ABSTRACT
Root exudates contribute to shaping the root-associated microbiomes, but it is unclear which of the many exudate compounds are important in this process. Here, we focused on understanding the influence of sugars and jasmonic acid (JA) concentrations in maize root exudates on the rhizobacterial communities. Twelve maize genotypes were identified with variable concentrations of sugars and JA based on a screening of 240 maize genotypes grown in a semihydroponic system. These twelve maize genotypes were grown in a replicated field experiment in which samples were collected at three maize developmental stages. The 16S rRNA gene (V4 region) was amplified and sequenced. Sugars and JA concentrations from rhizosphere soils were also quantified. The results indicated that the maize genotypic variability in sugars and JA concentration in root exudates, measured in the semihydroponic system, significantly affected the rhizosphere bacterial community composition at multiple stages plant development. In contrast, the root endosphere and bulk soil bacterial communities were only affected at specific growth stages. Sugars and JA concentration as quantified in rhizosphere soil samples confirmed that these two compounds affected the rhizobacterial communities at all developmental stages analyzed. The effects of specific sugars on the composition of the rhizobacterial communities were also measured, with larger effects of sucrose at earlier developmental stages and trehalose at later developmental stages. Our results indicate that JA and sugars are important root exudate compounds that influence the composition of the maize rhizobacterial communities.
IMPORTANCE Roots secrete exudates that are important in interactions with soil microbes that promote plant growth and health. However, the exact chemical compounds in root exudates that participate in these interactions are not fully known. Here, we investigated whether sugars and the phytohormone jasmonic acid influence the composition of the rhizobacterial communities of maize, which is an important crop for food, feed, and energy. Our results revealed that both compounds contribute to the assemblage of rhizobacterial communities at different maize developmental stages. Knowledge about the specific compounds in root exudates that contribute to shape the rhizobiome will be important for future strategies to develop sustainable agricultural practices that are less dependent on agrochemicals.
KEYWORDS: agricultural microbiology, phytohormones, rhizobacteria, rhizosphere microbiome, soil ecology, sugars
INTRODUCTION
It is estimated that up to 40% of total carbon (C) fixed by plant photosynthesis is destined for rhizodeposition, which significantly increases microbial population densities in the rhizosphere compared to the bulk soil (1, 2). In exchange for the higher C availability in the root zone, many microbes support plant growth and health in a mutualistic relationship, although other members of the root-associated microbiomes are commensals or even opportunistic pathogens (3–5). The rhizosphere microbiome is dominated by bacteria, and many strains perform beneficial activities such as nitrogen (N) fixation, phosphate solubilization, iron chelation, phytohormone production, alleviation of abiotic stresses, and biocontrol of phytopathogens, supporting plant growth and health (2, 4, 6–8). Therefore, understanding the ecological plant-microbe interactions and how to manipulate the rhizobiome in order to enhance their beneficial activities to increase plant productivity is an important strategy for developing more sustainable agricultural systems (9–11).
Many factors contribute to the rhizosphere microbiome assemblage of cultivated plants, including soil type, management practices, climate, plant species, and even plant genotype (6, 12, 13). Among the several substances present in rhizodeposits, including root mucilage, mucigel, border cells, cell lysates, and exudates, the latter contain the most labile C sources and therefore are assumed to be the main compounds participating in the microbiome selection (14, 15). However, many of the specific exudate compounds that affect the microbiome composition and select the most beneficial microbial communities are not clearly known. Sugars are considered the main C sources used by root-associated microbes and are the product of plant photosynthesis (14, 16). Some studies have analyzed the effects of sugars added to soil on the microbial community composition (17–19) or the correlation between temporal changes in sugar exudation and the corresponding changes in the rhizobiome along developmental stages of Arabidopsis (20, 21). However, it is unclear whether plants with distinct concentrations of sugars in the root exudates assemble different rhizosphere microbiomes.
Secondary metabolites comprise another class of compounds that have strong effects on shaping the root-associated microbiomes, e.g., benzoxazinoids, coumarins, camalexins, and triterpenes (22). Despite the fact that some secondary metabolites were shown to affect the structure of microbial communities, less attention was given to the effect of plant produced and root exuded hormones on the microbiomes (14, 22). Phytohormones such as auxins, gibberelins, and cytokinins were shown to be produced by many rhizobacteria to promote plant growth (5, 15), but the effects of phytohormones exuded by plant roots on shaping the associated microbiomes are largely unknown. Some studies characterized changes in the root-associated microbiome due to added phytohormones or changes in the root signaling of phytohormones. For example, salicylic acid (SA) signaling mutants and added SA were used to show that this immune-signaling hormone strongly modulates the root microbiome colonization of Arabidopsis (23). Jasmonic acid (JA) is another plant hormone involved in plant defense signaling that has been suggested to be important in shaping the root-associated microbiomes (24–28). However, the studies on JA did not investigate changes due to root exudation of JA but rather mutations in the plant JA signaling pathways or activation of these pathways by application of methyl jasmonate (24–27). Therefore, it is unknown whether the exudation of different concentrations of JA affects the composition of root-associated microbiomes.
Our study used maize lines naturally exuding different concentrations of sugars and JA to test the hypothesis that the structure and diversity of maize rhizosphere bacterial communities are affected by exudation of these compounds. We screened 240 maize inbred lines from the Goodman and Buckler association panel (29) using a semihydroponic system to collect root exudates in vitro (30) and analyzed the concentrations of sugars and JA through targeted metabolomics. We then selected 12 maize genotypes exuding variable concentrations of JA and sugars to conduct a field experiment. The field experiment allowed for the use of native soils under realistic conditions. We collected samples from the root endosphere, rhizosphere, and two bulk soil (within rows and between rows) regions at three maize developmental stages. The soil and root-associated bacterial communities were analyzed using 16S rRNA amplicon sequencing. We previously used this approach to show that the concentrations of the secondary metabolites 2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one (DIMBOA) and γ-aminobutyric acid (GABA) in root exudates affected the maize rhizobiome (31). In that study, we validated the use of the semihydroponic system to collect and analyze root exudates, as well as the use of natural variation in the concentration of root exudate compounds between genotypes within a plant species to investigate changes in the belowground microbiomes (31). Here, we extended this investigation to other exudate compounds and soil compartments. Our results indicated that the concentrations of JA and sugars in root exudates had a significant effect on the rhizosphere bacterial communities at all developmental stages analyzed and on the root endosphere and bulk soil bacterial communities at specific developmental stages, showing the importance of these compounds for maize root-soil microbe interactions.
RESULTS
Differences between maize genotypes in total sugars and jasmonic acid concentration in the root exudates collected from a semihydroponic system.
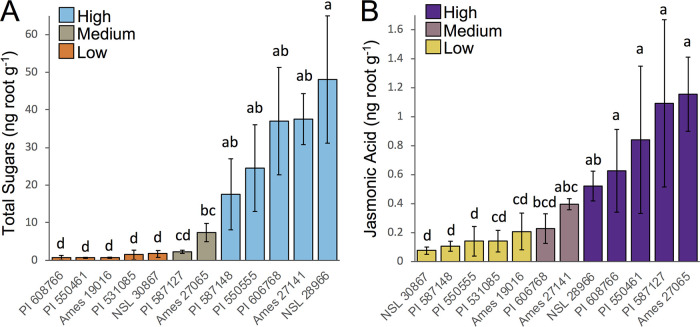
The results of targeted metabolomics of total sugars and JA indicated a large amount of natural variation in the concentration of these compounds in the root exudates among the 12 maize genotypes. According to the statistical results, we classified the genotypes as low, medium, or high exuders of sugars and JA (Fig. 1). All genotypes classified as low were significantly different from all genotypes classified as high sugars or JA exuders (Fig. 1). A total of 10 genotypes (five in each category) were classified as low or high sugars and JA exuders. For both exudate compounds (sugars and JA), two genotypes were classified as medium in their exudate levels because they were not significantly different from most genotypes in any of the two main groups (Fig. 1). Different genotypes were classified in the high, medium, and low categories for JA or sugars exudation, allowing the differentiation of the effects of JA and sugar levels on the belowground bacterial communities described in the next sections. The genotypes Ames19016, PI531085, and NSL30867 were classified as low sugars and low JA exuders; PI608766 and PI550461 were classified as low sugars and high JA exuders; PI587127 and Ames27065 were classified as medium sugars and high JA exuders; PI587148 and PI550555 were classified as high sugars and low JA exuders; PI606768 and Ames27141 were classified as high sugars and medium JA exuders; and NSL28966 was classified as a high sugar and a high JA exuder.
FIG 1.
