Abstract
ADP-glucose synthesis through ADP-glucose pyrophosphorylase defines the major rate-controlling step of storage polysaccharide synthesis in both bacteria and plants. We have isolated mutant strains defective in the STA6 locus of the monocellular green alga Chlamydomonas reinhardtii that fail to accumulate starch and lack ADP-glucose pyrophosphorylase activity. We show that this locus encodes a 514-amino-acid polypeptide corresponding to a mature 50-kDa protein with homology to vascular plant ADP-glucose pyrophosphorylase small-subunit sequences. This gene segregates independently from the previously characterized STA1 locus that encodes the large 53-kDa subunit of the same heterotetramer enzyme. Because STA1 locus mutants have retained an AGPase but exhibit lower sensitivity to 3-phosphoglyceric acid activation, we suggest that the small and large subunits of the enzyme define, respectively, the catalytic and regulatory subunits of AGPase in unicellular green algae. We provide preliminary evidence that both the small-subunit mRNA abundance and enzyme activity, and therefore also starch metabolism, may be controlled by the circadian clock.
Starch accumulation defines a distinctive feature of the photosynthetic eukaryotic cell. Bacteria, fungi, and animal cells all synthesize glycogen, a simpler form of α-1,4-linked and α-1,6-branched storage polysaccharides. Starch and glycogen can be easily distinguished by a number of structural features. Glycogen granules are water soluble and are composed of a single homogeneous highly branched polysaccharide fraction (20). Starch consists of large semicrystalline insoluble granules containing at least two distinct polysaccharide fractions (7). Amylopectin defines the major branched fraction of starch while amylose consists of smaller molecules with less than 1% of its glucosidic linkages as α-1,6 branches. It is believed that the asymmetrical distribution of the branches of amylopectin is responsible for the crystallization of this polysaccharide within the plant plastids. Despite these major differences, the pathway of starch biosynthesis shares a number of common features with glycogen biosynthesis in photosynthetic bacteria (3, 24). Both bacteria and plants use ADP-glucose as a nucleotide sugar donor for polysaccharide biosynthesis while fungi and other eukaryotes synthesize glycogen from UDP-glucose. In yeasts and animal cells, elongation of the glycogen polymer through glycogen synthase defines the major rate-controlling step of glycogen biosynthesis. The enzyme sensitivity to a number of allosteric effectors is finely tuned through a complex series of posttranslational modifications involving protein kinases and phosphatases. In bacteria and plants, the flux of carbon into the pathway is mainly regulated at the level of ADP-glucose synthesis (24, 26). ADP-glucose pyrophosphorylase catalyzes the formation of the glucosyl nucleotide from ATP and glucose-1-phosphate. In cyanobacteria and plants, this enzyme is activated by 3-phosphoglyceric acid (3-PGA) and inhibited by orthophosphate (for review, see reference 26). However, the pathway of polysaccharide synthesis in plants can be distinguished from that in bacteria through the multiplicity of enzyme forms that are present for each step of the biosynthetic pathway. While bacteria, with few exceptions, contain one subunit for the homotetramer AGPase, one glycogen synthase, and one branching enzyme, plants always contain two related subunits for their heterotetramer AGPase (26), a minimum of four distinct starch synthases, and two branching enzymes (7). All of these proteins display some sequence homology with the corresponding cyanobacterial enzymes and are only very distantly related to the fungal or animal glycogen pathway enzymes. Chlamydomonas reinhardtii is the only starch-synthesizing unicellular organism intensively studied by geneticists. It therefore offers a unique opportunity to understand the basic mechanisms of starch biosynthesis (1, 6). We have previously reported that strains with mutations in the STA1 locus accumulate restricted amounts of starch because of a lowered sensitivity of AGPase to 3-PGA activation (2, 27). We have further shown that STA1 encodes a 53-kDa protein that displays homology to both large subunits of vascular plants and cyanobacterial homotetrameric subunits (27). The mutants retained between 5 and 10% of the normal starch amount. However, the remaining polysaccharide displayed major structural alterations that came as immediate consequences of the limitation in ADP-glucose supply (27). The wild-type enzyme was purified to near-homogeneity and displayed a 53-kDa band with an N-terminal sequence identical to that deduced from the STA1 gene sequence (15, 27). The pure enzyme preparation also contained a 50-kDa band that cross-reacted with antibodies directed against the spinach leaf enzyme and therefore could be defined as a heterotetramer (15). We now report the selection and characterization of a starchless (<0.01% of the wild-type amount of starch) mutant of C. reinhardtii lacking ADP-glucose pyrophosphorylase activity. We demonstrate that the wild-type STA6 locus encodes a 50-kDa protein with homology to the small subunit of vascular plant AGPase. We bring suggestive evidence for circadian clock regulation of the small-subunit mRNA levels and of the corresponding enzyme activity.
MATERIALS AND METHODS
Materials.
[α-32dCTP] was purchased from Amersham (Little Chalfont, United Kingdom). The starch determination kit, phosphoglucomutase, and glucose-6-phosphate dehydrogenase were purchased from Boehringer (Mannheim, Germany). Percoll was from Pharmacia LKB Biotechnology (Uppsala, Sweden). ADP-glucose and glucose-1-phosphate were from Sigma Chemical Co. (St. Louis, Mo.).
Strains, media, incubation, and growth conditions.
Our reference strains are 137C (mt− nit1 nit2), 37 (mt+ pab2 ac14), NV314 (mt− pab2 ac14 sta1-1), and 330 (mt+ arg7-7 cw15 nit1 nit2). BAFJ3 (mt+ cw15 arg7-7 nit1 nit2 sta1-2::ARG7) and BAFJ5 (cw15 arg7-7 nit1 nit2 sta6-1::ARG7) were derived from strain 330 by random integration of pARG7 into the nuclear genome as described below. DP1 was generated by somatic fusion between gametolysin-generated protoplasts of strain NV314 and the cell wall-defective strain BAFJ5 as described below. Tris-acetate phosphate (TAP) liquid medium, Sueoka liquid medium, and solid minimum medium were as fully detailed previously (12) while nitrogen-starved medium (TAP-N) was as previously described (4, 8). Growth conditions were also previously detailed (2, 4). For the study of the expression of the small-subunit ADP-glucose pyrophosphorylase transcript and the measurement of the enzyme activity in a 12-h day and continuous 24-h night period, cells were grown with a high level of CO2 (4%) bubbling in Sueoka medium. Cells were previously trained through eight generations grown under a 12-h day–12-h night cycle in Sueoka medium under a high CO2 level.
