Abstract
Nine homologous proteins, CspA to CspI, constitute the CspA family of Escherichia coli. Recent studies are aimed at elucidating the individual cellular functions of these proteins. Two members of this family, CspC and CspE, are constitutively produced at 37°C. In the present study, these two proteins were evaluated for their cellular role(s). The expression of three stress proteins, OsmY, Dps, and UspA, is significantly affected by the overexpression and deletion of CspC and CspE. RpoS is a regulatory element for osmY and dps. Further analysis showed a larger amount and greater stability of the rpoS mRNA as well as a higher level of RpoS itself with the overexpression of CspC and CspE. This suggests that CspC and CspE upregulate the expression of OsmY and Dps by regulating the expression of RpoS itself. Indeed, this upregulation is lost in the ΔrpoS strain. Other RpoS-controlled proteins such as ProP and KatG, are also upregulated by the overexpression of CspC. The present study suggests that CspC and CspE are the important elements involved in the regulation of the expression of RpoS, a global stress response regulator, and UspA, a protein responding to numerous stresses. In the light of these observations, it seems plausible that CspC and CspE function as regulatory elements for the expression of stress proteins in the complex stress response network of E. coli.
When exponentially growing Escherichia coli cells are transferred to a low temperature of 15°C, there is a growth lag period, during which the synthesis of cold shock proteins is induced transiently. Among these, CspA is a major cold shock protein. The CspA family of E. coli comprises nine homologous proteins, CspA to CspI. These are of similar sizes, and the corresponding genes are scattered on the E. coli chromosome (for reviews, see references 26, 28, and 37). Of these proteins, only CspA, CspB, CspG, and CspI are cold shock inducible. However, their induction patterns are different. Maximum induction of CspA is at 10 to 24°C, while that of CspB and CspG is at 15°C (7). CspI is produced at 10 to 15°C (36). CspD is induced upon nutritional starvation (38). CspC and CspE are produced mainly at 37°C. These two proteins were originally identified as multicopy supressors of a temperature-sensitive chromosome-partitioning mutant (39). CspE is constitutively produced throughout the growth stages except the lag period, when a 1.5-fold increase in its production is observed (3). It has been shown that CspE inhibits phage lambda Q-mediated transcriptional antitermination in vitro (11). CspE has also been shown to negatively regulate the expression of CspA by increasing the promoter-proximal pausing efficiency of the RNA polymerase (3). CspA has been proposed to act as an RNA chaperone that facilitates translation at low temperature by blocking the formation of secondary structures in mRNA (16). E. coli CspA-family RNA chaperones are shown to be transcription antiterminators (4). CspA has 43% identity to the cold shock domain of the eukaryotic Y-box protein family. This protein family is implicated in various cellular functions such as transcription, DNA replication and repair, and masking of maternal mRNAs (5, 30). CspA binds to RNA with low sequence specificity and low binding affinity. In contrast, CspB, CspC, and CspE are able to selectively bind RNA and single-stranded DNA sequences (27). However, it remains to be seen if this selectivity has any in vivo significance.
In spite of the recent extensive studies (for reviews, see references 26, 28, and 37) on the CspA family of E. coli, the cellular functions of these proteins are not fully elucidated and it is not clear why E. coli has so many CspA-like proteins. Yamanaka et al. (37) had suggested that this large Csp family probably originated from a number of gene duplications and, after subsequent adaptation, resulted in specific groups of genes responding to different environmental stresses. In the present study, using two-dimensional gel electrophoresis, the effect of overproduction of CspC and CspE on the overall protein pattern of the cell was tested. The proteins that were significantly and consistently upregulated by the overexpression of CspC and CspE were identified. This result was analyzed by studying the effect of CspC on the amount and stability of mRNAs for these proteins by primer extension. Interestingly, these three proteins, OsmY, Dps, and UspA, are induced in response to different stresses and also upon stationary phase (2, 13, 14, 17, 24). RpoS is a regulatory element for osmY and dps (19, 42). Further analysis showed that the amount and stability of the rpoS mRNA, as well as the level of RpoS itself, are greatly enhanced by the overexpression of CspC and CspE and that this may result in the upregulation of OsmY and Dps. This possibility was confirmed by using the rpoS deletion strain. The other RpoS-regulated proteins such as ProP and KatG were also upregulated by the overexpression of CspC. The significance of this observation for a possible role of CspC and CspE in the regulation of expression of stress response proteins is discussed.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains used in this study are listed in Table 1. The bacterial cultures were grown in Luria broth or in M9 medium supplemented with glucose (0.02 to 0.4%) and 0.4% Casamino Acids. The cultures were supplemented with antibiotics (50 μg ml−1) such as ampicillin, kanamycin, or spectinomycin as required. The E. coli wild-type strain used in this study was JM83 (40).
TABLE 1.
Bacterial strains used in this study
| E. coli strain | Relevant genotype | Reference or source |
|---|---|---|
| JM83 | F araΔ (lac-proAB) rpsL(Strr) | 40 |
| AR137 | (Δ(argF-lac)U169) but pcnB80 | 12 |
| WB023 | JM83 but ΔcspE::Kanr | 3 |
| WBC | JM83 but ΔcspC::Spcr | This study |
| WBCE | JM83 but ΔcspC::Spcr ΔcspE::Kanr | This study |
| KNJ114 | JM83 but ΔrpoS::Kanr | Personal communication |
Construction of the ΔcspE strain (WB023) has been reported previously (3). The ΔcspC strain (WBC) was constructed in a similar manner, in which the chromosomal ΔcspC coding sequence was replaced by the Spcr gene. Further details will be described elsewhere. The ΔcspC ΔcspE strain (WBCE) was constructed by using phage P1vir-mediated transduction of the ΔcspE strain (21). The ΔcspC ΔcspE strain (AR137) was constructed in a similar manner. The ΔrpoS strain (KNJ114) was a gift from K. Yamanaka.
The isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible pINIIIA3 plasmid and the pINIII-cspC and pINIII-cspE expression vectors containing cspC and cspE, respectively, have been described previously (4, 15).
Using the total E. coli genome as a template, the PCR fragment encompassing the region from upstream of codon 13 of osmY (bases −88 to +284) (42) was generated and inserted into the EcoRI and BamHI sites of pUC19. The PCR fragment for dps contained the region from bases −150 to +81 (codon 14) (2). The sequence of each construct was confirmed. Plasmids pUC19-osmY and pUC19-dps were digested with EcoRI and BamHI, and this was followed by gel purification of the inserts and subcloning in the pRS414 vector to create the respective translational lacZ fusion constructs.
Assay for β-galactosidase activity.
The lacZ constructs of interest were transformed into AR137 strain. The cells were grown in Luria broth, and the β-galactosidase activity was measured as described by Miller (21). To check the expression of CspC and CspE, the cells were exposed to various stresses such as 0.5 M NaCl, 0.5 M KCl, 5% ethanol, pH 10, pH 4, 15°C, 50°C, and anaerobiosis for 15 min and the β-galactosidase activities were determined.
