Abstract
Genetic testing for SMA diagnosis, newborn screening, and carrier screening has become a significant public health interest worldwide, driven largely by the development of novel and effective molecular therapies for the treatment of spinal muscular atrophy (SMA) and the corresponding updates to testing guidelines. Concurrently, understanding of the underlying genetics of SMA and their correlation with a broad range of phenotypes and risk factors has also advanced, particularly with respect to variants that modulate disease severity or impact residual carrier risks. While testing guidelines are beginning to emphasize the importance of these variants, there are no clear guidelines on how to utilize them in a real-world setting. Given the need for clarity in practice, this review summarizes several clinically relevant variants in the SMN1 and SMN2 genes, including how they inform outcomes for spinal muscular atrophy carrier risk and disease prognosis.
Keywords: spinal muscular atrophy, carrier screening, diagnosis, SMN1, SMN2
1. Spinal Muscular Atrophy Disease Etiology
Spinal muscular atrophy (SMA) is an autosomal recessive neuromuscular disease caused by loss of survival motor neuron 1 (SMN1) gene function and is a primary genetic cause of infant death [1]. SMA is a rare disease with a pan-ethnic incidence of ~1/11,000 live births and a high carrier rate of ~1/54 [2]. SMA is divided into clinical types based on the age of onset and maximum motor milestone achievement, with a gradient of phenotypes ranging from never sitting unassisted, with onset prior to six months of age, to adult-onset mild muscular weakness. Most SMA patients are classified into three main types in order of decreasing severity: type 1 (~60% of patients), type 2 (~30% of patients), and type 3 (~10% of patients). Rarer SMA types, such as type 0 and type 4, also exist [3,4,5].
Bi-allelic loss of the SMN1 gene is the cause of disease in ~95% of patients with SMA. The remaining 5% of patients are compound heterozygotes, with an SMN1 deletion on one chromosome and a loss-of-function point mutation in SMN1 on the other chromosome. The vast majority (~98%) of SMA patients inherit the SMN1 alterations from their parents [6,7]. SMA carriers lack a functional SMN1 copy on a single chromosome and frequently have one functional copy on the other (1 + 0). However, a cis carrier genotype with two SMN1 copies on a single chromosome (2 + 0), commonly referred to as a silent carrier, is also well-documented [8]. In one study examining a large North American population, the detection rate of SMA carriers using SMN1 copy number alone varied from ~71% to 95% depending on ethnicity [9]. Most of the missed carriers were due to silent carriers (2 + 0) that cannot be resolved from wild-type (1 + 1) individuals solely based on copy number, since results would be 2 SMN1 copies for both genotypes [9]. While gene conversion from SMN2 to SMN1 is known to occur and is one potential cause for the silent carrier (2 + 0) genotype [8], the clinical significance of gene conversions is not fully understood. Recent studies have shown that variants c.*3+80T>G and c.*211_*212del in SMN1 (Figure 1A) are associated with SMN1 duplication in many ethnic groups and their presence informs the risk of silent carrier SMN1 genotypes (2 + 0) to varying degrees depending on ethnicity [10,11].
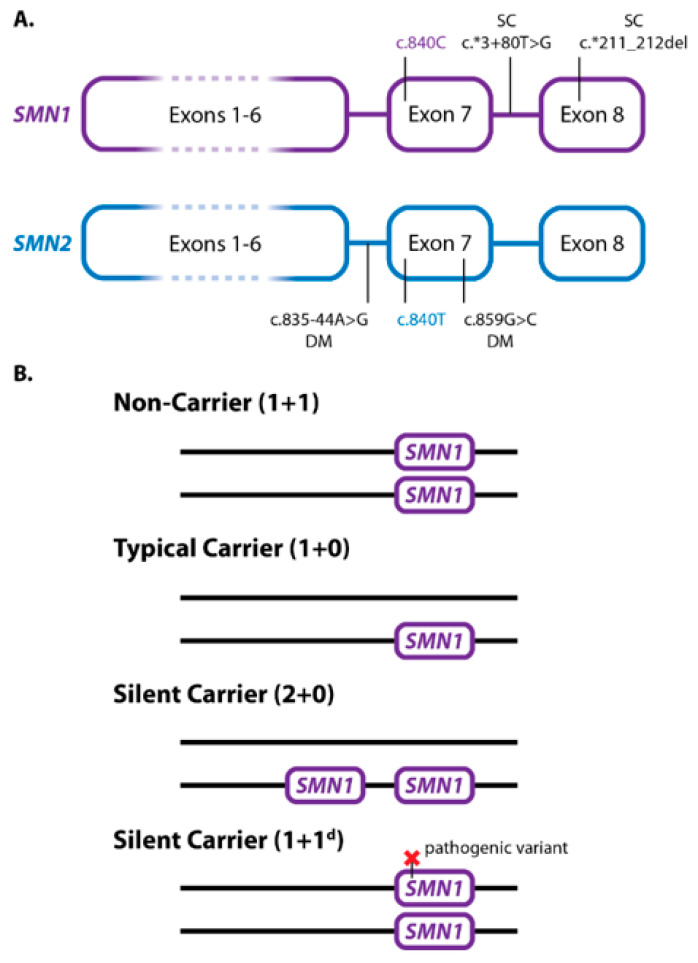
Figure 1.
