ABSTRACT
Pseudomonas aeruginosa is a notorious pathogen that causes various nosocomial infections. Several prophage genes located on the chromosomes of P. aeruginosa have been reported to contribute to bacterial pathogenesis via host phenotype transformations, such as serotype conversion and antibiotic resistance. However, our understanding of the molecular mechanism behind host phenotype shifts induced by prophage genes remains largely unknown. Here, we report a systematic study around a hypothetical recombinase, Pg54 (RecT), located on a 48-kb putative prophage (designated PP9W) of a clinical P. aeruginosa strain P9W. Using a ΔrecT mutant (designated P9D), we found that RecT promoted prophage PP9W excision and gene transcription via the inhibition of the gene expression level of pg40, which encodes a CI-like repressor protein. Further transcriptomic profiling and various phenotypic tests showed that RecT modulated like a suppressor to some transcription factors and vital genes of diverse cellular processes, providing multiple advantages for the host, including cell growth, biofilm formation, and virulence. The versatile functions of RecT hint at a strong impact of phage proteins on host P. aeruginosa phenotypic flexibility.
IMPORTANCE Multidrug-resistant and metabolically versatile P. aeruginosa are difficult to eradicate by anti-infective therapy and frequently lead to significant morbidity and mortality. This study characterizes a putative recombinase (RecT) encoded by a prophage of a clinical P. aeruginosa strain isolated from severely burned patients, altering prophage lifestyle and host core cellular processes. It implies the potential role of RecT in the coevolution arm race between bacteria and phage. The excised free phages from the chromosome of host bacteria can be used as weapons against other sensitive competitors in diverse environments, which may increase the lysogeny frequency of different P. aeruginosa subgroups. Subsequent analyses revealed that RecT both positively and negatively affects different phenotypic traits of the host. These findings concerning RecT functions of host phenotypic flexibility improve our understanding of the association between phage recombinases and clinical P. aeruginosa, providing new insight into mitigating the pathogen infection.
KEYWORDS: RecT, P. aeruginosa, prophage, excision, phenotypic flexibility
INTRODUCTION
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen that can be readily isolated from living sources and various environments (1). In recent decades, P. aeruginosa has been predominant in nosocomial and severe infections with significant morbidity and mortality (2). The mortality rate of burn patients infected with P. aeruginosa is much higher than those noninfected due to the subsequent septic complications triggered by bacterial virulence and antibiotic resistance. Considering a certain proportion of inevitable infections, how to reduce the cost of nosocomial diseases via restraining the antibiotic resistance and virulence of P. aeruginosa is the core problem for specialized burn centers (3). The problematic anti-pseudomonas therapy failure is mainly caused by comprehensive features of clinical strains, such as the intrinsic resistance to multiple drugs, acquired resistance through mutations, versatile roles in severe infections, and carrying multidrug resistance plasmids (horizontal gene transfer) or prophages (transduction and lysogenic conversion) (4).
Phages are viruses specific to bacteria that play a core role in bacterial abundance, diversity, and metabolism. The replication of phages primarily includes lytic and lysogenic processes (5). In the lytic cycle, the phage replicates and releases new progeny via host cell lysis (6). While in the lysogenic cycle, the phage genome integrates into the host chromosome (prophage) or is maintained as a plasmid. Many different induction factors can lead to the excision of prophages, such as temperature, pH, or bacterial growth rate (7). The temperate phages can replicate via the lytic or lysogenic cycle, which seems to be in a dynamic balance (8). A study of 2,110 sequenced bacterial genomes indicates that nearly half were lysogen with a highly variable number of prophages (9). These cryptic prophages facilitate bacterial survival in adverse environments by providing various benefits (10). Some phage proteins play a crucial role in diverse aspects resulting in clinical pathogen infection (11–13), including serotype conversion, cell motility, host cell predation escape, virulence factors, cell competitive fitness, acid, and antibiotic resistance (14–19).
RecT is a known single-strand annealing protein, which interacts with RecE (exonuclease VIII) to promote RecA-independent recombination and double-strand break repair by a DNA strand invasion mechanism in Escherichia coli (20–23). Nevertheless, the performance of “RecT” in P. aeruginosa is still unclear. Here, we identified a gene pg54 that encodes a putative recombinase containing a RecT domain on the genome of a prophage PP9W. The function of Pg54 (RecT) was investigated by reverse transcription-quantitative real-time (RT-qPCR), electrophoretic mobility shift assay (EMSA), transcriptome sequencing (RNA-seq), and a series of phenotypic tests between indicated strains. Our data show that RecT affects prophage excision and gene transcription and various cellular processes of the host by repressing several transcription factors and essential genes. These findings are important to improving our understanding of the association between phage proteins and clinical P. aeruginosa, highlighting the perspective to mitigate the pathogen infection.
RESULTS
Phage PP9W isolated from P. aeruginosa P9W can infect P8W.
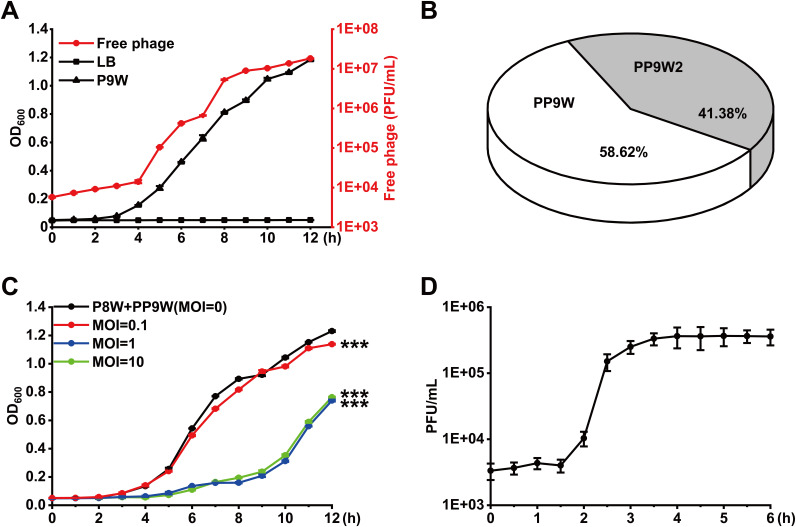
We isolated 5 P. aeruginosa strains (Table 1) from severely burned patients and used a spot assay to investigate the ecological interactions between them (Table S1 in the supplemental material). P8W and P9W were antagonism paired isolates, which were whole-genome sequenced and further investigated in this study. An integrative active prophage hunting tool, Prophage Hunter (24), was used to identify the location and the number of active prophages on the genomes of P8W and P9W. Compared to P8W, P9W harbors two additional prophages, which were designated PP9W and PP9W2 (Fig. S1). To determine their activities, we performed a growth-curve experiment and quantified the corresponding number of free phages in the supernatant of P9W using P8W as the indicator strain. The number of free phages was increased as the bacterial biomass of P9W (Fig. 1A). After serial dilution, we selected the double agar whose PFU was around 30 to dig out the central zone of plaques and used as PCR templates (Table 2). We found that both phage PP9W and PP9W2 were active and mixed in the supernatant of P9W, and their proportions were nearly half (Fig. 1B). The isolated excised phage PP9W from the plaques was further whole-genome sequenced.
TABLE 1.
Bacterial strains, phages, and plasmids used in this study
| Category | Genotype or description | Source |
|---|---|---|
| P. aeruginosa | ||
| P7 | Wild type P. aeruginosa clinical isolate | This work |
| P8W | Wild type P. aeruginosa clinical isolate | This work |
| P9W | Wild type P. aeruginosa clinical isolate | This work |
| P18 | Wild type P. aeruginosa clinical isolate | This work |
| P27 | Wild type P. aeruginosa clinical isolate | This work |
| P9D | P9W ΔrecT | This work |
| E. coli | ||
| DH5α | F− ϕ80 lacZΔM15 Δ(lacZYA-argF) U169 endA1 recA1 endA1 hsdR17(rK−, mK+) supE44λ− thi-1 gyrA96 relA1 phoA | TianGen |
| S17-1 | RP4-2 Tc::Mu Km::Tn7 Tpr Smr Pro Res− Mod+ | TransGen |
| JM109 | endA1 recA1 gyrA96 thi hsdR17(rk−, mk+) relA1 supE44D(lac-proAB) [F’traD36 proAB laq lqZΔM15] | Biomed |
| BL21(DE3) | F-ompT hsdSB(rB−, mB−) gal dcm(DE3) | Biomed |
| Phages | ||
| PP9W | A excised prophage isolated from P9W | This work |
| PP9W2 | Another excised prophage isolated from P9W | This work |
| PP9D | The recT gene deletion derivative of PP9W | This work |
| Plasmids | ||
| pEX18Tc | Gene replacement vector; Tcr oriT+ sacB+ | W. Wu |
| pEX18Tc::U + D | The recT gene knockout vector | This work |
| pUCP18 | Broad host range shuttle vector, Apr | W. Wu |
| pCT2 | The recT gene complementation vector | This work |
| pUC19 | Commonly used cloning vector; lac lacZ Apr | NEB |
| pMD19 (Simple) | A linearized T-vector for cloning promoter fragments, Apr | TaKaRa |
| pEDS40 | pg40 gene promoter cloned in pMD19 (Simple), Apr | This work |
| pEDS50 | pg50 gene promoter cloned in pMD19 (Simple), Apr | This work |
| pEDS55 | pg55 gene promoter cloned in pMD19 (Simple), Apr | This work |
| pEDS57 | pg57 gene promoter cloned in pMD19 (Simple), Apr | This work |
| pEDS72 | pg72 gene (IntR) promoter cloned in pMD19 (Simple), Apr | This work |
| pET32a(+) | Fusion vector for N-terminal His tag, Apr | NovaGen |
| pET32a-T | his-recT fusion in pET32a(+) vector, Apr | This work |
| pEASY-T1 | Cloning vector for ploting the standard curves, Apr Kanr | TransGen |
| pEASY-I | T-I fusion in pEASY-T1 vector, Apr Kanr | This work |
| pEASY-A | T-pg47 fusion in pEASY-T1 vector, Apr Kanr | This work |
FIG 1.
Characterization of phage PP9W isolated from the supernatant of P9W. (A) Growth curve of P9W during 12 h (the black triangle line) and LB is the control group (the black square line). The number of free phages in the supernatant of P9W (the red dot line) is shown. (B) The ratio of PP9W/PP9W2 in the supernatant of P9W was determined by plaque PCR. (C) Growth-inhibition curve of PP9W on P8W was monitored. Multiplicity of infection (MOI) represents the initial PP9W/P8W dose ratio (the black dot line: P8W + PP9W, MOI = 0, the control group; the red dot line: MOI = 0.1; the blue dot line: MOI = 1; the green dot line: MOI = 10). (D) One-step growth curve of phage PP9W during 6 h. P8W was used as the indicator strain and MOI = 1. The experiments were independently replicated three times and triplicates of each sample were tested. Data were analyzed by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons (α < 0.05) to examine the mean differences between the end points of the data groups. ***, P < 0.001, by Student's t test. Error bars show standard deviations.
TABLE 2.
