Abstract
Listeria monocytogenes serotype 4b strains account for about 40% of sporadic cases and many epidemics of listeriosis. Mutations in a chromosomal locus resulted in loss of reactivity with all three monoclonal antibodies (MAbs) which were specific to serotype 4b and the closely related serotypes 4d and 4e. Here we show that this locus contains a serotype 4b-4d-4e-specific gene cassette (3,071 bp) which consists of two genes, gltA and gltB, and is flanked by palindromic sequences (51 and 44 nucleotides). Complete loss of reactivity with the three serotype-specific MAbs resulted from insertional inactivation of either gltA or gltB. The gltA and gltB mutants were characterized by loss and severe reduction, respectively, of glucose in the teichoic acid, whereas galactose, the other serotype-specific sugar substituent in the teichoic acid, was not affected. Within L. monocytogenes, only strains of serotypes 4b, 4d, and 4e harbored the gltA-gltB cassette, whereas coding sequences on either side of the cassette were conserved among all serotypes. Comparative genomic analysis of a serotype 1/2b strain showed that the 3,071-bp gltA-gltB cassette was replaced by a much shorter (528-bp) and unrelated region, flanked by inverted repeats similar to their counterparts in serotype 4b. These findings indicate that in the evolution of different serotypes of L. monocytogenes, this site in the genome has become occupied by serotype-specific sequences which, in the case of serotype 4b, are essential for expression of serotype-specific surface antigens and presence of glucose substituents in the teichoic acids in the cell wall.
Numerous serotypes of Listeria monocytogenes have been identified using the antigenic scheme of Seeliger and Hoehne (16). However, three serotypes, 1/2a, 1/2b, and 4b, account for more than 95% of clinical isolates (5). Serotype 4b is of special interest, as it is implicated in about 40% of sporadic cases and the majority of epidemics of food-borne listeriosis reported in Europe and North America during the past 20 years (1, 7, 15). This may reflect relatively high virulence of serotype 4b strains for humans, although unique pathogenesis attributes of this serotype have not yet been identified.
The somatic component of the serotypic designation in Listeria resides primarily in the anionic polymer, teichoic acid (TA), which consists of polyribitol phosphate and is covalently linked to peptidoglycan (4, 6, 18). Glycosidic substitution(s) of the ribitol phosphate units render the TA variable, structurally and antigenically, among different serotypes. In serogroup 1/2 (e.g., serotypes 1/2a and 1/2b), N-acetylglucosamine and rhamnose are present as substituents on the ribitol, whereas in serogroup 4, N-acetylglucosamine is integral to the TA chains. A unique glycosidic substitution pattern is present in serotype 4b, where the integral N-acetylglucosamine bears both galactose and glucose substituents (4, 18).
In an effort to develop tools useful for the identification of antigenic and genetic attributes unique to serotype 4b bacteria, we have used monoclonal antibodies (MAbs) (c74.22, c74.33, and c74.180) which reacted with strains of serotypes 4b, 4d, and 4e (referred to collectively as serotype 4b-4d-4e) (8) to identify serotype-specific genomic regions. One such region was shown to harbor the serogroup 4-specific gene gtcA, which has been recently described (14). Insertional inactivation of gtcA resulted in loss of reactivity with one of the MAbs (c74.22), loss of galactose, and marked reductions in the glucose in the TA of the cell (14). A different genomic region was found to be specific to serotypes 4b, 4d, and 4e, and mutants in this region lacked reactivity with all three MAbs (10). Here we report the cloning and characterization of the genes composing this region and provide genetic evidence for their involvement in serotype-specific surface antigen expression and TA glycosylation in L. monocytogenes serotype 4b.
MATERIALS AND METHODS
Bacterial strains and media.
Listeria and Escherichia coli strains were grown and preserved as described before (14). Antibiotics used for Listeria and for E. coli were as described before (14). Generation of transposon mutants of the serotype 4b strain 4b1 and screening of the mutants with the MAbs have been described elsewhere (10).
Biochemical analysis of cell wall composition.
Cell wall composition was determined as described by Fiedler et al. (4). TA from Listeria was prepared and analyzed as previously described (4, 6).
Molecular procedures.
Procedures for extraction of plasmid DNA from E. coli and genomic DNA from Listeria and for nonradioactive labeling and detection of DNA were previously described (10). Fragment XL7-1, which flanks the single transposon insertion in mutant XL7, has been described elsewhere (10). This fragment was sequenced, and inverse PCR (13) was employed to obtain genomic fragments on either side, using as template genomic DNA of the wild-type strain 4b1 digested with EcoRI or Sau3A, purified from low-melting-point agarose with phenol-chloroform extractions (2), and self-ligated. Amplified fragments were cloned in pCR2.1 (Invitrogen) and sequenced. Sequence information was used to design new primers at the end of the known sequence for additional inverse PCRs. Transposon-flanking fragments from other mutants were amplified using the Tn916 terminal primer OTL (5′-CGG AAT TCC GTG AAG TAT CTT CCT ACA G-3′) with a 5′-end EcoRI site (underlined) and primer cP1 (5′-CAC AGA AGC GAT ACG ATG A-3′).
Probe construction.