Total sugar and jasmonic acid concentrations in the root exudates of 12 maize genotypes grown in the semihydroponic system. Exudates were collected from the semihydroponic system and analyzed with gas chromatography- or liquid chromatography-mass spectrometry to measure sugars and jasmonic acid concentration (ng g−1 fresh root weight) among the genotypes. Three to five replicates were used for each genotype. (A) Total sugar concentrations for the 12 genotypes. All individual sugars were summed to quantify total sugar concentration. (B) Jasmonic acid concentrations for the 12 genotypes. Different letters indicate significant differences (P < 0.05) according to Kruskal-Wallis tests. The error bars indicate the standard deviation between replicates. Five genotypes were classified as low and five genotypes were classified as high exuders of sugars and jasmonic acid according to statistical results. Two genotypes had intermediate values and were not clearly different from the high and low groups, so they were classified as medium exuders of sugars (PI587127 and Ames27065) and jasmonic acid (PI606768 and Ames27141).
Shifts in α- and β-diversity between sampling compartments, growth stages, and maize genotypes from the field experiment.
When comparing the differences between the four sampling compartments, we observed significantly lower species richness (observed amplicon sequence variants [ASVs]) and species diversity (Shannon index) in the root endosphere compared to the rhizosphere and bulk soil bacterial communities (Fig. S1A). The rhizosphere bacterial communities showed similar species richness compared to the two bulk soil compartments but intermediate species diversity between the bulk soil and root endosphere compartments (Fig. S1A). The sampling compartment was also the main factor shaping the β-diversity of the bacterial communities (Fig. S1B). Permutational multivariate analysis of variance (PERMANOVA) indicated that compartment, growth stage, and maize genotype were all significant factors (P < 0.001) affecting the bacterial communities, with 29.2% of the variation explained by sampling compartment, 1.5% of the variation explained by growth stage, and 1.2% of the variation explained by maize genotype. The bacterial community structure of all four compartments were different from each other according to pairwise PERMANOVA (P < 0.001). The soil-within-rows samples were located between the rhizosphere, and the soil-between-rows samples on the ordination (Fig. S1B). The bacterial community composition was dominated by Proteobacteria (22.7%) and Acidobacteria (13.9%) in the rhizosphere, Proteobacteria (46.9%) and Firmicutes (19.4%) in the root endosphere, Planctomycetes (21.3%) and Acidobacteria (20.0%) in the soil within rows, and Actinobacteria (17.4%) and Planctomycetes (16.4%) in the soil between rows (Fig. S1C).
Changes in the bacterial communities associated with different exudation levels of total sugars and jasmonic acid according to the semihydroponics data.
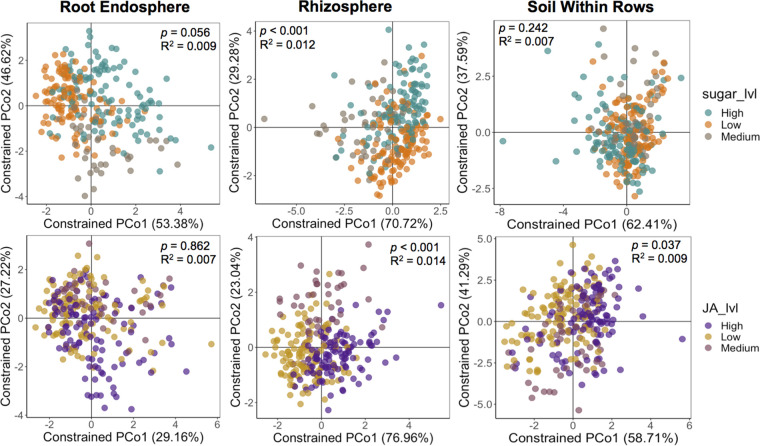
We then separated samples according to compartment (endosphere, rhizosphere, and soil within and between rows) to verify whether the different concentration levels of sugars and JA in root exudates according to the semihydroponics data (Fig. 1) affected the bacterial communities. For this initial analysis, each individual compartment was analyzed, comprising the samples from all three growth stages together. PERMANOVA indicated that both JA (P < 0.001; R2 = 0.014) and total sugar (P < 0.001; R2 = 0.012) concentration levels in exudates significantly affected the rhizosphere bacterial community structure (Fig. 2). The root endosphere bacterial communities were not significantly affected by the two compounds, but samples showed some separation according to sugar levels (Fig. 2). The bacterial communities in the soil within rows were not affected by sugar levels but were significantly affected by different levels of JA concentration in root exudates (P = 0.037; R2 = 0.009) (Fig. 2). We also observed a significant interaction effect between JA and sugar levels on the root endosphere (P = 0.034; R2 = 0.005) and rhizosphere (P = 0.012; R2 = 0.006) bacterial communities (Table S1). The bacterial community structure of the soil between rows was not affected by sugars or JA concentration levels in root exudates (Table S1).
FIG 2.
Bacterial community structure according to different exudation levels of sugars or jasmonic acid based on the hydroponics data for each plant and soil compartment. Shown are the constrained principal coordinate analysis (CAP) results based on the Bray-Curtis distance matrices showing the separation of samples from the different levels of sugars or jasmonic acid (JA) concentration in root exudates. Samples from the 12 maize genotypes were classified as low, medium, or high sugars/jasmonic acid exuders according to Fig. 1. Analyses include samples from the three maize developmental stages. The values shown in each graph indicate whether the factor tested (jasmonic acid or sugar levels) significantly affected (P < 0.05) the bacterial communities according to permutational multivariate analysis of variance (PERMANOVA). R2 indicates how much of the sample variability is explained by that factor. The graphs showing the separation of samples according to the different levels of total sugars or jasmonic acid in root exudates were constrained for the factors sugar levels and jasmonic acid levels. The soil within rows refers to the bulk soil collected in the rows where the plants were growing and close to plant roots. The other bulk soil compartment sampled (between rows and further from roots) was not affected by JA and sugar levels in root exudates, and for this reason, it was not included in this figure. PCo, principal coordinate analysis.
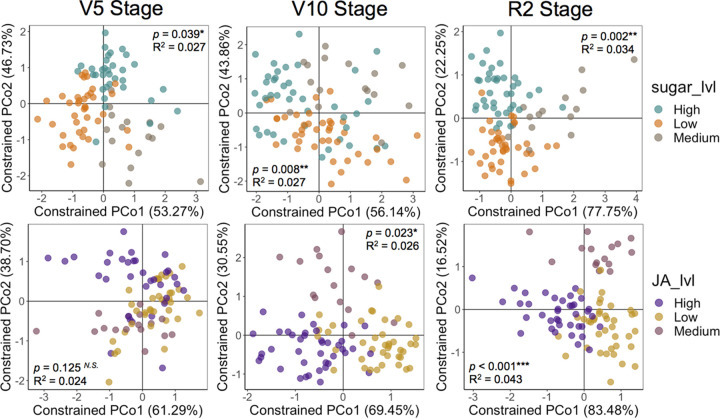
Next, the data of each specific growth stage were analyzed separately. This was done to understand in which specific developmental stages the concentration levels of sugars and JA in the root exudates had the largest effect on the rhizosphere bacterial communities. To do this analysis, the data were split into the different growth stages within each compartment. The different sugar exudation levels significantly affected the rhizosphere bacterial community structure at the three developmental stages analyzed (Fig. 3). The rhizosphere β-diversity was more affected by sugar concentration levels at the R2 reproductive stage (P = 0.002; R2 = 0.034) than at the V5 (P = 0.008; R2 = 0.027) and V10 (P = 0.039; R2 = 0.027) vegetative stages (Fig. 3). The impact of JA exudation levels on rhizosphere β-diversity was dependent on plant development (Fig. 3). Similar to what was observed for sugars, JA concentration levels in root exudates had a larger impact on the rhizosphere bacterial communities at the reproductive R2 stage (P < 0.001; R2 = 0.043) than at the V5 (not significant) and V10 (P = 0.023; R2 = 0.026) vegetative stages (Fig. 3). Together, sugars and JA concentration levels in root exudates explained 7.7% of the variation on the rhizosphere bacterial communities at the R2 stage (Fig. 3). We also observed significant interaction effects between sugars and JA levels on the rhizosphere bacterial communities at the V5 (P = 0.039; R2 = 0.014) and V10 (P = 0.004; R2 = 0.018), but not at the R2 stage (P = 0.147; R2 = 0.012). With respect to the bacterial communities inhabiting the other compartments, the root endosphere was significantly affected by sugar concentration levels only at the V10 stage (P < 0.001; R2 = 0.036), while the soil within rows was affected by JA levels only at the R2 stage (P = 0.029; R2 = 0.026) (Fig. S2). Sugar exudation level did not affect the bulk soil (within and between rows) bacterial communities at any plant growth stage analyzed. JA exudation level also did not affect the root endosphere bacterial communities at any growth stage. The root endosphere α-diversity was significantly affected by sugar levels at the V10 stage (Fig. S3). The only additional change in α-diversity was observed in the rhizosphere at the R2 stage due to different JA concentration in root exudates (Fig. S3).
