Abstract
Several DNA regions containing genes involved in poly-β-hydroxybutyrate (PHB) biosynthesis and degradation and also in fatty acid degradation were identified from genomic sequence data and have been characterized in the serine cycle facultative methylotroph Methylobacterium extorquens AM1. Genes involved in PHB biosynthesis include those encoding β-ketothiolase (phaA), NADPH-linked acetoacetyl coenzyme A (acetyl-CoA) reductase (phaB), and PHB synthase (phaC). phaA and phaB are closely linked on the chromosome together with a third gene with identity to a regulator of PHB granule-associated protein, referred to as orf3. phaC was unlinked to phaA and phaB. Genes involved in PHB degradation include two unlinked genes predicted to encode intracellular PHB depolymerases (depA and depB). These genes show a high level of identity with each other at both DNA and amino acid levels. In addition, a gene encoding β-hydroxybutyrate dehydrogenase (hbd) was identified. Insertion mutations were introduced into depA, depB, phaA, phaB, phaC, and hbd and also in a gene predicted to encode crotonase (croA), which is involved in fatty acid degradation, to investigate their role in PHB cycling. Mutants in depA, depB, hbd, and croA all produced normal levels of PHB, and the only growth phenotype observed was the inability of the hbd mutant to grow on β-hydroxybutyrate. However, the phaA, phaB, and phaC mutants all showed defects in PHB synthesis. Surprisingly, these mutants also showed defects in growth on C1 and C2 compounds and, for phaB, these defects were rescued by glyoxylate supplementation. These results suggest that β-hydroxybutyryl-CoA is an intermediate in the unknown pathway that converts acetyl-CoA to glyoxylate in methylotrophs and Streptomyces spp.
Many prokaryotes, including aerobic methylotrophic bacteria, accumulate poly-β-hydroxybutyrate (PHB) intracellularly as a carbon and energy reserve material. Methylotrophic bacteria employing the serine cycle for formaldehyde assimilation are able to accumulate up to 80% of PHB by dry weight, while methylotrophic bacteria with the Calvin-Benson-Bassham cycle of C1 assimilation can synthesize up to 20% of PHB by dry weight (14). In obligate methanol utilizers, which use the ribulose monophosphate pathway for assimilation of reduced C1 compounds, PHB has not been detected (14).
Two pathways for PHB synthesis are known in bacteria. In Ralstonia eutropha, Methylobacterium extorquens, Zoogloea ramigera, and Azotobacter beijerinckii PHB is synthesized from acetyl coenzyme A (acetyl-CoA) as a result of sequential action of three enzymes: β-ketothiolase, NADPH-linked acetoacetyl-CoA reductase, and PHB synthase (5, 15, 19, 33, 34) (Fig. 1). In Rhodospirillum rubrum and Methylobacterium rhodezianum PHB synthesis is catalyzed by five enzymes: β-ketothiolase, NADH-linked acetoacetyl-CoA reductase, l-(+) and d-(−)-specific crotonyl-CoA hydratases (crotonases), and PHB synthase (27, 28, 29).
FIG. 1.
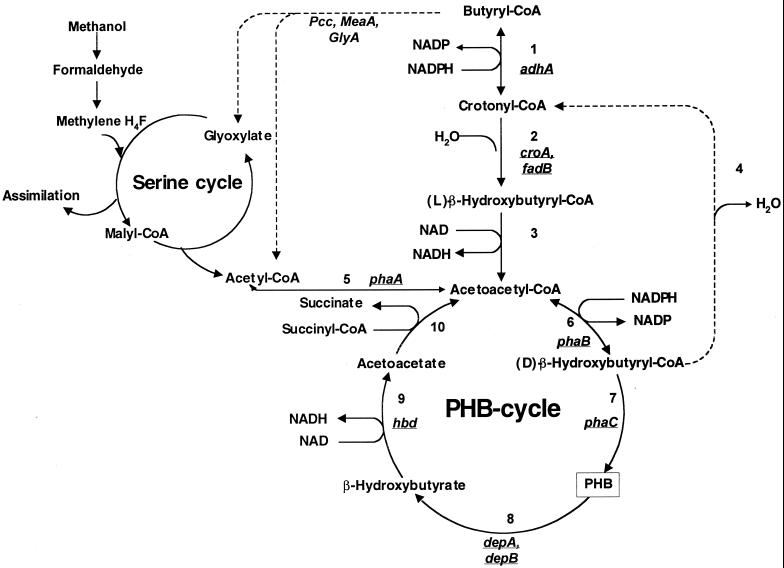
Pathways of PHB metabolism and fatty acid degradation in M. extorquens AM1 and their connection to methylotrophy. Enzymes of the fatty acid degradation pathway: 1, crotonyl-CoA reductase; 2, l-crotonase; 3, NADH-linked acetoacetyl-CoA reductase; 4, d-crotonase. Enzymes of PHB synthesis and degradation: 5, β-ketothiolase; 6, NADPH-linked acetoacetyl-CoA reductase; 7, PHB synthase; 8, PHB depolymerase; 9, β-hydroxybutyrate dehydrogenase; 10, acetoacetate–succinyl-CoA transferase. Dotted lines reflect enzymes not yet known or detected.
The degradation of PHB in most bacteria is catalyzed by PHB depolymerase, β-hydroxybutyrate dehydrogenase, acetoacetate–succinate-CoA transferase and β-ketothiolase (Fig. 1). β-Ketothiolase, NADH-linked acetoacetyl-CoA reductase, l-crotonase, and crotonyl-CoA reductase are also involved in the fatty acid degradation pathway (1) (Fig. 1).
Genes responsible for polyhydroxyalkanoic acid (PHA) synthesis have been isolated and characterized from several bacteria. In R. eutropha the genes encoding β-ketothiolase (phaA), acetoacetyl-CoA reductase (phaB), and PHB synthase (phaC) are closely linked on the chromosome (33, 38, 42). In Rhizobium meliloti, Paracoccus denitrificans, and Z. ramigera the two genes, phaA and phaB, are organized in an operon (32, 44, 48). In P. denitrificans phaC is located on the chromosome together with genes encoding PHB depolymerase (orf1), PHB granule-associated protein (phaP), and a regulator for PHB granule-associated protein (phaR) (23). The PHB synthase gene (phaC) was isolated and characterized from M. extorquens strain IBT no.6. In this strain, it has been shown that phaC is located separately from other genes encoding enzymes for PHB biosynthesis (45).
In order to understand carbon flow during methylotrophic growth of M. extorquens AM1, it is necessary to understand PHB synthesis and degradation, since cells growing exponentially on methanol accumulate a significant proportion of their biomass as PHB (42). The goal of the current study was to characterize genes participating in PHB cycling in M. extorquens AM1 and to determine their roles in methylotrophic metabolism.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Escherichia coli DH5α (Bethesda Research Laboratories) and S17-1 (40) were used in the study. They were grown in the Luria-Bertani medium in the presence of appropriate antibiotics as described by Maniatis et al. (24). M. extorquens AM1 was grown in a minimal medium described previously (18). Succinate (20 mM), methanol (100 mM), ethanol (50 mM), ethylamine (20 mM), formate (40 mM), pyruvate (40 mM), or β-hydroxybutyrate (20 mM) were used as substrates. The following antibiotic concentrations were used for M. extorquens AM1: tetracycline, 10 μg ml−1; kanamycin, 100 μg ml−1; and rifamycin, 50 μg ml−1. The growth responses of mutants were tested on plates containing the substrates listed above in the presence or absence of supplements of glyoxylate (5 mM), glycolate (20 mM), acetoacetate (0.1%), β-hydroxybutyrate (0.1%), or butyrate (0.04%). The following cloning vectors were used: pUC19 (Pharmacia) for cloning and subcloning, pAYC61 (8) as a suicide vector, pRK2013 (12) as a helper plasmid, and pCR2.1 (Invitrogen) for cloning of PCR products. In addition, a recently developed small IncP plasmid, pCM80 (C. Marx and M. Lidstrom, unpublished data), was used. pCM80 is an expression vector designed for use in M. extorquens AM1 that utilizes the strong promoter upstream of the methanol dehydrogenase gene cluster (26).
