Abstract
Lactobacillus strains are considered safe and healthy probiotics for manufacturing “natural food” products; this is due to their ability to produce bioactive compounds that reduce the incidence of various human diseases. Lactobacillus paracasei TK1501 is a novel probiotic strain isolated from naturally fermented congee; and can produce a high yield of genistein, one of the most widely studied isoflavone aglycones with plenty of physiological functions. To better understand the molecular basis of isoflavone aglycones biosynthesis, the complete 2,942,538 bp genome of L. paracasei TK1501 was sequenced and assembled; a group of genes that are involved in isoflavone aglycones production were identified. Of note, a β-glucosidase was analyzed in the L. paracasei TK1501. Moreover, we also found that L. paracasei TK1501 could be used in soymilk fermentation; which would remarkably increase the contents of genistein, daidzein, and glycitein. This work was meaningful to the application of L. paracasei TK1501 and the molecular mechanism analysis of isoflavone aglycones biosynthesis in Lactobacillus strains.
Keywords: complete genome sequence, Lactobacillus paracasei TK1501, isoflavone aglycones, β-glucosidase
1. Introduction
Soy isoflavone aglycones are a class of phytoestrogens with highly bioactive properties on lots of cellular processes and human diseases [1,2], including osteoporosis, cardiovascular disease [3], type 2 diabetes [4,5], and cancers [6]. Compared to the related glycosides, the isoflavone aglycones are with higher bioactivity due to their lower molecular weight and unimpeded gut absorption [7]. Therefore, there is a growing interest in the production of “natural food” products containing high levels of isoflavone aglycones. Glycosylated isoflavones can be converted to isoflavone aglycones according to a hydrolysis reaction using β-glycosidases. In addition, β-glycosidases commonly exist in plants and microorganisms, especially in lactic acid bacteria. Lactic acid bacteria are normally found in the gastrointestinal tract, and are frequently used as probiotics in fermented dairy products such as soybean milk [8]. Lactobacillus paracasei (L. paracasei) is a member of the normal gut microbiota and has the ability to produce genistein. Therefore, it is one of the most promising candidates for the manufacture of safe and healthy natural food such as fermented soymilk [9,10,11]. L. paracasei strains have already been used for in situ fortification of certain foods in the dairy industry. Thus, it would be beneficial to identify novel L. paracasei strains with higher yields of isoflavone aglycones. Herein, we have isolated a novel L. paracasei TK1502 from naturally fermented congee. Moreover, L. paracasei TK1502 could produce a high yield of genistein, an important isoflavone aglycone with many functions. In order to gain a better insight into the molecular mechanism of the isoflavone aglycones biosynthesis, the complete genome of L. paracasei TK1502 was sequenced; and a comparative β-glucosidase gene analysis was performed between L. paracasei TK1502 and several other bacterial strains. We also probed the beneficial effects of L. paracasei TK1502 in the application of fermented soymilk.
2. Materials and Methods
2.1. Strains and Data Storage
L. paracasei TK1501 was stored in the China General Microbiological Culture Collection (CGMCC) with an accession number of CGMCC13130. The complete genome sequences of L. paracasei TK1501 were deposited in the NCBI database under the accession number of CP017716.
2.2. DNA Extraction
The lactobacillus strains in this study were isolated from traditional fermented congee obtained from Inner Mongolia, China. These bacteria were cultured at 37 °C in brain-heart infusion broth under anaerobic conditions (80% N2, 10% H2, and 10% CO2) for 24 h. Lactobacillus selected for identification by a sequence analysis of 16S rDNA were collected by centrifugation (12,000× g, 5 min) from cultures that were incubated for 24 h at 37 °C; and their DNA was extracted using a previously described method, with some modifications [12].
2.3. PCR Amplification and Sequencing
The polymerase chain reaction was used to amplify the 16S rDNA gene using the primers (16S-S Forward:5′-AGAGTTTGATCCTGGCTCAG-3′; 16S-R Reverse:5′-TACGGCTACCTTGTTACGACTT-3′). The polymerase chain reaction conditions were as follows: 95 °C for 5 min, followed by 30 cycles of 1 min at 95 °C, 1 min at 55 °C, and 1 min at 72 °C, followed by 5 min at 72 °C. The complete genome was sequenced using a whole genome shotgun strategy combining next-generation sequencing and third-generation sequencing based on Illumina MiSeq paired-end and PacBio standard sequencing technology.
