Abstract
Several pathogenic bacteria have evolved a specialized protein secretion system termed type III to secrete and deliver effector proteins into eukaryotic host cells. Salmonella enterica serovar Typhimurium uses one such system to mediate entry into nonphagocytic cells. This system is composed of more than 20 proteins which are encoded within a pathogenicity island (SPI-1) located at centisome 63 of its chromosome. A subset of these components form a supramolecular structure, termed the needle complex, that resembles the flagellar hook-basal body complex. The needle complex is composed of a multiple-ring cylindrical base that spans the bacterial envelope and a needle-like extension that protrudes from the bacterial outer surface. Although the components of this structure have been identified, little is known about its assembly. In this study we examined the effect of loss-of-function mutations in each of the type III secretion-associated genes encoded within SPI-1 on the assembly of the needle complex. This analysis indicates that the assembly of this organelle occurs in discrete, genetically separable steps. A model for the assembly pathway of this important organelle is proposed that involves a sec-dependent step leading to the assembly of the base substructure followed by a sec-independent process resulting in the assembly of the needle portion.
Several important animal and plant pathogenic bacteria have evolved a specialized protein secretion system to interact with their hosts (11, 15). This protein secretion system, termed type III or contact dependent, is designed to deliver bacterial proteins to the host cell cytoplasm to modulate cellular functions for the pathogen's benefit. Salmonella enterica, encodes two such systems. One system, located at centisome 63, is required for the initial interaction of Salmonella with the intestinal epithelium (10). The other, located at centisome 31, is essential for the establishment of systemic infection (26, 29).
Type III secretion systems are composed of more than 20 proteins that are essential for the secretion and delivery of effector proteins into the host cell. Core components of type III secretion systems are localized and/or exert their function in the bacterial cytoplasm, the bacterial envelope, or the extracellular environment (11, 15). For example, a set of low-molecular-weight, acidic polypeptides are thought to function within the confines of the bacterial cytoplasm as chaperones, secretion pilots, or translational regulators of cognate secreted proteins (31). A group of secreted proteins required for the translocation of bacterial effectors into eukaryotic cells are thought to exert their function at the host cell membrane (6). Yet another group of type III secretion-associated proteins function at the bacterial envelope. Among them, two distinct groups with presumably different functions can be recognized. One subset is composed of several highly conserved inner membrane proteins that form the equivalent of what has been described as the “export machinery” in the related flagellar system (11, 15). Although the actual function of the export machinery is poorly understood, it is thought that it facilitates the engagement and subsequent transport through the inner membrane of the type III secreted proteins. The other subset is composed of a group proteins that form a supramolecular structure termed the needle complex (21, 23).
The needle complex was first identified in S. enterica serovar Typhimurium but has also been detected in other bacterial species encoding type III secretion systems (3, 30). This supramolecular complex spans both the inner and outer membranes and resembles the flagellar hook-basal body complex. The most salient features of this organelle are the presence of a four-ring hollow and cylindrical base that is anchored to both the inner and the outer membranes and a slender, needle-like structure that protrudes outward from the outer membrane (21). The protein components of the base and the needle substructures have been recently identified (23). PrgH, PrgK, and InvG make up the base substructure. The PrgH and PrgK proteins exhibit signature features of lipoproteins, while InvG belongs to the secretin family of outer membrane exporter proteins. These three proteins are unique among components of the type III secretion system in that they exhibit typical sec-dependent signal sequences. The major component of the needle substructure is PrgI, a low-molecular-weight protein also encoded within the type III secretion-associated cluster of genes in SPI-1 (23). The length of the needle portion is controlled by the function of InvJ, a protein previously shown to be secreted via the type III secretion system (5, 23). Absence of InvJ results in abnormally long needles and the complete absence of type III secretion (5, 23).
Little is known about the assembly of the needle complex on the bacterial envelope and the potential role that other type III secretion-associated gene products may play in this process. In this study, we have undertaken a genetic analysis of the assembly of the needle complex in serovar Typhimurium. Using electron microscopy and biochemical fractionation, we have examined the contribution of each one of the type III-secretion associated gene products encoded within the serovar Typhimurium pathogenicity island-1 (SPI-1). This analysis has allowed us to begin to build a model for the assembly of this very important organelle.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains used in this study were derivatives of the serovar Typhimurium strain SJW2941 (32) and are listed in Table 1. The construction of nonpolar mutations in invA (12), invC (9), invE (14), invG (17), invH (1), invI and invJ (5), spaO, spaP, spaQ, spaR, and spaS (4); prgH and prgK (21), prgI (23), sipB and sipC (19), and sipD (18) have been described elsewhere. Mutations in iacP, iagB, prgJ, orgA, and orgB were constructed by inserting a copy of the terminator-less aphT gene cassette, which confers kanamycin resistance, into unique sites within these genes. The mutated alleles were introduced into the serovar Typhimurium chromosome by allelic exchange as previously described (17). Mutations were moved into the serovar Typhimurium SJW2941 background strain by P22 HTint-mediated transduction (28). In all cases, the phenotypes associated with the introduction of the mutations could be complemented by the introduction of a wild-type copy of the gene. Strains were grown on L agar or in L broth supplemented with 0.3 M sodium chloride to allow optimal expression of the components of the invasion-associated type III secretion system. When required, the following antibiotics were added at the concentrations indicated: kanamycin, 50 μg/ml; ampicillin, 100 μg/ml; streptomycin, 100 μg/ml; and tetracycline, 10 μg/ml.
TABLE 1.