Genetics of SMN1, SMN2, and SMA Carriers. (A) Silent carriers and disease-modifying variants in SMN1 and SMN2. Nucleotides at position c.840 in exon 7, typically used to distinguish SMN1 and SMN2, are indicated by color (PSVs). Gene duplication variants in SMN1 associated with 2 + 0 silent carriers are indicated by the letters SC. Common disease modifier variants in SMN2 are indicated by the letters DM. (B) SMA carrier genetics. Non-carriers typically have one copy of SMN1 on each chromosome. Typical carriers have only one SMN1 copy, lacking SMN1 on the other chromosome. Silent carriers (2 + 0) often have two copies of SMN1 on a single chromosome, lacking SMN1 on the other chromosome. Silent carriers can also have one copy of SMN1 on both chromosomes but with a pathogenic variant in one copy.
SMA disease severity inversely correlates with SMN2 copy number, meaning the more copies of SMN2, the less severe the phenotype [5]. SMN1 and SMN2 differ in 16 paralogue sequence variants (PSVs) [12]. One PSV, c.840C>T, disrupts a splice enhancer that decreases the number of exon 7 containing mRNAs to 10–20%, which results in a significantly reduced amount of functional SMN protein compared to that made from a functional SMN1 gene. However, due to complete homology with the SMN1-associated SMN protein sequence, SMN2-generated SMN protein levels offer a compensatory effect, thus resulting in lessened disease severity with increased SMN2 copies. Though the SMN2 copy number is vital for assessing disease severity, there are also a few variants known to be SMA disease modifiers. Specifically, c.859G>C in SMN2 (Figure 1A) is linked to improved splicing efficiency of SMN2 by 20%, which also leads to reduced disease severity [13,14]. Indeed, 44 SMA patients carrying the c.859G>C variant have been described, all of whom presented a milder phenotype than expected according to their SMN2 copies. This variant has been described in various populations, showing a common haplotype that points towards a common ancestral origin [12]. Thus, SMN1 is associated with molecular SMA diagnosis and carrier status, whereas SMN2 is associated with the severity of the disease.
2. SMA Diagnostic and Carrier Screening Testing
Copy number analysis for SMN1 and SMN2 genes associated with SMA can be difficult, as the copy number of these varies much more than other regions within the genome. Furthermore, rapid turnaround time for SMA diagnostic testing is important for timely administration of therapies which halt neuron degeneration [15,16]. SMA genetic testing for SMN1 and SMN2 exon 7 copy numbers is accomplished using a variety of methods, including PCR followed by capillary electrophoresis (PCR/CE), quantitative PCR (qPCR), digital droplet PCR (ddPCR), multiplex ligation-dependent probe amplification (MLPA), and next-generation sequencing (NGS). These methods have recently been extensively described, including the strengths and weaknesses of each approach [17]. PCR-based systems are generally the fastest and simplest methods, though qPCR and ddPCR assays require separate reactions for each gene, and qPCR requires the generation of a standard curve, which can limit throughput. MLPA and PCR/CE both provide copy numbers for SMN1 and SMN2, but MLPA has a longer and more complex workflow, requiring at least 24 h to complete as compared to PCR/CE, which can be completed in a few hours [17]. While PCR/CE is restricted to quantifying exon 7 and intron 7 from SMN1 and SMN2, MLPA quantifies all exons in these genes, which can reveal partial gene deletions. NGS provides the most comprehensive analysis for variants, hybrid genes, and partial deletions, but the workflow can be laborious, time-intensive, and requires complex instrumentation. Furthermore, NGS analysis and interpretation requires significant hardware resources and bioinformatics expertise, especially for SMN1 and SMN2 analysis, given the high homology between the genes and high variability in potential copy numbers [18]. Recently, a more focused NGS method to analyze these genes provides full characterization of the SMN region in an affordable manner [19].
Traditionally, testing for SMN1 exon 7 copy number alone is used for SMA diagnosis. However, a deletion of exon 8 alone has been reported in milder SMA types in two patients [20]. In addition, exon 8 information may have utility for the detection of hybrid genes, depending on the testing methodology [21,22]. Although typical SMN1/2 hybrids involving exon 7 and exon 8 are the most common reported in the literature [6,23], hybrid genes may also be detected using other loci that differentiate SMN1 and SMN2, for example, by comparing exon 7 and intron 7 [24] or involving intron 6 to exon 8 [19].
In addition to copy numbers, some methods are also able to detect variants in the SMN1 and SMN2 genes associated with silent carrier risk and disease severity, as detailed in the following sections. In short, the test methodology should balance the need for the right information to guide clinical care in the shortest possible timeframe with practical constraints such as the availability of instrumentation, personnel, and other resources.
3. SMA Carrier Genotypes, Testing, and Reporting
An SMA carrier is an asymptomatic individual lacking a functional copy of SMN1 on one chromosome. Most SMA carriers have an SMN1 deletion on one chromosome and one functional SMN1 copy on the other (1 + 0), representing a heterozygous deletion (Figure 1B). Silent carriers, in contrast, have a (2 + 0) genotype, whereas others may have another type of pathogenic variant in SMN1 on one chromosome and two SMN1 copies (1d + 1), or rarer genotypes with higher SMN1 copy numbers (1d + 2, 3 + 0) [8]. Due to these multiple genotypes, the detection rate of SMA carriers using the SMN1 copy number alone to detect (1 + 0) genotypes varies from ~71% up to 95% depending on ethnicity [9]. Thus, there is a proportion of false-negative results for carrier status when reporting only the SMN1 copy number. Residual carrier risk estimations based on the SMN1 copy number alone have been calculated by compiling results across multiple studies and ethnicities (Table 1, first four columns) [25]. Since the total SMN1 copy number is used to assess carrier risk, the limitations of such testing, specifically the inability to detect silent carriers using SMN1 copy number alone, should be described when reporting results [8].
