Abstract
Using reporter gene (lacZ) transcriptional fusions, we examined the transcriptional dependencies of the bgl promoter (Pbgl) and the entire operon regulatory region (Pbgl-bglG) on eight transcription factors as well as the inducer, salicin, and an IS5 insertion upstream of Pbgl. Crp-cAMP is the primary activator of both Pbgl and the bgl operon, while H-NS is a strong dominant operon repressor but only a weak repressor of Pbgl. H-NS may exert its repressive effect by looping the DNA at two binding sites. StpA is a relatively weak repressor in the absence of H-NS, while Fis also has a weak repressive effect. Salicin has no effect on Pbgl activity but causes a 30-fold induction of bgl operon expression. Induction depends on the activity of the BglF transporter/kinase. IS5 insertion has only a moderate effect on Pbgl but causes a much greater activation of the bgl operon expression by preventing the full repressive effects of H-NS and StpA. While several other transcription factors (BglJ, RcsB, and LeuO) have been reported to influence bgl operon transcription when overexpressed, they had little or no effect when present at wild type levels. These results indicate the important transcriptional regulatory mechanisms operative on the bgl operon in E. coli.
Keywords: bgl operon, β-glucosides, H-NS, StpA, Crp, Fis, DNA loop, insertion sequences (IS)
1. Introduction
The E. coli bgl operon encodes BglG, BglF, and BglB that are involved in operon regulation and the utilization of aromatic β-glucosides, salicin, arbutin, and esculin, as well as non-aromatic β-glucosides such as cellobiose, as carbon sources [1,2]. The first gene bglG, formerly named bglC [3], flanked by two Rho-independent terminators, encodes an antiterminator protein whose function is to prevent the formation of terminator structures, stabilizing the 5′ end of the bgl mRNA and enabling operon transcription [4,5]. The second gene, bglF, codes a phosphoenol pyruvate-dependent phosphotransferase system (PTS)-dependent enzyme/transporter essential for β-glucoside uptake and phosphorylation, but it also plays a regulatory role, controlling the antitermination process [3,6,7,8]. The third gene in the operon, bglB, codes for a phospho-β-glucosidase that is responsible for hydrolyzing phosphorylated aromatic β-glucosides, such as salicin-P, arbutin-P, and esculin-P, allowing it to release glucose-6-P and the aglycone [2,9].
Although the bgl operon is found to be expressed in E. coli infecting mouse livers [10], the operon is transcriptionally silent (cryptic) and uninducible by β-glucosides in wild-type E. coli strains under standard lab conditions due to the presence of two terminators flanking the bglG gene [11] and strong repression by H-NS, the Histone-like Nucleoid-Structuring protein [12,13,14]. A variety of mutations [12,14,15,16,17,18,19,20] can occur in wild type cells after prolonged incubation with β-glucosides, thereby suppressing the silencing state, a process activating transcription. The activated bgl operon gives rise to a Bgl+ phenotype (which is able to use β-glucosides as the sole carbon source for growth). The most common types of Bgl+ mutations are due to insertions of IS (insertion sequence) elements such as IS1 and IS5 upstream of the bgl promoter region [2,21], a region carrying a SIDD (Superhelical stress-Induced DNA Duplex Destabilization) structure [22]. IS insertional mutations are found to be enhanced by the presence of the operon substrates salicin or arbutin and positively regulated by BglG [23]. Once the bgl operon is activated by a mutation, it becomes inducible. In the absence of β-glucosides, the Crp-dependent promoter initiates transcription, but it subsequently terminates at one of those two terminators flanking bglG. In the presence of operon inducers, transcription partially bypasses both terminators and yields mature operon transcripts.
BglG and BglF comprise a sensory system that dictates the termination and antitermination processes of bgl operon transcription [24]. The BglF sensor phosphorylates BglG in the absence of β-glucosides, thus inactivating it (operon silencing), while BglF dephosphorylates BglG in the presence of β-glucosides, thus activating it (operon expression). After dephosphorylation by BglF, BglG is phosphorylated by HPr (or FPr) and subsequently forms homodimers that bind to a site in the bgl mRNA that partially overlaps the Rho-independent terminators, impeding the formation of terminator hairpin structures and enabling transcriptional readthrough [4,5].
In addition to two intra-operon regulators, BglG and BglF, a number of transcription factors are thought to play roles in regulating bgl operon expression. Crp, the cyclic AMP (cAMP) receptor protein, is a global regulator in E. coli that binds to cAMP to regulate (usually to activate) the expression of genes involved in carbon utilization [25]. Crp-cAMP regulates more than 180 genes by responding to the changing amounts of intracellular cAMP [26]. As a major activator of the bgl operon, the Crp-cAMP complex binds to an upstream site near the promoter to activate it [14,27]. A point substitution within the Crp operator, yielding a more favorable binding site, results in a Bgl+ phenotype [15]. BglJ, a LysR-type transcriptional regulator carrying a C-terminal helix-turn-helix motif [17,20], when overexpressed, causes the activation of the H-NS-repressed bgl operon [17,28,29]. In wild-type E. coli cells, bglJ expression is negligible due to the strong repression by H-NS [30]. RcsB, a LuxR-type transcriptional regulator, is a response regulator involved in the regulation of colonic acid capsule synthesis, cell division, motility, and biofilm formation [31,32]. RcsB and BglJ, harboring similar DNA binding domains at their C-termini, can form heterodimers, binding to an upstream promoter region and relieving H-NS-mediated repression of the bgl operon [29,33]. LeuO is a global transcription factor that not only regulates the leucine biosynthesis operon of E. coli but is also involved in the regulation of stress responses [34,35]. Its overexpression interferes with the silencing of bgl by H-NS and thereby activates bgl operon transcription, although it is not required for bgl activation upon IS insertional mutation or the absence of H-NS [16]. Alternatively, LeuO may indirectly regulate the bgl promoter by increasing bglJ expression [30]. Similar to bglJ, leuO expression is subject to repression by H-NS [30].