Oligonucleotides used in this study
| Primer name | 5′–3′ sequence | Source or function |
|---|---|---|
| recTU-Fa | CCGGAATTCGGCGTACACCTCCTGAAGAT | A 1-kb fragment upstream of the recT gene |
| recTU-R | CGCGGATCCGCTGCTGTCTCCTTGTAACC | |
| recTD-F | CGCGGATCCCATGCCCAGCCGAACCATT | A 1-kb fragment downstream of the recT gene |
| recTD-R | CCCAAGCTTATGGCTCACTCCAGGTCGA | |
| recTA-F | GCGGTAGGTGTTCAGTTCG | A 3-kb fragment includes recT |
| recTA-R | TGAACGGCGAGCCTATCAG | |
| recT-F | CCGGAATTCCGGATTCACCACGAATACGG | Cloning the recT gene into pUCP18 |
| recT-R | CGCGGATCCATGAGCGAACCCACCCAAC | |
| PP9-1F | CTTCTGGAACTGCCGACTACT | Plaque PCR identification of the fragment of PP9W2 |
| PP9-1R | CGCTGGATTATCAACGTGAACA | |
| PP9-2F | CAACTGAGCTGAAGAAGGA | Plaque PCR identification of the fragment of PP8-1 or PP9-2 |
| PP9-2R | ATCTTGAACAGGAACGACAT | |
| PP8-2F | CGCAACTGCATACATTCTGGT | Plaque PCR identification of the fragment of PP8-2 |
| PP8-2R | AGTTCTTCGTCGTCCTCGTC | |
| PP9-3F | GTGGACAACGCTCAGAACAG | Plaque PCR identification of the fragment of PP9W |
| PP9-3R | AAGTGCGGCTGGCAGTAA | |
| PP9-4F | CATGGCAAGACCACTCTGAC | Plaque PCR identification of the fragment of PP8-3 or PP9-4 |
| PP9-4R | TGACGACCACCTTCTTCCTT | |
| PP9-5F | TCCAATCAGAACCGCCTAGC | Plaque PCR identification of the fragment of PP8-4 or PP9-5 |
| PP9-5R | CGCTTGCTCCGTTGATGGT | |
| P9L-F | CACGACAGGTTGAGGACGAT | Linear form in P9W when PP9W is excised |
| P9L-R | GCGAAGGTGCGGATCATCA | |
| P9I-F | AGCAGGCACGACAGGTTGA | I fragment in P9W when PP9W is integrated |
| P9I-R | CGTTCTTGCATCCTCCCTCTG | |
| PP9C-F | CGCTGAGATGGGCAGATATTG | Circular form in phage PP9W |
| PP9C-R | AATCCATCACATCGGGCATG | |
| PP9T-F | AACCCACCCAACTGGAGCA | Identification of the existence of the recT gene |
| PP9T-R | TTTCGCCCGTCTCGGTGTT | |
| pg47-F | GCATATCGGCGACAACATCA | Cloning pg47 into pEASY-T1 |
| pg47-R | GATTCCAGGACTCGACAAGATC | |
| rpoD-F | CGTCCTCAGCGGCTATATCG | Internal reference gene in RT-qPCR assay |
| rpoD-R | TCTTCCTCGTCGTCCTTCTCT | |
| pg10-F | AGACGAACCAAGGACCACATT | RT-qPCR assay for pg10 |
| pg10-R | CGGCATGGCACAGTATCATC | |
| pg26-F | CGAGGAGATGAAGGGCTTGT | RT-qPCR assay for pg26 |
| pg26-R | CGGAGAGCGGTTGTGACTT | |
| pg29-F | GGTGTGGTTCTCGTTCTCC | RT-qPCR assay for pg29 |
| pg29-R | GTTCCAGGTGATCCAGACTTC | |
| pg30-F | GACATCACATCCGCCATCCA | RT-qPCR assay for pg30 |
| pg30-R | TCGAAGCCGCTGAGGTACT | |
| pg40-F | TGATGCCGACCAAGAAGTGA | RT-qPCR assay for pg40 |
| pg40-R | GCCAAACGATACGACCGATAA | |
| pg48-F | TTCAGCAACGCGCAACTC | RT-qPCR assay for pg48 |
| pg48-R | TCGTCGTCCTCCTGCTCTT | |
| pg49-F | GCGGTTCGGATGATTGAGG | RT-qPCR assay for pg49 |
| pg49-R | TGTTCGTGCTCCTGAGTTCT | |
| pg50-F | AGCTCGACGACCTGGTGAT | RT-qPCR assay for pg50 |
| pg50-R | CGCTTGATGGTCAGTGCCA | |
| pg55-F | CCGAACCATTGAAGAGCAGTT | RT-qPCR assay for pg55 |
| pg55-R | GGACTCGCTGAGGAACATC | |
| pg56-F | ACAAGGATTGCCTGCTCATCAT | RT-qPCR assay for pg56 |
| pg56-R | TCGCTGTCGTCTGCTGGTT | |
| pg57-F | CGCTGTTCGAGAAGGTGAAG | RT-qPCR assay for pg57 |
| pg57-R | GCCGATTCCAGTTCCTCCT | |
| pg66-F | GGCAGTAGTAGAGGAATCCATC | RT-qPCR assay for pg66 |
| pg66-R | CGTTACGCAGATACAGCACTA | |
| fliM-F | GCGGTGCTGGAGATGAACT | RT-qPCR assay for fliM |
| fliM-R | TCGTGGTCGGACTGGAAAC | |
| pilB-F | AGTCGTATCTCTGCTCGTCTC | RT-qPCR assay for pilB |
| pilB-R | TCCTTCTGGTCCTCCTCGTA | |
| pctA-F | ACAAAGAGCAGGTGATGAAGAC | RT-qPCR assay for pctA |
| pctA-R | GGATGGCGACGATGGAGAT | |
| rmd-F | TGACTCAGCGTCTGTTCGT | RT-qPCR assay for rmd |
| rmd-R | GTGCCAAGGAGGTTGATCTG | |
| mexT-F | TCTGAACCTGCTGATCGTGTT | RT-qPCR assay for mexT |
| mexT-R | ATGGCGGTGGAGATGGAATC | |
| arnB-F | CCTGGCACCTGTTCATCCT | RT-qPCR assay for arnB |
| arnB-R | GAGTTCCACTCGCTGTTGG | |
| 19.1Sb | cggtaactatcgtcttgagtccaacccggtaagacacgacttatcgccac | 50-bp single-strand DNA of pUC19 |
| 19.2S | gtggcgataagtcgtgtcttaccgggttggactcaagacgatagttaccg | Reverse fragment to 19.1S |
| 19.3S | ttcggtcctccgatcgttgtcagaagtaagttggccgcagtgttatcact | Another 50-bp single-strand DNA of pUC19 |
| CIP-F | ATTCATACGGCTGCCTCCAT | The promoter region of pg40 for EMSA dsDNA binding assay |
| CIP-R | CGCTGGTTGAGTCTGTCTGA | |
| 50P-F | AGAACTCAGGAGCACGAACA | The promoter region of pg50 for EMSA dsDNA binding assay |
| 50P-R | AGATTCACCACCAGGGTCTC | |
| 55P-F | GTCAACGACTTCATCGAACCG | The promoter region of pg55 for EMSA dsDNA binding assay |
| 55P-R | GGCTCGTCTCCATCGTCTTC | |
| 57P-F | CGAAATCACCAATTTCCAAGGG | The promoter region of pg57 for EMSA dsDNA binding assay |
| 57P-R | CCATAGCCACGCCATCAAG | |
| 59P-F | ACCCTGCTGGACCTCTTCA | The promoter region of pg59 for EMSA dsDNA binding assay |
| 59P-R | GCTGATGAACACGGCATCC | |
| M13-47 | CGCCAGGGTTTTCCCAGTCACGAC | Amplification of the promoter regions |
| RV-M | GAGCGGATAACAATTTCACACAGG |
The underlined sequences represent the recognition sites of different restriction enzymes.
The primers 19.1S, 19.2S, and 19.3S are also used for EMSA ssDNA binding assay.
To characterize PP9W, we performed a growth inhibition experiment (25) at different multiplicity of infection (MOI) to determine the inhibitory effect of PP9W on P8W (Fig. 1C). Here, the remarkable MOI was “1” since the final bacterial biomass of P8W (12 h) significantly decreased when MOI increased from 0.1 to 1. However, no more obvious inhibitory effect was observed when more phage particles were added (MOI = 10). Therefore, the MOI was set at 1 in the subsequent one-step growth curve experiment (26). The typical triphasic curve showed the latent period was about 1.5 to 2 h, and the burst size was approximately 109 (Fig. 1D). These data suggested that the excised active phage PP9W of P9W leads to the lysis of P8W.
A recombinase RecT improves the host cell growth rate.
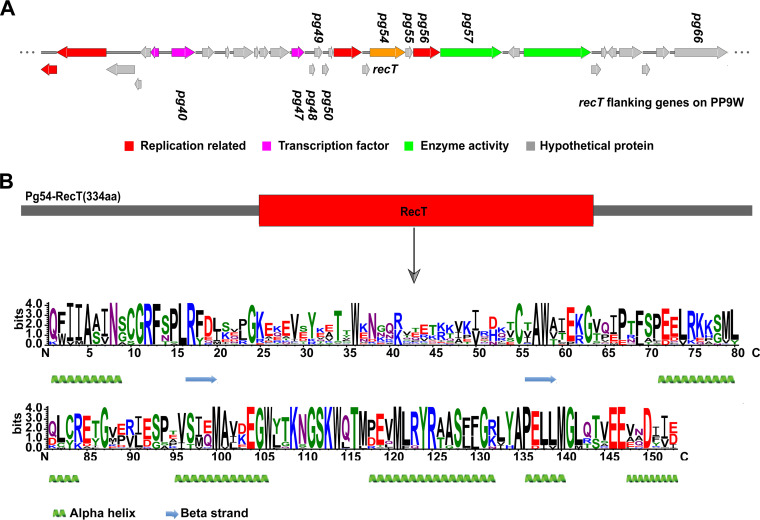
Many phage proteins have been reported to contribute to the phenotypic flexibility of P. aeruginosa (27). Moreover, phage-encoded recombinases are crucial factors in a high evolution rate among temperate phages with a remarkable level of horizontal gene transfer based on genomic comparisons (28). We screened the coding sequences of the phage PP9W (Fig. S2 to S3). We found that the pg54 gene encodes a putative recombinase containing a RecT domain (Fig. 2), which implied it might participate in DNA binding and metabolic process. An electrophoretic mobility shift assay (EMSA) demonstrated that RecT could bind to both single-strand DNA (ssDNA) and double-strand DNA (dsDNA) in a sequence-independent fashion (Fig. S4A to B). Similar results were obtained when different oligonucleotides (Table 2) were tested (data not shown). We further performed a d-loop formation assay to identify DNA strand invasion mediated by RecT. Three light bands showed that both three different ssDNA substrates (Table 2) homologous to the plasmid pUC19 could initiate the formation of joint molecules when incubated with the RecT protein (Fig. S4C). These results hinted that the Pg54 protein (RecT) is indeed a recombinase of the phage PP9W.
FIG 2.
Characterization of the RecT protein. (A) Gene context of pg54 (recT) in the phage PP9W genome. Different colors represent diverse proteins, as indicated above. Some related genes in the subsequent analysis are displayed. (B) A structure diagram of the Pg54 protein. RecT motif is indicated under the gray arrow by WebLogo, in which larger letters represent higher conservative amino acid residues in multiple alignments. The secondary structure analysis of RecT was performed using an online website tool, Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index).
