Abstract
Streptococcus thermophilus strain CNRZ 302 is unable to ferment galactose, neither that generated intracellularly by lactose hydrolysis nor the free sugar. Nevertheless, sequence analysis and complementation studies with Escherichia coli demonstrated that strain CNRZ 302 contained structurally intact genes for the Leloir pathway enzymes. These were organized into an operon in the order galKTE, which was preceded by a divergently transcribed regulator gene, galR, and followed by a galM gene and the lactose operon lacSZ. Results of Northern blot analysis showed that the structural gal genes were transcribed weakly, and only in medium containing lactose, by strain CNRZ 302. However, in a spontaneous galactose-fermenting mutant, designated NZ302G, the galKTE genes were well expressed in cells grown on lactose or galactose. In both CNRZ 302 and the Gal+ mutant NZ302G, the transcription of the galR gene was induced by growth on lactose. Disruption of galR indicated that it functioned as a transcriptional activator of both the gal and lac operons while negatively regulating its own expression. Sequence analysis of the gal promoter regions of NZ302G and nine other independently isolated Gal+ mutants of CNRZ 302 revealed mutations at three positions in the galK promoter region, which included substitutions at positions −9 and −15 as well as a single-base-pair insertion at position −37 with respect to the main transcription initiation point. Galactokinase activity measurements and analysis of gusA reporter gene fusions in strains containing the mutated promoters suggested that they were gal promoter-up mutations. We propose that poor expression of the gal genes in the galactose-negative S. thermophilus CNRZ 302 is caused by naturally occurring mutations in the galK promoter.
After its discovery almost 40 years ago, the Escherichia coli lactose operon, encoding enzymes of lactose metabolism, became the first model for gene regulation (reviewed in reference 4). The key component of this system is the lac repressor (LacI), the product of the lacI gene. The lac operon contains a primary operator (O1), which is the major element of repression by LacI, and two pseudo-operators, which enhance repressor binding to O1 by cooperativity. Control of the lac operon also involves activation by the cyclic AMP receptor protein. Many other paradigm systems of negative control have since been described, including GalR, one of the two repressors of the gal regulon encoding enzymes of galactose transport and metabolism in E. coli. Regulation of the gal regulon is mediated through GalR, GalS (Gal isorepressor), and the cyclic AMP receptor protein. GalR and GalS negatively regulate transcription of the two promoters of the gal operon, although GalS is not as efficient as GalR (57).
The bioconversion of lactose, which is the primary carbon and energy source in milk, into lactic acid is an essential process in industrial dairy fermentations carried out by lactic acid bacteria. Genetic studies of the metabolic pathways for lactose utilization in these gram-positive bacteria have revealed a variety of lac operons that differ from the paradigm known in E. coli (13). The thermophilic yogurt bacteria Streptococcus thermophilus and Lactobacillus bulgaricus contain a highly homologous lacSZ operon in which the β-galactosidase (lacZ) gene is located downstream from the lacS gene encoding a lactose permease (LacS), which belongs to the galactoside-pentose-hexuronide translocators (27, 41, 48, 49).
Although lactose is efficiently transported and hydrolyzed intracellularly, many strains of S. thermophilus and L. bulgaricus do not grow on galactose and ferment only the glucose portion of lactose, while the galactose is excreted into the medium in amounts stoichiometric with the uptake of lactose (20, 22). Kinetic studies indicated that LacS mediates both galactoside exchange (e.g., lactose-galactose) and movement of galactosides and protons (15). The exchange reaction is highly favored with excess galactosides on either side of the membrane and may account for the galactose-negative (Gal−) phenotype of S. thermophilus in milk which contains an excess of lactose (40). Another explanation for the Gal− phenotype may be the absence of functional Leloir pathway enzymes, including galactokinase (GalK), galactose-1-phosphate uridylyltransferase (GalT), and UDPglucose 4-epimerase (GalE), products of the galK, galT, and galE genes, respectively. Remarkably, under appropriate selective conditions, such as limiting lactose and excess galactose, Gal+ derivatives of S. thermophilus were obtained which fermented galactose and contained Leloir enzyme activities (21, 50). However, no molecular explanation was given, and the genetics of the Leloir pathway has only been poorly investigated in S. thermophilus. The lacSZ operon of strain A147 was found to be preceded by galE and galM (42). The galM gene appeared to be constitutively expressed and could encode a mutarotase that, similar to the homologous enzyme of E. coli, forms the galactokinase substrate α-d-galactose from β-d-galactose pyranose generated from lactose by β-galactosidase (LacZ) (7). However, S. thermophilus A147 is not a Gal+ strain.
The present study was undertaken to gain insight into the presence and regulation of the gal genes of S. thermophilus and the mechanism by which the genes, in particular the galK gene, are prevented from being expressed. Here we describe the characterization of the gal operon, consisting of the galK, galT and galE genes, and its promoter from S. thermophilus CNRZ 302, for which galactose-fermenting (Gal+) revertants have been reported (5). A regulatory gene, galR, was identified that is divergently transcribed from the gal operon. Analysis of mRNA for the gal metabolic genes from a Gal+ fermenting derivative of CNRZ 302 indicated that regulation occurred at the transcriptional level. In contrast, the gal metabolic genes of the original Gal− strain were not sufficiently transcribed to allow galactose metabolism. Furthermore, we demonstrate that GalR acts as a transcriptional activator of both the gal and lac operons and negatively regulates its own expression. To the best of our knowledge, this is the first report describing the mechanisms regulating galactose utilization in S. thermophilus at the molecular level.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. S. thermophilus strains were subcultured in M17 broth (Oxoid, Basingstoke, England), containing either 1% lactose, glucose, or galactose as necessary, at 42°C unless stated otherwise. The taxonomic position of strain CNRZ 302 was confirmed by 16S rRNA sequence analysis and corresponded to Streptococcus thermophilus (GenBank accession number X68418). E. coli strains HB101 or LE392 and TG1 were used for the isolation of pACYC184- and pUC19-derived plasmids and for the propagation of bacteriophage M13 chimeras, respectively. E. coli was routinely grown in TY medium (45) or brain heart infusion (Difco) broth with aeration at 37°C. MacConkey agar base (Difco Laboratories) supplemented with 1% galactose was used to detect galactose-positive (Gal+) E. coli strains. Agar media were prepared by adding 1.5% agar to broth. The antibiotics used for selection in media were chloramphenicol at 4 μg/ml and erythromycin at 2.5 μg/ml for S. thermophilus and chloramphenicol at 15 μg/ml, tetracycline at 12 μg/ml, and ampicillin at 100 μg/ml for E. coli. Emr E. coli strains were selected on brain heart infusion agar containing 150 μg of erythromycin per ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain | Relevant features | Reference or source |
|---|---|---|
| E. coli strains | ||
| HB101 | F−hsdS20(rB− mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20(Smr) xyl-5 mtl-1 supE44 λ− | 46 |
| LE392 | F−hsdR514(rK− mK+) supE44 supF58 lacY1 galK2 galT22 metB1 trpR55 λ− | 46 |
| TG1 | Δ(lac-pro) supE thi hsdD5 F′[traD36 proA+B+ lacIqlacZΔM15] | 46 |
| S. thermophilus strains | ||
| CNRZ 302 | Wild-type Gal− strain | 5 |
| ST11 | Gal− strain | 32 |
| NZ302G | Gal+ class I mutant | This study |
| SS1 | Gal+ class I mutant | This study |
| SS2 | Gal+ class II mutant | This study |
| SS3 | Gal+ class I mutant | This study |
| SS4 | Gal+ class I mutant | This study |
| SS5 | Gal+ class II mutant | This study |
| SS6 | Gal+ class I mutant | This study |
| SS7 | Gal+ class III mutant | This study |
| SS8 | Gal+ class I mutant | This study |
| SS9 | Gal+ class II mutant | This study |
| NZ302GΔR | NZ302G carrying a disruption in galR | This study |
| ST11ΔR | ST11 carrying a disruption in galR | This study |
| Plasmids | ||
| pACYC184 | Tcr | 9 |
| pUC19 | Apr | Gibco-BRL |
| pNZ680 | 4.9-kb gal insert in pACYC184; Tcr | This study |
| pNZ273 | Contains gusA reporter gene; Cmr | 39 |
| pNZ6871 | Contains galR gene, and galK promoter of strain CNRZ 302 fused to gusA; Cmr | This study |
| pNZ6872 | Contains galR gene, and galK promoter of strain SS2 fused to gusA; Cmr | This study |
| pG+host9 | Temperature-sensitive shuttle vector; Emr | 28 |
| pNZ684 | pG+host9 with internal fragment of galR gene; Emr | This study |
| pNZ6811 | Derived from pNZ6871, carries galR gene of strain CNRZ 302; Cmr | This study |
| Phage | ||
| M13mp18/19 | 58 |
DNA isolation and manipulations.
