Abstract
The genes encoding ammonia monooxygenase (amoCAB), hydroxylamine oxidoreductase (hao), and the c-type cytochrome c-554 (hcy) are present in multiple copies in the genome of Nitrosomonas europaea. The upstream regions of the two copies of amoC, the three copies of hao, and one copy of hcy were cloned and sequenced. Primer extension reactions were done to identify transcription start sites for these genes, as well as for amoA. Putative ς70 promoter sequences were found associated with all but one of the mapped transcription start sites. Primer extensions were done with amoC primers using RNA harvested from cells incubated with and without ammonium. The experiments suggested that N. europaea cells may be able to use different promoters in the presence and absence of ammonium.
Nitrosomonas europaea is a chemolithoautotrophic soil bacterium which oxidizes ammonia to nitrite in the process of nitrification. Ammonia is oxidized to nitrite by the sequential actions of ammonia monooxygenase and hydroxylamine oxidoreductase, providing reductant for the cell (22). In N. europaea, the primary nitrification genes are present in multiple copies. Ammonia monooxygenase is encoded by the genes amoC, amoA, and amoB, which are arranged contiguously and are cotranscribed as a 3.5-kb RNA (12, 19) (Fig. 1). The amoCAB operon is found in two copies in the N. europaea genome (12). These two copies of amoC have only 10 bp differences in 813 bp. In addition, there is a third copy of amoC, which is not associated with amoA or amoB (19), with about 60% sequence similarity to the other copies of amoC. The two copies of amoA only have 1 bp difference in 828 bp, and the two copies of amoB are identical. The gene for hydroxylamine oxidoreductase (hao) is present in three copies in the N. europaea genome (13, 20), and these genes are transcribed monocistronically (20). The three copies of hao only have 1 bp difference in 1,701 bp. The gene encoding cytochrome c-554 (hcy or cyc), an electron acceptor from hydroxylamine oxidoreductase, is also present in three copies which are located 950 bp downstream from each copy of hao (1, 9, 13, 20). The sequences of two copies of the gene for c-554 are known and differ by only 1 bp in 708 bp.
FIG. 1.
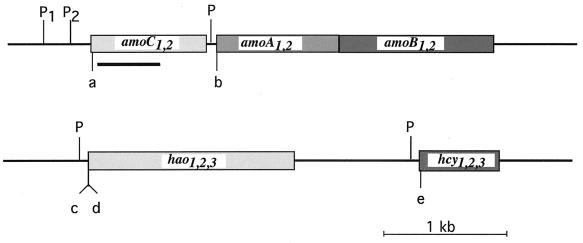
Physical map of the amoCAB operon, hao, and hcy. The locations of the putative promoters for amoC (P1 and P2), amoA, hao, and hcy are indicated. The locations of the oligonucleotide primers used in primer extension experiments are indicated: a, C23 (amoC); b, A5 (amoA); c, ph10 (hao3); d, HB5 (hao1,2); and e, pc12 (hcy). The dark line beneath amoC indicates the location of the amoC probe used in Northern blots.
The physiological significance of multiple copies of the nitrification genes is still unknown. Mutagenesis experiments have shown that no single copy of amo or hao is essential (7, 8). In the case of amoA, when one copy is inactivated, transcription of the other operon increases to compensate for the loss of expression of the first copy during growth. However, while amoA1 can completely compensate for the inactivation of amoA2, amoA2 can only compensate to about 65% of full transcription activity when amoA1 is inactivated (7). This reduced level of amo transcript in the amoA1 mutant may be the cause for the reduced growth rate observed in this mutant strain in laboratory cultures (7). Mutagenesis experiments on amoA suggest that under some conditions transcription of the two copies of amo is not identical. To better understand the basis for differences in transcription of the two copies of amoCAB, experiments were done to map the putative promoters for amoCAB and to examine the expression of amo in the presence and absence of ammonium (NH4+). Transcript mapping was also done for hao and hcy.