FIG 3.
Specific growth stages in which the rhizosphere bacterial communities were affected by total sugar or jasmonic acid concentration levels in root exudates based on the hydroponics data. Shown are the constrained principal coordinates analysis (CAP) results based on the Bray-Curtis distance matrices showing the separation of samples according to the different levels of total sugars and jasmonic acid concentrations in root exudates. Samples from the 12 maize genotypes were classified as low, medium, or high sugars/jasmonic acid exuders according to Fig. 1. PERMANOVA P values and R2 values are shown in each graph to indicate the significance and effect size of sugars and jasmonic acid concentration in root exudates on the rhizosphere bacterial community structure at each growth stage. The graphs showing the separation of samples according to the different levels of total sugars or jasmonic acid in root exudates were constrained for the factors sugar levels and jasmonic acid levels, respectively. PCo, principal coordinate analysis.
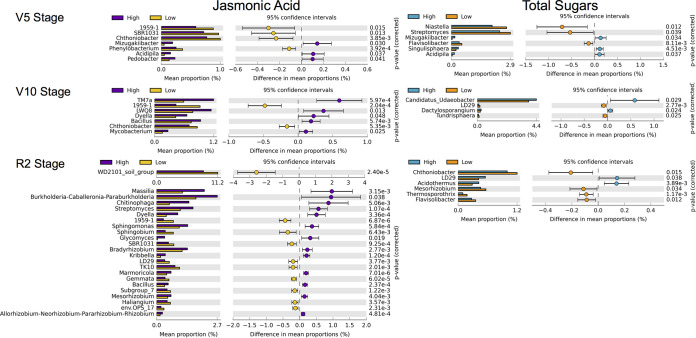
We also investigated which specific rhizosphere bacterial taxa changed in relative abundance between genotypes exuding high and low levels of JA or total sugars. Results indicated that at all developmental stages analyzed, there were specific bacterial genera that changed in relative abundance between genotypes exuding high and low levels of the two compounds studied (Fig. 4). At the V5 stage, a total of seven and six bacterial genera changed in relative abundance due to JA and sugar concentration levels in root exudates, respectively. At the V10 stage, a total of seven and four bacterial genera changed in relative abundance between the genotypes exuding different levels of JA and sugars, respectively. At the R2 stage, a total of 23 and 6 bacterial genera changed in relative abundance between genotypes exuding different levels of JA and sugars, respectively. Therefore, the greatest effect on the relative abundance of specific bacterial genera was observed at the R2 stage due to different JA exudation levels (Fig. 4). This is consistent with the results of β-diversity, for which the highest percentage of explanation of the changes in bacterial community structure was also observed at the R2 stage due to JA levels (Fig. 3). Specific bacterial genera also changed in relative abundance between genotypes exuding distinct sugars or JA levels in the other compartments. In the root endosphere, four bacterial genera changed in relative abundance between sugar exudation levels at the V10 stage, while in the soil within rows, five bacterial genera changed in relative abundance genotypes with associated with genotypic variation in JA exudation at the R2 stage (Fig. S4).
FIG 4.
Rhizobacteria changing in relative abundance between genotypes exuding contrasting levels of sugars or jasmonic acid at each growth stage. Bacterial genera showing significant changes in relative abundance between genotypes with high or low concentrations of jasmonic acid and sugars in root exudates in the rhizosphere compartment at the three growth stages analyzed. The genera displayed in the graphs showed P < 0.05 in the Welch’s t test after the Benjamini-Hochberg false discovery rate (FDR) correction, and the differences between proportions of sequences were >0.1%. Unclassified genera were filtered out.
Shifts in the rhizobacterial communities due to different concentrations of sugars and jasmonic acid as quantified in rhizosphere soil samples from the field.
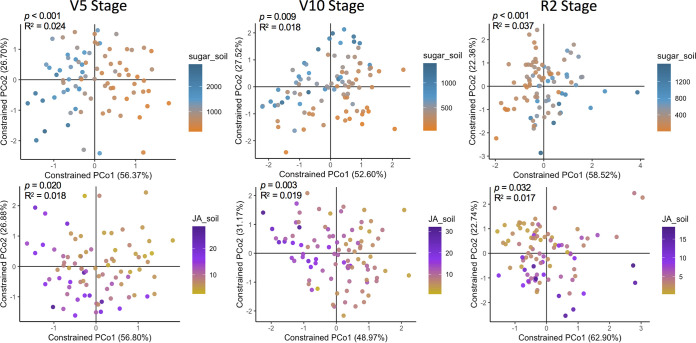
We next analyzed the concentration of sugars and JA in the rhizosphere samples collected in the field (Table S2) with the aim of testing whether the analysis with these data confirms the β-diversity results obtained with the previous analysis using the semihydroponics data to classify maize genotypes. In this case, the actual concentrations of sugars and JA quantified in each rhizosphere sample were used instead of the low, medium, and high categories used before. The results of PERMANOVA indicated that the concentration of sugars in the rhizosphere field samples significantly affected the rhizobacterial communities at the three developmental stages analyzed (V5: P < 0.001, R2 = 0.024; V10: P = 0.009, R2 = 0.018; R2: P < 0.001, R2 = 0.037) with the largest impact also observed at the R2 stage (Fig. 5). The concentration of JA in the rhizosphere field samples also significantly affected the rhizobacterial communities (V5: P = 0.02, R2 = 0.018; V10: P = 0.003, R2 = 0.019; R2: P = 0.032, R2 = 0.017) at all developmental stages (Fig. 5), including the V5 stage that was not significant in the previous analysis using the hydroponics data (Fig. 3). However, no significant interaction effects between the concentration of sugars and JA on the rhizosphere bacterial communities were observed at any growth stage, which is in contrast to the analysis with the hydroponics data. Another difference between the two approaches was that we measured the largest changes in rhizobacterial communities associated with JA concentration at the R2 stage based on the hydroponics data (Fig. 3), while the V10 stage was the most affected according to the concentration of JA in the rhizosphere field samples (Fig. 5). Nevertheless, the two approaches demonstrate the importance of sugars and JA concentration in root exudates in shaping rhizobacterial communities across maize development.
FIG 5.
Specific growth stages in which the rhizobacterial communities were affected by different jasmonic acid and sugar concentrations quantified in the rhizosphere field samples. Shown are the constrained principal coordinates analysis (CAP) results based on the Bray-Curtis distance matrices showing the separation of samples according to the concentrations of sugars and jasmonic acid quantified from rhizosphere samples collected in the field. PERMANOVA P values and R2 values are shown in each graph to indicate the significance and effect size of sugars and jasmonic acid concentration on the rhizosphere bacterial community structure at each growth stage. Sugars and jasmonic acid concentration were used as numeric factors in the multivariate analyses. Each sample had a corresponding value of sugars and jasmonic acid concentration as quantified in the rhizosphere field samples. The variation range of sugars and jasmonic acid concentration (ng soil g−1) is shown in the legends of each graph. The graphs showing the separation of samples according to the different concentrations of total sugars or jasmonic acid were constrained for the factors sugar and jasmonic acid concentration. PCo, principal coordinate analysis.
