was expressed by plasmid pDM1.21, while the expression vector pDM1.2 was used to replace it for the no-repressor control.
Repression ratio, calculated as XylE units with repressor/XylE units without repressor.
TrbA was expressed by plasmid pMZT24, while the expression vector pGBT30 was used to replace it for the no-repressor control.
For clarity, the standard deviation values, which were generally in the same range as shown in the other tables, were omitted.
In all constructs where OB9 was present a strong repression was observed in both overnight (19- to 39-fold) and logarithmic (38- to 112-fold) cultures (Table 3). We also included two other plasmids, pMZT39 and pMZT14. pMZT39 contains a 578-bp segment (RK2 coordinates 18337 to 18915) that covers trbBp amplified from a template in which the putative degenerate KorB operator has been mutated (8). Previously, the effect of this mutation was tested in the context of the whole trfA-trbA-trbBp region (RK2 coordinates 18007 to 18915), but the presence of the upstream sequences including the trfA and trbA promoters complicated the interpretation of the results. Plasmid pMZT14 was included to determine whether the changes in the −10 region of trbBp affect regulation by KorB. We found that mutation in the putative degenerate KorB operator does not influence regulation by KorB even in the absence of the upstream trfA and trbA promoters. Similarly, the mutations in the −10 region of trbBp do not change sensitivity to KorB regulation.
Mapping the TrbA binding site.
Although the ability of TrbA to repress trbBp is well documented (7, 26), the binding site for TrbA has not been mapped. Therefore, we used deletion analysis to determine the approximate location of the target for TrbA. The set of derivatives of the trbBp reporter plasmid (pGBT63) already described above, which included transposon insertions (pGBT66 and pMZT37), an internal deletion (pMZT38), site-directed mutations (pMZT14 and pMZT39), and deletions from the 5′ end (pMZT40, pMZT41, and pMZT42), were tested for in vivo repression by trbA provided in trans under the control of the tac promoter. All these derivatives were repressed by TrbA at least 13-fold in overnight cultures, which is similar to the repression of the full-length promoter fragment (pGBT63), which was repressed 18-fold. Even pMZT42, which includes only a 50-nucleotide-long trbBp region, was still 17-fold repressed by TrbA. The results obtained in logarithmic cultures were similar, except that the repression was much lower (Table 3). This indicates that the TrbA target is adjacent to or within the promoter sequence. Further definition was provided by the fact that pMZT14, which has two nucleotides changed within the trbBp −10 region (Fig. 2) (26), is not sensitive to TrbA repression (Table 3).
Cooperation between KorB and TrbA in repression of trbBp depends on the presence of their binding sites.
Since there is some hint that KorB may still repress trbBp very weakly when OB9 is deleted, it was possible that the cooperativity between TrbA and KorB might not require OB9. Therefore we tested all the plasmids previously used for mapping of TrbA and KorB binding sites for cooperativity in the three-plasmid system, as described above. As shown in Fig. 3, cooperative repression by KorB and TrbA is only possible when both OB9 and an intact −10 region of trbBp are present. If OB9 is removed or if the putative target for TrbA in the −10 region is mutated, KorB and TrbA no longer give cooperative repression of trbBp. Therefore, both KorB and TrbA must bind their normal targets to achieve cooperative repression.
FIG. 3.
Genetic analysis to determine whether the presence of both TrbA and KorB binding sites are necessary for cooperative repression of trbBp. Arrows indicate trbBp which is fused to xylE. M1, mutations in the putative degenerate KorB operator (Fig. 2); M2, mutation in the −10 region of trbBp which also reduces sensitivity to trbA repression (Fig. 2); OB9, the known KorB operator; 103 and 104, EcoRI sites in a 35-bp segment left as a result of Tn1723 transposon insertion followed by deletion of the internal EcoRI fragment which is defined by EcoRI sites 15 bp from each end of the transpsoson (20). c.i., cooperativity in vivo index when KorB was expressed from pDM1.21 (pDM1.2 as control) and TrbA was expressed from pMZT24 (pGBT30 as control). Repressor expression was induced with IPTG as indicated in Table 2.
The C terminus of TrbA is essential for cooperativity to occur.