Table 1.
Residual SMA carrier risk estimates by ethnicity based on SMN1 copy number and gene duplication variant status. Carrier frequency represents carrier risk without testing by ethnicity. Subsequent columns estimate residual risk based on SMN1 copy number alone. The last two columns estimate the residual risk with two copies of SMN1 with additional information on the presence of SMN1 gene duplication variants (SMN1 c.*3+80T>G and c.*211_*212del), where “positive” indicates presence of one or both variants, and “negative” indicates absence of both variants. Values are rounded to the nearest integer. Asian includes groups with South Asian and East Asian ancestry.
| Ethnicity | Carrier Frequency | 2 Copies SMN1 Exon 7 | 3 Copies SMN1 Exon 7 | 2 Copies SMN1, Variant Status “Negative” | 2 Copies SMN1, Variant Status “Positive” |
|---|---|---|---|---|---|
| Ashkenazi Jewish | 1:56 a | 1:514 a | 1:5899 a | 1:580 b | ~1 b |
| Asian | 1:50 a | 1:719 a | 1:5185 a | 1:779 c | 1:57 c |
| African American/Black | 1:71 a | 1:132 a | 1:6997 a | 1:375 d | 1:39 d |
| Caucasian/European | 1:45 a | 1:604 a | 1:4719 a | 1:814 c | 1:12 c |
| Hispanic | 1:83 a | 1:641 a | 1:7574 a | 1:906 d | 1:99 d |
| Spanish | 1:40 e | 1:781 e | Not Reported | 1:888 e | ~1 e |
| Israeli Jewish | 1:38 a | 1:450 a | 1:4004 a | Not Reported | Not Reported |
| Asian Indian | 1:50 a | 1:428 a | 1:5252 a | Not Reported | Not Reported |
| Iranian | 1:16 a | 1:96 a | 1:1604 a | Not Reported | Not Reported |
In addition to the SMN1 copy number, data has shown that the presence of SMN1 gene duplication variants c.*3+80T>G in intron 7 and c.*211_*212del in exon 8 (Figure 1A) can be indicative of the silent carrier (2 + 0) genotype in many ethnicities [10,11]. Several test methods can detect these variants, including MLPA (P-460), NGS, PCR/Sanger, and PCR/CE [12,18,19,24,26,27]. Typically, these variants co-occur [10]; however, individuals with only one of the two variants have been identified [11]. Detection of either c.*3+80T>G or c.*211_*212del alone is generally considered indicative of SMN1 gene duplication, and thus associated with increased silent carrier risk [10]. However, c.*211_*212del in exon 8 has been detected in SMN2 hybrid genes in SMA patients with no copies of SMN1, indicating that it is possible that an isolated occurrence of either can be associated with a hybrid gene [11].
In response to characterization of the SMN1 gene duplication variants across multiple ethnicities, guidelines have been updated to reflect that these variants improve residual risk estimates [28]. Table 1 (last two columns) summarizes these results across several studies, which can be used to provide an estimate of residual risk based on ethnicity. The impact of these variants has not been evaluated in all ethnicities, and some studies show varying residual risk levels within an ethnicity [10,18,29]. This is likely due to both the broad range of ethnic backgrounds included in each category and the fact that ethnicities are often self-reported, which creates ambiguity in how these groups are classified and reported [30]. Consequently, the numbers shown here represent risk estimations from studies with the largest number of individuals analyzed for each ethnicity, recognizing that while these are the best estimations available, they are not exact figures. Continued research is needed to further refine both diagnostic interpretations and residual risk values for different genetic ancestries, so literature should be reviewed regularly [31].
The absence of these gene duplication variants does not rule out the possibility of a carrier (2 + 0) genotype, nor does their presence definitively diagnose silent carriers across different ancestries. In these cases, the analysis of copy number in the progenitors of the carrier under study would help to determine the cis or trans configuration of SMN1 genes, though this implies extra testing that is not always possible [11]. Nevertheless, resolution of SMN1 gene duplication variants modifies the residual risk of SMA carrier status in all ethnicities studied to date (Table 1). Therefore, co-occurrence of these variants with two copies of SMN1 indicates increased carrier risk, while absence of the variants with two copies of SMN1 indicates reduced carrier risk compared to using SMN1 copy number alone, regardless of ethnicity [10,11,18,28,29].
For reporting purposes, SMN1 gene duplication variant information is relevant only when two copies of SMN1 are present; variant interpretation is not necessary when a one SMN1 copy carrier genotype (1 + 0) is identified through SMN1 copy number testing. Furthermore, when three or more copies of SMN1 are present, interpretation of these variants is unnecessary given the extremely low likelihood of being a carrier [25]. In cases where ethnicity is unknown, uncertain, or unreported, a range of possible risk values may be provided and discussed in counseling patients, while noting that risk varies depending on ethnicity and, more specifically, ancestry [30]. To clarify potential reporting, examples of SMN1 copy number and gene duplication variant status results in a carrier screening setting are provided in Table 2 based on available guidelines [8,28]. See also Prior et al. 2011 for an example report [8].