Probe locations are shown in Fig. 1. Probe XL7-1 (1.1 kb), which flanks the transposon insertion in mutant XL7, has been described elsewhere (10). Probe XL7-4 (0.6 kb) is internal to open reading frame Z (ORFZ) and consists of a 0.6-kb Sau3A fragment cloned into pUC19. Probe XL7-5 (1.6 kb), which includes ORFP and part of ORFO, was obtained as a PCR fragment with primers −1F5 (5′ CCG ACT GTA TCT TCT TTT CC 3′) and −1R9 (5′ TTT GCT ACT CAA CGG AGC CAC 3′) and 4b1 DNA as the template. The XL7-AB probe (2.9 kb), which includes both gltA and gltB, was obtained as a PCR fragment using primers 2P3 (5′-GTA ACG TCT CAT ATA GGG AG-3′) and −1R5 (5′-GTA GAA CAA TTG TAG TAC CG-3′). DNA fragments were isolated from low-melting-point agarose gels, purified by phenol-chloroform extractions (2), and labeled with a Genius kit.
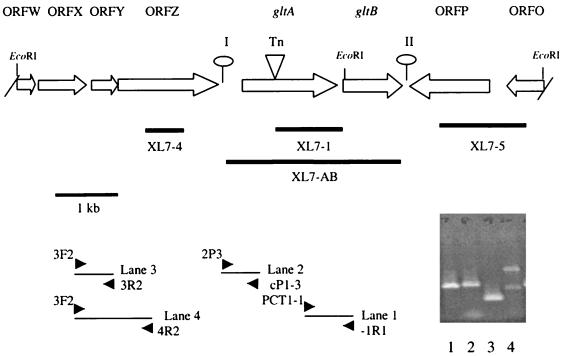
FIG. 1.
Genomic organization of the region harboring the transposon insertion in mutant XL7. Open arrows indicate ORFs and predicted direction of transcription. Slashes at the borders indicate that ORFW and ORFO are partial; lollipops represent putative stem-loop structures. The location of the Tn916ΔE insertion in gltA (Tn) is indicated by a triangle. Thick lines represent DNA fragments used as probes in Southern blots, and arrowheads at the bottom indicate primers used in RT-PCR. RT-PCR was done as described in Materials and Methods with -1R1 as the primer for cDNA synthesis. The gel shows products of PCRs with cDNA as the template and the primer pairs -1R1–PCT1-1 (lane 1), cP1-3–2P3 (lane 2), 3R2-3F2 (lane 3), and 4R2-3F2 (lane 4). Negative controls (using RNA instead of cDNA as the template and the same pairs of primers) were devoid of any product (data not shown).
RT-PCR.
Procedures for RNA extraction from Listeria, construction of cDNA, and reverse transcription PCR (RT-PCR) were as described elsewhere (14).
Construction of integration mutant in gltB.
To construct an integration mutant in gltB, an internal fragment of the gene was cloned in the temperature-sensitive shuttle vector pKSV7 (17), and integrants were selected by growth at the restrictive temperature (43°C) in medium containing chloramphenicol (CM medium) as follows. The internal fragment was amplified with primers −1F2 (5′-TTG GTA ACT CAC TAG TAC GT) and −1R4 (5′-ACA AGC ACA AAC AAA GAC GC), cloned in pCR2.1, recovered by EcoRI digestion, and subcloned into EcoRI-digested and dephosphorylated pKSV7. The resulting recombinant was electroporated into electrocompetent cells of the parental strain 4b1 as previously described (14), and transformants were isolated on CM medium after 48 h at 30°C. Integrants were isolated following four consecutive passages in CM medium at the restrictive temperature (43°C) and confirmed by Southern blotting. For colony immunoblots, the cultures were grown at room temperature.
Construction of pKA and pKAB.
Listeria DNA fragments harboring gltA and gltA-gltB were amplified from DNA of the parental strain 4b1 by PCR using High Fidelity enzyme (Roche). Fragment A (containing gltA) was obtained by PCR using primers 2P3 and −1R1 (5′-CAA GGC AAG AGT ACA GCT AC-3′). Fragment AB (containing gltA-gltB) was amplified using primers 2P3 and −1R5 (described above for construction of the gltA-gltB probe XL7-AB), which had a HindIII site and a BamHI site, respectively, at the 5′ end. The PCR fragments were excised from low-melting-point agarose gels, purified with phenol-chloroform, and cloned into pCR2.1. Fragment A was isolated following digestion of the recombinant plasmid with EcoRI and was subcloned into pKSV7 which had been digested by EcoRI and dephosphorylated. Fragment AB was obtained following digestion of the plasmid with BamHI and HindIII and directionally cloned into pKSV7 digested with the same enzymes. The resulting plasmids, consisting of pKSV7 with inserts of gltA and gltA-gltB, were named pKA and pKAB, respectively. Upon electroporation, 100 μl of the cells was plated on CM medium, and the plates were incubated at 30°C for 3 to 4 days.
Cloning of serotype 1/2b sequences.
Primers 2P2 (5′- GAC CAT ATC GTC GTG CTA CA-3′) and −1R65 (5′-CGA GCA TAC AAG TGC TCG TT-3′) were used to amplify a 1.1-kb DNA fragment with DNA of strain F4242 (serotype 1/2b) as the template. The 1.1-kb PCR product was directly cloned into pCR2.1 and sequenced on both strands.
DNA sequencing and sequence analysis.
Nested deletions were generated using the Erase-a-Base system (Promega) as suggested by the vendor. DNA was sequenced and analyzed as previously described (14).
Nucleotide sequence accession number.