Cloning of PHB cycle genes.
Data from the M. extorquens AM1 genome project (http://faculty.washington.edu/lidstrom/genome.html) were used for designing primers specific for putative genes of M. extorquens AM1 encoding the following enzymes: intracellular PHB depolymerases (phb1, 5′-TCGCCGACCGCCGTGAAC-3′, and phb2 reverse, 5′-GCGAGATACTCGTCGTAGAAG-3′, complementary to nucleotides 608 to 625 and nucleotides 861 to 841, respectively, with respect to the translation start site of gene encoding first depolymerase; phb3, 5′-GTGATGTCGTCCTTCTCGCC-3′, and phb4 reverse 5′-GCTTGATTCAAGCGAGCTGC-3′, complementary to nucleotides 1024 to 1005 and nucleotides −38 to −19, respectively, with respect to the translation start site of gene encoding second depolymerase), acetoacetyl-CoA reductase (re1, 5′-GAACGCGTCGCCCTCGTCACG-3′, and re2, 5′-GCCGTTGATCGACGAGATG-3′, complementary to nucleotides 10 to 30 and nucleotides 416 to 398, respectively, with respect to the translation start site of gene encoding acetoacetyl-CoA reductase), β-ketothiolase (th1, 5′-CGCACTGGAGCGCGCAGGCG-3′, and th2, 5′-CGAAAGCCTCGTTCGCCTCG-3′, complementary to nucleotides 116 to 135 and nucleotides 960 to 941, respectively, with respect to the translation start site of gene encoding β-ketothiolase), β-hydroxybutyrate dehydrogenase (hbd1, 5′-CATCGCCAAGCACTTCGCC-3′, and hbd2, 5′-CCGGTGATCTGGCTGGCGCTG-3′, complementary to nucleotides 59 to 77 and nucleotides 745 to 725, respectively, with respect to the translation start site of gene encoding β-hydroxybutyrate dehydrogenase), and crotonase (kr1, 5′-CATATAGGTCATTACGATCAG-3′, and kr2, 5′-GCCTGAGACGCTTGACGAGGAAC-3′, complementary to nucleotides 755 to 736 and nucleotides −29 to −7, respectively, with respect to the translation start of gene encoding crotonase). GenBank data (http://www.ncbi.nlm.nih.gov) were used for the design of primers specific to the gene encoding PHA synthase of M. extorquens AM1. The primers used were as follows: s1, 5′-GCAACGATCTGATGGAGTTG-3′, and s2, 5′-GATCCTCGACGATCAGCCAG-3′, which is complementary to nucleotides 721 to 740 and nucleotides 2417 to 2398, respectively, with respect to the translation start of gene encoding PHA synthase.
Cloning of transhydrogenase genes.
A 3.05-kb PCR fragment containing pntA and pntB of E. coli (GenBank accession no. AAC7467) was cloned into pCR2.1, the resulting plasmid was digested with HindIII-XbaI, and this fragment was cloned into pCM80 in the correct orientation with respect to the mxaF promoter. The resulting plasmid was transferred into phaA and phaB mutants of M. extorquens AM1 by conjugation.
Cloning of isocitrate lyase gene.
A 1.31-kb PCR product containing icl of E. coli (GenBank accession no. X12431) was cloned into pCR2.1, restricted by HindIII-XbaI, and cloned into pCM80 in the correct orientation with respect to the mxaF and lacZ promoters. The resulting plasmid was transferred into phaA and phaB mutants of M. extorquens AM1.
DNA-DNA hybridization.
Preparation of nitrocellulose filters and DNA probes for DNA-DNA hybridizations was carried out as described previously (16).
DNA manipulations.
Plasmid isolation, E. coli transformation, restriction enzyme digestion, ligation, blunting ends with T4 DNA polymerase, or filling in ends with Klenow enzyme were carried out as described by Maniatis et al. (24). The chromosomal DNA of M. extorquens AM1 was isolated by the procedure of Saito and Miura (36).
DNA sequencing.
DNA sequencing from both strands was carried out with an Applied Biosystems automated sequencer by the Department of Biochemistry, University of Washington Sequencing Facility.
Computer analysis.
Translation and analyses of DNA and DNA-derived polypeptide sequences were carried out using Genetics Computer Group and NCBI ORF Finder programs (http://www.ncbi.nlm.nih.gov).
Enzyme assays.
Enzyme activities were determined in M. extorquens AM1 crude extracts obtained by passing cells through a French pressure cell at 1.2 × 108 Pa, followed by centrifugation for 10 min at approximately 15,000 × g. All measurements were done at room temperature (26°C) in a total volume of 1 ml. Pyridine nucleotide transhydrogenase was assayed as described by Zahl et al. (49). β-Ketothiolase (EC 2.3.1.16), acetoacetyl-CoA reductase (EC 1.1.1.36), and β-hydroxybutyrate dehydrogenase were determined by the methods of Senior and Dawes (39). l=(+)- and d-(−)-specific enoyl-CoA hydratases (EC 4.2.1.17) were measured as described by Shuto et al. (40). Isocitrate lyase was assayed according to the method of Copeland et al. (11). Spectrophotometric methods (21, 47) were used for protein determination.
Matings.
Triparental or biparental matings between E. coli and M. extorquens AM1 were performed overnight on nutrient agar at 30°C. Cells were then washed with sterile medium and plated on selective medium at appropriate dilutions. In triparental matings, pRK2013 (12) was used as a helper plasmid. Rifamycin was used for E. coli counterselection.
PHB analysis.
The PHB concentration in bacterial cells was determined by a gas chromatographic method (6). A model GC-14A capillary gas chromatograph (Shimadzu, Kyoto, Japan) with an AT-WAX capillary column (0.53 mm by 10 m; 1.2-μm film thickness) (Alltech, Deerfield, Ill.) and a flame ionization detector were used. The flow rate of helium carrier gas was 2.7 ml/min. The initial column temperature of 60°C was held for 2 min, and then the temperature was increased by 5°C/min up to 160°C.
Nucleotide sequence accession number.
The sequences of 1,206 nucleotides containing depA and of 6,100 nucleotides containing phaA and phaB have been deposited with GenBank under accession numbers AF287908 and AF287907, respectively.
RESULTS
Nucleotide sequence analysis.
