Abstract
The expression of sll1689, an open reading frame from the cyanobacterium Synechocystis sp. strain PCC 6803 putatively encoding a member of the ς70 family of sigma factors, appears to be regulated by the nitrogen control transcription factor NtcA. Disruption of sll1689 had no noticeable effect on exponential growth, identifying its product as a member of the group 2, nonessential class of ς70-like sigma factors; however, this disruption decreased the viability of the cells after long periods of nitrogen starvation. We have named this gene rpoD2-V. The expression of glnN, encoding a type III glutamine synthetase, was impaired in strains bearing an inactivated copy of the rpoD2-V gene.
Eubacterial RNA polymerase consists of a core enzyme and a sigma factor that directs the complex to a specific class of promoter sequences. Most eubacterial genomes encode a number of different sigma factors that recognize different subsets or groups of promoters. This variety of sigma factors allows basal expression of certain genes as well as regulated expression of other genes in response to environmental or developmental signals (44). Two families of sigma factors have been defined on the basis of sequence similarity, the ς70 and the ς54 families, named after the primary sigma factor and the factor involved in nitrogen regulation in Escherichia coli, respectively. All eubacteria contain one or more sigma factors belonging to the ς70 family (44). Sigma factors that are responsible for the transcription of most genes in exponentially growing cells and that are essential for survival constitute group 1 or primary sigma factors; group 2 includes “primary-like” sigma factors that are nonessential for exponential cell growth (secondary sigma factors); and group 3 includes the so-called alternative sigma factors (30). A single group 2 sigma factor, stationary-phase ς38, has been found in E. coli, but multiple group 2 sigma factors are present in high-GC-content gram-positive bacteria (e.g., Streptomyces spp.), cyanobacteria, and Chloroflexus aurantiacus (26, 44).
Cyanobacteria are a widely distributed group of phototrophic bacteria that carry out oxygenic photosynthesis and are considered the precursors of chloroplasts. In cyanobacteria and chloroplasts, the core of RNA polymerase consists of five subunits (α2ββ′γ in cyanobacteria and α2ββ′β" in chloroplasts) (28, 39) and thus is slightly different from that in other eubacteria (α2ββ′ tetramer) (9). The β′ subunit of eubacterial RNA polymerase is considered to be split into two subunits in cyanobacteria and chloroplasts, β′γ and β′β", respectively (5). Multiple sigma factors of the ς70 family (groups 1 and 2) have been found in cyanobacteria of the genera Anabaena (7, 8), Nostoc (11), Synechococcus (12, 13, 27, 40, 41), Microcystis (2, 3) and Synechocystis (29). The chromosome of Synechocystis sp. strain PCC 6803 contains five open reading frames (ORFs) that would correspond to sequences encoding primary or primary-like sigma factors (groups 1 and 2) and three ORFs that would correspond to sequences encoding alternative sigma factors (29). The sequences of the genome of Anabaena sp. strain PCC 7120 available to date (http://www.kazusa.or.jp /cyano/anabaena) contain at least eight ORFs similar to sequences encoding primary or primary-like sigma factors. No sigma factor belonging to the ς54 family has yet been identified for cyanobacteria. Phylogenetic analysis of cyanobacterial sigma factor sequences known to date indicates that group 1 sigma factors are tightly clustered (cluster I) and that group 2 sigma factors form a separate, coherent clade (25, 26) which appears to be further divided into four clusters, II to V (25). Each cyanobacterial strain analyzed to date bears members belonging to each group 2 cluster (25).
Cyanobacteria are able to use a number of different nitrogen sources (17). The regulation of the use of these sources is mediated by the nitrogen control, CAP (catabolite activator protein)-family transcription factor NtcA. In the absence of ammonium, NtcA activates the expression of genes required for the assimilation of nitrogen sources alternative to ammonium (42). A consensus sequence needed for NtcA binding to DNA (GTAN8TAC) has been defined (31). The NtcA-activated promoters bear this sequence, located at about position −40.5, and a −10 sequence of the form TAN3T (18, 31). The ntcA gene is widely distributed in cyanobacteria (21). In all strains analyzed to date, the sequence of the NtcA protein is highly conserved, as is the sequence of the promoters activated by NtcA in those strains. Insertional mutants of ntcA have been obtained for Synechococcus sp. strain PCC 7942 and Anabaena sp. strain PCC 7120. However, attempts to isolate a completely segregated ntcA mutant of Synechocystis sp. strain PCC 6803 have been unsuccessful (23).