The nucleotide sequence data for L. monocytogenes serotypes 4b and 1/2b have been deposited in GenBank under accession numbers AF033015 and AF033016, respectively.
RESULTS
Mutants negative for serotype-specific MAbs.
The single-insertion Tn916ΔE mutant XL7 lacked reactivity with all three serotype-specific MAbs (C74.22, C74.33, and C74.180) but had no readily detectable phenotypic differences from its wild-type counterparts in terms of growth at 20 and 35°C, motility, sensitivity to serotype-specific phage 2671 or Listeria-specific phage A511, hemolytic activity, and colony or cellular morphology. Furthermore, four additional independent transposon mutants (33N1, 33N2, 33N3, and 8A3) phenotypically identical to XL7 were found to harbor transposon insertions in the same EcoRI and HindIII genomic fragment as XL7 (10), suggesting that the MAb-negative phenotype of XL7 was associated with the Tn916ΔE insertion. DNA sequence analysis of XL7-1 and of the additional fragments derived by inverse PCR showed that the transposon was inserted in an ORF termed gltA (for glucose in teichoic acid). The transposon insertion sites in mutants 33N1, 33N2, 33N3, and 8A3 were within a 10-nucleotide (nt) region in gltA, which also harbored the insertion in XL7. The target sequence for the transposon insertions conformed to the consensus target sequence (T[T/A]TTTTNNNNNNAAAAA[A/T]A) for Tn916 (11).
Genomic organization and ORF analysis of the gltA-gltB region.
Sequence analysis revealed six complete ORFs (ORFX, ORFY, ORFZ, gltA, gltB, and ORFP) and two partial ORFs (ORFW and ORFO) in this region (Fig. 1). ORFW (partial), ORFX, ORFY, ORFZ, gltA, and gltB were transcribed in the same direction and convergently to ORFP and ORFO (partial). Two palindromic sequences with the potential to form pronounced stem-loop structures flanked the gltA-gltB region. The palindrome for putative stem-loop I (51 nt; calculated free energy of formation, −46 kcal/mol) was in the region between ORFZ and gltA, 55 nt downstream of ORFZ and 279 nt upstream of gltA, whereas that for putative stem-loop II (44 nt; calculated free energy of formation also −46 kcal/mol) was 8 and 27 nt downstream of gltB and ORFP, respectively (Fig. 1). The organization of the region suggests that stem-loops I and II may serve as transcription terminators for ORFZ and ORFP, respectively.
The G+C contents of gltA and gltB were 34 and 34.8%, respectively, lower than is typical for L. monocytogenes (38%). In contrast, the other ORFs in this region had G+C contents noticeably higher than those of gltA and gltB: ORFW, ORFX, ORFY, and ORFZ had G+C contents of 38.8, 39.7, 41.5, and 39.8%, respectively, whereas the values for ORFP and ORFO were 39.6 and 40.6%, respectively.
gltA-gltB region.
The transposon-harboring ORF (gltA) (1,647 bp) was 386 nt downstream of ORFZ. We were unable to identify sequences upstream of gltA with detectable similarity to the canonical Shine-Dalgarno ribosome recognition sequences. A putative −10 promoter element (TATTAT) was identified 92 nt upstream of the putative start codon of gltA. The coding sequence of gltA appears to be novel, as screens of the nucleotide and protein databases failed to identify sequences with significant homology to either the gene or the deduced gene product. The latter (548 amino acids, calculated Mr of 62,755, pI 9.0) may be membrane associated in L. monocytogenes, as hydrophobicity analysis of the deduced polypeptide revealed 11 putative transmembrane segments (data not shown).
Immediately downstream of gltA was gltB (948 nt). The gltA-gltB intergenic space was only 10 nt, and the putative Shine-Dalgarno site preceding gltB (AGGAGAGA) included the last nucleotide of the gltA ochre codon, suggesting that the two ORFs may be translationally coupled. gltB had 57% identity over its entire length with rfbj (ORF10X5) and ORF10X9, which are adjacent to each other on the genome of Shigella flexneri (accession no. X71970). In S. flexneri this region has been shown to be involved in polymerization of lipopolysaccharide (12), although the exact functions of these two ORFs are unknown.
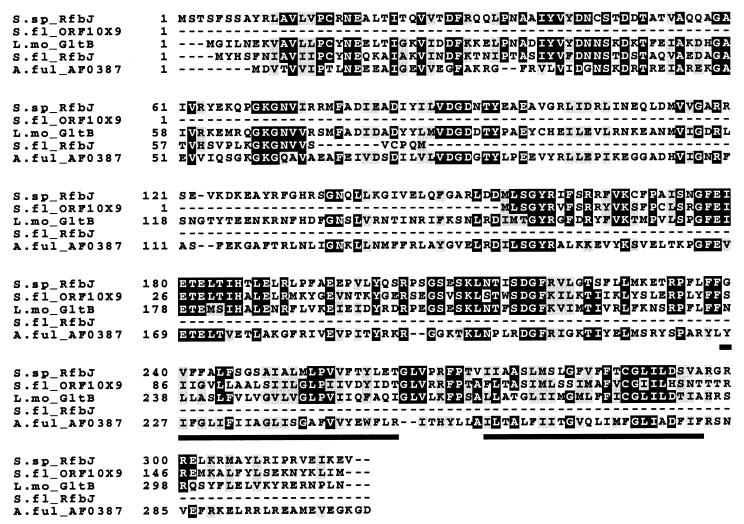
The deduced gltB gene product (315 amino acids, calculated Mr of 36,223, pI 6.04) contained two putative transmembrane domains (underlined in Fig. 2). Protein database searches showed significant similarity between the putative GltB and the deduced products of the S. flexneri rfbj and ORF10X9 (56 and 42% identity, respectively). The putative GltB also had 48% identity over its entire length with RfbJ of Synechocystis sp. strain PCC6803 (accession no. S77381) and lower (25 to 35%) identity with numerous glycosyltransferases and dolichol phosphate mannosyltransferases from bacteria and archaea. Figure 2 shows alignment of the deduced gltB gene product sequence with selected sequences.