Relevant genotypes and phenotypes of the strains used in these studies
| Strain | Relevant genotype | EMa detection of:
|
Detection of:
|
Host cell invasion | ||||
|---|---|---|---|---|---|---|---|---|
| Base | Needle | InvG | PrgH | PrgK | PrgI | |||
| SB905 | Wild type | + | + | + | + | + | + | + |
| SB906 | prgH | − | − | + | − | − | − | − |
| SB907 | prgK | − | − | + | − | − | − | − |
| SB1171 | invG | − | − | − | + | − | +c | − |
| SB1073 | prgI | + | − | + | + | + | − | − |
| SB1074 | prgJ | + | − | + | + | + | − | − |
| SB912 | invA | + | − | + | + | + | − | − |
| SB917 | invC | + | − | + | + | + | − | − |
| SB1174 | spaO | + | − | + | + | + | − | − |
| SB1202 | spaP | + | − | + | + | + | − | − |
| SB1175 | spaQ | + | − | + | + | + | − | − |
| SB1176 | spaR | + | − | + | + | + | − | − |
| SB1201 | spaS | + | − | + | + | + | − | − |
| SB1081 | orgA | + | − | + | + | + | − | − |
| SB1082 | orgB | + | − | + | + | + | − | − |
| SB1076 | invI | + | − | + | + | + | − | − |
| SB1077 | invJ | + | +b | + | + | + | + | − |
| SB1075 | invH | +c | +c | +c | +c | +c | +c | +c |
| SB1180 | iagB | + | + | + | + | + | + | + |
| SB1203 | iacP | + | + | + | + | + | + | + |
| SB1205 | invE | + | + | + | + | + | + | − |
| SB1210 | sipB | + | + | + | + | + | + | − |
| SB1211 | sipC | + | + | + | + | + | + | − |
| SB1212 | sipD | + | + | + | + | + | + | − |
EM, electron microscopy.
The needle substructures of the invJ mutant strain were abnormally long.
Reduced in comparison to the wild type.
Isolation and analysis of the needle complex.
The isolation and analysis of the serovar Typhimurium needle complex was carried out as described elsewhere (21, 23). Briefly, bacterial cultures were grown in L broth containing 0.3 M NaCl at 37°C to an optical density at 600 nm of ∼0.8. The cells were pelleted and resuspended in 0.5 M sucrose–0.15 M Tris. Lysozyme (0.2 mg/ml, final concentration) and EDTA (1 mM, final concentration) were added, and the cells were incubated on ice for 1 h. Bacteria were then lysed by the addition of a 3% solution of lauryldimethylamine oxide (LDAO). Lysates were cleared of debris by low speed centrifugation (10,000 × g for 15 min at 4°C), the pH was adjusted to 10.5 and, after incubation for 1 h at 4°C, the lysates were centrifuged again at 10,000 × g for 15 min. The cleared lysates were then subjected to high-speed centrifugation (250,000 × g for 1 h at 4°C), and the pellets were resuspended in 0.5 M sucrose–0.1 M Tris–0.03% LDAO (pH 10.5) and spun briefly (10,000 × g for 10 min) to remove any particulate matter. Samples were centrifuged again at 250,000 × g for 1 h at 4°C, and the pellets were resuspended in 0.01 M Tris–0.02 M EDTA–0.03% LDAO (pH 8.0) and loaded onto 30% (wt/vol) CsCl density gradients. The gradients were centrifuged at 15,000 × g for 15 h at 20°C in a swinging-bucket rotor in a Beckman (model L 80) ultracentrifuge. Gradient fractions containing needle complexes were pooled and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western immunoblotting using an antibody specific to the base structure as previously described (21, 23). Needle complex base-specific antiserum was prepared in New Zealand White rabbits immunized with highly purified preparations of base structures isolated as described elsewhere (23). The resulting antiserum was absorbed with an acetone powder preparation of a serovar Typhimurium strain not expressing the components of the needle complex. This protocol is optimal for the isolation of base substructures, since under these conditions the fragile needle substructures tend to break from the bases. To isolate needle complexes containing both the base and needle substructures, a slight modification to the isolation protocol was required. Bacterial cells were grown and pelleted as indicated above and resuspended in 0.5 M sucrose–0.15 M Tris (pH 8). Lysozyme (0.2 mg/ml, final concentration) and EDTA (1 mM, final concentration) was added, and the cells were incubated on ice for 1 h, followed by incubation at 37°C for 15 min. Cells were lysed by the addition of a 3% LDAO as indicated above, and cell lysates were subjected to a low-speed centrifugation step (10,000 × g for 15 min at 4°C), followed by high-speed centrifugation (250,000 × g for 1 h at 4°C) but without raising the pH in between the centrifugation steps. The pellets were resuspended in Laemmli buffer and analyzed by Western immunoblotting with an antibody directed to a peptide derived from the PrgI sequence.
The preparation of osmotically shocked bacterial cells for electron microscopy was carried out as previously described (21). Samples were negatively stained with 2% phosphotungstic acid (pH 7.0) and observed under transmission electron microscope (EM410; Philips). Micrographs were taken at an accelerating voltage of 80 kV.
RESULTS AND DISCUSSION
The assembly of the needle complex and its subsequent protein delivery activities are highly regulated. This regulation involves both transcriptional as well as posttranscriptional mechanisms that control, in a temporal and spatial manner, the delivery of effector proteins into host cells. Little is known about the mechanisms that control the actual assembly of the needle complex. In order to gain insight into this process, we carried out an extensive analysis of the effect of mutations in each of the type III secretion-associated genes encoded within the SPI-1 on the assembly process (10). We examined the effect of loss-of-function mutations in genes that encode structural components of the needle complex, other components of the type III secretion and translocation system, and several accessory proteins, as well as proteins encoded within SPI-1 whose role in type III secretion has not been previously characterized. We evaluated the phenotypes of these mutants using biochemical and electron microscopy assays for the assembly of the needle and the base substructures of the needle complex.
