Abstract
The Escherichia coli SOS-regulated umuDC gene products participate in a DNA damage checkpoint control and in translesion DNA synthesis. Specific interactions involving the UmuD and UmuD′ proteins, both encoded by the umuD gene, and components of the replicative DNA polymerase, Pol III, appear to be important for regulating these two biological activities of the umuDC gene products. Here we show that overproduction of the ɛ proofreading subunit of Pol III suppresses the cold sensitivity normally associated with overexpression of the umuDC gene products. Our results suggest that this suppression is attributable to specific interactions between UmuD or UmuD′ and the C-terminal domain of ɛ.
The Escherichia coli umuDC operon encodes a DNA polymerase, DNA Pol V, with the remarkable ability to replicate over DNA lesions, such as abasic sites (27, 35) and cyclobutane dimers (34), that act as strong blocks for the replicative DNA polymerase, DNA Pol III holoenzyme (Pol III). UmuC protein contains the catalytic DNA polymerase activity (27), while the umuD gene encodes two related gene products, UmuD and UmuD′ (3, 21, 28), that collectively act to manage the activity of UmuC (21, 31). Reflecting the universality of how all organisms must deal with damage to their genetic material, it has recently become apparent that the UmuC protein is the founding member of a diverse and ubiquitous family of novel DNA polymerases that collectively possess the ability to replicate imperfect DNA templates (reviewed in references 7, 11, 36, and 37). Importantly, members of this family of UmuC-like DNA polymerases, referred to as the UmuC-DinB-Rad30-Revl superfamily, are found in all three kingdoms of life (4, 9, 16).
In the gram-negative bacterium E. coli, DNA damage induces a highly regulated response referred to as the SOS response (10, 32, 38). This response involves the coordinated expression of approximately 30 unlinked genes whose products function primarily in DNA damage tolerance and repair (8). In the absence of DNA damage, the expression of these genes is efficiently repressed, albeit to varying degrees, by the LexA protein (8). However, following treatments that damage the bacterial genome, such as exposure to UV light or chemical carcinogens, RecA protein nucleates onto the single-stranded DNA (ssDNA) generated by the cell's failed attempts to replicate over damaged bases (8). These RecA/ssDNA nucleoprotein filaments, in addition to serving an essential role in homologous recombination, also act to facilitate the latent ability of both LexA and UmuD to autodigest (15). Autodigestion of LexA leads to its inactivation as a transcriptional repressor of the SOS regulon (17, 18), while autodigestion of UmuD leads to the production of UmuD′ (3, 21, 28). Functionally, UmuD and UmuD′ differ in that UmuD together with UmuC act as part of a DNA damage checkpoint control (20, 24), while UmuD′ together with UmuC functions in translesion DNA synthesis (26, 27, 35). Thus, RecA/ssDNA facilitated cleavage of UmuD to yield UmuD′ acts as a molecular switch that releases the UmuD2C-dependent DNA damage checkpoint control while enabling UmuD′2C-dependent translesion DNA synthesis that serves as the mechanistic basis for SOS mutagenesis in E. coli (21, 24, 31).
In addition to acting as part of a DNA damage checkpoint control and in translesion DNA synthesis, overproduction of the umuDC gene products confers a cold-sensitive growth phenotype (19, 25). This cold sensitivity correlates with a rapid inhibition of DNA synthesis at the nonpermissive temperature (19, 20). Although overproduction of uncleaved UmuD together with UmuC confers a greater degree of cold sensitivity than does overproduction of a synthetically engineered form of UmuD′ together with UmuC, overproduction of umuD′C nonetheless confers a significant degree of cold sensitivity (25). Thus, the biological activity (or activities) of the umuDC gene products responsible for their ability to confer cold sensitivity has not yet been defined. In this report, we describe experiments designed to address whether interactions between the umuDC gene products and components of Pol III are involved in umuDC-mediated cold sensitivity.
Overproduction of the ɛ subunit of Pol III suppresses umuDC-mediated cold sensitivity.