Radioactive labelling of the cells and two-dimensional gel electrophoresis.
Cells were grown in M9 medium supplemented with glucose, 19 amino acids (without methionine), and thiamine at 37°C to an optical density at 600 nm (OD600) of 0.5. Portions (1 ml) of the cultures were labeled with [35S]methionine (1,092 Ci mol−1, 53 Ci ml−1 [Amersham]) for 5 min and then were chased for 3 min by addition of nonradiactive methionine to a final concentration of 0.2 M. To examine the effect of overexpression of CspC and CspE, the pINIIIA3, pINIII-cspC, and pINIII-cspE plasmids were transformed into strain JM83. The exponentially growing cells at an OD600 of 0.5 were induced with 1 mM IPTG for 30 min and then labeled with [35S]methionine. Cell lysates were prepared and were analyzed by two-dimensional gel electrophoresis (34).
For identification of proteins from the two-dimensional gel pattern, the cells were not labeled; instead, the gels were strained with a silver stain (25) and the protein spots of interest were cut out from the gels and identified by peptide mass spectrophotometric fingerprinting (Protein Core Facility, Columbia University).
Isolation of RNA and primer extension.
The respective cultures were grown at 37°C to an OD600 of 0.5. Total RNA was extracted by the hot-phenol method described previously (29). Primer 750077 (5′-TACAGCCAGCAGAGTTTTCGAAAT-3′), which corresponds to the sequence from codons 15 to 8 of osmY (41), primer 750078 (5′-ATAAAGCAGATTGGTCGCTTTTGA-3′), which corresponds to the sequence from codons 16 to 9 of dps (2), and primer 750079 (5′-GCTTTCCGGGGAGAGGTCGACCGC-3′), which corresponds to the sequence from codons 16 to 9 of uspA (22), were labeled with [γ-32P]ATP (DuPont-New England Nuclear) by using T4 polynucleotide kinase (Gibco BRL). For detection of ompF, primer 7018 (5′-ACGGGATCCTTCATCATTATTTATTA-3′), which includes the region encompassing from codon 1 of ompF (accession numbers, JO1655, M10311, and M10312) was used. Primer 979747 (5′-CAGCGTATTCTGACTCATAAGGTG-3′), which corresponds to the region from codon 6 of rpoS (31), primer 956661 (5′-ACGAAGGGTAATCGGTTTTACTTT-3′), which corresponds to the region from codons 13 to 6 of proP (accession number, M83089), and primer 956660 (5′-AGTGGCTGTGGTGTTATGGATATC-3′), which corresponds to the region from codons 13 to 6 of katG (33), were used. Primer extension was carried out with 5 μg of RNA at 42°C for 1 h in a final reaction volume of 10 μl containing 50 mM Tris-HCl (pH 8.5), 8 mM MgCl2, 30 mM KCl, 1 mM dithiothreitol, 0.4 pmol of 32P-labeled primer, 0.5 mM each deoxynucleoside triphosphate, 10 U of RNase inhibitor (Boehringer Mannheim), and 6.25 U of reverse transcriptase (Boehringer Mannheim). The products were analyzed on 6% polyacrylamide gel under denaturing conditions. Quantitation of primer extension products was carried out by direct radioactive measurements.
For measuring the stability of the mRNAs, the cells were grown at 37°C to the mid-log phase and rifampin (Sigma) was added at a final concentration of 200 μg ml−1 to stop the transcription. Samples (1.5 ml) were removed at each time point, and RNA was isolated as described above.
Western blot analysis.
To examine the effect of CspC and CspE overproduction on the RpoS levels by Western blot analysis, wild-type cells containing pINIIIA3, pIN-cspC, or pIN-cspE, grown at 37°C and induced as described above, were concentrated by centrifugation at 13,000 × g, the resulting cell pellets were suspended in sodium dodecyl sulfate (SDS) loading buffer, and the proteins were resolved by SDS-polyacrylamide gel electrophoresis. The Western blots were prepared as described by the antibody manufacturer (Neoclone). The blots were probed with a 1:1,000 dilution of the anti-RpoS monoclonal antibodies (Neoclone) and then with a 1:1,000 dilution of sheep anti-mouse alkaline phosphatase conjugate (Chemicon).
RESULTS AND DISCUSSION
Expression pattern of CspC.
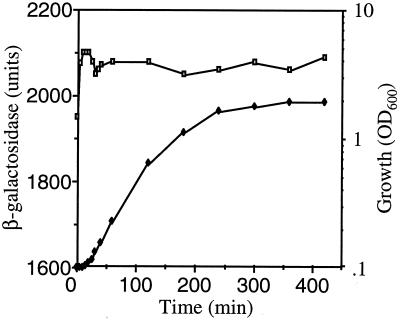
Using the translational cspE-lacZ fusion, it was shown previously that CspE is induced 1.5-fold after dilution of the overnight culture into fresh medium. However, apart from this, its expression is constitutive throughout growth (3). The cspC-lacZ fusion, in which the lacZ gene is translationally fused at codon 13 of cspC, was described previously (18). The cspC-lacZ fusion construct was introduced into strain AR137 which maintains plasmids at low copy number (12), and the β-galactosidase activity was measured during various stages of growth. As seen in Fig. 1, CspC was also expressed constitutively during growth at 37°C but there was no significant induction during the lag phase. We also examined whether CspC and CspE are induced by any stress by using the respective translational lacZ constructs as described in Materials and Methods. CspC and CspE were not significantly induced by 0.5 M NaCl, 0.5 M KCl, 5% ethanol, pH 10, pH 4, temperature stress of 15 or 50°C, or anaerobiosis (data not shown).
FIG. 1.
Expression pattern of the cspC-lacZ translational fusion. The cspC-lacZ expression pattern in different growth stages at 37°C is shown. Open squares, β-galactosidase activities; solid diamonds, OD600 of cells.
Effect of overproduction of CspC and CspE on the overall protein pattern of the cell.