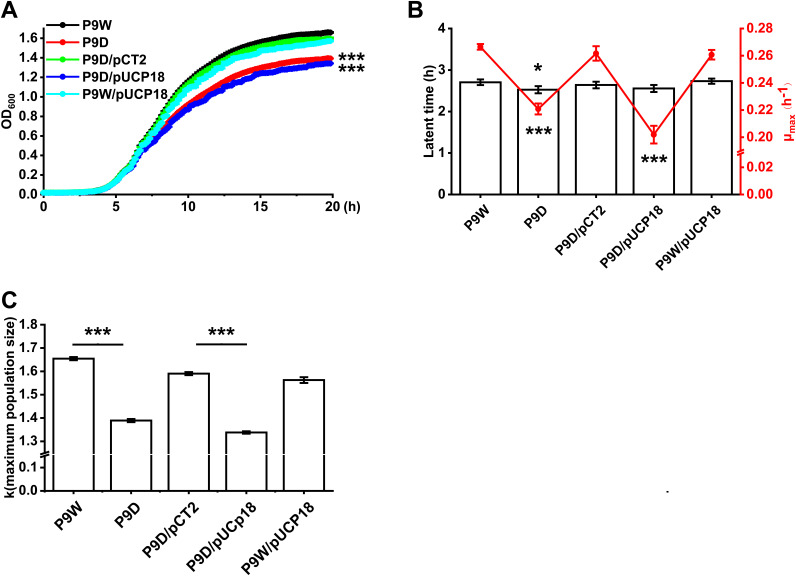
To further investigate the effect of RecT on host bacterial growth, we constructed the recT gene deletion strain, P9D. The growth-curve experiment data showed that RecT improves the cell growth of P9W compared to P9D (P < 0.001, Fig. 3A). Introduction of the empty vector pUCP18 into P9W or P9D did not cause an additional growth burden since no difference was observed in the indicated curves. Meanwhile, complementation of the recT gene (on the plasmid pCT2, Table 2) in P9D restored its final biomass compared to P9D/pUCP18 (P < 0.001). Further nonlinear curve fitting results suggested that the maximum growth rate of P9W was significantly higher than P9D (P < 0.001), although its latent period (29) was a little longer (P < 0.05, Fig. 3B). Similarly, a significant difference of the maximum growth rate between P9D/pCT2 and P9D/pUCP18 was found (P < 0.001). R package analysis also revealed a bigger maximum population size of P9W than P9D and the same results for P9D/pCT2 in comparison with P9D/pUCp18 (P < 0.001, Fig. 3C). These results demonstrated that RecT improves the cell growth rate of P9W.
FIG 3.
Assays for cell growth in different strains. (A) Growth curves of P9W (black line), P9D (red line), P9D/pCT2 (green line, pCT2 carries recT), P9D/pUCP18 (blue line, pUCP18 is the empty vector), and P9W/pUCP18 (cyan line). Overnight cultured strains were inoculated into 200 μl fresh LB medium for each well of 96-well plates at a 1/1,000 dilution ratio. OD600nm value was measured at 5-min intervals for 20 h. (B) Nonlinear curve fitting results correspond to panel A. The blank bar represents the latent time and the red line stands for μmax (maximum growth rate). (C) Maximum population size corresponding to panel A was analyzed by R package (Growthcurve). The experiments were independently replicated three times and each sample was tested in triplicate. Error bars show standard deviations. Data were analyzed by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons (α < 0.05) to examine the mean differences between the data groups. *, P < 0.05; ***, P < 0.001.
Comprehensive effects of RecT on phage excision and gene expression.
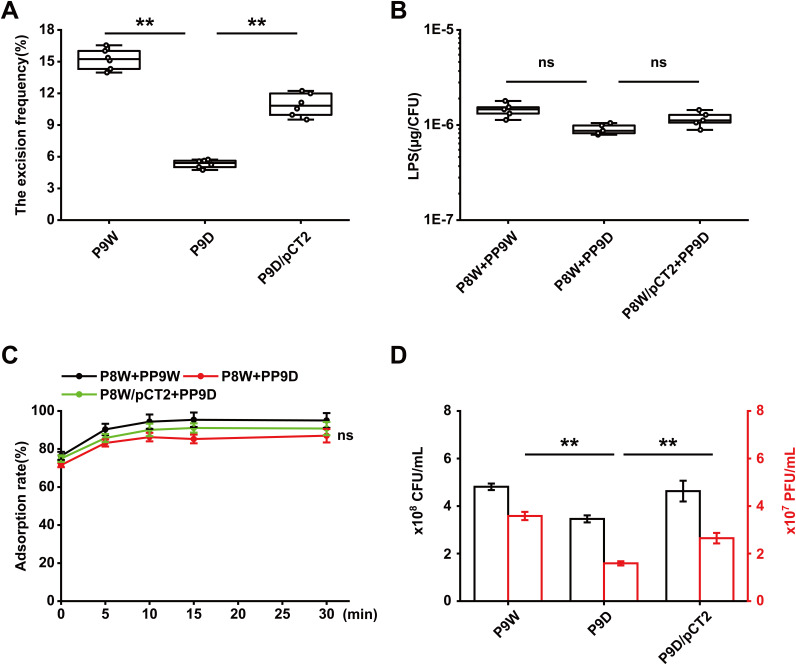
Prophage excision has been shown to provide extra benefits for host bacteria and affect various pathways (30–32). We then asked whether the deletion of recT would interfere with the excision process of PP9W. Several primer pairs (Table 2) were applied to evaluate the excision activity of PP9W (Fig. S5A). Both PP9W and PP9D (without recT) could spontaneously excise from the bacterial chromosome and circularize (Fig. S5B). DNA sequencing confirmed that PP9W and PP9D share the same 59-bp attachment site sequence (5′-AAAACCCCGTAACCCGCGCCTTCCAAGCCCCTCCTTGTGTTTCCGCGACATTTCCTTAT-3′) with the identical L and C forms (Fig. S5C). To further explore the impacts of RecT on the excision frequency of PP9W, we used a qPCR method (Fig. S6) to determine the relative copies of specific DNA fragments (33). In the mid-log phase, about a 1.9-fold increase of the excision frequency in P9W compared to that in P9D was observed. At the same time, the complementation group restored its excision activity by a nearly 1-fold increase (P < 0.01, Fig. 4A). These data indicated that RecT affects the excision frequency rather than the excision process of the prophage.
FIG 4.
RecT impacts the excision frequency of PP9W rather than the relative adsorption rate. (A) Excision frequency analysis in different strains. Each sample was tested in sextuplicate. (B) LPS content quantification in different groups after adsorption with related phages. Each sample was tested in quintuplicate. (C) Adsorption rate assay for PP9W (black line) and PP9D (red line) on P8W. The plasmid pCT2 carries the recT gene. P8W/pCT2+PP9D represents the complementation group (green line). (D) Evaluation of the impacts of recT on the excised phages in the supernatant of P9W. The black border bar represents the bacterial biomass of different strains after growth for 12 h. The red border bar shows the corresponding number of free phages in the supernatant. The experiments were independently replicated three times (A to D), and each sample was tested in triplicate (C and D). Error bars show standard deviations. Data were analyzed by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons (α < 0.05) to examine the mean differences between the data groups. **, P < 0.01; ns, no significance.
Phylogenetic analysis revealed that the phage tail protein (Pg10) of PP9W is highly homologous to D3 (coverage, 100%; identity, 97.47%; data not shown), which is a Pseudomonas phage, using LPS (lipopolysaccharide) as a receptor in the adsorption process. We then quantified the LPS content after adsorption, and there was no difference between the related groups (Fig. 4B). We measured the relative adsorption rate based on the purified phage stocks. The adsorption rate of all the indicated groups peaked at 15 min and remained stable after another 15 min. When P8W was infected with PP9D, the adsorption rate was virtually the same as that of the control group (P > 0.05, Fig. 4C). We also controlled the identical initial bacterial load of P9W and P9D to count the free phages in the supernatant. The final contents of phages were quite different (P < 0.01) due to the different excision frequencies of the prophages (Fig. 4D). These results implied that RecT is probably essential for the efficient excision of PP9W rather than the adsorption process.
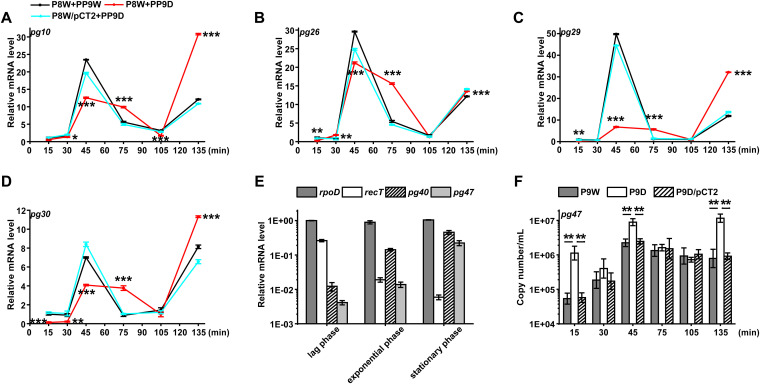
After adsorption, the injected phage genome may initiate a series of processes, such as transcription and translation, which consume the extra energy from the host. The expression of the phage genes encoded structural proteins was assessed by the RT-qPCR method (Fig. 5A to D). According to the one-step growth curve of PP9W (Fig. 1D), we cocultured P8W with the indicated phages to investigate the expression dynamics of these genes. In the first 45 min, the transcription levels of pg10 (encodes the phage tail tip protein) and pg26 (encodes a head decoration protein) were continuously increasing. The red line group (P8W + PP9D) was significantly weaker than the control and the complementation group (the cyan line) at the same time (Fig. 5A and B). These results could be attributed to phage structural protein synthesis. Terminase is required for the maturation of λ-phage DNA and the morphogenesis of the phage head (34). It might explain the similar trends of pg29 (encodes the terminase, large subunit) and pg30 (encodes the terminase, small subunit) (Fig. 5C and D). These results indicated that RecT possibly promotes the transcriptional activity of some functional phage genes when PP9W infected P8W. Besides, we investigated the expression of most recT neighboring genes (Fig. 2A). The curve trends were also obviously different between the red line and the rest groups. Notably, pg55 (encodes a hypothetical protein) was upregulated in all the time points, while the expression pattern of pg56 (encodes a cell division protein) was contrasting (Fig. S7). Both of them were right downstream of the recT gene in PP9W. These results might be explained by polarity on downstream transcripts since the knockout recT fragment contains a predicted promoter around 210 bp upstream of pg55 and pg56.
FIG 5.
RT-qPCR assay. (A to D) for different gene transcription curves of P8W or P8W/pCT2 infected with phage PP9W or PP9D (without recT). The black line indicates the control group, and the red line represents the treatment group. P8W/pCT2 + PP9D (cyan line) stands for the complementation group. (A) pg10 encodes the phage tail tip protein. (B) pg26 encodes the head decoration protein. (C) pg29 encodes terminase large subunit. (D) pg30 encodes terminase small subunit. (E) Gene expression levels of P9W in different growth phases. (lag phase: 1 h; exponential phase: 8 h; stationary phase: 20 h). The dark gray bar indicates rpoD (the host reference gene). The blank bar stands for recT. The striate-filled bar represents pg40 (encodes a CI-like repressor protein C). The light gray bar shows pg47 (encodes a protein containing a CsrA domain). (F) Absolute quantification of pg47 in different strains at lag phase indicated by the corresponding bars above. The experiments were independently replicated three times, and each sample was tested in triplicate. Data were analyzed by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons (α < 0.05) to examine the mean differences between the data groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars show standard deviations.