N. europaea cells (ATCC 19178) were grown in defined medium (3). For the primer extension experiments, N. europaea cells were grown for 3 days to the late exponential growth phase. Cells were harvested, washed three times, and stored as a pellet at 4°C for at least 24 h to allow the amo mRNA to be degraded (21). The cells were then resuspended in 150 ml of fresh medium (∼1.3 × 1010 cells/ml) in the presence or absence of 25 mM (NH4)2SO4. The cells were shaken at 30°C for 3 h before harvesting for RNA extraction.
Following the numbering of Hirota et al. (6), the copy of hao which we had previously obtained as a genomic lambda clone (8, 20) is hao3. The upstream regions of hao1 and hao2 and of the two copies of amoC were cloned using inverse PCR (18). The PCR-amplified fragments were cloned into the pGEMT-Easy cloning vector (Promega Corp, Madison, Wis.), and the DNA sequences of the promoter regions of these genes were obtained (Center for Gene Research and Biotechnology, Oregon State University). Total RNA was isolated as described (16). DNA probes were labeled by random priming using the Prime-a-Gene kit from Promega Corporation with [α-32P]dCTP (3,000 Ci/mmol; DuPont NEN Products, Wilmington, Del.). The hybridization signals were analyzed using a PhosphorImager and ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.).
Oligonucleotide primers were synthesized by the Center for Gene Research and Biotechnology (Table 1). The amoC primers C23 and C21 were specific to amoC1,2 and did not bind to the related gene amoC3. The primers were 5′-end labeled with T4 polynucleotide kinase using the Prime-a-Gene kit (Promega) with [γ-32P]dATP (6,000 Ci/mmol; DuPont NEN Products). Primer extensions were done using 1 μg of total RNA and 5 × 105 cpm of labeled primer as described (18). Primer extensions were done using reverse transcriptase (Perkin-Elmer, Branchburg, N.J.) following the manufacturer's directions. The resulting labeled cDNA fragments were resolved on a sequencing gel (8 M urea, 8% acrylamide) at 60 W for 2 h. An M13 sequencing ladder labeled with [α-35S]dATP was used as a size standard. The labeled fragments were analyzed by phosphorimagery as above.
TABLE 1.
Primers used in primer extension experiments
| Primer | Position relative to first nucleotide of protein coding region | Sequence (5′-3′) |
|---|---|---|
| C23 (amoC) | 54 to 30 | ATAGCCTCTCGACGAGACTGATGAG |
| C21 (amoC) | 78 to 55 | GGAGTCATACCACAGTGACATGTC |
| A5 (amoA) | 21 to 1 | CCGTTCTAAATATACTCACCC |
| ph10 (hao3) | −9 to −27 | CTCCGTAAATATGCGGGTCA |
| HB5 (hao1,2) | −9 to −34 | CTCCGTAATCTACCTGTTCGTCGGCTT |
| pc12 (hcy) | 83 to 64 | CGGGGCGTCTGCTGCCAGAC |
The following sequences are available in GenBank under the indicated accession numbers: amoC1, AF058691; amoC2, AF058692; hao1, U04053; hao2, U04053; hao3, U04053; and hcy, U05951.
DNA sequencing.
Using inverse PCR, the upstream flanking DNA for amoC1, amoC2, hao1, hao2, and hcy was obtained and sequenced. The nucleotide sequence of the upstream region of hao3 was reported previously (20). The DNA sequences upstream of the two copies of amoC were found to be identical for 334 bp upstream of the gene. The sequences upstream of hao1 and hao2 were found to be similar for 160 bp upstream of the genes, having 3 nucleotide (nt) differences in that span. Further upstream, the sequences had no significant similarity. For hao3, a different upstream sequence was obtained that diverged from the other hao copies 15 bp upstream of the start codon (Fig. 2).
FIG. 2.
Sequence alignment of the upstream regions of amoC1, amoC2, hao1,2,3 (hao3 is from reference (20)), amoA1 (12) (GenBank no. AF058691), and amoA2 (GenBank no. AF058692), hcy, and the E. coli consensus ς70 promoter sequence (5). The sequences are aligned to their mapped transcriptional start sites (underlined). The −10 and −35 regions of the putative promoters are also underlined. Two promoters for amoC (P1 and P2) are indicated. In the E. coli consensus sequence, nucleotides found in more than 65% of the sequences are in uppercase letters, and nucleotides found in less than 65% are in lowercase (5). The numbering is relative to the start codon of the gene.