Genetic techniques.
Gametogenesis and crosses were as previously described (13). Vegetative diploids were always selected from microcolonies growing after 4 days on minimal medium with nitrate as a sole nitrogen source. The diploid strain DP1 was constructed as follows. Because of the sterility displayed by BAFJ5, we performed somatic fusions between it and NV314. Protoplasts of NV314 were prepared with gametolysin (14). Fresh cells of NV314 were suspended in the gametolysin extract that was prepared as previously detailed (14) at a concentration of 5 × 106 cells ml−1 and were incubated at 34°C for 30 min. This mixture was centrifuged for 10 min at 2,000 × g, and the pellet was subjected to another round of the same treatment and then was finally washed once in TAP medium. Each somatic fusion partner (5 × 107 cells) was gently spread on a 9-cm-diameter petri dish containing solid minimal medium. A 0.1-ml volume of the fusion solution containing 85 mM polyethylene glycol (PEG) 8000, 20 mM CaCl2, and 20 mM glycine (pH 8.0, adjusted by NaOH) was added. The cells were gently mixed and spread on the agar. Fusion products were selected from microcolonies that were growing vigorously after 5 days of incubation. All fusion products produced by this technique have been previously proven to be diploid and equivalent to vegetative diploid strains produced by standard crossing (21).
After checking the somatic cell fusion products for starch content, cellular volume, and mating type, we crossed one of these diploid strains (DP1) with the haploid wild-type strain 37 to obtain the sta1-2::ARG7/sta6-1::ARG7/+ triploid zygotes. The aneuploid segregants were analyzed at random after meiosis. Previous work (9) on C. reinhardtii triploid zygotes has established that this technique could be used routinely despite the lower viability of the aneuploid progeny.
Crude extract preparation, ADP-glucose pyrophosphorylase assay.
Soluble crude extracts were prepared from late-log-phase cells (2 × 106 cells ml−1) grown in TAP medium under continuous light (250 μmol of photons m−1 s−1) except for the study on the expression of the small-subunit ADP-glucose pyrophosphorylase. For this study, crude extracts were prepared from cells grown in 5 liters of Sueoka medium and were subjected to different illumination regimes that included bubbling of CO2 (4%). The samples (200 ml) were taken every 3 h and were pelleted at 5,000 × g for 10 min at 4°C. The pellet was then frozen in liquid nitrogen. The cells were disrupted in liquid nitrogen and 2 ml of the extraction buffer (HEPES–100 mM NaOH [pH 7.6], 20 mM MgCl2 · 6H2O, 5 mM NaF, 5 mM dithiothreitol, 2 mM CaCl2 · 2H2O, 10% glycerin, 3% PEG 6000) was added. The extract was centrifuged at 17,500 × g for 10 min at 4°C to remove the cell fragments, and the supernatant was kept at −20°C for further use. After thawing, the latter was centrifuged at 17,500 × g for 10 min at 4°C and the ADP-glucose pyrophosphorylase assay was performed as previously described (2).
Measures of starch levels, starch purification, and spectral properties of the iodine-starch complex.
Amyloglucosidase assays, starch purification on Percoll gradient, and λmax, the wavelength of the maximal absorbance of the iodine-polysaccharide complex, were as previously described (8).
Chlamydomonas DNA purification, RNA purification, cloning, and sequencing.
A description of algal DNA extraction can be found elsewhere (25). Total RNA extraction and purification were fully described previously (19).
Cloning of the small subunit of ADP-glucose pyrophosphorylase was performed using a λzap (titer, 109 PFU ml−1) cDNA library (Stratagene, La Jolla, Calif.). The oligonucleotide 5′-GAGAAGCCGTACATCGCCTCCATGGGC-3′ and the antisense oligonucleotide 5′-CCAACGCGGGCGTTCTTGTCAATG-3′ generated a 550-bp PCR fragment from genomic DNA of strain BAFJ3. This fragment was used to screen the cDNA library. Five positive clones were then partially or fully sequenced. All clones displayed extensive sequence overlap. The rapid amplification of cDNA ends PCR kit from Gibco BRL (Gaithersburg, Md.) was used to generate the 5′ end of the cDNA sequence. The amplification was performed using the antisense oligonucleotides 5′-GGTGCTTGCGGACAAA-3′ and 5′-GTCGCCCGACAGGATGAGGA-3′ corresponding, respectively, to the FVRKH and LILSGD peptides. Sequencing was performed by the dideoxy chain termination method using the Sequenase Version 2.0 DNA sequencing kit (Amersham).
Northern blot analysis.
Total RNAs (50 μg) were separated on a 1.1% formaldehyde agarose gel containing electrophoresis buffer (20 mM MOPS [morpholinepropanesulfonic acid], 8 mM sodium acetate, 1 mM EDTA, 2.2 M formaldehyde) and were subsequently transferred to a nylon membrane (Gene Screen Plus; Dupont). The RNA was cross-linked to the membrane by UV exposure for 2 min and prehybridized at 42°C for 1 h in a solution containing 30% (wt/vol) formamide, 5% (wt/vol) sodium dodecyl sulfate (SDS), 1 mM NaCl, and 250 μg of sonicated herring sperm DNA ml−1. Hybridization was performed for 16 to 18 h in the prehybridization solution with the biotin-labeled probe. After washing at 42°C with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate; pH 7.0)–0.5% SDS for 15 min, the membrane was washed under high-stringency conditions for 30 min at 60°C with 0.5× SSC–0.5% (wt/vol) SDS and subsequently at 65°C for 30 min with 0.2× SSC–0.5% (wt/vol) SDS. Southern-Star (Tropix, Bradford, Mass.) was used for the detection of the biotin-labeled probes. The probe was generated by PCR amplification using the primers 5′-ATGGCCCTGAAGATGCGGGT-3′ and 5′-AGAGGATACAGACGGGTGCC-3′ that correspond, respectively, to the MALKMRV and GTRLYP peptides.