To further explore whether the expression levels of recT and other prophage genes are impacted by the growth phase. We sampled at 1 h (lag phase), 8 h (exponential phase), and 20 h (stationary phase) in the lysogeny broth (LB) medium of P9W. The pg40 gene encodes a CI-like repressor containing Cro/CI-type HTH and peptidase s24 domains (35). Besides, pg47 is a single copy gene integrated on PP9W that was selected as the marker gene to calculate the total copies (Fig. S6). As shown in Fig. 5E, the transcription level of the host reference gene rpoD remained stable, while that of recT reduced with time. The pg40 and pg47 gene expression levels negatively correlated with recT. Moreover, elevated transcription levels of pg40 and pg47 in a specific period were observed in Fig. S7H and Fig. 5F (P9D versus P9W). These results suggested that a high transcription level of recT depends on the initial stage of host growth, which might repress the expression of pg40 and pg47 uncertainly.
RecT alters bacterial motility, chemotaxis, and biofilm formation.
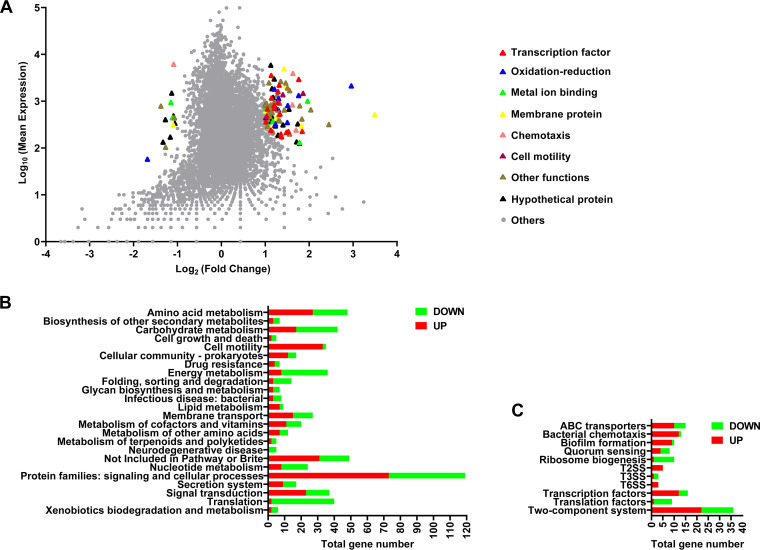
To further figure out the potential role of RecT in the phenotypic flexibility of host bacteria, we conducted RNA-seq analysis to compare gene expression differences between P9D and P9W. Differential expression gene (DEG; P9D versus P9W) data included 350 upregulated and 239 downregulated genes (Data set S1), of which highly expressed genes were classified into several terms by Gene Ontology annotation (Fig. 6A). We also classified these DEGs into 24 central metabolic pathways based on KEGG_B_class (Fig. 6B) and further divided them into 11 crucial groups based on KEGG_C_class (Fig. 6C). Since so many related pathways gene expression had been affected in P9D, we performed a series of tests to verify the potential phenotypic variations.
FIG 6.
Transcriptome analysis of the strain P9D compared to P9W. (A) x axis represents log2-transformed value of gene expression change folds, and y axis represents the logarithm-transformed value of gene mean expression levels. Different colored triangles indicate various pathway genes with significant expression differences. Gray dots represent other genes that do not belong to the above pathways. (B) Enrichment histogram of the genes belong to KEGG_B_class referred to P. aeruginosa PAO1. Green color represents the genes significantly downregulated, while red color represents the genes significantly upregulated. (C) Genes belonging to the different metabolic pathways in KEGG_C_class correspond to panel B.
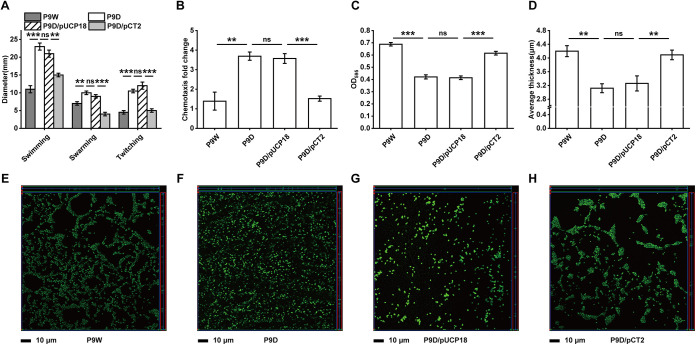
Cell motility is involved in the competition for nutrients and survival space among bacteria in different environments (36). Bacterial flagella have been reported to be associated with swimming and swarming motilities, while type IV pili are required for twitching motility (37). Our transcriptome data showed that more than 30 related genes were significantly upregulated, such as fliM, pilM, and pilB. It was further verified by motility assay, with an increased average migration zone diameter of P9D than P9W in three specific categories. Similar results were obtained when P9D/pUCP18 compared to P9D/pCT2 (P < 0.01 and P < 0.001, Fig. 7A). The subsequent RT-qPCR results of fliM and pilB also identify the RNA-seq data (P < 0.05, Fig. S8A and B). These results suggested that there is a limitation of RecT to the bacterial motility of P9W.
FIG 7.
Assays for bacterial motility, chemotaxis, and biofilm formation. (A) Motility measurement by the migration zone diameter on plates. (B) Chemotaxis activity analysis of cell response to the induction by casein hydrolysates. (C) Microtiter dish biofilm formation assay, the extent of biofilm formation (24 h) was measured using the dye crystal violet in 96-well plates. (D) Average thickness in biofilm formation of the indicated strains corresponding to panels E to H. (E to H) Representative orthogonal image renderings of biofilm formation by the indicated strains grown at 37°C for 16 h. Different groups with an appropriate mixture of the SYTO 9 and propidium iodide stained were then analyzed by a confocal laser scanning microscopy. (E) P9W. (F) P9D. (G) P9D/pUCP18. (H) P9D/pCT2. The scale bar represents 10 μm. The experiments were independently replicated three times and each sample was tested in triplicate. Error bars show standard deviations. Data were analyzed by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons (α < 0.05) to examine the mean differences between the data groups. **, P < 0.01; ***, P < 0.001.
Chemotaxis is positively correlated with the number of chemoreceptors based on genome analysis (38). Three paralogous receptors (PctA, PctB, and PctC) have been reported to regulate P. aeruginosa PAO1 chemotaxis to proteinogenic amino acids and chlorinated hydrocarbons (39). We evaluated several bacteria chemotaxis activity by casein hydrolysates induction. The migrating cell number of P9D increased 1.7-fold compared to that of P9W (P < 0.01). Meanwhile, a 1.3-fold increased migrating cell number of P9D/pUCP18 than P9D/pCT2 was observed (P < 0.001, Fig. 7B). Transcription levels of numerous chemotaxis-related genes were upregulated in P9D, such as pctC, pctA, aer, and cttP (Fig. 6C), except for the only one downregulated gene PA4915 (encodes a probable chemotaxis transducer). We also found that the transcription level of pctA in P9D increased nearly 1.5-fold more than in P9W, while there was a 1.7-fold elevated transcription level of pctA in P9D/pUCp18 than in P9D/pCT2 by RT-qPCR assay (P < 0.01, Fig. S8C). These data indicated that RecT negatively influences the chemotaxis activity of P9W since P9D cells migrated more rapidly and efficiently.
The regulation of biofilm formation in P. aeruginosa includes three most important factors: quorum sensing (QS), cyclic diguanylate (c-di-GMP), and small RNAs (sRNAs) (40). Apart from these factors, a two-component system, adhesins, motility, rhamnolipids, and extracellular polymeric substances (EPS) also play a role in biofilm formation (41). The transcriptome analysis showed that many related genes of P9D expressed significantly differently, such as rhlR, lasR (QS system), flgM, algR, algZ (two-component system), wspA, fleS, and fliM (motility). We thus quantify the extent of biofilm formation by a crystal violet staining method. The biofilm biomass of P9D decreased by 38.8% (versus P9W), and that of P9D/pUCp18 is decreased by 32.7% (versus P9D/pCT2), respectively, without the presence of RecT (P < 0.001, Fig. 7C). To further observe the morphology differences, we stained the inchoate biofilms of the indicated strains (16 h). The P9D group showed a more dispersed bacterial distribution (Fig. 7E and F) and a thinner biofilm in the z axis (P < 0.01, Fig. 7D) than P9W group. Similar results were observed when P9D/pUCP18 group compared to P9D/pCT2 group (Fig. 7D, G, and H). These results demonstrated that RecT promotes the biofilm formation in P9W.
RecT impacts host catalase activity, antibiotic resistance, and virulence.
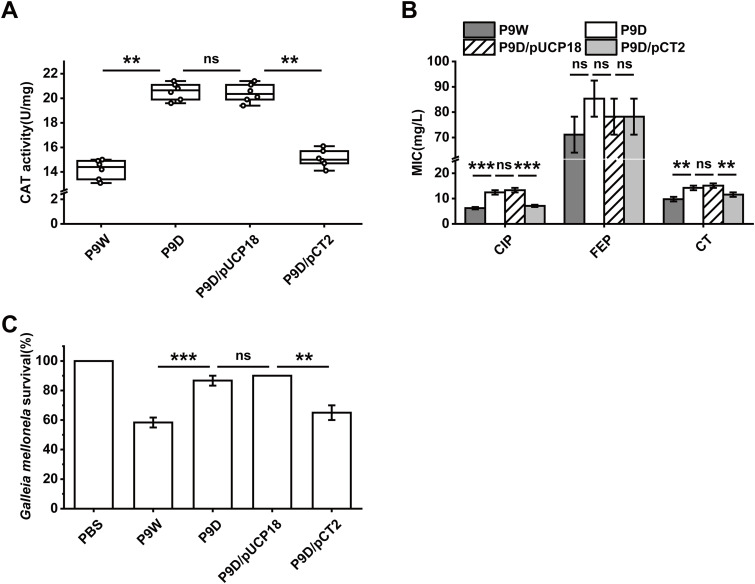
The SOS regulatory system was found to be involved in the repair of damaged DNA or DNA replication inhibition, which is in response to reactive oxygen species (ROS) in E. coli (42). Oxidation-reduction-related gene expression was significantly altered for 10 genes, of which nine were upregulated, such as ahpB, folM, rmd, xdhA, and hmgA, except for the only 1 downregulated gene, PA4515 (Fig. 6A). The catalase (CAT) activity of P9D increased by 44.7% (versus P9W), while that of P9D/pUCP18 increased by 35.2% (versus P9D/pCT2, P < 0.01, Fig. 8A). The transcription level of rmd in P9D increased almost 1.9-fold than P9W. There was an about 1.5-fold enhanced expression level of rmd in P9D/pUCP18 compared to P9D/pCT2 (P < 0.001, Fig. S8D). These data implied that RecT inhibits the catalase activity of the host strain.
FIG 8.