In addition to those positive regulators described above, the bgl operon is subject to the repression of several negative regulators. As the main silencer of the bgl operon, the histone-like heat-stable nucleoid structural protein (H-NS) is a major nucleoid protein that is involved in chromosomal stability and transcriptional regulation [36]. H-NS preferentially binds to A/T rich and curved DNA [37,38]. When bound to the DNA, H-NS can self-oligomerize [39] and create a nucleoprotein complex that often represses transcription, either by blocking RNA polymerase binding or by trapping the polymerase [40,41,42]. Therefore, H-NS is often an important negative regulator of transcription, and, as expected, it decreases the transcriptional readthrough of the bgl operon. H-NS represses the bgl operon by binding to the upstream promoter region and a site within the bglG gene [13] in an apparently synergistic fashion [43]. However, thus far, a DNA looping mechanism mediated by H-NS when bound to its two binding sites within the bgl operon has not been demonstrated, although the repression of transcriptional initiation by such DNA looping has been reported in several other E. coli promoters [40,44,45,46].
H-NS is also involved in regulating physiological adaptation to the environment [47] and it can play a role in regulating transposon-mediated directed mutation, determining how mutations can occur at higher frequencies when beneficial to the organism under stressful environmental conditions ([23]; Lam et al., manuscript in preparation).
StpA, an H-NS paralog, can form heterodimers with H-NS [48,49] and plays a role in gene regulation and silencing [50]. This small nucleoid protein can function as a DNA-binding adaptor that is necessary for repression by a C-terminally truncated H-NS or an H-NS carrying the I119T mutation (both defective in DNA binding) [28,51]. One report suggests that StpA alone does not repress the bgl promoter [28].
Fis is a small, abundant nucleoid-associated protein primarily expressed in the exponential phase [52]. Similar to other DNA structural proteins, Fis is capable of binding to and bending the DNA and acts as a global regulator that participates in essential cell processes such as rRNA and tRNA gene transcription [53,54]. In vitro assays showed that Fis is a repressor of the bgl promoter, contributing to promoter silencing and antagonizing Crp for promoter activation [55]. Scant research has demonstrated the characterization of the in vivo interaction of these two proteins in bgl regulation. Lastly, the stress/stationary phase-response sigma factor RpoS can be involved in bgl operon repression [56,57]. RpoS-dependent repression requires the presence of Crl [58], an RNA polymerase holoenzyme assembly factor [59]. In an rpoS mutant background, increased levels of BglG conferred a growth advantage to Bgl+ cells during the stationary phase [60,61].
In this paper, we examine the transcriptional activities of the bgl promoter (Pbgl) alone (with no terminators) and the entire operon regulatory region (Pbgl-bglG) (the promoter plus the first gene flanked by two terminators) by comprehensively exploring the effects of each proposed regulator introduced above, either singly or in combination. The possible H-NS-mediated DNA looping and the effects of StpA and in vivo Crp/Fis antagonism on bgl operon regulation were examined as well. Deletion mutants were constructed for each of these genes, and they were analyzed by measuring their effects on Pbgl and Pbgl-bglG using a single copy lacZ reporter gene located at the lac locus (with an intact native bgl operon simultaneously present), with and without salicin as the inducer of bgl operon expression. Our results show that Crp is essential for both the promoter’s and the operon’s activities. As expected, H-NS is the major silencer of the operon, but it is a weaker repressor of the promoter. H-NS may exert its inhibitory effect by binding to the promoter and the bglG gene and looping the DNA. IS insertions dramatically increase bgl operon transcription in the presence of β-glucosides, but they only slightly enhance promoter activity. StpA moderately represses the bgl operon in the absence of H-NS, while Fis exerts a more recessive effect in the absence of Crp. β-glucosides have no effect on the promoter but significantly induce activated bgl operon transcription. At the wild type levels, BglJ, RcsB, the BglJ/RcsB combination, and LeuO have negligible effects, although their overexpression leads to a Bgl+ phenotype [16,17,29]. The results provide further insight into the importance and possible mechanism that each gene product plays in regulating the expression of the bgl operon.