Effect of mutations in prgH, prgK, and invG.
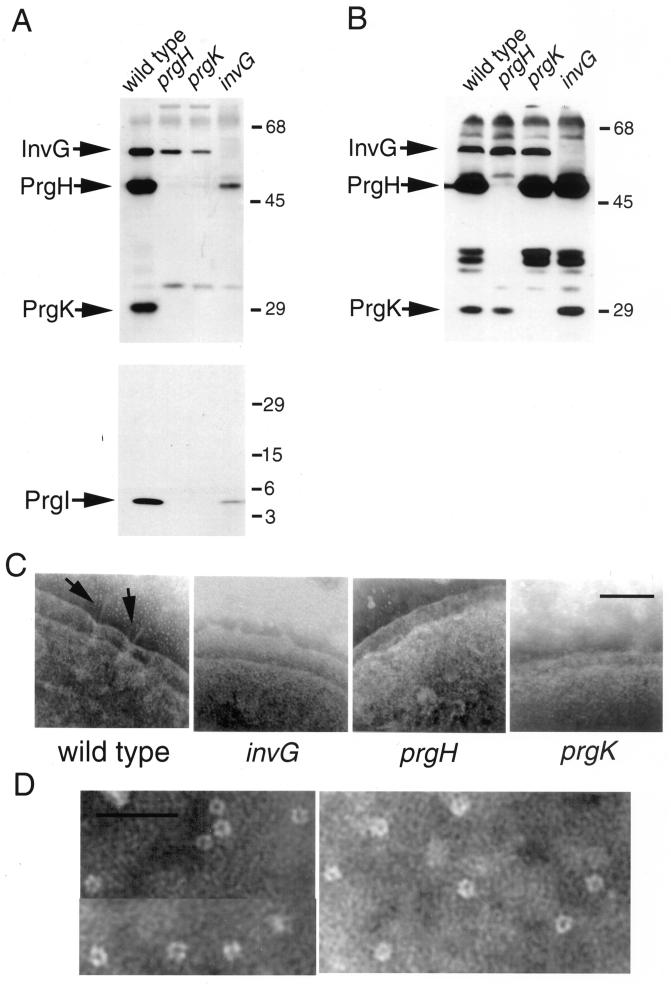
prgH, prgK, and invG encode three essential components of the base substructure of the needle complex (23). In an effort to identify intermediate structures, we examined the effect of loss-of-function mutations in each one of theses genes on the assembly of the needle complex. Needle complex base and needle preparations were obtained from strains carrying loss-of-function mutations in prgH, prgK, or invG and analyzed by Western immunoblotting with an antiserum specific for either PrgH, PrgK, and InvG or the needle protein PrgI, respectively. Both the prgH and prgK mutant strains were capable of assembling InvG into a stable complex that was able to withstand the harsh conditions of the needle complex isolation protocol (Fig. 1A, upper panel). However, the amount of InvG in the complexes isolated from these mutant strains was significantly lower than that isolated from the wild type (Fig. 1A, upper panel), even though the total amount of InvG in whole-cell lysates was comparable in all the strains (Fig. 1B, upper pannel). These results indicate that InvG can form a complex in the absence of PrgH and PrgK but that the formation of such a complex is enhanced by the presence of these two proteins. PrgH and PrgK may therefore assist in the outer membrane localization of InvG or stabilize its multimeric form. Consistent with this observation, no structures resembling the needle complex base substructure were observed under the electron microscope in preparations of osmotically shocked cells from the prgH and prgK mutant strains (Fig. 1C). However, structures which appeared identical to the previously described InvG multimers (7) were observed in preparations obtained from the prgH and prgK (but not invG) mutant strains following the needle complex isolation protocol (Fig. 1D and data not shown).
FIG. 1.
Effect of mutations in prgH, prgK, and invG on the assembly of the needle complex. (A) Needle complex preparations were obtained from wild-type serovar Typhimurium and the isogenic prgH, prgK, and invG mutant strains and then examined by Western immunoblotting using an antibody specific to InvG, PrgH, and PrgK or the needle protein PrgI. The positions of the InvG, PrgH, PrgK, and PrgI proteins, and the molecular mass standards (in kilodaltons) are indicated. (B) Whole-cell lysates of the same strains were examined for the presence of InvG, PrgH, and PrgK by Western immunoblotting. (C) Electron micrographs of negatively stained osmotically shocked preparations of wild-type and the invG, prgH, and prgK isogenic mutant strains are also shown (bar, 100 nm). (D) Electron micrographs of negatively stained preparations of InvG ring structures (bar, 100 nm)
A loss-of-function mutation in prgH abolished the formation of a PrgK complex (Fig. 1A, upper panel) without affecting the total amount of PrgK (Fig. 1B, upper panel). The same results were obtained with a prgK mutant strain which abolished PrgH complex formation without affecting its total level (Fig. 1A and B, upper panels). These results indicate that these proteins require the presence of each other to assemble into stable complexes. Needle complex base preparations isolated from the invG mutant strain exhibited a small but significant amount of PrgH but no detectable PrgK (Fig. 1A, upper panel). However, less-stringent needle complex preparations of the invG mutant strain contained both PrgH and PrgK, indicating that these proteins can form a complex, albeit one incapable of withstanding the more-stringent isolation protocol (data not shown). Comparison of the relative abundance in wild-type bacteria of PrgH, PrgK, and InvG in purified needle complexes versus total cell lysates indicated that there is a significant proportion of PrgH that is not associated with the needle complex (compare Fig. 1A versus Fig. 1B). Whether this pool of PrgH free from the needle complex is assembled into a multimeric structure is not known. Unlike the InvG ring, which is expected to be located in the outer membrane, the PrgH-PrgK intermediate would be most likely located in the inner membrane. In any case, our results indicate that the presence of the three proteins is necessary for the stability of the entire complex.