Primer extensions on amoC, amoA, hao, and hcy.
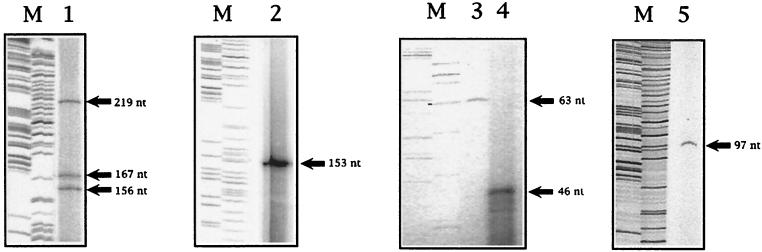
Primer extension experiments from amoC revealed three fragments 219, 167, and 156 nt in size (Fig. 3, lane 1). These fragments indicate three potential transcription start sites located at −166, −114, and −103 bp upstream relative to the start codon of amoC (Fig. 2). The −166 and −103 sites both have putative promoter sequences associated with them (designated P1 and P2, respectively; Fig. 4C) that are similar to consensus ς70-type Escherichia coli promoters (Fig. 2). These results were confirmed with a second amoC primer (C21, Table 1) located downstream from C23 (data not shown). Both promoter sequences were identical for each of the two copies of amoC. The origin of the 167-nt fragment, which mapped to bp −114, is unclear. The −114 site did not have an identifiable match with the E. coli consensus ς70-type sequence (5). It may be a secondary transcription start site for P2, or alternatively, the 167-nt fragment may represent a partially degraded RNA derived from P1.
FIG. 3.
Primer extensions. Lane 1, amoC using primer C23; lane 2, amoA using primer A5; lane 3, hao using primer ph10; lane 4, hao using primer HB5; lane 5, hcy using primer pc12. Lanes M, M13 sequence as size standards.
FIG. 4.
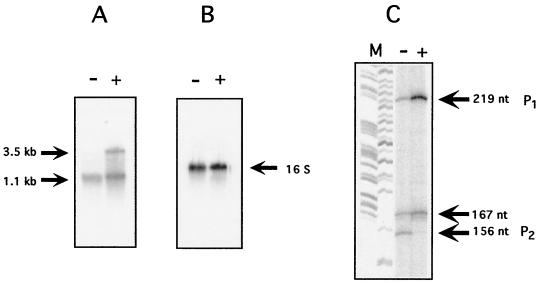
Primer extensions for amoC with and without NH4+. N. europaea cells whose amo mRNA was depleted by incubation in the absence of NH4+ were resuspended in fresh medium either without (−) or with (+) 25 mM (NH4)2SO4. (A) Northern blots using an amoC probe. (B) The same blot as in A, stripped and reprobed with a 16s rRNA probe. (C) Primer extensions were done using an amoC primer (C23). The 219-, 167-, and 156-nt fragments correspond to transcription initiation at −166, −114, and −103 bp upstream of the start codon of amoC, respectively. Lane M, M13 sequencing ladder was used as size standards.
Northern blots had indicated a transcript consisting of amoAB (19). Therefore we searched for a promoter upstream of amoA. Primer extensions done from amoA produced a 153-nt fragment which mapped to bp −155 relative to the amoA start codon located in the intergenic region between amoC and amoA. (Fig. 3, lane 2). This site was associated with a possible ς70-type promoter sequence. Both copies of amoA are known to be expressed (7), and the intergenic region between amoC and amoA where the transcription start site and putative promoter are located have identical DNA sequences. Experiments have shown that the intergenic region between amoC and amoA from another nitrifying bacterium, Nitrosospira NpAV, which also has the amoCAB operon, could drive expression of amoA in E. coli, lending credence to the idea that this is a viable promoter (15).