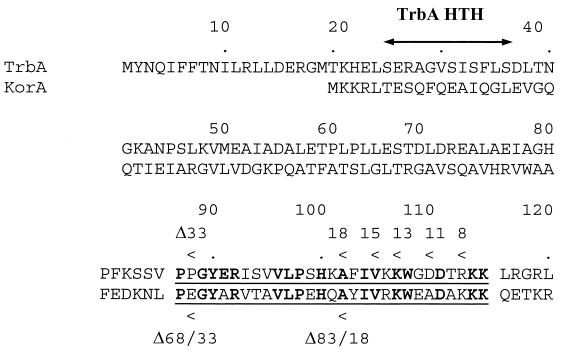
Cooperativity between KorB and another RK2 repressor, KorA, has recently been described (11). The C-terminal, basic region of KorA was found to be essential for this interaction. Since the C-terminal regions of KorA and TrbA are highly conserved, showing 76% similarity (55% identity) over a 29-residue overlap (Fig. 4), we determined whether the C-terminal region of TrbA is also important for cooperative interaction with KorB. For this purpose, a series of C-terminal deletions of TrbA was prepared (Fig. 4). pMZT48, which has 33 amino acids removed from its C terminus, lacks all of the basic, conserved region. pMZT47, which has 18 amino acids removed, lacks about half of the conserved region. pMZT46, pMZT45, pMZT44, and pMZT43 lack 15, 13, 11, and 8 amino acids, respectively.
FIG. 4.
Alignment of TrbA and KorA proteins of RK2. The underlining indicates the conserved C-terminal region. Identical amino acids are shown in bold. Arrowheads indicate the end points of deletions. For TrbA, the numbers indicate the number of amino acids removed from the C terminus. For the KorA deletions, the numbers indicate the distance from the N terminus, as described by Kostelidou et al. (11), as well as the number of amino acids removed from the C terminus (N-terminal distance/C-terminal residues removed). The other main feature is the TrbA helix-turn-helix (HTH) motif as predicted by Jagura-Burdzy et al. (7).
We tested the ability of C-terminally truncated TrbA proteins to repress trbBp by introducing each of the deletion derivatives in trans to the pGBT63 reporter plasmid (Table 4). XyIE assays showed that all TrbA deletion derivatives were able to repress trbBp. TrbA with 8 amino acids removed showed almost wild-type repression levels (4.1-fold in logarithmic cultures, 15.8-fold in overnight cultures), whereas the deletion of 15 amino acids showed the weakest repression (1.7-fold in logarithmic cultures, 3.5-fold in overnight cultures). All the other deletions tested showed intermediate levels of repression (Table 4).
TABLE 4.
Repression of trbBp-xylE fusion in pGBT63 by deletion derivatives of TrbA (in pMZT24 and derived plasmids)a
| Plasmid | No. of amino acids deleted from CTD | Logarithmic culture (+ 0.5 mM IPTG)
|
Overnight culture (+ 0.05 mM IPTG)
|
||||
|---|---|---|---|---|---|---|---|
| XylE units ± SD | Rb | CI with KorBc | XylE units ± SD | Rb | CI with KorBc | ||
| pMZT24 | 0d | 0.15 ± 0.02 | 5.8 | 10.7 | 0.57 ± 0.09 | 17.8 | 6.19 |
| pMZT43 | 8 | 0.21 ± 0.04 | 4.1 | 9.6 | 0.64 ± 0.01 | 15.8 | 5.16 |
| pMZT44 | 11 | 0.29 ± 0.08 | 3.0 | 1.23 | 2.45 ± 0.14 | 4.1 | 1.36 |
| pMZT45 | 13 | 0.40 ± 0.11 | 2.2 | 0.70 | 1.20 ± 0.15 | 8.5 | 0.34 |
| pMZT46 | 15 | 0.51 ± 0.10 | 1.7 | 0.89 | 3.47 ± 0.31 | 2.9 | 0.65 |
| pMZT47 | 18 | 0.35 ± 0.06 | 2.5 | 0.73 | 1.83 ± 0.49 | 5.5 | 0.57 |
| pMZT48 | 33 | 0.32 ± 0.05 | 2.7 | 0.58 | 1.58 ± 0.15 | 6.4 | 0.39 |
| pGBT30 | 0.87 ± 0.07 | 10.15 ± 1.51 | |||||
Results come from at least 3 experiments.