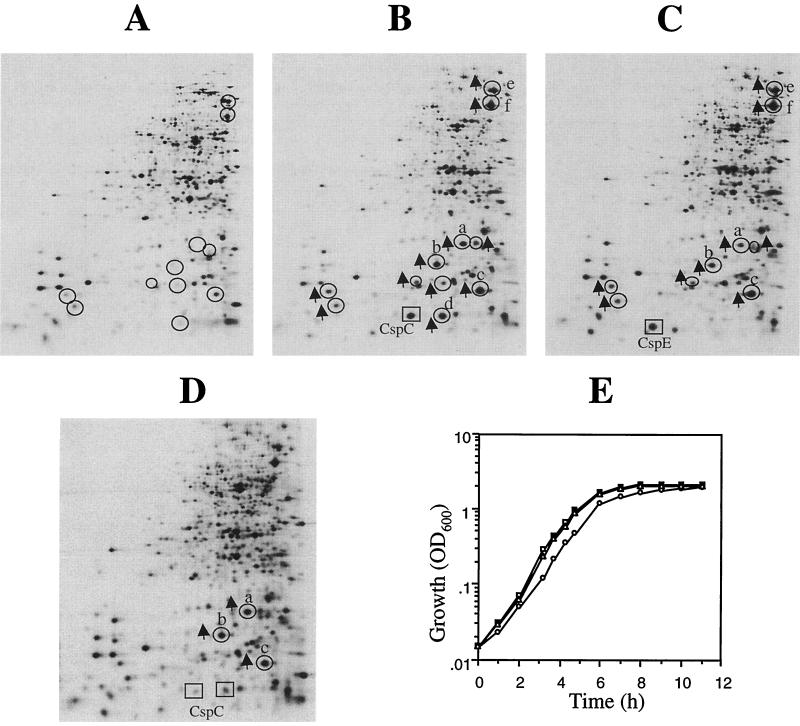
CspC and CspE were overexpressed in wild-type E. coli JM83 using the IPTG-inducible pINIIIA3 vector system. Overexpression of both CspC (10-fold) (Fig. 2B) and CspE (8-fold) (Fig. 2C) seemed to have similar effects on the overall protein pattern of E. coli. The expression of some proteins was upregulated by the overexpression of CspC and CspE. These proteins are circled in Fig. 2. However, we observed that the intensity of the same protein spot sometimes varied in different electrophoretic runs. Hence, we carried out several independent electrophoretic runs of different cell extracts and found that only four of these proteins (labeled a to d in Fig. 2B) were consistently and significantly (more than fourfold) affected by the overexpression of CspC. The intensities of the spots labeled a, b, c, and d were 10-, 8-, 4-, and 10-fold higher, respectively, than those of the corresponding spots in the control (Fig. 2A). The intensities of other circled spots varied in different runs, and in some cases less than a twofold increase was observed with overexpression of CspC and CspE.
FIG. 2.
Effect of overexpression of CspC and CspE in the exponentially growing cells at 37°C. (A to C) The wild-type cells (JM83) transformed with pINIIIA3 (A), pINIII-cspC (B), and pINIII-cspE (C) were induced with 1 mM IPTG for 30 min. (D) The cells transformed with pINIII-cspC were also induced with 0.25 mM IPTG. The cells were labeled with [35S]methionine, and the protein synthesis patterns were compared by two-dimensional gel electrophoresis. Electrophoresis in the first-dimension (isoelectric focusing) gel was carried out between pH 3.5 (right side) and pH 10 (left side). The proteins whose expression is upregulated by CspC and CspE are circled and marked with arrows. The position of CspC is marked with a square in panels B and D, while the position of CspE is marked with a square in panel C. Four independent sets of experiments were carried out, and the proteins consistently showing significant upregulation are designated a to d. (E) Growth curves of the wild-type cells transformed with pINIIIA3 (squares), pINIII-cspC (circles), and pINIII-cspE (triangles) induced with 1 mM IPTG.
The protein designated c was identified as universal stress protein, UspA, from the E. coli database for two-dimensional gel electrophoresis protein patterns (35). Other proteins were identified by peptide mass spectrophotometric fingerprinting. Spot a is an osmotically inducible protein, OsmY, and spot b is the DNA protection protein during starvation, Dps. These three proteins were also upregulated by overexpression of CspE. The fourth protein, designated spot d in Fig. 2B, was identified to be another form of CspC. As expected, this protein was not upregulated in the strain overexpressing CspE (Fig. 2C). We have previously reported that UspA was downregulated in the ΔcspE strain and that when cspE was reintroduced in trans using a plasmid carrying cspE (pKX714) to complement the ΔcspE strain, the level of UspA was restored (3). Downregulation of UspA was also seen in the ΔcspC strain (data not shown). Two more proteins, DnaK and GroEL (spots e and f) were also upregulated by CspC and CspE overexpression; however, the levels were significantly less strongly induced (3-fold) than those seen with their heat shock induction (>10-fold) (data not shown).
Since 1 mM IPTG induced massive production of the two Csp proteins, we also checked the effect of a lower level of CspC induction by using 0.25 mM IPTG. Figure 2D shows that a two times induction of CspC resulted in significant induction of OsmY, Dps, and UspA (8-, 6-and 2.5-fold, respectively). This result suggests that even lower induction of CspC is sufficient for significant induction of stress proteins and supports our conclusion that in spite of being induced constitutively, these proteins are important for regulation of expression of stress response proteins. To determine if overproduction of CspC and CspE has any effect on the growth rate, we checked the growth rates of the wild-type strain containing pINIIIA3, pINIII-cspC, or pINIII-cspE. The cells were induced with 1 mM IPTG at an OD600 of 0.5 as described above. Figure 2E shows that overexpression of CspE had no effect on growth rate. The growth was slower with CspC overproduction; however, the maximum cell density was similar to that without the overexpression. Since CspC and CspE have similar effects on the induction of RpoS and UspA and since CspE does not have any effect on the growth rate, it seems that induction of RpoS and UspA is not due to reduction in the growth rate by any stress created by overproduction of the Csps.
Effect of CspC overexpression and deletion on the amount and stability of mRNAs for osmY, dps, and uspA.