Putative genes for the two intracellular PHB depolymerases, acetoacetyl-CoA reductase, β-ketothiolase, β-hydroxybutyrate dehydrogenase, crotonase, and PHA synthase were identified in the course of an ongoing M. extorquens AM1 genome sequencing project, by performing automated BLAST searches on raw single-read sequences from the M. extorquens AM1 genomic database (http://faculty.washington.edu/lidstrom/genome.html) by the high level of identity of the translated polypeptide sequences to, respectively, the intracellular PHB depolymerase of R. eutropha, the β-ketothiolase and acetoacetyl-CoA reductase of Z. ramigera, the β-hydroxybutyrate dehydrogenase of S. meliloti, the crotonase of Thermoanaerobacterium thermosaccharolyticum, and the PHA synthase of M. extorquens (3, 25, 32, 45). Primers were designed complementary to the coding regions of these sequences and used to amplify seven corresponding fragments by PCR. After the identities of the PCR products were confirmed by sequencing, three PCR fragments encoding PHB depolymerase, acetoacetyl-CoA reductase, and β-ketothiolase were used as probes against a cosmid library of M. extorquens AM1 chromosomal DNA (16). Two types of cosmids were identified by DNA-DNA (Southern) hybridization in filters: one that hybridized with both β-ketothiolase and acetoacetyl-CoA reductase probes and one that hybridized with the probe for PHB depolymerase. HindIII fragments from selected cosmids that hybridized to the appropriate probes were cloned into pUC19 and partially sequenced.
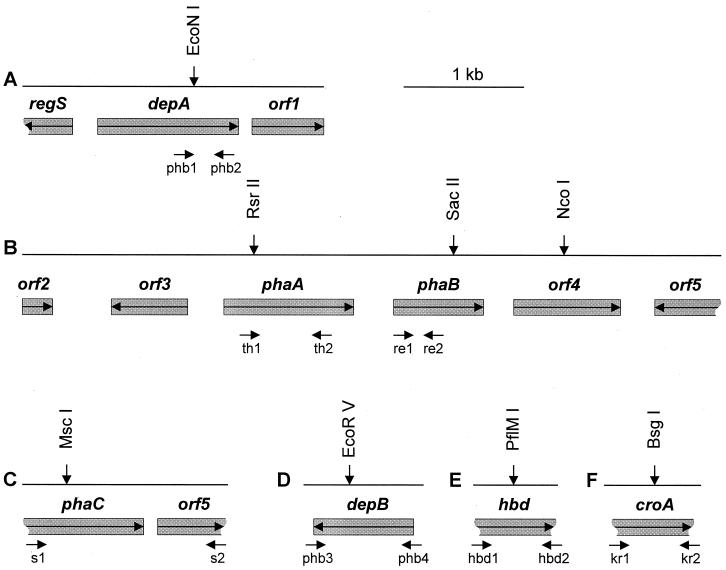
The complete PHB depolymerase gene was sequenced and designated depA (Fig. 2, top line). The amino acid sequence translated from depA showed the highest level of identity (47%) with the intracellular PHB depolymerase from R. eutropha (GenBank accession no. BAA33394). The two surrounding open reading frames (ORFs) showed identities (54 and 29%, respectively) with a putative histidine kinase from Bradyrhizobium japonicum (GenBank accession no. CAA06858) and a hypothetical protein from Deinococcus radiodurans (GenBank accession no. AAF12030). These were designated regS and orf1, respectively (Fig. 2).
FIG. 2.
Physical map of the M. extorquens AM1 chromosomal regions analyzed in this study. Only restriction sites discussed here are shown. Arrows within the boxes indicate the direction of transcription, vertical arrows indicate the sites of insertion mutations, and the small arrows underneath indicate the primers used to generate PCR products.
The physical map of the DNA region containing genes for β-ketothiolase and acetoacetyl-CoA reductase is shown in Fig. 2 in the center line. The polypeptide translated from the first ORF showed the highest level of identity (60%) with the ribosomal protein L32 of Neisseria meningitidis (GenBank accession no. CAB83837 [31]). This gene was designated orf2. The polypeptide translated from the second ORF showed the highest identities with an unknown protein located in the phb locus of S. meliloti (GenBank accession no. AAA90981; 52% identity [3]) and also with a regulator for granule-associated protein (phaR) in P. denitrificans (GenBank accession no. BAA77259; 32% identity [23]) and phbF of R. eutropha (GenBank accession no. AAC3819; 41% identity [42]). This gene was designated orf3. The third ORF encodes a polypeptide that is similar to β-ketothiolase (PhaA) of Z. ramigera (GenBank accession no. 7766963; 67% identity [25]). This gene was designated phaA. The next ORF located downstream from phaA and its translated product showed a high degree of identity with acetoacetyl-CoA reductase (PhaB) from Z. ramigera (GenBank accession no. P23238; 67% identity [32]). This gene was designated phaB. The polypeptides translated from the two last ORFs showed identities with lauroyl acyltransferase (htrB) of Rickettsia prowazekii (GenBank accession no. CAA15149; 26% identity [2]) and aminopeptidase of Thermus aquaticus (GenBank accession no. P42778; 57% identity [30]), respectively. These genes were designated orf4 and orf5.
The remaining genes of interest were completely or partially sequenced from the corresponding PCR fragments. The complete gene for the second PHB depolymerase (depB) was identified within the 1,060-bp PCR fragment (Fig. 2, bottom line). The polypeptide translated from depB showed high identity with the intracellular PHB depolymerase of R. eutropha (GenBank accession no. BAA33394; 49% identity) and depA of M. extorquens AM1 described above (72% identity). The gene for β-hydroxybutyrate dehydrogenase (hbd) was identified as a partial ORF within the 700-bp PCR fragment. The translated product of hbd showed high identity with β-hydroxybutyrate dehydrogenase (Hbd) of S. meliloti (GenBank accession no. AAD12733; 60% identity in the 229-amino-acid overlap [3]). The gene for crotonase (croA) was identified as a partial ORF in the 791-bp PCR product. The product translated from croA showed identity with crotonase (CroA) of T. thermosaccharolyticum (GenBank accession no. CAB07495; 36% identity in a 164-amino-acid overlap). The last PCR fragment contained 1,700 bp encoding two partial ORFs. The partial ORFs showed 100% identity at the nucleotide sequence level to polyhydroxyalkanoic acid synthase (PhaC) from M. extorquens strain IBT no.6 and a hypothetical protein in the phaC 3′ region of the same strain (GenBank accession no. AAA72330 [45]), respectively. These genes were designated phaC and orf6, respectively (Fig. 2).
Construction of insertion mutations.
Insertion mutations in depA, depB, orf4, phaA, phaB, phaC, hbd, and croA were constructed in vitro using the kanamycin resistance (Kmr) gene cartridge as described earlier (8). The mutation sites for each gene are indicated in Fig. 2. Mutants were selected in the presence of Kanamycin, and Kmr colonies were checked for their resistance to tetracycline. Tcs colonies were chosen as potential double-crossover recombinants, while Tcr colonies were assumed to be single-crossover recombinants. The identity of the double-crossover mutants was confirmed by diagnostic PCR with primers specific to the insertion sites.
Phenotypic analysis of insertion mutants. (i) Growth substrates.