Repression ratio compared to activity in the presence of pGBT30.
KorB was expressed by plasmid pDM1.21, while the expression vector pDM1.2 was used to replace it for the no-repressor control.
Wt TrbA.
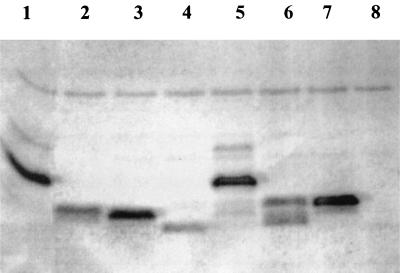
To determine whether the variation in effect of these TrbA derivatives was due to the concentration of each protein after induction we checked the protein levels by Western blotting on cleared lysates of cultures previously used in XylE assays. Antibodies were prepared, and Western blotting was performed as described in Materials and Methods. The results are presented in Fig. 5. Expression levels of wild-type (wt) TrbA (pMZT24), TrbA-CΔ8 (pMZT43), TrbA-CΔ13 (pMZT45), and TrbA-CΔ18 (pMZT47) were very similar, whereas expression of TrbA-CΔ15 (pMZT46) and TrbA-CΔ33 (pMZT48) was weaker. TrbA-CΔ11 (pMZT44) seems to be quite unstable since it is detected as two bands on the Western blot.
FIG. 5.
Western blot analysis of C-terminally deleted TrbA derivatives, carried out as described in Materials and Methods. Lanes: 1, pMZT24 (TrbA wt); 2, pMZT46 (C-Δ15); 3, pMZT47 (C-Δ18); 4, pMZT48 (C-Δ33); 5, pMZT43 (C-Δ8); 6, pMZT44 (C-Δ11); 7, pMZT45 (C-Δ13); 8, pGBT30 (no insert).
We tested all TrbA deletion derivatives for the ability to cooperate with KorB for the repression of trbBp. Cooperativity was tested in vivo in the three-plasmid system, and the CI was calculated as described for wild-type TrbA. Deletion of the last eight amino acids still allows for cooperativity to occur, and the CI is very similar to that of wt TrbA (Table 4). Deletion of a further three amino acids (pMZT44) almost completely abolished cooperativity (CI of about 1.0, indicating that the repression is at the level expected for noncooperating repressors). However, this protein seems to be quite unstable. Deletion of 13 amino acids (or more) resulted in complete loss of cooperativity (CI < 1, which indicates that the repression is smaller than predicted for noninteracting repressors), despite the fact that this protein is expressed at a level comparable with that of wt TrbA and is still able to repress trbBp (8.5-fold in overnight and 2.2-fold in logarithmic cultures).
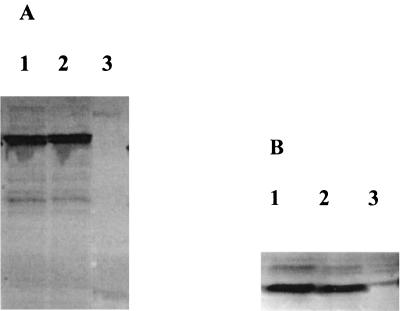
The effect of enhanced repression in the presence of both repressors could come from simple stablization of one protein by another. Therefore, we used Western blotting to compare the level of TrbA in the presence and absence of KorB and the level of KorB in the presence and absence of TrbA. As shown in Fig. 6A, the level of KorB was the same in the presence and absence of TrbA, and similarly, the level of TrbA was the same in the presence and absence of KorB (Fig. 6B). Thus, the effect of enhanced repression in the presence of both proteins does not come from better expression or stability. We propose that it is the result of cooperative interaction between KorB and TrbA, which most probably involves direct interaction between the two proteins, and that the C-terminal domain of TrbA is essential for this interaction (see Discussion).
FIG. 6.
Western blot analysis of the relative amounts of TrbA and KorB alone and in combination with the second repressor, carried out as described in Materials and Methods. Cleared lysates used for cooperativity testing were run on SDS-polyacrylamide gel electrophoresis gels. (A) Rabbit anti-KorB as primary antibody. Lanes: 1, KorB and TrbA; 2, KorB only; 3, TrbA only. (B) Rabbit anti-TrbA as primary antibody. Lanes: 1, KorB and TrbA; 2, TrbA only; 3, KorB only. Antibodies were prepared as described in Materials and Methods.
