Abstract
Gladiolus is an important ornamental plant that is one of the world’s four most-grown cut flowers. Gladiolus gandavensis has only been found in the Cangnan County (Zhejiang Province) of China, which is recorded in the “Botanical”. To explore the origin of G. gandavensis, chloroplast genome sequencing was conducted. The results indicated that a total of 151,654 bp of circular DNA was obtained. The chloroplast genome of G. gandavensis has a quadripartite structure (contains a large single-copy (LSC) region (81,547 bp), a small single-copy region (SSC) (17,895 bp), and two inverted repeats (IRs) (IRa and IRb, 52,212 bp)), similar to that of other species. In addition, a total of 84 protein-coding genes, 8 rRNA-encoding genes, and 38 tRNA-encoding genes were present in the chloroplast genome. To further study the structural characteristics of the chloroplast genome in G. gandavensis, a comparative analysis of eight species of the Iridaceae family was conducted, and the results revealed higher similarity in the IR regions than in the LSC and SSC regions. In addition, 265 simple sequence repeats (SSRs) were detected in this study. The results of the phylogenetic analysis indicated that the chloroplast genome of G. gandavensis has high homology with the Crocus cartwrightianus and Crocus sativus chloroplast genomes. Genetic analysis based on the rbcl sequence among 49 Gladiolus species showed that samples 42, 49, 50, and 54 had high homology with the three samples from China (64, 65, and 66), which might be caused by chance similarity in genotypes. These results suggest that G. gandavensis may have originated from South Africa.
Keywords: Gladiolus gandavensis, chloroplast genome sequence, comparative analysis, evolutionary genetics
1. Introduction
Gladilous comprises approximately 265 species, which is one of the largest genera in the family Iridaceae [1]. In addition, Gladiolus is a valuable ornamental plant with beautiful colors, and is one of the world’s four most-grown cut flowers [2]. Gladiolus is native to Africa and southern Europe. Currently, G. gandavensis is found in Cangnan County and Zhejiang Province, China. In Cangnan County, G. gandavensis is distributed in Xiaguan town, Mazhan town, and Beiguang Island [3]. G. gandavensis likes warm and sunny environments with good ventilation. In its environment, it displays red and yellow flowers and leaves shaped like swords [4]. Therefore, G. gandavensis has a high ornamental value, and is mainly used for flower arrangements, bouquets and baskets, as well as in flower beds and as potted plants [5].
The nuclear genome is biparentally inherited, while the chloroplast genome is maternally inherited [6]. In addition, the nuclear genome can be spread by pollen and seeds, and the chloroplast genome can be spread only by seeds in most angiosperm species [7]. Therefore, the chloroplast genome is suitable for identifying plants because of its special characteristics, such as its small size and conservation. Now, the chloroplast genome plays an irreplaceable role in evolution [8], migration [9], and identification [10].
The chloroplast is an important organelle used for photosynthesis and metabolic activities in higher plants and a few algae and prokaryotes [11]. In addition, chloroplasts also play important roles in other aspects of plant physiology and development, which are important for plant responses to light [12,13], heat [14], drought [15,16], salt [17], and other stress [18].
With the popularization and development of NGS technology, chloroplast genome databases are becoming increasingly abundant. Chloroplast genomes of increasing numbers of species have been sequenced, including Nicotiana tabacum [19], Oryza sativa [20], Zea mays [21], Pinus massoniana [22], and many other species. In general, there is a typical double-linked loop structure in the chloroplast genome of higher plants, whose sizes range between 120 and 180 kb. The chloroplast genome usually contains a small single-copy (SSC) region, a large single-copy (LSC) region, and an inverted repeat (IR) sequence [23,24].
G. gandavensis was only distributed in Cangnan County, China. However, the origin of G. gandavensis is unclear. The chloroplast genome was sequenced, and then the cpDNA rbcl sequence was used to identify 46 samples of Gladilous, to explore the origin of G. gandavensis.
2. Materials and Methods
2.1. Sequencing, Assembly, and Annotation
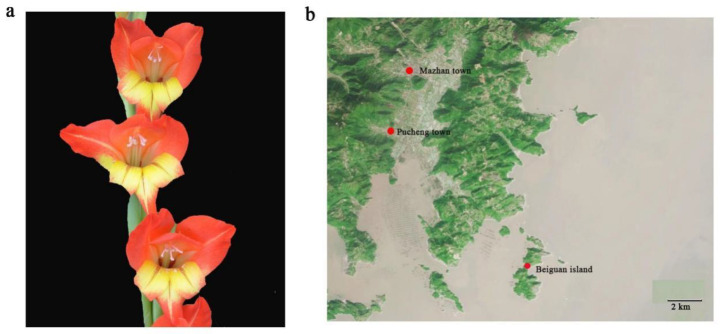
The G. gandavensis flowers showed red and yellow pigment, and their distribution was found in Cangnan county, Zhejiang Province, China. In Cangnan county, G. gandavensis was distributed in Xiaguan town, Mazhan town, and Beiguang Island (Figure 1). The G. gandavensis leaves were collected from Mazhan town (Zhejiang, China, N 27.29°, E 120.43°) for sequencing. An improved extraction method was used to isolate cpDNA from fresh leaves of G. gandavensis [25]. A library was constructed with 1 μg of DNA, and a Covaris M220 ultrasonic instrument was used to break the DNA into 300~5500 bp fragments. Afterward, the 3′ ends were polyadenylated and connected to index fragments (TruSeq™ Nano DNA Sample Prep Kit). Library enrichment and PCR amplification for 8 cycles were performed with a 2% agarose gel recycling destination bar (Certified Low Range Ultra Agarose), and then TBS380 (picogreen) was used for quantitation; the materials were mixed according to the data ratio. Then, the generated clusters were subjected to bridge PCR amplification on a CBOT solid phase, and 2 × 150 bp sequencing was performed with an Illumina HiSeq sequencing platform [26].
Figure 1.
(a) The flower with red and yellow colors of G. gandavensis. (b) The distribution of G. gandavensis in Cangnan County (Manzhan town, Pucheng town, and Beiguan Island).