Nucleotide sequence accession number.
The nucleotide sequence of the C. reinhardtii small-subunit ADP-glucose pyrophosphorylase cDNA has been deposited in the GenBank-EMBL database under accession no. AF193431.
RESULTS
Selection of starchless mutants lacking AGPase.
Over 12,000 colonies were isolated after transformation of strain 330 with the pARG7 plasmid. Mutant phenotypes were screened after staining nitrogen-starved cell patches with iodine vapors (Fig. 1). The yellow-staining BAFJ5 mutant strain accumulated less than 0.05 μg of starch per 106 cells, which was the sensitivity limit of our assay, with no detectable oligosaccharide or water-soluble polysaccharide production. These parameters thus define the most severe starch defect phenotype recorded to date for Chlamydomonas. Despite the fact that the mutant grew normally in all conditions tested, BAFJ5 had become sterile and nonmotile. The paralyzed phenotype is probably due to the presence of abnormal flagellae appearing as short stubs in the BAFJ5 mutant. Cell-free extracts of the mutant were used to monitor all enzyme activities suspected to be involved in starch biosynthesis. BAFJ5 proved selectively defective in ADP-glucose pyrophosphorylase activity (Table 1). In contrast with the STA1 locus mutants that contained residual amounts of enzyme activity and starch (2), BAFJ5 lacked both.
FIG. 1.
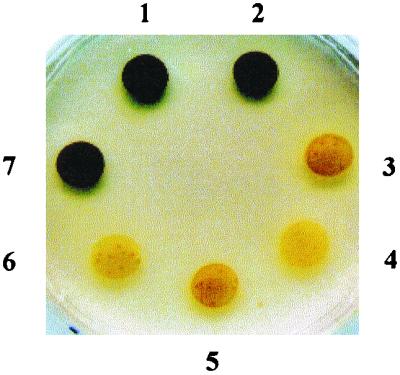
Phenotypes of wild-type, mutant haploid, and diploid strains. Strains 330 (1) and 137C (2) are wild-type haploid references. DP1 (7) is a vegetative diploid strain generated by somatic fusion between the strains NV314 (sta1-1) (6) and BAFJ5 (sta6-1::ARG7) (4). Strains 17 (sta1-1) (3) and BAFJ3 (sta1-2::ARG7) (5) have allelic mutations in the locus STA1. Cell patches were incubated for 5 days on solid nitrogen-deprived medium and sprayed with iodine vapors.
TABLE 1.
ADP-glucose pyrophosphorylase activity and starch accumulation in 137C (wild-type reference), NV314 (sta1-1), BAFJ5 (sta6-1::ARG7), and DP1 (vegetative diploid generated from fusion between NV314 and BAFJ5) strains
| Strains | Activity (10−2)a | Activation factorb | Starch amtc |
|---|---|---|---|
| 137C | 12.8 ± 3.4 | 8 | 45 ± 8 |
| NV314 | 1.8 ± 0.7 | 1 | 2.5 ± 1.3 |
| BAFJ5 | NDd | ND | ND |
| DP1 | 10.7 ± 2.3 | 10 | 55 ± 7 |
The assay was performed in the degradation direction by measuring the glucose-1-phosphate formed in the presence of 1 mM ADP-glucose, 1 mM pyrophosphate and 1.5 mM activator 3-PGA. The activity is expressed in nanomoles of glucose-1-phosphate formed per milligram of protein per minute.
The activation factor of ADP-glucose pyrophosphorylase represents the ratio between the activities measured with 1.5 mM 3-PGA and without 3-PGA.
Amount of starch expressed (in micrograms) per 106 cells. Cells were grown in a nitrogen-deprived medium that allows a maximum accumulation of starch.
ND, not detectable.
Somatic fusion of the sterile sta6-1::ARG7 mutant with protoplasts carrying the sta1-1 mutation.
Cell wall-defective mutants derived from strain 330 very often display reduced fertility when mated with many wild-type strains. We have, however, never failed to generate enough zygotes to achieve random spore analysis. In addition, we were always able to produce a large number of vegetative diploid clones that grew vigorously after 5 days on selective medium. Despite intensive efforts, we never managed to generate vegetative diploid clones or standard zygotes from BAFJ5. To overcome the sterility problem, we fused the cell wall-defective BAFJ5 mutant to protoplasts of the NV314 strain containing the sta1-1 mutation. The fusion products proved to contain the expected Southern blot patterns upon hybridization with appropriate probes which are specific for both BAFJ5 or NV314 (Fig. 2). It's been previously proven that under the experimental conditions used, all PEG-induced fusion products defined stable diploid clones (21). The doubling of the mean cellular volume and the segregation obtained after subsequent crossing confirmed the diploid nature of the fused somatic hybrids. The diploids accumulated wild-type amounts of starch and ADP-glucose pyrophosphorylase activities (Table 1), suggesting that BAFJ5 contained a recessive mutation (sta6-1::ARG7) at a novel Chlamydomonas locus that was tentatively named STA6.
FIG. 2.
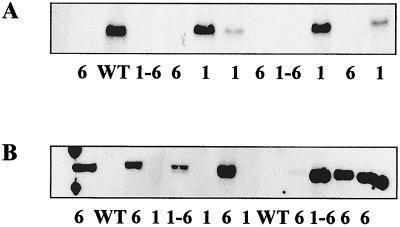
Southern blot analysis of PstI-digested chromosomal DNA extracted from 137C (wild-type reference), NV314 (sta1-1), BAFJ5 (sta6-1::ARG7), and DP1 (fusion diploid between NV314 and BAFJ5) strains. (A) The probe covers the 356-bp PvuII fragment from the coding region of the 52.3-kDa ADP-glucose pyrophosphorylase large-subunit cDNA. (B) The probe covers the 323-bp NruI-SalI fragment from the prokaryotic part of the pARG7.8 plasmid that was used for insertional mutagenesis. Lanes 1, 2, 3, and 4 represent about 15 μg of DNA extracted from, respectively, strains 137C, NV314, and BAFJ5 and the somatic hybrid diploid DP1.