Table 2.
Carrier Results Interpretation Examples. The examples provided here are interpretations based on relevant guidelines [8,11] and literature [10,11,18,25,29]. When interpreting and presenting results, all relevant local guidelines and regulations should be followed.
| Example Results | SMN1 Copies | c.*3+80T>G | c.*211_ *212del |
Interpretation |
|---|---|---|---|---|
| Case 1 | 1 | Not indicated | Not indicated | Carrier The SMN1 copy number indicates a carrier of SMA. Genetic counseling is recommended and carrier testing should be made available to other at-risk family members. |
| Case 2 | 2 | Positive | Negative | Increased Carrier Risk The SMN1 copy number is two, ruling out a typical carrier genotype (1 + 0). However, the presence of one or more variants indicates an increased risk of being a silent carrier. The residual risk of SMA carrier status based on genotype alone is between 1:99 to ~1 depending on ethnicity. Ethnic-specific risk values based on these results are provided (see Table 1, last column). Parental testing should be considered to elucidate the presence of a silent carrier (2 + 0). Genetic counseling is recommended and carrier testing should be made available to other at-risk family members. |
| Case 3 | 2 | Positive | Positive | Increased Carrier Risk Refer to Case 2 for example language. |
| Case 4 | 2 | Negative | Negative | Reduced Carrier Risk The SMN1 copy number and variant status indicate reduced, but not eliminated, carrier risk. The residual risk of SMA carrier status based on genotype alone is between 1:375 and 1:906 depending on ethnicity. Ethnic-specific risk values based on these results are provided (see Table 1, 2nd to last column). Genetic counseling is recommended. |
| Case 5 | 3 | At genetic counselor’s discretion | At genetic counselor’s discretion | Reduced Carrier Risk The SMN1 copy number indicates a significantly reduced, but not eliminated, carrier risk. The residual risk of SMA carrier status based on genotype is low. Ethnic-specific risk values based on these results are provided (see Table 1, Column 4). Genetic counseling is recommended. |
Since gene conversions are another mechanism that can lead to silent carriers [8], evidence of conversion from SMN2 to SMN1 (SMN1/2 hybrids) could inform silent carrier risk. However, this possibility has not been sufficiently investigated clinically, and hybrid genes have a variable gene architecture [32]. As a result, there is insufficient evidence to determine carrier risk based on hybrid genes.
4. Disease Prognosis Genotypes, Testing, and Reporting
While the SMN2 copy number is not relevant for the diagnosis of SMA, guidelines recommend that SMN2 copy number results be reported to inform prognosis and treatment decisions [17,33,34,35]. The SMN2 copy number is strongly correlated with SMA type, but the copy number alone is not sufficient to predict SMA type. This limitation should be clearly communicated when reporting SMN2 copy number results.
Additionally, the c.859G>C variant is a positive disease modifier associated with reduced disease severity and improved prognosis. Several test methods can detect this variant, including NGS, specific PCR/Sanger, and PCR/CE [19,24]. Evidence indicates that c.859G>C improves SMN2 splicing, exon 7 inclusion, and full-length SMN protein production, leading to improved phenotypic outcomes [13,14]. For instance, while 90% of individuals with SMA and two copies of SMN2 exon 7 typically have SMA type 1, individuals with SMA that have two copies of SMN2 exon 7 and the c.859G>C variant typically have SMA type 2 or type 3, with no known cases of SMA type 1 in individuals where this variant is present [13,14,33]. A similar effect has been observed in patients with three copies of SMN2 exon 7 and the c.859G>C variant, typically resulting in SMA type 3 [12,33]. The number of SMN2 copies with c.859G>C also correlates with phenotype, with multiple copies leading to milder phenotypes [12]. While the c.859G>C variant has not been reported in patients with one or four copies of SMN2, available evidence suggests that any individual with this variant would have a milder phenotype than expected based on SMN2 copy number alone.
In addition to c.859G>C, another positive modifier known as c.835-44A>G has been described (Figure 1A), albeit with limited investigation in SMA patients to date. This variant is one of the PSV differentiating SMN1 from SMN2, and its presence in intron 6 of SMN2 increases the inclusion of exon 7 [36]. This modifier can be detected with specific PCR/Sanger or NGS methods [12,19]. Other putative positive and negative disease modifiers have been described [15,17,32]. However, these variants have been identified only in a small number of patients without a clear genotype-phenotype correlation [19].
Aside from SNP and INDEL variants that impact disease prognosis, several recent publications have mentioned SMN1/2 hybrids as another positive disease modifier [15,37,38,39]. These hybrid genes arise when SMN1 is partially converted to SMN2 or vice versa. Since they retain elements of SMN1, some hybrids can increase exon 7 inclusion in SMN mRNAs compared to typical SMN2, producing greater quantities of full length SMN protein that lead to a milder phenotype [37,38]. However, SMN1/2 hybrids are heterogeneous, and their impacts on full-length SMN transcript and protein quantity are likely dependent on which SMN1 elements are retained [37]. More data are needed to inform the interpretation of hybrid genotypes beyond the general observation that SMN1/2 hybrids can be associated with milder phenotypes.