Original reads were filtered before assembly. SOAPdenovo (version: 2.04, http://soap.genomics.org.cn/soapdenovo.html, accessed on 1 May 2022) was used to assemble the clean data and obtain the optimal assembly results after multiple adjustment parameters [27]. The contigs were obtained by the assembly. The results were partially assembled and optimized according to the reads’ paired ends and overlapping relationships. Then, GapCloser (version: 1.12, http://soap.genomics.org.cn/soapdenovo.html, accessed on 1 May 2022) was used to repair the internal gaps in the sequences, and the redundant sequences were removed to obtain the final assembly sequence [28].
2.2. Sequence Analysis
MISA (http://pgrc.ipk-gatersleben.de/misa/misa.html, accessed on 1 May 2022) was used to identify the microsatellite motif [29]. MAFFT v7.310 (https://mafft.cbrc.jp/alignment/software/, accessed on 1 May 2022), which is a multiple sequence alignment software, was used to align the IR sequences between some species in Gladiolus [30]. MAUVE was used to locate structural differences among whole-genome alignments [31]. The codon usage bias of (RSCU) was analyzed by (number of codons encoding one amino acid/number of all codons encoding the amino acid)/(1/type of codon encoding the amino acid), namely, the actual frequency of codon usage/frequency of theoretical usage of the codon. Vmatch v2.3.0 (http://www.vmatch.de/, accessed on 1 May 2022) was used to identife the repeat sequence, and the parameter was set to: minimum length (minimum length) = 30 bp, hamming distance (hamming distance) = 3 [32]. KaKs_Calculator v2.0 (https://sourceforge.net/projects/kakscalculator2/, accessed on 1 May 2022) was used to calculate the Ka/Ks, while dnasp5 was used to calculate the pi of every gene [33].
2.3. Phylogenetic Analysis with Other Species
For in-depth research, the 8 chloroplast genome sequences of Iridaceae were aligned by MAFFT version 7 [30]. The Iridaceae species and sequences used included Iris lactea (MT740331), Iris sanguinea (KT626943), Iris loczyi (MT254070), Iris missourensis (MH251636), Crous sativus (MH542233), Crous cartwrightianus (MH542231), and Iris domestica (MW039136), which was downloaded from https://www.ncbi.nlm.nih.gov/ (accessed on 1 May 2022). Then, the maximum likelihood (ML) method was used to construct the phylogenetic tree [34].
2.4. Genetic Evolutionary Analysis
The total DNA of 49 Gladiolus (Table 1) samples was extracted using a Plant Genprep DNA Kit (Tiangen, Beijing, China) and quantified using a NanoDrop 2000c instrument (ThermoFisher Scientific, Wilmington, DE, USA) [35,36]. The DNA templates were detected via 1% agarose gel electrophoresis. The ABI-2720 PCR instrument (Applied Biosystems, Waltham, MA, USA) was for PCR amplification. PCR was carried out according to the manufacturer’s protocol. The primers of the chloroplast genome rbcl sequence used were ATGTCACCACAAACAGAAAC (forward primer) and TCGCATGTACCTGCAGTAGC (reverse primer). The PCR products were sent to Shanghai Suny Biotechnology Co., Ltd., Shanghai, China, for sequencing. The original sequence data were obtained with Sequencing Analysis 5.2 software. MAFFT was used to align all the sequences. The arithmetic means (UPGMA) method was used to construct the phylogenetic tree [37].
Table 1.
Sample geographic information on Gladiolus.
| Number | Specific Name |
Latin Name | Origin |
|---|---|---|---|
| 1 | 1 | Gladiolus abbreviatus | South Africa |
| 2 | 2 | Gladiolus alatus | South Africa |
| 3 | 3 | Gladiolus angustus | South Africa |
| 4 | 4 | Gladiolus brevifolius var brevifolius | South Africa |
| 5 | 5 | Gladiolus carinatus | South Africa |
| 6 | 6 | Gladiolus carmineus | South Africa |
| 7 | 7 | Gladiolus carneus | South Africa |
| 8 | 9 | Gladiolus carneus (macowanianus) | South Africa |
| 9 | 11 | Gladiolus ceresianus | South Africa |
| 10 | 12 | Gladiolus crassifolius | South Africa |
| 11 | 14 | Gladiolus dalenii | South Africa |
| 12 | 15 | Gladiolus densiflorus | South Africa |
| 13 | 18 | Gladiolus exiguus | South Africa |
| 14 | 19 | Gladiolus ferrugineus | South Africa |
| 15 | 21 | Gladiolus floribundus ssp floribundus | South Africa |
| 16 | 23 | Gladiolus griseus | South Africa |
| 17 | 25 | Gladiolus hirsutus | South Africa |
| 18 | 26 | Gladiolus hyalinus | South Africa |
| 19 | 29 | Gladiolus liliaceus | South Africa |
| 20 | 30 | Gladiolus longicollis | South Africa |
| 21 | 31 | Gladiolus macelatus | South Africa |