Transformation and isolation of plasmid DNA from E. coli were performed essentially by established protocols (46). Chromosomal DNA was extracted from exponentially growing cells of S. thermophilus by the procedure of Hayes et al. (19). The Anderson and McKay (3) lysis procedure was used to detect plasmid DNA in S. thermophilus. Restriction enzymes, T4 DNA ligase, and other DNA-modifying enzymes were used as recommended by the supplier (Gibco-BRL, Life Technologies, Gaithersburg, Md.). DNA fragments were recovered from agarose gels with the GlassMatrix DNA isolation system (Gibco-BRL). Electroporation of S. thermophilus was performed by the procedure of Mollet et al. (32) with the modification that the harvested cells were incubated in the electroporation buffer at 4°C for at least 4 h prior to electroporation. PCR was performed under the conditions described previously (25) using Taq polymerse (Life Technologies) or Pwo polymerase (Boehringer Mannheim). Oligonucleotides were synthesized by Eurogentech (Gent, Belgium).
Cloning of gal genes.
S. thermophilus CNRZ 302 total genomic DNA was digested with EcoRI, and the DNA fragments were separated by agarose gel electrophoresis (0.7% agarose). The DNA was transferred to a GeneScreen Plus (Dupont, Boston, Mass.) membrane by established methods (46). The membrane was hybridized with a 700-bp AccI fragment, containing part of the S. thermophilus F140 galK gene kindly provided by B. Hutkins (34). The labeling of this fragment with horseradish peroxidase and the hybridization and detection methods were as described in the manufacturer's manual for the ECL system (Amersham, Little Chalfont, United Kingdom). Fragments of approximately 5 kb were recovered, ligated with EcoRI-linearized calf intestinal alkaline phosphatase-treated pACYC184, and used to transform E. coli HB101 and LE392. Gal+ clones were selected as red Tcr colonies on McConkey galactose agar at a frequency of approximately 1%. Analysis of the plasmid content of all 10 Gal+ colonies indicated that they contained a recombinant plasmid with a 4.9-kb insert that showed an identical restriction pattern. One of the clones, designated HB101(pNZ680), was used in further experiments.
Nucleotide sequence analysis.
DNA fragments were subcloned in the phage vectors M13mp18 and M13mp19 with TG1 as a host by using standard techniques (46). Nucleotide sequences of both strands were determined by the dideoxy-chain termination method (47), adapted for Sequenase version 2.0 (U.S. Biochemical Corp., Cleveland, Ohio) with either the M13 universal primer or specifically synthesized primers. The gal promoter regions of S. thermophilus CNRZ 302 and its 10 Gal+ derivatives were isolated as 350-bp PCR fragments from agarose gels. The purified fragments and primers were annealed by boiling for 5 min and rapidly freezing in liquid nitrogen, and sequencing proceeded as described above. The sequence data were assembled and analyzed with PC/GENE version 6.6 (Genofit, Geneva, Switzerland). Amino acid sequence comparisons were performed with the EMBL (release 31.0), SwissProt (release 28.0), and NBRF/PIR (release 40.0) databases using the FASTA program (36), through the facilities of the CAOS/CAMM Center (Nijmegen, The Netherlands). The curvature of DNA was predicted as described by Munteau et al. (33).
Isolation of Gal+ S. thermophilus strains.
S. thermophilus CNRZ 302 cultures grown in M17 broth supplemented with 1% lactose were diluted 100-fold into M17 broth containing 1% galactose and 0.01% glucose and incubated for 24 h. Cultures that exhibited growth were transferred to M17 broth containing 1% galactose (Gal-M17) and incubated for 16 to 20 h. Ten Gal+ single-colony isolates were obtained by plating 10 cultures treated as described above on M17 agar with 1% galactose, and these were designated NZ302G and SS1 to SS9.
RNA isolation, Northern blotting, and primer extension analysis.
S. thermophilus strains were grown in M17 broth (50 ml) containing 1% lactose, glucose, or galactose to an optical density (600 nm) of 0.6 to 1.0. Total RNA was isolated from the harvested cells as described by Kuipers et al. (26) with the following modification: before being subjected to bead beating, the cells were treated with 2 mg of lysozyme per ml for 2 min on ice, which increased the RNA yield. The RNA was either fractionated on a 1.0% formaldehyde gel (46) or glyoxylated and fractionated on a 1.2% agarose gel as described previously (52). RNA size markers were obtained from Bethesda Research Laboratories. RNA was transferred to GeneScreen Plus membranes by following the protocols outlined by the manufacturers. Hybridizations were performed at 65°C in a 0.5 M sodium phosphate buffer (pH 7.2) containing 1.0% bovine serum albumin (fraction V), 1.0 mM EDTA, and 7.0% sodium dodecyl sulfate, and the blots were washed at 55 to 65°C in 1.0 to 0.1× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 1% sodium dodecyl sulfate (46).
Gel-purified restriction fragments and PCR products that had been labeled by nick translation with α-32P (Amersham) were used as hybridization probes (46). These included a galK-specific probe isolated as a 0.6-kb HpaI-HindIII fragment from pNZ680, a galTE probe consisting of a 1.2-kb PstI-EcoRI fragment, a galR-specific probe amplified from pNZ680 with primers 5′-GCC CAA TGA GTA GGC C-3′ and 5′-CGG ATA TTA ACT ATC GCT G-3′, and a 1.6-kb lacS-specific probe generated with primers 5′-TAA CAC AGG TGA TCC AAA GCA-3′ and 5′-GGT GAC CAG AAC TCA AGA AG-3′. The primer GALRAS (5′-GTT GAA ATA GAT ACA CCT GC-3′), which is complementary to the 5′ end of the sense strand of the galR gene, was end labeled with γ-32P using polynucleotide kinase (Bethesda Research Laboratories).
Primer extension was performed by annealing 5 ng of oligonucleotide 3′-ACT AAC CAC TCG TAT GCC TGA T-5′ and 5 ng of oligonucleotide 5′-GTA TCC TCT GTT ACG G-3′ complementary to the mRNA for galK and galR, respectively, to 20 μg of S. thermophilus RNA and performing complementary DNA synthesis as previously described (52). The reaction products were separated by electrophoresis on a 5% sequencing gel, together with a sequencing reaction product obtained using the same primers.
Construction of plasmids for analysis of galK promoters.
The promoters from the CNRZ 302 and a class II Gal+ mutant, strain SS2, were amplified by PCR using primers 5′-CGG GAT CCT GCT AAT TTT GCG ATA TCT G-3′ and 5′-CGG AAT TCC TTT AAA CTT TTC TCT TAA C-3′, with built-in BamHI and EcoRI sites (underlined), respectively. The 210-bp products were cloned into BamHI-EcoRI-digested pUC19, generating pNZ680.1 and pNZ680.2. A 1.5-kb NsiI-EcoRV fragment from pNZ680 containing the CNRZ 302 galR gene and a potential transcription terminator (Fig. 1A) was attached in frame to the galR promoters (this step was necessary for the stability of further constructs), using the PstI and EcoRV sites in the pUC19 derivatives, generating plasmids pNZ6861 and pNZ6862. The “gal promoter and galR gene cassettes” were removed as 1.7-kb EcoRI-HindIII fragments (the 3′ recessed terminus of the HindIII sites were first filled) and subsequently ligated into the EcoRI-ScaI-digested pNZ273. Plasmid pNZ273 contains the gusA reporter gene that encodes the β-glucuronidase enzyme. The plasmids, designated pNZ6871 and pNZ6872 for the CNRZ 302 and SS2 mutants, respectively, contain the galK promoter fused to the gusA gene and the galR gene under its own promoter. The integrity of the amplified promoter regions was confirmed by sequence analysis. The constructs were initially made in E. coli and were subsequently used to transform S. thermophilus ST11 and selected on M17 agar containing 1% sucrose and chloramphenicol. Histochemical screening for β-glucuronidase activity by selecting for blue colonies with 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc) (Research Organics Inc., Cleveland, Ohio) was performed as previously described (11).