For reporting purposes, likely prognosis can be interpreted using SMN2 copy number alone when disease-modifying variants are not detected, noting that the correlation between genotype and phenotype is not absolute [8,34,35]. A positive result for c.859G>C may be reported as a marker associated with reduced severity and/or improved prognosis in comparison with the typical presentation based on the SMN2 copy number genotype. To clarify probable SMA types based on SMN2 copy number and c.859G>C, a summary of published treatment guidelines and peer-reviewed studies is provided in Table 3. This prognostic information is relevant only for individuals diagnosed with SMA. Examples for reporting SMN2 copy number and c.859G>C status when providing test results are provided. Other disease modifier variants such as c.835-44A>G or the presence of SMN2 hybrids can be reported when further research genetic studies are performed, mainly in discordant patients [15,17].
Table 3.
Likely SMA prognosis based on SMN2 copy number and variant status. SMN1 copy numbers are presumed to be 0, consistent with diagnosis. Genotypes not referenced below (e.g., 3 copies SMN2 with two or more c.859G>C alleles) have not yet been reported. The reporting examples provided here are interpretations based on consensus recommendations published by the American College of Medical Genetics (ACMG), Cure SMA, and the SMA Care group [8,34,35], as well as other relevant guidelines and literature [13,14,17,33]. For recommendations on follow-up testing and management of SMA cases as well as probability estimations of SMA type based on results, see [17]. When interpreting and presenting results, all relevant local guidelines and regulations should be followed.
| SMN2 Copy Number | c.859G>C Variant Status | Interpretation and Reporting Example |
|---|---|---|
| 1 | Negative | SMA (Type 0 probable) a Most individuals with SMA and one SMN2 copy present with Type 0 congenital disease. While the relationship between SMN2 copy number and disease outcomes is strongly correlated, it is not absolute, and individual exceptions do occur. Genetic counseling is recommended. |
| 2 | Negative | SMA (Type 1 probable) a Most individuals with SMA and two SMN2 copies present with Type 1 SMA. Refer to other examples with Negative c.859G>C Variant Status for example language. |
| 2 | Detected in one copy | SMA (Type 2/3 probable) b,c Whereas most individuals with SMA and two SMN2 copies present with Type 1 SMA, the presence of the c.859G>C variant in one SMN2 copy is associated with reduced severity consistent with SMA Type 2/3. Genetic counseling is recommended. |
| 2 | Detected in two copies | SMA (Type 3/4 probable) c,d Whereas most individuals with SMA and two SMN2 copies present with Type 1 SMA, the presence of the c.859G>C variant in two SMN2 copies is associated with reduced severity consistent with SMA Type 3/4. Genetic counseling is recommended. |
| 3 | Negative | SMA (Type 2/3 probable) a Refer to other examples with negative c.859G>C variant status for an example language. |
| 3 | Detected in one copy | SMA (Type 3 probable) c,e Whereas most individuals with SMA and three SMN2 copies present with Type 2/3 SMA, the presence of the c.859G>C variant in one SMN2 copy is associated with reduced severity consistent with SMA Type 3. Genetic counseling is recommended. |
| ≥4 | Negative | SMA (Type 3/4 probable) a Refer to other examples with negative c.859G>C variant status for example language. |
5. Newborn Screening for SMA
With multiple treatment options available and compelling data showing the value of early treatment to maximize patient benefit, SMA newborn screening (NBS) has become an increasing priority. In the US, this screening is included in the RUSP (Recommended Uniform Screening Panel) and other NBS recommendations [34]. In the same line, the SMA NBS Alliance promotes the implementation of NBS in all of Europe by 2025 (www.sma-screening-alliance.org/ (accessed on 12 September 2022)).
In SMA NBS, SMN1 is the primary indicator of disease status. Given the throughput and cost restrictions necessary for NBS, testing is often limited to the presence or absence of SMN1 exon 7 using DNA isolated from dried blood spots (DBS) and is frequently combined with testing for severe combined immunodeficiency (SCID) in a single assay [34,40]. When positive screening results are identified, follow-up testing is performed to confirm diagnosis and obtain SMN2 copy number results to infer disease prognosis. However, recent studies have provided data supporting the reporting of SMN2 copy numbers along with initial screening results, as it is beneficial for SMA patients with two copies of SMN2 where treatment timing is most crucial [16]. Others have suggested that disease modifier variant testing is also important to further refine the likely prognosis for SMA patients identified through NBS with two or three copies of SMN2 [17]. As NBS programs and our understanding of the intersection of screening and treatment continue to expand, it is likely that NBS testing will move toward providing as much genetic information as possible to maximize treatment benefits in newborns with SMA [41]. As the complexity of NBS is increasing, genetic programs in newborns should come along with adequate pre-test genetic counseling to provide more precise information to the families.