We also prepared two N-terminal deletion derivatives of TrbA, pMZT49 (N-Δ18) and pMZT50 (N-Δ23), to check if the presence of the intact CTD is sufficient for cooperativity to occur. Unfortunately, both derivatives were very unstable: neither of them was detected by Western blot analysis, and neither showed any repression of trbBp in the reporter assay. The lack of ability to repress trbBp could have resulted from instability as well as from removal of the putative DNA recognition domain. Therefore, we could not use them in our test for cooperativity.
DISCUSSION
The topic of this paper is the cooperativity between KorB and TrbA at the trbB promoter. For the background to this, we explored the targets necessary for KorB and TrbA to repress trbBp. We mapped the TrbA binding site to a 50-nucleotide fragment spanning the trbBp −10 sequence. Such a small fragment is still repressed by TrbA 17-fold in stationary-phase cultures and 7.3-fold in log-phase cultures. We explored further the idea that there may be specific secondary binding sites for KorB that may be necessary for its repression of trbBp. Our result with an internal deletion rules out a specific role for the region between transposon insertions 103 and 104, which was suggested as a region potentiating KorB binding in vitro (8). If there is a specific secondary interaction our results point to one in the trbBp region: a 50-nucleotide fragment spanning the trbBp −10 sequence still showed slight sensitivity to KorB repression (about twofold in log-phase cultures [KorB generally represses this promoter better in logarithmic cultures than in overnight cultures]). However, mutation of the best candidate for secondary interaction with KorB, a putative degenerate binding site based on similarity to the OB consensus, did not support the idea (8). This would point to RNA polymerase (RNAP) as the direct target for KorB rather than a specific DNA sequence, as was already observed for the φ29 repressor protein (14).
If KorB and RNAP interact at a distance they may create a loop between them in the DNA. The cooperativity with TrbA could be due to a parallel looping interaction between KorB and TrbA which favors the interaction between KorB and RNAP as well as increasing the steric hindrance, which blocks RNAP access to the promoter in the first place. This is consistent with the fact that cooperativity depends on the presence of both binding sites OB9 for KorB and OT1 for TrbA. An alternative to looping would be the idea that KorB spreads along the DNA from OB9, silencing flanking genetic functions as suggested for the ParB protein of P1 (18). In vitro footprints for KorB at and around OB9 (8) are not consistent with this hypothesis, but the in vivo situation may be different from the in vitro situation.
The C-terminal region of TrbA is essential for cooperativity but not for repression. This is consistent with the prediction that a helix-turn-helix motif is located close to the N-terminal domain (7). However, the different deletion derivatives vary in their stability: Δ11, Δ15, and Δ33 are shown by Western blotting to be less stable than the others, suggesting that removal of critical residues results in changes in folding and unmasking of sequences sensitive to proteases. The last eight amino acids, which constitute a very basic C terminus, RKKLRGRL, are not necessary for cooperative interaction, which is surprising since KorB is an acidic protein. Removal of the next three amino acids, DDT, resulted in almost complete loss of the ability to cooperate, but it also conferred instability. Removal of a further two residues, WG, restored stability but made the protein completely unable to cooperate. Thus, the patch WGDDT marks the start of the region essential for TrbA to cooperate with KorB. An alternative explanation, that this patch is responsible for additional contact with DNA which changes the structure in such a way that it allows KorB bound at OB9 to be brought into the vicinity of the promoter, does not seem very likely.
The ability of KorB and TrbA to cooperate for the repression of trbBp provides a second example of cooperativity between repressors which control RK2 regulatory circuits, the other being KorB-KorA interactions (11), and suggests that the interaction of KorB with other regulators is key to the behavior of these circuits. However, there are clear differences between the interactions described so far. The most important difference is the spacing between operators: OA1 and OB1 in korAp are separated by 22 bp, while OT1 and OB9 at trbBp are separated by approximately 165 bp. In addition, the deletion analysis suggests that different patches may be responsible for cooperative interaction of KorB with KorA and TrbA, despite the high similarity in their C termini. For KorA, the patch involved in cooperativity maps between residues 68 and 83 (Fig. 4), and the region downstream of residue 83 does not seem to be important. The analogous region of TrbA is still essential for cooperativity. These differences may reflect the spatial organization of KorA and KorB on korAp and TrbA and KorB at trbBp. Interaction between KorB and the second protein when they are side by side or separated by a DNA loop may impose different ways of interacting, in addition to the differences caused by the nonidentical N-terminal domains in TrbA and KorA.