Triploid genetic analysis.
To confirm that we had indeed defined a novel Chlamydomonas locus conditioning ADP-glucose pyrophosphorylase activity, we crossed the somatic hybrid diploid clone DP1 with a wild-type strain of opposite mt+ mating type. DP1 behaved as a standard vegetative diploid Chlamydomonas strain. It displayed the dominant mt− mating type and was fully fertile and motile. We calculated the expected trisomic segregation patterns and compared these to the phenotypes recorded after meiosis of the triploid zygote. The genes segregated as expected from the inferred triploid genotypes in a fashion similar to that previously reported for such zygotes (9). However, the calculated segregation ratios could not distinguish between one or two unlinked STA loci. We therefore performed complementation tests between those low-starch recombinants carrying suitable markers and standard sta1 mutants of mt+ or mt− mating type. From this analysis, we found starchless recombinants that fully complemented the sta1-1 or the sta1-2::ARG7 mutations. Most of these recombinants were fertile and enabled us to isolate sta6-1::ARG7 strains for further complementation analysis. We were thus able to deduce the genotypes of 16 recombinants from a total of 52 low-starch recombinants which displayed one of the following three genotypes: sta1-1 (3), sta6-1::ARG7 (3), and sta1-1 sta6-1::ARG7 (6). These results, obtained from a restricted number of starch-defective recombinants, were nevertheless sufficient to establish that STA1 and STA6 defined two independent loci required for normal ADP-glucose pyrophosphorylase activity. In addition, they proved that paralysis and sterility segregated independently from the STA1 and STA6 loci. Finally, the occurrence of the sta1 sta6 double mutant class enabled us to establish full epistasis of sta6 on sta1. The double mutants contained no detectable ADP-glucose pyrophosphorylase activity and were consequently starchless. To further rule out possible misinterpretations due to the presence of chromosome imbalances in the aneuploid progeny, we crossed a recombinant sta6-1::ARG7 with a standard wild-type strain and confirmed the starchless phenotype of all mutant sta6-1::ARG7 recombinants. No sterile or nonmotile strains were found in the progeny of this cross. During this triploid genetic analysis and the subsequent crosses, we found linkage between STA6 and NIT2 on chromosome III.
Molecular cloning of the 50-kDa small subunit of ADP-glucose pyrophosphorylase.
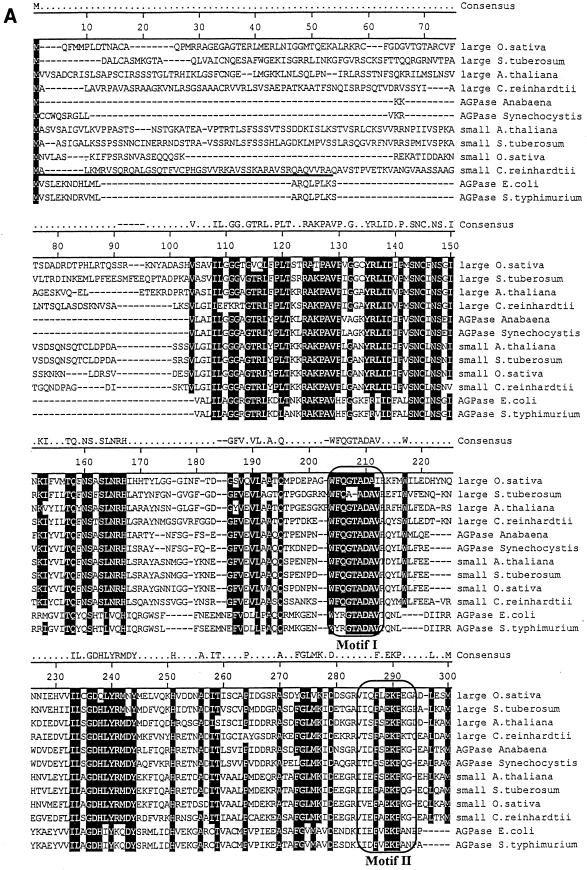
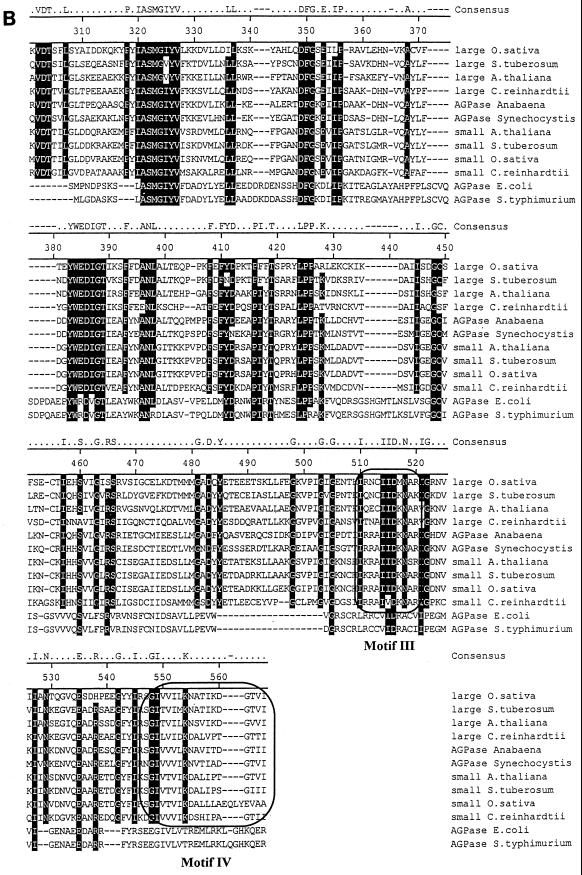
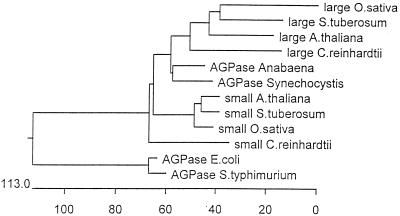
We have previously described the molecular cloning of the ADP-glucose pyrophosphorylase large-subunit cDNA (27). The initial cloning step consisted of PCR amplification of sequences present in cDNA libraries by using oligonucleotides derived from peptide sequences that are conserved from cyanobacteria to vascular plants. Despite repeated attempts, we never selected fragments coding for a second ADP-glucose pyrophosphorylase subunit. We therefore concentrated our efforts on PCR amplification of gDNA using a particular mutant strain (BAFJ3) that carried a deletion for most of the large-subunit genomic sequences (27). We were able to select a 550-bp PCR fragment that contained an open reading frame encoding a product bearing homologies to ADP-glucose pyrophosphorylase subunits. The sequence was clearly different from that previously cloned and was therefore used to select for appropriate cDNA clones. A combination of cDNA cloning and multiple rapid amplification of cDNA ends PCR (see Materials and Methods) yielded a 2,741-bp sequence. The complete cDNA coded for a 514-amino-acid protein displayed in Fig. 3. The N-terminal sequence coded for a putative transit peptide with some sequence organization similarity to many such peptides identified in Chlamydomonas. A hypothetical transit peptide cleavage sequence which yielded a 50-kDa mature protein was found at position 47. The mass of the product matched precisely that which was described for the second subunit of the pure Chlamydomonas enzyme (15). This 50-kDa