2. Results
2.1. Crp Strongly Activates the bgl Promoter While H-NS Weakly Represses It
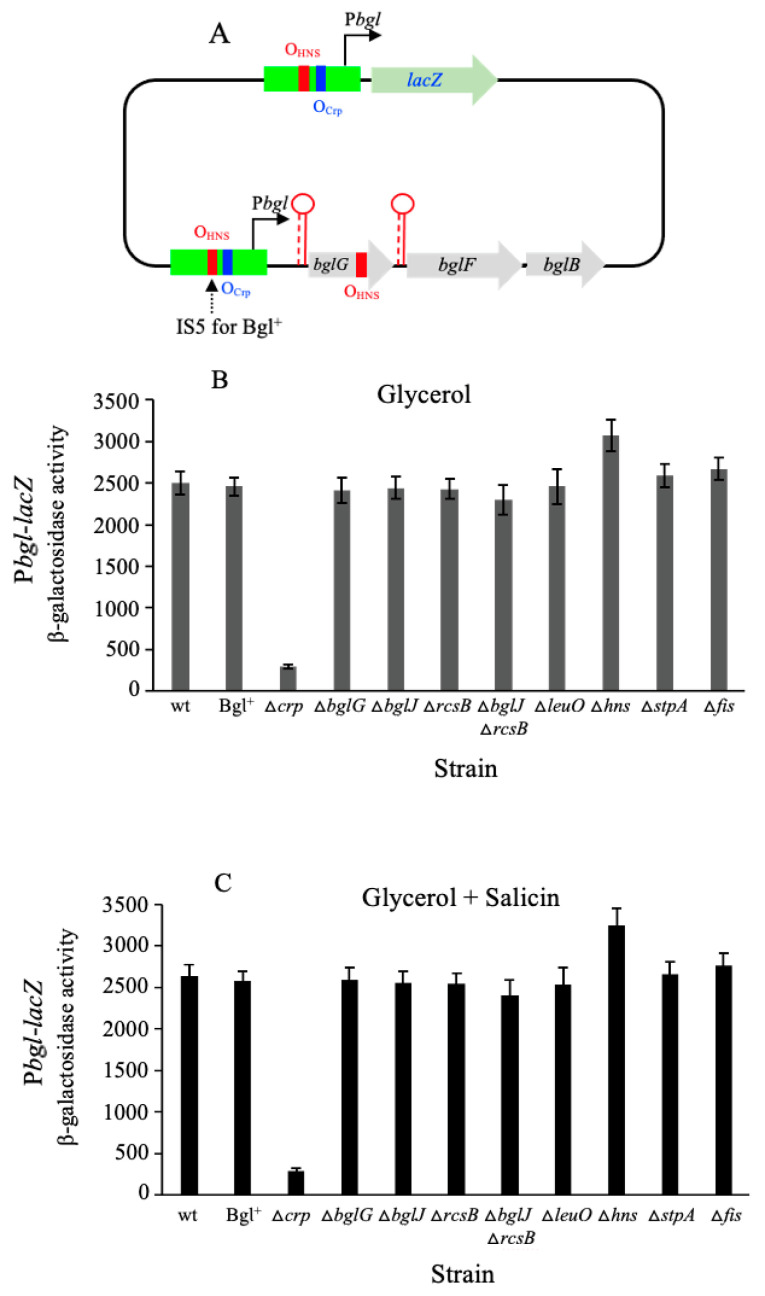
We began our studies on bgl operon expression by examining the effects of the genetic deletion of eight transcription factors (previously reported to exert influences on the transcription of this operon) on the bglGFB promoter (Pbgl) activity. Using a Pbgl-lacZ transcriptional reporter integrated within the E. coli lac locus while the native bgl operon remained intact (Figure 1A), we measured the promoter activities in the presence and absence of each of these eight transcriptional factors. The cells were first cultured with glycerol as the carbon source; only two transcription factors exerted appreciable effects on Pbgl activity: Crp, an activator, and H-NS, a repressor (Figure 1B). While the dependency on Crp was great (a 10-fold reduction in the absence of Crp; see Figure 1B columns 1 and 3 from the left), the dependency on H-NS was small (only a 20% increase in the absence of H-NS; see Figure 1B, columns 1 and 9). This result presumably reflects the fact that the H-NS binding site in the bglG gene is not present as this gene was replaced by the lacZ reporter gene (see Section 3). All other transcription factors had negligible effects on the promoter activity in their presence versus their absence. This was also true when glycerol plus salicin served as the carbon sources during bacterial growth (compare Figure 1B with Figure 1C). This result showed that salicin, a potent inducer of bgl operon expression, had essentially no effect on bgl promoter activity under the conditions used in this study (see the Section 4).
Figure 1.
The bgl promoter (Pbgl) activities in the wild type and various isogenic genetic backgrounds. Cells were grown in M63 minimal media with shaking at 37 °C. At least four samples were collected at OD600 values of 0.2 to 1.0 during the exponential growth phase. Bacterial samples were subject to β-galactosidase assays as described in Section 4, and the enzyme activities were calculated using the equation [(OD420−1.75 × OD550)/(sample volume in mL × time in min)] × 1000. For a given test strain, the slope of OD600 values versus β-galactosidase activities was referred to as the promoter activity. (A) Diagram showing the lacZ transcriptional reporter for the bgl promoter (Pbgl-lacZ). Pbgl (−205 to + 54 relative to the transcriptional start site) with no terminators was fused to the upstream region of the lacZ’s RBS (that is, to TTTCACACAGGAAACAGCT) at the lac locus, replacing lacI and the lacI/lacZ intergenic region. The native bgl operon remained intact. However, for strain Bgl+, there is an IS5 element oriented in the inverse direction and inserted at −207.5 upstream of the bglG translation start site. For both the promoter reporter and the native bgl operon, the blue bars represent the Crp binding sites (OCrp) while the red bars represent the proposed H-NS binding sites (OHNS). (B) Pbgl activities in cells grown with glycerol as the primary carbon source. (C) Pbgl activities in cells grown with glycerol and salicin as carbon sources.