As expected, none of the mutants exhibited needle substructures when observed under the electron microscope (Fig. 1C). Consistent with this observation, the needle protein PrgI was not detected in needle complex preparations from the prgH or the prgK mutant strains (Fig. 1A, lower panel). However, a small but reproducible amount of PrgI was consistently detected in needle complex preparations of the invG mutant strain (Fig. 1A, lower panel). Since PrgI could not be detected in similar preparations from strains carrying mutations in components of the export machinery (see below), these results indicate that PrgH and PrgK must be able to associate with the export machinery in the absence of InvG and that this complex must be competent for the export of the PrgI needle protein. In the absence of InvG, PrgI remains associated with PrgH and PrgK, presumably in the periplasmic space, but is unable to traverse the outer membrane and assemble into needle substructures.
Effect of a mutation in invH.
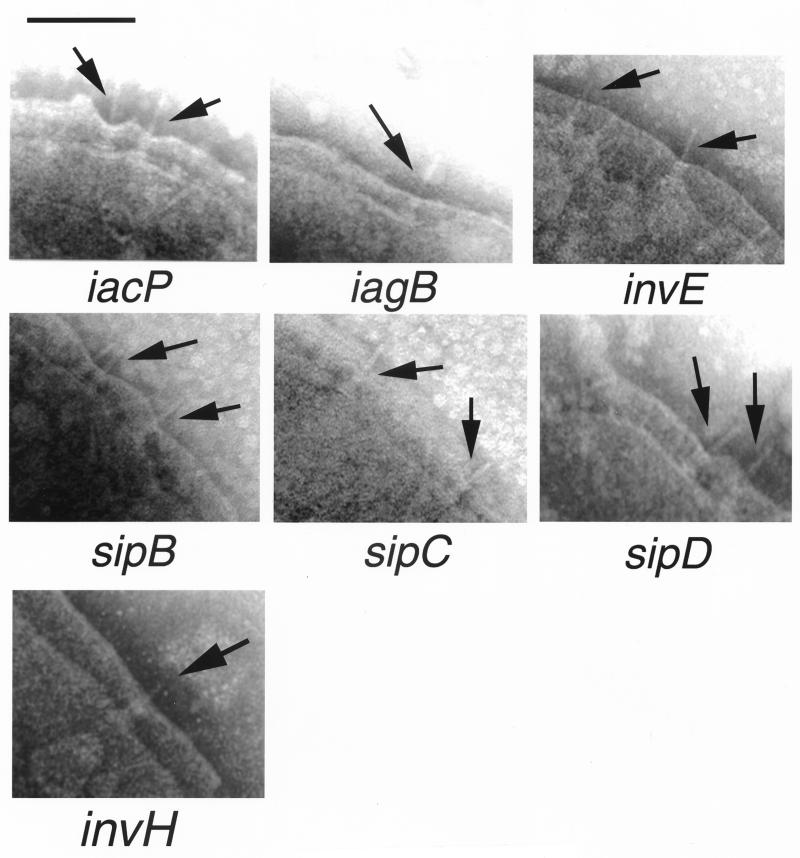
It has been suggested previously that InvH is required for the multimeric assembly and insertion of InvG into the bacterial outer membrane (7, 8). Since InvH does not appear to be a structural component of the needle complex (21, 23), it must aid the assembly process through a transient interaction with InvG. Introduction of a loss-of-function mutation in invH resulted in a marked reduction in the number of needle complexes on the bacterial envelope as observed by electron microscopy of osmotically shocked cells (data not shown). However, unlike the case of the invG mutant (Fig. 1), complete needle complexes were distinctly detected on the envelope of the invH mutant strain (Fig. 2). Consistent with the electron microscopy observations, biochemical analysis of purified base and needle substructures from the serovar Typhimiurium invH mutant strain showed the presence of reduced amounts of InvG-PrgH-PrgK complexes (Fig. 3A, upper panel) and the needle protein PrgI (Fig. 3A, lower panel). These results indicate that InvH is not essential for the assembly of the needle complex. However, InvH significantly enhances the efficiency of needle complex formation, presumably through its stabilizing activity on InvG. These results are also consistent with our previous observation that invH mutants of S. enterica retain significant invasion capacity, an indication of the presence of a functional type III secretion system (1).
FIG. 2.
Electron micrographs of negatively stained osmotically shocked preparations of strains of serovar Typhimurium carrying mutations in type III secretion-associated genes that do not affect the assembly of the needle complex. The identity of the different mutant strains is indicated below each panel. The arrows point to needle complex structures (bar, 100 nm).
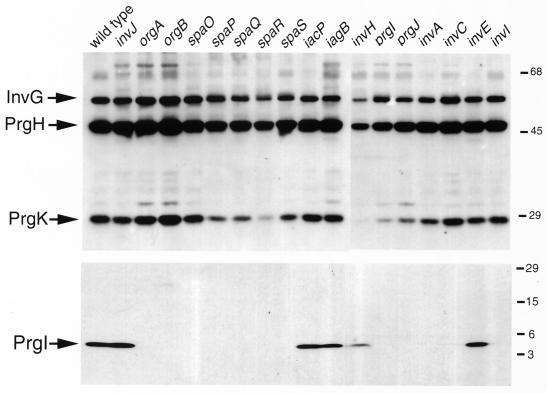
FIG. 3.