| 22 | 32 | Gladiolus martleyi | South Africa |
| 23 | 33 | Gladiolus meliusculus | South Africa |
| 24 | 35 | Gladiolus mostertiae | South Africa |
| 25 | 36 | Gladiolus ochroleucus | South Africa |
| 26 | 37 | Gladiolus oppositiflorus ssp salmoneus | South Africa |
| 27 | 38 | Gladiolus papilio | South Africa |
| 28 | 39 | Gladiolus pardalinus | South Africa |
| 29 | 41 | Gladiolus patersoniae | South Africa |
| 30 | 42 | Gladiolus permeabilis ssp permeabilis | South Africa |
| 31 | 43 | Gladiolus phoenix | South Africa |
| 32 | 44 | Gladiolus pole-evansii | South Africa |
| 33 | 45 | Gladiolus quadrangularis | South Africa |
| 34 | 46 | Gladiolus recurvus | South Africa |
| 35 | 47 | Gladiolus rudis | South Africa |
| 36 | 48 | Gladiolus saccatus | South Africa |
| 37 | 49 | Gladiolus saundersii | South Africa |
| 38 | 50 | Gladiolus scabridus | South Africa |
| 39 | 51 | Gladiolus sericeovillosus | South Africa |
| 40 | 52 | Gladiolus splendens | South Africa |
| 41 | 53 | Gladiolus stellatus | South Africa |
| 42 | 54 | Gladiolus sufflavus | South Africa |
| 43 | 55 | Gladiolus teretifolius | South Africa |
| 44 | 59 | Gladiolus venustus | South Africa |
| 45 | 60 | Gladiolus vernus | South Africa |
| 46 | 61 | Gladiolus virescens | South Africa |
| 47 | 64 | G. gandavensis | China/Beiguan Island |
| 48 | 65 | G.gandavensis | China/Mazhan |
| 49 | 66 | G.gandavensis | China/Pucheng |
3. Results
3.1. Characteristics of Chloroplast Genomes
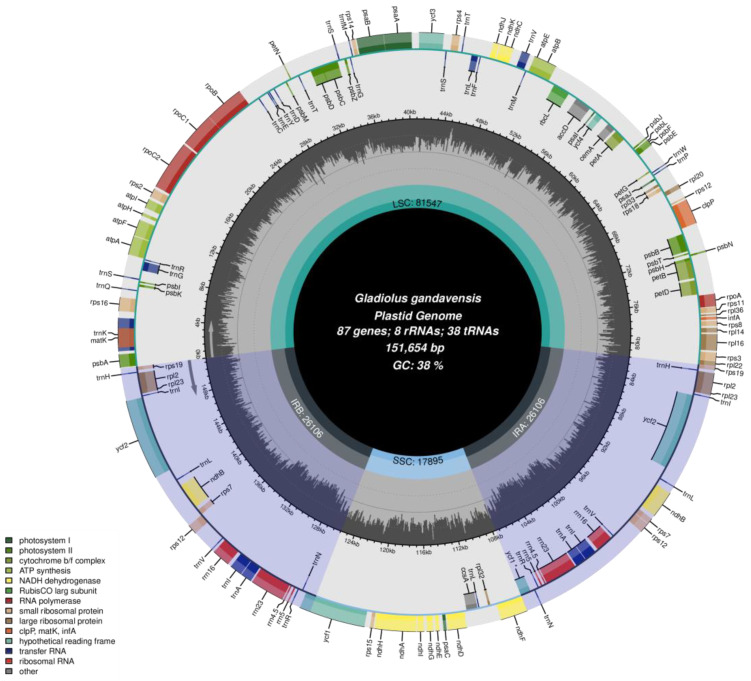
A circular chloroplast genome with 151,654 bp from G. gandavensis was assembled (GenBank accession number: OM304631). The chloroplast genome locations of 1–81,547 bp, 81,548–107,653 bp, 107,654–125,548 bp, and 125,549–151,654 bp were large single-copy (LSC) regions, first inverted repeat (IRa) regions, small single-copy (SSC) regions, and second inverted repeat (IRb) regions, respectively. The contents of the LSCs, IRs, and SSCs in the chloroplast genome were 53.78%, 29.01%, and 17.21%, respectively (Figure 2). In addition, the A, T, C, and G of the nucleotide compositions in the chloroplast genome were 30.79%, 30.88%, 19.2%, and 19.13%, respectively, and the total GC content was 38.34%. Additionally, the chloroplast genome of G. gandavensis contains 84 protein-coding genes, 38 transfer-RNA genes (tRNA), and 8 ribosomal RNA genes (rRNA) (Table 2).
Figure 2.
The chloroplast genome of G. gandavensis. Genes drawn outside are presented in the clockwise direction, while those inside the circle are presented in the counterclockwise direction. In addition, genes with different functions are represented with different colors.
Table 2.
Gene contents of chloroplast genome in G. gandavensis.
| Category | Gene Group | Gene Contents |
|---|---|---|
| Photosynthesis | Subunits of photosystem I | psaA,psaB,psaC,psaI,psaJ |
| Subunits of photosystem II | psbA,psbB,psbC,psbD,psbE,psbF, psbH,psbI,psbJ,psbK,psbL,psbM,psbN,psbT,psbZ |
|
| Subunits of cytochrome b/f complex | petA,petB*,petD*,petG,petN | |
| Subunits of ATP synthase | atpA,atpB,atpE,atpF*,atpH,atpI | |
| Subunits of NADH-dehydrogenase | ndhA*,ndhB*(2),ndhC,ndhD,ndhE,ndhF,ndhG,ndhH, ndhI,ndhJ,ndhK |
|
| Subunit of rubisco rbcL | rbcL | |
| Self-replication | Small subunit of ribosome | rps11,rps12**(2),rps14,rps15,rps16*,rps18,rps19(2), rps2,rps3,rps4,rps7(2),rps8 |
| Large subunit of ribosome | rpl14,rpl16*,rpl2*(2),rpl20,rpl22,rpl23(2),rpl32,rpl33, rpl36 |
|
| DNA-dependent RNA polymerase | rpoA,rpoB,rpoC1*,rpoC2 | |
| Protease clpP | clpP | |
| Maturase | matK | |
| Envelope membrane protein cemA | cemA | |
| Translation initiation factor infA | infA | |
| Cytochrome c biogenesis ccsA | ccsA | |
| Subunit Acetyl-CoA-Carboxylate | accD | |
| Ribosomal RNAs | rrn16(2), rrn23(2), rrn4.5(2), rrn5(2) | |
| Transfer RNA | trnA-UGC*(2), trnC-GCA, trnD-GUC, trnE-UUC,trnF-GAA,trnG-GCC*,trnG-UCC, trnH-GUG(2),trnI-CAU(2),trnI-GAU*(2), trnK-UUU*,trnL-CAA(2),trnL-UAA*, trnL-UAG,trnM-CAU,trnN-GUU(2), trnP-UGG,trnQ-UUG,trnR-ACG(2), trnR-UCU,trnS-GCU,trnS-GGA,trnS-UGA, trnT-GGU,trnT-UGU,trnV-GAC(2),trnV-UAC*, trnW-CCA,trnY-GUA,trnfM-CAU |
|
| other genes | Maturase | matK |
| Protease | clpP** | |
| Envelope membrane protein | cemA | |
| Acetyl-CoA carboxylase | accD | |
| c-type cytochrome synthesis gene | ccsA | |
| Translation initiation factor | infA | |
| Genes of unknown function | Conserved open reading frames | ycf1, ycf2, ycf2-D2, ycf3, ycf4, |
*: one intron; **: two intron.