product displayed more homology to the vascular plant small-subunit sequences than to the large-subunit sequences of both higher plants and Chlamydomonas. In addition, both large and small subunits of Chlamydomonas displayed more homology to unique cyanobacterial subunits than between themselves. The sequence comparisons displayed in Fig. 3 enabled us to build the phylogenetic tree shown in Fig. 4 by using the Clustal method with the PAM250 residue weight table. It is clear from this analysis that divergence of small (catalytic) subunits from the large regulatory subunits within the ADP-glucose pyrophosphorylase structure occurred at a very early stage during the evolution of photosynthetic eukaryotes. In addition, the predicted sequence suggests that we are probably dealing with the catalytic subunit of the algal enzyme. Mutation in a catalytic subunit is expected to yield a severe depletion in starch such as that witnessed in the sta6-deficient mutants. We have therefore set to investigate the relation existing between the small subunit of ADP-glucose pyrophosphorylase and the STA6 gene.
FIG. 3.
Protein sequence of the small subunit of Chlamydomonas ADP-glucose pyrophosphorylase. Shown is a comparison of the algal protein sequence to those of ADP-glucose pyrophosphorylase subunits of Oryza sativa, Solanum tuberosum, A. thaliana, E. coli, and Salmonella enterica serovar Typhimurium. Large and small correspond to the large and small subunits of ADP-glucose pyrophosphorylase, respectively. Accession numbers (top to bottom) are as follows: AAB58473, P55242, BAA76362, X91736, P30521, BAA18822, AAB09585, P23509, P15280, AF193431, P00584, and P05415. The putative transit peptide of the Chlamydomonas small ADP-glucose pyrophosphorylase subunit is underlined. Its cleavage site remains hypothetical since it varies from that of the large subunit, and we have been unable to sequence the N terminus of the purified small subunit (15). The framed boxes marked Motif I and Motif II (A) represent, respectively, the regions considered to be involved in the binding to ATP and glucose-1-phosphate. The domains contained within the framed boxes marked Motif III and Motif IV (B) are considered to be involved in the binding to 3-PGA in plants. The consensus amino acids shaded in black are those identical in at least 10 of 12 sequences. The figure was constructed using the Clustal method with the PAM250 residue weight table.
FIG. 4.
Phylogenetic tree of selected ADP-glucose pyrophosphorylase subunits. The accession numbers of the proteins are the same as those presented in Fig. 3. This phylogenetic tree was built using the Clustal method with the PAM250 residue weight table.
STA6 defines the small-subunit structural gene.
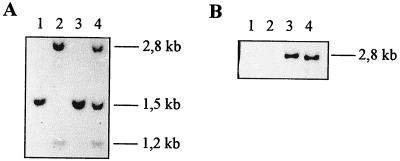
An internal 322-bp HincII-BstXI fragment corresponding to the small-subunit sequence was used in Southern hybridization experiments against restricted gDNA corresponding to a wild-type strain or to mutant strains lacking, respectively, the STA1 or STA6 locus. From the results displayed in Fig. 5, it is clear that this probe recognizes a 2.8-kbp PstI fragment that is absent from all sta6-1::ARG7 mutant recombinants (n = 34). This analysis proves that the sta6-1::ARG7 deletion spans a large part of the coding sequence corresponding to the small subunit of AGPase. The sta6-1::ARG7 recombinants always contained a 2.7-kbp PstI fragment that hybridized with a 323-bp NruI-SalI restriction fragment corresponding to the prokaryotic part of the pARG7.8 plasmid that was used for insertional mutagenesis. These recombinants were all arginine prototrophs while 4 out of 10 of the sta1-1 recombinants required arginine for growth. This establishes linkage between sta6-1::ARG7 and an integrated copy of pARG7.8. We never found sta1-1 segregants carrying the 323-bp NruI-SalI restriction fragment corresponding to the prokaryotic part of the pARG7.8 plasmid. We would have expected such strains to appear in the aneuploid progeny if both STA1 and STA6 were located on separate linkage groups. This can be taken as suggestive evidence for the presence of both loci on chromosome III.
FIG. 5.