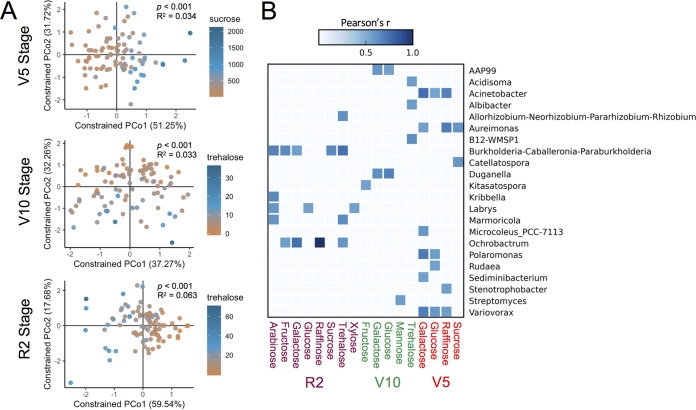
In addition to analyzing the effect of total sugar concentration, we also investigated which specific sugars affected the rhizobacterial communities at the three growth stages. The PERMANOVA results indicated that sucrose, glucose, and mannose significantly affected the rhizobacterial communities at the V5 stage; trehalose, arabinose, mannose, and glucose significantly affected the rhizobacterial communities at the V10 stage; and trehalose, arabinose, galactose, and glucose significantly affected the rhizobacterial communities at the R2 stage (Fig. 6A; Table S3; Fig. S5). Therefore, a different set of sugars affected the bacterial communities at each stage, but glucose was always significant (Fig. S5). However, sucrose was the sugar with the largest impact at the V5 stage, while trehalose had the largest impact at the V10 and R2 stages (Fig. 6A). We also analyzed the linear correlations between the relative abundance of individual bacterial genera and the concentration of specific sugars quantified in the rhizosphere (ng g soil−1). The results indicated a total of 41 significant correlations, of which 16 were observed at the V5 stage, 9 were observed at the V10 stage, and 16 were observed at the R2 stage (Fig. 6B). A total of 22 bacterial genera were significantly correlated with specific sugars. Glucose and galactose were correlated with at least one bacterial genus at all of the stages analyzed (Fig. 6B).
FIG 6.
Influence of specific sugars on the rhizobacteria. (A) Constrained principal coordinates analyses (CAPs) based on the Bray-Curtis distance matrices showing the specific sugars that had the largest effects on the rhizobacterial communities at each growth stage. PERMANOVA P values and R2 values are shown in each graph to indicate the significance and effect size of sucrose (V5 stage) and trehalose (V10 and R2 stages). The concentrations of the specific sugars were used as numeric factors in the multivariate analyses. Each sample had a corresponding value of sucrose or trehalose as quantified in the rhizosphere field samples, the variation of which (ng soil g−1) is shown in the legend of each graph. The CAP ordinations were constrained according to the factors sucrose concentration or trehalose concentration in each respective graph. (B) Correlations between specific sugars (ng g soil−1) and the relative abundance of specific bacterial genera. The heat map shows all individual bacterial genera that were significantly correlated (P < 0.05 after the Bonferroni P-value correction) to specific sugars at each growth stage according to Pearson’s correlation test. Significant correlations are shown in blue squares, while white squares indicate no correlation. The darker the blue is, the greater is the correlation (higher r) between a bacterial genus and a specific sugar.
DISCUSSION
In this study, we tested whether the natural variation in the root exudate concentrations of sugars and JA between maize genotypes affected the rhizosphere bacterial communities. The natural variation approach has some advantages compared to studies using plant mutants defective in the biosynthesis or exudation of a targeted compound (19, 23, 24, 26). Generating mutants is generally more time consuming due to the difficulty of identifying the genes associated with the production and exudation of specific compounds, especially in crop plants. In addition, using multiple genotypes increases the applicability of the study for the plant species level instead of making conclusions restricted to one or few genotypes and enhances the statistical power and confidence in the results. However, there are also possible disadvantages, such as the introduction of more uncontrolled factors associated with unrelated genotypes that can decrease the statistical power and the possibility of having low natural variation between genotypes for a particular compound. We validated the natural variation approach in a previous study that confirmed the influence of DIMBOA exudation on the maize root-associated microbiomes and showed for the first time the impact of GABA on rhizobiome composition and diversity (31).
Here, our results showed that the differences in the exudate concentrations of sugars and JA among maize genotypes significantly affected the composition of the rhizosphere bacterial communities. We used two different strategies to reach this conclusion: through analysis of bacterial community changes according to sugars and JA concentration as quantified in the semihydroponic system and in the rhizosphere soil samples collected in the field. The two strategies are complementary in their strengths and weaknesses. For example, hydroponics avoids contaminant microbes that may bias the real concentration of root exudates by consuming and/or synthesizing the compounds under study, which may occur in the rhizosphere soil extracts (16, 32, 33). On the other hand, the hydroponics uses artificial glass beads to mimic soil that may distort plant exudation compared to realistic field conditions. Therefore, it is important to consider the results of both strategies for a more accurate interpretation, as we did in our previous work (31). The combined effect of JA and sugar concentration in root exudates explained a maximum of 7.7% of the bacterial community variation in our study. The reason for the relatively low explanation of variation provided by the two compounds is probably because many other exudate compounds and environmental factors also affect the rhizobiome simultaneously (6). In our previous study, the maximum explanation of the rhizobiome variation provided by GABA and DIMBOA was similar (10.6%), which was observed at the V10 stage (31). Similar to our results, other studies also indicated that growth stage and plant genotype explain much of the variation in the root-associated bacterial communities (13, 21, 34–39). Therefore, individual exudate compounds may not explain a large amount of the rhizobacterial community structure variability, but their statistical significance indicates that they have a contribution to the rhizobiome assemblage, which was the case for sugars and JA in the present study.
Jasmonic acid is a phytohormone important to plants because it regulates many growth/developmental processes and mediates responses to abiotic and biotic stresses such as mechanical damage, herbivore insect attack, and pathogen infection (28, 40–42). Our study suggested that JA in maize root exudates also contributes to shape the rhizosphere bacterial community composition. In addition, the soil within rows bacterial community was affected by different JA exudate concentrations, indicating that this compound can influence the microbes inhabiting the soil beyond the rhizosphere (bulk soil). On the other hand, both JA and total sugar exudate concentrations did not affect the soil between rows bacterial community composition at any stage, suggesting that root exudates may affect only the bulk soil that is in closer proximity to plant roots. Several bacterial genera that have strains known to promote plant growth were enriched in the rhizosphere of genotypes exuding higher JA levels at the maize reproductive R2 stage, such as Massilia, Bacillus, Burkholderia, Sphingomonas, Streptomyces, and rhizobia (19, 43–46). Massilia was also enriched in the soil within rows of genotypes exuding higher JA levels at the R2 stage. The enrichment of Streptomyces in the rhizosphere of genotypes exuding higher JA levels at the R2 stage was particularly interesting because this genus is known to alleviate several types of plant abiotic stresses (47). Therefore, selection of Streptomyces might be a potential method mediated by plants that exude larger amounts of JA to enhance their ability to cope with environmental stress (28). Further studies under stressful conditions such as drought will be required to test this possibility.
Although no previous studies have characterized the impact of JA in plant root exudates on the rhizobiome, previous studies analyzed shifts in the rhizosphere microbiome of Arabidopsis (24, 25) and in the root endosphere and rhizosphere microbiomes of wheat (27) associated with the activation of internal JA signaling pathways by the application of exogenous methyl jasmonate. Some studies also compared the microbiomes of Arabidopsis wild type and mutants defective in the JA response (24, 26). Our study clearly showed changes in the rhizobacterial community composition due to variation in JA concentrations in root exudates. Some of the previous studies in which the JA signaling pathways were activated or used mutant plants defective in the JA response also detected changes in the rhizobacterial community composition (25, 26), while others did not (24, 27). However, those studies did not measure the actual jasmonic acid concentration in root exudates, which seems to be of primary importance for shaping soil bacterial communities outside the root. Therefore, one explanation for the divergence between our results and some of the previous studies is that activating the JA signaling pathways by addition of methyl jasmonate and mutation in JA-response pathways might not change the exudate concentrations of JA, despite other exudate compounds being affected (26). Our study confirmed the direct effects of JA concentration in root exudates on the rhizobacterial communities. On the other hand, in our study, no changes in the root endosphere bacterial communities were detected due to different concentrations of JA in root exudates, while differences in the wheat root endosphere microbiome composition were detected after activating the JA signaling pathways, which were associated with a higher transcription of 10 JA-signaling-related genes (27). Therefore, the endogenous JA signaling that was activated may alter multiple cellular processes that affect the microbiome inside root tissues but not necessarily the microbiome outside roots (27). Future studies are needed to test whether the concentration of JA in root exudates is also changed when the endogenous JA signaling pathways are activated.
Sugars are derived from photosynthesis in plants and are transported via the phloem to various sinks, including roots, and also play a role in the regulation of growth and developmental processes (48, 49). Sugars have been found in abundance in root exudates and have been suggested to shape the rhizosphere microbiome (17–19), but studies analyzing the effects of the actual sugar concentration in root exudates on the belowground microbiomes are not available in the literature. Our study analyzing multiple maize genotypes indicated that the maize root-associated bacterial communities are shaped by the concentration of sugars in root exudates. The largest impact was on the rhizosphere, but even the root endophytic bacterial community was affected at a specific growth stage (V10). On the other hand, sugars did not affect the bulk soil bacterial communities at any growth stage, probably because they are mostly metabolized by the rhizosphere microbes and barely reach the bulk soil, since sugars are the most labile compounds from root exudation and considered the main nutritional source for rhizosphere microbes (14, 16).