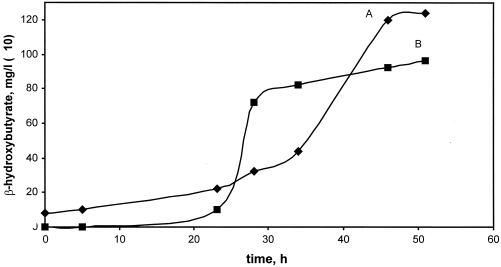
Mutations in all the genes produced double-crossover recombinants (null mutants) on succinate plates with a frequency of 10 to 50% relative to the total number of recombinants. These results demonstrated that none of these genes were required for growth on succinate. Growth responses were determined for double-crossover recombinant insertion mutants. All mutants grew on both succinate and pyruvate on plates, and all grew normally on a combination of methanol and succinate, demonstrating that none were methanol sensitive. In addition, depA, depB, and croA mutants grew normally on all compounds tested, including C1 compounds (methanol and formate), C2 compounds (ethylamine and ethanol), and β-hydroxybutyrate. The mutant in hbd grew normally on methanol, formate, ethylamine, and ethanol but did not grow on β-hydroxybutyrate. During growth on methanol, this mutant accumulated β-hydroxybutyrate (Fig. 3).
FIG. 3.
Characteristics of cell growth (A) and 3-hydroxybutyrate accumulation (B) of the hbd mutant grown on methanol. Cell growth is measured by optical density at 600 nm (values shown on y axis have been multiplied by 100).
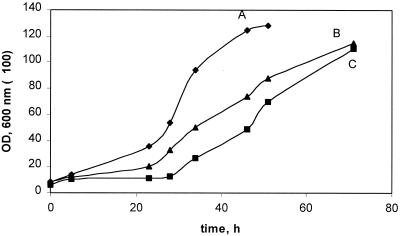
In contrast to the other mutants, the phaA, phaB, and phaC mutants showed multiple growth defects (Table 1). phaA and phaB mutants grew fairly well on plates on succinate. However, in liquid they both grew more slowly on succinate than wild-type M. extorquens AM1, with a longer growth lag and about half the growth rate of the wild type (Fig. 4). The phaA mutant also was unable to grow on methanol, formate, ethylamine, ethanol, or β-hydroxybutyrate, indicating that the product of phaA is required for both C1 and C2 metabolism. The phaB mutant grew poorly on methanol and formate compared to wild-type M. extorquens AM1 and did not grow on ethylamine, ethanol, or β-hydroxybutyrate (Table 1). The phaC mutant grew more slowly on methanol, ethylamine, ethanol, and β-hydroxybutyrate than wild-type M. extorquens AM1. However, this mutant produced with high frequency second-site suppressor mutants on methanol and ethylamine media. These grew normally on methanol, ethylamine, ethanol, and β-hydroxybutyrate, but were not revertants, as they still contained the Km insertion in the correct site.
TABLE 1.
Growth responses of pha mutants on plates
| Mutant | Growth responsea
on:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Succinate | Pyruvate | Methanol plus succinate | Methanol | Ethanol | Ethylamine | Methanol plus glyoxylate or glycolate | Ethylamine plus glyoxylate or glycolate | Formate | β-Hydroxybutyrate | |
| phaA | ++ | ++ | ++ | − | − | − | − | − | − | − |
| phaB | ++ | ++ | ++ | + | − | − | ++ | ++ | + | − |
| phaCb | ++ | ++ | ++ | + | + | + | NT | NT | + | + |
++, Similar to wild type; +, decrease compared to wild type; −, no growth; NT, not tested.
Generates second site suppressor mutants at high frequency.
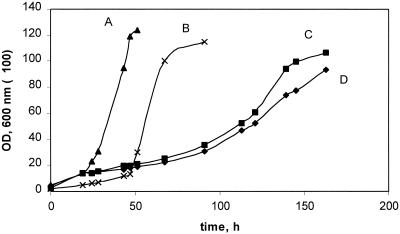
FIG. 4.
Growth of M. extorquens AM1 (A) and the phaA (C) and phaB (B) mutants on succinate.
The simultaneous loss of ability to grow on C1 and C2 compounds is associated with defects in an unknown portion of the serine cycle for formaldehyde assimilation that functions to oxidize acetyl-CoA to glyoxylate (10, 13). Therefore, the phenotypes of the phaA and phaB mutants suggested that these genes might be involved in this unknown acetyl-CoA oxidation pathway. Mutants defective in this pathway are characterized by the ability to grow on C1 and C2 compounds when supplemented with glyoxylate or glycolate (13). These compounds were tested for the ability to restore the growth of the phaA and phaB mutants on methanol, ethylamine, and ethanol. Both supplements were able to restore growth of the phaB mutant on C1 and C2 compounds but did not affect the growth of the phaA mutant. However, β-hydroxybutyrate or acetoacetate were able to partially rescue growth of the phaA mutant on methanol (Fig. 5). It was not possible to test the growth of the phaC mutants in liquid culture due to the outgrowth of the suppressor mutants.
FIG. 5.
Growth of M. extorquens AM1 on methanol (A) and on methanol in the presence of 0.1% β-hydroxybutyrate (B) and of phaA mutant on methanol in the presence of 0.1% acetoacetate (C) and in the presence of 0.1% β-hydroxybutyrate (D).
(ii) Enzyme activities.
The activities of β-ketothiolase and NADPH- and NADH-linked acetoacetyl-CoA reductase were measured in wild type and in phaA and phaB mutants grown on succinate (Table 2). β-Ketothiolase activity was not detectable in the phaA mutant, while it was present in wild-type M. extorquens AM1 and the phaB mutant, confirming that phaA is necessary for β-ketothiolase activity. NADPH-linked acetoacetyl-CoA reductase activity was not found in the phaB mutant, while it was present at a low level in wild-type M. extorquens AM1 and the phaA mutant. Low levels of NADH-linked acetoacetyl-CoA reductase were found in the phaA and phaB mutants and in wild-type M. extorquens AM1. These results show that phaB is necessary for NADPH-linked acetoacetyl-CoA reductase and that a separate enzyme must be present that accounts for NADH-linked acetoacetyl-CoA reductase activity.
TABLE 2.
Activity of PHB cycling enzymes in extracts of succinate-grown M. extorquens AM1 wild-type and mutants.
| Enzyme | Activity (nmol/min/mg of
protein)a
|
||||
|---|---|---|---|---|---|
| AM1 | PhaA− | PhaB− | Cro− | Hbd− | |
| β-Ketothiolase | 392 | 0 | 400 | NT | NT |
| Acetoacetyl-CoA reductase | |||||
| NADH-dependent | 17 | 12 | 11 | NT | NT |
| NADPH-dependent | 13 | 10 | 0 | NT | NT |
| l-(+)-Crotonase | 25 | NT | NT | 17 | NT |
| NAD-dependent β-hydroxybutyrate dehydrogenase | 9 | NT | NT | NT | 0 |
Values are the average of three to four determinations; the range was ±10%. AM1, wild-type M. extorquens AM1; 0, not detectable; NT, not tested.
Further proof of the identity of phaB was obtained by overexpressing this gene in E. coli and M. extorquens AM1. A 1.39-kb PCR product containing phaB was cloned into pCR2.1 and subsequently subcloned into pCM80 in the correct orientation with respect to the mxaF and lacZ promoters. The plasmid was transferred into M. extorquens AM1. The activity of acetoacetyl-CoA reductase with NADH and NADPH were measured in E. coli and M. extorquens AM1 containing phaB and also in plasmid-free strains. In E. coli carrying phaB, high NADPH-linked acetoacetyl-CoA reductase activity (71 mU/mg) was found, while no activity was detected in the plasmid-free strain. The NADH-linked activity was at the same low level (about 1 mU/mg) in the control and plasmid-bearing strains. In M. extorquens AM1 containing the construct with phaB, the activity of the NADPH-linked acetoacetyl-CoA reductase increased to 42 mU/mg of protein in M. extorquens AM1. These results identify phaB as the gene encoding the NADPH-specific acetoacetyl-CoA reductase.