6. Conclusions
While understanding of the impact of SMN1 and SMN2 variants on SMA carrier status and disease prognosis continues to evolve, a solid foundation of clinical studies demonstrates the utility of identifying several variants in addition to copy numbers. More specifically, when variants predicting SMN1 copies in cis are present, it is possible to adjust the risk of silent carrier status, which can help inform reproductive decisions for couples. Additionally, disease modifier testing can improve prognostic predictions in individuals diagnosed with SMA, explaining some of the discrepancies between observed SMN2 copy numbers and expected SMA disease progression. The information provided by these variants can benefit laboratories and clinicians interested in providing more accurate SMA carrier screening and prognostic predictions.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
John N. Milligan is an employee of Asuragen, a Bio-Techne Brand.
Funding Statement
This work was partially supported by Grants from Biogen ESP-SMG-17-11256 (to E.F.T. supporting L.B.-P.), Roche and Spanish Instituto de Salud Carlos III, Fondo de Investigaciones Sanitarias and co-funded with ERDF funds (Grant No. FIS PI18/000687) (to E.F.T.).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stabley D.L., Harris A.W., Holbrook J., Chubbs N.J., Lozo K.W., Crawford T.O., Swoboda K.J., Funanage V.L., Wang W., Mackenzie W., et al. SMN1 and SMN2 copy numbers in cell lines derived from patients with spinal muscular atrophy as measured by array digital PCR. Mol. Genet. Genom. Med. 2015;3:248–257. doi: 10.1002/mgg3.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sugarman E.A., Nagan N., Zhu H., Akmaev V.R., Zhou Z., Rohlfs E.M., Flynn K., Hendrickson B.C., Scholl T., Sirko-Osadsa D.A., et al. Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: Clinical laboratory analysis of >72,400 specimens. Eur. J. Hum. Genet. 2012;20:27–32. doi: 10.1038/ejhg.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Talbot K., Tizzano E.F. The clinical landscape for SMA in a new therapeutic era. Gene Ther. 2017;24:529–533. doi: 10.1038/gt.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sugarman E., Nagan N., Zhu H., Akmaev V.R., Zhou Z., Rohlfs E.M., Flynn K., Hendrickson B.C., Scholl T., Sirko-Osadsa D.A., et al. Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet. Med. 2002;4:20–26. doi: 10.1097/00125817-200201000-00004. [DOI] [PubMed] [Google Scholar]
- 5.Wadman R.I., Stam M., Gijzen M., Lemmink H.H., Snoeck I.N., Wijngaarde C.A., Braun K.P.J., Schoenmakers M.A.C.G., van den Berg L.H., Dooijes D., et al. Association of motor milestones, SMN2 copy and outcome in spinal muscular atrophy types 0–4. J. Neurol. Neurosurg. Psychiatry. 2017;88:365–367. doi: 10.1136/jnnp-2016-314292. [DOI] [PubMed] [Google Scholar]
- 6.Alías L., Bernal S., Fuentes-Prior P., Barceló M.J., Also E., Hernandez R.M., Rodríguez-Alvarez F.J., Martín Y., Aller E., Grau E., et al. Mutation update of spinal muscular atrophy in Spain: Molecular characterization of 745 unrelated patients and identification of four novel mutations in the SMN1 gene. Hum. Genet. 2009;125:29–39. doi: 10.1007/s00439-008-0598-1. [DOI] [PubMed] [Google Scholar]
- 7.Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA) Hum. Mutat. 2000;15:228–237. doi: 10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 8.Prior T.W., Nagan N., Sugarman E.A., Batish S.D., Braastad C. Technical standards and guidelines for spinal muscular atrophy testing. Genet. Med. 2011;13:686–694. doi: 10.1097/GIM.0b013e318220d523. [DOI] [PubMed] [Google Scholar]
- 9.Hendrickson B.C., Donohoe C., Akmaev V.R., Sugarman E.A., Labrousse P., Boguslavskiy L., Flynn K., Rohlfs E.M., Walker A., Allitto B., et al. Differences in SMN1 allele frequencies among ethnic groups within North America. J. Med. Genet. 2009;46:641–644. doi: 10.1136/jmg.2009.066969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo M., Liu L., Peter I., Zhu J., Scott S.A., Zhao G., Eversley C., Kornreich R., Desnick R.J., Edelmann L. An Ashkenazi Jewish SMN1 haplotype specific to duplication alleles improves pan-ethnic carrier screening for spinal muscular atrophy. Genet. Med. 2014;16:149–156. doi: 10.1038/gim.2013.84. [DOI] [PubMed] [Google Scholar]