3.2. Bias of Codon Usage
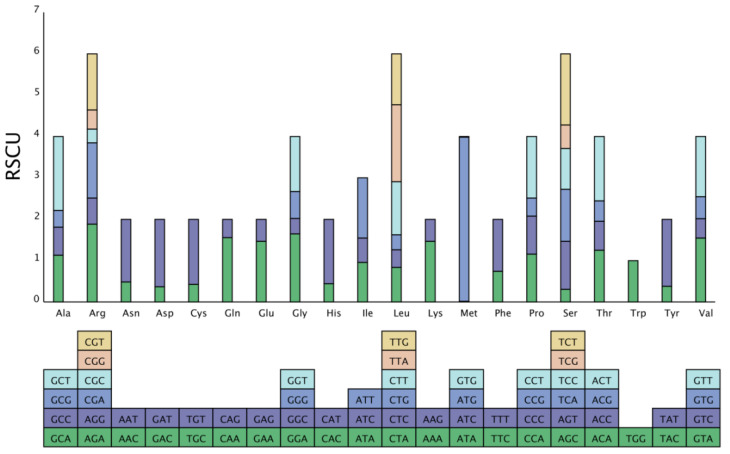
Genes from different species or within the same species show different codon usage bias modes, and the current oscillating use of unbalanced codons in biology is called codon usage bias, which helps in better understanding the environmental adaptability and molecular evolution of organisms. In this study, 26,108 codons were identified in all protein-coding sequences. Ile had the highest number (2276) of amino acids, while Met had the lowest number (85) of amino acids. Sixty-eight codons were identified with an RSCU > 1 (Figure 3).
Figure 3.
Codon content of 20 amino acids and stop codons in all protein-coding genes of the G. gandavensis chloroplast genome. The codons are represented by different colors in the histogram.
3.3. Microsatellite Polymorphisms
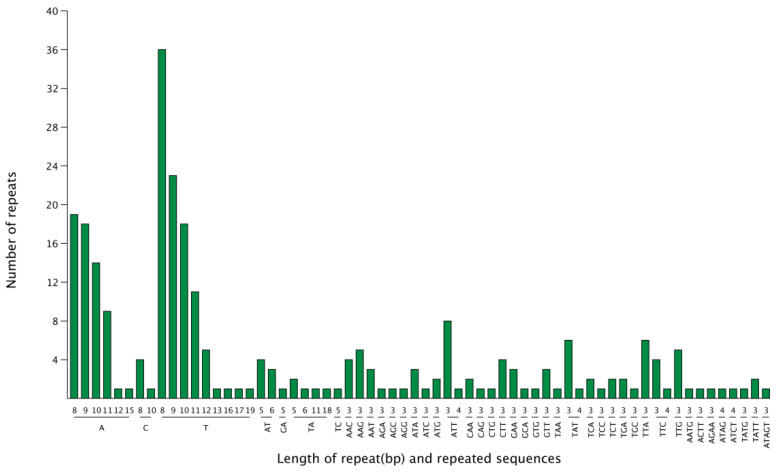
Microsatellite polymorphisms (i.e., simple sequence repeats (SSRs)) were identified in the chloroplast genome of G. gandavensis, and distributed in the two different types of regions. There were 171 SSRs located in the LSC regions (64.6%), while 47 (17.7%) and 44 (16.6%) SSRs were located in the SSC regions and IR regions, respectively. In this study, 265 SSRs were identified in the G. gandavensis chloroplast genome. Among them, 164 were mononucleotides, 14 were dinucleotides, 78 were trinucleotides, 8 were tetranucleotides, and 1 was a pentanucleotide (Figure 4).
Figure 4.
The number of SSRs repeats in the chloroplast genome of G. gandavensis. The number of repeats is represented by the green bar charts, and the x-ray represents the length of the repeat (bp) and repeated sequence.
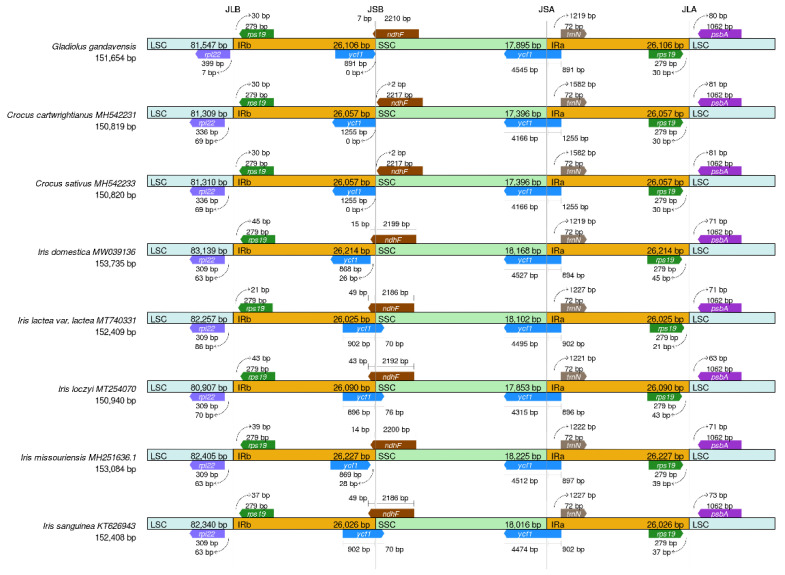
3.4. IR Expansion and Contraction
The IRs serve as integral components of maintaining the stability of the chloroplast genome, as loss of IRs could result in changes in the chloroplast genome [38]. Previous reports have indicated that IR expansion and contraction occur in many plant species [39]. In this study, the IR regions and the junction sites of the LSC and SSC regions in the chloroplast genomes of eight Iridaceae family members (including G. gandavensis) were analyzed (Figure 5). The results showed that the IR regions ranged from 150,819 bp in C. sativus to 153,735 bp in I. domestica. In addition, the ycf1 gene was located at the SSC/IRa junction in all the chloroplast genomes of different species; however, the ycf1 gene was missing in the Iris missouriensis and I. domestica, but was located at the SSC/IRb junction in other chloroplast genomes. Notably, the coding region of rpl22 was located at the LSC/IRb junction of all the chloroplast genomes, which resulted in the generation of 7, 69, 69, 63, 86, 70, 63, or 63 bp at the LSC/IRb border, respectively.