Effect of mutations in type III-associated genes on the assembly of the needle complex. Needle complex preparations were obtained from wild-type serovar Typhimurium and the indicated isogenic mutant strains and then examined by Western immunoblotting using an antibody specific to InvG, PrgH, and PrgK or to the needle protein PrgI. The positions of InvG, PrgH, PrgK, and PrgI proteins and the molecular mass standards (in kilodaltons) are indicated.
Effect of mutations in prgI and prgJ.
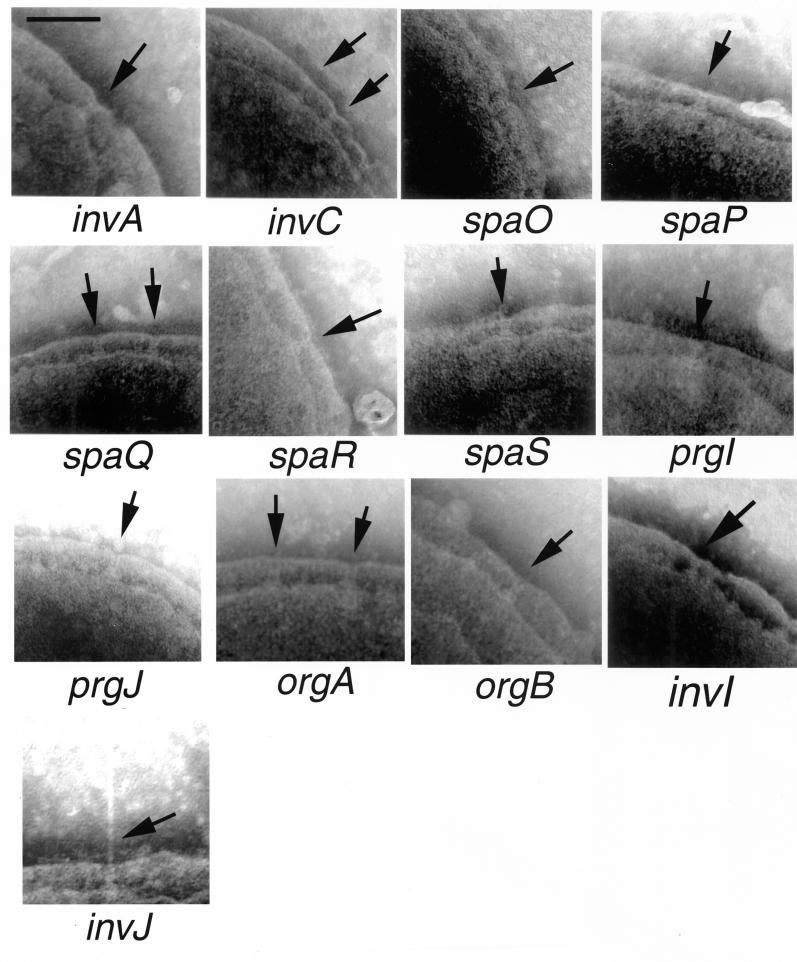
We have recently identified PrgI as the main component of the needle substructure of the needle complex (23). The prgI and prgJ genes are located between the prgH and prgK genes and are in the same transcriptional unit (2). The organization of these genes suggests a functional relationship. We therefore examined the role of PrgI and PrgJ in the assembly of the needle complex. Introduction of loss-of-function mutation in prgI or prgJ resulted in strains that are capable of assembling the base but lack the needle substructure (Fig. 4). Biochemical analysis of needle complexes obtained from these mutant strains confirmed the absence of the needle protein PrgI (Fig. 3, lower panel) and showed slightly reduced levels of PrgK (Fig. 3, upper panel). It is possible that this reduction was at least in part due to a slight polar effect of the prgI and prgJ mutations on the expression of prgK since transcomplementation of these mutants did not restore wild-type levels of PrgK (data not shown). These results indicate that PrgI and PrgJ are largely dispensable for the assembly of the base substructure but are essential for the assembly of the needle. The mechanisms by which PrgJ contributes to needle assembly are not understood. It is possible that PrgJ is a minor component of the needle or is required for its assembly, perhaps serving as a scaffolding protein in a manner analogous to FlgD in the flagellar assembly pathway (27). Consistent with this hypothesis, we have found that PrgJ is secreted into the culture medium (A. Sukhan and J. E. Galan, unpublished observations).
FIG. 4.
Electron micrographs of negatively stained, osmotically shocked preparations of strains of serovar Typhimurium carrying mutations in genes that affect the assembly of the needle portion of the needle complex without affecting the integrity of the base substructure. The identity of the different mutant strains is indicated below each panel. The arrows point to base structures (bar, 100 nm).
Effect of mutations in genes encoding components of the export machinery.
We have recently shown that InvA and InvC are required for the assembly of the needle substructure of the needle complex but are dispensable for the assembly of the base (23). These two proteins are components of the export machinery of the SPI-1 type III secretion system that are conserved in other type III secretion systems (9, 12). We therefore extended our analysis and examined the role of other conserved components of the export apparatus in needle complex assembly. Serovar Typhimurium strains carrying loss-of-function mutations in invA, invC, spaP, spaQ, spaR, and spaS (4, 9, 12) were examined for the presence of needle complexes in osmotically shocked cells. As shown in Fig. 4, all of these mutant strains exhibited base structures indistinguishable in shape and number from those formed by the wild type. However, all of these strains lacked the needle portion of the needle complex. Consistent with the electron microscopy observations, biochemical analysis of needle complexes isolated from these strains showed the absence of the PrgI needle protein (Fig. 3, lower panel) but wild-type levels of the base proteins PrgH, PrgK and InvG (Fig. 3, upper panel). These results indicate that all of the components of the export machinery are required for the assembly of the needle substructure of the needle complex but are not required for the assembly and/or stability of the base.