We have previously suggested that interactions of the umuDC gene products with components of the replicative DNA polymerase might serve as the mechanistic basis for the rapid inhibition of DNA synthesis we have observed following overproduction of umuDC (24, 31). Indeed, we have shown that both UmuD and UmuD′ interact physically with the α (catalytic), ɛ (proofreading), and β (processivity) subunits of Pol III (31). In this study, we reasoned that if these interactions involving the umuDC gene products and components of Pol III were in fact important for umuDC-mediated cold sensitivity, we might then be able to observe an effect on the extent of the cold sensitivity conferred by umuDC by the simultaneous overproduction of certain components of Pol III using the previously described quantitative transformation assay (25). In this analysis, we transformed plasmids that overproduced the individual subunits of Pol III into a lexA(Def) E. coli strain that contained a compatible plasmid that expressed elevated levels of the umuDC gene products from their native LexA-regulated promoter because of the absence of LexA. We then plated aliquots of these transformation reactions onto plates that either lacked or were supplemented with 0.05 or 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) to induce the expression of the Pol III subunits of interest. Transformants were counted after incubation overnight at 30 or 42°C.
This analysis indicated that overproduction of ɛ, the 3′→5′ exonuclease proofreading subunit of Pol III, suppressed the cold sensitivity for growth conferred by elevated levels of the umuDC gene products (Table 1). In addition to its proofreading activity, ɛ also acts to enhance the DNA polymerase activity of the α (catalytic) subunit when in the form of the αɛ complex (30). With the exception of β, whose overexpression exacerbated the cold sensitivity (M. D. Sutton et al., unpublished data), elevated levels of the other polymerase subunits had no apparent effect on umuDC-mediated cold sensitivity (data not shown).
TABLE 1.
Overexpression of ɛ suppresses umuDC-mediated cold sensitivity
| Straina | IPTG (mM) | Transformants/mlb |
|---|---|---|
| GW8111(pSU38-DC)(pBR322kan) | 0 | 0.0023 |
| 0.05 | 0.0028 | |
| 0.1 | 0.0023 | |
| GW8111(pSU38-DC)(pHN1) | 0 | 0.0035 |
| 0.05 | 0.18 | |
| 0.1 | 0.52 |
GW8111 [lexA300(Def)::spec recA441 xyl-5 mtl-1 galK2 rpsL31 kdgK51 Δ(argF-lacU)169 lacY1 tsx-33 supE44 thi-1 leuB6 hisG4(Oc) mgl-51 argE3(Oc) rfbD1 ara-14 thr-1 qsr′ qin-111 sulA11 ilvD(Ts) λ− Rac−/F′ lacIq] is a GW8018 derivative (25) containing the F′ lacIq from XL-1 Blue (Stratagene). pSU38-DC is a pSU38 derivative (1) containing the complete umuDC operon on a PstI fragment and is similar to pSU18-DC (31). GW8111(pSU38-DC) was transformed with either pBR322kan, a pBR322 derivative containing the kanamycin resistance cassette from Tn5 inserted into its tet gene (25), or pHN1, a pBR322 derivative bearing the structural gene for ɛ under Ptac control (14). Aliquots of the transformation reactions were then plated onto selective media containing the indicated level of IPTG to induce expression of ɛ.
Expressed as the total number of CFU obtained at 30°C divided by the total number of CFU obtained at 42°C. Transformation efficiencies were on the order of ∼1 × 105 to 5 × 105 CFU/μg of DNA.
The ability of elevated levels of ɛ to suppress umuDC-mediated cold sensitivity is consistent with our previous observation that ɛ physically interacts with UmuD and UmuD′ in vitro (31) and suggests that these interactions also occur in vivo. Further previously published evidence that interactions involving ɛ and the umuDC gene products occur in vivo, and that these interactions affect that ability of Pol III to replicate DNA, includes the following observations: (i) the umuDC gene products enhance the mutator phenotype of a dnaQ49 strain (5); (ii) a plasmid expressing the umuDC gene products cannot be maintained by a dnaQ49 strain at any temperature (22); and (iii) overexpression of the umuDC gene products in a dnaQ+ strain leads to an inhibition of SOS mutagenesis (6, 12, 13).
The suppression of umuDC-mediated cold sensitivity by overproduction of ɛ is due to the release of the replication block conferred by the umuDC gene products.