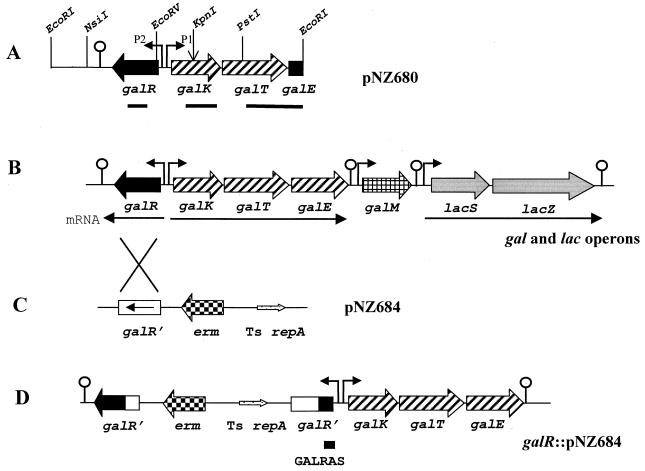
FIG. 1.
Organization of the gal and lac genes in their operons, and construction of a galR gene disruption. (A) Illustration of the gal genes on the 4.9-kb EcoRI fragment of S. thermophilus CNRZ 302 cloned in pNZ680. The positions of relevant restriction enzyme cleavage sites are indicated, and the frameshift mutation in the KpnI site is marked with a vertical arrow. The galK and galR promoters are indicated by arrows P1 and P2, respectively. The black lines below the genes represent the DNA probes used in the Northern blot experiments. (B) Organization of the gal and lac genes in the chromosome of S. thermophilus CNRZ 302. The mRNA transcripts for the gal and lac genes are indicated by arrows. Positions of promoters and potential terminators are indicated. (C) Plasmid pNZ684 carrying the thermosensitive replicon (Ts repA) of pG+host9 (small arrow) is shown in a linear form for convenience; the directions of the sense strand in the 750-bp PmlI-NlaIV galR fragment in pNZ684 and the Emr gene (erm) are indicated. (D) Orientation of integration of pNZ684 in the galR gene. The two partially deleted copies of galR are designated galR', and the location of the primer GALRAS is indicated by the bar.
Construction and use of integrating and complementing plasmids.
A 750-bp PmlI-NlaIV fragment of the galR gene from pNZ680 was ligated into the calf intestinal alkaline phosphatase-treated EcoRV site of the thermosensitive pG+host9 vector, generating pNZ684 (Fig. 1C). Electrotransformation of pNZ684 into S. thermophilus NZ302G resulted in four Emr transformants, all of which contained the expected plasmid at 30°C. To obtain integration of pNZ684, cultures grown overnight in M17 sucrose broth with erythromycin at 28°C were diluted 100-fold into fresh medium and reincubated at 28°C to allow the exponential phase of growth to resume. The cultures were shifted to 42°C and grown until they reached stationary phase. Dilutions of the cultures were plated at 42°C, and integrants appeared as Emr colonies after 24 to 48 h of incubation. Correct integration within the galR gene in the chromosome (Fig. 1D) was confirmed by both Southern hybridization and PCR for integrant NZ302GΔR (data not shown). The galR gene of strain ST11 was disrupted in the same manner.
To construct a plasmid with the galR gene without gusA, the gusA gene in pNZ6871 was removed by digesting with EcoRI-HindIII and the recessed 3′ termini of the remaining 4.9-kb fragment were filled in and ligated to generate pNZ6811.
Enzyme and protein assays and chromatography.
The S. thermophilus strains were grown in M17 broth containing either 1% lactose, galactose, glucose, or sucrose with the appropriate antibiotics to an optical density at 600 nm (OD600) of 1.0. For the preparation of extracts, cells were disrupted with zirconium glass beads in a Bead Beater (Biospec Products, Bartlesville, Okla.) for 3-min treatments with intervals of 1 min on ice between treatments and cellular debris was removed by centrifugation. The extracts were kept on ice, and enzyme assays were performed within 4 h. Galactose 1-phosphate uridylyltransferase activity was assayed in the resultant extracts with 30 to 350 μg of protein per assay by the spectrophotometric method of Isselbacher (23). β-Galactosidase was assayed at 37°C by the method of Miller (31) using 1 to 6 μg of protein per assay and galactokinase assays by the method of Ajdic et al. (2). All enzyme activity measurements presented were the mean of at least two independent experiments. Proteins concentrations were estimated by a dye binding assay (8).
Lactose and galactose were detected by high-performance liquid chromatography with a refractive index detector (M410; Waters) using a Polyspher CHPb18 column (Merck). The separations were carried out on a M6000 isocratic pumping system (Perkin-Elmer) in combination with an automatic sample injector (717+; Waters), and water was used as the eluent.
Nucleotide sequence accession number.
The GenBank accession number assigned to the nucleotide sequence encoding S. thermophilus galR, galK, galT, and the partial galE gene is U61402.
RESULTS
Isolation and localization of the S. thermophilus galK and galT genes.
Southern hybridizations of genomic DNA of S. thermophilus strain CNRZ 302 identified an EcoRI fragment of approximately 5 kb that hybridized with a probe consisting of a 0.7-kb internal fragment of the galK gene of S. thermophilus F140 (34). A minibank of fragments including the hybridizing fragment was constructed in the chloramphenicol resistance (cat) gene of pACYC184, and the putative galK gene of S. thermophilus CNRZ 302 was isolated by functional complementation of the galK2 mutation of E. coli HB101. The complementing plasmid, designated pNZ680 (Fig. 1A), contained a 4.9-kb insert that allowed HB101 to form red colonies on McConkey galactose agar and utilize galactose as the sole carbon source in minimal M9 medium. Introduction of a frameshift mutation in the KpnI site on pNZ680 resulted in a plasmid which could not restore a Gal+ phenotype to HB101, indicating that the galK gene was overlapping this site (data not shown). Moreover, pNZ680 also complemented both the galK2 and galT22 mutations of E. coli LE392, indicating that it also contained the galT gene of S. thermophilus. Although these gal genes are hardly expressed in S. thermophilus, the promoter upstream of the cat gene in pACYC184 is likely to be responsible for their expression in E. coli. The presence of a functional galT gene was further confirmed by assaying for GalT enzyme activity. Cell extracts of LE392(pNZ680) contained 119 nmol of GalT activity per min per mg, whereas no activity was detected for the LE392 strain alone.
Organization and similarity studies of the S. thermophilus gal region.
Commencing at the KpnI site on pNZ680, nucleotide sequence analysis in both directions revealed the galK open reading frame (ORF), 1,164 bp in length (Fig. 1A). The deduced galK sequence had the strongest similarity to the GalK proteins of several gram-positive bacterial species including Streptococcus mutans (83%) and Lactobacillus casei (70%) (2, 6). The S. thermophilus CNRZ 302 GalK was also 79% similar to the GalK of the Gal+ S. thermophilus F410 strain (34). A potential ribosome binding site (5′-GAGA-3′), complementary to the 3′ end of the 16S rRNA of lactic acid bacteria (12), was located 8 bp upstream from the first translational initiation codon at nucleotide (nt) 1483.
Upstream of galK located in a divergent orientation, a 1,014-bp ORF was designated galR on the basis of the similarity of its deduced amino acid sequence to proteins of the LacI-GalR family of transcriptional regulators (55) (Fig. 1A). The translational initiation site at nt 1340 is proposed on the basis of the position of the putative ribosome binding site (5′-AGGAGGA-3′, nt 1351 to 1345) and the similarity between related proteins (see also below). The S. thermophilus GalR had the greatest similarity to the GalR repressor of S. mutans (75%; 57% identity) and the potential GalR repressor of L. casei (59%; 40% identity) (2, 6). There was also significant similarity, 53 and 48% (35 and 27% identity), to the evolved β-galactosidase (EbgR) and galactose (GalR) repressors, respectively, of E. coli (18, 54). An inverted-repeat structure and a stretch of five T nucleotides (nt 95 to 56) that could function as a rho-independent transcriptional terminator (38). followed the galR gene sequence.
DNA sequence analysis downstream of galK revealed the galT gene (1,482 bp), whose deduced sequence was similar (67 to 74%) to the GalT proteins from several gram-positive bacteria including S. mutans, L. casei, and Lactococcus lactis (Fig. 1A) (2, 6, 53). The stop codon for the S. thermophilus galK gene and the start codon of the galT gene were separated by just 19 bp. The translational initiation site for galT was preceded by a putative ribosome binding site (nt 2654 to 2660). Finally, a fourth ORF was present immediately downstream of the galT gene and reading beyond the pNZ680 clone (Fig. 1A). The nucleotide and predicted amino acid sequence were identical to the aminoterminus of the previously characterized galE gene from S. thermophilus A147 (42).