ACKNOWLEDGMENTS
We thank Kalliope Kostelidou and Grazyna Jagura-Burdzy for useful discussions.
M.Z. was supported by Wellcome Trust grant 046356/Z/95, A.C.J. was supported by Wellcome Trust grant 048040/Z/96, and L.B. was supported by Wellcome Trust grant 056000. DNA sequencing was performed by AltaBioscience using an ABI automated sequencer partly funded by a shared equipment grant from the Wellcome Trust (038654/Z/93). The phosphorimager was purchased with shared equipment grants from the Wellcome Trust (037160/Z/92) and the Medical Research Council (G9216078MB).
REFERENCES
- 1.Balzer D, Ziegelin G, Pansegrau W, Kruft V, Lanka E. KorB protein of promiscuous plasmid RK2 recognises inverted sequence repetitions in regions essential for conjugative transfer. Nucleic Acids Res. 1992;20:1851–1858. doi: 10.1093/nar/20.8.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gornall A G, Bardawill C J, David M M. Determination of serum proteins by means of the biuret reaction. J Biol Chem. 1949;177:751–766. [PubMed] [Google Scholar]
- 4.Jagura-Burdzy G, Thomas C M. KorA protein of promiscuous plasmid RK2 controls a transcriptional switch between divergent operons for plasmid replication and conjugative transfer. Proc Natl Acad Sci USA. 1994;91:10571–10575. doi: 10.1073/pnas.91.22.10571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jagura-Burdzy G, Thomas C M. Purification of KorA protein from broad host range plasmid RK2: definition of a hierarchy of KorA operators. J Mol Biol. 1995;253:39–50. doi: 10.1006/jmbi.1995.0534. [DOI] [PubMed] [Google Scholar]
- 6.Jagura-Burdzy G, Ibbotson J P, Thomas C M. The korF region of broad-host-range plasmid RK2 encodes two polypeptides with transcriptional repressor activity. J Bacteriol. 1991;173:826–833. doi: 10.1128/jb.173.2.826-833.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jagura-Burdzy G, Khanim F, Smith C A, Thomas C M. Cross-talk between plasmid vegetative replication and conjugative transfer: regulation of the trfA operon by trbA of broad-host-range plasmid RK2. Nucleic Acids Res. 1992;20:3939–3944. doi: 10.1093/nar/20.15.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jagura-Burdzy G, Macartney D P, Zatyka M, Cunliffe L, Cooke D, Huggins C, Westblade L, Khanim F, Thomas C M. Repression at a distance by the global regulator KorB of promiscuous IncP plasmids. Mol Microbiol. 1999;32:519–532. doi: 10.1046/j.1365-2958.1999.01365.x. [DOI] [PubMed] [Google Scholar]
- 9.Kahn M, Kolter R, Thomas C, Figurski D, Mayer R, Remaut E, Helinski D R. Plasmid cloning vehicle derived from ColE1, F, R6K and RK2. Methods Enzymol. 1979;68:268–280. doi: 10.1016/0076-6879(79)68019-9. [DOI] [PubMed] [Google Scholar]
- 10.Kostelidou K, Thomas C M. The hierarchy of KorB binding at its twelve binding sites on the broad host range plasmid RK2 and modulation of this binding by IncCl protein. J Mol Biol. 2000;295:411–422. doi: 10.1006/jmbi.1999.3359. [DOI] [PubMed] [Google Scholar]
- 11.Kostelidou K, Jones A C, Thomas C M. Conserved C-terminal region of global repressor KorA of broad-host-range plasmid RK2 is required for co-operativity between KorA and a second RK2 global regulator, KorB. J Mol Biol. 1999;289:211–221. doi: 10.1006/jmbi.1999.2761. [DOI] [PubMed] [Google Scholar]
- 12.Macartney D P, Williams D R, Stafford T, Thomas C M. Divergence and conservation of the partitioning and global regulation functions in the central control region of the IncP plasmids RK2 and R751. Microbiology. 1997;143:2167–2177. doi: 10.1099/00221287-143-7-2167. [DOI] [PubMed] [Google Scholar]