Previous studies found a correlation between Arabidopsis root exudate composition and the temporal shifts in the rhizosphere microbiome across developmental stages. Their results suggested that sugars may have a greater effect on the microbiomes at earlier stages, while phenolics and amino acids may be more important at later stages (20, 21). In contrast, our data indicated that sugar concentration affects the rhizobacterial communities at different stages of maize development, with the largest impact at the reproductive stage according to multiple approaches (Fig. 3, 4, and 6). One hypothesis for the different results is that plants with a very distinct genetic background such as Arabidopsis and maize may exude sugars differently across development, which reflects in the rhizobiome shaping. Another possible reason is that when a single plant genotype is analyzed across time (21), the contribution of sugars for the rhizobiome shaping may be relatively lower at later stages in that specific genotype, but when multiple genotypes are analyzed as we did here, the differences in sugar exudation between genotypes may be more detectable or obvious at later stages, causing a greater microbiome divergence.
We also identified specific sugars affecting the rhizobiome. Glucose affected the rhizobacterial communities and was correlated with specific bacterial genera at all developmental stages analyzed. Glucose is one of the most abundant organic molecules in the biosphere, and most organisms are able to consume this sugar (16), which may explain its influence on the rhizobiome across maize development. However, other sugars showed a greater impact than glucose at each growth stage analyzed, e.g., sucrose at the earlier vegetative stage and trehalose at later stages. Sucrose may be the second most used sugar by soil microbes after glucose, since plants exude large amounts of this disaccharide (19). Sucrose may be more important at earlier stages of plant development due to its potential to attract microbes as a nutritional source, since plant roots recruit microbes from the surrounding soil (3, 5). In addition, sucrose was shown to act as a signaling molecule and trigger the root colonization of Bacillus subtilis and potentially other bacteria (19).
Trehalose functions as a signal and homeostatic regulator of sucrose levels in plants (50), but its role in shaping the rhizobiome is unclear. Many soil microbes accumulate trehalose to resist abiotic stresses (33, 51), which may be one of the possible reasons trehalose had a larger impact on the rhizobacterial communities later in the season when soil nutrients and water content may be reduced. Trehalose alone explained 6.3% of the variation in the rhizobacterial communities at the R2 stage (Fig. 6A), which was larger than the percentage of variation explained by JA, total sugars, and other individual sugars at any growth stage. Therefore, the reason for the largest impact of sugars at the maize R2 stage may be connected to a larger effect of trehalose. Previous studies showed that trehalose-6-phosphate (T6P) downregulation in internal tissues during the maize flowering period increases plant yield and drought resistance (52, 53). When the endogenous levels of T6P decrease, the levels of sucrose and secondary metabolites increase, which are important to confer drought stress tolerance (52, 53). It is possible that genotypes with great amounts of endogenous T6P also exude more trehalose and may use this sugar to select plant growth-promoting rhizobacteria (PGPR) in order to compensate for their metabolic disadvantage. Trehalose was indeed correlated with some genera containing PGPR strains such as rhizobia and Burkholderia (46, 51) at the R2 stage, which supports this hypothesis. In our previous study, we observed that maize genotypes with higher DIMBOA concentrations in root exudates were enriched in metabolic pathways associated with plant disease, indicating that exuding larger concentrations of a particular compound is not necessarily positive for plants (31). Our results indicate that trehalose exudation has an important role in the plant-microbiome interactions at later developmental stages.
Other bacterial genera containing PGPR strains also responded to increased concentrations of specific sugars such as Acinetobacter to raffinose, glucose, and galactose at the V5 stage (54, 55); Streptomyces to mannose at the V10 stage (47); and Burkholderia to trehalose, sucrose, galactose, fructose, and arabinose at the R2 stage (46). It is interesting to note that the abundance of some bacterial genera was correlated with the concentrations of different sugars at each growth stage, suggesting that they may respond to specific sugars across the season (Fig. 6B). This result may reflect a variable availability of each sugar molecule in the rhizosphere soil due to changes in root exudation over time and/or new ecological interactions with other microbes such as competition for the same C source as the microbiome changes across growth stages (21). It is also noteworthy that in some cases more than one sugar was correlated with the same bacterial genera within a growth stage, which suggests a combined influence of different sugars to select specific bacteria or a greater metabolic flexibility for C utilization. Finally, we observed that some individual sugars such as glucose and galactose were correlated with several bacterial genera across multiple plant developmental stages, suggesting that certain sugars are more universally used by rhizosphere bacteria. In sum, our results suggest that sugars are important for the maize rhizobiome selection across the growing season, but different blends of sugars may be used at each growth stage to select for specific bacteria important for crop growth and development.
Conclusion.
This study indicated that the concentrations of JA and total sugars in root exudates affected the rhizosphere bacterial communities across multiple stages of plant development with the largest impact later in the season. Total sugar concentration in root exudates also affected the root endosphere bacterial communities at the maize V10 stage, while JA concentration affected the bacterial community composition of the soil within rows at the reproductive stage. The rhizosphere of maize genotypes exuding higher JA levels was enriched in several bacterial genera potentially able to promote plant growth and increase stress tolerance, suggesting that in addition to being important as an endogenous signal molecule in stress response, JA may also be used in root exudates to select beneficial rhizobacteria to help plants thrive under stress. Different sugars affected the rhizobacterial communities at each developmental stage, despite glucose being effective at all stages. Sucrose and trehalose were the sugars with the largest impacts on the rhizobacterial communities at the earlier and later stages, respectively. Different bacterial genera containing strains known to promote plant growth were stimulated by specific sugars at each growth stage. Therefore, our results suggest that the concentration of sugars in root exudates are important for the maize rhizobiome composition across the growing season, but the blend of specific sugars influences the rhizobacteria shifts across plant development. In sum, this study contributed to advancing our understanding of the influence that specific exudate compounds have on the rhizobiome.
MATERIALS AND METHODS
Collection and analysis of root exudates from a semihydroponic system.
A semihydroponic system developed for collecting root exudates (30) was used to screen 240 maize inbred lines (n ≥ 3 replications per line) belonging to the Goodman and Buckler association panel (29), with the aim of identifying maize genotypes differing in the concentrations of total sugars and JA in root exudates. For that, seeds were surface disinfected with Cl2 gas, germinated on plates with sterile 1 mM CaCl2, and planted in sterile tubes filled with glass beads and 1× Hoagland’s solution. In this semihydroponic system, the nutrient solution was initially held in the tubes for short periods of time and constantly replaced. Six days after planting, the solution was allowed to flow freely through the glass beads using peristaltic pumps to ensure proper oxygenation and moisture levels of the plant root system. The system was always manipulated in aseptic conditions. After a total of 2 weeks, the tubes were drained and then filled with sterile 1 mM CaCl2 solution to collect root exudates for 2 h, which was collected in sterile jars and frozen at −80°C (30).
The solutions with exudates were then freeze-dried (LabConco Freezone Bulk Tray Drier), resuspended in 2% formic acid, desalted using solid-phase extraction cartridges (Oasis MCX 1 cc Vac Cartridge, Waters), and analyzed by targeted liquid chromatography mass spectrometry (LC-MS) (QTRAP6500, Thermo Fisher Scientific) using standards to determine the concentration of JA in the exudates. The solutions were also analyzed using an Agilent gas chromatography (GC)-MS with an Agilent HP-5MS column to quantify the concentrations of sugars. The concentration of the compounds was normalized by dividing by the root weight in each sample. For an estimation of total sugar concentration in the exudates, the values of the 10 sugars analyzed (arabinose, xylose, fructose, mannose, galactose, glucose, sucrose, lactose, trehalose, and raffinose) were summed in each sample. This approach was previously used to identify maize genotypes differing in the concentration of other exudate compounds (GABA and DIMBOA) and to test their impact on the root-associated microbiome (31).
Field experiment with selected maize genotypes, soil/plant sampling, and quantification of sugars and jasmonic acid from the rhizosphere soil.
Twelve maize genotypes with variable concentrations of JA and total sugars in root exudates according to the semihydroponic data were selected for the field experiment. The experiment was performed near Mead, Nebraska (USA) in the summer of 2020 using a randomized block design with eight replicates. Each of the eight replicate blocks were planted with the 12 maize genotypes previously selected, generating a total of 96 plots. Each plot contained two rows (5.8 m) of each genotype spaced by 76 cm. Each row was planted with 45 seeds at a distance of 13 cm between seeds on May 15. Before planting, the field was disked and fertilized with 134.5 kg ha−1 of N. A subsoil irrigation system was installed at 20 cm below the ground. The herbicide Acuron was applied at the V2 growth stage to control weeds in the field and hand cultivated after that.