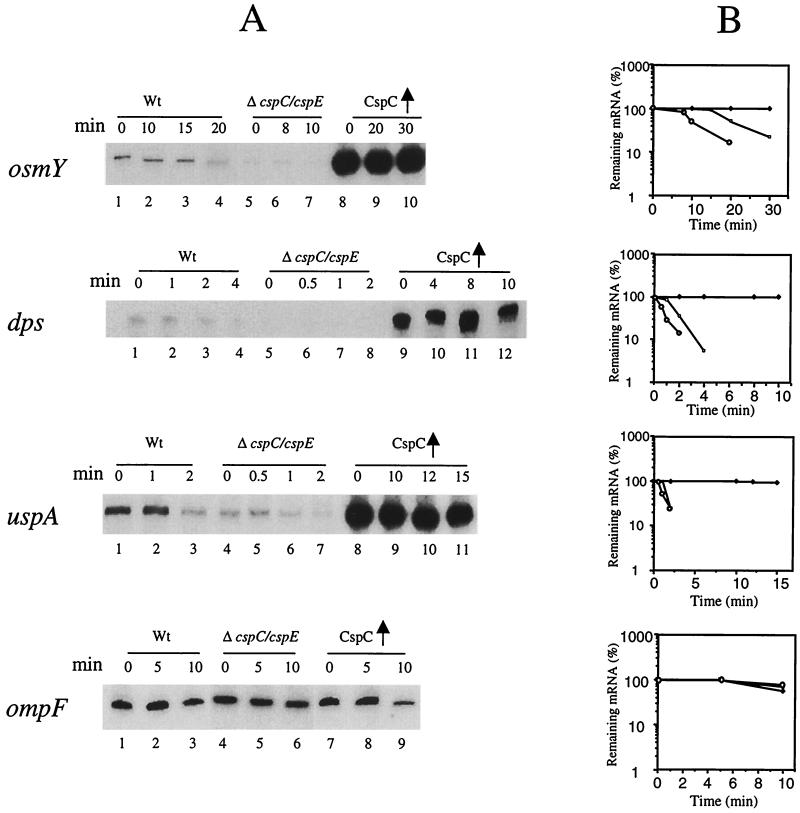
RNAs were isolated from the wild-type strain with and without the overexpression of CspC and from the ΔcspC ΔcspE strain in the exponential phase. Since CspE also influences the expression of these three proteins, a cspC cspE double-deletion strain was used for this study. As seen in Fig. 3A, in the strain overexpressing CspC, the level of osmY mRNA was much higher (15-fold) (lane 8; 0 min) than in the wild-type strain (lane 1; 0 min). On the other hand in the ΔcspC ΔcspE strain, osmY mRNA was significantly downregulated (lanes 5 to 7; 0, 8, and 10 min, respectively). Also, the half-life of RNA in the wild-type strain and in the CspC-overexpressing strain was 20 min and more than 30 min, respectively, while in the ΔcspC ΔcspE strain it was reduced to 10 min (Fig. 3B).
FIG. 3.
Stability and amount of the osmY, dps, and uspA mRNAs in the wild-type, wild-type overexpressing CspC, and ΔcspC ΔcspE strains. The respective cultures were grown at 37°C to an OD600 of 0.5. Total RNA was extracted by the hot-phenol method. For measuring the stability of the mRNAs, rifampin was added at a final concentration of 200 μg ml−1 to stop the transcription. Samples (1.5 ml) were removed at each time point, RNA was isolated, and primer extension analysis was carried out with oligonucleotides corresponding to osmY, dps, or uspA. (A) For osmY mRNA, the wild-type (Wt) strain is lanes 1 to 4 (0, 10, 15, and 20, min, respectively), the ΔcspC ΔcspE strain is in lanes 5 to 7 (0, 8, and 10 min, respectively), and the wild-type strain overexpressing CspC is in lanes 8 to 10 (0, 20, and 30 min, respectively). For dps mRNA, the wild-type strain is in lanes 1 to 4 (0, 1, 2, and 4 min, respectively), the ΔcspC ΔcspE strain is in lanes 5 to 8 (0, 0.5, 1, and 2 min, respectively), and the wild-type strain overexpressing CspC is in lanes 9 to 12 (0, 4, 8, and 10 min, respectively). For uspA mRNA, the wild-type strain is in lanes 1 to 3 (0, 1, and 2 min, respectively), the ΔcspC ΔcspE strain is in lanes 4 to 7 (0, 0.5, 1, and 2 min, respectively), and the wild-type strain overexpressing CspC is lanes 8 to 11 (0, 10, 12, and 15 min, respectively). For ompF mRNA, the wild-type strain is in lanes 1 to 3 (0, 5, and 10 min, respectively), the ΔcspC ΔcspE strain is in lanes 4 to 6 (0, 5, and 10 min, respectively), and the wild-type strain overexpressing CspC is in lanes 7 to 9 (0, 5, and 10 min, respectively). (B) Graphical representation of the results in panel A, which shows the half-lives of the osmY, dps, uspA, and ompF mRNAs. The respective mRNA at the zero time point was takes as 100% in each case. Solid squares, mRNA (i.e., osmY, dps, uspA, or ompF) from cells overexpressing CspC; open squares, mRNAs from wild-type cells; open circles, mRNAs from ΔcspC ΔcspE cells.
As seen in Fig. 3A, the level of dps mRNA was 12 times higher in the CspC-overexpressing strain (lane 9; 0 min) than in the wild-type strain (lane 1; 0 min). Its half-life was 1.5 min in the wild-type strain whereas in the CspC-overexpressing strain it was stable even after 10 min. In the ΔcspC ΔcspE strain, the amount of dps mRNA was significantly reduced (lanes 5 to 8; 0, 0.5, 1, and 2 min, respectively) and the half-life was decreased to 0.5 min (Fig. 3B). The half-life of uspA mRNA was 1.8 min in the wild-type strain, which is consistent with the previous report (22). It was stable for up to 15 min in the CspC-overexpressing strain. The level of uspA mRNA was four times higher in the CspC-overexpressing strain (lane 8; 0 min) than in the wild-type strain (lane 1; 0 min); it was reduced by more than three fold in the ΔcspC ΔcspE strain (lane 4; 0 min), and the half-life was reduced to 1 min (lane 6). Hence, the overexpression and deletion of CspC appears to have a significant effect on the amount and stability of osmY, dps, and uspA mRNAs.
To check if the effect of CspC overexpression is specific, primer extension was carried out using the ompF mRNA. The reason for using OmpF is that it is not induced by any of the above stresses. As seen from Fig. 3A, the amount and stability of ompF mRNA were not affected by the deletion of cspC and cspE (lanes 4 to 6; 0, 5, and 10 min, respectively or by the overexpression of CspC (lanes 7 to 9; 0, 5, and 10 min, respectively) compared to the values in the wild-type strain (lanes 1 to 3; 0, 5, and 10 min, respectively). This suggests that osmY, dps, and uspA are specifically regulated by CspC. We also carried out these experiments using the wild-type strain containing pINIIIA3 vector and found that results were same as those with the wild-type strain alone. The pINIIIA3 vector had no effect on stability of the tested mRNAs or the transcript levels.
Effect of overexpression of CspC and CspE on the amount and stability of rpoS mRNA.
While OsmY is produced in response to osmotic stress and during the stationary phase, its function is unclear. Its expression is transcriptionally regulated by a stationary-phase sigma factor, RpoS, in both osmotic stress and stationary phase (42). Dps is induced by osmotic, oxidative stress and upon stationary phase and is subject to complex regulation including that by OxyR in the exponential phase and by RpoS and integration host factor in the stationary phase (1, 2, 19, 20). UspA is produced under the stresses that lead to growth arrest, and its expression is independent of the global regulators such as RpoS, PhoB, AppY, OmpR, Lrp, and RpoH (8, 9, 22).