Effect of mutations in orgA and orgB.
A recent study has shown that an open reading frame located immediately downstream of prgK and originally incorrectly identified as encoding a single protein, OrgA, actually encodes two proteins, OrgA and OrgB (16, 20). Both of these proteins were shown to be required for serovar Typhimurium invasion of epithelial cells (20). We therefore investigated the role of these two proteins in the assembly of the serovar Tyhimurium needle complex. Strains carrying loss-of-function nonpolar mutations in orgA and orgB were examined by electron microscopy for the presence of needle complexes in osmotically shocked cells. Both the orgA and orgB mutants displayed apparently normal base structures on the bacterial envelope but lacked the needle substructure (Fig. 4). Analysis of isolated complexes from these mutants showed a total absence of the needle protein PrgI (Fig. 3, lower panel) and wild-type levels of PrgH, PrgK, and InvG assembled into base structures (Fig. 3, upper panel). Therefore the orgA and orgB mutations appear to have a similar phenotype to the loss-of-function mutations in components of the export machinery.
Effect of mutations in invI, invJ, and spaO.
InvI, InvJ, and SpaO are essential for type III secretion but, unlike other components of the export machinery, share only limited sequence similarity to proteins in other type III secretion systems (4, 5). Two of these proteins, InvJ and SpaO, are themselves secreted into the culture supernatant through the type III secretion system (4, 5). Therefore, these proteins have the potential to exert a function distinct from that of the conserved components of the type III secretion-associated export machinery. As previously shown (23), a strain carrying a loss-of-function mutation in invJ displayed abnormally long needle structures (Fig. 2). In contrast, strains carrying mutations in either spaO or invI lacked needle substructures (Fig. 4). All three strains exhibited base structures that were indistinguishable from those of the wild type (Fig. 2). Consistent with the electron microscopy observations, biochemical analysis of needle complexes isolated from these mutants showed the presence of PrgI in the needle preparation obtained from the invJ mutant strain but not in those obtained from the invI and spaO mutants (Fig. 3, lower panel). In addition, base preparations obtained from all these mutants showed the presence of PrgH, PrgK, and InvG at levels that were indistinguishable from those observed in the wild type (Fig. 3, upper panel). It was previously suggested that InvI may function as a chaperone for InvJ secretion (5). This hypothesis was based on at least three observations: (i) the immediate proximity of the genes encoding these two proteins, a frequently observed arrangement in type III secreted chaperones and their cognate substrates; (ii) the secondary structure of InvI that resembles that of other type III secretion associated chaperones; and (iii) the requirement of InvI for InvJ secretion (5). Since a loss-of-function mutation in InvI did not lead to elongated needle structures, these results indicate that either InvI is not a chaperone for InvJ, the secretion of InvJ is not required for its needle-length control function, and/or that InvI is also required for PrgI secretion.
Effect of a mutation in invE.
InvE was one of the first proteins identified as essential for serovar Typhimurium entry into host cells (14). However, unlike other components of the type III secretion system, InvE does not seem to be required for the assembly of the invasomes on the surface of serovar Typhimurium (13). The role of InvE in type III secretion has not been investigated. We examined under the electron microscope osmotically shocked cells from a strain carrying a loss-of-function mutation in invE. As shown in Fig. 4, the morphology of the needle complexes of the invE-null strain was indistinguishable from that of the wild type. Furthermore, the amount of the base and needle proteins obtained from needle complex preparations from this strain were also indistinguishable from that of the wild type (Fig. 3, upper and lower panels). These results indicate that InvE must exert its essential role in type III secretion-associated function independently of needle complex assembly.
Effect of mutations in iagB, iacP, and components of the translocation machinery.
IagB and IacP are encoded within the type III secretion-associated genes in SPI-1 and are thought to play accessory roles in type III secretion and/or the phenotypes associated with this system. The IagB protein exhibits amino acid sequence similarity to a family of muramidases that are associated with the assembly of large membrane protein complexes such as flagella and the type III and type IV protein secretion systems (24). It has been proposed that the function of this family of proteins may be to form a hole in the peptidoglycan layer to allow the insertion of these large complexes into the bacterial envelope. The IacP protein belongs to a family of acyl-carrier proteins, and it is thought that this protein may serve to modify either components of the needle complex or perhaps the secreted targets of the SPI-1 system (18). Translocation of bacterial effector proteins through the eukaryotic host cell membrane requires the function of SipB, SipC, and SipD, which are themselves proteins secreted through the type III system. Mutations in genes encoding any of these three proteins completely abolish protein translocation into host cells but do not affect protein secretion. To assess the potential role of these genes in needle complex assembly, strains carrying loss-of-function mutations in iacP, iagB, sipB, sipC, or sipD were examined under the electron microscope. These mutant strains exhibited normal needle complexes as examined both by electron microscopy (Fig. 4) or biochemical fractionation analysis (Fig. 3, upper and lower panels). These results indicate that IagB, IacP, and the Sip proteins are not essential for the assembly of the needle complex. In contrast, in the flagellar system FliJ, which is the homolog of IagB, has been shown to be essential for flagellar assembly (22, 25). Either the type III secretion complex can assemble without the assistance of these muramidases or an unidentified functionally redundant protein can substitute for IagB.
Assembly pathway of the type III secretion-associated needle complex.