FIG. 2.
Multiple sequence alignment (CLUSTAL) of the deduced sequences of, from top to bottom, RfbJ of Synechocystis sp. strain PCC6803 (accession no. S77381), ORF10X9 of S. flexneri (accession no. P37787), GltB of L. monocytogenes (accession no. AF033015), RfbJ of S. flexneri (accession no. P37786), and a putative glycosyltransferase of Archaeoglobus fulgidus (accession no. AE001078). The underlined segments represent predicted transmembrane regions.
Coding sequences upstream of gltA-gltB (ORFW to ORFZ).
BLAST and motif search analysis of the deduced amino acid sequences of ORFW (partial), ORFX, ORFY, and ORFZ suggested that all had characteristics of ABC (ATP-binding cassette) transporters (3). A putative ATP/GTP-binding site motif A (P loop) was identified in the deduced sequences of ORFX (residues 217 to 225) and ORFZ (residues 368 to 375). The ORFW-ORFX and ORFX-ORFY intergenic spaces were 2 and 21 nt, respectively, whereas the stop codon of ORFY overlapped by one nucleotide with the putative start codon of ORFZ, suggesting that ORFY and ORFZ are translationally coupled.
Coding sequences downstream of gltA-gltB (ORFP and ORFO).
FASTA and BLAST analysis of ORFP, located downstream of gltB and transcribed convergently, suggested that the deduced product may be a penicillin-binding protein (PBP), having 34 to 55% identity over the entire amino acid sequence with PBPs from numerous other bacteria. Highest similarity (55% identity) was observed with the d-alanyl-d-alanine carboxypeptidase, PBP5, of Bacillus subtilis (accession no. P08750). ORFP was preceded by ORFO (partial), transcribed in the same orientation as ORFP and separated from it by 214 nt. The deduced ORFO product had 49 and 46% identity over its entire available length (189 amino acids) with the 7-β-(4-carboxybutanamido)cephalosporanic acid acylase (glutaryl 7-amino cephalosporanic acid [7-ACA] acylase precursor) of Bacillus laterosporus and with the cocaine esterase of Rhodococcus sp. strain MB1, respectively. These similarities are difficult to evaluate at this time, as such enzymatic activities have not been detected before in L. monocytogenes.
Transcriptional studies.
The quantitative levels of gltA-gltB transcripts were too low for reliable detection and size determination by Northern blotting (data not shown), and RT-PCR was used for transcriptional studies. When primer −1R1 (located in gltB) was used for reverse transcription, a PCR product of the expected size was obtained using primers −1R1 and PCT1-1 (spanning gltB and gltA) (Fig. 1, lane 1), suggesting that gltA and gltB were cotranscribed. Furthermore, the transcript contained the 386-nt region between gltA and ORFZ, which includes the palindromic sequence, as suggested by a product of the expected size with primers cP1-3 and 2P3 (located in the region between ORFZ and gltA) (Fig. 1, lane 2). Surprisingly, cDNA produced by primer −1R1 could be amplified by primer 3R2 (located in the 3′ region of ORFX) and either 3R2 or 4R2 (Fig. 1, lanes 3 and 4), suggesting that ORFY and ORFZ were included in the transcript as well. These and additional RT-PCR data (not shown) suggest the presence of transcripts harboring not only gltA-gltB but also extending through the relatively long (386-nt) region between gltA and ORFZ, to at least 2,287 nt upstream of gltA. gltB appears to be the last ORF in this transcriptional unit.
Insertional inactivation of either gltA or gltB results in absence of glucose in the TA, whereas galactose is not affected.
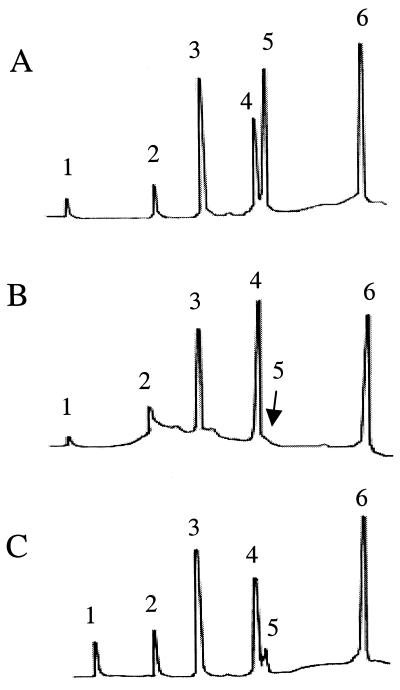
Transposon mutants in gltB were not identified, and an integration mutant (4b1-INTB) was constructed, using the temperature-sensitive plasmid pKSV7. Similarly to XL7, the mutant had normal growth and other phenotypic characteristics but lacked reactivity with all three MAbs (data not shown). Biochemical analysis of TA from XL7 and 4b1-INTB showed that both mutants were severely deficient in glucose. In contrast to the wild-type parental strain 4b1, which had both galactose and glucose as substituents on the N-acetylglucosamine of the TA, as is typical of serotype 4b (4), glucose was undetectable in the TA of XL7 and present in only trace amounts in the TA of 4b1-INTB (Fig. 3). Interestingly, the other serotype-specific substituent, galactose, was present in normal amounts in the TA of the mutants, as were the integral components of TA (ribitol phosphate and N-acetylglucosamine) (Fig. 3). The loss of glucose in the TA of XL7 was also seen with the independently obtained gltA mutants 33N1, 33N2, 33N3, and 8A3 (data not shown).