- 13.McKenney K, Shimatake H, Court D, Schmeissner U, Brady C, Rosenberg M. A system to study promoter and termination signals recognized by E. coli RNA polymerase. In: Chirikjian J C, Papas T S, editors. Gene amplification and analysis. Vol. 2. Amsterdam, The Netherlands: Elsevier/North Holland; 1981. pp. 383–415. [PubMed] [Google Scholar]
- 14.Monsalve M, Calles B, Mencia M, Rojo F, Salas M. Binding of phage φ29 protein p4 to the early A2c promoter: recruitment of a repressor by the RNA polymerase. J Mol Biol. 1998;283:559–569. doi: 10.1006/jmbi.1998.2084. [DOI] [PubMed] [Google Scholar]
- 15.Motallebi-Veshareh M, Balzer D, Lanka E, Jagura-Burdzy G, Thomas C M. Conjugative transfer functions of broad-host-range plasmid RK2 are coregulated with replication. Mol Microbiol. 1992;6:907–920. doi: 10.1111/j.1365-2958.1992.tb01541.x. [DOI] [PubMed] [Google Scholar]
- 16.Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harbor Symp Quant Biol. 1986;51:263–273. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- 17.Pansegrau W, Lanka E, Barth P T, Figurski D H, Guiney D G, Haas D, Helinski D R, Schwab H, Stanisich V A, Thomas C M. Complete nucleotide sequence of Birmingham IncPα plasmids: compilation and comparative analysis. J Mol Biol. 1994;239:623–663. doi: 10.1006/jmbi.1994.1404. [DOI] [PubMed] [Google Scholar]
- 18.Rodionov O, Lobocka M, Yarmolinsky M. Silencing of genes flanking the P1 plasmid centromere. Science. 1999;283:546–549. doi: 10.1126/science.283.5401.546. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Shingler V, Thomas C M. Analysis of the trfA region of broad host range plasmid RK2 by transposon mutagenesis and identification of polypeptide products. J Mol Biol. 1984;175:229–250. doi: 10.1016/0022-2836(84)90346-2. [DOI] [PubMed] [Google Scholar]
- 21.Shingler V, Thomas C M. Transcription in the trfA region of broad host range plasmid RK2 is regulated by trfB and korB. Mol Gen Genet. 1984;195:523–529. doi: 10.1007/BF00341457. [DOI] [PubMed] [Google Scholar]
- 22.Thompson V J, Jovanovic O S, Pohlman R F, Chang C-H, Figurski D H. Structure, function, and regulation of the kilB locus of promiscuous plasmid RK2. J Bacteriol. 1993;175:2423–2435. doi: 10.1128/jb.175.8.2423-2435.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorsted P B, Shah D S, Macartney D, Kostelidou K, Thomas C M. Conservation of the genetic switch between replication and transfer genes of IncP plasmids but divergence of the replication functions which are major host range determinants. Plasmid. 1996;36:95–111. doi: 10.1006/plas.1996.0037. [DOI] [PubMed] [Google Scholar]
- 24.Williams D R, Motallebi-Veshareh M, Thomas C M. Multifunctional repressor KorB can block transcription by preventing isomerisation of RNA-polymerase promoter complexes. Nucleic Acids Res. 1993;21:1141–1148. doi: 10.1093/nar/21.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zatyka M, Jagura-Burdzy G, Thomas C M. Regulation of transfer genes of promiscuous IncPα plasmid RK2: repression of Tra1 region transcription both by relaxosome proteins and by the Tra2 regulator TrbA. Microbiology. 1994;140:2981–2990. doi: 10.1099/13500872-140-11-2981. [DOI] [PubMed] [Google Scholar]
- 26.Zatyka M, Jagura-Burdzy G, Thomas C M. Transcriptional and translational control of the genes for the mating pair apparatus of promiscuous IncP plasmids. J Bacteriol. 1997;179:7201–7209. doi: 10.1128/jb.179.23.7201-7209.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zukowski M M, Gaffney D F, Speck D, Kauffman M, Findelli A, Wisecup A, Lecocq J-P. Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned Pseudomonas gene. Proc Natl Acad Sci USA. 1983;80:1101–1105. doi: 10.1073/pnas.80.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]