- 11.Alías L., Bernal S., Calucho M., Martínez E., March F., Gallano P., Fuentes-Prior P., Abuli A., Serra-Juhe C., Tizzano E.F. Utility of two SMN1 variants to improve spinal muscular atrophy carrier diagnosis and genetic counselling. Eur. J. Hum. Genet. 2018;26:1554–1557. doi: 10.1038/s41431-018-0193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blasco-Pérez L., Costa-Roger M., Leno-Colorado J., Bernal S., Alias L., Codina-Solà M., Martínez-Cruz D., Castiglioni C., Bertini E., Travaglini L., et al. Deep Molecular Characterization of Milder Spinal Muscular Atrophy Patients Carrying the c.859G>C Variant in SMN2. Int. J. Mol. Sci. 2022;23:8289. doi: 10.3390/ijms23158289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prior T.W., Krainer A.R., Hua Y., Swoboda K.J., Snyder P.C., Bridgeman S.J., Burghes A.H., Kissel J.T. A positive modifier of spinal muscular atrophy in the SMN2 gene. Am. J. Hum. Genet. 2009;85:408–413. doi: 10.1016/j.ajhg.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vezain M., Saugier-Veber P., Goina E., Touraine R., Manel V., Toutain A., Fehrenbach S., Frébourg T., Pagani F., Tosi M., et al. A rare SMN2 variant in a previously unrecognized composite splicing regulatory element induces exon 7 inclusion and reduces the clinical severity of spinal muscular atrophy. Hum. Mutat. 2010;31:E1110–E1125. doi: 10.1002/humu.21173. [DOI] [PubMed] [Google Scholar]
- 15.Costa-Roger M., Blasco-Pérez L., Cuscó I., Tizzano E.F. The Importance of Digging into the Genetics of SMN Genes in the Therapeutic Scenario of Spinal Muscular Atrophy. Int. J. Mol. Sci. 2021;22:9029. doi: 10.3390/ijms22169029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee B.H., Deng S., Chiriboga C.A., Kay D.M., Irumudomon O., Laureta E., Delfiner L., Treidler S.O., Anziska Y., Sakonju A., et al. Newborn Screening for Spinal Muscular Atrophy in New York State: Clinical Outcomes from the First 3 Years. Neurology. 2022 doi: 10.1212/WNL.0000000000200986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuscó I., Bernal S., Blasco-Pérez L., Calucho M., Alias L., Fuentes-Prior P., Tizzano E.F. Practical guidelines to manage discordant situations of SMN2 copy number in patients with spinal muscular atrophy. Neurol. Genet. 2020;6:e530. doi: 10.1212/NXG.0000000000000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X., Sanchis-Juan A., French C.E., Connell A.J., Delon I., Kingsbury Z., Chawla A., Halpern A.L., Taft R.J., Bentley D.R., et al. Spinal muscular atrophy diagnosis and carrier screening from genome sequencing data. Genet. Med. 2020;22:945–953. doi: 10.1038/s41436-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blasco-Pérez L., Paramonov I., Leno J., Bernal S., Alías L., Fuentes-Prior P., Cuscó I., Tizziano E.F. Beyond copy number: A new, rapid, and versatile method for sequencing the entire SMN2 gene in SMA patients. Hum. Mutat. 2021;42:787–795. doi: 10.1002/humu.24200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gambardella A., Mazzei R., Toscano A., Annesi G., Pasqua A., Annesi F., Quattrone F., Oliveri R.I., Valentino P., Bono F., et al. Spinal muscular atrophy due to an isolated deletion of exon 8 of the telomeric survival motor neuron gene. Ann. Neurol. 1998;44:836–839. doi: 10.1002/ana.410440522. [DOI] [PubMed] [Google Scholar]
- 21.Scheffer H., Cobben J.M., Matthijs G., Wirth B. Best practice guidelines for molecular analysis in spinal muscular atrophy. Eur. J. Hum. Genet. 2001;9:484–491. doi: 10.1038/sj.ejhg.5200667. [DOI] [PubMed] [Google Scholar]
- 22.Niba E.T.E., Nishio H., Wijaya Y.O.S., Lai P.S., Tozawa T., Chiyonobu T., Yamadera M., Okamoto K., Awano H., Takeshima Y., et al. Clinical phenotypes of spinal muscular atrophy patients with hybrid SMN gene. Brain Dev. 2021;43:294–302. doi: 10.1016/j.braindev.2020.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Cusco I., Barceló M., Del Rio E., Martín Y., Hernández-Chico C., Bussaglia E., Baiget M., Tizzano E. Characterisation of SMN hybrid genes in Spanish SMA patients: De novo, homozygous and compound heterozygous cases. Hum. Genet. 2001;108:222–229. doi: 10.1007/s004390000452. [DOI] [PubMed] [Google Scholar]
- 24.Milligan J.N., Larson J.L., Filipovic-Sadic S., Laosinchai-Wolf W., Huang Y.-W., Ko T.-M., Abbott K.M., Lemmink H.H., Toivonen M., Schleutker J., et al. Multisite Evaluation and Validation of a Sensitive Diagnostic and Screening System for Spinal Muscular Atrophy that Reports SMN1 and SMN2 Copy Number, along with Disease Modifier and Gene Duplication Variants. J. Mol. Diagn. 2021;23:753–764. doi: 10.1016/j.jmoldx.2021.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macdonald W.K., Hamilton D., Kuhle S. SMA carrier testing: A meta-analysis of differences in test performance by ethnic group. Prenat. Diagn. 2014;34:1219–1226. doi: 10.1002/pd.4459. [DOI] [PubMed] [Google Scholar]
- 26.Park J.E., Yun S.A., Roh E.Y., Yoon J.H., Shin S., Ki C.S. Carrier Frequency of Spinal Muscular Atrophy in a Large-scale Korean Population. Ann. Lab. Med. 2020;40:326–330. doi: 10.3343/alm.2020.40.4.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez-Lopez D., Loucera C., Carmona R., Aquino V., Salgado J., Pasalodos S., Miranda M., Alonso Á., Dopazo J. SMN1 copy-number and sequence variant analysis from next-generation sequencing data. Hum. Mutat. 2020;41:2073–2077. doi: 10.1002/humu.24120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prior T.W., Nagan N., Sugarman E.A., Batish S.D., Braastad C. ADDENDUM: Technical standards and guidelines for spinal muscular atrophy testing. Genet. Med. 2016;18:752. doi: 10.1038/gim.2016.76. Addendum to Genet. Med. 2011, 13, 686–694. [DOI] [PubMed] [Google Scholar]