The organization of lac genes in relation to the galE gene, which have been characterized for S. thermophilus A147 (41, 42), was demonstrated to be identical in CNRZ 302. Long-range PCR was performed using primers based on the sequences of the galK gene of CNRZ 302 and the lacS gene (41) of A147, which resulted in the expected 5.7-kb product. Restriction enzyme analysis of the PCR product showed an identical pattern to that of A147, confirming the presence of the galE, galM, and lacS genes downstream of galK-galT in strain CNRZ 302 (Fig. 1B).
Transcriptional analysis of the gal genes.
The transcription of the gal genes was analyzed in the wild-type S. thermophilus strain CNRZ 302, and in strain NZ302G, an isogenic spontaneous Gal+ mutant strain. Strain NZ302G has a doubling time of 58 min in M17 medium containing 1% galactose at 42°C, in contrast to the wild-type CNRZ 302 parental strain which does not grow at all on galactose. The Gal+ phenotype of NZ302G was stably maintained even after several subcultures in M17 containing lactose.
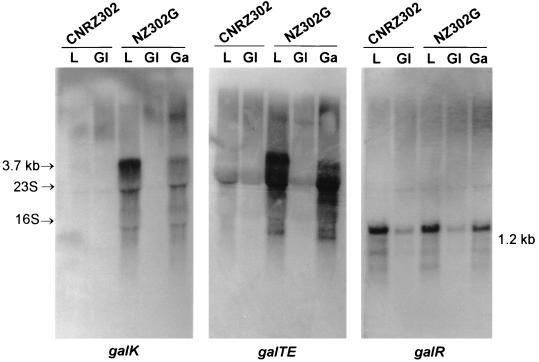
Northern analysis failed to detect hybridization signals for galK or galTE from the glucose-grown wild-type and Gal+ strains (Fig. 2). Only after prolonged exposure, were weak signals obtained for lactose-grown wild-type cells (data not shown). However, mRNA was detected for the lactose- and galactose-grown NZ302G cells (Fig. 2), in accordance with the Gal+ phenotype. The weaker signal obtained for the galactose-grown cells may be due to the poorer-quality RNA obtained as a result of their slower growth. The transcripts were stronger when hybridized with the larger galTE probe than when hybridized with the galK probe. The size of the predominant mRNA hybridizing to the galK and galTE probes was approximately 3.7 kb, indicating that the galK, galT, and galE genes, which are 1.2, 1.4, and 1.0 kb, respectively, are transcribed together as a single mRNA. In conclusion, sufficient induction of galKTE mRNA for galactose metabolism occurred only in the Gal+ NZ302G strain when it was cultured in lactose- or galactose-containing M17 medium.
FIG. 2.
Northern blot analysis of RNA isolated from S. thermophilus strains CNRZ 302 (Gal−) and NZ302G (Gal+) grown on glucose (Gl), lactose (L), or galactose (Ga). The probes used for the hybridization are indicated below the blot. The 23S and 16S rRNAs and the mRNA transcript sizes are indicated by arrows.
A major transcript of approximately 1.2 kb was identified for galR from both the wild-type strain grown in lactose medium and the mutant strain grown in lactose or galactose medium (Fig. 2). However, only a weak signal for this galR mRNA was detected in glucose-grown cells for both strains. The size of the transcript for the 1.0-kb galR gene suggests that it is transcribed alone and supports the functional role of the terminator following galR. These data indicate that the expression of the galR gene is also regulated at the level of transcription.
Mapping and characterization of the galR and galK promoters.
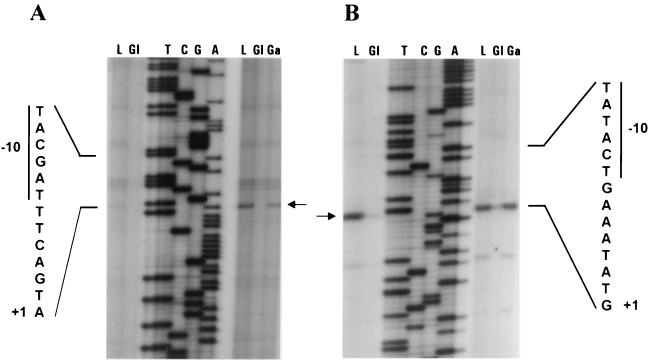
The transcriptional start points of the galK and galR genes were determined by primer extension analysis. Total RNA was isolated from lactose- and glucose-grown S. thermophilus CNRZ 302 cells and from lactose-, glucose-, and galactose-grown NZ302G cells. A major transcriptional start site was observed for the galK gene of strain NZ302G that mapped at an A residue (nt 1452), 6 bp downstream of the inferred −10 sequence (TACGAT) (Fig. 3A). The latter was separated by 17 bp from a −35 sequence (TTGATT) that conforms well to the E. coli and S. thermophilus promoter consensus sequences (Fig. 4A). Essentially no clear signal for the galK gene of CNRZ 302 was detected, in accordance with its Gal− phenotype (Fig. 3A).
FIG. 3.
Primer extension analysis of the 5′ ends of the RNA transcripts for the galK (A) and galR (B) genes of S. thermophilus CNRZ 302 (two left-hand lanes) and NZ302G (three right-hand lanes) grown in lactose (L), glucose (Gl) or galactose (Ga). The ladder obtained from pNZ680 sequenced with the relevant primer is in the middle of each panel. Relevant nucleotide sequences of the promoter regions are presented on either side of the figure. The −10 region is denoted by a vertical bar, and the transcription initiation sites are indicated by +1. The arrows point to the major primer-extended products.
FIG. 4.
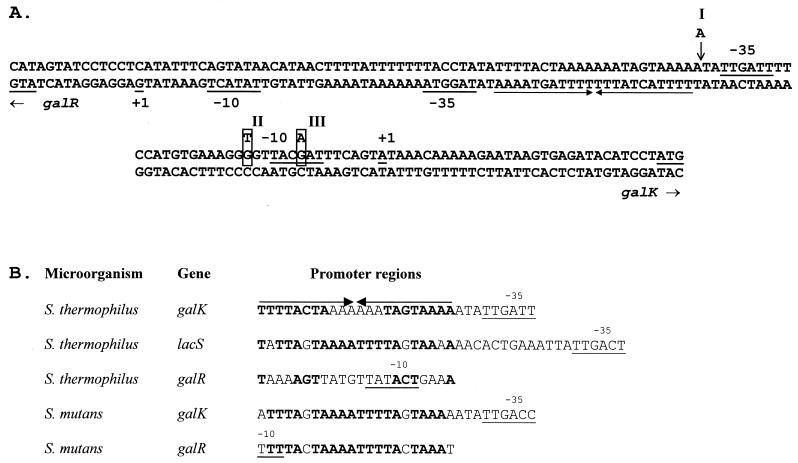
(A) Intergenic region containing the promoter sequences for the galK and galR genes. The −10 and −35 regions are underlined, and the transcriptional start sites are indicated as +1. The point mutations for the three classes (I, II, and III) of S. thermophilus Gal+ mutants are as described in the text. Inverted repeats are indicated by arrows. (B) Alignment of similar regions in the promoters of lac and/or gal genes of S. thermophilus and S. mutans. Inverted repeats in the S. thermophilus galK promoter are indicated by arrows. Nucleotides that are complementary in each arm of the inverted repeats are in boldface type.
For galR, a strongly labeled extension product was detected that initiated at a G residue (nt 1352) 7 bp upstream of a putative −10 sequence (TATACT) (Fig. 3B) for both CNRZ 302 and NZ302G. A putative −35 sequence (TAGGTA) could be found 18 bp upstream of the −10 box (Fig. 4A). The presence of the primer extended products for galK and galR matched exactly the results obtained in the Northern experiments and confirmed control at the transcriptional level.
Effect of the galR gene disruption on galactose utilization.