The plant and soil samplings were performed at two vegetative (V5 and V10) and one reproductive (R2) maize developmental stages. The samplings of the V5, V10, and R2 stages were performed on June 17, July 8, and August 3, respectively. Four compartments were sampled: root endosphere, rhizosphere, and the two bulk soil compartments of soil within rows and soil between rows. The reason for sampling two bulk soil compartments was to investigate whether root exudates can also affect bulk soil bacterial communities and whether the distance from the plant roots is an important factor. The soil between rows was collected between the two rows planted with the same genotype (~38 cm distant from plants) at a 0- to 10-cm depth in two random points, which were combined into a composite sample in each plot. The other three compartments were sampled by excavating two random plants in each plot. We considered the soil within rows as the soil that fell from the root ball of the two plants after shaking. Both soil-between-rows and soil-within-rows samples were placed in Ziploc bags (~500 g) and stored on ice. The roots were randomly cut from the two plants and placed in 50-mL tubes containing 35 mL of phosphate buffer (6.33 g liter−1 NaH2PO4, 8.5 g liter−1 Na2HPO4 anhydrous, 200 μL liter−1 Silwet L-77). The tubes were vigorously shaken to detach the soil in close and tight connection with the roots, i.e., the rhizosphere soil. The washed roots were blotted dry and transferred to a clean 50-mL tube. Tubes containing the roots and the rhizosphere soil with phosphate buffer were placed on ice. The root and rhizosphere samples were processed in the laboratory according to McPherson et al. (56). Briefly, the roots were surface sterilized in 5.25% sodium hypochlorite, 0.01% Tween 20, and 70% ethanol; rinsed in sterile ultrapure water; ground with liquid N; and stored at −20°C. The rhizosphere samples were filtered (100-μm mesh) to remove coarse organic material such as root hairs. Then, the rhizosphere samples were pelleted (3,000 × g for 10 min) to separate the rhizosphere soil and supernatant. The rhizosphere soil was stored at −20°C, while the rhizosphere supernatant was stored at −80°C in 2-mL tubes. Portions of the soil-within-rows and soil-between-rows samples were also saved in 2-mL microtubes at −20°C, and the remaining soil was stored at 4°C.
The rhizosphere supernatant from the field samples was used to assess the concentration of sugars through GC-MS, as well as the concentration of JA through LC-MS. The frozen supernatant was freeze-dried at −50°C (Labconco FreeZone Dryer), desalted, and analyzed as described previously. The weight of the rhizosphere soil pellet was used to normalize the metabolite data. For an estimation of total sugar concentration in the rhizosphere samples, the values of all the sugars detected (arabinose, xylose, fructose, mannose, galactose, glucose, sucrose, lactose, trehalose and raffinose) were summed. The same methods used for analysis and preparation of the samples from the semihydroponic system was used for the sugar and JA analysis of the rhizosphere supernatant.
16S rRNA gene sequencing and bioinformatics analysis of reads.
The PowerSoil-htp 96-well Soil DNA isolation kit (MoBio, Carlsbad, CA) was used to extract the DNA from the rhizosphere and bulk soil (within and between rows) samples, while the PowerPlant-htp 96-well plant DNA isolation kit (MoBio, Carlsbad, CA) was used to extract the DNA from the root endosphere samples. The resulting DNA was quantified using QuantiFluor ONE dsDNA dye (Promega Corporation, Madison, WI), and PCR amplifications were targeted to the V4 region of the 16S rRNA gene with the primer pair V4_515F/V4_806R according to Caporaso et al. (57). The sequencing libraries were prepared using a dual-index primer system consisting of the Illumina adapter, an 8-nucleotide index sequence, a 10-nucleotide pad-sequence, a 2-nucleotide linker sequence, and the primer sequence (58). A total of four different libraries were prepared, one for each compartment studied. Amplicons were purified and normalized using SequalPrep normalization plates (Invitrogen), pooled in an equimolar mixture, concentrated using a SpeedVac, and size-selected within a range of 200 to 700 bp using the SPRIselect beads (Beckman Coulter, Brea, CA). Then, sequencing libraries were quantified and quality checked using a high-sensitivity DNA kit on an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA). Libraries were spiked with 20% PhiX prior to loading the Illumina flow cell. Sequencing of the four libraries were performed on the University of Nebraska Medical Center (UNMC) using the Illumina MiSeq platform with the MiSeq 600 cycles v3 kit (59). Each library contained negative controls represented by blank DNA extraction control and water blank control (same water used in the library preparation), as well as positive controls represented by the amplification of DNA from microbial mock community B v5.1L (BEI Resources, Manassas, VA).
The sequencing reads were analyzed with the QIIME 2.0 software (60) using the DADA2 algorithm for denoising (61), which includes filtering sequences by quality, trimming reads in the positions of choice, merging the paired-end reads, removing chimeric sequences, and generating the ASV table. The ASVs reflect individual sequences with 100% nucleotide identity (62). The reads were taxonomically classified using the SILVA database version 138 (63). Mitochondria and chloroplast sequences were removed from the ASV table. Initially, reads from the four compartments were analyzed together. All samples were rarified to 4,000 reads for α- and β-diversity analyses. The Shannon index and observed ASVs were calculated to infer on species diversity and richness, respectively. The Bray-Curtis distance matrix was generated and exported for statistical analyses. The same process was performed after splitting the ASV table for each compartment and then for each growth stage within each compartment. For diversity analysis, the root endosphere, rhizosphere, soil-within-rows, and soil-between-rows samples were rarified to 4,000, 10,000, 8,000, and 15,000 reads, respectively.
Statistical analyses.
The Bray-Curtis distance matrices were exported to the R software, where all multivariate analyses were performed using the VEGAN package v. 2.5-6 (64). Unconstrained and constrained principal coordinate analyses (PCoA and CAP) were generated using the capscale function and visualized with the ggplot function in the ggplot package version 3.3.0 (65). Permutative multivariate analysis of variance (PERMANOVA, 999 permutations) was performed to test for significant effects of the diverse factors studied (compartments, growth stage, genotype, sugar level, and JA level) and their interaction using the adonis function. The factors sugar level and jasmonic acid level referred to the classification based on the quantification of these compounds from exudates collected in the semihydroponic system. Data from each growth stage within each compartment were also individually analyzed with CAP and PERMANOVA to identify the specific developmental stages in which the bacterial communities were affected by different JA or sugar levels in root exudates. The function strata was always used in the PERMANOVAs to account for the spatial variation associated with different blocks in the field. For the graphs showing the separation of samples according to different categories of sugar or JA levels (hydroponics classification), the CAP ordinations were constrained for the factors sugar level and JA level, respectively.
In addition to analyzing changes in bacterial community structure using the hydroponic data to classify the genotypes in low, medium, or high JA/sugars exuders, we also quantified the concentration of JA and sugars in the rhizosphere samples collected from field soils and used those data to test their effects on the bacterial communities (Bray-Curtis matrices) using CAP (capscale), and PERMANOVA (adonis). For that, sugars and JA concentration were used as numeric instead of categorical factors where each microbiome sample from the lines grown in 96 plots had an unique concentration of JA and sugars rather than grouping samples in low, medium, or high categories. For the graphs showing the separation of samples according to different concentrations of sugar or JA, the CAP ordinations were constrained for the factors sugar concentration and JA concentration, respectively. In addition to analyzing the effect of total sugar concentration, we also assessed the effect of the 10 specific sugars on the rhizobacterial communities at each growth stage using PERMANOVA (adonis).
Linear correlations between the concentration of specific sugars and the relative abundance of individual bacterial genera were identified using the Pearson’s test with the Bonferroni P-value correction in the PAST 4.0 software (66). Differences in total sugars and jasmonic acid concentration in root exudates between maize genotypes, as well as differences in species diversity (Shannon index) and richness (observed ASVs) were assessed using the Kruskal-Wallis test in the PAST 4.0 software (66). Differences in the relative abundance of bacterial genera between genotypes with distinct levels of sugars and jasmonic acid were analyzed with the Welch’s t test and the Benjamini-Hochberg false discovery rate (FDR) P-value correction in the STAMP software (67). Differences were considered significant when P < 0.05 and differences between proportions of sequences were >0.1%.
Data availability.
The sequences used in this article were submitted to NCBI-SRA under the BioProject accession number PRJNA759602.