2.4. Phylogenetic Analysis and Gene Annotation
The reads were de novo assembled using Newbler and HGAP 2.3.0; and integration of the assembled contigs was carried out using Mummer and Pilon. Automated gene prediction and annotation of the whole genome sequences were carried out using Glimmer v.3.0 and the NCBI Prokaryotic Genome Annotation Pipeline, respectively. Ribosomal RNA genes were identified using RNAmmer v.1.2, and tRNA genes were predicted using a tRNA scan-SE v.1.3.1 [13]. Each gene was functionally assigned to a category using the following databases: Evolutionary Genealogy of Genes: Non-supervised Orthologous Groups (eggNOG) [14]; Kyoto Encyclopedia of Genes and Genomes (KEGG) [15]; Swiss-Prot, Gene Ontology (GO) [16]; and the NCBI Non-Redundant Protein Database (NR) [17]. Cycle maps of the genomes were made with cgview and compiled by Photoshop CS [18]. The phylogenetic tree was constructed using MEGA 6.0. HMMER 3.0 software [19], which was used to predict the carbohydrate-active enzyme (CAZy) genes from the genome sequence. The critical E-value was set as 1 × 106 when the sequence was ≥ 80 amino acids; and was set as 1 × 103 when the sequence was <80 amino acids.
2.5. β-Glucosidase Activity Analysis of L. paracasei TK1502
We analyzed the β-glucosidase catalytic capacity of L. paracasei TK1502 after fermentation for 24 h, following our previously reported method [20]. One unit (U) of β-glucosidase activity was defined as the amount of enzyme that generated 1 μM p-nitrophenol (pNP) from the substrate of p-nitrophenyl-β-D-glucopyranoside (pNPG) per min. After being fermented at 37 °C for 24 h, the supernatant of L. paracasei TK1502 was removed at 15,000× g for 10 min. The cell pellet was suspended in 10 mM PBS buffer (pH 7.4) and sonicated for 20 min (on 1 s, off 1 s); the supernatant was obtained after being centrifuged at 4 °C, 15,000× g for 10 min. Then, 5 mM pNPG was mixed with a 5 mL L. paracasei TK1502 cell lysis solution and incubated at 37 °C for 30 min. The reaction was stopped by adding 250 μM 0.2 M Na2CO3. Then, the absorbance of samples at 410 nm was tested using a UV–vis spectrophotometer.
2.6. Application of L. paracasei TK1502 in the Fermentation of Soymilk
L. paracasei TK1502 or other bacterial strains were cultured at 37 °C in 10 mL of sterile soymilk for 24 h to prepare the seeds. After calculating the concentration of the bacteria, 1 × 107 CFU/mL of strains seed was transferred into an 80 mL fresh sterile soymilk medium and cultured at 37 °C for 24 h. The sterile soymilk medium without any bacterial strains was defined as the blank control; it was also treated in the same incubation conditions as the experimental group. The fermented solution was lyophilized, and stored at −80 °C until use. In total, 0.5 g of the lyophilized samples were dissolved in a small amount of DMSO, and re-solved in 5 mL of 80% methanol. The sample solutions were incubated by shaking at 60 °C for 2 h, and then centrifuged at 12,000 rpm, 4 °C for 20 min. The supernatant of each sample was filtered using a 0.22 μm membrane for measurement.
High-performance liquid chromatography (HPLC) was used to test the changes of the soy isoflavones and isoflavone glycosides. Herein, we mainly measured three soy isoflavones, including genistin, daidzin, and glycitin; and their hydrolysis products of genistein, daidzein, and glycitein, respectively. All of the standard substances were purchased from Sigma-Aldrich (St. Louis, MO, USA). A NovaPak C18 column (250 × 4.6 mm, 5 μm, Waters, Milford, MA, USA) was used for the separation, and a photodiode array detector (PDA-100, Dionex) was used for the detection process. Mobile phase A was water with 0.1% acetic acid, and phase B was methanol with 0.1% acetic acid. The flow rate was 1 mL/min and the other parameters were as follows: 0 min, 20% phase B; 8 min, 60% phase B; 18 min, 95% phase B; 19 min, 20% phase B; 24 min, 20% phase B; the column temperature was 27 °C; and the wavelength was 260 nm.