To determine the function of the galR gene in the Gal+ S. thermophilus NZ302G, the gene was disrupted using pNZ684, consisting of the temperature-sensitive pG+ host9 vector that carried an internal fragment of the galR gene (Fig. 1C). The disruption caused by pNZ684 resulted in two partially deleted copies of the galR gene, one of which lacks the DNA for the 17 N-terminal amino acids including most of the conserved DNA binding motif while the other suffers from a 200-bp deletion that can encode 67 amino acids of the C-terminal region (Fig. 1D). In contrast to the NZ302G strain, the isogenic NZ302GΔR integrant could no longer grow in M17 broth containing 1% galactose. To determine whether NZ302GΔR could utilize the galactose moiety of lactose, its growth was compared with that of the parental strain NZ302G in medium containing 0.4% lactose. Although NZ302G is Gal+, when the strain is grown in medium containing excess lactose (1%), the glucose moiety is preferentially metabolized while galactose is excreted, and presumably acid inhibits growth and prevents subsequent metabolism of the galactose portion of lactose. Reduction of the concentration of lactose to 0.4% eliminates this imbalance while supporting normal growth of strain NZ302G (Table 2). High-performance liquid chromatography analysis of the spent medium indicated that the galactose moiety of lactose was completely metabolized by strain NZ302G while, in contrast, a substantial amount of galactose (72% of the amount that could be hydrolyzed from lactose) was not utilized by the NZ302GΔR integrant. Moreover, the doubling time of NZ302GΔR on lactose increased to approximately 1 h in comparison to that of NZ302G, which is 25 min, and the final OD600 was less than half that of NZ302G (Table 2).
TABLE 2.
Growth and galactose utilization of S. thermophilus NZ302G and derivatives in medium containing 0.4% lactose
| Strain | Final OD600 | % Galactosea |
|---|---|---|
| NZ302G | 1.84 | NDb |
| NZ302GΔR | 0.76 | 0.143 |
| NZ302GΔR1(pNZ6811) | 1.37 | ND |
| NZ302GΔR2(pNZ6811) | 1.30 | ND |
| NZ302GΔR3(pNZ6811) | 1.43 | ND |
Percentage of galactose remaining in medium following growth.
ND, not detectable (<0.01%)
To exclude any possible polar effects of the integration of pNZ684 in the NZ302GΔR integrant, complementation of its galR mutation was studied. Plasmid pNZ6811 contains the galR gene under its own promoter on a high-copy-number vector based on pNZ123 (12). Three transformants of NZ302GΔR harboring pNZ6811 grew to an OD600 of approximately 1.4 in M17 medium containing 0.4% lactose broth, and no residual galactose was detected in the cell-free supernatant (Table 2). Furthermore, the transformants could again utilize galactose as the sole carbon source (data not shown). Thus, GalR is necessary for the ability to utilize galactose.
Effect of galR disruption on transcription of the gal and lac genes.
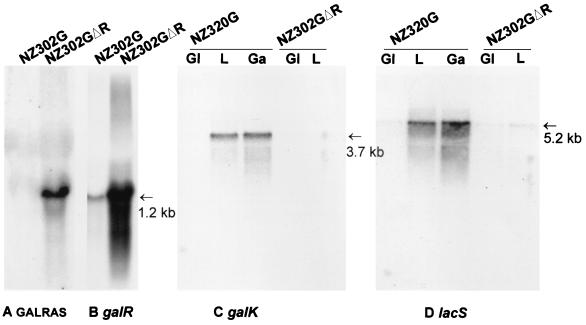
Northern hybridizations were performed to determine the effect of the galR disruption on the transcription of the galR gene and of the gal and lac operons. The primer GALRAS, which is complementary to the 5′ end of the sense strand of galR, was chosen since it can hybridize to the single copy of the 3′-deleted galR (galR′) in the chromosome that is under control of the galR promoter (Fig. 1D). The 1.2-kb galR transcript, which was only weakly visible for NZ302G growing on glucose, gave a signal of much greater intensity for strain NZ302GΔR (Fig. 5A). The 5′-deleted copy of galR did not appear to be transcribed since the same result was obtained by probing with a 600-bp fragment internal to the galR gene (Fig. 5B). This constitutive overexpression of the 3′-truncated galR′ was also observed for lactose-grown cells (data not shown) and suggests that the product of the galR gene is a negative regulator of its own expression. It should be noted that termination of transcription of galR′ in NZ302GΔR is located at an unknown point within the integration plasmid pNZ684, and therefore it is coincidental that the galR and galR′ transcripts appear to have similar sizes.
FIG. 5.
(A and B) Northern blot analysis of RNA from S. thermophilus NZ302G and NZ302GΔR grown on glucose hybridized with the GALRAS primer (A) and galR probe (B). (C and D) Northern blot analysis of RNA from S. thermophilus NZ302G and NZ302ΔR grown on glucose (Gl), lactose (L), or galactose (Ga) hybridized with the galK probe (C) and the lacS probe (D). The mRNA transcript sizes are indicated by arrows.
A galK probe was used to monitor the transcription of the galKTE operon in NZ302G and the mutant NZ302GΔR (Fig. 5C). While the 3.7-kb transcript of the galKTE operon was observed when NZ302G was grown on lactose, no transcript was detected for NZ302GΔR. This indicates that GalR is an activator of transcription for the galKTE operon and explains why the galactose moiety of lactose is not effectively metabolized by strain NZ302GΔR.
LacS is the sole galactoside transporting activity in S. thermophilus and therefore is essential for both lactose and galactose transport (15). We speculated that GalR may also play a role in the regulation of the lac operon (lacS-lacZ). Northern hybridization of NZ302G probed with an internal fragment of lacS showed that a 5.2-kb transcript, corresponding to the sizes of the lacS and lacZ genes, was expressed weakly when the strain was grown on glucose (barely visible on the blot) and induced when the strain was grown on lactose or galactose (Fig. 5D). The lac operon was still transcribed in NZ302GΔR when the strain was grown on glucose, but there was no longer induction of transcription when it was grown on lactose. This indicates that in addition to being the positive regulator of the gal operon, GalR functions as an inducer of transcription of the lac operon.
Effect of galR disruption on GalT and LacZ enzyme activities.
To further substantiate the effect of the galR gene disruption on the expression of the gal and the lac operons, enzyme assays were performed. Since the galT gene is the central one of the three genes in the gal operon, GalT activity was used as a measure of the activity of the Leloir pathway enzymes (Table 3). In strain NZ302G, GalT activity is induced three- to fourfold in lactose- and galactose-grown cultures in comparison to growth in glucose-containing medium. In contrast, very low activity was detected in the galR disruption strain NZ302GΔR when grown on glucose and no significant increase was observed following growth on lactose, indicating a lack of induction of the enzyme. However, the levels of activity of the NZ302GΔR mutant complemented by pNZ6811 were similar to those of NZ302G on all the sugars tested and thus were also induced on lactose and galactose.
TABLE 3.
Enzyme activity measurements of S. thermophilus strains NZ302G, ST11, and derivatives
| Carbohydrate | Strain | Gal-1-phosphate uridylyltransferase activity (nmol/min/mg of protein)a | β-Galactosidase activity (μmol/min/mg of protein)a |
|---|---|---|---|
| Glucose | NZ302G | 59 ± 6 | 4.2 ± 0.9 |
| NZ302GΔR | 19 ± 3 | 3.1 ± 0.5 | |
| NZ302GΔR(pNZ6811) | 70 ± 12 | 2.5 ± 0.4 | |
| Lactose | NZ302G | 178 ± 73 | 7.7 ± 0.8 |
| NZ302GΔR | 23 ± 9 | 3.1 ± 0.4 | |
| NZ302GΔR(pNZ6811) | 149 ± 35 | 6.7 ± 1.8 | |
| Galactose | NZ302G | 230 ± 70 | 15.6 ± 2.8 |
| NZ302GΔR(pNZ6811) | 393 ± 21 | 9.9 ± 0.8 | |
| Sucrose | ST11 | NDb | 4.8 ± 1.0 |
| ST11ΔR | ND | 2.2 ± 0.3 | |
| Lactose | ST11 | ND | 13.3 ± 0.9 |
| ST11ΔR | ND | 5.5 ± 0.6 |
Results are expressed as mean ± standard deviation. The enzyme assays were performed on the same extracts within 4 h.
ND, not determined.
LacZ activity was measured to determine the expression of the lac operon genes. In NZ302G, LacZ activity was induced about two- and fourfold during growth on lactose and galactose, respectively, in comparison to growth on glucose (Table 3). In contrast, LacZ levels in NZ302GΔR were not induced during growth on lactose. LacZ was again inducible in NZ302GΔR carrying the galR-expressing plasmid pNZ6811, although the activity was not as high as that of the original strain. Thus, the enzyme activity measurements confirmed the transcriptional analysis results, demonstrating the role of GalR as an activator of the gal and lac operons.