FIG. 3.
TA composition of wild-type strain 4b1 (A), gltA mutant XL7 (B), and gltB mutant 4b1-INTB (C). TA preparation and analysis were done as described previously (4, 6). Peaks: 1, glycerol; 2, anhydroribitol; 3, ribitol; 4, galactose; 5, glucose; 6, glucosamine. The arrow indicates the position of the missing glucose peak in panel B.
The MAb-negative phenotype of XL7 can be partially complemented by gltA alone or in combination with gltB.
The recombinant plasmids pKA and pKAB, harboring gltA alone and together with gltB, respectively, were electroporated into mutant XL7. Both plasmids included 219 nt upstream of the start codon of gltA, since a promoter may be contained within this region. The resulting strains were grown in the presence of chloramphenicol at 30°C, a temperature which permits both replication of the temperature-sensitive plasmid (17) and optimal expression of the serotype-specific surface antigens (8). Reactivity of the mutant with c74.22, c74.33, and c74.180 was restored partially and to the same levels by both plasmids, whereas XL7 harboring the shuttle vector pKSV7 alone remained negative with the MAbs (data not shown). Although pKA and pKAB partially restored reactivity with the MAbs, glucose in the TA of the mutant was not restored to detectable levels (data not shown).
gltB is needed for heterologous expression of the serotype-specific surface antigens in strains of serotypes 4a and 4c.
Strains of serotypes 4a and 4c lacked reactivity with the gltA-derived probe XL7-1 (10). When transformed with pKAB, strains ATCC 19114 (serotype 4a) and ATCC 19116 (serotype 4c) were rendered reactive with at least two of the MAbs, c74.22 and c74.33 (data not shown). When transformed by pKA these strains remained MAb negative, suggesting that gltB was required for expression of c74.22- and c74.33-specific surface antigens in these heterologous hosts.
Within L. monocytogenes, only strains of serotype 4b-4d-4e harbored sequences with homology to gltA and gltB, whereas ORFP and ORFZ were conserved among different serotypes.
Hybridizations using probe XL7-AB, which contains both gltA and gltB, showed that the genes were unique to L. monocytogenes serotype 4b-4d-4e and could not be detected in DNA from strains of other serotypes. The genes have also been detected in a unique lineage (lineage I) of L. innocua (9). EcoRI restriction fragment length polymorphisms using XL7-AB as the probe could differentiate between L. monocytogenes serotype 4b-4d-4e and L. innocua lineage I (Table 1). Southern blot and PCR data suggest that the gltA-gltB cassette was flanked by ORFZ and ORFP in L. innocua lineage I, as in L. monocytogenes serotype 4b (data not shown). No hybridization was observed with DNA from other L. innocua strains or other Listeria species (Table 1).
TABLE 1.
Southern blot hybridization data using probes from the gltA-gltB genomic region and EcoRI-digested DNAs from different strains of Listeria
| Strain (serotype) | Hybridization with indicated DNA probea
|
|||
|---|---|---|---|---|
| XL7-4 (ORFZ) | XL7-1 (gltA) | XL7-AB (gltA-gltB) | XL7-5 (ORFP-PRFO) | |
| L. monocytogenes | ||||
| 4b1 (4b) | 5.0 | 5.0 | 5.0, 3.0 | 3.0 |
| F2381 (4b) | 4.5 | 4.5 | 4.5, 3.0 | 3.0 |
| G2228 (1/2a) | 2.6 | 0 | 0 | 6.0 |
| F4242 (1/2b) | 2.6 | 0 | 0 | 2.6 |
| F4245 (1/2b) | 2.6 | 0 | 0 | 5.0 |
| LM103 (1/2c) | 2.6 | 0 | 0 | 2.6 |
| ATCC 19113 (3a) | 2.6 | 0 | 0 | 2.6 |
| G3331 (3b) | 2.6 | 0 | 0 | 2.6 |
| ATCC 2540 (3b) | 3.0 | 0 | 0 | 2.6 |
| G4315 (3c) | 2.6 | 0 | 0 | 2.6 |
| SLCC 2479 (3c) | 2.6 | 0 | 0 | 2.6 |
| ATCC 19114 (4a) | 2.8 | 0 | 0 | 6.5 |
| ATCC 19116 (4c) | 6.5 | 0 | 0 | 6.5 |