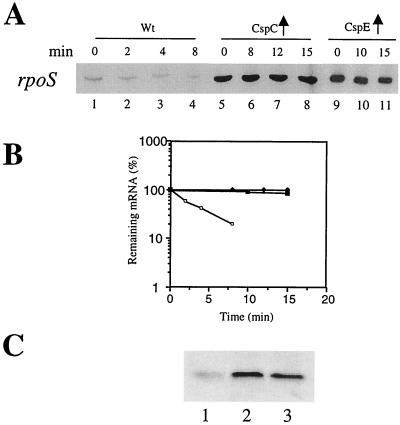
Next we examined if the expression of osmY and dps is influenced by CspC and CspE through RpoS itself. RNAs were isolated from the wild-type strain with and without the overexpression of CspC and CspE, and the stability was measured as described previously. As seen in Fig. 4A, the amount of rpoS mRNA was approximately fourfold higher in the strain overexpressing CspC (lane 5; 0 min) than in the wild-type strain (lane 1; 0 min). The half-life was 2.5 min (lane 2) and more than 15 min (lane 8) in the wild-type strain and the strain overexpressing CspC, respectively. Similar results were observed with CspE overexpression (lanes 9 to 11). The amount of rpoS mRNA was almost undetectable in the ΔcspC ΔcspE strain (data not shown). The upregulation of RpoS by the overexpression of CspC and CspE was also examined at the protein level by Western blot analysis using anti-RpoS monoclonal antibodies. Figure 4C shows RpoS levels in the wild-type strain without (lane 1) and with the overexpression of CspC (lane 2) or CspE (lane 3). The RpoS level was significantly higher in the latter, consistent with its correspondingly higher transcript levels. This raises the interesting possibility that the upregulation of osmY and dps may also be caused by transcriptional activation by RpoS, which in turn is regulated by CspC and CspE.
FIG. 4.
Stability and amount of the rpoS mRNA and RpoS levels in the wild-type strain and the wild-type strains overexpressing CspC and CspE. The cultures were grown at 37°C to an OD600 of 0.5. Total RNA was extracted by the hot-phenol method. For measuring stability of the mRNAs, rifampin was added at a final concentration of 200 μg ml−1 to stop the transcription. Samples (1.5 ml) were removed at each time point, RNA was isolated, and primer extension analysis was carried out with oligonucleotides corresponding to rpoS. (A) The wild-type (wt) strain is in lanes 1 to 4 (0, 2, 4, and 8 min, respectively), the wild-type strain overexpressing CspC is in lanes 5 to 8 (0, 8, 12, and 15 min, respectively), and the wild-type strain overexpressing CspE is in lanes 9 to 11 (0, 10, and 15 min, respectively). (B) Graphical representation of the results in A, which shows the half-life of the rpoS mRNA. The mRNA at the zero time point was considered 100% in each case. Solid diamonds, rpoS mRNA from cells overexpressing CspC; solid squares, rpoS mRNA from cells overexpressing CspE; open squares, rpoS mRNA from wild-type cells. (C) Western blot analysis of RpoS levels in the wild-type strain without (lane 1) and with the overproduction of CspC (lane 2) and CspE (lane 3). Equal numbers of cells were applied to each lane. The Blots were probed with anti-RpoS monoclonal antibodies.
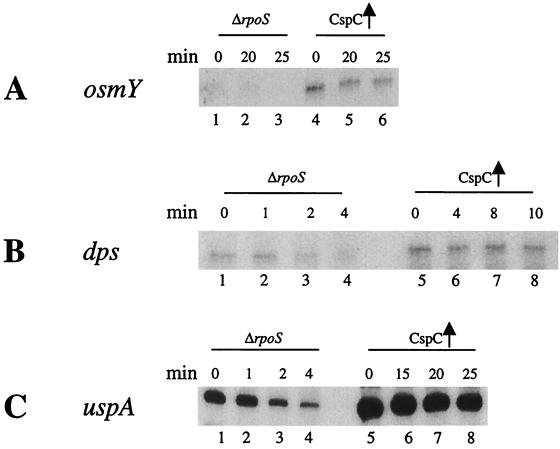
Effect of CspC overexpression on osmY, dps, and uspA mRNAs in the ΔrpoS strain.
To confirm the above possibility, RNAs were isolated from the ΔrpoS strain with and without the overexpression of CspC and the effect of this overexpression on the osmY and dps mRNAs was investigated as described above. As seen in Fig. 5A (lane 1; 0 min), the amount of osmY mRNA was drastically reduced in the ΔrpoS strain, which is consistent with the previous report by Hengge-Aronis et al. (13). In the ΔrpoS strain overexpressing CspC, osmY mRNA could be detected (lane 4; 0 min), even though its level was dramatically lower than that in the wild-type strain overexpressing CspC (Fig. 3A). The amount of dps mRNA in the strain overexpressing CspC (lane 5; 0 min) was twice as high as that in the ΔrpoS strain (lane 1; 0 min). However, as with osmY mRNA, this level was significantly lower than that in the wild-type strain overexpressing CspC (Fig. 3A). These results support the possibility that CspC regulates the expression of OsmY and Dps by influencing the expression of RpoS itself. On the other hand, the level of uspA mRNA in the ΔrpoS strain was unaffected (Fig. 4C, lane 1; 0 min) compared to that in the wild-type strain (Fig. 3A), which is consistent with the fact that expression of UspA is independent of RpoS (22). The stability and amount of uspA mRNA were increased by the overexpression of CspC in the ΔrpoS strain (lanes 5 to 8; 0, 15, 20, and 25 min, respectively).
FIG. 5.
Stability and amount of the osmY, dps, and uspA mRNA in the ΔrpoS strain and the ΔrpoS strain overexpressing CspC. The respective cultures were grown at 37°C to an OD600 of 0.5. Total RNA was extracted by the hot-phenol method. For measuring the stability of the mRNAs, rifampin was added at a final concentration of 200 μg ml−1 to stop the transcription. Samples (1.5 ml) were removed at each time point, RNA was isolated, and primer extension analysis was carried out with oligonucleotides corresponding to osmY, dps, or uspA. (A) osmY mRNA. The ΔrpoS strain is in lanes 1 to 3 (0, 20, and 25 min, respectively), and the ΔrpoS strain overexpressing CspC is in lanes 4 to 6 (0, 20, and 25 min, respectively). (B) dps mRNA. The ΔrpoS strain is in lanes 1 to 4 (0, 1, 2, and 4 min, respectively), and the ΔrpoS strain overexpressing CspC is in lanes 5 to 8 (0, 4, 8, and 10 min, respectively). (C) uspA mRNA. The ΔrpoS strain is in lanes 1 to 4 (0, 1, 2, and 4 min, respectively), and the ΔrpoS strain overexpressing CspC is in lanes 5 to 8 (0, 15, 20, and 25 min, respectively).
Effect of CspC overexpression on mRNAs for proP and katG.