The l-(+)- and d-(−)-specific crotonase activities were measured in wild-type M. extorquens AM1 and in the croA mutant grown on succinate or methanol. d-(−)-Specific crotonase was not detectable in wild-type M. extorquens AM1 or the mutant. l-(+)-specific crotonase was present in both wild-type M. extorquens AM1 and the croA mutant (Table 2). These data suggested that M. extorquens AM1 may have multiple genes responsible for crotonase activity. This idea is supported by the fact that a putative gene for the α-subunit of the fatty oxidation complex (which includes enoyl-CoA hydratase, β-hydroxyacyl-CoA dehydrogenase, and β-hydroxybutyryl-CoA epimerase) was also identified in the course of our M. extorquens AM1 genome sequencing project. The gene product showed identity (44% in a 231-amino-acid overlap) with the translated polypeptide sequence of fadB (putative fatty oxidation protein) of Mycobacterium tuberculosis (GenBank accession no. CAA17666).
The activity of NAD-linked β-hydroxybutyrate dehydrogenase was measured in wild-type M. extorquens AM1 and the hbd mutant grown on succinate. The activity was not detectable in the hbd mutant, while it was present in wild-type M. extorquens AM1 at a low level (Table 2), confirming the necessity of hbd for β-hydroxybutyrate dehydrogenase activity.
(iii) PHB accumulation.
PHB concentrations were determined as the percentage of dry weight in depA, depB, phaA, phaB, phaC, hbd, and croA mutants and also in wild-type M. extorquens AM1 grown on succinate or methanol (Table 3). depA, depB, hbd, and croA mutants accumulated normal levels of PHB (15 to 20% on succinate and 30 to 43% on methanol). phaA, phaB, and phaC mutants did not accumulate detectable PHB during growth on succinate. The phaB mutant accumulated a small amount (7 to 10%) of PHB during growth on methanol, demonstrating the presence of an alternate pathway for β-hydroxybutyryl-CoA synthesis. It is possible that this pathway uses enzymes participating in fatty acid degradation (Fig. 1). The second site suppressor phaC mutants showing wild-type growth characteristics (see above) also did not accumulate PHB during growth on methanol or ethylamine, confirming that they retained the PHB synthesis defect of the original phaC mutant.
TABLE 3.
Biosynthesis of PHB by M. extorquens AM1 and mutants during growth on methanol and succinate
| Strain | Carbon source | PHB (% [dry biomass wt])a |
|---|---|---|
| M. extorquens AM1 | Succinate | 18–20 |
| M. extorquens AM1 | Methanol | 34–42 |
| Mutantsb | Succinate | 15–20 |
| Mutantsb | Methanol | 30–43 |
| phaA mutant | Succinate | 02 |
| phaB mutant | Succinate | 0 |
| phaB mutant | Methanol | 7–9 |
| phaC mutant | Succinate | 0 |
| phaC suppressor mutant | Methanol | 0 |
Range of three determinations. 0, Not detectable.
One depA mutant, one depB mutant, one croA mutant, and one hbd mutant.
Expression of E. coli pyridine nucleotide transhydrogenase in phaA and phaB mutants.
It has been suggested that PHB can serve as a redox regulator, maintaining NADPH balance in the cell (1). It seemed possible that the pha defects might result in an increase in NADPH pools, which could in turn cause inhibition of key NADPH-dependent enzymes important for C1 and C2 metabolism and might cause the growth defects. To test this hypothesis, the pntA and pntB genes of E. coli, encoding the α and β subunits of pyridine nucleotide transhydrogenase were expressed in phaA and phaB mutants. Transhydrogenase expression should result in decreased NADPH levels in the mutants. The activity of pyridine nucleotide transhydrogenase was measured in phaA and phaB mutants carrying pntA and pntB genes transcribed from the mxaF promoter and shown to be present at higher levels (63 and 92 mU/mg in phaA and phaB mutants containing the construct with pntA and pntB, respectively) than in the mutants without the construct (13 mU/mg in phaA and phaB mutants). However, these transconjugants did not regain the ability to grow on methanol or ethylamine plates. These results show that while a significant increase in transhydrogenase activity was achieved, it had no effect on the growth of the mutants.
Expression of icl of E. coli in phaA and phaB mutants.
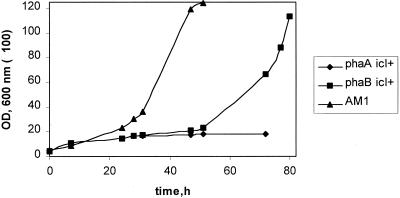
To verify the participation of phaA and phaB in the oxidation of acetyl-CoA to glyoxylate, the isocitrate lyase gene (icl) of E. coli was expressed in phaA and phaB mutants. Isocitrate lyase should allow the completion of the glyoxylate cycle in M. extorquens AM1 and should bypass the need for the native acetyl-CoA oxidation pathway. In phaA and phaB mutants carrying icl, the activity of isocitrate lyase was detected (89 and 242 mU/mg, respectively), while no activity was found in the plasmid-free mutants. The phaA and phaB transconjugants did not grow on ethylamine. However, the phaA transconjugants grew very slowly on methanol (Fig. 6). The phaB transconjugants grew more slowly on methanol than wild-type M. extorquens AM1 (Fig. 6). These data suggest that the NADPH-linked acetoacetyl-CoA reductase and β-ketothiolase are involved in the unknown pathway for glyoxylate regeneration in M. extorquens AM1.
FIG. 6.
Growth on methanol of M. extorquens AM1 wild type and of the phaA and phaB transconjugants containing the E. coli icl.
DISCUSSION
In this study, we have used preliminary genome sequence data to assess genes in M. extorquens AM1 involved in the biosynthesis and degradation of PHB and also in fatty acid degradation. Synthesis of PHB as a reserve material is a common feature among methylobacteria with the serine pathway for C1 assimilation (14). M. extorquens has been reported to produce PHB at up to 64% of the dry cell mass (43). Three enzymes are involved in PHB synthesis in M. extorquens: β-ketothiolase, NADPH-linked acetoacetyl-CoA reductase, and PHB synthase. Degradation of PHB in M. extorquens is catalyzed by PHB depolymerase, β-hydroxybutyrate dehydrogenase, acetoacetate–succinate-CoA transferase, and β-ketothiolase.
We have shown that the genes encoding β-ketothiolase and NADPH-linked acetoacetyl-CoA reductase (phaA and phaB) are closely linked on the M. extorquens AM1 chromosome and are transcribed in the same direction. It is not yet known whether the two are cotranscribed. Another gene was found in the region whose translated product showed identity with the regulator for a PHB granule-associated protein of P. denitrificans. In addition, M. extorquens AM1 has two genes, depA and depB, predicted to encode intracellular PHB depolymerase. These genes show high identity to each other and are located on different chromosomal fragments.
Mutants unable to synthesize PHB have been isolated from several bacterial species, i.e., R. eutropha, R. rubrum, Rhodobacter capsulatus, Rhodobacter sphaerodes, Rhizobium etli, and R. meliloti (7, 20, 22, 35, 37). Most mutants are defective in the PHB synthase gene (the phaC equivalent), and in most cases no obvious growth phenotype is associated with the inability to synthesize PHB. However, in R. etli, a phaC mutant showed reduced growth characteristics on minimal medium in comparison with wild-type strains and excreted large amounts of organic acids (7). In R. capsulatus a phaC mutant was sensitive to fatty acids (22). However, R. capsulatus phaA and phaAB mutants grew normally on tested substrates and were able to synthesize PHB, suggesting the presence of an alternative route for β-hydroxybutyryl-CoA synthesis (22).