2.2. H-NS Is the Strong Dominant Repressor of the bgl Operon
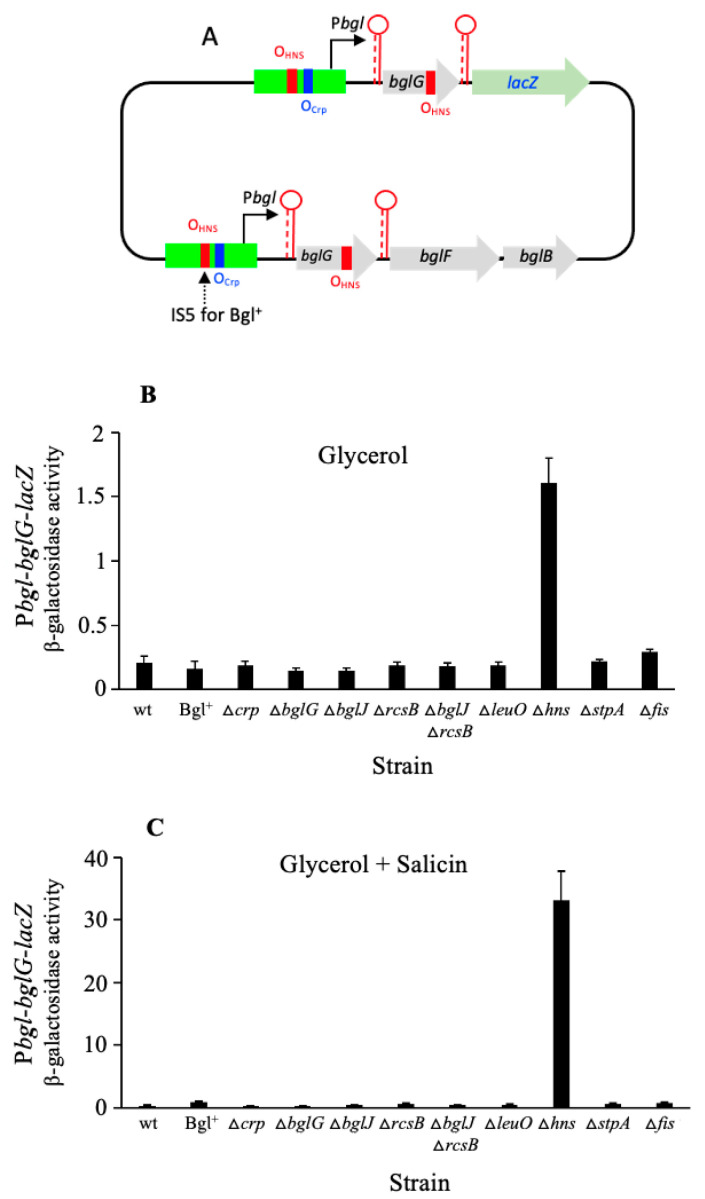
The above experiments were repeated using a bgl operon reporter (Pbgl-bglG-lacZ) integrated at the lac locus, in which the regulatory region of the bgl operon, including Pbgl plus the bglG gene, flanked by the two terminators, preceded the fused lacZ gene (Figure 2A). The results obtained using this construct are recorded in Figure 2B,C. In the absence of salicin, bgl operon expression was minimal due to the presence of both terminators that efficiently block transcriptional readthrough (Figure 2B). However, deletion of the hns gene still enhanced the expression of the operon about eightfold (see columns 1 and 9 of Figure 2B), indicating that with no H-NS binding, those two terminators flanking bglG are not sufficient for the complete abolition of bgl operon expression. In the presence of salicin, the deletion of hns dramatically enhanced bgl gene expression, up to 100-fold (compare columns 1 and 9 in Figure 2C). These results indicate that H-NS is the major repressor of the bgl operon, regardless of the presence of salicin.
Figure 2.
The bgl operon activities in the wild type and its various genetic backgrounds. Culture preparation, sample collection, and β-galactosidase assays were carried out as in Figure 1 (see Section 4). (A) Diagram showing the lacZ transcriptional reporter for the entire regulatory region of the bgl operon (Pbgl-bglG-lacZ). The region carrying Pbgl and bglG, including both terminators (−205 to + 1127 relative to the bglG transcriptional start site), was fused upstream of the lacZ’s RBS (that is, TTTCACACAGGAAACAGCT) at the lac locus. Strain Bgl+ carries an IS5 element oriented in the inverse direction and inserted at −207.5 upstream of the bglG translation start site. For all other strains, the native bgl operon remains unchanged. For both the operon reporter and the native bgl operon, the blue bars represent the Crp binding sites (OCrp) while the red bars represent the proposed H-NS binding sites (OHNS). (B) bgl operon activities in cells grown in M63 with glycerol as the primary carbon source. (C) bgl operon activities in cells grown with glycerol and salicin as carbon sources.
All other transcription factors examined had only a small effect or were essentially without an effect (Figure 2B,C). For Bgl+ cells, the native bglGFB operon should have a significantly increased expression by allowing for the entry of salicin into the cell cytoplasm. However, the operon reporter, Pbgl-bglG-lacZ, at the lac locus, remained silent in these Bgl+ cells grown with salicin since there is no IS insertion present in the reporter construct (Figure 2A and column 2 of Figure 2C). The results obtained clearly suggest that salicin enhances the expression of the operon, although it had no effect on the promoter strength or H-NS repression. Moreover, salicin did not promote regulation by any of the other transcription factors examined.