Assessments of catalase activity, antibiotic resistance, and virulence of the indicated strains. (A) Catalase activity test by a CAT enzyme activity kit with the microplate reader. (B) Antibiotic resistance analysis, ciprofloxacin (CIP), cefepime (FEP), and colistin sulfate (CT). (C) Virulence evaluation by Galleia mellonella survival rate injected with the indicated strains after 24 h. PBS stands for the control group (no treated). The experiments were independently replicated three times and each sample was tested in triplicate (B and C) or sextuplicate (A). Data were analyzed by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons (α < 0.05) to examine the mean differences between the data groups. **, P < 0.01; ***, P < 0.001. Error bars show standard deviations.
An increasing threat of chronic and hospital-acquired infections is associated with the prevalence of multidrug-resistant (MDR) P. aeruginosa (43). All the P. aeruginosa strains in this study were isolated from severely burned patients, and a routine test for MIC was conducted (Table S2). RNA-seq data revealed seven drug resistance genes’ expression distinctly altered in P9D (Fig. 6B). We used a disk diffusion test, including 11 antibiotics to screen the possible targets (data not shown) and further reduced the MIC variation range progressively in 96-well plates. Enhanced resistance of P9D to ciprofloxacin (2-fold) and colistin sulfate (1.5-fold) was observed (versus P9W). Analogous to the improved resistance of P9D/pUCP18 (versus P9D/pCT2) to ciprofloxacin (1.9-fold) and colistin sulfate (1.3-fold), respectively (P < 0.01 and 0.001, Fig. 8B). These results were consistent with the RT-qPCR tests of mexT and arnB (P < 0.01, Fig. S8E and F), suggesting that RecT blocks the multidrug resistance of P9W.
To further investigate the role of RecT in vivo, a Galleria mellonella killing assay was performed. In the absence of RecT, the survival rate of the P9D-infected group significantly elevated by 28.3% (versus the P9W-infected group, P < 0.001), while that of the P9D/pUCP18-infected group increased by 25% (versus the P9D/pCT2-infected group, P < 0.01, Fig. 8C), respectively. The transcriptome analysis showed that the expression of several bacterial infectious disease (KEGG_B_class) genes was dramatically changed (Fig. 6B), which might explain the hypo toxicity of P9D in Galleria mellonella acute infection model.
DISCUSSION
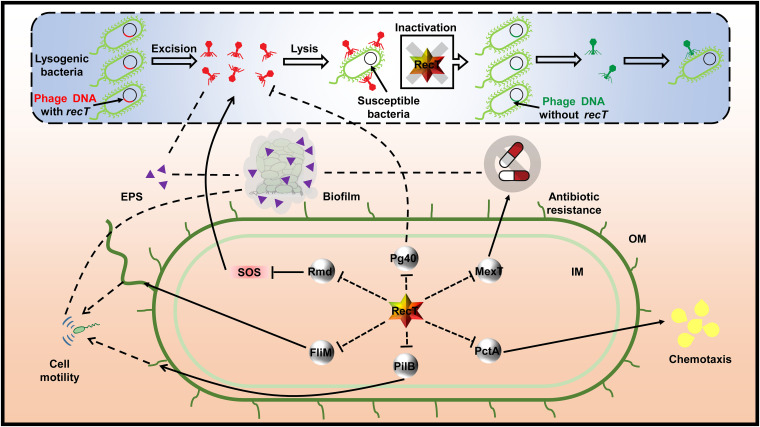
Many phage proteins have been shown to improve host growth rate, mediate spontaneous phage induction (SPI), and play crucial roles in bacterial physiological phenotypes via various mechanisms (27). There are still difficulties in comprehensively defining those functionally unclear genes in phage genomes due to the high variation of phages, the limitations of classification based on alignment, and the reference phage sequences in databases (44). In this study, the Pg54 protein (RecT) located on a prophage PP9W of P. aeruginosa P9W is identified to function in prophage excision and gene transcription and phenotypic traits of the host. Here, we proposed a schematic model of RecT functions combined with some identified research findings (Fig. 9).
FIG 9.
Schematic of RecT’s role in modulation of prophage excision and omnifarious host core phenotypic traits. Solid lines show explicit regulatory relationships that have been established previously. Dotted lines indicate cryptic hints (direct or indirect) that need further investigation. Arrowheads stand for a specific extent of stimulation or enhancement, while bars represent inhibition of a sort. RecT is depicted in gradient-colored hexagon textboxes. Related structural proteins or enzymes are depicted in gray circular textboxes. EPS, extracellular polymeric substances; SOS, stress response; IM, inner membrane; OM, outer membrane.
The lysogenic state of E. coli phage λ is generally stable. However, spontaneous phage induction (SPI) is readily implemented, attributed to two widely accepted hypotheses (45). The first is the self-cleavage of the phage repressor CI stimulated by RecA nucleoprotein filaments, which then activates the lytic promoters, leading to the prophage excision (42). Another route is that RecA nucleoprotein filament triggers the autocatalytic cleavage of LexA, resulting in the derepression of SOS genes expression and the formation of antirepressor. It finally promotes the inactivation of the repressor CI (46, 47). In this study, a CI-like protein (48) is encoded by pg40 in prophage PP9W, whose expression level was negatively correlated with recT in the RT-qPCR assay (Fig. 5E). In addition, we observed that the prophage excision frequency of P9D distinctly reduced compared to that of P9W (Fig. 4A). These results indicated that the increased transcription level of pg40 might suppress the excision of the prophage in the absence of recT.
Phage lytic development and oxidative stress are two direct and indirect triggers of the SOS response, a global regulatory system in response to DNA damage modulated by LexA and RecA (49). Interestingly, numerous oxidation-reduction-related gene expression levels were significantly elevated in P9D (Fig. 6A). The catalase activity assay of P9D (Fig. 8A) and related RT-qPCR data (Fig. S8D) further support the transcriptome results. The oxidoreductase such as Rmd probably reduces the highly intracellular oxidative stress and further alleviates the SOS response, eventually leading to the downregulated prophage excision frequency (Fig. 9). To sum up, RecT likely promotes prophage excision of the host cell by a dual inhibition analogous to RecA (Fig. 9). More experiments are required to clarify the relationship between RecT and RecA in P. aeruginosa and reveal the underlying mechanisms.
Many bacteria use swimming and swarming motility to achieve surface colonization in liquid medium, which is associated with bacteria flagellum (50). Numerous flagella-related genes were significantly upregulated by the transcriptome analysis (Fig. 6B and C), such as flhA, flhF (flagellar biosynthesis protein), fliM (flagellar motor switch protein), fliE (flagellar hook-basal body complex protein), fliD (flagellar capping protein), fliC (flagellin type B), flgB, flgF, and flgG (flagellar basal-body rod protein), etc. Twitching motility allows cells to move forward and backward on a moist surface at specific speeds, which is involved in polar pili (51). We found the increased transcription levels of some pili-related genes, including pilB and pilM (type 4 fimbrial biogenesis protein). P9D showed a noticeable enhancement in cell motility (Fig. 7A). The RNA-seq analysis indicated the upregulation of chemotaxis-related gene expression in P9D, such as pctA, pctC (chemotactic transducer), aer (aerotaxis receptor), and cttP (chemotactic transducer for trichloroethylene). The subsequent chemotaxis test (Fig. 7B) and RT-qPCR assay results (Fig. S8C) are consistent. These data revealed that RecT exhibits adverse effects on host motility and chemotaxis by various factors (Fig. 9).
We found that the biofilm formation of P9D was significantly decreased (Fig. 7C to H). Many factors might affect biofilm biosynthesis in P9D, such as a lower content of EPS (owing to the lower excision frequency) and more vigorous cell motility (which leads to a more dispersed planktonic state). Biofilms often derive several emergent features, one of which is antibiotic resistance (52). However, the resistance of P9D to ciprofloxacin (fluoroquinolones) or colistin sulfate (lipopeptide antibiotics) was enhanced (Fig. 8B). It may be attributed to the upregulation of mexT and arnB in RNA-seq (Fig. 6B) and RT-qPCR assay (Fig. S8E and F). MexT is a regulator of the MexEF-OprN efflux pump in P. aeruginosa, which can accelerate the excretion of intracellular fluoroquinolones (53). Addition of the 4-amino-4-deoxy-l-arabinose (l-Ara4N) moiety to the phosphate group(s) of lipid A can confer polymyxin resistance. The relevant aminotransferase is encoded by ArnB (54). These results indicated that RecT perhaps plays multiple roles in biofilm formation and antibiotic resistance via complicated mechanisms (Fig. 9).
Our results demonstrate that the inactivation of RecT (P9D) results in increases in host cell motility (Fig. 7A), chemotaxis (Fig. 7B), catalase activity (Fig. 8A), and antibiotics resistance (Fig. 8B). At the same time, it also leads to decreases in host growth rate (Fig. 3B), prophage excision frequency (Fig. 4A), biofilm formation (Fig. 7C to H), and virulence (Fig. 8C). In a nutshell, RecT probably acts as a suppressor to several crucial transcription factors and essential genes in host diverse core cellular processes (Fig. 9). These data improve our understanding of bacteria-phage interplays, given that the phage recombinase-like RecT plays a pivotal role in host phenotypic flexibility, which may be speculated by the adaptive balance under continuous evolutionary pressure. There are still many unclear molecular mechanisms underlying the complicated modulation web of RecT. Further study is warranted to illustrate its detailed performance in P. aeruginosa, and more attention should be paid to unclear prophage genes in many other bacteria.
MATERIALS AND METHODS
Bacterial strains, phages, plasmids, and culture conditions.
Bacterial strains, phages, and plasmids used in this study are listed in Table 1. Primers utilized in this work are listed in Table 2. We routinely cultured our strains in lysogeny broth (LB) medium at 37°C supplemented with appropriate antibiotics unless otherwise stated.
Spot test for the interactions of five clinical P. aeruginosa isolates.
One milliliter of the overnight culture medium of five P. aeruginosa isolates was first centrifuged. The supernatant was stocked by sterile disposable syringes with 0.22-μm filters. Cell pellets were resuspended in 0.9% sterile saline, and this was repeated three times. Then, these pellets were used to adjust optical density at 600 nm (OD600nm) to 0.5, and we inoculated 100 μl of each strain with 3.5 ml of cooled soft top (0.75%) agar and poured the mixture onto LB agar (1.5%) plates with gentle sloshing to produce an overlay of the indicator strain lawn. Next, we pipetted 10-μl supernatant stocks of individual test strains and spotted them onto the overlay agar, incubating them inverted at 37°C overnight when the spots were dried.
PCR conditions.
All primers we used here were designed by Primer Premier 6 using P9W or PP9W Genome as a reference (Table 2). Our PCR volume of 25 μl comprised 12.5 μl 2× Taq master mix, 11 μl of deionized water, and 0.5 μl each of the primers (10 mM) and templates. PCR conditions generally used in this work were 94°C for 5 min, followed by 34 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 60 s/kb, followed by a final extension at 72°C for 7 min.
Counting the ratio of PP9W/PP9W2 in the supernatant of P9W.