3. Results and Discussion
3.1. Genome Organization and Base Composition
In this present study, L. paracasei TK1501 was isolated from naturally fermented congee. This strain is a facultative anaerobic, rod-shaped and Gram-positive bacteria; the colonies of which are milky white, round, opaque, and the surface of which is smooth and with a convex elevation. D-glucose, fructose, lactose, maltose, D-mannose, and cellose can be used as carbon source; in addition, peptone, beef extract, yeast extract, ammonium nitrate, ammonium sulfate, and ammonia chloride can be used as nitrogen source for the L. paracasei TK1501. Furthermore, L. paracasei TK1501 is catalase-negative and protease-positive; also, it cannot digest gelatin or produce sulfuretted hydrogen. L. paracasei TK1501 showed a ≥99% nucleotide sequence similarity to other Lactobacillus strains deposited in the GenBank database based on 16S rRNA gene analysis. Phylogenetic analysis showed that TK1501 formed a distinct branch with L. paracasei strains (Figure 1), and was most closely related to L. paracasei N1115. TK1501; it was therefore preliminarily designated as a L. paracasei strain. Furthermore, the capacity of L. paracasei TK1501 on the production of genistein by fermenting soymilk was compared with several other lactobacillus strains. After fermentation using L. paracasei TK1501, the contents of genistein in the soymilk was increased 25.34-fold compared with the blank control group that was fermented without any bacterial strains. Meanwhile, L. paracasei TK1501 had a much higher increase in the amount of times with respect to the genistein in the fermented soymilk than in any of the other bacterial strain groups (Table 1).
Figure 1.
Phylogenetic tree of L. paracasei TK1501. The phylogenetic tree was constructed using MEGA 6.0.
Table 1.
Comparison of genistein production in the soymilk after fermentation using the different lactobacillus strains.
| Strains | Increased Folds |
|---|---|
| Lactobacillus paracasei TK1501 | 25.34 |
| Lactobacillus rhamnosus CRL981 | 4.66 |
| Lactobacillus rhamnosus D7 | 17.06 |
| Lactobacillus paracasei BCRC 14023 | 1.10 |
| Lactobacillus delbrueckii 01181 | 8.36 |
| Lactobacillus plantarum 00144 | 7.15 |
| Streptococcus thermophilus CCRC14085 | 2.59 |
| Bifidobacterium breve K-101 | 3.01 |
| Lactobacillus acidophilus BCRC 10695 | 1.06 |
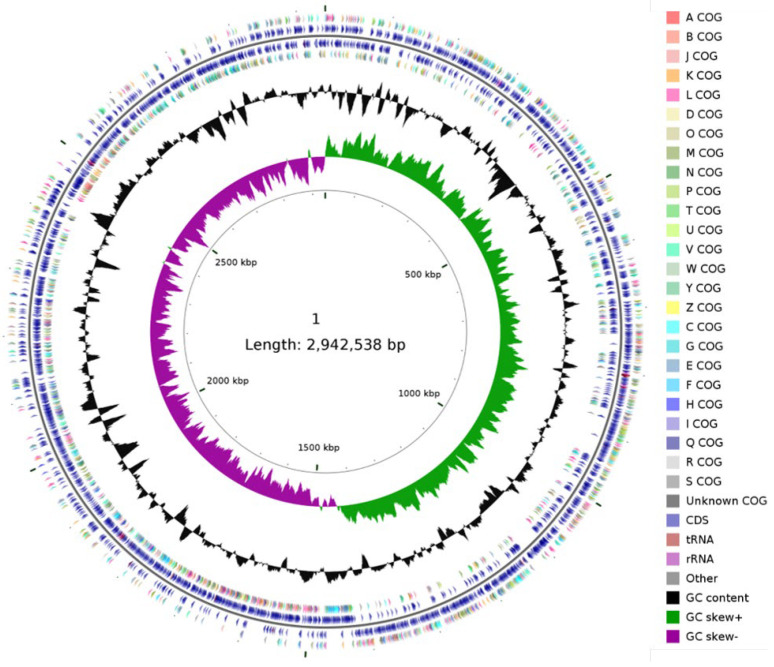
Complete genome sequencing and genomic analysis of bacterial strains can provide functional information. To gain insights into the genetic elements involved in the production of genistein, we sequenced the entire genome of L. paracasei TK1501. A total of 693,261 reads were generated, with an average length of 6768 nucleotides. As shown in Figure 2 and Table 2, the complete genome of TK1501 is composed of a single circular chromosome, 2,942,538 bp in length. There were 2947 protein-coding genes, 59 tRNA genes, and 15 rRNA genes, along with 40 other non-coding RNA regions. The predicted G + C content was 46.54%, and no plasmids were identified.