We also determined whether these transcriptional regulators significantly affect bgl operon expression in the absence of H-NS. These activators and repressors were individually deleted in a ∆hns background. As usual, all the double mutants were cultured with glycerol and salicin, and their operon activities were measured using the operon reporter, Pbgl-bglG-lacZ. As shown in Supplementary Figure S1, the deletion of crp almost abolished bgl operon expression (column 2), confirming that Crp is the primary positive regulator when the operon is activated by removing H-NS. However, the absence of all other transcription factors still had only small effects (≤10% changes) on operon expression. These results together with those described in Figure 2 indicate that except for Crp and H-NS, all other transcription factors, when expressed at their wild-type levels, play minor roles in regulating the bgl operon, both in the presence and in the absence of H-NS.
Our studies were extended by examining the consequences of the increased expression of bglG by using additional strong promoters (Supplementary Figure S2), comparing the transcriptional rates when the wild type bgl promoter was used, versus the stronger lacIq promoter, or the very strong tet promoter. As it can be seen, the overexpression of bglG only increased the operon expression two to threefold, in either the absence (left panel) or the presence (right panel) of salicin. This is consistent with our previous observation that such low levels of residual operon expression due to bglG overexpression remains Bgl− [23].
The above-described measurements were conducted using cells from the exponential growth phase. We next examined bgl operon expression during the stationary phase with or without RpoS, the stationary phase sigma factor. Supplementary Figure S3 (left panel) shows that using the wild-type cells, bgl operon expression remains silent from the middle to the late stationary growth phase. This is also true for the cells deleted for rpoS (right panel of Supplementary Figure S3), although it recognizes RpoD (sigma 70) promoters [62,63]. The loss of RpoS had a negligible impact on the bgl operon-silencing state (when comparing the left panel and the right panel). This is not consistent with the previously reported literature [56,57].
2.3. StpA Represses the bgl Operon Only in the Absence of H-NS
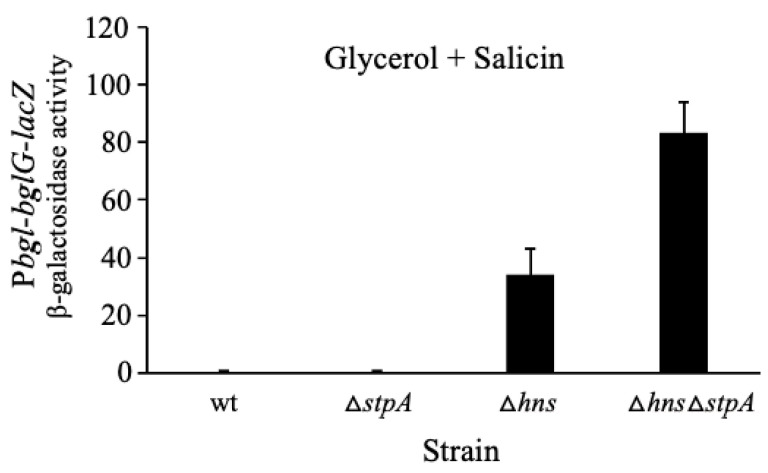
We have shown that the deletion of the stpA gene did not exert an effect on bgl operon expression when the hns gene was intact (Figure 2; also see the first two columns of Figure 3). However, in the absence of H-NS, the deletion of the stpA gene (that is, strain ∆hns∆stpA) more than doubled the expression of the operon (see columns 3 and 4 of Figure 3) in the cells cultured in the presence of salicin. It is possible that H-NS and StpA bind to the same site(s) and exert their repressive effects by similar or overlapping mechanisms. The effects of StpA on bgl operon expression documented here had not been examined in previous publications.
Figure 3.
Inhibitory effect of StpA on bgl operon expression in the absence of H-NS. Using the operon reporter, Pbgl-bglG-lacZ (at the lac locus), the bgl operon transcriptional activities were assayed, comparing the stpA single mutant (∆stpA), the hns single mutant (∆hns), and the hns/stpA double mutant (∆hns∆stpA). Cells were cultured in M63 with glycerol and salicin. Sample preparation and β-galactosidase assays were carried out as in Figure 1.
2.4. IS Insertions Promote Both the bgl Promoter and Operon Expression
Figure 4A,B show a new Pbgl reporter and a new operon reporter, respectively. They are similar to the two previously described reporters used for Figure 1, Figure 2 and Figure 3, except that an IS5 insertion was present upstream of these reporter constructs (see Section 4). For the strain Bgl+, the same IS5 insertion as that for the new reporter construct was present upstream of the native bgl operon, while for all the other test strains there was no change to the native operon.