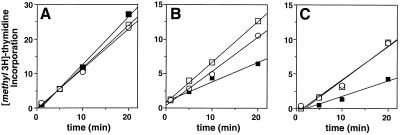
To confirm that suppression of umuDC-mediated cold sensitivity by overproduction of ɛ was due to the inability of the umuDC gene products to interfere with DNA synthesis and not to some other aspect of the unique physiology associated with the cold sensitivity (25), we measured the rate of DNA replication in an E. coli strain that was overexpressing umuDC with or without simultaneous overproduction of ɛ (Fig. 1). This was done by measuring the extent of [methyl-3H]thymidine (Dupont, NEN) incorporated into the DNA of growing cultures as previously described (2). Briefly, three cultures of E. coli GW8025 [relevant genotype, lexA300(Def)::spec ΔumuDC595::cat] were grown in liquid culture at the permissive temperature of 42°C. Two of these cultures bore plasmid pSU38-DC, which expresses the umuDC gene products from their native promoter, in addition to pHN1, which overproduces ɛ in response to IPTG. The third culture, containing only pBR322kan, did not express umuDC and served as a positive control. When the cultures reached mid-exponential phase, they were each split in two. One half of each was maintained at 42°C, and the other half was shifted to 30°C. At this time, IPTG (0.1 mM) was added to one pair of the cultures bearing the ɛ-overproducing plasmid pHN1. At times corresponding to both 10 min and 2 h after addition of IPTG, aliquots of each culture received a pulse of [methyl-3H]thymidine. Incorporation of [methyl-3H]thymidine into the bacterial DNA was then quantitated by trichloroacetic acid precipitation and collection of the acid-insoluble material onto glass fiber filters (Schleicher & Schuell) followed by liquid scintillation spectroscopy.
FIG. 1.
Effect of overproduction of ɛ on the ability of elevated levels of the umuDC gene products to interfere with DNA replication at the nonpermissive temperature of 30°C in E. coli GW8025 [lexA300(Def)::spec ΔumuDC595::cat recA441 xyl-5 mtl-1 galK2 rpsL31 kdgK51 Δ(argF-lacU)169 lacY1 tsx-33 supE44 thi-1 leuB6 hisG4(Oc) mgl-51 argE3(Oc) rfbD1 ara-14 thr-1 qsr′ qin-111 sulA11 ilvD(Ts) λ− Rac−] (25). [methyl-3H]thymidine incorporation (counts per minute of [methyl-3H]thymidine per unit of optical density at 600 nM × 10−4) was measured as described elsewhere (2) after incubation for 10 min at 42°C (A) or either 10 min (B) or 2 h (C) following a shift in the incubation temperature from 42°C to 30°C. Note the differences in scales for [methyl-3H] thymidine incorporation at 30 and 42°C. Symbols: □, GW8025(pBR322kan); ■, GW8025(pSU38-DC)(pHN1) (minus IPTG); ○, GW8025(pSU38-DC)(pHN1) (plus 0.1 mM IPTG).
Although DNA replication in the strain overexpressing the umuDC gene products and bearing the ɛ overproducer (pHN1) in the absence of IPTG was unaffected at 42°C (Fig. 1A), its rate was significantly reduced after just 10 min at 30°C (Fig. 1B and 1C), consistent with previous observations concerning the effects of overexpression of umuDC (20, 24). However, addition of IPTG to induce expression of ɛ led to a partial reduction in the extent of the interference of replication after 10 min at 30°C (Fig. 1B). Two hours after the shift of the cultures to 30°C and addition of IPTG, the rate of DNA synthesis in the strain overexpressing the umuDC gene products was completely restored relative to that observed for the control strain bearing pBR322kan that did not express umuDC (Fig. 1C). The ability of elevated levels of ɛ to alleviate the replication block at 30°C indicates that the suppression of umuDC-mediated cold sensitivity provided by overexpression of ɛ occurs via its ability to prevent the umuDC gene products from interfering with DNA replication. Taken together, these results indicate that interactions involving the umuDC gene products with ɛ likely constitute an important component of umuDC-mediated cold sensitivity.
Deletion of dnaQ suppresses umuDC-mediated cold sensitivity.
To further test the possibility that the umuDC gene products interact with the replisome, in part through contacts with ɛ, we investigated whether or not deletion of the structural gene for ɛ (dnaQ) would similarly affect umuDC-mediated cold sensitivity. Although strains bearing a ΔdnaQ allele are viable, they grow poorly (29). However, this poor growth is efficiently suppressed by a gain-of-function suppressor mutation, called spq-2, that encodes a V832G substitution in the α (catalytic) subunit of Pol III that serves to modestly increase its polymerase activity (29).