Figure 2.
Circular genome map of L. paracasei TK1501.
Table 2.
Features of the L. paracasei TK1501 genome.
| Attributes | Values |
|---|---|
| Genome size (bp) | 2,942,538 |
| GC content (%) | 46.54% |
| Plasmid | 0 |
| rRNAs | 15 |
| tRNAs | 59 |
| Other ncRNAs | 40 |
| Proteins | 2947 |
3.2. Analysis of Carbohydrate-Active Enzymes
The carbohydrate-active enzymes (CAZy) database mainly contains glycoside hydrolases (GHs), glycosyl transferases (GTs), polysaccharide lyases (PLs), carbohydrate esterases (CEs), and auxiliary activities (AAs) [21]. Moreover, carbohydrate-binding modules (CBMs) are also included. Genes of CAZys in the genome sequences were predicted with HMMER (version 3.0). The analysis results were shown in Table 3; the numbers of CAZy genes are 77, which is 2.61 percent of the whole genome. In addition, GHs and GTs were the main enzymes of the predicted CAZy genes.
Table 3.
Analysis of CAZy in L. paracasei TK1501.
| Property | Numbers of Genes | Percentage (%) |
|---|---|---|
| GHs | 36 | 1.22 |
| GTs | 26 | 0.88 |
| PLs | 0 | 0.00 |
| CEs | 9 | 0.31 |
| AAs | 4 | 0.14 |
| CBMs | 2 | 0.07 |
| Total | 77 | 2.61 |
3.3. Comparative Analysis of β-Glucosidase from L. paracasei TK1501
β-glucosidases are the key enzyme during genistein biosynthesis, and belong to GHs; in addition, they have been reported to have the function to hydrolyse isoflavones [22]. Firstly, we analyzed the β-glucosidase catalytic capacity of L. paracasei TK1502 using pNPG as the substrate. The results showed that the β-glucosidase activity of L. paracasei TK1502 was 17.45 U/mL; this was remarkably higher than many probiotic stains or other microorganisms, such as L. perolens FI10842 (49.10 mU/mL) [23], L. bulgaricus CFR2028 (~160 mU/mL) [24], A. niger NRRL 3112 (9.30 U/mL) [25], T. atroviride TUB F-172 (11.70 U/mL) [26], and so on. Therefore, L. paracasei TK1502 was a good source of β-glucosidases that possessed an excellent β-glucosidase catalytic capacity.
In order to analyze the genomic properties of β-glucosidase in L. paracasei TK1501, the β-glucosidase amino acid sequence from L. paracasei TK1501 was compared with the corresponding sequences from several other available bacterial strains of Lactobacillus casei ATCC334, Lactobacillus rhamnosus GG, and Bifidobacterium B.b. The TK1501 sequence showed a 99.8% similarity to that of L. casei ATCC334; however, it had only a 76.4% and 36.6% similarity to the sequences from L. rhamnosus GG and Bifidobacterium B.b, respectively (Figure 3). Together, our findings suggest that the production of genistein is potentially related to several important genetic differences between TK1501 and closely related strains.
Figure 3.
Comparison of β-glucosidase amino acid sequences from L. paracasei TK1501 (TK) with L. casei ATCC334 (334), L. rhamnosus GG (GG), and Bifidobacterium B.b (B.b). The navy-blue means consistent residues in all four enzymes, purple means consistent residues in three enzymes, and light blue means consistent residues in two enzymes.