Figure 5.
The results of comparing the borders of the LSC, SSC, and IR regions among eight Iridaceae chloroplast genomes: I. lactea (MT740331), I. sanguinea (KT626943), I. loczyi (MT254070), I. missou-riensis (MH251636), C. sativus (MH542233), C. cartwrightianus (MH542231), and I. domestica (MW039136). Genes located at the IRa/b junctions are represented by colored boxes above (sense), while gene segments represent the boxes below (antisense) the horizontal line.
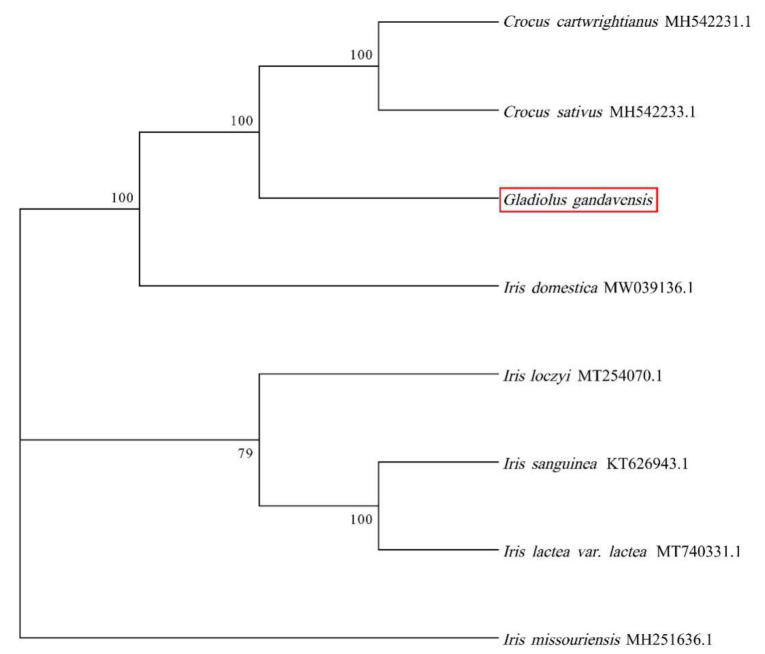
3.5. Phylogenetic Analysis
A phylogenetic tree of eight Iridaceae species was constructed by the GTRGAMMA model. The results showed that G. gandavensis had high homology with C. cartwrightianus and C. sativus, followed by I. domestica and other Iris species. Therefore, we speculate that Gladiolus has high homology with Crocus (Figure 6).
Figure 6.
The phylogenetic tree was constructed based on the eight monocotyledons chloroplast genomes: I. lactea (MT740331), I. sanguinea (KT626943), I. loczyi (MT254070), I. missou-riensis (MH251636), C. sativus (MH542233), C. cartwrightianus (MH542231), and I. domestica (MW039136). The red box represented the chloroplast genome of G. gandavensis which was sequenced in this study. The GTRGAMMA model of NJ was used in this study. Bootstrap replicates = 1000.
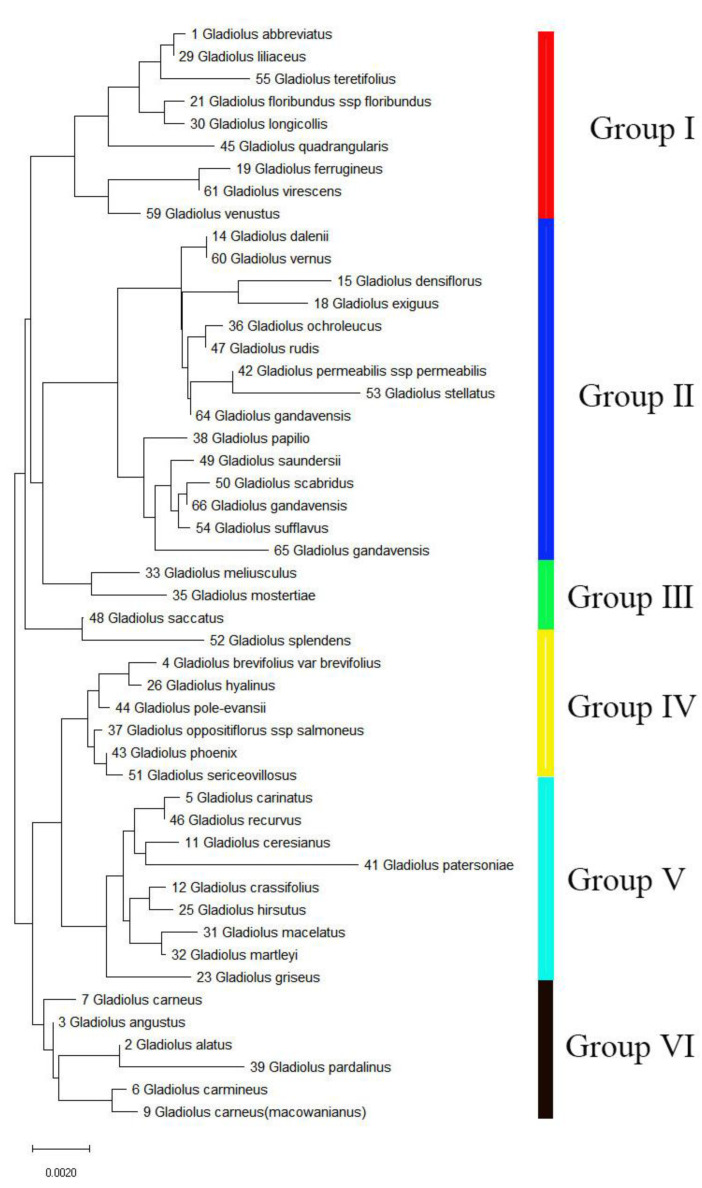
3.6. Genetic Relationship Analysis of Gladiolus
For further study, we amplified and sequenced the rbcl segment (cpDNA region) of the 49 Gladiolus species to analyze the genetic relationship in Gladiolus. Then, we obtained a phylogenetic tree by using the neighbor-joining (NJ) approach (Figure 7). The results showed that the three Chinese species (64, 65, and 66) all clustered into Group II. Specifically, species 66 had high homology with 50 and 54, 65 had high homology with the three species (50, 54, and 65), and 49, and 64 had high homology with 42 and 53. The results indicated that these species may be closely related to the three species.