Using the quantitative transformation assay described above (25), we investigated the effect of deletion of dnaQ on umuDC-mediated cold sensitivity with three isogenic strains that differ in their dnaE and/or dnaQ alleles. As shown in Table 2, the ΔdnaQ903::tet allele partially (∼33-fold) suppressed the cold sensitivity conferred by overexpression of umuDC. In contrast, the spq-2 allele had no apparent effect on umuDC-mediated cold sensitivity. The reduced efficiency of suppression by the ΔdnaQ903::tet allele compared to overexpression of ɛ (compare Tables 1 and 2) suggests that the umuDC gene products interact with more than one component of the replisome, as we have previously demonstrated in vitro (31). Taken together, these results suggest that removal of ɛ from the replisome by deletion of dnaQ reduces the affinity of the umuDC gene products for Pol III, but to a lesser extent than do elevated levels of ɛ, which presumably acts to titrate the umuDC gene products away from Pol III.
TABLE 2.
Deletion of dnaQ suppresses umuDC-mediated cold sensitivity
| Straina | dnaE allele | dnaQ allele | Plasmidb | Transformants/mlb |
|---|---|---|---|---|
| GW8018 | dnaE+ | dnaQ+ | pSU18 | 0.95 |
| pSU18-DCc | 0.00012 | |||
| GW8112 | spq-2 | dnaQ+ | pSU18 | 0.77 |
| pSU18-DC | 0.00017 | |||
| GW8113 | spq-2 | ΔdnaQ903::tet | pSU18 | 0.97 |
| pSU18-DC | 0.0056 |
The three strains used are isogenic except for the indicated dnaE and/or dnaQ alleles and were constructed by P1vir-mediated transduction. The presence of the spq-2 allele was confirmed by automated DNA sequence analysis (data not shown). The resultant genotype is identical to that of GW8111 lacking the F′ lacIq from XL-1 Blue (25). The spq-2 allele used in GW8112 was linked to zae-3905::Tn10kan (29). The ΔdnaQ903::tet allele is a marker replacement that is missing the coding sequence corresponding to the C-terminal half of the gene (29).
Expressed as the total number of CFU obtained at 30°C divided by the total number of CFU obtained at 42°C. Transformation efficiencies were on the order of ∼1 × 105 to 5 × 105 CFU/μg of DNA. The differences in relative plating efficiencies between pSU18-DC and pSU38-DC are presumably related to the difference in antibiotics used (pSU18-DC confers chloramphenicol resistance, while pSU38-DC confers kanamycin resistance).
A pSU18 derivative containing the complete umuDC operon on a PstI fragment (31).
UmuD and UmuD′ each interact with the small C-terminal domain of ɛ.
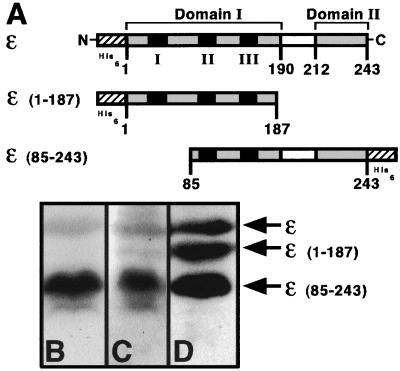
It has recently been suggested that ɛ is comprised of two separate domains, a large N-terminal domain that interacts with the Θ subunit of Pol III and contains the three exonuclease motifs, and a small C-terminal domain that interacts with the α subunit of Pol III (33). To determine which domain of ɛ is involved in interaction with UmuD and UmuD′, we analyzed the abilities of 32P-labeled UmuD and UmuD′ to interact with the different domains of ɛ by Far Western blotting as previously described (31). For this, we used PCR to clone two ɛ derivatives, one that corresponds to the proposed N-terminal domain [ɛ(1–187)] and a second that corresponds to a portion of the N-terminal domain together with the small C-terminal domain [ɛ(85–243)] (Fig. 2A). Attempts to overexpress an ɛ derivative corresponding to just the small C-terminal domain (residues 212 to 243), with or without the proposed Q linker that constitutes residues 190 to 212 (33), were unsuccessful. All of the derivatives used in this study contain a His6 tag at either the N- or C-terminal end that allowed for their one-step purification under denaturing conditions via nickel chromatography (Qiagen).
FIG. 2.