To analyze whether the activating role of GalR was specific for the galactose-utilizing strain NZ302G, the galR gene of S. thermophilus ST11, a well-characterized Lac+ Gal− strain (32), was also disrupted using the pNZ684 construct. PCR analysis indicated an identical organization of the gal genes in this strain and in CNRZ 302 (data not shown). Since strain ST11 grows very poorly on glucose, LacZ activities were compared using medium containing sucrose (Table 3). Both ST11 and ST11ΔR showed an induction of LacZ of approximately 2.5- to 3-fold after growth on lactose in comparison to growth on sucrose, but the levels of the LacZ activity of ST11ΔR were at least half those produced by ST11 on both carbon sources. The failure to fully induce LacZ in ST11ΔR strongly suggests that GalR also plays a role in activating the lac operon of this strain.
Characterization of the galK promoters for the Gal+ mutants.
To gain an understanding of the ability of NZ302G to transcribe the gal metabolic genes, in contrast to the parent strain CNRZ 302, a series of hybridization experiments using specific galR and galK probes on the gal region and PCR amplification of the galKTE gene cluster were performed on the Gal+ mutant. However, no DNA structural rearrangements were detected within galR or the metabolic gal gene cluster compared to the parent CNRZ 302 strain (data not shown). The galR and galK promoter regions of CNRZ 302 and NZ302G were amplified by PCR in duplicate with the same primers as those used in the primer extension experiments, and both strands of each product were sequenced. The analysis revealed that an extra A residue was inserted in a stretch of adenines preceding the −35 region (nt 1410 to 1414 [Fig. 4A]) of the galK promoter in the NZ302G DNA sequence, resulting in 6 A residues in the mutant in comparison to 5 in the Gal− parent. To determine whether other Gal+ mutants of CNRZ 302 contained similar mutations in the promoter region, nine more independently isolated Gal+ mutants of CNRZ 302 were investigated. DNA sequence analysis of these nine promoters showed that the galK promoter of each Gal+ mutant also contained a point mutation. The mutants could be divided into three classes based on their mutations: class I consisted of five mutants with an A insertion, as described above for NZ302G; in class II, the three mutants contained a G-to-T substitution 3 bp preceding the −10 box; and the one mutant in the third class had a G-to-A substitution in the −10 box (Fig. 4A).
galK promoter activity.
The GalK activity of the mutant strains was used as a reporter for comparing the expression of the mutated promoters with that of the wild-type. A low level of GalK activity, 10 nmol/min/mg, was detected for the Gal− S. thermophilus CNRZ 302 grown on glucose-containing medium, and it increased to 37 nmol/min/mg on lactose. For the Gal+ NZ302G and the other five class I mutants, the GalK activity increased from an average 46 nmol/min/mg on glucose to 264 and 305 nmol/min/mg on lactose and galactose, respectively. The activity increased from 53 nmol/min/mg on glucose to 193 and 323 nmol/min/mg on lactose and galactose, respectively, for the class III mutant. The highest activity was observed with the three class II mutants, an average of 68 nmol/min/mg on glucose to 400 and 458 nmol/min/mg on lactose and galactose, respectively. Thus, it is likely that the basal GalK activity has increased in the mutant strains due to promoter-up mutations, which allows sufficient expression of the galK gene on induction for galactose utilization.
To support our hypothesis that point mutations in the promoters were largely responsible for the Gal+ phenotype of the mutants, the galK promoters of CNRZ 302 and SS2 (class II mutant) were cloned in front of the gusA reporter gene in plasmids pNZ6871 and pNZ6872, respectively. Both plasmids were stable in E. coli, but the frequency of transformation for pNZ6872 in S. thermophilus ST11 was consistently 10-fold lower than for pNZ6871. Since ST11 grows very poorly on glucose, the strength of the promoters was examined in medium containing sucrose or lactose. Expression of the gusA gene results in β-glucuronidase activity, which is indicated by the development of a blue color in colonies on plates containing the substrate X-Gluc. Surprisingly, growth of S. thermophilus ST11 transformants harboring pNZ6871 resulted in blue colonies on X-Gluc plates containing sucrose, while on lactose plates very small and bluer colonies developed, suggesting increased activity from the galK promoter. Colonies of ST11 harboring pNZ6872 developed as very small blue colonies on both sucrose and lactose plates, indicating strong activity of the class II mutant promoter on both carbon sources. However, ST11 transformed with pNZ6872 resulted in colonies that were smaller than those obtained with pNZ6871, and some white colonies also appeared. Analysis of the plasmid content of these colonies demonstrated that the pNZ6871 construct remained intact while deletion derivatives of the pNZ6872 construct were present in ST11 (data not shown). Thus, pNZ6872 is unstable, probably due to toxic effects of high β-glucuronidase activity.
DISCUSSION
This study demonstrates conclusively that S. thermophilus CNRZ 302 possesses the full complement of genes necessary for galactose metabolism despite its Gal− phenotype. In many organisms, the galK, galT, and galE genes, which constitute the Leloir pathway of galactose metabolism, may be clustered or organized in a single operon, and the order of these genes within the operon may be highly variable. While the similarity between the deduced primary sequences of the enzymes is very high within the lactic acid bacteria, the genomic organization of the gal clusters and gene order is species specific (17). The gene order for the S. thermophilus galKTE operon and the divergent galR gene is identical to that in S. mutans (2), which reflects the evolutionary relationship between these bacteria. A potential transcriptional regulatory gene, galR, has also been identified in L. casei and is transcribed in the L. casei gal operon galKETRM (6). In contrast, the putative homologues in E. coli, galR and galS, are not linked to the gal operon (56), and regulatory genes have as yet not been identified for other gal operons in lactic acid bacteria such as L. lactis, Leuconostoc lactis, and Lactobacillus helveticus.
The effect of glucose, lactose, and galactose on the galR mRNA levels strongly suggested that GalR was a transcriptional regulator of the S. thermophilus gal operon. Transcriptional analysis of the two galR copies generated following disruption of galR in the Gal+ NZ302G indicated that the copy resulting in a C-terminally truncated GalR protein is driven by the galR promoter. The second copy, which lacks most of the DNA binding motif, was not transcribed. This was expected based on the orientation of the galR gene fragment in pNZ684. Since the substantial deletion from the C terminus includes residues contributing to inducer binding and dimerization (55), the truncated S. thermophilus GalR proteins are no longer functional. Northern analysis of NZ302GΔR confirmed that GalR functions as an activator of transcription for the gal operon and explained the inability of the disruption mutant to use galactose or the galactose moiety of lactose as a carbon source. The very low activity still detected for the GalT enzyme in NZ302GΔR might be due to a basal transcription level in the absence of GalR. Alternatively, it might reflect a low level of reversion to wild type. When the galR gene was provided in trans, the Gal+ phenotype was restored.
The constitutive transcription of the nonfunctional galR gene in the absence of GalR indicates that the latter normally functions as a negative regulator of its own expression. Autoregulation is a common feature in prokaryotic gene regulation strategies. Negative autoregulation has been reported for other members of the LacI-GalR family such as GalS, PurR, and CytR (16, 30, 56). It is noteworthy that the majority of LacI-GalR members function as negative regulators while some positive regulators also belong to this family and some (like CcpA) perform both functions (43, 45, 51).
The expression of the lac operon genes of both S. thermophilus NZ302G and ST11 is induced by growth on lactose- or galactose-containing medium. In both strains, β-galactosidase activity could no longer be induced to the usual level when the galR gene was disrupted, confirming that GalR also functions as a transcriptional activator of the lac operon. β-Galactosidase is more strongly induced in NZ302G in galactose-containing medium than in lactose-containing medium. This differential gene expression of the lac genes may be due to catabolite repression by the glucose moiety of lactose on the lac operon promoter (51), which will result in repression of the lac operon by lactose but not by galactose. Furthermore, the excretion of galactose by LacS in lactose medium would effectively reduce the availability of inducer in the cell.