The complete sequence of the chromosome of Synechocystis sp. strain PCC 6803 is available (29) (http://www.kazusa.or.jp /cyano/cyano.html). In the context of our attempts to identify genes controlled by the global nitrogen regulator NtcA, we have carried out a search for putative NtcA-activated promoters in the genomic sequence of strain PCC 6803. In this report, the identification of sll1689, encoding a protein homologous to ς70-like sigma factors, as a nitrogen-regulated gene as well as the effects of disruption of sll1689 are described.
Search for putative NtcA-activated promoters.
A computer search for sequences of the genome of Synechocystis sp. strain PCC 6803 that could correspond to putative NtcA-activated promoters was carried out using overlapping 30-kb segments of the genome sequence and MatInspector software (34). The consensus sequence needed for NtcA binding, GTAN8TAC, was found in a total of 367 positions, 44 of which were followed, at a distance of 20 to 23 nucleotides, by a sequence matching the consensus for a putative −10 box, TAN3T. Only in 31 of those cases was the NtcA box located less than 2 kb upstream from an ORF, that is, in a position that might be considered compatible with a role in transcription activation. Although some of the identified sequences were located upstream from ORFs encoding hypothetical proteins of unknown function, 22 of them were located upstream from either known genes or ORFs whose predicted products were similar to some proteins in the databases. Among those, as expected, we could identify the NtcA-regulated promoter of the glnA gene, encoding glutamine synthetase (35), and the region upstream from nirA, encoding nitrite reductase, a gene whose expression has been shown to be NtcA dependent in Synechococcus sp. strain PCC 7942 (31) and Anabaena sp. strain PCC 7120 (20). A sequence, GTAN8TACN21TAN3T, matching the structure of NtcA-regulated promoters was found 270 nucleotides upstream from sll1689, one of the five ORFs identified in the genome of strain PCC 6803 as rpoD-like genes encoding members of the ς70 family of sigma factors (29). We refer to this ORF as rpoD2-V because its product belongs to cluster V of the phylogenetic tree of cyanobacterial primary or primary-like sigma factors (25).
Regulation of expression of rpoD2-V.
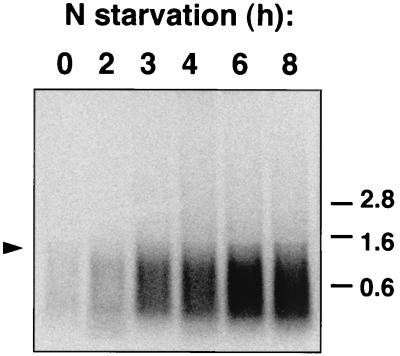
In order to analyze whether the expression of rpoD2-V was in fact under nitrogen control, Northern hybridization analysis was carried out with RNA isolated from Synechocystis sp. strain PCC 6803 cells that had been subjected to nitrogen starvation. Standard molecular biology procedures were performed by published methods (4, 37). Synechocystis cells were grown in BG110 medium (medium BG11 [36] without NaNO3) supplemented with 0.84 g of NaHCO3 liter−1 (BG110C medium) and 15 mM NH4Cl and bubbled with a mixture of CO2 (1% [vol/vol]) and air. Twice as much N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES)–NaOH buffer (pH 7.5) as NH4Cl was added to all NH4Cl-containing media. For RNA isolation, cells growing exponentially in NH4Cl-containing BG110C medium were harvested at room temperature and either used directly or washed with and resuspended in BG110C medium (nitrogen free) and further incubated under culture conditions for various times. RNA was isolated as previously described (22). Northern hybridization was carried out using Hybond N+ membranes according to manufacturer recommendations. Northern blot experiments performed using the insert of pCSAM82 (a plasmid containing sll1689 and flanking sequences; see below) as a probe indicated that the expression of rpoD2-V was indeed induced in cells subjected to nitrogen deprivation (Fig. 1). As has also been observed for the sigC transcript of Synechococcus sp. strain PCC 7002 (13), the transcript hybridizing to the rpoD2-V probe appeared as a smear. Control hybridization experiments with different probes showed that the RNA in the filters was not degraded (data not shown).
FIG. 1.
Northern blot analysis of the expression of rpoD2-V in Synechocystis sp. strain PCC 6803. RNA was isolated from ammonium-grown cells (lane 0) or from ammonium-grown cells incubated in nitrogen-free medium for 2, 3, 4, 6, or 8 h. Hybridization to a probe for rpoD2-V was carried out as described in the text. Samples contained 40 μg of RNA. Sizes of standards (in kilobases) are indicated on the right. Arrowhead indicates transcripts of about 1.5 kb.