(A) Structures of the ɛ derivatives used in this study. Proposed domains of ɛ are indicated (33). Amino acid residues are indicated, as are the three exonuclease motifs (black boxes), the proposed N- and C-terminal domains (gray boxes) and Q linker (white boxes), and the hexa-histidine tags (cross-hatched boxes). Note that the figures are not to scale. Polyvinylidene difluoride membranes (Millipore) containing the indicated ɛ derivatives (1 μg each) after fractionation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis were processed either as a Far Western blot (31) by probing with 32P-labeled UmuD (B) or UmuD′ (C) or as a standard Western blot (D) using antipolyhistidine tag antibodies as described elsewhere (23, 31). The relative position of each ɛ derivative is indicated.
As shown in Fig. 2, probing a membrane bearing these ɛ derivatives with 32P-labeled UmuD or UmuD′ as described elsewhere (31) indicated that both UmuD and UmuD′ interact specifically with the C-terminal domain of ɛ. In fact, both UmuD and UmuD′ appear to interact better with ɛ(85–243) than they do with wild-type ɛ. We observed no interaction of UmuD with the N-terminal domain of ɛ and only a very weak interaction between this domain and UmuD′. Based on the characterization of the dnaQ101 allele that contains a deletion of the exonuclease III motif but retains the C-terminal 30 residues intact, it was suggested that the ability of overproduced ɛ to suppress SOS mutagenesis was due to its ssDNA binding activity (13). However, our finding that the umuD gene products interact with the C-terminal domain of ɛ suggests the alternate possibility that suppression of SOS mutagenesis by overproduction of ɛ (6, 12) may in fact be due to its ability to titrate the umuDC genes away from the replisome, as was originally suggested (5, 6, 12).
Summary and conclusions.
The results discussed in this report indicate that the interaction of the umuDC gene products with the replisome involves, at least in part, a specific interaction between the umuD gene products and the C-terminal domain of ɛ. These findings, although a result of studies that used strains expressing higher than normal levels of the umuDC and ɛ gene products from multicopy plasmids, provide further evidence that interactions of the umuDC gene products with components of Pol III help to determine which biological role the umuDC gene products will play and are consistent with a direct role for Pol III in translesion DNA synthesis (31). Our observations that the domain of ɛ involved in interaction with UmuD and UmuD′ corresponds to the same region of ɛ that interacts with α raises some interesting questions. For example, do the umuDC gene products interact with ɛ and α simultaneously, forming a ternary complex that acts to regulate the activity of Pol III, or do they compete with α for binding to ɛ? Also, does interaction of UmuD and/or UmuD′ with ɛ serve to inhibit its proofreading exonuclease activity in response to DNA damage? Further work will be necessary to establish whether UmuD and UmuD′ exhibit similar affinities for ɛ, or whether they possess different affinities for ɛ, as the umuD gene products do for α and β (31). In addition, further biochemical and physiological studies will be required to better define the precise physiological role of the interactions involving ɛ and the umuDC gene products.
Acknowledgments
M.D.S., S. M., and T. O. contributed equally to this work.
We thank Charles McHenry for the Pol III-overproducing plasmids, Russell Maurer for the ΔdnaQ903::tet and spq-2 zae-3905::Tn10kan E. coli strains, and members of our laboratory, in particular Brad Smith, for helpful discussions.
This work was supported by Public Health Service grant CA21615 to G.C.W. from the National Cancer Institute. M.D.S. was supported by a fellowship (5 F32 CA79161-02) from the National Cancer Institute. C. K. carried out her research as part of the Undergraduate Research Opportunities Program (UROP) at the Massachusetts Institute of Technology.