Transcript mapping for hao was done using primers ph10 and HB5, which are located upstream of where the DNA sequences upstream of hao1,2 and hao3 diverge. In this way, the primers were targeted to different hao copies. The primer extensions produced a 46-nt and a 63-nt fragment with primers ph10 and HB5, respectively (Fig. 3, lanes 4 and 3). For hao3 (ph10), the transcription start site mapped to 54 bp upstream of the start codon. For hao1 and hao2 (HB5), having identical upstream sequences, the transcription start sites mapped to 71 bp upstream of the start codon. We could not distinguish fragments formed by primer extension from these two copies of hao. All three copies of hao had ς70-type promoter sequences associated with their putative transcription start sites.
A potential transcription start site was also found for hcy, a gene encoding the cytochrome c554 and located downstream of hao. The start site for hcy was at −97 bp relative to the start codon for hcy and was also associated with a ς70-type promoter sequence (Fig. 3, lane 5). However, in this experiment it was not possible to distinguish which copy of hcy was being used as the template for the primer extension. The putative promoter sequences from different N. europaea genes were compared and have no apparent conserved sequences (Fig. 2).
Effect of NH4+ on primer extensions from amoC.
The production of multiple fragments in primer extension reactions could result from tandem promoters for amoC. This possibility was investigated by comparing the relative amounts of the 219-, 167-, and 156-nt fragments produced in primer extension reactions when transcription of amo was induced by the addition of NH4+ (Fig. 4). N. europaea cells which had their endogenous RNA levels depleted were transferred to fresh medium either lacking NH4+ or containing 25 mM (NH4)2SO4. The amount of amo mRNA present after 3 h was estimated by primer extensions and by Northern blots. Primer extensions showed three fragments in samples grown with and without NH4+ which responded differently to the addition of NH4+ (Fig. 4C). Ratios were calculated for the amount of cDNA produced in primer extension reactions in the plus-NH4+ sample relative to the minus-NH4+ sample. In this experiment (Fig. 4C), the ratio for the 219-nt fragment was 4.36, the ratio for the 167-nt fragment was 2.0, and the ratio for the 156-nt fragment was 0.6. When the average ratios were calculated from four independent experiments, the ratio for the 219-nt fragment was 3.3 ± 1.6, the ratio for the 167-nt fragment was 1.3 ± 0.5, and the ratio for the 156-nt fragment was 0.42 ± 0.28.
Northern blots.
In the absence of NH4+, Northern blots probed with an amoC probe showed a single fragment of about 1.1 kb (Fig. 4A). Previous work has shown that this fragment is derived from amoC1,2 but not amoC3 (19). Northern blots from samples incubated in medium containing NH4+ highlighted a 3.5-kb fragment (transcription of the full amoCAB operon) as well as the 1.1-kb fragment seen in the minus-NH4+ sample. The same blot stripped and reprobed with a 16S rRNA probe indicated that the relative loading concentrations of the two samples were the same (Fig. 4B). These results are consistent with the results of the primer extensions. In the sample without NH4+, the 1.1-kb RNA fragment was likely the source of the three fragments produced in the primer extensions from the sample without NH4+. Although the Northern blot only showed one fragment in the sample without NH4+, the size resolution of fragments visualized on Northern blots was not sufficient to detect a 70-nt difference. Therefore, the fragments seen in the Northern blots were apparently a mixed population containing the different 5′ ends shown by the primer extensions. When RNA from the sample with NH4+ was used, the same three fragments were observed in the primer extensions as in the sample without NH4+, albeit in different relative proportions. Since both the 3.5-kb fragment in the Northern and the 219-nt fragment in the primer extension are induced in the presence of NH4+, it may be that the 219-nt fragment is primarily transcribed from P1 on the 3.5-kb RNA, while the 167- and 156-nt fragments derive mainly from the 1.1-kb RNA. However, the results presented here do not allow us to ascribe primer extension products to specific RNAs on Northern blots.