Figure 7.
The phylogenetic tree of 49 Gladiolus samples was constructed based on the rbcl segment. The neighbor-joining (NJ) method was used to constructed the phylogenetic tree. Bootstrap replicates = 1000.
4. Discussion
4.1. The Chloroplast Genome of G. gandavensis
The complete chloroplast genome of G. gandavensis was assembled. Then, the sequence data were submitted to the NCBI database, under the GenBank number (OM304631). The structure and characteristics of the G. gandavensis chloroplast genome were analyzed in this study, and the results were consistent with the traits of most angiosperms. In this study, only the clpP and rps12 genes included two introns. Research on the clpP gene indicated that it plays an important role in plant chloroplasts, which are the proteolytic subunits of the ATP-dependent Clp protease [40,41,42]. The rps12 gene is the most unique of all the chloroplast genes and is composed of two parts that are far apart in the genome [43]. Therefore, the study of these two genes would help in understanding the evolutionary process of genes and the genomic characteristics of G. gandavensis.
The sequence analysis among G. gandavensis and Iridaceae speciesCodon usage bias could avert transcriptional errors in the chloroplast genome by affecting the amino acid functions [44,45,46]. This research showed that the content of bases A and T in mononucleotide SSRs in the chloroplast genome of G. gandavensis was 96.95% on average, which was the most frequent. This result is consistent with that of previous reports for most angiosperm chloroplast genomes [47]. Notably, repetitive sequences could be used in phylogenetic studies and genome rearrangements [48]. A comparative analysis of eight Iridaceae plants was conducted. The expansion or contraction of the IR regions in this study indicates that the ycf1 gene was located at the SSC/IRa and SSC/IRb junctions, and this result will help in exploring the evolution of the chloroplast genome in G. gandavensis. Additionally, the phylogenetic tree of eight Iridaceae species indicated that G. gandavensis had higher homology with Crocus than with Iris species. These results show that G. gandavensis is closely related to the Crocus, which would cause similar molecular evolutionary mechanisms.
4.2. The Evolutionary Genetics of G. gandavensis
In addition, we obtained the rplc sequence from the 49 Gladiolus samples to analyze the evolution of Gladiolus, and then, constructed a phylogenetic tree by the NJ approach in this study. The results showed that 42, 49, 50, and 54 samples had high homology with the three Chinese species (64, 65, and 66), which may indicate a close evolutionary relationship. Regain was used with AFLP markers to analyze the genetic relationship of 54 Gladiolus cultivars, and the results showed that most of the exotic cultivars as well as indigenous cultivars were closely related to each other. This might be due to a chance similarity in their genotypes [49]. Therefore, we speculate that G. gandavensis originated from South Africa. Our results are similar to the previously mentioned studies; however, the specific relationship still needs further exploration. In the future, a study on population genetics, species identification, and conservation biology of Gladiolus may be conducted.
5. Conclusions
The chloroplast genome of G. gandavensis was assembled by Illumina sequencing technology. The sequence information was deposited into the NCBI database under the GenBank number (OM304631). By comparing its structure with that of other Iridaceae species, we found that Gladiolus had a higher homology with Crocus than with Iris. A study on the theoretical relationship among the Gladiolus species based on the rbcl chloroplast genome sequence will provide reference information for relationship homology, germplasm resource preservation, and sustainable use of these Gladiolus species.