REFERENCES
- 1.Bartolome B, Jubete Y, Martinez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 2.Bukau B, Walker G C. ΔdnaK52 mutants of Escherichia coli have defects in chromosome segregation and plasmid maintenance at normal growth temperatures. J Bacteriol. 1989;171:6030–6038. doi: 10.1128/jb.171.11.6030-6038.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burckhardt S E, Woodgate R, Scheuermann R H, Echols H. UmuD mutagenesis protein of Escherichia coli: overproduction, purification, and cleavage by RecA. Proc Natl Acad Sci USA. 1988;85:1811–1815. doi: 10.1073/pnas.85.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elledge S J, Walker G C. Proteins required for ultraviolet light and chemical mutagenesis. Identification of the products of the umuC locus of Escherichia coli. J Mol Biol. 1983;164:175–192. doi: 10.1016/0022-2836(83)90074-8. [DOI] [PubMed] [Google Scholar]
- 5.Foster P L, Sullivan A D. Interactions between epsilon, the proofreading subunit of DNA polymerase III, and proteins involved in the SOS response of Escherichia coli. Mol Gen Genet. 1988;214:467–473. doi: 10.1007/BF00330482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foster P L, Sullivan A D, Franklin S B. Presence of the dnaQ-rnh divergent transcriptional unit on a multicopy plasmid inhibits induced mutagenesis in Escherichia coli. J Bacteriol. 1989;171:3144–3151. doi: 10.1128/jb.171.6.3144-3151.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedberg E C, Gerlach V L. Novel DNA polymerases offer clues to the molecular basis of mutagenesis. Cell. 1999;98:413–416. doi: 10.1016/s0092-8674(00)81970-4. [DOI] [PubMed] [Google Scholar]
- 8.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 9.Gerlach V L, Aravind L, Gotway G, Schultz R A, Koonin E V, Friedberg E C. Human and mouse homologs of Escherichia coli DinB (DNA polymerase IV), members of the UmuC/DinB superfamily. Proc Natl Acad Sci USA. 1999;96:11922–11927. doi: 10.1073/pnas.96.21.11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodman M F. Coping with replication ‘train wrecks’ in Escherichia coli using Pol V, Pol II and RecA proteins. Trends Biochem Sci. 2000;25:189–195. doi: 10.1016/s0968-0004(00)01564-4. [DOI] [PubMed] [Google Scholar]
- 11.Johnson R E, Washington M T, Prakash S, Prakash L. Bridging the gap: a family of novel DNA polymerases that replicate faulty DNA. Proc Natl Acad Sci USA. 1999;96:12224–12226. doi: 10.1073/pnas.96.22.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jonczyk P, Fijalkowska I, Ciesla Z. Overproduction of the epsilon subunit of DNA polymerase III counteracts the SOS mutagenic response of Escherichia coli. Proc Natl Acad Sci USA. 1988;85:9124–9127. doi: 10.1073/pnas.85.23.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanabus M, Nowicka A, Sledziewska-Gojska E, Jonczyk P, Ciesla Z. The antimutagenic effect of a truncated epsilon subunit of DNA polymerase III in Escherichia coli cells irradiated with UV light. Mol Gen Genet. 1995;247:216–221. doi: 10.1007/BF00705652. [DOI] [PubMed] [Google Scholar]
- 14.Kim D R, McHenry C S. In vivo assembly of overproduced DNA polymerase III. Overproduction, purification, and characterization of the alpha, alpha-epsilon, and alpha-epsilon-theta subunits. J Biol Chem. 1996;271:20681–20689. doi: 10.1074/jbc.271.34.20681. [DOI] [PubMed] [Google Scholar]
- 15.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulaeva O I, Koonin E V, McDonald J P, Randall S K, Rabinovich N, Connaughton J F, Levine A S, Woodgate R. Identification of a DinB/UmuC homolog in the archeon Sulfolobus solfataricus. Mutat Res. 1996;357:245–253. doi: 10.1016/0027-5107(96)00164-9. [DOI] [PubMed] [Google Scholar]
- 17.Little J W. Control of the SOS regulatory system by the level of RecA protease. Biochimie. 1982;64:585–589. doi: 10.1016/s0300-9084(82)80092-8. [DOI] [PubMed] [Google Scholar]