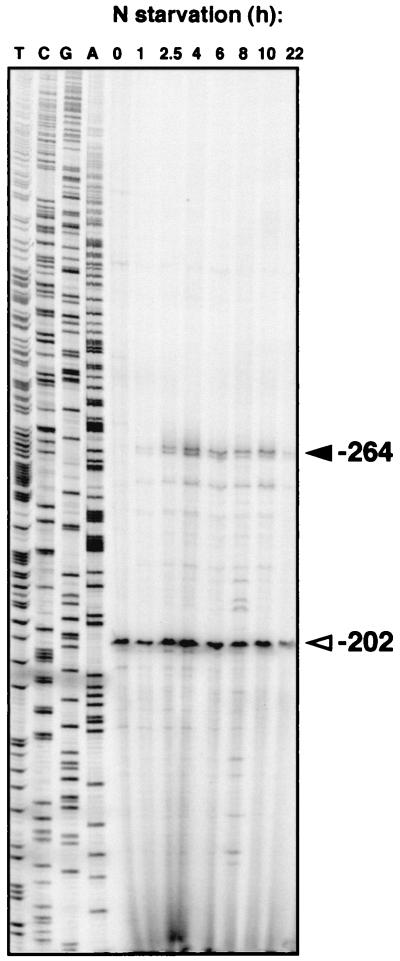
Primer extension experiments with 32P-labeled primers were carried out in order to determine the possible transcription start point(s) (tsp) from which the observed transcription originated. These experiments were performed as previously described (33) using the above-mentioned RNA samples and oligonucleotide RD5 (5′-TTA TTC AAT CGG CCA GAG C-3′, complementary to positions −89 to −107 with respect to the putative translational start site of rpoD2-V) (Fig. 2). In addition to a constitutive putative tsp located at position −202 with respect to the translational start site of rpoD2-V, an inducible transcript which corresponded to a putative tsp located at position −264 was observed. The sequences located upstream from the inducible tsp corresponded to the consensus NtcA-activated promoter sequence (GTAN8TACN21TAN3TN5 tsp) identified in the computer search described above. The identification of these two putative tsp was confirmed by using oligonucleotides RD4 (5′-AGA GCA TCA TCC AGA TAG ACC-3′, complementary to positions −103 to −123 with respect to the putative translational start site of rpoD2-V) and RD6 (5′-TCA GCG AGG CCA TCC AAA GCC-3′, complementary to positions +77 to +57 with respect to the putative translational start site of rpoD2-V) (data not shown). The apparent inconsistency between the Northern and primer extension results could be due to different stabilities of the constitutive and inducible transcripts. In any case, the tsp detected at −264 is the one which reproduces the regulatory pattern observed by Northern analysis for the rpoD2-V gene.
FIG. 2.
Primer extension analysis of the expression of the rpoD2-V gene in Synechocystis sp. strain PCC 6803. Primer extension assays were carried out with RNA isolated from cells grown on ammonium (lane 0) or grown on ammonium and incubated in nitrogen-free medium for 1, 2.5, 4, 6, 8, 10, or 22 h. Assays were carried out using oligonucleotide RD5 (see the text). The sequencing ladders shown were generated with the same oligonucleotide and plasmid pCSAM82. Arrowheads indicate the putative tsp identified at positions −264 and −202.
Band shift assays were carried out with a 402-bp DNA fragment containing the putative inducible promoter of rpoD2-V. This fragment was amplified by PCR using oligonucleotides RD1 (5′-ACG CTT GGA ATG GCA ACA GG-3′, corresponding to positions −545 to −526 with respect to the putative translational start site of rpoD2-V) and RD2 (5′-CCA CTT TCA GCT ATG CGC ACT GCG G-3′, complementary to positions −144 to −168 with respect to the putative translational start site of rpoD2-V) and plasmid pCSAM82 (see below) as a template. Binding assays with purified NtcA protein were carried out as described previously (33). The results shown in Fig. 3 indicate that NtcA specifically bound to the fragment containing the putative inducible promoter of rpoD2-V. The binding could be competed by the addition of the same, unlabeled fragment or a fragment containing the NtcA-regulated promoter of the glnA gene from Anabaena sp. strain PCC 7120 (19; see the fragment used in reference 33).