3.4. The Isoflavone Aglycones in Soymilk Were Increased after Fermentation Using L. paracasei TK1502
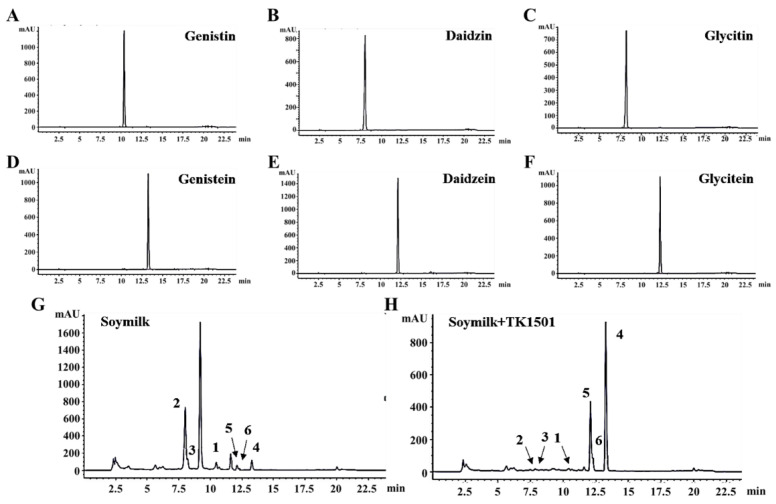
Since β-glucosidases have a broad catalytic capacity for the hydrolysis of many isoflavones to produce isoflavone aglycones, we further measured the changes of three important isoflavones, including genistin, daidzin, and glycitin; and their corresponding aglycones of genistein, daidzein, and glycitein after fermentation using L. paracasei TK1501 by HPLC. The HPLC data of the standard molecules of genistin, daidzin, and glycitin; and their aglycones of genistein, daidzein, and glycitein are shown in Figure 4A–F; herein, all the retention times of the isoflavones were less than their aglycones. The degradation rates of genistin, daidzin, and glycitin in the soymilk after fermentation using L. paracasei TK1501 were (94.87 ± 0.06), (83.57 ± 0.14), and (97.05 ± 0.08)%, respectively (Table 4). Compared to the soymilk without L. paracasei TK1501, the contents of genistein, daidzein, and glycitein were remarkably increased from (8.04 ± 0.06), (2.24 ± 0.09), and (2.43 ± 0.08) μg/g to (211.65 ± 1.27), (25.39 ± 0.13), and (109.10 ± 1.19) μg/g, respectively. The results proved that the L. paracasei TK1501 could be applied in the fermentation of soymilk, and could increase the contents of several important isoflavone aglycones; this could be easily absorbed and beneficial to ameliorate many human conditions, including diabetes, obesity, cancers, hypertension, and so forth [2,7,27].
Figure 4.
Analysis of isoflavones and related isoflavone aglycones changes in the fermentation of soymilk by L. paracasei TK1501 using HPLC. Standard molecules of isoflavones, including (A) genistin, (B) daidzin, and (C) glycitin; and their corresponding aglycones of (D) genistein, (E) daidzein, and (F) glycitein. Fermented soymilk (G) without and (H) with L. paracasei TK1501; in which compounds 1–6 were genistin, daidzin, and glycitin; and genistein, daidzein, and glycitein, respectively.
Table 4.
The degradation rate of isoflavones in the L. paracasei TK1501 fermented soymilk.
| Isoflavones | Degradation Rate (%) |
|---|---|
| Genistin | 94.87 ± 0.06 |
| Daidzin | 83.57 ± 0.14 |
| Glycitin | 97.05 ± 0.08 |
4. Conclusions
In this study, we reported the complete sequence of a novel probiotic strain isolated from the naturally fermented congee, L. paracasei TK1501. The complete genome of L. paracasei TK1501 was 2,942,538 bp. A group of isoflavone aglycone production-related genes, including the β-glucosidase gene, were analyzed and identified in the L. paracasei TK1501. Moreover, the contents of isoflavone aglycones, including genistein, daidzein, and glycitein in the soymilk, were remarkably increased after fermentation using L. paracasei TK1501. The complete genome sequence of L. paracasei TK1501 will enrich its application, and help us to better understand the molecular basis of isoflavone aglycones biosynthesis in the lactobacillus.