Southern blot analysis of PstI-digested chromosomal DNA of segregants from the cross between DPI and NV314. (A) The probe covers the 322-bp HincII-BstXI fragment from the coding region of the 50-kDa ADP-glucose pyrophosphorylase small-subunit cDNA. (B) The probe covers the 323-bp NruI-SalI fragment from the prokaryotic part of the pARG7.8 plasmid that was used for insertional mutagenesis. For each segregant, 15 μg of DNA was loaded. WT, wild type; 1, sta1-1 mutants; 6, sta6-1::ARG7 mutants; 1-6, double mutants sta1-1/sta6-1::ARG7.
Small-subunit ADP-glucose pyrophosphorylase mRNA abundance is suggestive of a circadian clock control mechanism.
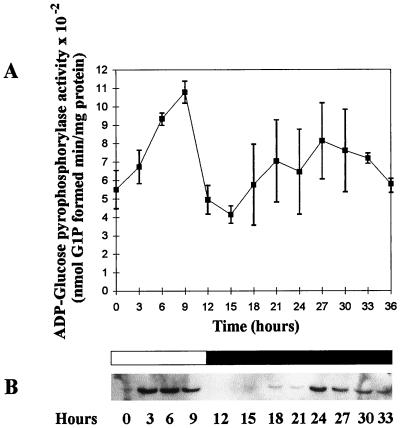
We measured the amount of ADP-glucose pyrophosphorylase and small-subunit mRNA levels in cultures grown under high or low CO2 levels experiencing a 12-h day and 12-h night cycle of growth. The enzyme activity and mRNA levels were correlated and peaked at 6 h after dawn while decreasing during the night (for example, the first 24 h in the three distinct experiments detailed in Fig. 6). The presence of high or low amounts of CO2 was not found to modify the transcript or activity levels. The amount of enzyme activity decreased at most by 60% while the mRNA levels decreased by over 90%. To test the possibility that the enzyme activity and mRNA levels might be subjected to regulation by the circadian clock in addition to or in place of a light-dependent induction mechanism, we measured both after switching the cultures to continuous darkness. A summary of the results obtained in three different sets of experiments is presented in Fig. 6. The rhythm of enzyme activity dampened somewhat in continuous darkness. Nevertheless, the enzyme specific activity did show a significant increase during the virtual day, suggesting that there could be a circadian component in enzyme activity regulation.
FIG. 6.
(A) Activity of ADP-glucose pyrophosphorylase in wild-type cells grown in a 12-h day and a prolonged 24-h night period under 4% CO2. Cells were previously trained through eight cycles of 12-h day–12-h night under 4% CO2. The assay was performed in the pyrophosphorolysis direction by measuring the glucose-1-phosphate produced from 1 mM ADP-glucose in the presence of 1 mM pyrophosphate (2). The concentration of 3-PGA used was 1.5 mM. The results displayed are the means ± standard deviations of three entirely distinct and independent experiments. (B) Northern analysis of ADP-glucose pyrophosphorylase small-subunit gene expression in wild-type cells grown in the same conditions. Cells were previously trained as above. An amount of 50 μg of total RNA was loaded in each lane, and the blot was probed with a 355-bp fragment from the N-terminal coding region of the ADP-glucose pyrophosphorylase small-subunit cDNA. That probe was generated by PCR amplification and was biotin labeled.
DISCUSSION
The appearance of starch coincides with the acquisition of photosynthesis by unicellular eukaryotes. The storage of glucose in an insoluble semicrystalline form provided a powerful osmotically inert intraplastidial carbon sink for the emerging plant cells. It has been known for years that starch metabolism in plants could be distinguished from that of cyanobacterial glycogen by the presence of multiple forms of enzymes at each step of the pathway (1, 7, 24, 26). It was thus thought that the interplay between multiple forms of branching enzymes and starch synthases would be sufficient to explain the complexity of starch granule biogenesis. Vascular plant ADP-glucose pyrophosphorylase can also be distinguished from that of cyanobacteria by the presence of two distinct subunits of related sequence in the tetrameric enzyme composition. Genetic dissection of plant mutants and expression of either or both subunits in Escherichia coli have clearly established the small subunit as required for catalysis and the large subunits as necessary for normal allosteric regulation of the enzyme (1; reviewed in references 7, 24, and 26). Most vascular plants contain several genes corresponding to each of the subunits which are, in some cases, expressed in a tissue-specific manner. In some instances, the same catalytic subunit can be found coexpressed with distinct regulatory subunits, thereby expanding the range of different activities and providing each tissue with an enzyme with optimal kinetics and allosteric regulations (16). One could thus better understand the appearance of these specialized subunits in multicellular organisms. Such speculations, however, do not apply to Chlamydomonas, and the advantages given to this organism by the presence of a heterotetrameric ADP-glucose pyrophosphorylase remain elusive. It is striking to note that while the primary enzyme structures of the algal enzyme fits the evolutionary position of green algae between vascular plants and cyanobacteria, C. reinhardtii displays exactly the same enzymatic makeup as that of plants. Furthermore, the structure of the algal starch is identical to that of vascular plants (6) and the phenotypic consequences of the absence of enzyme activities through gene mutations remain the same (reviewed in reference 1). Reductive activation or inhibition through the ferredoxin-thioredoxin system defines another important feature distinguishing chloroplasts from cyanobacteria. Reductive activation of potato tuber ADP-glucose pyrophosphorylase has recently been demonstrated to enhance the enzyme's sensitivity to the allosteric activator 3-PGA (5, 10). It would thus be tempting to speculate that this mechanism of enzyme activation might have favored the appearance of two specialized subunits. In this respect, it is worth noting that Cys12, the small-subunit residue which has been demonstrated to be involved in the activation process (5), is absent from the Chlamydomonas sequence. It must be stressed that while green algae also contain ferredoxin-thioredoxin systems of enzyme activation, they do not contain exactly the same plastidial components. It therefore remains possible that the Chlamydomonas enzyme is sensitive to another mechanism of ferredoxin-thioredoxin activation, or another Cys residue may be involved.