FIG. 3.
Band shift assays of a DNA fragment from the rpoD2-V promoter with purified histidine-tagged NtcA. Assays were carried out as described in the text using a fragment from the region upstream of rpoD2-V. Assay mixtures contained 5 pmol of purified histidine-tagged Anabaena NtcA without competitor DNA or with a 25-fold molar excess of the same unlabeled fragment (rpoD2) or with an unlabeled fragment from the NtcA-regulated promoter of the glnA gene of Anabaena sp. strain PCC 7120 (glnA).
The expression of rpoD2-V in an ntcA background could not be tested, since no completely segregated ntcA mutant of strain PCC 6803 is currently available. However, the two pieces of evidence presented here, the location of sequences corresponding to an NtcA-activated promoter sequence in front of the inducible tsp and the specific binding of purified NtcA to those sequences, suggest that the expression of rpoD2-V is directly regulated by the global nitrogen regulator NtcA. Thus, induction of the expression of rpoD2-V appears to be part of the transcriptional changes that are induced by NtcA and take place in response to nitrogen starvation in Synechocystis sp. strain PCC 6803. Interestingly, the expression of Synechococcus sp. strain PCC 7002 SigC (a sigma factor also belonging to cluster V) has also been found to increase under nitrogen limitation (13). One of the three possible tsp defined for strain PCC 7002 sigC, located at position −255 (13), could in fact correspond to an NtcA-regulated promoter, since it exhibits a sequence (GTAN8AAC) that resembles that of the NtcA box, separated by 22 nucleotides from a putative −10 box of the form TAN3T.
Disruption of rpoD2-V.
A 2-kb DNA fragment comprising rpoD2-V and flanking sequences was amplified by PCR using oligonucleotides RD1 (see above) and RD3 (5′-GAT GCG AGC GAA GAT TTC TG-3′, complementary to positions +342 to +323 with respect to the putative translational stop site of rpoD2-V) and total DNA (isolated as described in reference 10) from strain PCC 6803 as a template. The PCR product was cloned in vector pGEM-T (Promega), generating plasmid pCSAM82. This plasmid was digested with BamHI and BglII (both sites are internal to the rpoD2-V gene), and the 1.3-kb Kmr gene cassette C.K1 excised from pRL161 (S.A1/L.HEH1/C.K1; nomenclature as in reference 16) with BamHI was inserted between the BamHI and BglII sites of rpoD2-V, rendering plasmid pCSAM89 (a or b, depending on the orientation of C.K1 with respect to the rpoD2-V gene).
After transformation of Synechocystis sp. strain PCC 6803 with plasmids pCSAM89a and pCSAM89b (14), Kmr transformants were selected and maintained on solid BG110C medium supplemented with 4 mM NH4Cl and 50 μg of kanamycin ml−1. To test whether the resulting mutant strains were homozygous for the mutant chromosomes, PCR amplification with primers RD1 and RD3 and genomic DNA from the mutants as templates and Southern hybridization analysis were carried out. Clones homozygous for the mutated chromosomes were chosen and named CSAM4 (rpoD2-V::C.K1, which carries the gene cassette in the same orientation as rpoD2-V) and CSAM5 (rpoD2-V::C.K1, which carries the gene cassette in the orientation opposite that in rpoD2-V).
No noticeable effect on cell growth was detected in the mutant strains with respect to the wild-type strain in nitrate- or ammonium-containing media. The growth rate constant was determined for single cultures of the wild-type and mutant strains in 4 mM NH4Cl-containing medium, and values of 0.62 day−1 (wild type), 0.57 day−1 (CSAM4), and 0.64 day−1 (CSAM5) were obtained. Thus, rpoD2-V is not essential for cell viability under nutrient-replete conditions and can be classified as a group 2 sigma factor gene.