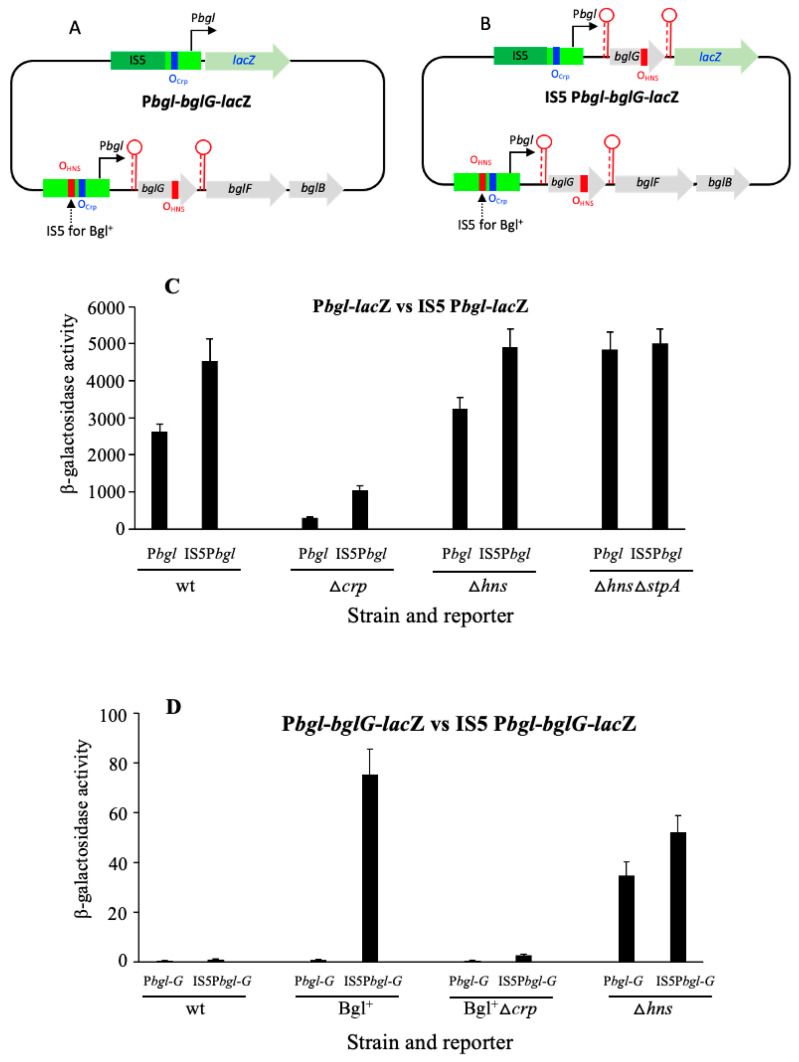
Figure 4.
IS5 insertion stimulates both promoter and operon activities. (A) Diagram showing IS5 insertion at the Pbgl reporter (IS5Pbgl-lacZ). An IS5 element in the reverse direction is located at -207.5 upstream of the bglG translation start site. The native bgl operon is unchanged. (B) Diagram showing IS5 insertion at the bgl operon reporter (IS5Pbgl-bglG-lacZ). IS5 orientation and location are the same as in Figure 4A. In both (A) and (B), the blue bars represent the Crp binding sites (OCrp) while the red bars represent the proposed H-NS binding sites (OHNS). (C) Effects of IS5 insertion on Pbgl activities. (D) Effects of IS5 insertion on bgl operon activities. In both (C) and (D), test strains were cultured in M63 with glycerol and salicin as carbon sources. Sample collections and β-galactosidase assays were carried out as in Figure 1.
In Figure 4C, it is evident that the presence of IS5 enhanced the reporter gene expression <2-fold in the wild type background (see the first two columns). The absence of crp decreased gene expression as expected, but LacZ activity was enhanced to a greater degree (about 4x) due to the IS5 insertion (see columns 3 and 4 from the left). These results suggest that with no Crp, H-NS binding is more repressive to Pbgl since both proteins bind upstream of Pbgl, and their binding sites may partially overlap. On the other hand, in the absence of H-NS, the IS5 insertion only moderately increased operon expression (see columns 5 and 6). However, when both hns and stpA were simultaneously deleted, there was almost no effect of IS5 on the reporter gene expression. These results suggest that with no H-NS, StpA exerts some inhibitory effects on Pbgl.
Figure 4D shows a similar series of experiments except that the bgl operon was examined instead of just the promoter. In the wild type cells, the expression of lacZ was hardly detected, regardless of the presence or absence of the inserted IS5. This is not surprising since the native bgl operon remains silent and the anti-terminator protein, BglG, remains inactivated, even in the presence of salicin, the operon inducer. However, in the Bgl+ cells, the IS5 insertion dramatically elevated β-galactosidase activity >100 fold (see columns 3 and 4 of Figure 4D), confirming that the IS element activated the operon and rendered it highly inducible. This activity was abolished by deletion of the crp gene (columns 5 and 6), indicating that Crp is still essential for operon expression in the presence of salicin, even when the operon is activated by IS insertion. In other words, there are three requirements for full bgl operon expression: IS insertion (activating the operon), Crp (activating Pbgl), and the operon inducer (activating BglG). Furthermore, the loss of H-NS facilitated expression with a moderate increase upon the rate of insertion of IS5 in front of the operon reporter gene construct (the last two columns of Figure 4D). This increase in activity is presumably due to StpA, which appears to exert a mild repressive effect when the hns gene is not present.
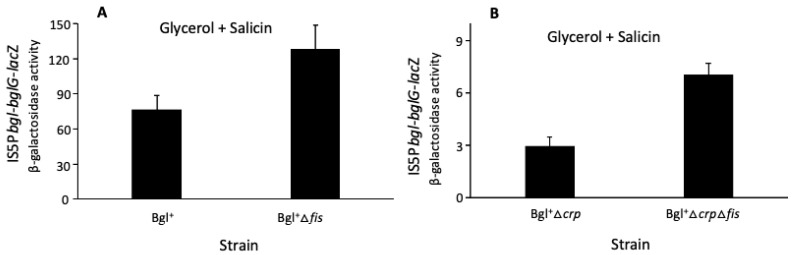
2.5. Fis Represses the Activated bgl Operon, Possibly by Interacting with Crp
Figure 5 reveals a previously unrecognized phenomenon, namely, that Fis has a moderate repressive effect on expression of the bgl operon. Thus, in Figure 5A, in a Bgl+ background, when IS5 activates the native bgl operon, the deletion of fis gives rise to an approximately 50% increase in LacZ activity, indicating that when the bgl operon is activated by IS insertion, Fis is inhibitory to the operon. In the same Bgl+ strain, in which the crp gene was deleted, there is over a 2-fold increase in bgl operon expression when fis is deleted (Figure 5B), even though the total activity has decreased 20-fold compared to when Crp was present (compare Figure 5A,B). These results show that Fis exerts a repressive effect on the bgl operon, and the repression is even stronger when Crp is absent. Thus, while H-NS is the major repressor of the bgl operon, Fis and StpA are minor repressors of this operon. The nature of the degree of increase observed when Crp is absent compared with when it is present (again compare Figure 5A,B) suggests (tentatively) that Crp and Fis may exert partially, but not fully, antagonist effects.