Author Contributions
Design of the experimental approach and writing of the original draft, R.Q.; main experiments, data analysis and writing of the original draft, Y.Y.; image preparation, X.M.; article modification, Q.H.; provision of materials, J.Z. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
Zhejiang Forestry Science and Technology Promotion Project (2021B06); Wenzhou Major Scientific and Technological Innovation Projects (ZS2020002).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Myouga F. A Heterocomplex of Iron Superoxide Dismutases Defends Chloroplast Nucleoids against Oxidative Stress and is Essential for Chloroplast Development in Arabidopsis. Plant. Cell. 2008;20:11. doi: 10.1105/tpc.108.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar P.N., Raju D.V.S. Dormancy in Gladiolus: The Cause and Remedy—A Review. Agric. Rev. 2007;28:309–312. [Google Scholar]
- 3.Wang D., Qi Y., Li B., Zhang G. A New Anthraquinone from Gladiolus Gandavensis. Nat. Prod. Res. 2003;17:365–368. doi: 10.1080/14786410310001605940. [DOI] [PubMed] [Google Scholar]
- 4.Tai Z., Yang X., Le C., Sun W., Yang Y. Studies on the Chemical Constituents from The Aerial Parts of Gladiolus gandavensis. J. Chin. Med. Mater. 2010;33:1257–1259. [PubMed] [Google Scholar]
- 5.Yamagami T., Mine Y., Ishiguro M. Complete Amino Acid Sequence of Chitinase-a from Bulbs of Gladiolus (Gladiolus gandavensis) Biosci. Biotech. Bioch. 2014;62:386–389. doi: 10.1271/bbb.62.386. [DOI] [PubMed] [Google Scholar]
- 6.Zhao L., Jiang X.W., Zuo Y.J., Liu X.L., Chin S.W., Haberle R., Wen J. Multiple Events of Allopolyploidy in The Evolution of the Racemose Lineages in Prunus (Rosaceae) Based on Integrated Evidence from Nuclear and Plastid Data. PLoS ONE. 2016;11:e0157123. doi: 10.1371/journal.pone.0157123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schubert M., Petersson U., Haas B. Proteome Map of the Chloroplast Lumen of Arabidopsis thaliana. J. Biol. Chem. 2002;277:8354–8365. doi: 10.1074/jbc.M108575200. [DOI] [PubMed] [Google Scholar]
- 8.Ohyama K. Chloroplast and Mitochondrial Genomes from A Liverwort, Marchantia polymorpha--Gene Organization and molecular evolution. J. Agric. Chem. Soc. Jpn. 1996;60:16–24. doi: 10.1271/bbb.60.16. [DOI] [PubMed] [Google Scholar]
- 9.Ferris C., Oliver R., Davy A., Hewitt G. Using Chloroplast DNA to Trace Postglacial Migration Routes of Oaks into Britain. Mol. Ecol. 2010;4:731–738. doi: 10.1111/j.1365-294X.1995.tb00273.x. [DOI] [PubMed] [Google Scholar]
- 10.James C., Gibby M., Barrett J. Molecular Studies in Pelargonium (Geraniaceae). A Taxonomic Appraisal of Section Ciconium and the Origin of the ‘Zonal’ and ‘Ivy-leaved’ cultivars. Plant Syst. Evol. 2004;243:131–146. doi: 10.1007/s00606-003-0074-2. [DOI] [Google Scholar]
- 11.Heath R., Packer L. Photoperoxidation in Isolated Chloroplast I. Kinetics and Stoichiometry ofFatty Acid Peroxidation. Arch. Biochem. Biophys. 1968;125:189–198. doi: 10.1016/0003-9861(68)90654-1. [DOI] [PubMed] [Google Scholar]
- 12.Kasahara M., Swartz T.E., Olney M.A., Onodera A., Mochizuki N., Fukuzawa H., Briggs W.R. Photochemical Properties of the Flavin Mononucleotide-Binding Domains of the Phototropins from Arabidopsis, rice, and Chlamydomonas reinhardtii. Plant. phys. 2002;129:762–773. doi: 10.1104/pp.002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kagawa T. Arabidopsis NPL1: A Phototropin Homolog Controlling the Chloroplast High-Light Avoidance Response. Science. 2001;291:2138–2141. doi: 10.1126/science.291.5511.2138. [DOI] [PubMed] [Google Scholar]
- 14.Heckathorn S.A. The Small, Methionine-Rich Chloroplast Heat-Shock Protein Protects Photosystem II Electron Transport during Heat Stress. Plant Phys. 1998;116:439–444. doi: 10.1104/pp.116.1.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyer J., Potter J. Chloroplast Response to Low Leaf Water Potentials: IV. Quantum Yield is Reduced. Plant Physiol. 1973;51:989–1092. doi: 10.1104/pp.51.6.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamburino R., Vitale M., Ruggiero A. Chloroplast Proteome Response to Drought Stress and Recovery in Tomato (Solanum lycopersicum L.) BMC Plant Biol. 2017;17:40. doi: 10.1186/s12870-017-0971-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mittova V., Tal M., Volokita M. Salt Stress Induces Up-Regulation of an Efficient Chloroplast Antioxidant System in the Salt-tolerant Wild tomato Species Lycopersicon pennellii but not in theCultivated Species. Physiol. Plant. 2010;115:3. doi: 10.1034/j.1399-3054.2002.1150309.x. [DOI] [PubMed] [Google Scholar]
- 18.Siddhartha D. Role of Temperature Stress on Chloroplast Biogenesis andProtein Import in Pea. Plant Physiol. 2009;150:1050–1061. doi: 10.1104/pp.109.137265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T. The Complete Nucleotide Sequence of the Tobacco Chloroplast Genome: Its Gene Organization and Expression. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiratsuka J., Shimada H., Whittier R., Ishibashi T., Sakamoto M., Mori M. The Complete Sequence of the rice (Oryza sativa) Chloroplast Genome: Intermolecular Recombination between Distinct tRNA Genes Accounts for a Major Plastid DNA Inversion during the Evolution of the Cereals. Mol. Gen. Genet. 1989;217:185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- 21.Maier R., Kai N., Igloi G. Complete Sequence of the Maize Chloroplast Genome: Gene Content, Hotspots of Divergence and Fine Tuning of Genetic Information by Transcript Editing. J. Mol. Biol. 1995;251:614. doi: 10.1006/jmbi.1995.0460. [DOI] [PubMed] [Google Scholar]
- 22.Ni Z., Zhou P., Xin Y., Xu M., Xu L.A. Parent–offspring variation transmission in full-sib families revealed predominantly paternal inheritance of chloroplast DNA in Pinus massoniana (Pinaceae) Tree Genet. Genomes. 2021;17:36. doi: 10.1007/s11295-021-01519-6. [DOI] [Google Scholar]
- 23.Palmer J., Stein D. Conservation of chloroplast genome structure among vascular plants. Curr. Genet. 1986;10:823–833. doi: 10.1007/BF00418529. [DOI] [Google Scholar]