The mutation to a Gal+ phenotype does not result in constitutive expression of the gal genes that are induced in the presence of lactose and galactose, which strongly suggests that S. thermophilus was Gal+ but became Gal− in the recent past. While the advantages of the exchange reaction of the lactose transport protein offer a rationale for the observed excretion of galactose (40), the precise mechanism by which the enzymes of the Leloir pathway are suppressed has not been determined. Characterization of the 10 Gal+ mutants of CNRZ 302 revealed that a point mutation had occurred in the galK promoter region of every isolate, the majority of which were single-base insertions in a homopolymeric run of adenine residues. Interestingly, a study of the molecular basis for the adaptive response of E. coli populations to conditions of nonlethal selection such as nutrient deprivation also identified single-base variations mainly in short mononucleotide repeats (14, 44), and slipped-strand mispairing was proposed as the responsible mechanism. In the S. thermophilus galK promoter, the G-to-A substitution in the class III mutant results in a −10 box (TACAAT) with greater homology to the −10 consensus (TATAAT) sequence (29). In class II mutants, the G-to-T substitution gives a TG doublet 1 bp upstream of the −10 sequence, which is a feature present in the promoters of gram-positive bacteria (12). This may correspond to the “extended −10” sequence that functions as a −35-independent promoter and requires the TG motif for efficient initiation at such promoters (24). Thus, these substitutions that resemble promoter-up mutations may increase the level of transcription of the gal genes and allow metabolism of galactose. The A insertion in class I mutants may also be a promoter-up mutation, although the reason for the enhanced activity in this case is not so apparent. The extra A increases the size of the inverted repeat preceding the −35 box from 11 to 15 nt (see below). In particular, the intrinsic DNA curvature that is predicted in this region is enhanced by the A insertion (data not shown), and this may result in increased promoter strength. The presence of curved DNA upstream of promoters, of which A tracts appear to be a major determinant, is associated with increased transcription (37).
The CNRZ 302 and SS2 galK promoter fusions to the gusA gene support the hypothesis that mutations in the galK promoter of S. thermophilus CNRZ 302 suppress the expression of the gal genes. Although β-glucuronidase activity was not expected from the ST11(pNZ6871) strain since galactokinase activity is barely detectable in CNRZ 302, factors such as the high copy number of this plasmid (12) and the gene dosage effect of the GalR activator are likely to be responsible. The only difference between the pNZ6871 and pNZ6872 plasmids was the G-to-T mutation in the galK promoter of the latter. Very high β-glucuronidase activity as a consequence of this promoter-up mutation would result in lethal effects on the host (P. G. G. A. de Ruyter, personal communication). This would explain the reduced frequency of transformation for pNZ6872, the small size of the blue colonies on medium containing X-Gluc, and the instability observed for this plasmid in the S. thermophilus host.
The intergenic region between galR and galK of S. thermophilus consists of 142 bp, which contains the promoter sequences of both genes in a back-to-back configuration (Fig. 4A). An 11-bp inverted-repeat sequence (IR) (nt 1392 to 1413; 5′-TTTTACTA-3′, 8 out of 11 matching nt) was detected in this region that could be an operator for GalR. The potentially global regulation by GalR prompted us to search for homologous sequences in the promoters of the lac operon and galR gene of CNRZ 302. Similar 11-bp IRs were found 13 bp upstream of the −35 box in the lac promoter and also overlapping the −10 box of the galR promoter (Fig. 4B). A consensus sequence could be deduced, with the 11-bp half of the IR consisting of a central 3 bp highly conserved portion, A(C/G)T, flanked on either side by four predominantly adenine and thymine bases. It is usually the −40 to −35 region of a promoter that is approached by an activator site; exceptions include the MerR family regulators (10, 35). In contrast, sites for repressors may be located from the −35 to −10 region. When activator proteins are used for repression, which generally occurs in cases of autoregulation, the operators often appear in positions for repression rather than activation. The potential operator sites for GalR conform to these general rules.
The gal genes of S. mutans are homologous to those of S. thermophilus and are organized in a similar divergent orientation (2). Alignment of the nucleotide sequences of the S. thermophilus and S. mutans promoter regions revealed homologous palindromic sequences, as described above (Fig. 4B). It is noteworthy, however, that a single-crossover disruption of the S. mutans galR gene resulted in constitutive expression of galactokinase, indicating that GalR functions as a repressor of the gal operon in this species. In contrast, GalR of S. thermophilus activates the gal and lac operons while repressing its own expression. The presence of these potential operators gives further credence to the hypothesis that repression of the gal operon was caused by recent mutations in the galK promoter.
ACKNOWLEDGMENTS
We thank R. Hutkins for the gift of the galK gene fragment from S. thermophilus F410, and we thank B. Poolman for communicating the unpublished galT gene sequence and for helpful discussions. We thank R. Holleman for performing the high-performance liquid chromatography assays and E. Maguin of INRA, Jouy en Josas, France, for the kind gift of pG+ host9. We are grateful to M. Kleerebezem and R. Siezen for their interest in this work and for critical reading of the manuscript.
Part of this work was funded by the BIOTECH Programmes of the European Union (contracts ERBB102-CT92-5123 and ERBBIO04-CT96-0439) and by a grant from Ancona University for a specialization abroad to Pasquale Catzeddu.
REFERENCES
- 1.Ajdic D, Ferretti J J. Transcriptional regulation of the Streptococcus mutans gal operon by the GalR repressor. J Bacteriol. 1998;180:5727–5732. doi: 10.1128/jb.180.21.5727-5732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ajdic D, Sutcliffe I C, Russel R R B, Ferretti J J. Organization and nucleotide sequence of the Streptococcus mutans galactose operon. Gene. 1996;180:137–144. doi: 10.1016/s0378-1119(96)00434-9. [DOI] [PubMed] [Google Scholar]
- 3.Anderson D G, McKay L L. Simple and rapid method method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beckwith J. The lactose operon. In: Neindardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 1444–1452. [Google Scholar]
- 5.Benataya A, Bracquart P, Linden G. Galactose-fermenting mutants of Streptococcus thermophilus. Can J Microbiol. 1991;35:136–140. [Google Scholar]
- 6.Bettenbrock K, Alpert C-A. The gal genes for the Leloir pathway of Lactobacillus casei 64H. Appl Environ Microbiol. 1998;64:2013–2019. doi: 10.1128/aem.64.6.2013-2019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouffard G G, Rudd K E, Adhya S L. Dependence of lactose metabolism upon mutarotase encoded in the gal operon in Escherichia coli. J Mol Biol. 1994;244:269–278. doi: 10.1006/jmbi.1994.1728. [DOI] [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Chang A C, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collado-Vides J, Magasanik B, Gralla J D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Ruyter P G G A, Kuipers O P, Beerthuyzen M M, van Alen-Boerrigter I J, de Vos W M. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J Bacteriol. 1996;178:3434–3439. doi: 10.1128/jb.178.12.3434-3439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Vos W M, Simons G. Gene cloning and expression systems in lactococci. In: Gasson M G, de Vos W M, editors. Genetics and biotechnology of lactic acid bacteria. London, United Kingdom: Chapman & Hall; 1994. pp. 52–105. [Google Scholar]
- 13.de Vos W M, Vaughan E E. Genetics of lactose utilization in lactic acid bacteria. FEMS Microbiol Rev. 1994;15:217–237. doi: 10.1111/j.1574-6976.1994.tb00136.x. [DOI] [PubMed] [Google Scholar]
- 14.Foster P L, Trimarchi J M. Adaptive reversion of a frameshift mutation in Escherichia coli by simple base deletions in homopolymeric runs. Science. 1994;265:407–409. doi: 10.1126/science.8023164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foucaud C, Poolman B. Lactose transport system of Streptococcus thermophilus. J Biol Chem. 1992;267:22087–22094. [PubMed] [Google Scholar]
- 16.Gerlach P, Valentin-Hansen P, Bremer E. Transcriptional regulation of the cytR repressor gene of Escherichia coli: autoregulation and positive control by the cAMP/CAP complex. Mol Microbiol. 1990;4:479–488. doi: 10.1111/j.1365-2958.1990.tb00614.x. [DOI] [PubMed] [Google Scholar]