Three milliliters of overnight culture media of strain P9W was harvested by centrifugation. The pellet was washed twice and resuspended in an equal volume of fresh LB medium. We then pipetted 200 μl to inoculate into 20 ml LB medium and sampled 100 μl every hour to measure the OD600nm value for 12 h (triplicates of each sample were tested, and LB was the control group). Meanwhile, we collected the related supernatant after centrifugation by further filtration with a 0.22-μm filter. The P8W strain lawn was used as an indicator to quantify the number of plaques caused by free phages in the supernatant of P9W every hour. Through serial dilution, we selected appropriate double agar (PFU around 30) to dig out the central zone of plaques and use as PCR templates with two pair primers, which amplify specific fragments of PP9W and PP9W2, respectively (Table 2). The ratio of PP9W/PP9W2 depends on the corresponding number of positive bands after gel electrophoresis.
Prediction of the number and location of prophages in P8W and P9W.
The number and location of prophages in genomes of P8W and P9W were predicted by an online tool, Prophage Hunter (https://pro-hunter.genomics.cn/). Then, we screened the potential active prophages by the scores and further identified their activity in the supernatant of P8W and P9W by PCR using different paired primers listed in Table 2. We found that the P8W chromosome likely integrated four active prophages and one more for the P9W chromosome (data not shown). Spot assay and PCR results revealed that only PP9W and PP9W2 could excise spontaneously. Moreover, PP8-1/PP9-2, PP8-3/PP9-4, and PP8-4/PP9-5 were three predicted paired prophages, highly homologous to each other (>99%). Although PP8-2 seems to be a specific prophage integrated into P8W, the corresponding fragment in P9W was indeed highly homologous to PP8-2 (>75%).
Growth inhibition experiment.
Five milliliters of overnight culture medium of strain P8W was harvested by centrifugation. The pellet was washed twice and resuspended in 1 ml fresh LB medium. It was further incubated with the phage PP9W at different MOI (0.1, 1, and 10). Then, the mixtures were subjected to 50 ml fresh LB medium with violent shaking (180 rpm) at 37°C for 12 h. The optical densities of samples were measured three times at a wavelength of 600 nm at 1-h intervals.
One-step growth experiment.
Five milliliters of culture medium of P8W was incubated to an OD600nm value at 0.4 to 0.6 and centrifuged. The pellet was resuspended in an equal volume of fresh LB medium, and we used 100 μl to mix with 1 ml purified phage PP9W solution (1 × 107 PFU/ml). Then, 10 mM CaCl2 was also added according to the previous study (26). After adsorption for 15 min at 37°C, the mixture was immediately subjected to 100 ml fresh LB medium. The culture was continuously incubated at 37°C for 6 h, which we sampled (100 μl) every 30 min, and the phage titer was determined by a double-layer agar plate method.
Construction of the recT gene deletion derivate.
A homologous recombination method was used to knock out the recT gene in P9W. In brief, we first amplified the upstream and downstream fragments (about 1 kb) of the recT gene and ligated the molecules by T4 ligase. Then, the complex was amplified and cloned into the plasmid pEX18Tc. The new fusion plasmid pEX18Tc::U+D was first transformed in E. coli DH5α, then transformed in E. coli S17-1, and finally, introduced into the strain P9W by conjugation. The recT gene mutant derivates were first screened by LB agar supplemented with 100 mg/liter tetracycline and 50 mg/liter kanamycin. The second-round screening was conducted by LB agar supplemented with 7.5% sucrose. The result ΔrecT mutants (P9D) were identified by PCR and DNA sequencing. Bacteria and plasmids used here are listed in Table 1, and primers are listed in Table 2.
Complementation test.
The fragment of the recT gene was amplified and cloned into the plasmid pUCP18, and we named the resultant plasmid pCT2. It was electroporated into the competent cells of P9D. The complemented derivate was selected by LB agar supplemented with 100 mg/liter ampicillin and further verified by PCR and DNA sequencing. The bacteria, plasmids, and primers used are listed in Table 1 and 2.
Prophage excision frequency quantification.
The frequency of prophage excision in different strains was quantified by a modified quantitative PCR method (31). In brief, the number of total PP9W copies was quantified rely on the single-copy reference gene pg47. The number of integrated PP9W copies was quantified using primer pair P9I-F/P9I-R (Table 2), which only results in PCR products when the prophage is integrated. The binding efficiency of the primers used to quantify the excision frequency was evaluated by various proportions of genomic DNA from wild-type versus ΔrecT mutant type (ranging from 1/105 to 1/1010) as a template to generate the standard curves.
Lipopolysaccharide quantification.
LPS of the related groups was extracted following the instructions of an LPS extraction kit (Abcam, ab239718). Briefly, bacteria were grown overnight in LB medium, and 1 ml culture was resuspended in an equal volume of PBS. Then, it was mixed with related phages for 15 min, centrifuged and resuspended in extraction buffer, sonicated, and incubated on ice for 10 min. After centrifugation, the supernatant was treated with proteinase K at 60°C for 1 h. Any unlysed debris was removed by centrifugation. Total carbohydrate content (g/CFU) in the supernatant was quantified following a total carbohydrate quantification assay (Abcam, ab155891).
Adsorption rate assay.
A previously described method (55) was used with some modifications. Briefly, the indicated overnight cultured cells were centrifuged (12,000 rpm, 3 min), washed twice, and resuspended at a final concentration of 1 × 107 CFU/ml (determined by plate count) in fresh LB medium supplemented with 10 mM CaCl2. We added purified phage stocks at a multiplicity of infection (MOI) of 1, and the mixture was incubated without shaking at 37°C for 30 min. At time intervals, an aliquot was centrifuged (12,000 rpm, 3 min), and the number of free phages in the supernatant was determined by the double-layer agar plate method. In contrast, the total phage particle count was determined directly without centrifugation. The phage adsorption rate was quantified by (total − free)/total × 100%.
Purification of the RecT protein.
We first cloned the recT gene fragment into the plasmid pET32a(+) to produce a fusion plasmid pET32a-T and transformed it into E. coli BL21(DE3). The positive clones were selected by antibiotic resistance agar plates (100 mg/liter ampicillin), PCR, and DNA sequencing validation. Then, we inoculated the strain E. coli BL21(DE3)/pET32a-T and cultured it overnight. On the second day, we pipetted a 1-ml bacteria solution and centrifuged (12,000 rpm, 3 min) and resuspended it in 1 ml fresh LB medium three times. Then, 200 μl of that solution was inoculated into 20 ml LB medium supplemented with 100 mg/liter ampicillin. After 2 h, 1.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to induce the expression of recT, and the mixture was further grown for 3 h. Cells were harvested by centrifugation, and we first used One Step Bacterial Active Protein Extraction kit (Sangon Biotech) to obtain soluble proteins. Ni-NTA-Sefinose Column (Bio basic, Canada) was further used to extract the His-tagged RecT protein according to the manufacturer’s protocol. Finally, a DeSalting Gravity Column (Sephadex) was used to purify the target protein, and the concentration was measured by Implen Nano Photometer (Version 3.1).
Electrophoretic mobility shift assay.
We performed both ssDNA and dsDNA binding tests for the RecT protein here. For ssDNA, we directly synthesized three different homologous fragments (50 bp) to the plasmid pUC19 through Sangon Biotech (Table 2). As for dsDNA, we amplified several promoters of the indicated genes (Table 2), which are close to the recT gene in PP9W. The PCR fragments (200 to 300 bp) were first cloned into pMD19 (Simple, TaKaRa), and the ligated fragments were again amplified using the primers M13-47 and RV-M (Table 2). The electrophoretic mobility shift assay (EMSA) kit (Molecular Probes) was used here. Briefly, we first mixed various concentrations of purified His-tagged RecT protein (0 to 5.28 μM) with ssDNA (6.00 μM) or dsDNA (1.21 μM) in Component E (750 mM KCl, 0.5 mM dithiothreitol, 0.5 mM EDTA, 50 mM Tris, pH 7.4) at 25°C for 30 min. At the end of the incubation period, we added 6× EMSA gel-loading solution (Component D) into reaction mixtures and mixed gently but thoroughly. Finally, we separated the DNA-protein complexes by electrophoresis using a 5% nondenaturing polyacrylamide gel. The gel was visualized with the Automatic Digital Gel Image Analysis System (Tanon 1600R).
D-loop formation assay.
The test was conducted as previously described (21) with minor modifications. Briefly, the reaction mixtures (30 μl) contained 2 μM single-stranded oligonucleotide, 25 mM NaCl, 20 mM Tris-HCl (pH 7.5), 100 mg/ml BSA (bovine serum albumin), 0.5 mM DTT (dithiothreitol), and 2 μM purified RecT protein (Treated group, Fig. S4, lanes 3 to 5). The single-stranded oligonucleotide was replaced by an equal volume of ddH2O in the control group (Fig. S4, lane 2). Mixtures were incubated at room temperature for 20 min, and 15 μM supercoiled DNA (pUC19) was added. Mixtures were further incubated at 37°C for 30 min. The final concentrations of 50 mM EDTA (pH 8.0), 0.2% SDS, and 0.5 mg/ml proteinase K were added to stop the reactions. Then, the incubation was continued at 37°C for an additional 20 min, and the mixtures were finally separated by 0.8% agarose gel electrophoresis.
Growth curve experiment.
Overnight cultured indicated strains were inoculated at a 1/1,000 dilution ratio using 96-well plates. Two hundred microliters of fresh LB medium was added to each well, and each sample was tested in triplicate. An automatic microplate reader (BioScreen, Finland) was used to quantify the OD600nm value at 5-min intervals for 20 h. The latent time and maximum growth rate were analyzed by nonlinear curve fitting, while the maximum population size was analyzed by R package (Growthcurve).
Reverse transcription-quantitative real-time PCR.
In part 1 (Fig. 5A to D), we incubated the strain P8W or P8W/pCT2 with the phage PP9W or PP9D in 100 ml LB medium at an MOI of 1 according to growth inhibition experiment results. We took multiple 1-ml samples every 15 min or 30 min that were immediately subjected to total RNA extraction using a Total RNA Extraction kit (Promega). Then, 1 μg total RNA was subjected to reverse transcription using the FastKing gDNA Dispelling RT SuperMix (TianGen) according to the manufacturer’s protocol. The cDNA was finally subjected to RT-qPCR analysis, and the RT-qPCR volume of 20 μl comprised 10 μl TB Green Premix Ex TaqII (TaKaRa Bio), 8.2 μl of deionized water, 0.4 μl each of the primers (10 mM), and a 1-μl template. The RT-qPCR conditions in this study were 94°C for 30 s; followed by 40 cycles of 94°C for 5 s, 57.4°C for 12 s, and 72°C for 12 s; followed by 95°C for 30 s, 45°C for 30 s, and final melt curve at 60°C to 95°C (increment of 0.5°C) for 5 s. A typical 2−ΔΔCT method was used to perform relative quantification (56). All samples were tested in triplicate, and the primers used here are listed in Table 2.
In part 2 (Fig. 5F and Fig. S7), we incubated the strain P9W, P9D, and P9D/pcT2 directly in 100 ml LB medium, and the remaining steps were the same as above. Particularly in Fig. 5E, we only incubated the strain P9W and sampled at 1 h, 8 h, and 20 h.
RNA-seq analysis.