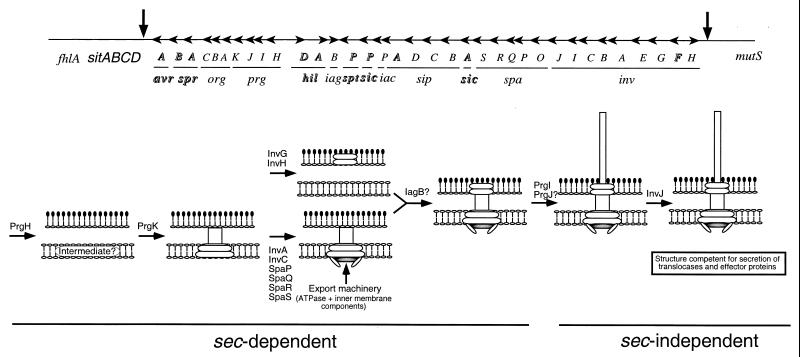
Our analysis suggests a model for the assembly pathway of the type III secretion-associated needle complex (Fig. 5). The assembly of the base substructure does not require the function of any of the components of the type III secretion export machinery. However, this machinery is absolutely required for the assembly of the needle substructure. Since all the components of the base substructure possess typical sec-dependent signal sequences, the first step in the assembly process must therefore be the sec-dependent export of PrgH, PrgK, InvG, and the accessory protein InvH (see below). Upon secretion through the inner membrane, these proteins may form intermediate structures that eventually lead to the assembly of the complete and stable base structure which, in conjunction with the export machinery, functions as a type III secretion machinery. This incomplete type III secretion machinery must be able to recognize only a limited number of substrates which are required for the assembly of the needle portion of the needle complex. No bacterial translocases or effectors of type III-mediated cellular responses can be secreted until completion of the assembly of the needle substructure (4). Once the entire needle complex is assembled, the type III secretion machinery switches specificity to secrete bacterial effector proteins. This specificity switch is governed by InvJ since loss-of-function mutations in this protein result in a machinery unable to secrete any other protein but PrgI, the main subunit of the needle substructure (5, 23), and PrgJ, a putative minor component or scaffold of this substructure (A. Sukhan and J. E. Galán, unpublished observations). As a result, the invJ mutant strain exhibit needle complexes with grossly elongated needles (23).
FIG. 5.
Model for the assembly pathway of the needle complex. In the upper panel a scheme representing SPI-1 is shown. Highlighted genes encode transcription factors, effector proteins, or their cognate chaperones, and therefore their potential contribution to the assembly of the needle complex was not investigated.
Previous studies and our own results indicate that InvG is capable of forming a ring-like structure (Fig. 1D) (7). Formation of this structure is aided by the presence of the accessory protein InvH (7, 8). However, our results show that InvH is not essential for needle complex assembly since a strain carrying a loss-of-function mutation in InvH retained the capacity to form wild-type needle complexes, albeit less efficiently. These results are consistent with our previous observation that strains carrying mutations in invH retained significant signaling capacity as measured by their ability to induce actin cytoskeleton rearrangements and enter into host cells (1). Therefore, although not essential for needle complex assembly, InvH increases the efficiency of this process, presumably through its function as a facilitator of InvG multimerization. Our results also argue that the formation of the InvG substructure can take place in the absence of either PrgK or PrgH. However, in the absence of PrgK or PrgH, the amount of InvG complex is significantly reduced, indicating that these proteins may help to stabilize the InvG ring or aid InvG's multimerization. We postulate that PrgH and PrgK form intermediate structures before becoming associated with the InvG ring substructure. Consistent with this hypothesis, PrgH-containing complexes were isolated in the absence of InvG. However, these complexes were not detected in the absence of PrgK, indicating that the stability of this intermediate depends on the presence of this protein. Our results suggest that a PrgH-PrgK intermediate complex must be able to associate with the inner-membrane components of the export machinery even in the absence of InvG. This conclusion is based on the detection of the PrgI needle protein in needle complexes obtained from an invG mutant strain but not from strains carrying mutations in any of the component of the export machinery. Nevertheless, this PrgH-PrgK export machinery intermediate complex most likely becomes associated with InvG very rapidly and therefore may be virtually undetectable in wild-type bacteria. Our results also indicate that in the absence of any of the three main components of the base structure, the stability of the base is seriously compromised.
In summary, we have shown that the assembly of the needle complex occurs in discrete independent steps and requires the coordinated function of several gene products. More studies will be required to define additional intermediate structures and to better understand the assembly of the needle substructure.
ACKNOWLEDGMENTS
We thank Sumati Murli, Alessandra Pancetti and Carol Peña for critical review of this manuscript.
T. K. was supported by a fellowship from the Human Frontiers Science Program. This work was supported by Public Health Service Grant AI30492 from the National Institutes of Health.