ACKNOWLEDGMENTS
We thank Ellen Marsh, Allyn Pella, Bertrand Devilbiss, Jingjie Hao, Yen Ning Chai, Mari Butler, Ibrahim Alnajem, Maddie Holland, and Ngoc Pham for help with field sampling, sample processing, and laboratory analyses.
L.D.L. contributed to the study design and selection of maize genotypes and field sampling; performed laboratory, bioinformatics, and statistical analyses; interpreted the results; and wrote the manuscript. P.W. contributed to the study design and the screening of maize genotypes using the semihydroponics system. S.L.F. prepared the sequencing libraries and contributed to the field sampling. D.P.S. designed the study; supervised the field experiment, sampling, and laboratory analyses; interpreted the results; and edited the manuscript. All authors read and approved the final manuscript.
We declare no conflict of interest.
This work was supported by a grant to Daniel P. Schachtman from the National Science Foundation EPSCOR to fund the Center for Root and Rhizobiome Innovation (award OIA-1557417) and USDA-NIFA award 2020-67019-31796.
Footnotes
Supplemental material is available online only.
Contributor Information
Daniel P. Schachtman, Email: daniel.schachtman@unl.edu.
Gladys Alexandre, University of Tennessee at Knoxville.
REFERENCES
- 1.Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. 2006. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57:233–266. 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- 2.Berendsen RL, Pieterse CMJ, Bakker PAHM. 2012. The rhizosphere microbiome and plant health. Trends Plant Sci 17:478–486. 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Hirsch PR, Mauchline TH. 2012. Who’s who in the plant root microbiome? Nat Biotechnol 30:961–962. 10.1038/nbt.2387. [DOI] [PubMed] [Google Scholar]
- 4.Mendes R, Garbeva P, Raaijmakers JM. 2013. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37:634–663. 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- 5.Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P. 2013. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838. 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 6.Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH. 2013. Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 11:789–799. 10.1038/nrmicro3109. [DOI] [PubMed] [Google Scholar]
- 7.Compant S, Samad A, Faist H, Sessitsch A. 2019. A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J Adv Res 19:29–37. 10.1016/j.jare.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Vries FT, Griffiths RI, Knight CG, Nicolitch O, Williams A. 2020. Harnessing rhizosphere microbiomes for drought-resilient crop production. Science 368:270–274. 10.1126/science.aaz5192. [DOI] [PubMed] [Google Scholar]
- 9.Sessitsch A, Pfaffenbichler N, Mitter B. 2019. Microbiome applications from lab to field: facing complexity. Trends Plant Sci 24:194–198. 10.1016/j.tplants.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Singh BK, Trivedi P, Egidi E, Macdonald CA, Delgado-Baquerizo M. 2020. Crop microbiome and sustainable agriculture. Nat Rev Microbiol 18:601–602. 10.1038/s41579-020-00446-y. [DOI] [PubMed] [Google Scholar]
- 11.Arif I, Batool M, Schenk PM. 2020. Plant microbiome engineering: expected benefits for improved crop growth and resilience. Trends Biotechnol 38:1385–1396. 10.1016/j.tibtech.2020.04.015. [DOI] [PubMed] [Google Scholar]
- 12.Berg G, Smalla K. 2009. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol 68:1–13. 10.1111/j.1574-6941.2009.00654.x. [DOI] [PubMed] [Google Scholar]
- 13.Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE. 2013. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci USA 110:6548–6553. 10.1073/pnas.1302837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasse J, Martinoia E, Northen T. 2018. Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci 23:25–41. 10.1016/j.tplants.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Hassan MK, McInroy JA, Kloepper JW. 2019. The interactions of rhizodeposits with plant growth-promoting rhizobacteria in the rhizosphere: a review. Agriculture 9:142. 10.3390/agriculture9070142. [DOI] [Google Scholar]
- 16.Gunina A, Kuzyakov Y. 2015. Sugars in soil and sweets for microorganisms: review of origin, content, composition and fate. Soil Biol Biochem 90:87–100. 10.1016/j.soilbio.2015.07.021. [DOI] [Google Scholar]
- 17.Shi S, Richardson AE, O’Callaghan M, DeAngelis KM, Jones EE, Stewart A, Firestone MK, Condron LM. 2011. Effects of selected root exudate components on soil bacterial communities. FEMS Microbiol Ecol 77:600–610. 10.1111/j.1574-6941.2011.01150.x. [DOI] [PubMed] [Google Scholar]
- 18.Badri DV, Chaparro JM, Zhang R, Shen Q, Vivanco JM. 2013. Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J Biol Chem 288:4502–4512. 10.1074/jbc.M112.433300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian T, Bingbing S, Shi H, Gao T, He Y, Li Y, Liu Y, Li X, Zhang L, Li S, Wang Q, Yunrong C. 2021. Sucrose triggers a novel signaling cascade promoting Bacillus subtilis rhizosphere colonization. ISME J 15:2723–2737. 10.1038/s41396-021-00966-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaparro JM, Badri DV, Bakker MG, Sugiyama A, Manter DK, Vivanco JM. 2013. Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS One 8:e55731. 10.1371/journal.pone.0055731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaparro JM, Badri DV, Vivanco JM. 2014. Rhizosphere microbiome assemblage is affected by plant development. ISME J 8:790–803. 10.1038/ismej.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacoby RP, Koprivova A, Kopriva S. 2021. Pinpointing secondary metabolites that shape the composition and function of the plant microbiome. J Exp Bot 72:57–69. 10.1093/jxb/eraa424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lebeis SL, Paredes SH, Lundberg DS, Breakfield N, Gehring J, McDonald M, Malfatti S, del Rio TG, Jones CD, Tringe SG, Dangl JL. 2015. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349:860–864. 10.1126/science.aaa8764. [DOI] [PubMed] [Google Scholar]
- 24.Doornbos RF, Geraats BPJ, Kuramae EE, Loon LCV, Bakker PAHM. 2011. Effects of jasmonic acid, ethylene, and salicylic acid signaling on the rhizosphere bacterial community of Arabidopsis thaliana. Mol Plant Microbe Interact 24:395–407. 10.1094/MPMI-05-10-0115. [DOI] [PubMed] [Google Scholar]
- 25.Carvalhais LC, Dennis PG, Badri DV, Tyson GW, Vivanco JM, Schenk PM. 2013. Activation of the jasmonic acid plant defence pathway alters the composition of rhizosphere bacterial communities. PLoS One 8:e56457. 10.1371/journal.pone.0056457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carvalhais LC, Dennis PG, Badri DV, Kidd BN, Vivanco JM, Schenk PM. 2015. Linking jasmonic acid signaling, root exudates, and rhizosphere microbiome. Mol Plant Microbe Interact 28:1049–1058. 10.1094/MPMI-01-15-0016-R. [DOI] [PubMed] [Google Scholar]
- 27.Liu H, Carvalhais LC, Schenk PM, Dennis PG. 2017. Effects of jasmonic acid signaling on the wheat microbiome differ between body sites. Sci Rep 7:41766. 10.1038/srep41766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruan J, Zhou Y, Zhou M, Yan J, Khurshid M, Weng W, Cheng J, Zhang K. 2019. Jasmonic acid signaling pathway in plants. Int J Mol Sci 20:2479. 10.3390/ijms20102479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flint-Garcia SA, Thuillet AC, Yu J, Pressoir G, Romero SM, Mitchell SE, Doebley J, Kresovich S, Goodman MM, Buckler ES. 2005. Maize association population: a high-resolution platform for quantitative trait locus dissection. Plant J 44:1054–1064. 10.1111/j.1365-313X.2005.02591.x. [DOI] [PubMed] [Google Scholar]
- 30.Lopez-Guerrero MG, Wang P, Phares F, Schachtman DP, Alvarez S, van Dijk K. 2022. A glass bead semi-hydroponic system for intact maize root exudate analysis and phenotyping. Plant Meth 18:25. 10.1186/s13007-022-00856-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang P, Lopes LD, Lopez-Guerrero M, van Dijk K, Alvarez S, Riethoven JJ, Schachtman DP. 2022. Natural variation in root exudation of GABA and DIMBOA impacts the maize root endosphere and rhizosphere microbiomes. J Exp Bot 73:5052–5066. 10.1093/jxb/erac202. [DOI] [PubMed] [Google Scholar]