| ATCC 19117 (4d) | 5.0 | 5.0 | 5.0, 3.0 | 3.0 |
| ATCC 19118 (4e) | 4.5 | 4.5 | 4.5, 3.2 | 3.2 |
| G2940 (4ab) | 7.5 | 0 | 0 | 1.5 (weak) |
| SLCC 2480 (7) | 2.6 | 0 | 0 | 2.6 |
| L. innocua | ||||
| 120A1 | 4.2 | 0 | 0 | 1.5 |
| F8596b | 3.0 | 2.2 | 2.2, 1.7 | 1.5 |
| G6882 | 4.2, 3.0 | 0 | 0 | 5.0, 1.5 |
| G803 | 4.2, 3.0 | 0 | 0 | 5.0, 1.5 |
| L. grayi | 0 | 0 | 0 | 0 |
| L. ivanovii | 4.0 | 0 | 0 | 1.5 |
| L. seeligeri | 7.5 (weak) | 0 | 0 | 7.5 (weak) |
| L. welshimeri | 0 | 0 | 0 | 0 |
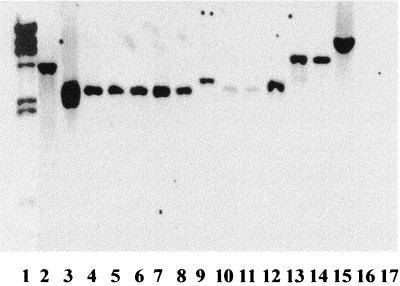
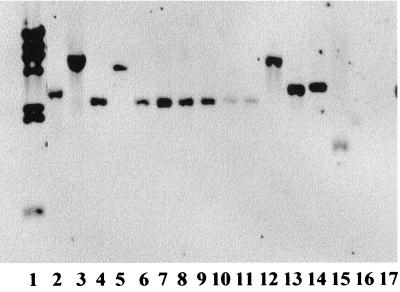
Southern blots using probes derived from ORFP-ORFO and ORFZ hybridized with all screened serotypes of L. monocytogenes suggesting that, in contrast to gltA and gltB, these sequences were conserved among different serotypes (Fig. 4 and 5). EcoRI restriction fragment length polymorphisms could be detected with probes derived from ORFZ and ORFP (Table 1). Sequences homologous to the ORFP- and ORFZ-derived probes were detected in other Listeria species as well, except for L. grayi and L. welshimeri (Table 1).
FIG. 4.
Southern blot of EcoRI-digested genomic DNAs from L. monocytogenes of different serotypes, using the ORFZ-derived fragment XL7-4 (Fig. 1) as the probe. Lane 1, λ HindIII-digested molecular size markers (fragment sizes [from the top to bottom], 23, 9.4, 6.6, 4.4, 2.3, 2.0, and 0.56 kb); lanes 2 to 15, L. monocytogenes F2381 (4b), G2228 (1/2a), F4242 (1/2b), F4254 (1/2b), LM103 (1/2c), ATCC 19113 (3a), G3331 (3b), SLCC 2540 (3b), G4315 (3c), SLCC 2479 (3c), ATCC 19114 (4a), ATCC 19117(4d), ATCC 19118 (4e), and G2940 (4ab), respectively; lanes 16 and 17, B. subtilis 168 and B. subtilis W23, respectively. Lanes 10 and 11 contained relatively low amounts of DNA.
FIG. 5.
Southern blot of EcoRI-digested genomic DNAs from L. monocytogenes of different serotypes, using the ORFP-ORFO-derived fragment XL7-5 (Fig. 1) as the probe. Lanes are identical to those in Fig. 4. The membrane used for the Southern blot in Fig. 4 was stripped of its probe and reprobed with XL7-5.
Serotype 1/2b L. monocytogenes harbors a novel locus genomically equivalent to the gltA-gltB cassette of serotype 4b-4d-4e.
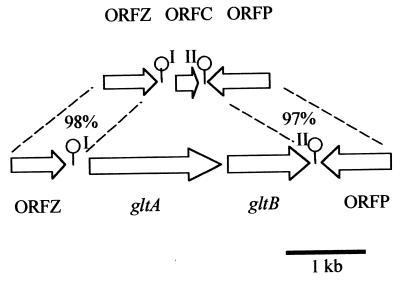
The genomic equivalent of the region flanked by the conserved ORFZ and ORFP was amplified from strain F4242 (serotype 1/2b) as described in Materials and Methods. In serotype 1/2b, ORFP and ORFZ flanked a region of 528 bp, in contrast to 3,071 bp in serotype 4b (Fig. 6). Interestingly, the region in serotype 1/2b was flanked by palindromic sequences with significant sequence identity (72 and 84%) to their counterparts in serotype 4b (Fig. 7). The remainder of the 528-bp region, however, showed no detectable homology with the serotype 4b sequences. The 1/2b sequence contained only a small potential coding sequence (ORFC, 75 amino acids), which was preceded by a putative Shine-Dalgarno sequence 7 nt upstream of the putative start codon. The palindromic sequence between ORFC and ORFP was followed by a string of eight T's, suggesting that it may function as a rho-independent terminator. The G+C content of ORFC was unusually low (26%), and no homologous sequences were identified in searches of the DNA and protein databases.
FIG. 6.
Comparison between the gltA-gltB region of serotype 4b (bottom) and the genomically equivalent region in serotype 1/2b (top). Putative stem-loops I and II (lollipops) represent the boundaries of unique cassettes of different size (3,071 bp in serotype 4b and 528 bp in serotype 1/2b). The percentages indicate nucleotide sequence identities between the corresponding conserved ORFs on either side of the cassettes.