The NH4+ induction experiment results are consistent with the presence of tandem promoters that are not functionally equivalent, since they respond differently to the addition of NH4+. While a number of examples of tandem promoters are known (10, 11, 14, 17), a situation such as with N. europaea, where tandem ς70-type promoters are regulated differently, appears to be unusual. However, Grafe et al. (4) reported that the streptokinase gene (skc) in Streptococcus equisimilis had tandem overlapping functional promoters. A 200-bp region located 100 bp upstream of the transcription start site enhanced transcription from one promoter but not the other.
An alternative explanation for the pattern of fragments seen in the amoC primer extension experiments is that the 5′- ends represented by the 167- and 156-nt fragments in primer extensions are degradation products of the RNA represented by the 219-nt fragment. In this scenario, the relative abundance of the three fragments observed in primer extensions in the plus- and minus-NH4+ samples reflects changes in the rates of transcription and degradation. During induction, the increased rates of transcription of amo from P1 allow the 219-nt fragment to accumulate. When NH4+ is removed, synthesis rates decrease, allowing degradation to the smaller fragment sizes to occur. Examination of the sequence upstream of amoC reveals an 11-bp inverted repeat starting 67 bp upstream from the start codon. Secondary structures in RNAs are known to affect their resistance to degradation (2). A structure such as this may be relevant to the continued presence of the 156- and 167-nt fragments in the absence of NH4+.
In this work we identified transcript start sites and putative promoters for copies of amo, hao, and hcy. Previous studies on the mutagenesis of amoA in N. europaea (7) have shown that the two operons of amo are expressed at different levels. We expected that these differences in transcription would be reflected in differences in promoter sequences between the copies. Surprisingly, the DNA sequences of the putative amoC1 and amoC2 promoters were found to be identical and therefore cannot account for the observed transcriptional differences in amo in the mutant strains. Perhaps the differential expression of the two amo copies involves sequences upstream of the promoters in the region where the DNA sequences in the two copies diverge. We are continuing to explore the regulation of amo and hao under different growth conditions, particularly regarding the contribution of each of the copies to the overall expression of the genes.
Acknowledgments
This work was supported by DOE grant DE-FG03-97ER20266 to D. J. Arp and L. A. Sayavedra-Soto.
REFERENCES
- 1.Bergmann D J, Arciero D M, Hooper A B. Organization of the hao gene cluster of Nitrosomonas europaea: genes for two tetraheme c cytochromes. J Bacteriol. 1994;176:3148–3153. doi: 10.1128/jb.176.11.3148-3153.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coburn G, Mackie G. Degradation of mRNA in Escherichia coli: an old problem with some new twists. In: Moldave K, editor. Progress in nucleic acid research and molecular biology. Vol. 62. San Diego, Calif: Academic Press; 1999. pp. 55–108. [DOI] [PubMed] [Google Scholar]
- 3.Ensign S A, Hyman M R, Arp D J. In vitro activation of ammonia monooxygenase from Nitrosomonas europaea by copper. J Bacteriol. 1993;175:1971–1980. doi: 10.1128/jb.175.7.1971-1980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grafe S, Ellinger T, Malke H. Structural dissection and functional analysis of the complex promoter of the streptokinase gene from Streptococcus equisimilis H46A. Med Microbiol Immunol. 1996;185:11–17. doi: 10.1007/s004300050009. [DOI] [PubMed] [Google Scholar]