P. aeruginosa P9W and P9D cells were grown to OD600nm of 0.8 and harvested by centrifugation. The collected cells were immediately subjected to freezing in liquid nitrogen and delivered to Novogene (Beijing) in dry ice for transcriptome analysis. In brief, total RNA of each sample was extracted and subsequently subjected to rRNA removal. Fragmentation was carried out using purified mRNA before cDNA synthesis. The cDNA libraries were sequenced in triplicate on an Illumina Hiseq platform, and paired-end reads were generated. Reference genome, P. aeruginosa P9W (CP081202.1), and gene model annotation files were downloaded from the genome website directly. Both building index of the reference genome and aligning clean reads to the reference genome were by Bowtie2-2.2.3 (57). The total mapped reads number of the P9W sample was 25,665,718 (97.09%), while there was a total of 18,766,508 (96.96%) reads of the P9D sample. Differential expression analysis of two groups was performed using the DESeq R package (1.18.0) (58). The resulting P values were adjusted using Benjamini and Hochberg’s approach to controlling the false discovery rate (FDR). Genes with an adjusted P value of <0.05 found by DESeq were assigned as differentially expressed, in which the DEGs with FDR ≤ 0.001 and fold change |log2Ratio| ≥1 were considered significantly changed. The KEGG database (http://www.genome.jp/kegg/) was used to classify the DEGs based on their functions (59). P. aeruginosa PAO1 was selected as the reference genome to convert gene IDs (http://kobas.cbi.pku.edu.cn/kobas3/annotate/).
Cell motility assay.
In the swimming test, bacteria were grown to an OD600nm of 0.5, and 1 μl of the culture was dropped onto the semisolid medium containing 0.2% (wt/vol) agar, 1% (wt/vol) tryptone, and 0.5% (wt/vol) NaCl as described previously (27). The diameter of the swimming zones was measured when the plates were incubated at 37°C for 24 h. The swarming assay is similar to the above except for another semisolid medium with 0.3% (wt/vol) agar. For twitching motility assay, bacteria were inoculated onto 1% L-agar by stabbing isolated fresh colonies to the bottom layer. The plates were incubated for 24 h at 37°C, then the agar was gently removed, and the biomass that adhered to the bottom was stained with 1% crystal violet for 1 min, washed three times with water (60). The plates were further dried inverted overnight, and the diameter of the stained zone was measured.
Chemotaxis evaluation.
We first prepared a pipette tip filled with the 100 μl indicated strain culture (OD600nm = 0.5). Then, 1 ml 1% casein hydrolysates inducer was transferred into a 5-ml sterile syringe. Then, the needle was dipped into the culture in the prepared pipette tip. After incubation at room temperature for 30 min, the mixture in the syringe was plated onto LB agar to enumerate the migrating cell numbers.
Biofilm formation assay.
Overnight cultured bacteria were diluted at 1:100 and resuspended in fresh LB medium, of which 100 μl was spotted onto a glass bottom cell culture dish (15 mm, polystyrene nonpyrogenic, sterile; Sigma, USA) and incubated at 37°C without shaking in dark for 16 h. Then, the planktonic residual liquid was carefully removed, and the biomass adhered to the bottom dish was stained with a mixture of SYTO 9/propidium iodide in dark for 30 min according to the Live/Dead Bac Light Viability kit. After that, the excess dye was also removed and the dishes were subjected to image analysis by a confocal laser scanning microscopy (LSM880, Zeiss). Both orthogonal images and the average thickness of the z axis were analyzed by an application program, Zen. In the microtiter dish biofilm formation assay, we used a typical method described previously (61). Briefly, we first grew cultures of the indicated strains overnight in LB medium, diluted the cultures at 1:100 into fresh M63 medium, and then added 100 μl of the dilution to each well in a 96-well dish(six replicate wells per treatment). The plates were incubated for 24 h at 37°C. Then, the planktonic cells were dumped out by turning the plate over and washing three times. Next, 125 μl of 0.1% crystal violet (CV) solution was added to each well of the plate and incubated at room temperature for 15 min. Then, the plate was rewashed three to four times. Another 125 μl of 30% acetic acid solution was added to each well until the plate was dried and incubated at room temperature for another 15 min. We finally transferred 100 μl of the solubilized CV to a new dish and measured absorbance in a plate reader at 595 nm using 30% acetic acid solution as the blank.
Catalase activity assay.
A catalase activity detection kit (mbio) was used here. In brief, overnight cultured bacterial cells were harvested by centrifugation and resuspended in PBS (1:100 diluted). About 5 × 106 cells were collected by centrifugation and removed from the supernatant. Then, 1 ml extracting solution was added, followed by ultrasonic disruption, and the supernatant was placed onto the ice after centrifugation (8,000 × g, 4°C, 10 min). The spectrophotometer was preheated for 30 min, the wavelength was adjusted to 240 nm, and the distilled water was used for zero setting. CAT detection solution was heated in the water bath for 10 min. Then, 1 ml CAT detection solution was added to a 1-ml quartz colorimetric dish, followed by a mix with a 35-μl sample for 5 s. The initial absorbance value A1 at 240 nm and the absorbance value A2 after 1 min were both measured immediately at room temperature. The catalase activity was determined by CAT (1 U = nmol/min/104 cell) = 1.356 × (A1 − A2).
Antibiotics resistance experiment.
According to the MIC of wild-type P. aeruginosa P9W (Table S2), we first performed a disk diffusion test following the manufacturer’s instructions to screen the target antibiotics. Further MIC in detail was determined by increasing amounts of ciprofloxacin, cefepime, and colistin sulfate to the indicated strains in 96-well plates (100-μl volumes) gradually. Samples were incubated at 37°C without shaking for 18 h (62) and measured an optical density at 600 nm.
Galleria mellonella acute infection model.
Galleria mellonella killing assay was conducted following a previous study (63). Briefly, overnight grown P. aeruginosa in LB medium at 37°C was diluted at 1:100 into fresh media at 37°C until OD600nm reached 0.6 to 0.8. Then, the cells were collected by centrifugation, washed twice, and resuspended to a final concentration of 1 × 105 CFU/ml (plate count) using 10 mM MgSO4. A microsyringe (10 μl; Hamilton, Germany) was used to squeeze a 10-μl volume diluted strain solution or MgSO4 (aq) into Galleria mellonella, and the larvae were grown at 37°C without shaking in dark for 24 h. Five experimental groups of Galleria mellonella are described as follows: (i) not treated; (ii) injected with P9W; (iii) injected with P9D; (iv) injected with P9D/pUCP18; and (v) injected with P9D/pCT2. Each group comprised 20 larvae and three replicates.
Statistical analysis.
One-way ANOVA with Tukey’s multiple comparisons (α < 0.05) was used to compare the mean differences between the data groups in this study.
Data availability.
The whole-genome sequencing data of P8W and P9W were deposited at Sequence Read Archive of NCBI with accession no. SRA: PRJNA753640 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA753640). The whole-genome sequencing data of PP9W have been made available at the National Center for Biotechnology Information with GenBank no.: MZ773939.1 (https://www.ncbi.nlm.nih.gov/nuccore/2095887126). The transcriptome data have been deposited at Sequence Read Archive of NCBI with accession no. SRA: PRJNA774287 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA774287). Other related data are available in this article or from the corresponding authors upon request.
ACKNOWLEDGMENTS
This work was supported by Key projects of the National Natural Science Foundation of China (41831287), the National Key R&D Program of China (2020YFC1806904), the China National Science Fund for Distinguished Young Scholars (41525013), and National Natural Science Foundation of China (31870351).
We thank W. Wu (College of Life Sciences, Nankai University) for kindly providing the plasmids in this study.
Footnotes
Supplemental material is available online only.
Contributor Information
Weihui Wu, Email: wuweihui@nankai.edu.cn.
Yi Luo, Email: luoy@nankai.edu.cn.
Maia Kivisaar, University of Tartu.
REFERENCES
- 1.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morita Y, Tomida J, Kawamura Y. 2014. Responses of Pseudomonas aeruginosa to antimicrobials. Front Microbiol 4:422. 10.3389/fmicb.2013.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fournier A, Voirol P, Krahenbuhl M, Bonnemain CL, Fournier C, Pantet O, Pagani JL, Revelly JP, Dupuis-Lozeron E, Sadeghipour F, Pannatier A, Eggimann P, Que YA. 2016. Antibiotic consumption to detect epidemics of Pseudomonas aeruginosa in a burn centre: a paradigm shift in the epidemiological surveillance of Pseudomonas aeruginosa nosocomial infections. Burns 42:564–570. 10.1016/j.burns.2015.10.030. [DOI] [PubMed] [Google Scholar]
- 4.Livermore DM. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis 34:634–640. 10.1086/338782. [DOI] [PubMed] [Google Scholar]
- 5.Mirzaei MK, Maurice CF. 2017. Ménage à trois in the human gut: interactions between host, bacteria and phages. Nat Rev Microbiol 15:397–408. 10.1038/nrmicro.2017.30. [DOI] [PubMed] [Google Scholar]
- 6.Li Z, Li W, Ma W, Ding Y, Zhang Y, Yang Q, Wang J, Wang X. 2021. Characterization and application of a lytic phage D10 against multidrug-resistant Salmonella. Viruses 13:1626. 10.3390/v13081626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loś JM, Loś M, Wegrzyn G, Wegrzyn A. 2009. Differential efficiency of induction of various lambdoid prophages responsible for production of Shiga toxins in response to different induction agents. Microb Pathog 47:289–298. 10.1016/j.micpath.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Liu X, Tang K, Wang P, Zeng Z, Guo Y, Wang X. 2019. Excisionase in Pf filamentous prophage controls lysis-lysogeny decision-making in Pseudomonas aeruginosa. Mol Microbiol 111:495–513. 10.1111/mmi.14170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Touchon M, Bernheim A, Rocha EP. 2016. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J 10:2744–2754. 10.1038/ismej.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Kim Y, Ma Q, Hong SH, Pokusaeva K, Sturino JM, Wood TK. 2010. Cryptic prophages help bacteria cope with adverse environments. Nat Commun 1:147. 10.1038/ncomms1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor VL, Fitzpatrick AD, Islam Z, Maxwell KL. 2019. The diverse impacts of phage morons on bacterial fitness and virulence. Adv Virus Res 103:1–31. 10.1016/bs.aivir.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Newton GJ, Daniels C, Burrows LL, Kropinski AM, Clarke AJ, Lam JS. 2001. Three-component-mediated serotype conversion in Pseudomonas aeruginosa by bacteriophage D3. Mol Microbiol 39:1237–1247. 10.1111/j.1365-2958.2001.02311.x. [DOI] [PubMed] [Google Scholar]