Author Contributions
Conceptualization, Y.X., F.L. and L.J.; methodology, Y.X., Y.W. and Y.H.; validation, Y.X.; investigation, Y.X., Y.W., Y.H. and J.Z.; supervision, S.L., H.W. and F.L.; funding acquisition, Y.X., S.W. and J.Z.; writing—original draft preparation, Y.X. and L.J.; writing—review and editing, L.J. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Funding Statement
This research was funded by the Higher Education Teaching Reform Research Project in Heilongjiang Province (grant number SJGY20210525); the National Natural Science Foundation of China (grant number 31801516); the Innovation and Entrepreneurship Project for College Students in Heilongjiang Province (grant number 202210234020); and the Zibo City Integration Development Project (grant numbers 2020SNPT0003 and 2021SNPT0069).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ren B., Liu Y., Zhang Y., Cai Y., Gong X., Chang Y., Xu L., Zheng J. Genistein: A Dual Inhibitor of Both Amyloid beta and Human Islet Amylin Peptides. ACS Chem. Neurosci. 2018;9:1215–1224. doi: 10.1021/acschemneuro.8b00039. [DOI] [PubMed] [Google Scholar]
- 2.Hsiao Y.H., Ho C.T., Pan M.H. Bioavailability and health benefits of major isoflavone aglycones and their metabolites. J. Funct. Foods. 2020;74:104164. doi: 10.1016/j.jff.2020.104164. [DOI] [Google Scholar]
- 3.Omoni A.O., Aluko R.E. Soybean foods and their benefits: Potential mechanisms of action. Nutr. Rev. 2005;63:272–283. doi: 10.1111/j.1753-4887.2005.tb00141.x. [DOI] [PubMed] [Google Scholar]
- 4.Babu P.V., Liu D., Gilbert E.R. Recent advances in understanding the anti-diabetic actions of dietary flavonoids. J. Nutr. Biochem. 2013;24:1777–1789. doi: 10.1016/j.jnutbio.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller N.T., Odegaard A.O., Gross M.D., Koh W.P., Yu M.C., Yuan J.M., Pereira M.A. Soy intake and risk of type 2 diabetes in Chinese Singaporeans [corrected] Eur. J. Nutr. 2012;51:1033–1040. doi: 10.1007/s00394-011-0276-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adlercreutz H. Phyto-oestrogens and cancer. Lancet Oncol. 2002;3:364–373. doi: 10.1016/S1470-2045(02)00777-5. [DOI] [PubMed] [Google Scholar]
- 7.Harada T., Masuda S., Arii M., Adachi Y., Nakajima M., Yadomae T., Ohno N. Soy isoflavone aglycone modulates a hematopoietic response in combination with soluble beta-glucan: SCG. Biol. Pharm. Bull. 2005;28:2342–2345. doi: 10.1248/bpb.28.2342. [DOI] [PubMed] [Google Scholar]
- 8.Marazza J.A., Garro M.S., de Giori G.S. Aglycone production by Lactobacillus rhamnosus CRL981 during soymilk fermentation. Food Microbiol. 2009;26:333–339. doi: 10.1016/j.fm.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Chun J., Kim G.M., Lee K.W., Choi I.D., Kwon G.H., Park J.Y., Jeong S.J., Kim J.S., Kim J.H. Conversion of isoflavone glucosides to aglycones in soymilk by fermentation with lactic acid bacteria. J. Food Sci. 2007;72:M39–M44. doi: 10.1111/j.1750-3841.2007.00276.x. [DOI] [PubMed] [Google Scholar]
- 10.Mangiapane E., Mazzoli R., Pessione A., Svensson B., Riedel K., Pessione E. Ten years of subproteome investigations in lactic acid bacteria: A key for food starter and probiotic typing. J. Proteom. 2015;127 Pt. B:332–339. doi: 10.1016/j.jprot.2015.04.028. [DOI] [PubMed] [Google Scholar]
- 11.Otieno D.O., Shah N.P. A comparison of changes in the transformation of isoflavones in soymilk using varying concentrations of exogenous and probiotic-derived endogenous beta-glucosidases. J. Appl. Microbiol. 2007;103:601–612. doi: 10.1111/j.1365-2672.2006.03245.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhang N., Liu J.H., Li J.J., Chen C., Zhang H.T., Wang H.K., Lu F.P. Characteristics and Application in Food Preservatives of Lactobacillus plantarum TK9 Isolated from Naturally Fermented Congee. Int. J. Food Eng. 2016;12:377–384. doi: 10.1515/ijfe-2015-0180. [DOI] [Google Scholar]