Figure 5.
Fis represses bgl operon expression when it is activated by IS insertion. (A) Fis repression of bgl operon expression after activation by IS insertion. (B) Antagonism between Fis and Crp in regulating the bgl operon. In both (A) and (B), IS5 insertion is present both in the native operon and the operon lacZ reporter. Cells were cultured in M63 with glycerol and salicin as carbon sources. Sample collection and β-galactosidase assays were carried out as in Figure 1.
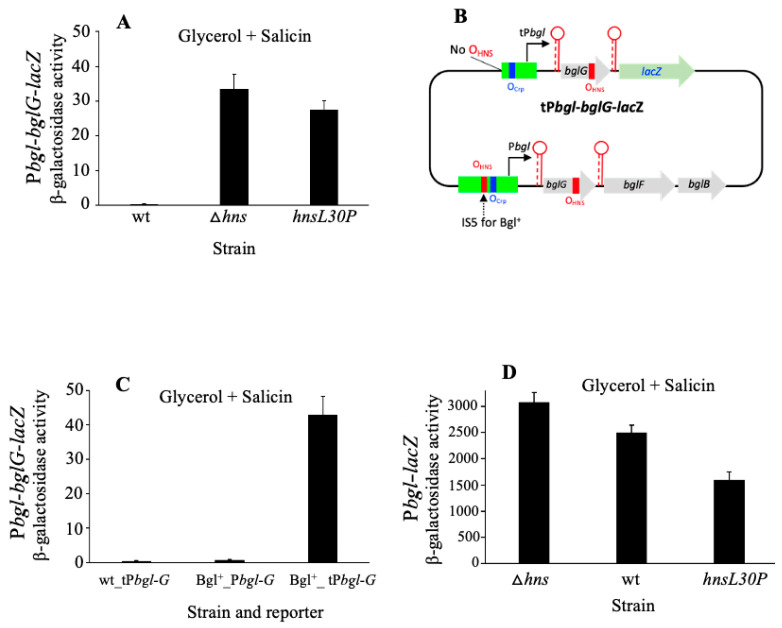
2.6. DNA Looping Mediated by H-NS May Be Essential for Full bgl Operon Silencing
The results presented in Figure 6A lead to preliminary mechanistic suggestions regarding the mode of action of H-NS as a primary repressor of bgl operon transcription. In a wild type genetic background (with wild type H-NS), there is essentially no detectable operon expression. The deletion of the hns gene results in an increase in operon expression by more than 200-fold. When a mutant of H-NS is used with a single amino acid substitution at position 30 [i.e., changing a leucine to a proline (L30P)], thereby losing its self-oligomerization property and its DNA-looping activity [64,65], 15% of the repressive activity, observed in the absence of H-NS, is retained. The slightly lower operon expression seen for hnsL30P than for ∆hns is most likely due to binding of the mutant H-NS to both Pbgl and bglG. These results suggest that oligomerization of H-NS is important for H-NS-mediated repression. We speculate that this is because DNA looping between the two binding sites is required for the strong repressive activity of H-NS (see Section 3).
Figure 6.
Possible requirement of DNA looping for bgl operon silencing. (A) Effect of H-NSL30P on bgl operon expression. H-NSL30P is an H-NS derivative that carries a proline residue instead of leucine at residue 30 in the protein. This derivative is thought to maintain its DNA-binding capability but is deficient in oligomerization [64,65], thereby failing to bridge two or more DNA loci together. Using the operon reporter, Pbgl-bglG-lacZ, the effect of this mutant H-NS on bgl operon expression was determined by comparing the wild type H-NS and the absence of H-NS. (B) Diagram of a truncated bgl operon reporter (tPbgl-bglG-lacZ). It is the same as the regular operon reporter Pbgl-bglG-lacZ except that the regulatory region upstream of the Crp operator (believed to carry an H-NS binding site) in Pbgl has been removed. The blue bars represent the Crp binding sites (OCrp) while the red bars represent the proposed H-NS binding sites (OHNS). (C) The bgl operon activity using a reporter lacking the proposed H-NS binding site in the upstream regulatory region. (D) Effect of H-NSL30P on Pbgl. In (A,C,D), test strains were cultured in M63 with glycerol and salicin as carbon sources at 37 °C. Sample collection and β-galactosidase assays were carried out as in Figure 1.