- 24.Zhang X., Gu C., Zhang T., Tong B., Zhang H., Wu Y., Yang C. Chloroplast (Cp) Transcriptome of P. davidiana Dode× P. bolleana Lauch provides insight into the Cp drought response and Populus Cp phylogeny. BMC Evol. Biol. 2020;20:51. doi: 10.1186/s12862-020-01622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McPherson H., Merwe M., Delaney S., Edwards M., Henry R., McIntosh E. Capturing Chloroplast Variation for Molecular Ecology Studies: A Simple Next Generation Sequencing Approach Applied to a Rainforest Tree. BMC Ecol. 2013;13:8. doi: 10.1186/1472-6785-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruce B.D. Chloroplast Transit Peptides: Structure, Function and Evolution. Trends Cell. Biol. 2000;10:440–447. doi: 10.1016/S0962-8924(00)01833-X. [DOI] [PubMed] [Google Scholar]
- 27.Liu B., Xie Y., Li Z., Liu Y. SOAPdenovo2: An Empirically Improved Memory-Efficient Short-Read de novo Assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu M., Guo L., Gu S. TGS-GapCloser: A Fast and Accurate Gap Closer for Large Genomes with Low Coverage of Error-Prone Long Reads. GigaScience. 2020;9:9. doi: 10.1093/gigascience/giaa094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuravadi N., Tiwari P., Tanwar U. Identification and Characterization of EST-SSR Markers in Cluster Bean (Cyamopsis spp.) Crop. Sci. 2013;54:3. doi: 10.2135/cropsci2013.08.0522. [DOI] [Google Scholar]
- 30.Standley D. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013;30:772. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darling A., Mau B., Blattner F. Mauve: Multiple Alignment of Conserved Genomic Sequence with Rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurtz S. The Vmatch Large Scale Sequence Analysis Software-a Manual. Center. Bioinform. 2010;170:391–392. [Google Scholar]
- 33.Yu Z.J. KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinform. 2010;8:77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dempster A. Maximum-Likelihood from Incomlete Data via the Em Algorithm. J. R. Stat. Soc. Ser. B. 1977;39:1–38. [Google Scholar]
- 35.Huang B., Wang Z., Huang J., Li X., Zhu H., Wen Q., Xu L. Population Genetic Structure Analysis Reveals Significant Genetic Differentiation of the Endemic Species Camellia chekiangoleosa Hu. with a Narrow Geographic Range. Forests. 2022;13:234. doi: 10.3390/f13020234. [DOI] [Google Scholar]
- 36.Ni Z., Ye Y., Bai T., Xu L. Complete Chloroplast Genome of Pinus massoniana (Pinaceae): Gene Rearrangements, Loss of ndh Genes, and Short Inverted Repeats Contraction, Expansion. Molecules. 2017;22:1528. doi: 10.3390/molecules22091528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Backeljau T., de Bruyn L., de Wolf H., Jordaens K., van Dongen S., Winnepennincks B. Multiple UPGMA and Neighbor-Joining Trees, and the Performance of Some Computer Packages. Mol. Biol. Evol. 1966;13:2. doi: 10.1093/oxfordjournals.molbev.a025590. [DOI] [Google Scholar]
- 38.Lee E., Smith D., Fanwick P., Byrn S. Characterization and Anisotropic Lattice Expansion/Contraction of Polymorphs of Tenofovir Disoproxil Fumarate. Cryst. Growth. Des. 2010;10:2314–2322. doi: 10.1021/cg1000667. [DOI] [Google Scholar]
- 39.Dong W., Chao X., Tao C., Zhou S., Nicolas S. Complete Chloroplast Genome of Sedum sarmentosum and Chloroplast Genome Evolution in Saxifragales. PLoS ONE. 2013;8:e77965. doi: 10.1371/journal.pone.0077965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J., Hartling J., Flanagan J. The structure of ClpP at 2.3 AResolution Suggests a Model for ATP-Dependent Proteolysis. Cell. 1997;91:447. doi: 10.1016/S0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 41.Hricová A., Quesada V., Micol J.L. The SCABRA3 Nuclear Gene Encodes the Plastid RpoTp RNA Polymerase, which is Required for Chloroplast Biogenesis and Mesophyll Cell Proliferation in Arabidopsis. Plant Phys. 2006;141:942–956. doi: 10.1104/pp.106.080069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haynes C., Petrova K., Benedetti C., Yun Y., Ron D. ClpP Mediates Activation of a Mitochondrial Unfolded Protein Response in C. elegans. Dev. Cell. 2007;13:467–480. doi: 10.1016/j.devcel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 43.Liu X., Gillham N., Boynton J. Chloroplast Ribosomal Protein Gene rps12 of Chlamydomonas Reinhardtii. Wild-Type Sequence, Mutation to Streptomycin Resistance and Dependence, and Function in Escherichia coli. J. Biol. Chem. 1989;264:16100. doi: 10.1016/S0021-9258(18)71592-5. [DOI] [PubMed] [Google Scholar]
- 44.Kawabe A., Miyashita N. Patterns of codon usage bias in three dicot and four monocot plant species. Genes Genet. Syst. 2003;78:343–352. doi: 10.1266/ggs.78.343. [DOI] [PubMed] [Google Scholar]
- 45.Shackelton L., Parrish C., Holmes E. Evolutionary Basis of Codon Usage and Nucleotide Composition Bias in Vertebrate DNA Viruses. J. Mol. Evol. 2006;62:551–563. doi: 10.1007/s00239-005-0221-1. [DOI] [PubMed] [Google Scholar]
- 46.Dong F., Lin Z., Lin J. Chloroplast Genome of Rambutan and Comparative Analyses in Sapindaceae. Plants. 2021;10:283. doi: 10.3390/plants10020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J., Jin Z., Tan T. Genetic Diversity and Differentiation of Sinocalycanthus Chinensis Populations Revealed by Chloroplast Microsatellite (cpSSRs) Markers. Biochem. Syst. Ecol. 2012;41:48–54. doi: 10.1016/j.bse.2011.12.019. [DOI] [Google Scholar]
- 48.Cesare M., Hodkinson T., Barth S. Chloroplast DNA Markers (cpSSRs, SNPs) for Miscanthus, Saccharum and Related grasses (Panicoideae, Poaceae) Mol. Breed. 2010;26:539–544. doi: 10.1007/s11032-010-9451-z. [DOI] [Google Scholar]
- 49.Ranjan P., Bhat K.V., Misra R.L., Singh S.K., Ranjan J.K. GeneticRelationships of Gladiolus Cultivars Inferred from Fluorescence based AFLP Markers. Sci. Hortic. 2010;123:562–567. doi: 10.1016/j.scienta.2009.11.013. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.