Most ADP-glucose pyrophosphorylase-defective plant mutants or antisense constructs generated to date produce a reduced but significant amount of starch. The Arabidopsis adg1 mutant defective for the small subunit of ADP-glucose pyrophosphorylase remains the only exception to date (18). According to the authors, the mutant had no detectable activity and there were no measurable starch levels above the assay's background. The Chlamydomonas STA6 mutant reported in this work contained less than 0.05 μg per 106 cells in conditions favorable for maximal starch synthesis and in continuous light. This would amount to less than 0.1% of the wild-type starch content. As with the Arabidopsis mutant, we were never able to record any measure above the baseline afforded by the amyloglucosidase technique. This proves beyond a doubt that ADP-glucose synthesis through plastidial ADP-glucose pyrophosphorylase defines the sole route for starch biosynthesis in plants and in the unicellular green alga Chlamydomonas. The deletion of sequences corresponding to the small catalytic subunit of the Chlamydomonas ADP-glucose pyrophosphorylase gene within the sta6 mutants very strongly suggests that STA6 encodes this subunit. However, we have not fully ruled the possible presence of a tightly linked regulatory locus that would be codeleted in the mutant and responsible for the phenotype observed. In this improbable hypothesis, STA6 would not encode the small catalytic subunit of ADP-glucose pyrophosphorylase. Regulatory loci in contrast to structural genes are not expected to yield a strict correlation between the number of wild-type alleles and the amount of enzyme activity in heterozygous diploids or triploids. We have therefore performed gene dosage experiments and found heterozygous diploids (sta6/+) to display enzyme activities which did not always correlate with the amount of wild-type alleles in their genotype (data not shown), probably because of the complexity afforded by the quaternary organization of the heterotetrameric enzyme. A similar situation was precisely evidenced in the case of the Arabidopsis thaliana adg1 mutant lacking the small subunit of the enzyme (18, 28). However, in this case it was proven that ADG1 encoded the small subunit of the enzyme through a combination of molecular techniques, including complementation of the plant mutant with the corresponding wild-type Arabidopsis genomic sequences (28).
Finally, we found a 24-h period rhythm in the Chlamydomonas ADP-glucose pyrophosphorylase mRNA abundance. The pattern is somewhat followed but to a lesser extent by the enzyme activity. Because ADP-glucose pyrophosphorylase defines one of the major rate-controlling steps of starch biosynthesis, this important observation might explain part of the daily pattern of starch accumulation within algae. The fact that the enzyme activity rhythm dampens quickly suggests that there are other superimposed posttranslational controls on the ADP-glucose pyrophosphorylase itself. We are aware that the data presented in this paper define preliminary suggestive evidence for the regulation of starch metabolism by the circadian clock. To definitively prove that we are dealing with a circadian clock-dependent mechanism, other constant environmental conditions should be tested (for instance, continuous light). In addition, we should demonstrate the existence of both a temperature compensation mechanism and an entrainment mechanism that should set the period to exactly 24 h. Nevertheless, the data obtained clearly point in the direction of a circadian clock control mechanism, encouraging yet further work dealing with the rhythms of starch metabolism in unicellular green algae. Very few studies deal with the issue of circadian clock control of mRNA abundance for enzymes of starch metabolism. To our knowledge, the only case reported of possible circadian clock control is that reported for granule-bound starch synthase I mRNA abundance in snapdragon leaves (22). However, this fluctuation of the mRNA encoding the enzyme responsible for amylose biosynthesis was not shown to be paralleled by a similar pattern of granule-bound starch synthase I activity and amylose content. Moreover, it was not demonstrated either that the rhythm was temperature compensated or that it could be reset through the use of external 24-h-period stimuli. Diurnal starch accumulation patterns have been previously proven to be under circadian regulation both in sugar beet leaves and tobacco leaves (11, 17). We believe that our results concerning the major rate-controlling enzyme of starch biosynthesis should prompt more detailed studies on the potential circadian clock regulation of algal and leaf starch metabolism.
ACKNOWLEDGMENTS
This work was supported in part through the Ministère de l'Education Nationale, the Centre National de la Recherche Scientifique, and the Deutsche Forshungsgemeinschaft SFB 199 and in part through the Department of Energy (grant DE-FG02-93ER20121).
Special thanks to Silke Herdt and André Decq for their excellent technical assistance.
REFERENCES
- 1.Ball S. Regulation of starch biosynthesis. In: Rochaix J-D, Goldschmidt-Clermont M, Merchant S, editors. The molecular biology of chloroplasts and mitochondria in chlamydomonas. Dordrecht, The Netherlands: Kluwer; 1998. pp. 549–567. [Google Scholar]
- 2.Ball S, Marianne T, Dirick L, Fresnoy M, Delrue B, Decq A. A Chlamydomonas reinhardtiilow-starch mutant is defective for 3-phosphoglycerate activation and orthophosphate inhibition of ADP-glucose pyrophosphorylase. Planta. 1991;185:17–26. doi: 10.1007/BF00194509. [DOI] [PubMed] [Google Scholar]
- 3.Ball S, Guan H-P, James M, Myers A, Keeling P, Mouille G, Buléon A, Colonna P, Preiss J. From glycogen to amylopectin: a model explaining the biogenesis of the plant starch granule. Cell. 1996;86:349–352. doi: 10.1016/s0092-8674(00)80107-5. [DOI] [PubMed] [Google Scholar]