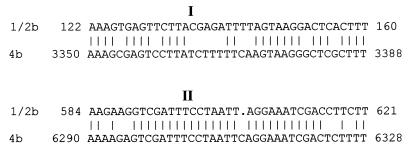
FIG. 7.
Comparison (BESTFIT) of the palindromic sequences corresponding to the putative stem-loops I and II in L. monocytogenes of serotypes 4b and 1/2b. Locations of putative stem-loops are as indicated in Fig. 6.
The sequenced 3′ portions of ORFP and ORFZ from serotype 1/2b were 97 and 98%, respectively, identical to their counterparts in serotype 4b. The corresponding C-terminal sequences of ORFP (144 residues) and ORFZ (31 residues) had 98 and 100%, respectively, identity, to their serotype 4b counterparts. Furthermore, Southern blots using probes derived from the regions outside the putative stem-loop structures (ORFP, ORFZ, and sequences distal to them) showed that the corresponding sequences and genomic organization were conserved between serotypes 4b and 1/2b (data not shown). The combined nucleotide sequence and Southern blot data suggest that the genomic organization of this region in serotypes 1/2b and 4b is as shown in Fig. 6.
DISCUSSION
The ca. 3-kb gene cassette described here represents a novel serotype-specific locus present in serotype 4b L. monocytogenes and the genetically closely related (albeit relatively rare) serotypes 4d and 4e but in no other serotypes of the species. In addition, a unique L. innocua lineage that reacts with the serotype 4b-4d-4e-specific MAbs (9) also harbors the cassette, in the same genomic location as serotype 4b L. monocytogenes. The distribution of the cassette in Listeria parallels precisely the pattern of reactivity of the serotype-specific MAbs (8).
The genes on either side of the cassette were found to be conserved among different serotypes of L. monocytogenes as well as other Listeria species (L. innocua, L. ivanovii, and L. seeligeri). On one side, at least one of these genes (ORFP) may be involved in cell wall biosynthesis, the deduced product being a putative PBP. On the other side, we identified four genes with homology to ABC transporters. It remains to be determined whether the products of these genes mediate transport of cell wall or TA precursors.
The serotype-specific distribution of the cassette and its unusually low (for Listeria) G+C content suggest the possibility that it may have been introduced to the L. monocytogenes serotype 4b-4d-4e lineage by horizontal transfer from some unidentified source. From there it could have been transferred to lineage I of L. innocua, as has been speculated for the gtcA locus recently identified in this lineage (9). The origin of the serotype-specific sequences may be elucidated by future identification of homologous sequences in other bacteria or bacteriophage. It is tempting to speculate that the inverted repeats flanking the serotype-specific sequences may represent remnants of a genetic system (e.g., a transposon or phage) that may have mediated this transfer. These palindromic sequences may have assumed novel functions in their current locations in serotype 4b L. monocytogenes, possibly related to transcriptional termination, message stability, or other regulatory mechanisms. Interestingly, these inverted repeats were similar to their counterparts in serotype 1/2b. In the latter, however, the genomic location of the ca. 3-kb serotype 4b-4d-4e-specific cassette was occupied by a much shorter (528-bp) region, which harbors a novel, unrelated ORF. Involvement of the 1/2b region (ORFC) in expression of surface antigen(s) in serotype 1/2b remains to be determined.
The integrated genetic, immunological, and biochemical results suggest that in L. monocytogenes serotype 4b, the gltA-gltB cassette is involved in expression of the surface antigens recognized by MAbs c74.22, c74.33, and c74.180 and in the addition of glucose substituents on the TA, but the precise biochemical functions of the two genes remain to be elucidated. The genes can be cotranscribed, and at this time we cannot exclude the possibility that the transposon insertion in gltA may have polar effects on gltB. The fact that the observed phenotypic complementation of XL7, albeit partial, was conferred equally by pKA and pKAB suggests that gltB was expressed to some extent in this mutant. Construction of alternative mutants in gltA (such as an in-frame deletion) and/or alternative complementation strategies will be needed to more precisely address the function(s) of gltA. Sequence analysis could not facilitate functional predictions in the case of gltA, as both the gene and the deduced gene product appeared to lack homologs in the databases. The deduced gltB product, however, had significant similarity with numerous glycosylases and dolichol phosphate mannosyltransferases, and a glycosylase function would be in agreement with the observed deficiency of glucose in the TA of the gltB mutant.
Complementation of MAb reactivity of XL7 by gltA or gltA-gltB was partial, for reasons that are not clear but may involve absence of possibly required cis elements or suboptimal copy number of the genes in the vector that was used. The low level of complementation may account for the lack of detectable restoration of glucose in the TAs. Such difficulties with complementation were not experienced with the previously studied gene gtcA, where both MAb reactivity and TA glycosylation were restored by the wild-type gene in trans (14). The mechanisms controlling regulation of expression of gltA and gltB are not understood but may be complicated, as suggested by the presence of the long and apparently transcribed region between ORFZ and gltA.