REFERENCES
- 1.Altmeyer R M, McNern J K, Bossio J C, Rosenshine I, Finlay B B, Galán J E. Cloning and molecular characterization of a gene involved in Salmonella adherence and invasion of cultured epithelial cells. Mol Microbiol. 1993;7:89–98. doi: 10.1111/j.1365-2958.1993.tb01100.x. [DOI] [PubMed] [Google Scholar]
- 2.Behlau I, Miller S J. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J Bacteriol. 1993;175:4475–4484. doi: 10.1128/jb.175.14.4475-4484.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blocker A, Gounon P, Larquet E, Niebuhr K, Cabiaux V, Parsot C, Sansonetti P. The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J Cell Biol. 1999;147:683–693. doi: 10.1083/jcb.147.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collazo C, Galán J E. Requirement of exported proteins for secretion through the invasion-associated Type III system in Salmonella typhimurium. Infect Immun. 1996;64:3524–3531. doi: 10.1128/iai.64.9.3524-3531.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collazo C M, Zierler M K, Galán J E. Functional analysis of the Salmonella typhimurium invasion genes invI and invJ and identification of a target of the protein secretion apparatus encoded in the inv locus. Mol Microbiol. 1995;15:25–38. doi: 10.1111/j.1365-2958.1995.tb02218.x. [DOI] [PubMed] [Google Scholar]
- 6.Cornelis G R, Boland A, Boyd A P, Geuijen C, Iriarte M, Neyt C, Sory M P, Stainier I. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev. 1998;62:1315–1327. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crago A M, Koronakis V. Salmonella InvG forms a ring-like multimer that requires the InvH lipoprotein for outer membrane localization. Mol Microbiol. 1998;30:47–56. doi: 10.1046/j.1365-2958.1998.01036.x. [DOI] [PubMed] [Google Scholar]
- 8.Daefler S, Russel M. The Salmonella typhimurium InvH protein is an outer membrane lipoprotein required for the proper localization of InvG. Mol Microbiol. 1998;28:1367–1380. doi: 10.1046/j.1365-2958.1998.00908.x. [DOI] [PubMed] [Google Scholar]
- 9.Eichelberg K, Ginocchio C, Galán J E. Molecular and functional characterization of the Salmonella typhimurium invasion genes invB and invC: homology of InvC to the F0F1 ATPase family of proteins. J Bacteriol. 1994;176:4501–4510. doi: 10.1128/jb.176.15.4501-4510.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gálan J E. Interaction of Salmonella with host cells through the centisome 63 type III secretion system. Curr Opin Microbiol. 1999;2:46–50. doi: 10.1016/s1369-5274(99)80008-3. [DOI] [PubMed] [Google Scholar]
- 11.Gálan J E, Collmer A. Type III secretion machines: bacterial devices for protein delivery into host cells. Science. 1999;284:322–328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 12.Gálan J E, Ginocchio C, Costeas P. Molecular and functional characterization of the Salmonella typhimurium invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol. 1992;17:4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginocchio C, Olmsted S B, Wells C L, Galán J E. Contact with epithelial cells induces the formation of surface appendages on Salmonella typhimurium. Cell. 1994;76:717–724. doi: 10.1016/0092-8674(94)90510-x. [DOI] [PubMed] [Google Scholar]
- 14.Ginocchio C, Pace J, Galán J E. Identification and molecular characterization of a Salmonella typhimurium gene involved in triggering the internalization of salmonellae into cultured epithelial cells. Proc Natl Acad Sci USA. 1992;89:5976–5980. doi: 10.1073/pnas.89.13.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones B D, Falkow S. Identification and characterization of a Salmonella typhimurium oxygen-regulated gene required for bacterial internalization. Infect Immun. 1994;62:3745–3752. doi: 10.1128/iai.62.9.3745-3752.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaniga K, Bossio J C, Galán J E. The Salmonella typhimurium invasion genes invF and invG encode homologues to the PulD and AraC family of proteins. Mol Microbiol. 1994;13:555–568. doi: 10.1111/j.1365-2958.1994.tb00450.x. [DOI] [PubMed] [Google Scholar]
- 18.Kaniga K, Trollinger D, Galán J E. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J Bacteriol. 1995;177:7078–7085. doi: 10.1128/jb.177.24.7078-7085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaniga K, Tucker S C, Trollinger D, Galán J E. Homologues of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J Bacteriol. 1995;177:3965–3971. doi: 10.1128/jb.177.14.3965-3971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein J R, Fahlen T F, Jones B D. Transcriptional organization and function of invasion genes within Salmonella enterica serovar Typhimurium pathogenicity island 1, including the prgH, prgI, prgJ, prgK, orgA, orgB, and orgC genes. Infect Immun. 2000;68:3368–3376. doi: 10.1128/iai.68.6.3368-3376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galán J E, Aizawa S-I. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 22.Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa S-I. Morphological pathway of flagellar assembly in Salmonella typhimurium. J Mol Biol. 1992;226:433–446. doi: 10.1016/0022-2836(92)90958-m. [DOI] [PubMed] [Google Scholar]
- 23.Kubori T, Sukhan A, Aizawa S-I, Galán J E. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc Natl Acad Sci USA. 2000;97:10225–102230. doi: 10.1073/pnas.170128997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mushegian A R, Fullner K J, Koonin E V, Nester E W. A family of lysozyme-like virulence factors in bacterial pathogens of plants and animals. Proc Natl Acad Sci USA. 1996;93:7321–7326. doi: 10.1073/pnas.93.14.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nambu T, Minamino T, Macnab R M, Kutsukake K. Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium. J Bacteriol. 1999;181:1555–1561. doi: 10.1128/jb.181.5.1555-1561.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ochman H, Soncini F C, Solomon F, Groisman E A. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohmishi K, Ohto Y, Aizawa S-I, Macnab R M. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J Bacteriol. 1994;176:2272–2281. doi: 10.1128/jb.176.8.2272-2281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:74–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 29.Shea J E, Hensel M, Gleeson C, Holden D W. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamano K, Aizawa S I, Katayama E, Nonaka T, Imajoh-Ohmi S, Kuwae A, Nagai S, Sasakawa C. Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO J. 2000;19:3876–3887. doi: 10.1093/emboj/19.15.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wattiau P, Woestyn S, Cornelis G R. Customized secretion chaperones in pathogenic bacteria. Mol Microbiol. 1996;20:255–262. doi: 10.1111/j.1365-2958.1996.tb02614.x. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi S, Fujita H, Ishihara A, Aizawa S, Macnab R M. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J Bacteriol. 1986;166:187–193. doi: 10.1128/jb.166.1.187-193.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]