- 18.Little J W, Mount D W, Yanisch-Perron C R. Purified lexA protein is a repressor of the recA and lexA genes. Proc Natl Acad Sci USA. 1981;78:4199–4203. doi: 10.1073/pnas.78.7.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsh L, Walker G C. Cold sensitivity induced by overproduction of UmuDC in Escherichia coli. J Bacteriol. 1985;162:155–161. doi: 10.1128/jb.162.1.155-161.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murli S, Opperman T, Smith B T, Walker G C. A role for the umuDC gene products of Escherichia coli in increasing resistance to DNA damage in stationary phase by inhibiting the transition to exponential growth. J Bacteriol. 2000;182:1127–1135. doi: 10.1128/jb.182.4.1127-1135.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nohmi T, Battista J R, Dodson L A, Walker G C. RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and posttranslational activation. Proc Natl Acad Sci USA. 1988;85:1816–1820. doi: 10.1073/pnas.85.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nowicka A, Kanabus M, Sledziewska-Gojska E, Ciesla Z. Different UmuC requirements for generation of different kinds of UV-induced mutations in Escherichia coli. Mol Gen Genet. 1994;243:584–592. doi: 10.1007/BF00284207. [DOI] [PubMed] [Google Scholar]
- 23.Ohta T, Sutton M D, Guzzo A, Cole S, Ferentz A E, Walker G C. Mutations affecting the ability of the Escherichia coli UmuD′ protein to participate in SOS mutagenesis. J Bacteriol. 1999;181:177–185. doi: 10.1128/jb.181.1.177-185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Opperman T, Murli S, Smith B T, Walker G C. A model for a umuDC-dependent prokaryotic DNA damage checkpoint. Proc Natl Acad Sci USA. 1999;96:9218–9223. doi: 10.1073/pnas.96.16.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Opperman T, Murli S, Walker G C. The genetic requirements for UmuDC-mediated cold sensitivity are distinct from those for SOS mutagenesis. J Bacteriol. 1996;178:4400–4411. doi: 10.1128/jb.178.15.4400-4411.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajagopalan M, Lu C, Woodgate R, O'Donnell M, Goodman M F, Echols H. Activity of the purified mutagenesis proteins UmuC, UmuD′, and RecA in replicative bypass of an abasic DNA lesion by DNA polymerase III. Proc Natl Acad Sci USA. 1992;89:10777–10781. doi: 10.1073/pnas.89.22.10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reuven N B, Arad G, Maor-Shoshani A, Livneh Z. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and is specialized for translesion replication. J Biol Chem. 1999;274:31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- 28.Shinagawa H, Iwasaki H, Kato T, Nakata A. RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis. Proc Natl Acad Sci USA. 1988;85:1806–1810. doi: 10.1073/pnas.85.6.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slater S C, Lifsics M R, O'Donnell M, Maurer R. holE, the gene coding for the theta subunit of DNA polymerase III of Escherichia coli: characterization of a holE mutant and comparison with a dnaQ (epsilon-subunit) mutant. J Bacteriol. 1994;176:815–821. doi: 10.1128/jb.176.3.815-821.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Studwell P S, O'Donnell M. Processive replication is contingent on the exonuclease subunit of DNA polymerase III holoenzyme. J Biol Chem. 1990;265:1171–1178. [PubMed] [Google Scholar]
- 31.Sutton M D, Opperman T, Walker G C. The Escherichia coli SOS mutagenesis proteins UmuD and UmuD′ interact physically with the replicative DNA polymerase. Proc Natl Acad Sci USA. 1999;96:12373–12378. doi: 10.1073/pnas.96.22.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutton M D, Smith B T, Godoy V G, Walker G C. The SOS response: recent insights into umuDC-dependent DNA damage tolerance. Annu Rev Genet. 2000;34:479–497. doi: 10.1146/annurev.genet.34.1.479. [DOI] [PubMed] [Google Scholar]
- 33.Taft-Benz S A, Schaaper R M. The C-terminal domain of dnaQ contains the polymerase binding site. J Bacteriol. 1999;181:2963–2965. doi: 10.1128/jb.181.9.2963-2965.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang M, Pham P, Shen X, Taylor J-S, O'Donnell M, Woodgate M, Goodman M F. Roles of E. coli DNA polymerase IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature. 2000;404:1014–1018. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- 35.Tang M, Shen X, Frank E G, O'Donnell M, Woodgate R, Goodman M F. UmuD′(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker G C. Skiing the black diamond slope: progress on the biochemistry of translesion DNA synthesis. Proc Natl Acad Sci USA. 1998;95:10348–10350. doi: 10.1073/pnas.95.18.10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woodgate R. A plethora of lesion-replicating DNA polymerases. Genes Dev. 1999;13:2191–2195. doi: 10.1101/gad.13.17.2191. [DOI] [PubMed] [Google Scholar]
- 38.Woodgate R, Levine A S. Damage inducible mutagenesis: recent insights into the activities of the Umu family of mutagenesis proteins. Cancer Surv. 1996;28:117–140. [PubMed] [Google Scholar]