In contrast to the results in R. capsulatus the phaA mutant of M. extorquens AM1 did not accumulate PHB, while the phaB mutant accumulated reduced amounts of PHB on methanol but did not accumulate PHB on succinate. These results indicate that both phaA and phaB are central to PHB biosynthesis in M. extorquens AM1. It is likely that a low-level alternate route of β-hydroxybutyryl-CoA synthesis is present that accounts for the small amount of PHB in methanol-grown cells.
Surprisingly, M. extorquens AM1 phaA, phaB, and phaC mutants showed growth defects on both C1 and C2 compounds. The defects are most severe in the phaA mutant, but the phaB mutant also exhibited very poor growth on C1 compounds and no growth on C2 compounds. The phaC mutant could grow on all tested substrates but at a slower rate on C1 and C2 compounds. Moreover, the phaB mutant phenotype on methanol and ethylamine was rescued by the addition of glyoxylate or glycolate, while the phaA mutant phenotype on methanol was partially rescued by the addition of β-hydroxybutyrate or acetoacetate.
A possible explanation for the growth phenotypes of the mutants is that the lack of PHB synthesis causes NADPH or NADH to accumulate to inhibitory levels by preventing NADPH consumption. NADH accumulation has been demonstrated in phaC mutants in R. etli, and inhibition of tricarboxylic acid cycle enzymes by NADH has been proposed as an explanation for the slow growth and organic acid excretion by this mutant when grown on glucose and succinate (7). Our results suggest this explanation is unlikely for M. extorquens AM1 because the mutants show growth defects on formate. Formate is a low-energy growth substrate and the only direct source of reducing equivalents in cells grown on formate is NADH produced via formate dehydrogenase. Therefore, NADPH should be limiting in this growth condition and NADH should not be in excess. Another line of evidence in this regard is the result from overexpression of pyridine nucleotide transhydrogenase. M. extorquens AM1 shows a low activity of transhydrogenase, and the main sink for NADPH in methanol- and ethylamine-grown cells is PHB synthesis. Therefore, it is possible that NADPH accumulates in the PHB− mutants, which might become inhibitory on these growth substrates. However, overexpression of the pyridine nucleotide transhydrogenase in these mutants did not affect their phenotypes. If NADPH accumulation were the main reason for the growth defect, then we might expect that some alleviation would occur under these conditions. In addition, it is not clear why glyoxylate or glycolate would rescue the growth phenotype of phaB mutants if the accumulation of reduced pyridine nucleotides were a major reason for the defects.
The alternative possibility is that β-ketothiolase and NADPH-linked acetoacetyl-CoA reductase are involved in the unknown pathway for glyoxylate regeneration from acetyl-CoA, since other mutants defective in this pathway show growth on C1 and C2 compounds in the presence of glyoxylate or glycolate. Serine cycle methylotrophs such as M. extorquens AM1 lack isocitrate lyase, and the pathway used for assimilation of acetate, which also operates during growth on C1 compounds and β-hydroxybutyrate, remains unknown. It has been shown previously using experiments with 14C-labeled C2 compounds that glyoxylate and glycolate are intermediates in this pathway (13).
Previously two M. extorquens AM1 chromosomal fragments have been identified as carrying genes encoding enzymes involved in assimilation of both C1 and C2 compounds and probably operating in the unknown pathway for glyoxylate regeneration. One of these, glyA, encodes serine hydroxymethyltransferase. This enzyme probably plays a dual role in M. extorquens AM1: both as the first enzyme in the serine cycle and as an enzyme in the glyoxylate regeneration pathway (9). The other chromosomal fragment is not linked to the fragment mentioned above and has been shown to contain three genes involved in the unknown pathway for glyoxylate regeneration, i.e., meaA, pccA, and adhA (10). The pccA gene product has been demonstrated to be a propionyl-CoA carboxylase (10). adhA has strong similarity to ccr, the gene encoding a NADPH-linked crotonyl-CoA reductase in Streptomyces collinus (17). This enzyme catalyzes the conversion of crotonyl-CoA to butyryl-CoA. We have recently shown that adhA of M. extorquens AM1 does encode the NADP-linked crotonyl-CoA reductase (enzyme 1 in Fig. 1; L. V. Chistoserdova and M. E. Lidstrom, unpublished results).
meaA encodes a novel CoB12-dependent mutase that is not methylmalonyl-CoA mutase or isobutyryl-CoA mutase (10, 17). A similar gene is present downstream of ccr in S. collinus. meaA- and ccr-blocked mutants of S. collinus exhibit dramatically reduced growth characteristics when acetate is the sole carbon source (17). This result suggests that both ccr and meaA are probably involved in the unknown pathway of acetate assimilation in S. collinus, similar to the one operating in M. extorquens AM1.
The phenotypes of the new mutants of M. extorquens AM1 described here indicate that the initial step of the glyoxylate regeneration pathway may be proceeding via intermediates of the PHB biosynthesis pathway (enzymes 5 and 6 in Fig. 1) and reversible reactions of fatty acid degradation (enzymes 1 and 4 in Fig. 1). It is not clear why the phaA mutants are not rescued for growth on C1 and C2 compounds by glyoxylate or glycolate. It is possible that these mutants accumulate an intermediate that has an inhibitory action or that they have low levels of an intermediate that is required for enzyme activation or for other pathways. These mutants are partially rescued for growth on C1 and C2 compounds by acetoacetate and β-hydroxybutyrate, pointing to acetoacetate or acetoacetyl-CoA as possible key intermediates in the unknown pathway.
As noted above, the M. extorquens AM1 phaB mutant accumulates low concentrations of PHB during growth on methanol. This phenotype indicates that an alternative mechanism for β-hydroxybutyryl-CoA production must be present in M. extorquens AM1. It is likely that NADH-linked acetoacetyl-CoA reductase (enzyme 3 in Fig. 1) and l- and d-crotonases (enzymes 2 and 4 in Fig. 1) supply PHB biosynthesis with a β-hydroxybutyryl-CoA pool and account for the slow growth of the mutants on methanol. Although we were unable to detect d-crotonase activity in cell extracts, it is possible that the activity is labile or requires special conditions to detect. Alternatively, another unknown pathway for d-β-hydroxybutyryl-CoA synthesis may exist in M. extorquens AM1.
The phaC mutant grows slowly on methanol and ethylamine but forms second site suppressor mutants on these substrates with high frequency. These suppressor mutants are not able to accumulate PHB, but they do regain the ability to grow normally on methanol or ethylamine, demonstrating that the growth defects are not due to the inability to synthesize PHB per se. These results suggest that PHB synthase is not required for assimilation of C1 and C2 compounds. Although more detailed studies will be necessary to determine the nature of the suppressors, it is possible that the phaC mutants accumulate an inhibitory intermediate or block the production of another key intermediate, as with the phaA mutants. In this case, the suppressor mutants may upregulate or downregulate a consumption or production pathway or alter a target of the inhibition. Since suppressor mutants do not accumulate at a significant frequency in the phaA mutants, the phenotypes must be due to different effects.