- 32.Eng F, Marin JE, Zienkiewicz K, Gutierrez-Rojas M, Favela-Torres E, Feussner I. 2021. Jasmonic acid biosynthesis by fungi: derivatives, first evidence on biochemical pathways and culture conditions for production. PeerJ 9:e10873. 10.7717/peerj.10873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanaporn M, Titball RW. 2020. Trehalose and bacterial virulence. Virulence 11:1192–1202. 10.1080/21505594.2020.1809326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen S, Waghmode TR, Sun R, Kuramae EE, Hu C, Liu B. 2019. Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome 7:136. 10.1186/s40168-019-0750-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edwards J, Cameron J, Santos-Medellin C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V. 2015. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci USA 112:E911–E920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edwards JA, Santos-Medellin CM, Liechty ZS, Nguyen B, Lurie E, Eason S, Phillips G, Sundaresan V. 2018. Compositional shifts in root-associated bacterial and archaeal microbiota track the plant life cycle in field-grown rice. PLoS Biol 16:e2003862. 10.1371/journal.pbio.2003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallart M, Adair KL, Love J, Meason DF, Clinton PW, Xue J, Turnbull MH. 2018. Host genotype and nitrogen form shape the root microbiome of Pinus radiata. Microb Ecol 75:419–433. 10.1007/s00248-017-1055-2. [DOI] [PubMed] [Google Scholar]
- 38.Zhalnina K, Louie KB, Hao Z, Mansoori N, da Rocha UN, Shi S, Cho H, Karaoz U, Loque D, Bowen BP, Firestone MK, Northen TR, Brodie EL. 2018. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol 3:470–480. 10.1038/s41564-018-0129-3. [DOI] [PubMed] [Google Scholar]
- 39.Veach AM, Morris R, Yip DZ, Yang ZK, Engle NL, Cregger MA, Tschaplinski TJ, Schadt CW. 2019. Rhizosphere microbiomes diverge among Populus trichocarpa plant-host genotypes and chemotypes, but it depends on soil origin. Microbiome 7:76. 10.1186/s40168-019-0668-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campos ML, Kang JH, Howe GA. 2014. Jasmonate-triggered plant immunity. J Chem Ecol 40:657–675. 10.1007/s10886-014-0468-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta A, Hisano H, Hojo Y, Matsuura T, Ikeda Y, Mori IC, Kumar MS. 2017. Global profiling of phytohormone dynamics during combined drought and pathogen stress in Arabidopsis thaliana reveals ABA and JA as major regulators. Sci Rep 7:4017. 10.1038/s41598-017-03907-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wasternack C, Hause B. 2013. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development: an update to the 2007 review in Annals of Botany. Ann Bot 111:1021–1058. 10.1093/aob/mct067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asaf S, Numan M, Khan AL, Al-Harrasi A. 2020. Sphingomonas: from diversity and genomics to functional role in environmental remediation and plant growth. Crit Rev Biotechnol 40:138–152. 10.1080/07388551.2019.1709793. [DOI] [PubMed] [Google Scholar]
- 44.Garrido-Oter R, Nakano RT, Dombrowski N, Ma KW, McHardy AC, Schulze-Lefert P; The AgBiome Team. 2018. Modular traits of the rhizobiales root microbiota and their evolutionary relationship with symbiotic rhizobia. Cell Host Microbe 24:155–167.e5. 10.1016/j.chom.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li C, Cao P, Du C, Zhang X, Bing H, Li L, Sun P, Xiang W, Zhao J, Wang X. 2021. Massilia rhizosphaerae sp. nov., a rice-associated rhizobacterium with antibacterial activities against Ralstonia solanacearum. Int J Syst Evol Microbiol 71:e05009. [DOI] [PubMed] [Google Scholar]
- 46.Mullins AJ, Murray JAH, Bull MJ, Jenner M, Jones C, Webster G, Green AE, Neill DR, Connor TR, Parkhill J, Challis GL, Mahenthiralingam E. 2019. Genome mining identifies cepacin as a plant-protective metabolite of the biopesticidal bacterium Burkholderia ambifaria. Nat Microbiol 4:996–1005. 10.1038/s41564-019-0383-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Viaene T, Langendries S, Beirinckx S, Maes M, Goormachtig S. 2016. Streptomyces as a plant’s best friend? FEMS Microbiol Ecol 92:fiw119. 10.1093/femsec/fiw119. [DOI] [PubMed] [Google Scholar]
- 48.Eveland AL, Jackson DP. 2012. Sugars, signalling, and plant development. J Exp Bot 63:3367–3377. 10.1093/jxb/err379. [DOI] [PubMed] [Google Scholar]
- 49.Lemoine R, La Camera S, Atanassova R, Dédaldéchamp F, Allario T, Pourtau N, Bonnemain J-L, Laloi M, Coutos-Thévenot P, Maurousset L, Faucher M, Girousse C, Lemonnier P, Parrilla J, Durand M. 2013. Source-to-sink transport of sugar and regulation by environmental factors. Front Plant Sci 4:272–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fichtner F, Lunn JE. 2021. The role of trehalose 6-phosphate (Tre6P) in plant metabolism and development. Annu Rev Plant Biol 72:737–760. 10.1146/annurev-arplant-050718-095929. [DOI] [PubMed] [Google Scholar]
- 51.Sharma MP, Grover M, Chourasiya D, Bharti A, Agnihotri R, Maheshwari HS, Pareek A, Buyer JS, Sharma SK, Schutz L, Mathimaran N, Singla-Pareek SL, Grossman JM, Bagyaraj DJ. 2020. Deciphering the role of trehalose in tripartite symbiosis among rhizobia, arbuscular mycorrhizal fungi, and legumes for enhancing abiotic stress tolerance in crop plants. Front Microbiol 11:509919. 10.3389/fmicb.2020.509919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nuccio ML, Wu J, Mowers R, Zhou H-P, Meghji M, Primavesi LF, Paul MJ, Chen X, Gao Y, Haque E, Basu SS, Lagrimini LM. 2015. Expression of trehalose-6-phosphate phosphatase in maize ears improves yield in well-watered and drought conditions. Nat Biotechnol 33:862–869. 10.1038/nbt.3277. [DOI] [PubMed] [Google Scholar]
- 53.Oszvald M, Primavesi LF, Griffiths CA, Cohn J, Basu SS, Nuccio ML, Paul MJ. 2018. Trehalose 6-phosphate regulates photosynthesis and assimilate partitioning in reproductive tissue. Plant Physiol 176:2623–2638. 10.1104/pp.17.01673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das S, Sultana KW, Chandra I. 2021. Isolation and characterization of a plant-growth promoting bacterium Acinetobacter sp. SuKIC24 from in vitro-grown Basilicum polystachion (L.) Moench. Curr Microbiol 78:2961–2969. 10.1007/s00284-021-02558-x. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki W, Sugawara M, Miwa K, Morikawa M. 2014. Plant growth-promoting bacterium Acinetobacter calcoaceticus P23 increases the chlorophyll content of the monocot Lemna minor (duckweed) and the dicot Lactuca sativa (lettuce). J Biosci Bioeng 118:41–44. 10.1016/j.jbiosc.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 56.McPherson MR, Wang P, Marsh EL, Mitchell RB, Schachtman DP. 2018. Isolation and analysis of microbial communities in soil, rhizosphere and roots in perennial grass experiments. J Vis Exp 137:e57932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108:4516–4522. 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gohl DM, Vangay P, Garbe J, MacLean A, Hauge A, Becker A, Gould TJ, Clayton JB, Johnson TJ, Hunter R, Knights D, Beckman KB. 2016. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat Biotechnol 34:942–949. 10.1038/nbt.3601. [DOI] [PubMed] [Google Scholar]
- 60.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, et al. 2019. Reproducible, interactive, scalable and extensive microbiome data science using QIIME2. Nat Biotechnol 37:852–857. 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Callahan BJ, McMurdie PJ, Holmes SP. 2017. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 11:2639–2643. 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2012. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930. 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 65.Wickham H. 2011. ggplot2. Comput Stat 3:180–185. 10.1002/wics.147. [DOI] [Google Scholar]
- 66.Hammer Ø, Harper DAT, Ryan PD. 2001. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron 4:1–9. [Google Scholar]
- 67.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. 2014. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30:3123–3124. 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S5. Download aem.00971-22-s0001.pdf, PDF file, 0.9 MB (941.2KB, pdf)
Table S1. Download aem.00971-22-s0002.xlsx, XLSX file, 0.01 MB (11.2KB, xlsx)
Table S2. Download aem.00971-22-s0003.xlsx, XLSX file, 0.02 MB (20.2KB, xlsx)
Table S3. Download aem.00971-22-s0004.xlsx, XLSX file, 0.01 MB (11.7KB, xlsx)
Data Availability Statement
The sequences used in this article were submitted to NCBI-SRA under the BioProject accession number PRJNA759602.