- 13.Li P., Gu Q., Zhou Q. Complete genome sequence of Lactobacillus plantarum LZ206, a potential probiotic strain with antimicrobial activity against food-borne pathogenic microorganisms. J. Biotechnol. 2016;238:52–55. doi: 10.1016/j.jbiotec.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 14.Powell S., Forslund K., Szklarczyk D., Trachana K., Roth A., Huerta-Cepas J., Gabaldon T., Rattei T., Creevey C., Kuhn M., et al. eggNOG v4.0: Nested orthology inference across 3686 organisms. Nucleic Acids Res. 2014;42:D231–D239. doi: 10.1093/nar/gkt1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moriya Y., Itoh M., Okuda S., Yoshizawa A.C., Kanehisa M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conesa A., Gotz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genom. 2008;2008:619832. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tatusov R.L., Galperin M.Y., Natale D.A., Koonin E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stothard P., Wishart D.S. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- 19.Finn R.D., Clements J., Eddy S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y., Hu X., Sang J., Zhang Y., Zhang H., Lu F., Liu F. An acid-stable beta-glucosidase from Aspergillus aculeatus: Gene expression, biochemical characterization and molecular dynamics simulation. Int. J. Biol. Macromol. 2018;119:462–469. doi: 10.1016/j.ijbiomac.2018.07.165. [DOI] [PubMed] [Google Scholar]
- 21.Lombard V., Ramulu H.G., Drula E., Coutinho P.M., Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki H., Takahashi S., Watanabe R., Fukushima Y., Fujita N., Noguchi A., Yokoyama R., Nishitani K., Nishino T., Nakayama T. An isoflavone conjugate-hydrolyzing beta-glucosidase from the roots of soybean (Glycine max) seedlings: Purification, gene cloning, phylogenetics, and cellular localization. J. Biol. Chem. 2006;281:30251–30259. doi: 10.1074/jbc.M605726200. [DOI] [PubMed] [Google Scholar]
- 23.Son S.H., Jeon H.L., Yang S.J., Sim M.H., Kim Y.J., Lee N.K., Paik H.D. Probiotic lactic acid bacteria isolated from traditional Korean fermented foods based on beta-glucosidase activity. Food Sci. Biotechnol. 2018;27:123–129. doi: 10.1007/s10068-017-0212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rekha C.R., Vijayalakshmi G. Isoflavone phytoestrogens in soymilk fermented with beta-glucosidase producing probiotic lactic acid bacteria. Int. J. Food Sci. Nutr. 2011;62:111–120. doi: 10.3109/09637486.2010.513680. [DOI] [PubMed] [Google Scholar]
- 25.Abdella A., Mazeed T.E.-S., El-Baz A.F., Yang S.-T. Production of β-glucosidase from wheat bran and glycerol by Aspergillus niger in stirred tank and rotating fibrous bed bioreactors. Process Biochem. 2016;51:1331–1337. doi: 10.1016/j.procbio.2016.07.004. [DOI] [Google Scholar]
- 26.Kovács K., Megyeri L., Szakacs G., Kubicek C.P., Galbe M., Zacchi G. Trichoderma atroviride mutants with enhanced production of cellulase and β-glucosidase on pretreated willow. Enzym. Microb. Technol. 2008;43:48–55. doi: 10.1016/j.enzmictec.2008.02.006. [DOI] [Google Scholar]
- 27.Ibrahim A.S., El-Shishtawy M.M., Pena A., Jr., Liou G.I. Genistein attenuates retinal inflammation associated with diabetes by targeting of microglial activation. Mol. Vis. 2010;16:2033–2042. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.