Glycosylated TA components have been shown to be important antigenic determinants in L. monocytogenes (6, 19), although their role in infection has not been elucidated. It is worthy of note that even though gltA or gltB mutants grew normally in the laboratory, our surveys of numerous serotype 4b field isolates (both food and clinical) failed to identify strains which had the XL7 or 4b1-INTB phenotype or which lacked gltA-gltB sequences. One may speculate that because of its surface exposure, abundance, and immunogenicity, properly decorated TA may be important in interactions between the bacteria and their host cells. Glycosylated TA may also affect physiological attributes of the bacteria in foods or in the environment, in response to environmental stresses, association with surfaces and with other organisms in biofilms, etc. Continuing studies in our laboratory aim toward further elucidation of the serotype-specific gene cassettes described in this report in terms of their evolution and potential roles in adaptive physiology and pathogenesis of the listerial lineages which harbor them.
ACKNOWLEDGMENTS
This research was partially supported by U.S. Department of Agriculture Competitive Research Initiative AAFS grant 99-35201-8183 and by ILSI North America.
We are grateful to Huyen Le Tran for assistance with graphics and to Vladimir Lazarevic for exchange of information related to teichoic acid biosynthesis genes. We thank Nattawan Promadej, Wei Zheng, Edward Lanwermeyer, and all other members of our laboratories for valuable feedback and support throughout the course of this work.
REFERENCES
- 1.Anonymous. Update: multistate outbreak of listeriosis—United States, 1998–1999. Morbid Mortal Wkly Rep. 1999;47:1117–1118. [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J D, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 3.Fath M J, Kolter R. ABC transporters: bacterial exporters. Microbiol Rev. 1993;57:995–1017. doi: 10.1128/mr.57.4.995-1017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiedler F, Seger J, Schrettenbrunner A, Seeliger H P R. The biochemistry of murein and cell wall teichoic acids in the genus Listeria. Syst Appl Microbiol. 1983;5:360–376. [Google Scholar]
- 5.Gellin B G, Broome C V. Listeriosis. JAMA. 1989;261:1313–1320. [PubMed] [Google Scholar]
- 6.Kaminsango K, Fujii H, Okumura H, Saiki I, Araki Y, Yamamura Y, Azuma I. Structural and immunochemical studies of teichoic acid of Listeria monocytogenes. J Biochem. 1983;93:1401–1409. doi: 10.1093/oxfordjournals.jbchem.a134275. [DOI] [PubMed] [Google Scholar]
- 7.Kathariou S. Pathogenesis determinants of Listeria monocytogenes. In: Cary J W, Linz J E, Bhatnagar D, editors. Microbial foodborne diseases. Lancaster, Pa: Technomics Publishing Co., Inc.; 2000. pp. 295–314. [Google Scholar]
- 8.Kathariou S, Mizumoto C, Allen R D, Fok A K, Benedict A A. Monoclonal antibodies with a high degree of specificity for Listeria monocytogenes serotype 4b. Appl Environ Microbiol. 1994;60:3548–3552. doi: 10.1128/aem.60.10.3548-3552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan Z, Fiedler F, Kathariou S. A sheep in wolf's clothing: Listeria innocua strains with teichoic acid-associated surface antigens and genes characteristic of Listeria monocytogenes serogroup 4. J Bacteriol. 2000;182:6161–6168. doi: 10.1128/jb.182.21.6161-6168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei X-H, Promadej N, Kathariou S. DNA fragments from regions involved in surface antigen expression specifically identify Listeria monocytogenes serovar 4 and a subset thereof: cluster IIB (serotypes 4b, 4d, and 4e) Appl Environ Microbiol. 1997;63:1077–1082. doi: 10.1128/aem.63.3.1077-1082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu F, Churchward G. Tn916 target DNA sequences bind the C-terminal domain of integrase protein with different affinities that correlate with transposon insertion frequency. J Bacteriol. 1995;177:1938–1946. doi: 10.1128/jb.177.8.1938-1946.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morona R, Mavris M, Fallarino A, Manning P A. Characterization of the rfc region of Shigella flexneri. J Bacteriol. 1994;176:733–747. doi: 10.1128/jb.176.3.733-747.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ochman H, Medhora M M, Garza D, Hartl D L. Amplification of flanking sequences by inverse PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. San Diego, Calif: Academic Press, Inc.; 1990. pp. 219–227. [Google Scholar]
- 14.Promadej N, Fiedler F, Cossart P, Dramsi S, Kathariou S. Cell wall teichoic acid glycosylation in Listeria monocytogenes serotype 4b requires gtcA, a novel, serotype-specific gene. J Bacteriol. 1999;181:418–425. doi: 10.1128/jb.181.2.418-425.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuchat A, Swaminathan B, Broome C V. Epidemiology of human listeriosis. Clin Microbiol Rev. 1991;4:169–183. doi: 10.1128/cmr.4.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seeliger H P R, Hoehne K. Serotypes of Listeria monocytogenes and related species. Methods Microbiol. 1979;13:31–49. [Google Scholar]
- 17.Smith K, Youngman P. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie. 1992;74:705–711. doi: 10.1016/0300-9084(92)90143-3. [DOI] [PubMed] [Google Scholar]
- 18.Uchikawa K, Sekikawa J, Azuma I. Structural studies on teichoic acids in cell walls of several serotypes of Listeria monocytogenes. J Biochem. 1986;99:315–327. doi: 10.1093/oxfordjournals.jbchem.a135486. [DOI] [PubMed] [Google Scholar]
- 19.Ullmann W W, Cameron J A. Immunochemistry of the cell walls of Listeria monocytogenes. J Bacteriol. 1969;98:486–493. doi: 10.1128/jb.98.2.486-493.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]