- 5.Harley C P, Roberts R P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirota R, Yamagata A, Kato J, A. K, Ikeda T, Takiguchi N, Ohtake H. Physical map location of the multicopy genes coding for ammonia monooxygenase and hydroxylamine oxidoreductase in the ammonia-oxidizing bacterium Nitrosomonas sp. strain ENI-11. J Bacteriol. 2000;182:825–828. doi: 10.1128/jb.182.3.825-828.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hommes N G, Sayavedra-Soto L A, Arp D J. Mutagenesis and expression of amo, which codes for ammonia monooxygenase in Nitrosomonas europaea. J Bacteriol. 1998;180:3353–3359. doi: 10.1128/jb.180.13.3353-3359.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hommes N G, Sayavedra-Soto L A, Arp D J. Mutagenesis of hydroxylamine oxidoreductase in Nitrosomonas europaea by transformation and recombination. J Bacteriol. 1996;178:3710–3714. doi: 10.1128/jb.178.13.3710-3714.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hommes N G, Sayavedra-Soto L A, Arp D J. Sequence of hcy, a gene encoding cytochrome c-554 in Nitrosomonas europaea. Gene. 1994;146:87–89. doi: 10.1016/0378-1119(94)90838-9. [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Matsumura P. Differential regulation of multiple overlapping promoters in flagellar class II operons in Escherichia coli. Mol Microbiol. 1996;21:613–620. doi: 10.1111/j.1365-2958.1996.tb02569.x. [DOI] [PubMed] [Google Scholar]
- 11.Marques S, Gallegos M T, Manzanera M, Holtel A, Timmis K N, Ramos J L. Activation and repression of transcription at the double tandem divergent promoters for the xylR and xylS genes of the TOL plasmid of Pseudomonas putida. J Bacteriol. 1998;180:2889–2894. doi: 10.1128/jb.180.11.2889-2894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McTavish H, Fuchs J A, Hooper A B. Sequence of the gene coding for ammonia monooxygenase in Nitrosomonas europaea. J Bacteriol. 1993;175:2436–2444. doi: 10.1128/jb.175.8.2436-2444.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McTavish H, LaQuier F, Arciero D, Logan M, Mundfrom G, Fuchs J A, Hooper A B. Multiple copies of genes coding for electron transport proteins in the bacterium Nitrosomonas europaea. J Bacteriol. 1993;175:2445–2447. doi: 10.1128/jb.175.8.2445-2447.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nielsen A K, Gerdes K, Murrell J C. Copper-dependent reciprocal transcriptional regulation of methane monooxygenase genes in Methylococcus capsulatus and Methylosinus trichosporium. Mol Microbiol. 1997;25:399–409. doi: 10.1046/j.1365-2958.1997.4801846.x. [DOI] [PubMed] [Google Scholar]
- 15.Norton J M, Low J M, Klotz M G. The gene encoding ammonia monooxygenase subunit A exists in three nearly identical copies in Nitrosospira sp. NpAV. FEMS Microbiol Lett. 1996;139:181–188. doi: 10.1111/j.1574-6968.1996.tb08200.x. [DOI] [PubMed] [Google Scholar]
- 16.Reddy K J, Gilman M. Preparation of bacterial RNA. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley and Sons; 1993. pp. 4.4.1–4.4.7. [Google Scholar]
- 17.Reitzer L J, Magasanik B. Expression of glnA in Escherichia coli is regulated at tandem promoters. Proc Natl Acad Sci USA. 1985;82:1979–1983. doi: 10.1073/pnas.82.7.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Sayavedra-Soto L A, Hommes N G, Alzerreca J J, Arp D J, Norton J M, Klotz M G. Transcription of the amoC, amoA, and amoB genes in Nitrosomonas europaea and Nitrosospira sp. NpAV. FEMS Microbiol Lett. 1998;167:81–88. doi: 10.1111/j.1574-6968.1998.tb13211.x. [DOI] [PubMed] [Google Scholar]
- 20.Sayavedra-Soto L A, Hommes N G, Arp D J. Characterization of the gene encoding hydroxylamine oxidoreductase in Nitrosomonas europaea. J Bacteriol. 1994;176:504–510. doi: 10.1128/jb.176.2.504-510.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sayavedra-Soto L A, Hommes N G, Russell S A, Arp D J. Induction of ammonia monooxygenase and hydroxylamine oxidoreductase mRNAs by ammonium in Nitrosomonas europaea. Mol Microbiol. 1996;20:541–548. doi: 10.1046/j.1365-2958.1996.5391062.x. [DOI] [PubMed] [Google Scholar]
- 22.Wood P M. Nitrification as a bacterial energy source. In: Prosser J I, editor. Nitrification. Oxford, U.K: Society for General Microbiology, IRL Press; 1986. pp. 39–62. [Google Scholar]