Since the expression of rpoD2-V increased under nitrogen-limiting conditions, a number of different phenotypes related to the assimilation of nitrogen sources in the mutant strains were tested. The induction of nitrate reductase and glutamine synthetase activities (measured as described previously [19]) took place in a similar fashion in the wild type and in the mutants (data not shown). Also, the decrease in phycobiliproteins that takes place in response to nitrogen deprivation (1, 15) was not altered in the mutants. The effect of rpoD2-V inactivation on the induction of nitrogen-regulated genes was also tested by Northern hybridization (Fig. 4). The genes whose expression was analyzed were amt1 (ammonium/methylammonium permease) (32), glnB (PII signaling protein) (22), and glnN (type III glutamine synthetase) (35). The probes used for amt1, glnB, and glnN were those indicated in the corresponding references. Quantification of the signals in the Northern blots shown in Fig. 4, normalized to the signal for the rnpB probe (43), used as a loading and transfer control, indicated that the expression of amt1 or glnB was not significantly altered in the rpoD2-V mutants. Activation of the expression of amt1 or glnB upon nitrogen deprivation took place earlier than the expression of the rpoD2-V gene itself (Fig. 1 and 4), making the participation of RpoD2-V in their expression unlikely. However, activation of the expression of glnN, which under our experimental conditions took place later than induction of the expression of amt1 and glnB in response to nitrogen starvation, was impaired in the mutants; a decrease of about 30% was observed after 4 h of nitrogen starvation (Fig. 4). Interestingly, glnN recently has been shown to influence the recovery of Synechococcus sp. strain PCC 7942 cells from long periods of nitrogen starvation (38).
FIG. 4.
Effect of the disruption of rpoD2-V on the expression of amt1, glnB, and glnN. RNA was isolated from ammonium-grown cells (lane 0) or from ammonium-grown cells incubated in nitrogen-free medium for 2, 4, 8, or 22 h. The filter was sequentially hybridized with probes for glnN, glnB, amt1, and rnpB. Samples contained 30 μg of RNA. Images of radioactive gels were obtained and quantified using a Cyclone storage phosphor system (Packard). The level of expression of each gene (defined as the ratio of the signal in each lane to the signal in the corresponding rnpB hybridization lane and expressed in arbitrary units) in the wild-type (WT) strain (solid circles) and in mutant strain CSAM4 (open circles) is depicted in the graphs on the right. Similar results were obtained with mutant strain CSAM5 (data not shown).
The survival of wild-type and mutant cells after different periods of nitrogen starvation was studied. For determination of survival after nitrogen deficiency, cells growing exponentially in liquid BG110C medium supplemented with 15 mM NH4Cl were harvested at room temperature, resuspended in BG110C medium (nitrogen free) at approximately 10 μg of chlorophyll ml−1, and further incubated under culture conditions. For the determination of CFU, appropriate dilutions of liquid cultures were plated as soft-agar overlays on solid BG110C medium supplemented with 4 mM NH4Cl. The viability of mutant cells after long periods of nitrogen starvation was severely altered with respect to that of wild-type cells. The survival of wild-type cells subjected to 25 days of nitrogen starvation was about 0.15, a value similar to that described for Synechococcus sp. strain PCC 7942 (24); however, survival dropped to 7.3 × 10−4 and 9.1 × 10−4 for mutant strains CSAM4 and CSAM5, respectively. Although differences were more dramatic after this long period of starvation, some differences in survival between the wild-type and mutant strains could be observed as soon as after 2 days of nitrogen deficiency.
In order to test whether the lower survival of the mutant strains was specific for nitrogen stress, we tested the survival of the cells after starvation for phosphorus, carbon, or sulfur. For this experiment, cells grown in 15 mM NH4Cl-containing medium were washed and resuspended in the corresponding nutrient-deficient medium (10 mM NH4Cl-containing BG110C medium lacking phosphorus in the case of phosphorus starvation, 10 mM NH4Cl-containing BG110C medium lacking sulfur in the case of sulfur starvation, and 4 mM NH4Cl-containing BG110 medium in the case of carbon starvation). All cultures except for the carbon-starved ones were bubbled with a mixture of CO2 (1% [vol/vol]) and air. N2 was bubbled through the carbon-starved cultures. The survival of wild-type PCC 6803 cells under phosphorus and carbon stress was similar to that under nitrogen stress, i.e., about 0.25 after 10 days of starvation. However, whereas the survival of rpoD2-V mutant cells after nitrogen starvation was decreased with respect to that of wild-type cells (0.01 after 10 days of nitrogen starvation), the mutant cells behaved like the wild-type cells under phosphorus and carbon stress. Survival under sulfur stress was much lower for both the wild type and the mutants, about 0.1 after 2 days of starvation. These experiments indicate that the sigma factor encoded by rpoD2-V is specifically involved in survival under severe nitrogen stress.
Concluding remarks.