- 13.Chung IY, Jang HJ, Bae HW, Cho YH. 2014. A phage protein that inhibits the bacterial ATPase required for type IV pilus assembly. Proc Natl Acad Sci USA 111:11503–11508. 10.1073/pnas.1403537111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pawluk A, Shah M, Mejdani M, Calmettes C, Moraes TF, Davidson AR, Maxwell KL. 2017. Disabling a type I-E CRISPR-Cas nuclease with a bacteriophage-encoded anti-CRISPR protein. mBio 8:e01751-17. 10.1128/mBio.01751-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmad AA, Stulberg MJ, Huang Q. 2017. Prophage Rs551 and its repressor gene orf14 Reduce virulence and increase competitive fitness of its Ralstonia solanacearum carrier strain UW551. Front Microbiol 8:2480. 10.3389/fmicb.2017.02480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su LK, Lu CP, Wang Y, Cao DM, Sun JH, Yan YX. 2010. Lysogenic infection of a Shiga toxin 2-converting bacteriophage changes host gene expression, enhances host acid resistance and motility. Mol Biol 44:54–66. 10.1134/S0026893310010085. [DOI] [PubMed] [Google Scholar]
- 17.Martin-Verstraete I, Peltier J, Dupuy B. 2016. The regulatory networks that control clostridium difficile toxin synthesis. Toxins (Basel) 8:153. 10.3390/toxins8050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sekulovic O, Meessen-Pinard M, Fortier LC. 2011. Prophage-stimulated toxin production in Clostridium difficile NAP1/027 lysogens. J Bacteriol 193:2726–2734. 10.1128/JB.00787-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wipf JR, Schwendener S, Perreten V. 2014. The novel macrolide-Lincosamide-Streptogramin B resistance gene erm(44) is associated with a prophage in Staphylococcus xylosus. Antimicrob Agents Chemother 58:6133–6138. 10.1128/AAC.02949-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iyer LM, Koonin EV, Aravind L. 2002. Classification and evolutionary history of the single-strand annealing proteins, RecT, Red beta, ERF and RAD52. BMC Genomics 3:8. 10.1186/1471-2164-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noirot P, Kolodner RD. 1998. DNA strand invasion promoted by Escherichia coli RecT protein. J Biol Chem 273:12274–12280. 10.1074/jbc.273.20.12274. [DOI] [PubMed] [Google Scholar]
- 22.Hall SD, Kolodner RD. 1994. Homologous pairing and strand exchange promoted by the Escherichia coli RecT protein. Proc Natl Acad Sci USA 91:3205–3209. 10.1073/pnas.91.8.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kusano K, Takahashi NK, Yoshikura H, Kobayashi I. 1994. Involvement of RecE exonuclease and RecT annealing protein in DNA double-strand break repair by homologous recombination. Gene 138:17–25. 10.1016/0378-1119(94)90778-1. [DOI] [PubMed] [Google Scholar]
- 24.Song W, Sun HX, Zhang C, Cheng L, Peng Y, Deng Z, Wang D, Wang Y, Hu M, Liu W, Yang H, Shen Y, Li J, You L, Xiao M. 2019. Prophage Hunter: an integrative hunting tool for active prophages. Nucleic Acids Res 47:W74–W80. 10.1093/nar/gkz380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui X, You J, Sun L, Yang X, Zhang T, Huang K, Pan X, Zhang F, He Y, Yang H. 2016. Characterization of Pseudomonas aeruginosa phage C11 and identification of host genes required for virion maturation. Sci Rep 6:39130. 10.1038/srep39130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang H, Liang L, Lin S, Jia S. 2010. Isolation and characterization of a virulent bacteriophage AB1 of Acinetobacter baumannii. BMC Microbiol 10:131. 10.1186/1471-2180-10-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsao Y-F, Taylor VL, Kala S, Bondy-Denomy J, Khan AN, Bona D, Cattoir V, Lory S, Davidson AR, Maxwell KL. 2018. Phage morons play an important role in pseudomonas aeruginosa phenotypes. J Bacteriol 200:e00189-18. 10.1128/JB.00189-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Paepe M, Hutinet G, Son O, Amarir-Bouhram J, Schbath S, Petit MA. 2014. Temperate phages acquire DNA from defective prophages by relaxed homologous recombination: the role of Rad52-like recombinases. PLoS Genet 10:e1004181. 10.1371/journal.pgen.1004181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prats C, Giro A, Ferrer J, Lopez D, Vives-Rego J. 2008. Analysis and IbM simulation of the stages in bacterial lag phase: basis for an updated definition. J Theor Biol 252:56–68. 10.1016/j.jtbi.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 30.Tao L, Wu X, Sun B. 2010. Alternative sigma factor sigmaH modulates prophage integration and excision in Staphylococcus aureus. PLoS Pathog 6:e1000888. 10.1371/journal.ppat.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng Z, Liu X, Yao J, Guo Y, Li B, Li Y, Jiao N, Wang X. 2016. Cold adaptation regulated by cryptic prophage excision in Shewanella oneidensis. ISME J 10:2787–2800. 10.1038/ismej.2016.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Kim Y, Wood TK. 2009. Control and benefits of CP4-57 prophage excision in Escherichia coli biofilms. ISME J 3:1164–1179. 10.1038/ismej.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong SH, Wang X, Wood TK. 2010. Controlling biofilm formation, prophage excision and cell death by rewiring global regulator H-NS of Escherichia coli. Microb Biotechnol 3:344–356. 10.1111/j.1751-7915.2010.00164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gold M, Becker A, Parris W. 1983. The bacteriophage lambda terminase enzyme. Methods Enzymol 100:183–191. 10.1016/0076-6879(83)00054-3. [DOI] [PubMed] [Google Scholar]
- 35.Little JW. 1990. Chance phenotypic variation. Trends Biochem Sci 15:138–138. 10.1016/0968-0004(90)90211-s. [DOI] [PubMed] [Google Scholar]
- 36.Harshey RM. 2003. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol 57:249–273. 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 37.Harshey RM. 1994. Bees aren’t the only ones: swarming in gram-negative bacteria. Mol Microbiol 13:389–394. 10.1111/j.1365-2958.1994.tb00433.x. [DOI] [PubMed] [Google Scholar]
- 38.Corral-Lugo A, Matilla MA, Martin-Mora D, Silva Jimenez H, Mesa Torres N, Kato J, Hida A, Oku S, Conejero-Muriel M, Gavira JA, Krell T. 2018. High-affinity chemotaxis to histamine mediated by the TlpQ chemoreceptor of the human pathogen Pseudomonas aeruginosa. mBio 9:e01894-18. 10.1128/mBio.01894-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rico-Jimenez M, Munoz-Martinez F, Garcia-Fontana C, Fernandez M, Morel B, Ortega A, Ramos JL, Krell T. 2013. Paralogous chemoreceptors mediate chemotaxis towards protein amino acids and the non-protein amino acid gamma-aminobutyrate (GABA). Mol Microbiol 88:1230–1243. 10.1111/mmi.12255. [DOI] [PubMed] [Google Scholar]
- 40.Fazli M, Almblad H, Rybtke ML, Givskov M, Eberl L, Tolker-Nielsen T. 2014. Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ Microbiol 16:1961–1981. 10.1111/1462-2920.12448. [DOI] [PubMed] [Google Scholar]
- 41.Valentini M, Filloux A. 2016. Biofilms and cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. J Biol Chem 291:12547–12555. 10.1074/jbc.R115.711507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mustard JA, Little JW. 2000. Analysis of Escherichia coli RecA interactions with LexA, lambda CI, and UmuD by site-directed mutagenesis of recA. J Bacteriol 182:1659–1670. 10.1128/JB.182.6.1659-1670.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oliver A, Mulet X, Lopez-Causape C, Juan C. 2015. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat 21–22:41–59. 10.1016/j.drup.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 44.Krishnamurthy SR, Wang D. 2017. Origins and challenges of viral dark matter. Virus Res 239:136–142. 10.1016/j.virusres.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 45.Nanda AM, Thormann K, Frunzke J. 2015. Impact of spontaneous prophage induction on the fitness of bacterial populations and host-microbe interactions. J Bacteriol 197:410–419. 10.1128/JB.02230-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nanda AM, Heyer A, Kramer C, Grunberger A, Kohlheyer D, Frunzke J. 2014. Analysis of SOS-induced spontaneous prophage induction in Corynebacterium glutamicum at the single-cell level. J Bacteriol 196:180–188. 10.1128/JB.01018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Little JW, Michalowski CB. 2010. Stability and instability in the lysogenic state of phage lambda. J Bacteriol 192:6064–6076. 10.1128/JB.00726-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsui H, Sano Y, Ishihara H, Shinomiya T. 1993. Regulation of pyocin genes in Pseudomonas aeruginosa by positive (prtN) and negative (prtR) regulatory genes. J Bacteriol 175:1257–1263. 10.1128/jb.175.5.1257-1263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erill I, Campoy S, Barbe J. 2007. Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol Rev 31:637–656. 10.1111/j.1574-6976.2007.00082.x. [DOI] [PubMed] [Google Scholar]
- 50.Nakamura S, Minamino T. 2019. Flagella-driven motility of bacteria. Biomolecules 9:279. 10.3390/biom9070279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henrichsen J, Blom J. 1975. Examination of fimbriation of some gram-negative rods with and without twitching and gliding motility. Acta Pathol Microbiol Scand B 83:161–170. 10.1111/j.1699-0463.1975.tb00088.x. [DOI] [PubMed] [Google Scholar]
- 52.Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. 2016. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14:563–575. 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 53.Maseda H, Saito K, Nakajima A, Nakae T. 2000. Variation of the mexT gene, a regulator of the MexEF-OprN efflux pump expression in wild-type strains of Pseudomonas aeruginosa. FEMS Microbiol Lett 192:107–112. 10.1111/j.1574-6968.2000.tb09367.x. [DOI] [PubMed] [Google Scholar]
- 54.Breazeale SD, Ribeiro AA, Raetz CRH. 2003. Origin of lipid A species modified with 4-amino-4-deoxy-L-arabinose in polymyxin-resistant mutants of Escherichia coli–an aminotransferase (ArnB) that generates UDP-4-amino-4-deoxy-L-arabinose. J Biol Chem 278:24731–24739. 10.1074/jbc.M304043200. [DOI] [PubMed] [Google Scholar]
- 55.Suarez V, Moineau S, Reinheimer J, Quiberoni A. 2008. Argentinean Lactococcus lactis bacteriophages: genetic characterization and adsorption studies. J Appl Microbiol 104:371–379. 10.1111/j.1365-2672.2007.03556.x. [DOI] [PubMed] [Google Scholar]
- 56.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 57.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang L, Feng Z, Wang X, Wang X, Zhang X. 2010. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26:136–138. 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 59.Mao X, Cai T, Olyarchuk JG, Wei L. 2005. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793. 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- 60.Semmler ABT, Whitchurch CB, Mattick JS. 1999. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology (Reading) 145:2863–2873. 10.1099/00221287-145-10-2863. [DOI] [PubMed] [Google Scholar]
- 61.Merritt JH, Kadouri DE, O'Toole GA. 2005. Growing and analyzing static biofilms. Curr Protoc Microbiol 1:Unit 1B 1. 10.1002/9780471729259.mc01b01s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wiegand I, Hilpert K, Hancock RE. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 63.Koch G, Nadal-Jimenez P, Cool RH, Quax WJ. 2014. Assessing Pseudomonas virulence with nonmammalian host: galleria mellonella. Methods Mol Biol 1149:681–688. 10.1007/978-1-4939-0473-0_52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S8 and Tables S1 and S2. Download aem.01068-22-s0001.pdf, PDF file, 0.9 MB (894.9KB, pdf)
Data Set S1. Download aem.01068-22-s0002.xls, XLS file, 0.9 MB (950.5KB, xls)
Data Availability Statement
The whole-genome sequencing data of P8W and P9W were deposited at Sequence Read Archive of NCBI with accession no. SRA: PRJNA753640 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA753640). The whole-genome sequencing data of PP9W have been made available at the National Center for Biotechnology Information with GenBank no.: MZ773939.1 (https://www.ncbi.nlm.nih.gov/nuccore/2095887126). The transcriptome data have been deposited at Sequence Read Archive of NCBI with accession no. SRA: PRJNA774287 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA774287). Other related data are available in this article or from the corresponding authors upon request.