- 4.Ball S G, Dirick L, Decq A, Martiat J C, Matagne R F. Physiology of starch storage in the monocellular alga Chlamydomonas reinhardtii. Plant Sci. 1990;66:1–9. [Google Scholar]
- 5.Ballicora M A, Frueauf J B, Fu Y, Schurmann P, Preiss J. Activation of potato tuber ADP-glucose pyrophosphorylase by thioredoxin. J Biol Chem. 2000;275:1315–1320. doi: 10.1074/jbc.275.2.1315. [DOI] [PubMed] [Google Scholar]
- 6.Buléon A, Gallant D-J, Bouchet B, Mouille G, D'Hulst C, Kossman J, Ball S G. Starches from A to C. Chlamydomonas reinhardtiias a model microbial system to investigate the biosynthesis of the plant amylopectin crystal. Plant Physiol. 1997;115:949–957. doi: 10.1104/pp.115.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buléon A, Colonna P, Planchot V, Ball S. Starch granules: structure and biosynthesis. Int J Biol Macromol. 1998;23:85–112. doi: 10.1016/s0141-8130(98)00040-3. [DOI] [PubMed] [Google Scholar]
- 8.Delrue B, Fontaine T, Routier F, Decq A, Wieruszeski J M, Van Den Koornhuyse N, Maddelein M L, Fournet B, Ball S. Waxy Chlamydomonas reinhardtii: monocellular algal mutants defective in amylose biosynthesis and granule-bound starch synthase activity accumulate a structurally modified amylopectin. J Bacteriol. 1992;174:3612–3620. doi: 10.1128/jb.174.11.3612-3620.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eves E M, Chiang K S. Genetics of Chlamydomonas reinhardtiidiploids. I. Isolation and characterization and meiotic segregation of a homozygous diploid. Genetics. 1982;100:35–60. doi: 10.1093/genetics/100.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Y, Ballicora M A, Leykam J F, Preiss J. Mechanism of reductive activation of potato tuber ADP-glucose pyrophosphorylase. J Biol Chem. 1998;273:25045–25052. doi: 10.1074/jbc.273.39.25045. [DOI] [PubMed] [Google Scholar]
- 11.Geiger D R, Shieh W J, Yu X M. Photosynthetic carbon metabolism and translocation in wild-type and starch-deficient mutant Nicotiana sylvestrisL. Plant Physiol. 1995;107:507–514. doi: 10.1104/pp.107.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris E H. Culture and storage methods. In: Harris E, editor. The Chlamydomonas sourcebook. A comprehensive guide to biology and laboratory use—1989. San Diego, Calif: Academic Press; 1989. pp. 25–63. [Google Scholar]
- 13.Harris E H. Genetic analysis. In: Harris E, editor. The Chlamydomonas sourcebook. A comprehensive guide to biology and laboratory use—1989. San Diego, Calif: Academic Press; 1989. pp. 399–446. [Google Scholar]
- 14.Harris E H. Procedures and resources. In: Harris E, editor. The Chlamydomonas sourcebook. A comprehensive guide to biology and laboratory use—1989. San Diego, Calif: Academic Press; 1989. pp. 575–641. [Google Scholar]
- 15.Iglesias A A, Charng Y Y, Ball S, Preiss J. Characterization of the kinetic, regulatory and structural properties of ADP-glucose pyrophosphorylase from Chlamydomonas reinhardtii. Plant Physiol. 1994;104:1287–1294. doi: 10.1104/pp.104.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.La Cognata U, Willmitzer L, Muller-Rober B. Molecular cloning and characterization of novel isoforms of potato ADP-glucose pyrophosphorylase. Mol Gen Genet. 1995;246:538–548. doi: 10.1007/BF00298960. [DOI] [PubMed] [Google Scholar]
- 17.Li B, Geiger D R, Shieh W J. Evidence for circadian regulation of starch and sucrose synthesis in sugar beet leaves. Plant Physiol. 1992;99:1393–1399. doi: 10.1104/pp.99.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin T P, Caspar T, Somerville C, Preiss J. Isolation and characterization of a starchless mutant of Arabidopsis thaliana(L.) Heynh lacking ADP-glucose pyrophosphorylase activity. Plant Physiol. 1988;86:1131–1135. doi: 10.1104/pp.86.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- 20.Manners D J. Recent developments in our understanding of glycogen structure. Carbohydr Polym. 1989;16:37–82. [Google Scholar]
- 21.Matagne R F, Deltour R, Ledoux L. Somatic fusion between cell wall mutants of Chlamydomonas reinhardtii. Nature. 1979;278:344–346. [Google Scholar]
- 22.Merida A, Rodriguez-Galan J M, Vincent C, Romero J M. Expression of granule-bound starch synthase I (waxy) gene from snapdragon is developmentally and circadian clock regulated. Plant Physiol. 1999;120:401–409. doi: 10.1104/pp.120.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myers A M, Morell M K, James M G, Ball S. Recent progress in understanding biosynthesis of the amylopectin crystal. Plant Physiol. 2000;122:989–998. doi: 10.1104/pp.122.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Preiss J, Romeo T. Molecular biology and regulatory aspects of glycogen synthesis in bacteria. In: Cohn W E, Moldave K, editors. Progress in nucleic acid research and molecular biology. Vol. 47. San Diego, Calif: Academic Press, Inc.; 1994. pp. 299–329. [DOI] [PubMed] [Google Scholar]
- 25.Rochaix J D, Mayfield S, Goldschmidt-Clermont M, Erickson J. Molecular biology of Chlamydomonas. In: Shaw C, editor. Plant molecular biology: a practical approach. Oxford, United Kingdom: IRL Press; 1991. pp. 253–275. [Google Scholar]
- 26.Smith-White B, Preiss J. Comparison of proteins of ADP-glucose pyrophosphorylase from diverse sources. J Mol Evol. 1992;34:449–464. doi: 10.1007/BF00162999. [DOI] [PubMed] [Google Scholar]
- 27.Van den Koornhuyse N, Libessart N, Delrue B, Zabawinski C, Decq A, Iglesias A, Preiss J, Ball S. Control of starch composition and structure through substrate supply in the monocellular alga Chlamydomonas reinhardtii. J Biol Chem. 1996;271:16281–16288. doi: 10.1074/jbc.271.27.16281. [DOI] [PubMed] [Google Scholar]
- 28.Wang S M, Lue W L, Yu T S, Long J H, Wang C N, Eimert K, Chen J. Characterization of ADG1, an Arabidopsis locus encoding for ADPG pyrophosphorylase small subunit, demonstrates that the presence of the small subunit is required for large subunit stability. Plant J. 1998;13:63–70. doi: 10.1046/j.1365-313x.1998.00009.x. [DOI] [PubMed] [Google Scholar]