The mutant in hbd has lost the ability to use β-hydroxybutyrate as sole carbon source confirming participation of hbd in the first step of β-hydroxybutyrate utilization. In addition, these mutants accumulate β-hydroxybutyrate without affecting growth on methanol, indicating that this intermediate is not inhibitory at these levels. The croA, depA, depB, and orf4 mutants had a wild-type phenotype. Thus, we have shown that β-hydroxybutyrate dehydrogenase and the croA, depA, depB, and orf4 products are not required for the assimilation of C1 and C2 compounds.
The details of the pathway for converting acetyl-CoA to glyoxylate in M. extorquens AM1 and other bacteria lacking isocitrate lyase are still not known. However, the results presented here strongly suggest that the first part of this pathway involves the conversion of two acetyl-CoA molecules to butyryl-CoA (Fig. 1).
ACKNOWLEDGMENTS
This work was supported by a joint grant from the EPA (R826729) and the NSF (BES9819957). The genome sequence data used in this project were obtained through support from the NIH (GM 98933-01).
REFERENCES
- 1.Anderson A J, Dawes E A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev. 1990;54:450–472. doi: 10.1128/mr.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson S G, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C, Podowski R M, Naslund A K, Eriksson A S, Winkler H H, Kurland C G. The genome sequence of Rickettsia prowazekiiand the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 3.Aneja P, Charles T C. Poly-3-hydroxybutyrate degradation in Rhizobium (Sinorhizobium) meliloti: isolation and characterization of a gene encoding 3-hydroxybutyrate dehydrogenase. J Bacteriol. 1999;181:849–857. doi: 10.1128/jb.181.3.849-857.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer E, Kaspar T, Fischer H M, Hennecke H. Expression of the fixR-nifA operon in Bradyrhizobium japonicum depends on a new response regulator, regR. J Bacteriol. 1998;180:3853–3863. doi: 10.1128/jb.180.15.3853-3863.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belova L L, Sokolov A P, Sidorov I A, Trotsenko Y A. Purification and characterization of NADPH-dependent acetoacetyl-CoA reductase from Methylobacterium extorquens. FEMS Microbiol Lett. 1997;156:275–279. [Google Scholar]
- 6.Braunegg G, Sonnleitner B, Lafferty R M. A rapid method for the determination poly-β-hydroxybutyric acid in microbial biomass. Eur J Appl Microbiol Biotechnol. 1978;6:29–37. [Google Scholar]
- 7.Cevallos M G, Encarnacion S, Leija A, Mora Y, Mora J. Genetic and physiological characterization of a Rhizobium etlimutant strain unable to synthesize poly-β-hydroxybutyrate. J Bacteriol. 1996;178:1646–1654. doi: 10.1128/jb.178.6.1646-1654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chistoserdov A Y, Chistoserdova L V, Mclntire W S, Lidstrom M E. Genetic organization of mau gene cluster in Methylobacterium extorquens AM1: complete nucleotide sequence and generation and characteristics of maumutants. J Bacteriol. 1994;176:4052–4065. doi: 10.1128/jb.176.13.4052-4065.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chistoserdova L V, Lidstrom M E. Cloning, mutagenesis and physiological effect of a hydroxypyruvate reductase gene from M. extorquensAM1. J Bacteriol. 1992;174:71–77. doi: 10.1128/jb.174.1.71-77.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chistoserdova L V, Lidstrom M E. Molecular characterization of a chromosomal region involved in the oxidation of acetyl-CoA to glyoxylate in the isocitrate-lyase-negative methylotroph Methylobacterium extorquensAM1. Microbiology. 1996;142:1459–1468. doi: 10.1099/13500872-142-6-1459. [DOI] [PubMed] [Google Scholar]
- 11.Copeland L, Quinnell R G, Day D A. Malic enzyme activity in bacteroids from soybean nodules. J Gen Microbiol. 1989;135:2005–2011. [Google Scholar]
- 12.Ditta G, Schmidhauser T, Yakobson F, Lu P, Liang X, Finlay D, Guiney D, Helinski D. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and monitoring gene expression. Plasmid. 1985;13:149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- 13.Dunstan P M, Anthony C, Drabble W T. Microbial metabolism of C1 and C2 compounds. The role of glyoxylate, glycollate and acetate in the growth of Pseudomonas AM1 on ethanol and on C1compounds. Biochem J. 1972;128:107–115. doi: 10.1042/bj1280107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Föllner C G, Madkour M, Mayer F, Babel W, Steinbüchel A. Analysis of the PHA granyle-associated proteins GA20 and GA11 in Methylobacterium extorquens and Methylobacterium rhodesianum. J Basic Microbiol. 1997;37:11–21. doi: 10.1002/jobm.3620370104. [DOI] [PubMed] [Google Scholar]
- 15.Fukui T, Ito M, Saito T, Kenkichi T. Purification and characterization NADP-linked acetoacetyl-CoA reductase from Zoogloea ramigeraI-16-M. Biochem Biophys Acta. 1987;917:365–371. [PubMed] [Google Scholar]
- 16.Fulton G F, Nunn D N, Lidstrom M E. Molecular cloning of a malyl coenzyme A lyase gene from Pseudomonassp. strain AM1, a facultative methylotroph. J Bacteriol. 1984;160:718–723. doi: 10.1128/jb.160.2.718-723.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han L, Reynolds K A. A novel alternative anaplerotic pathway to the glyoxylate cycle in streptomycetes. J Bacteriol. 1997;179:5157–5164. doi: 10.1128/jb.179.16.5157-5164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harder W, Attwood M, Quayle J R. Methanol assimilation by Hyphomicrobiumspp. J Gen Microbiol. 1973;78:155–163. [Google Scholar]
- 19.Haywood G W, Anderson A J, Chu L, Dawes E A. The role of NADH and NADPH-linked acetoacetyl-CoA reductase in the poly-3-hydroxybutyrate synthesizing organism Alcaligenes eutrophus. FEMS Microbiol Lett. 1988;52:259–264. [Google Scholar]
- 20.Hustede E, Steibuechel A, Schlegel H G. Relationship between the photoproduction of hydrogen and the accumulation of PHB in non-sulphur purple bacteria. Appl Microbiol Biotechnol. 1993;39:87–93. [Google Scholar]
- 21.Kalb V F, Bernlohr R W. A new spectrophotometric assay for protein in cell extracts. Anal Biochem. 1977;82:362–371. doi: 10.1016/0003-2697(77)90173-7. [DOI] [PubMed] [Google Scholar]