- 29.Feng Y., Ge X., Meng L., Scull J., Li J., Tian X., Zhang T., Jin W., Cheng H., Wang X., et al. The next generation of population-based spinal muscular atrophy carrier screening: Comprehensive pan-ethnic SMN1 copy-number and sequence variant analysis by massively parallel sequencing. Genet. Med. 2017;19:936–944. doi: 10.1038/gim.2016.215. [DOI] [PubMed] [Google Scholar]
- 30.Kaseniit K.E., Haque I.S., Goldberg J.D., Shulman L.P., Muzzey D. Genetic ancestry analysis on >93,000 individuals undergoing expanded carrier screening reveals limitations of ethnicity-based medical guidelines. Genet. Med. 2020;22:1694–1702. doi: 10.1038/s41436-020-0869-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davidson D., Kaseniit K., Haque I. Duplication Tag SNP g.27134T>G should not be considered diagnostic of SMA carrier status; Proceedings of the ACMG Annual Meeting; Phoenix, AZ, USA. 22–24 March 2017. [Google Scholar]
- 32.Ruhno C., McGovern V.L., Avenarius M.R., Snyder P.J., Prior T.W., Nery F.C., Muhtaseb A., Roggenbuck J.S., Kissel J.T., Sansone V.A., et al. Complete sequencing of the SMN2 gene in SMA patients detects SMN gene deletion junctions and variants in SMN2 that modify the SMA phenotype. Hum. Genet. 2019;138:241–256. doi: 10.1007/s00439-019-01983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calucho M., Bernal S., Alías L., March F., Venceslá A., Rodríguez-Álvarez F.J., Aller E., Fernández R.M., Borrego S., Millán H.M., et al. Correlation between SMA type and SMN2 copy number revisited: An analysis of 625 unrelated Spanish patients and a compilation of 2834 reported cases. Neuromuscul. Disord. 2018;28:208–215. doi: 10.1016/j.nmd.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Glascock J., Sampson J., Haidet-Phillips A., Connolly A., Darras B., Day J., Finkel R., Howell R.R., Klinger K., Kuntz N., et al. Treatment Algorithm for Infants Diagnosed with Spinal Muscular Atrophy through Newborn Screening. J. Neuromuscul. Dis. 2018;5:145–158. doi: 10.3233/JND-180304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mercuri E., Finkel R.S., Muntoni F., Wirth B., Montes J., Main M., Mazzone E.S., Vitale M., Snyder B., Quijano-Roy S., et al. Diagnosis and management of spinal muscular atrophy: Part 1, Recommendations for diagnosis, rehabilitation, orthopedic and nutritional care. Neuromuscul. Disord. 2018;28:103–115. doi: 10.1016/j.nmd.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 36.Wu X., Wang S.H., Sun J., Krainer A.R., Hua Y., Prior T.W. A-44G transition in SMN2 intron 6 protects patients with spinal muscular atrophy. Hum. Mol. Genet. 2017;26:2768–2780. doi: 10.1093/hmg/ddx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butchbach M.E.R. Genomic Variability in the Survival Motor Neuron Genes (SMN1 and SMN2): Implications for Spinal Muscular Atrophy Phenotype and Therapeutics Development. Int. J. Mol. Sci. 2021;22:7896. doi: 10.3390/ijms22157896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keinath M.C., Prior D.E., Prior T.W. Spinal Muscular Atrophy: Mutations, Testing, and Clinical Relevance. Appl. Clin. Genet. 2021;14:11–25. doi: 10.2147/TACG.S239603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maretina M.A., Zheleznyakova G.Y., Lanko K.M., Egorova A.A., Baranov V.S., Kiselev A.V. Molecular Factors Involved in Spinal Muscular Atrophy Pathways as Possible Disease-modifying Candidates. Curr. Genom. 2018;19:339–355. doi: 10.2174/1389202919666180101154916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor J.L., Lee F.K., Yazdanpanah G.K., Staropoli J.F., Liu M., Carulli J.P., Sun C., Dobrowolski S.F., Hannon W.H., Vogt R.F. Newborn blood spot screening test using multiplexed real-time PCR to simultaneously screen for spinal muscular atrophy and severe combined immunodeficiency. Clin. Chem. 2015;61:412–419. doi: 10.1373/clinchem.2014.231019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pane M., Donati M.A., Cutrona C., De Sanctis R., Pirinu M., Coratti G., Ricci M., Palermo C., Berti B., Leone D., et al. Neurological assessment of newborns with spinal muscular atrophy identified through neonatal screening. Eur. J. Pediatr. 2022;181:2821–2829. doi: 10.1007/s00431-022-04470-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Souza P.V.S., Pinto W.B.V.D.R., Ricarte A., Badia B.D.M.L., Seneor D.D., Teixeira D.T., Caetano L., Gonçalves E.A., Chieia M.A.T., Farias I.B., et al. Clinical and radiological profile of patients with spinal muscular atrophy type 4. Eur. J. Neurol. 2021;28:609–619. doi: 10.1111/ene.14587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.