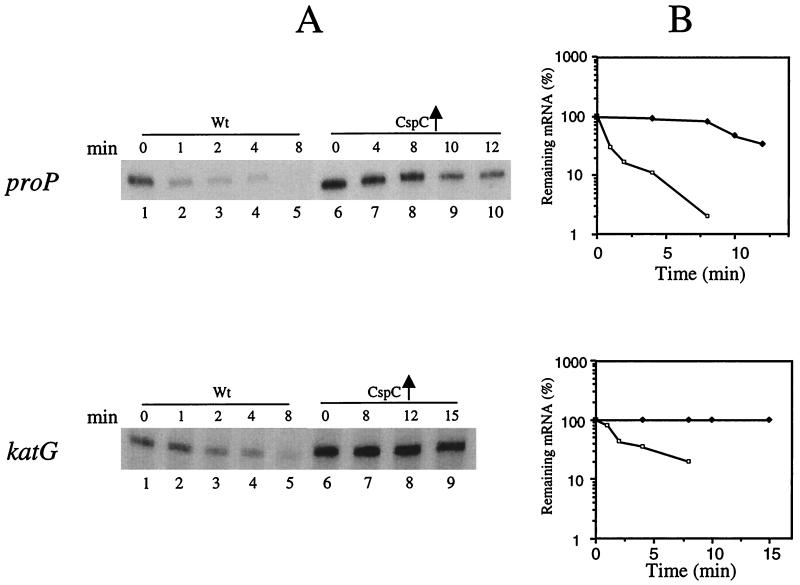
We also investigated if other stress proteins regulated by RpoS are upregulated by the overexpression of CspC and CspE. We tested the effect of CspC overexpression on the level and stability of mRNAs for proP and katG, which are regulated by RpoS (14). ProP is induced by osmotic stress (6), and KatG is induced by oxidative stress (14, 33). The half-life of proP mRNA increased from 0.5 min in the wild-type strain to 10 min in the strain overexpressing CspC (Fig. 6). The half-life of katG mRNA was 1.5 min and more than 15 min in the wild-type strain and the strain overexpressing CspC, respectively (Fig. 6). Neither of these proteins were identified by the two-dimensional polyacrylamide gel electrophoresis, possibly because they may not be resolved well by the system used and because some additional factors may also be responsible for regulation of their expression. Thus, the present results show clearly that CspC and CspE influence the expression of a number of stress proteins that are regulated by RpoS.
FIG. 6.
Stability and amount of the proP and katG mRNAs in the wild-type strain and the wild-type strain overexpressing CspC. The respective cultures were grown at 37°C to an OD600 of 0.5. Total RNA was extracted by the hot-phenol method. For measuring the stability of the mRNAs, rifampin was added at a final concentration of 200 μg ml−1 to stop the transcription. Samples (1.5 ml) were removed at each time point, RNA was isolated, and primer extension analysis was carried out with oligonucleotides corresponding to proP or katG. (A) For proP mRNA, the wild-type (Wt) strain was in lanes 1 to 5 (0, 1, 2, 4, and 8 min, respectively) and the wild-type strain overexpressing CspC was in lanes 6 to 10 (0, 4, 8, 10, and 12 min, respectively). For katG mRNA, the wild-type strain was in lanes 1 to 5 (0, 1, 2, 4, and 8 min, respectively) and the wild-type strain overexpressing CspC was in lanes 6 to 10 (0, 8, 12, and 15 min, respectively). (B) Graphical representation of the results is panel A, which shows the half-lives of the proP and katG mRNAs. The respective mRNA at the zero time point was taken as 100% in each case. Solid squares, respective mRNA (i.e., proP or katG) from cells overexpressing CspC; open squares, respective mRNA from wild-type cells.
Effect of cspC and cspE deletion on the osmotic induction of OsmY and Dps.
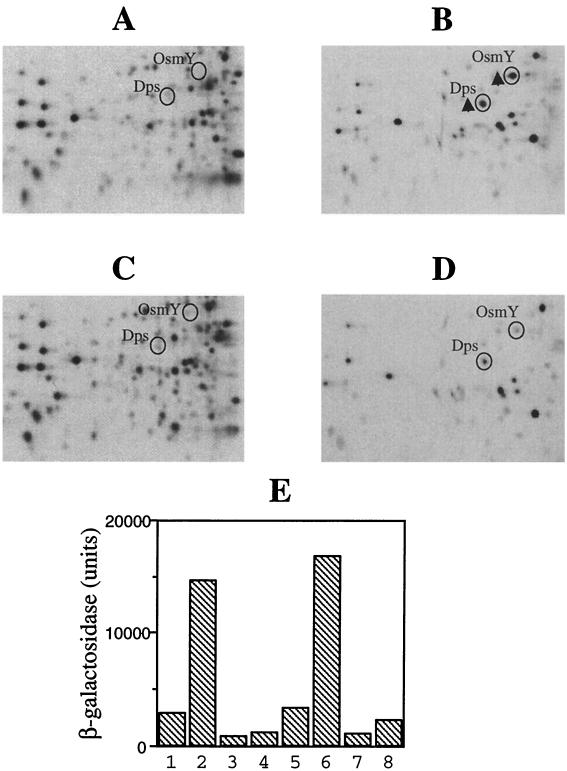
It has been reported that OsmY and Dps are induced by osmotic stress and that in the ΔrpoS strain, the induction of OsmY and Dps by the osmotic stress is reduced (13, 17, 19, 41). To check if deletion of cspC and cspE affects their osmotic induction, the wild-type and ΔcspC ΔcspE cells were treated with 0.3 M NaCl for 15 min and labeled with [35S]methionine and the protein patterns were analyzed by two-dimensional polyacrylamide gel electrophoresis. As seen in Fig. 7B, in the wild-type strain OsmY and Dps were induced by the osmotic stress (compare with the results for the untreated wild-type cells in Fig. 7A), while deletion of cspC and cspE resulted in reduced induction of these proteins by the osmotic stress (Fig. 7D). This may occur through the regulation of rpoS by CspC and CspE. UspA was not greatly induced under the conditions used, because complete growth inhibition by any stress is required for significant induction of UspA (24).
FIG. 7.
Effect of deletion of cspC and cspE on the induction of OsmY and Dps by osmotic stress. The wild-type and ΔcspC ΔcspE cells were exposed to 0.5 M NaCl for 15 min and labeled with [35S]methionine, and the protein synthesis patterns were compared by two- dimensional gel electrophoresis. (A) Wild-type cells. (B) Wild-type cells treated with NaCl. (C) ΔcspC ΔcspE cells. (D) ΔcspC ΔcspE cells treated with NaCl. The relevant parts of the gels are shown. (E) Comparison of the expression patterns of the osmY-lacZ and dps-lacZ translational fusion constructs in response to the osmotic stress in the AR137 and the AR137ΔcspCΔcspE strains. Exponentially growing cells of the respective strains were subjected to osmotic stress as in panels A to D, and β-galactosidase activities were determined in the pre- and postshift cells. The results are as follows: osmY-lacZ in the AR137 strain, preshift (lane 1) and postshift (lane 2); osmY-lacZ in the AR137ΔcspCΔcspE strain, preshift (lane 3) and postshift (lane 4). dps-lacZ in the AR137 strain, preshift (lane 5) and postshift (lane 6); dps-lacZ in the AR137ΔcspCΔcspE strain, preshift (lane 7) and postshift (lane 8).