- 17.Grossiord B, Vaughan E E, Leusink E, de Vos W M. Genetics of galactose utilisation via the Leloir pathway in lactic acid bacteria. Lait. 1998;78:77–84. [Google Scholar]
- 18.Hall B G, Betts P W, Wootton J C. DNA sequence analysis of artificially evolved ebg enzyme and ebg repressor genes. Genetics. 1989;123:635–648. doi: 10.1093/genetics/123.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayes F, Daly C, Fitzgerald G F. Identification of the minimal replicon of Lactococcus lactis subsp. lactis UC317 plasmid pCI305. Appl Environ Microbiol. 1990;56:202–209. doi: 10.1128/aem.56.1.202-209.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hickey M W, Hillier A J, Jago G R. Transport and metabolism of lactose, glucose, and galactose in homofermentative lactobacilli. Appl Environ Microbiol. 1986;51:825–831. doi: 10.1128/aem.51.4.825-831.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutkins R, Morris H A, McKay L L. Galactokinase activity in Streptococcus thermophilus. Appl Environ Microbiol. 1985;50:777–780. doi: 10.1128/aem.50.4.777-780.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutkins R W, Morris H A. Carbohydrate metabolism by Streptococcus thermophilus: a review. J Food Prot. 1987;50:876–884. doi: 10.4315/0362-028X-50.10.876. [DOI] [PubMed] [Google Scholar]
- 23.Isselbacher K J. Uridyl transferase. In: Bergmeyer H V, editor. Methods in enzymatic analysis. I. New York, N.Y: Academic Press, Inc.; 1974. pp. 802–805. [Google Scholar]
- 24.Keilty S, Rosenburg M. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J Biol Chem. 1987;262:6389–6395. [PubMed] [Google Scholar]
- 25.Kuipers O P, Boot H J, de Vos W M. Improved site-directed mutagenesis method using PCR. Nucleic Acids Res. 1991;19:4558. doi: 10.1093/nar/19.16.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuipers O P, Beerthuyzen M M, Siezen R J, de Vos W M. Characterization of the Tn5276-located nisin gene cluster nisABTCIPR of Lactococcus lactis and evidence for the involvement of expression of nisI and nisA in producer immunity. Eur J Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 27.Leong-Morgenthaler P, Zwahlen M C, Hottinger H. Lactose metabolism in Lactobacillus bulgaricus: analysis of the primary structure and expression of the genes involved. J Bacteriol. 1991;173:1951–1957. doi: 10.1128/jb.173.6.1951-1957.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maguin E, Prevost H, Ehrlich S D, Gruss A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClure W R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- 30.Meng L M, Kilstrup M, Nygaard P. Autoregulation of PurR repressor synthesis and involvement of purR in the regulation of purB, purC, purMN, and guaBA expression in Escherichia coli. Eur J Biochem. 1990;187:373–379. doi: 10.1111/j.1432-1033.1990.tb15314.x. [DOI] [PubMed] [Google Scholar]
- 31.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 32.Mollet B, Knol J, Poolman B, Marciset O, Delley M. Directed genomic integration, gene replacement, and integrative gene expression in Streptococcus thermophilus. J Bacteriol. 1993;175:4315–4324. doi: 10.1128/jb.175.14.4315-4324.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munteanu M G, Vlahovicek K, Parthasarathy S, Simon I, Pongor S. Rod models of DNA: sequence-dependent anisotropic elastic modelling of local bending phenomena. Trends Biochem Sci. 1998;23:341–347. doi: 10.1016/s0968-0004(98)01265-1. [DOI] [PubMed] [Google Scholar]
- 34.Mustapha A, Hutkins R W, Zirnstein W. Cloning and characterization of the galactokinase gene from Streptococcus thermophilus. J Dairy Sci. 1995;78:989–997. doi: 10.3168/jds.S0022-0302(95)76714-5. [DOI] [PubMed] [Google Scholar]
- 35.O'Halloran T V. Transition metals in control of gene expression. Science. 1993;261:715–725. doi: 10.1126/science.8342038. [DOI] [PubMed] [Google Scholar]
- 36.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez-Martin J, Rojo F, de Lorenzo V. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol Rev. 1994;58:268–290. doi: 10.1128/mr.58.2.268-290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- 39.Platteeuw C, Simons G, De Vos W M. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl Environ Microbiol. 1994;60:587–593. doi: 10.1128/aem.60.2.587-593.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poolman B. Energy transduction in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:125–148. doi: 10.1111/j.1574-6976.1993.tb00015.x. [DOI] [PubMed] [Google Scholar]
- 41.Poolman B, Royer T J, Mainzer S E, Schmidt B F. Lactose transport system of Streptococcus thermophilus: a hybrid protein with homology to the melibiose carrier and enzyme III of phosphoenolpyruvate-dependent phosphotransferase systems. J Bacteriol. 1989;171:244–253. doi: 10.1128/jb.171.1.244-253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poolman B, Royer T J, Mainzer S E, Schmidt B F. Carbohydrate utilization in Streptococcus thermophilus: characterization of the genes for aldose 1-epimerase (mutarotase) and UDPglucose 4-epimerase. J Bacteriol. 1990;172:4037–4047. doi: 10.1128/jb.172.7.4037-4047.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramseier T M, Bledig S, Michotey V, Feghali R, Saier M H., Jr The global regulatory protein FruR modulates the direction of carbon flow in Escherichia coli. Mol Microbiol. 1995;16:1157–1169. doi: 10.1111/j.1365-2958.1995.tb02339.x. [DOI] [PubMed] [Google Scholar]
- 44.Rosenberg S M, Longerich S, Gee P, Harris R S. Adaptive mutation by deletions in small mononucleotide repeats. Science. 1994;265:405–407. doi: 10.1126/science.8023163. [DOI] [PubMed] [Google Scholar]
- 45.Saier M H, Jr, Chauvaux S, Cook G M, Deutscher J, Paulsen I T, Reizer J, Ye J-J. Catabolite repression and inducer control in gram-positive bacteria. Microbiology. 1996;142:217–230. doi: 10.1099/13500872-142-2-217. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Sanger F, Coulson A R, Barell B G, Smith A J H, Roe B A. Cloning in single-stranded bacteriophages as an aid to rapid DNA sequencing. J Mol Biol. 1977;143:161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt B F, Adams R M, Requadt C, Power S, Minzer S E. Expression and nucleotide sequence of the Lactobacillus bulgaricus β-galactosidase gene cloned in Escherichia coli. J Bacteriol. 1989;171:625–635. doi: 10.1128/jb.171.2.625-635.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schroeder C J, Robert C, Lenzen G, McKay L L, Mercenier A. Analysis of the lacZ sequences from two Streptococcus thermophilus strains: comparison with the Escherichia coli and Lactobacillus bulgaricus β-galactosidase sequences. J Gen Microbiol. 1991;137:369–380. doi: 10.1099/00221287-137-2-369. [DOI] [PubMed] [Google Scholar]
- 50.Thomas T D, Crow V L. Selection of galactose-fermenting Streptococcus thermophilus in lactose-limited chemostat cultures. Appl Environ Microbiol. 1984;48:186–191. doi: 10.1128/aem.48.1.186-191.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van den Bogaard P T C, Kleerebezem M, Kuipers O P, de Vos W M. Control of lactose transport, β-galactosidase activity, and glycolysis by CcpA in Streptococcus thermophilus: evidence for carbon catabolite repression by a non-phosphoenolpyruvate-dependent phosphotransferase system sugar. J Bacteriol. 2000;182:5982–5989. doi: 10.1128/jb.182.21.5982-5989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Rooijen R J, de Vos W M. Molecular cloning, transcriptional analysis, and nucleotide sequence of lacR, a gene encoding the repressor of the lactose phosphotransferase system of Lactococcus lactis. J Biol Chem. 1990;265:18499–18503. [PubMed] [Google Scholar]
- 53.Vaughan E E, Pridmore R D, Mollet B. Transcriptional regulation and evolution of lactose genes in the galactose-lactose operon of Lactococcus lactis NCDO2054. J Bacteriol. 1998;180:4893–4902. doi: 10.1128/jb.180.18.4893-4902.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.von Wilcken-Bergmann B, Muller-Hill B. Sequence of galR gene indicates a common evolutionary origin of lac and gal repressor in Escherichia coli. Proc Natl Acad Sci USA. 1982;79:2427–2431. doi: 10.1073/pnas.79.8.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weickert M J, Adhya S. A family of bacterial regulators homologous to Gal and Lac repressors. J Biol Chem. 1992;267:15869–15874. [PubMed] [Google Scholar]
- 56.Weickert M J, Adhya S. Control of transcription of Gal repressor and isorepressor genes in Escherichia coli. J Bacteriol. 1993;175:251–258. doi: 10.1128/jb.175.1.251-258.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weickert M J, Adhya S. The galactose regulon of Escherichia coli. Mol Microbiol. 1993;10:245–251. doi: 10.1111/j.1365-2958.1993.tb01950.x. [DOI] [PubMed] [Google Scholar]
- 58.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]