Functions for cyanobacterial nonessential sigma factors have been assigned or suggested only in a few cases. The expression of sigB and sigC of Anabaena sp. strain PCC 7120 was found to be increased in the absence of nitrogen (8). However, single- and double-mutant strains bearing inactivated sigB and/or sigC genes were able to grow fixing nitrogen (8). The expression of sigB and sigC of Synechococcus sp. strain PCC 7002 is also induced under nutrient-limiting conditions (13), and sigE of this strain seems to be implicated in transcription in the stationary phase of growth (27). Mutations in rpoD2 of Synechococcus sp. strain PCC 7942 affect the circadian rhythm of this organism (41), whereas mutations in sigH of Nostoc punctiforme affect the symbiosis of this cyanobacterium with a plant host (11). On the other hand, mutations in sigF of Synechocystis sp. strain PCC 6803, the only cyanobacterial alternative (group 3) sigma factor gene that has been characterized so far, result in a pleiotropic phenotype that includes alterations in pilus formation and motility (6).
The low survival of the Synechocystis sp. strain PCC 6803 rpoD2-V mutants after nitrogen stress could have been due to decreased glnN expression. Unfortunately, we have been unable to analyze the expression of glnN after long periods of nitrogen starvation because of the poor quality of mRNA samples isolated from the starved cells. RpoD2-V could also be involved in the expression of other genes required for survival under conditions of severe nitrogen deficiency. The fact that NtcA-activated promoters bear a −10 box of the form TAN3T suggests that NtcA activates transcription mediated by RNA polymerase containing a ς70-like sigma factor. Therefore, NtcA-dependent rpoD2-V expression might have the effect of increasing the levels of an appropriate, ς70-like sigma factor to assist NtcA-activated gene expression under conditions of severe nitrogen starvation.
Acknowledgments
We thank Enrique Martínez-Force for help with the computer search, Ana Valladares for help in some primer extension experiments, and F. J. Florencio and J. C. Reyes for providing several DNA probes.
This work was supported by grants PB97-1137 and PB98-0481 from Ministerio de Ciencia y Tecnología (Madrid, Spain). A.M.M.-P. was the recipient of postdoctoral contracts from MEC and CSIC (Madrid, Spain) and a fellowship from the Universidad de Sevilla.
REFERENCES
- 1.Allen M M, Smith A J. Nitrogen chlorosis in blue-green algae. Arch Microbiol. 1969;69:114–120. doi: 10.1007/BF00409755. [DOI] [PubMed] [Google Scholar]
- 2.Asayama M, Suzuki A, Nozawa S, Yamada A, Tanaka K, Takahashi H, Aida T, Shirai M. A new sigma factor homolog in a cyanobacterium: cloning, sequencing, and light-responsive transcripts of rpoD2 from Microcystis aeruginosa K-81. Biochim Biophys Acta. 1997;1351:31–36. doi: 10.1016/s0167-4781(97)00019-5. [DOI] [PubMed] [Google Scholar]
- 3.Asayama M, Suzuki H, Sato A, Aida T, Tanaka K, Takahashi H, Shirai M. The rpoD1 gene product is a principal sigma factor of RNA polymerase in Microcystis aeruginosa K-81. J Biochem. 1996;120:752–758. doi: 10.1093/oxfordjournals.jbchem.a021475. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley-Interscience; 2000. [Google Scholar]
- 5.Bergsland K J, Haselkorn R. Evolutionary relationships among eubacteria, cyanobacteria, and chloroplasts: evidence from the rpoC1 gene of Anabaena sp. strain PCC 7120. J Bacteriol. 1991;173:3446–3455. doi: 10.1128/jb.173.11.3446-3455.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhaya D, Watanabe N, Ogawa T, Grossman A R. The role of an alternative sigma factor in motility and pilus formation in the cyanobacterium Synechocystis sp. strain PCC 6803. Proc Natl Acad Sci USA. 1999;96:3188–3193. doi: 10.1073/pnas.96.6.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brahamsha B, Haselkorn R. Isolation and characterization of the gene encoding the principal sigma factor of the vegetative cell RNA polymerase from the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1991;173:2442–2450. doi: 10.1128/jb.173.8.2442-2450.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brahamsha B, Haselkorn R. Identification of multiple RNA polymerase sigma factor homologs in the cyanobacterium Anabaena sp. strain PCC 7120: cloning, expression, and inactivation of the sigB and sigC genes. J Bacteriol. 1992;174:7273–7282. doi: 10.1128/jb.174.22.7273-7282.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgess R, Erickson B, Gentry D, Gribskov M, Hager D, Lesley S, Strickland M, Thompson N. Bacterial RNA polymerase subunits and genes. In: Reznikoff W S, Burgess R, Dahlberg J E, Gross C A, Record M T Jr, Wickens M P, editors. RNA polymerase and the regulation of transcription. New York, N.Y: Elsevier; 1987. pp. 3–16. [Google Scholar]