To examine if only the fold induction by the osmotic stress is affected by the cspC cspE deletion or if the steady-state levels are affected in both the preshift and postshift cells, translational lacZ fusion constructs of osmY and dps were transformed into strains AR137 and AR137ΔcspCΔcspE (compare lanes 1 and 3 for osmY and lanes 5 and 7 for dps). The preshift levels of osmY and dps were 3.5- and 3-fold lower, respectively, in the AR137ΔcspCΔcspE strain, while the corresponding postshift levels were 12- and 8-fold lower (compare lanes 2 and 4 for osmY and lanes 6 and 8 for dps). This suggests that in addition to maintaining the steady-state levels of RpoS, CspC and CspE are involved in regulation of the expression of RpoS and consequently of RpoS-regulated proteins during stress response.
The effect of CspC and CspE overexpression was studied in the exponentially growing cells, but upregulation of osmY and dps by the overexpression of these proteins was also observed in the stationary phase (data not shown). It has been suggested that RpoS may play a role in the exponentially growing cells as well (14). This theory is supported by the recent expression analysis of E. coli (32).
The expression of RpoS itself is subject to complex posttranscriptional and translational regulation. In the present case, CspC and CspE which are RNA-binding proteins (27), regulate the expression of rpoS, which results in the upregulation of the expression of at least some of the genes controlled by RpoS, such as osmY, dps, proP, and katG. The mRNAs for rpoS and uspA are dramatically stabilized by overexpression of CspC and CspE. In addition, transcriptional activation may be responsible for their upregulation. The exact mechanism of the stabilization is not clear, but it may be speculated that CspC and CspE protect these mRNAs from degradation by the virtue of physical binding. On the other hand, it is also possible that the effect of CspC and CspE on the expression of RpoS and UspA is indirect and that they act on an unknown factor(s), which in turn affects the expression of these stress proteins.
On the basis of the expression analysis of E. coli growing in minimal and rich media, it has been suggested that RpoS and UspA play important roles in the global control of carbon flow in cells growing in minimal media (32). The results suggested that RpoS plays a role in the regulation of carbon-metabolizing genes and that UspA may coordinate glucose and acetate metabolism. The involvement of the latter in the coupling of glucose and acetate metabolism is supported by the observation that mutants lacking uspA exhibited diauxic growth on minimal media. This is supposedly due to the failure to assimilate acetate until glucose becomes completely exhausted (23). This may give a new perspective to the role of CspC and CspE in the regulation of these two key proteins.
Conclusion.
The present study shows that RpoS is regulated by CspC and CspE, with the consequent regulation of some of the stress proteins controlled by the former. The cold shock domain family is considered to be one of the ancient protein families (10), and these proteins might have played an important role in the survival of the organism during evolution under various stress conditions (26, 28). It is significant that production of CspC and CspE at 37°C is constitutive. It is interesting that CspC and CspE are involved in the regulation of the expression of RpoS, a global stress response regulator, and UspA, a protein responding to numerous stresses, during normal growth as well as during the stress response. In the light of these observations, it seems plausible that CspC and CspE act as regulatory elements for the expression of the stress proteins in the complex stress response network of the cell.
ACKNOWLEDGMENTS
We appreciate the critical suggestions given by Kunitoshi Yamanaka. We thank Weonhye Bae for providing the ΔcspC and ΔcspC ΔcspE strains.
This work was supported by a grant from the National Institutes of Health (GM 19043).
REFERENCES
- 1.Almiron M, Link A J, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 2.Altuvia S, Almiron M, Huisman G, Kolter R, Storz G. The dps promoter is activated by OxyR during growth and by IHF and ςs in stationary phase. Mol Microbiol. 1994;13:265–272. doi: 10.1111/j.1365-2958.1994.tb00421.x. [DOI] [PubMed] [Google Scholar]
- 3.Bae W, Phadtare S, Severinov K, Inouye M. Characterization of Escherichia coli cspE, whose product negatively regulates transcription of cspA, the gene for the major cold shock protein. Mol Microbiol. 1999;31:1429–1441. doi: 10.1046/j.1365-2958.1999.01284.x. [DOI] [PubMed] [Google Scholar]
- 4.Bae W, Xia B, Inouye M, Serenov K. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc Natl Acad Sci USA. 2000;97:7784–7789. doi: 10.1073/pnas.97.14.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouvet P, Matsumoto K, Wolffe A P. Sequence-specific RNA recognition by the Xenopus Y-box proteins. J Biol Chem. 1995;270:28297–28303. doi: 10.1074/jbc.270.47.28297. [DOI] [PubMed] [Google Scholar]
- 6.Culham D E, Lasby B, Marangoni A G, Milner J L, Steer B A, van Nues R W, Wood J M. Isolation and sequencing of Escherichia coli gene proP reveals unusual structural features of the osmoregulatory proline/betaine transporter, ProP. J Mol Biol. 1993;229:268–276. doi: 10.1006/jmbi.1993.1030. [DOI] [PubMed] [Google Scholar]
- 7.Etchegaray J P, Jones P G, Inouye M. Differential thermoregulation of two highly homologous cold-shock genes, cspA and cspB, of Escherichia coli. Genes Cells. 1996;1:171–178. doi: 10.1046/j.1365-2443.1996.d01-231.x. [DOI] [PubMed] [Google Scholar]
- 8.Farewell A, Diez A A, DiRusso C C, Nystrom T. Role of the Escherichia coli FadR regulator in stasis survival and growth phase-dependent expression of the uspA, fad, and fab genes. J Bacteriol. 1996;178:6443–6450. doi: 10.1128/jb.178.22.6443-6450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freestone P, Nystrom T, Trinei M, Norris V. The universal stress protein, UspA, of Escherichia coli is phosphorylated in response to stasis. J Mol Biol. 1997;274:318–324. doi: 10.1006/jmbi.1997.1397. [DOI] [PubMed] [Google Scholar]
- 10.Graumann P, Marahiel M A. A case of convergent evolution of nucleic acid binding modules. Bioessays. 1996;18:309–315. doi: 10.1002/bies.950180409. [DOI] [PubMed] [Google Scholar]
- 11.Hanna M M, Liu K. Nascent RNA in transcription complexes interact with CspE, a small protein in E. coli implicated in chromatin condensation. J Mol Biol. 1998;282:227–239. doi: 10.1006/jmbi.1998.2005. [DOI] [PubMed] [Google Scholar]