- 22.Kranz R G, Gabbert K K, Locke T A, Madigan M T. Polyhydroxyalcanoate production in Rhodobacter capsulatus: genes, mutant, expression, and physiology. Appl Environ Microbiol. 1997;63:3003–3009. doi: 10.1128/aem.63.8.3003-3009.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maehara A, Ueda S, Nakano H, Yamane T. Analyses of a polyhydroxyalkanoic acid granule-associated 16-kilodalton protein and its putative regulator in the pha locus of Paracoccus denitrificans. J Bacteriol. 1999;181:2914–2921. doi: 10.1128/jb.181.9.2914-2921.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 25.Modis Y, Wierenga R K. Crystallographic analysis of the reaction pathway of Zoogloea ramigerabiosynthetic thiolase. J Mol Biol. 2000;297:1171–1182. doi: 10.1006/jmbi.2000.3638. [DOI] [PubMed] [Google Scholar]
- 26.Morris C J, Lidstrom M E. Cloning of methanol-inducible moxF promotor and its analysis in moxB mutant of Methylobacterium extorquensAM1rif. J Bacteriol. 1992;174:4444–4449. doi: 10.1128/jb.174.13.4444-4449.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moskowitz G J, Merrick J M. Metabolism of poly-β-hydroxybutyrate. II. Enzymatic synthesis of D(−)-β-hydroxybutyryl coenzyme A by enoyl hydratase from Rhodospirillum rubrum. Biochemistry. 1969;8:2748–2755. doi: 10.1021/bi00835a009. [DOI] [PubMed] [Google Scholar]
- 28.Mothes G, Babel W. Methylobacterium rhodesianumMB 126 possesses two acetacetyl-CoA reductases. Arch Microbiol. 1994;161:277–280. [Google Scholar]
- 29.Mothes G, Babel W. Methylobacterium rhodesianumMB 126 possesses two stereospecific crotonyl-CoA hydratases. Can J Microbiol. 1995;41:68–72. [Google Scholar]
- 30.Motoshima H, Minagawa E, Tsukasaki F, Kaminogawa S. Cloning of genes of the aminopeptidase T family from Thermus thermophilus. HB8 and Bacillus stearothermophilusNCIB8924: apparent similarity to the leucyl aminopeptidase family. Biosci Biotechnol Biochem. 1997;61:1710–1717. doi: 10.1271/bbb.61.1710. [DOI] [PubMed] [Google Scholar]
- 31.Parkhill J, Achtman M, James K D, Bentley S D, Churcher C, Klee S R, Morelli G, Basham D, Brown D, Chillingworth T, Davies R M, Davis P, Devlin K, Feltwell T, Hamlin N, Holroyd S, Jagels K, Leather S, Moule S, Mungall K, Quail M A, Rajandream M A, Rutherford K M, Simmonds M, Skelton J, Whitehead S, Spratt B G, Barrell B G. Complete DNA sequence of a serogroup A strain of Neisseria menigitidisZ2491. Nature. 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 32.Peoples O P, Sinskey A J. Fine structural analysis of the Zoogloea ramigera phaA-phaB locus encoding beta-ketothiolase and acetoacetyl-CoA reductase: nucleotide sequence of phaB. Mol Microbiol. 1989;3:349–357. doi: 10.1111/j.1365-2958.1989.tb00180.x. [DOI] [PubMed] [Google Scholar]
- 33.Peoples O P, Sinskey A J. Poly-β-hydroxybutyrate biosynthesis in Alcaligenes eutrophusH16. J Biol Chem. 1989;264:15293–15297. [PubMed] [Google Scholar]
- 34.Ritchie G A R, Senior P J, Dawes E A. The purification and characterization NADP-linked acetoacetyl-CoA reductase from Azotobacter beijerinckii. Biochem J. 1971;121:309–316. doi: 10.1042/bj1210309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Povolo S, Tombolini R, Morea A, Anderson A J, Casella S, Nuti M P. Isolation and characterisation of mutant of Rhizobium melilotiunable to synthesize poly-β-hydroxybutyrate. Can J Microbiol. 1994;40:823–829. [Google Scholar]
- 36.Saito H, Miura K-I. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochem Biophys Acta. 1963;72:619–629. [PubMed] [Google Scholar]
- 37.Schlegel H G, Lafferty R, Krauss I. The isolation of mutants not accumulating poly-β-hydroxybutyric acid. Arch Microbiol. 1970;71:283–294. doi: 10.1007/BF00410161. [DOI] [PubMed] [Google Scholar]
- 38.Schubert P, Krüger N, Steinbüchel A. Molecular analysis of the Alcaligenes eutrophuspoly(3-hydroxybutyrate) biosynthetic operon: identification of the N terminus of poly(3-hydroxybutyrate) synthase and identification of the promoter. J Bacteriol. 1991;173:168–175. doi: 10.1128/jb.173.1.168-175.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Senior P J, Dawes E A. The regulation of poly-3-hydroxybutyrate metabolism in Azotobacter beijerinckii. Biochem J. 1973;134:225–238. doi: 10.1042/bj1340225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shuto H, Fukui T, Saito T, Shirakura Y, Tomita K. An NAD-linked acetoacetyl-CoA reductase from Zoogloea ramigeraI-16-M. Eur J Biochem. 1981;118:53–59. doi: 10.1111/j.1432-1033.1981.tb05485.x. [DOI] [PubMed] [Google Scholar]
- 41.Simon R, Priefer U, Puhler A. Vector plasmids for in vivo manipulations of gram-negative bacteria. In: Puhler A, editor. Molecular genetics of the bacteria-plant interactions. Berlin, Germany: Springer-Verlag; 1983. pp. 98–106. [Google Scholar]
- 42.Slater S, Houmiel K L, Tran M, Mitsky T A, Taylor N B, Padgette S R, Gruys K J. Multiple beta-ketothiolases mediate poly(beta-hydroxyalkanoate) copolymer synthesis in Ralstonia eutropha. J Bacteriol. 1998;180:1979–1987. doi: 10.1128/jb.180.8.1979-1987.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki T, Yamane T, Schimizu S. Mass production of poly-β-hydroxybutyric acid by fed-batch culture with controlled carbon/nitrogen feeding. Appl Microbiol Biotechnol. 1986;24:370–374. [Google Scholar]
- 44.Tombolini R, Povolo S, Buson A, Squartini A, Nuti M P. Poly-beta-hydroxybutyrate (PHB) biosynthetic genes in Rhizobium meliloti41. Microbiology. 1995;141:2553–2559. doi: 10.1099/13500872-141-10-2553. [DOI] [PubMed] [Google Scholar]
- 45.Valentin H E, Steinbuechel A. Cloning and characterization of the Methylobacterium extorquenspolyhydroxyalkanoic-acid-synthase structural gene. Appl Microbiol Biotechnol. 1993;39:309–317. doi: 10.1007/BF00192084. [DOI] [PubMed] [Google Scholar]
- 46.Vrijbloed J W, Zerbe-Burkhardt K, Ratnatilleke A, Grubelnik-Leiser A, Robinson J A. Insertional inactivation of methylmalonyl coenzyme A (CoA) mutase and isobutyryl-CoA mutase genes in Streptomyces cinnamonensis: influence on polyketide antibiotic biosynthesis. J Bacteriol. 1999;181:5600–5605. doi: 10.1128/jb.181.18.5600-5605.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitaker J R, Granum P E. An absolute method for protein determination based on the difference in absorbance at 235 and 280 nm. Anal Biochem. 1980;109:156–159. doi: 10.1016/0003-2697(80)90024-x. [DOI] [PubMed] [Google Scholar]
- 48.Yabutani T, Maehara A, Ueda S, Yamane T. Analysis of β-ketothiolase and acetoacetyl-CoA reductase genes of methylotrophic bacterium, Paracoccus denitrificans and their expression in Escherichia coli. FEMS Microbiol Lett. 1995;133:85–90. doi: 10.1111/j.1574-6968.1995.tb07865.x. [DOI] [PubMed] [Google Scholar]
- 49.Zahl K J, Rose C, Hanson R L. Isolation and partial characterization of mutant of Escherichia colilacking pyridine nucleotide transhydrogenase. Arch Bichem Biophys. 1978;190:598–602. doi: 10.1016/0003-9861(78)90315-6. [DOI] [PubMed] [Google Scholar]