- 10.Cai Y, Wolk C P. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J Bacteriol. 1990;172:3138–3145. doi: 10.1128/jb.172.6.3138-3145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell E L, Brahamsha B, Meeks J C. Mutation of an alternative sigma factor in the cyanobacterium Nostoc punctiforme results in the increased infection of its symbiotic plant partner, Anthoceros punctatus. J Bacteriol. 1998;180:4938–4941. doi: 10.1128/jb.180.18.4938-4941.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caslake L F, Bryant D A. The sigA gene encoding the major ς factor of RNA polymerase from the marine cyanobacterium Synechococcus sp. strain PCC 7002: cloning and characterization. Microbiology. 1996;142:347–357. doi: 10.1099/13500872-142-2-347. [DOI] [PubMed] [Google Scholar]
- 13.Caslake L F, Gruber T M, Bryant D A. Expression of two alternative sigma factors of Synechococcus sp. strain PCC 7002 is modulated by carbon and nitrogen stress. Microbiology. 1997;143:3807–3818. doi: 10.1099/00221287-143-12-3807. [DOI] [PubMed] [Google Scholar]
- 14.Chauvat F, De Vries L, Van der Ende A, Van Arkel G A. A host-vector system for gene cloning in the cyanobacterium Synechocystis PCC 6803. Mol Gen Genet. 1986;204:185–191. [Google Scholar]
- 15.Collier J L, Grossman A R. Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC 7942: not all bleaching is the same. J Bacteriol. 1992;174:4718–4726. doi: 10.1128/jb.174.14.4718-4726.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elhai J, Wolk C P. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene. 1988;68:119–138. doi: 10.1016/0378-1119(88)90605-1. [DOI] [PubMed] [Google Scholar]
- 17.Flores E, Herrero A. Assimilatory nitrogen metabolism and its regulation. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 487–517. [Google Scholar]
- 18.Flores E, Muro-Pastor A M, Herrero A. Cyanobacterial nitrogen assimilation genes and NtcA-dependent control of gene expression. In: Peschek G A, Löffelhardt W, Schmetterer G, editors. The phototrophic prokaryotes. New York, N.Y: Plenum Publishing Corporation; 1999. pp. 463–477. [Google Scholar]
- 19.Frías J E, Flores E, Herrero A. Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120. Mol Microbiol. 1994;14:823–832. doi: 10.1111/j.1365-2958.1994.tb01318.x. [DOI] [PubMed] [Google Scholar]
- 20.Frías J E, Flores E, Herrero A. Nitrate assimilation gene cluster from the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1997;179:477–486. doi: 10.1128/jb.179.2.477-486.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frías J E, Mérida A, Herrero A, Martín-Nieto J, Flores E. General distribution of the nitrogen control gene ntcA in cyanobacteria. J Bacteriol. 1993;175:5710–5713. doi: 10.1128/jb.175.17.5710-5713.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.García-Domínguez M, Florencio F J. Nitrogen availability and electron transport control the expression of glnB gene (encoding PII protein) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol. 1997;35:723–734. doi: 10.1023/a:1005846626187. [DOI] [PubMed] [Google Scholar]
- 23.García-Domínguez M, Reyes J C, Florencio F J. NtcA represses transcription of gifA and gifB, genes that encode inhibitors of glutamine synthetase type I from Synechocystis sp. PCC 6803. Mol Microbiol. 2000;35:1192–1201. doi: 10.1046/j.1365-2958.2000.01789.x. [DOI] [PubMed] [Google Scholar]
- 24.Görl M, Sauer J, Baier T, Forchhammer K. Nitrogen-starvation-induced chlorosis in Synechococcus PCC 7942: adaptation to long-term survival. Microbiology. 1998;144:2449–2458. doi: 10.1099/00221287-144-9-2449. [DOI] [PubMed] [Google Scholar]