- 12.Harlocker S L, Rampersaud A, Yang W-P, Inouye M. Phenotypic revertant mutations of a new OmpR2 mutant (V203Q) of Escherichia coli lie in the envZ gene, which encodes the OmpR kinase. J Bacteriol. 1993;175:1956–1960. doi: 10.1128/jb.175.7.1956-1960.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hengge-Aronis R, Lange R, Henneberg N, Fischer D. Osmotic regulation of rpoS-dependent genes in Esherichia coli. J Bacteriol. 1993;175:259–265. doi: 10.1128/jb.175.1.259-265.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: ASM Press; 1996. pp. 1497–1512. [Google Scholar]
- 15.Inouye M. Multipurpose expression cloning vehicles in Escherichia coli. Experimental manipulation of gene expression. New York, N.Y: Academic Press, Inc.; 1983. [Google Scholar]
- 16.Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 17.Lange R, Barth M, Hengge-Aronis R. Complex transcriptional control of ςs-dependent stationary phase-induced and osmotically regulated osmY (csi-5) gene suggests novel roles for Lrp, cyclic AMP (cAMP) receptor protein-cAMP complex, and integration host factor in the stationary-phase response of Escherichia coli. J Bacteriol. 1993;175:7910–7917. doi: 10.1128/jb.175.24.7910-7917.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S J, Xie A, Jiang W, Etchegaray J P, Jones P G, Inouye M. Family of the major cold-shock protein, CspA (CS7.4), of Escherichia coli, whose members show a high sequence similarity with the eukaryotic Y-box binding proteins. Mol Microbiol. 1994;11:833–839. doi: 10.1111/j.1365-2958.1994.tb00361.x. [DOI] [PubMed] [Google Scholar]
- 19.Lomovskaya O L, Kidwell J P, Matin A. Characterization of the ς38-dependent expression of a core Escherichia coli starvation gene, pexB. J Bacteriol. 1994;176:3928–3935. doi: 10.1128/jb.176.13.3928-3935.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol. 1997;179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 22.Nystrom T, Neidhardt F C. Cloning, mapping and nucleotide sequencing of a gene encoding a universal stress protein in Escherichia coli. Mol Microbiol. 1992;6:3187–3198. doi: 10.1111/j.1365-2958.1992.tb01774.x. [DOI] [PubMed] [Google Scholar]
- 23.Nystrom T, Neidhardt F C. Isolation and properties of a mutant of Escherichia coli with an insertional inactivation of the uspA gene, which encodes a universal stress protein. J Bacteriol. 1993;175:3949–3956. doi: 10.1128/jb.175.13.3949-3956.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nystrom T, Neidhardt F C. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol Microbiol. 1994;11:537–544. doi: 10.1111/j.1365-2958.1994.tb00334.x. [DOI] [PubMed] [Google Scholar]
- 25.O'Connell K L, Stults J T. Identification of mouse liver proteins on two-dimensional electrophoresis gels by matrix-assisted laser desorption/ionization mass spectrometry of in situ enzymatic digests. Electrophoresis. 1997;18:349–359. doi: 10.1002/elps.1150180309. [DOI] [PubMed] [Google Scholar]
- 26.Phadtare S, Alsina J, Inouye M. Cold-shock response and cold-shock proteins. Curr Opin Microbiol. 1999;2:175–180. doi: 10.1016/S1369-5274(99)80031-9. [DOI] [PubMed] [Google Scholar]
- 27.Phadtare S, Inouye M. Sequence selective interactions with RNA by CspB, CspC and CspE, members of the CspA family of Escherichia coli. Mol Microbiol. 1999;33:1004–1014. doi: 10.1046/j.1365-2958.1999.01541.x. [DOI] [PubMed] [Google Scholar]
- 28.Phadtare S, Yamanaka K, Inouye M. The cold shock response. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: American Society for Microbiology; 2000. pp. 33–45. [Google Scholar]
- 29.Sarmientos P, Sylvester J E, Contente S, Cashel M. Differential stringent control of the tandem E. coli ribosomal RNA promoters from rrnA operon expressed in vivo in multicopy plasmids. Cell. 1983;32:1337–1346. doi: 10.1016/0092-8674(83)90314-8. [DOI] [PubMed] [Google Scholar]
- 30.Sommerville J, Ladomery M. Masking of mRNA by Y-box proteins. FASEB J. 1996;10:435–443. doi: 10.1096/fasebj.10.4.8647342. [DOI] [PubMed] [Google Scholar]
- 31.Takayanagi Y, Tanaka K, Takahashi H. Structure of the 5′ upstream region and the regulation of the rpoS gene of Escherichia coli. Mol Gen Genet. 1994;243:525–531. doi: 10.1007/BF00284200. [DOI] [PubMed] [Google Scholar]
- 32.Tao H, Bausch C, Richmond C, Blattner F R, Conway T. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Triggs-Raine B L, Doble B W, Mulvey M R, Sorby P A, Loewen P C. Nucleotide sequence of katG, encoding catalase HPI of Escherichia coli. J Bacteriol. 1988;170:4415–4419. doi: 10.1128/jb.170.9.4415-4419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.VanBogelen R A, Neidhardt F C. Ribosomes as sensors of heat and cold shock in Esherichia coli. Proc Natl Acad Sci USA. 1990;87:5589–5593. doi: 10.1073/pnas.87.15.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.VanBogelen R A, Abshire K Z, Pertsemldis A, Clark R L, Neidhardt F C. Gene-protein database of Escherichia coli K-12, edition 6. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: ASM Press; 1996. pp. 2067–2202. [Google Scholar]
- 36.Wang N, Yamanaka K, Inouye M. CspI, the ninth member of the CspA family of Escherichia coli, is induced upon cold shock. J Bacteriol. 1999;181:1603–1609. doi: 10.1128/jb.181.5.1603-1609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamanaka K, Fang L, Inouye M. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol Microbiol. 1998;27:247–255. doi: 10.1046/j.1365-2958.1998.00683.x. [DOI] [PubMed] [Google Scholar]
- 38.Yamanaka K, Inouye M. Growth-phase-dependent expression of cspD, encoding a member of the CspA family of Escherichia coli. J Bacteriol. 1997;179:5126–5130. doi: 10.1128/jb.179.16.5126-5130.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamanaka K, Mitani T, Ogura T, Niki H, Hiraga S. Cloning, sequencing and characterization of multicopy supressors of a mukB mutation in Escherichia coli. Mol Microbiol. 1994;13:301–312. doi: 10.1111/j.1365-2958.1994.tb00424.x. [DOI] [PubMed] [Google Scholar]
- 40.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 41.Yim H H, Villarejo M. osmY, a new hyperosmotically inducible gene, encodes a periplasmic protein in Escherichia coli. J Bacteriol. 1992;174:3637–3644. doi: 10.1128/jb.174.11.3637-3644.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yim H H, Brems R L, Villarejo M. Molecular characterization of the promoter of osmY, an rpoS-dependent gene. J Bacteriol. 1994;176:100–107. doi: 10.1128/jb.176.1.100-107.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]