- 25.Goto-Seki A, Shirokane M, Masuda S, Tanaka K, Takahashi H. Specificity crosstalk among group 1 and group 2 sigma factors in the cyanobacterium Synechococcus sp. PCC 7942: in vitro specificity and a phylogenetic analysis. Mol Microbiol. 1999;34:473–484. doi: 10.1046/j.1365-2958.1999.01608.x. [DOI] [PubMed] [Google Scholar]
- 26.Gruber T M, Bryant D A. Molecular systematic studies of eubacteria, using ς70-type sigma factors of group 1 and group 2. J Bacteriol. 1997;179:1734–1747. doi: 10.1128/jb.179.5.1734-1747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gruber T M, Bryant D A. Characterization of the alternative ς-factors SigD and SigE in Synechococcus sp. strain PCC 7002. SigE is implicated in transcription of post-exponential-phase-specific genes. Arch Microbiol. 1998;169:211–219. doi: 10.1007/s002030050563. [DOI] [PubMed] [Google Scholar]
- 28.Hudson G S, Holton T A, Whitfeld P R, Bottomley W. Spinach chloroplast rpoBC genes encode three subunits of the chloroplast RNA polymerase. J Mol Biol. 1988;200:639–654. doi: 10.1016/0022-2836(88)90477-9. [DOI] [PubMed] [Google Scholar]
- 29.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 30.Lonetto M, Gribskov M, Gross C A. The ς70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luque I, Flores E, Herrero A. Molecular mechanism for the operation of nitrogen control in cyanobacteria. EMBO J. 1994;13:2862–2869. doi: 10.1002/j.1460-2075.1994.tb06580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montesinos M L, Muro-Pastor A M, Herrero A, Flores E. Ammonium/methylammonium permeases of a cyanobacterium. Identification and analysis of three nitrogen-regulated amt genes in Synechocystis sp. PCC 6803. J Biol Chem. 1998;273:31463–31470. doi: 10.1074/jbc.273.47.31463. [DOI] [PubMed] [Google Scholar]
- 33.Muro-Pastor A M, Valladares A, Flores E, Herrero A. The hetC gene is a direct target of the NtcA transcriptional regulator in cyanobacterial heterocyst development. J Bacteriol. 1999;181:6664–6669. doi: 10.1128/jb.181.21.6664-6669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reyes J C, Muro-Pastor M I, Florencio F J. Transcription of glutamine synthetase genes (glnA and glnN) from the cyanobacterium Synechocystis sp. strain PCC 6803 is differently regulated in response to nitrogen availability. J Bacteriol. 1997;179:2678–2689. doi: 10.1128/jb.179.8.2678-2689.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rippka R, Deruelles J, Waterbury J B, Herdman M, Stanier R Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T, editors. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 38.Sauer J, Dirmeier U, Forchhammer K. The Synechococcus strain PCC 7942 glnN product (glutamine synthetase III) helps recovery from prolonged nitrogen chlorosis. J Bacteriol. 2000;182:5615–5619. doi: 10.1128/jb.182.19.5615-5619.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider G J, Tumer N E, Richaud C, Borbely G, Haselkorn R. Purification and characterization of RNA polymerase from the cyanobacterium Anabaena 7120. J Biol Chem. 1987;262:14633–14639. [PubMed] [Google Scholar]
- 40.Tanaka K, Masuda S, Takahashi H. Multiple rpoD-related genes of cyanobacteria. Biosci Biotechnol Biochem. 1992;56:1113–1117. doi: 10.1271/bbb.56.1113. [DOI] [PubMed] [Google Scholar]
- 41.Tsinoremas N F, Ishiura M, Kondo T, Andersson C R, Tanaka K, Takahashi H, Johnson C H, Golden S S. A sigma factor that modifies the circadian expression of a subset of genes in cyanobacteria. EMBO J. 1996;15:2488–2495. [PMC free article] [PubMed] [Google Scholar]
- 42.Vega-Palas M A, Flores E, Herrero A. NtcA, a global nitrogen regulator from the cyanobacterium Synechococcus that belongs to the Crp family of transcriptional regulators. Mol Microbiol. 1992;6:1853–1859. doi: 10.1111/j.1365-2958.1992.tb01357.x. [DOI] [PubMed] [Google Scholar]
- 43.Vioque A. The RNase P from cyanobacteria: short tandemly repeated repetitive (STRR) sequences are present within the RNase P RNA gene in heterocyst-forming cyanobacteria. Nucleic Acids Res. 1997;25:3471–3477. doi: 10.1093/nar/25.17.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wösten M M S M. Eubacterial sigma-factors. FEMS Microbiol Rev. 1998;22:127–150. doi: 10.1111/j.1574-6976.1998.tb00364.x. [DOI] [PubMed] [Google Scholar]