Figure 6B shows the diagram of a truncated bgl operon reporter (tPbgl-bglG-lacZ) that is essentially the same as the regular operon reporter, except that the region upstream of the Crp binding site has been removed. Conceivably, H-NS would not be able to bind to tPbgl. Figure 6C shows that wild type E. coli cells, with a wild type H-NS protein, blocks the transcription of the native bgl operon, and no activity was detected for the truncated operon reporter due to the presence of both terminators (column 1). Similarly, in Bgl+ cells, IS5 insertion activated the native bgl operon, but the operon reporter remained almost silent because IS5 was not present in this construct (column 2). However, using the equivalent Bgl+ cells carrying the truncated operon reporter (tPbgl-bglG-lacZ), a 60-fold increase in operon expression was seen (compare columns 2 and 3 of Figure 6C), suggesting that H-NS almost lost its repressive effect on the operon without binding, or with deficient binding, to the Pbgl region. Thus, we believe that the binding of H-NS to BOTH of its binding sites, in the promoter region and the bglG gene of the reporter construct, are required for effective repression. These observations suggest that H-NS exerts its repressive effect by looping the DNA between its two binding sites (see Section 3).
To determine if the H-NS derivative, H-NSL30P, still maintained its DNA-binding capacity (despite the loss of its oligomerization property), we measured the promoter activities in cells expressing either hns or hnsL30P. As seen from Figure 6D, the deletion of the hns gene yields the highest transcriptional activity (column 1), the restoration of H-NS function gives rise to about a 20% loss of LacZ activity (column 2), and the use of the H-NSL30P mutant protein results in the retention of the repressive activity, although it is decreased compared to the situation in which the wild type H-NS was present. These results demonstrated that both H-NS and H-NSL30P are capable of binding to the bgl promoter DNA, thereby repressing its activity. The decrease in transcriptional activity noted for the strain hnsL30P when comparing the activities reported in column 2 compared with column 3 (Figure 6D) can likely be attributed to the increased binding affinity to the H-NS binding site in Pbgl by H-NSL30P. Taken together, the significant loss of operon repression in the hnsL30P mutant (Figure 6A) is not due to a decrease in DNA-binding activity (the mutant H-NS seems to have a greater affinity for the Pbgl region). Instead, it is probably attributed to the failure to form a DNA loop. It seems likely that the DNA binding of H-NS gives rise to appreciable repression, but full repression is dependent on the DNA looping between the two binding sites, in the upstream promoter region and the downstream site in the bglG gene.
3. Discussion
As noted in the introductory section, the expression of the bgl operon has been subject to investigation by many different groups of researchers, suggesting that its regulation involves several operon-specific and global DNA-binding proteins in E. coli and other enteric bacteria. These studies have led to predictions as to many potential DNA-binding proteins that influence the rates of bgl operon transcription. However, several of these studies have reported the effects of gene overexpression on bgl operon activity, and consequently, some of these studies may not have physiological relevance in wild type E. coli cells. Thus, we have constructed reporter gene (lacZ) transcriptional fusions to (1) the promoter of the β-glucoside utilization operon, Pbgl, to identify the factors that influence promoter strength, and (2) the entire regulatory region of the bgl operon, Pbgl-bglG, including the two transcriptional terminator/anti-terminator structures flanking the bglG gene, in order to ascertain which factor(s) play roles in operon regulation, independently of the promoter.
The reporter gene constructs described above have been used to examine the effects of eight transcription factors (Crp, BglG, BglJ, RcsB, LeuO, H-NS, StpA, and Fis) by comparing the wild type levels of these factors under identical conditions except that each of the encoding genes had been deleted. Thus, under normal physiological conditions, we were able to gain relevant information about the involvement of the different DNA-binding proteins in the expression of Pbgl and Pbgl-bglG. The effects of the inducer, salicin, and of transposon IS5 insertion upstream of the promoter were also determined, and in several instances, we have combined the occurrence in a single strain of more than one of these factors on bgl transcription. For this purpose, the single bgl-lacZ constructs were expressed at one location on the chromosome while a native bgl operon was expressed at a distinct chromosomal location, both with and without the IS5 insertion, and with and without the bgl operon inducer, salicin.
The results of our studies can be summarized with the following primary conclusions. (1) The cAMP-Crp complex is the primary activator of both Pbgl and bgl operon expression. (2) H-NS is a strong dominant repressor of the operon, although it is only a weak repressor of Pbgl, irrespective of whether an inducer (salicin) is absent or present during growth. (3) The preliminary evidence suggests that H-NS exerts its repressive effect by binding to two sites and looping the DNA between these two sites. (4) StpA is a weak repressor of the bgl operon, but only in the absence of H-NS, suggesting that it exerts its effect independently of H-NS, contrary to a previous report [28]. (5) Fis also has a weak repressive effect on the bgl operon, but more so in the absence of Crp than in its presence, suggesting that there could be competition for DNA binding by these two proteins. (6) Salicin has no effect on Pbgl activity but causes a 30-fold induction of bgl operon expression, probably by counteracting transcriptional termination at the two terminators flanking the bglG gene. (7) While Pbgl is a strong promoter, strong transcriptional repression of the bgl operon occurs even under inducing conditions. (8) The inductive effect of salicin depends on the activity of the phosphoenolpyruvate-dependent BglF transporter/kinase. (9) The upstream IS5 insertion only has a moderate effect on Pbgl, but it causes a much greater activation of bgl operon expression by preventing the repressive effects of H-NS and StpA. (10) While several other transcription factors have been reported to influence bgl operon transcription when overexpressed, they have little or no effect when present at wild type levels. These results indicate the important transcriptional regulatory mechanisms operative on the E. coli bgl operon while confirming or refuting several previously published suggestions and conclusions.
In this paper, we present preliminary evidence that the mechanism of H-NS repression of bgl transcription involves the binding of this protein to two sites in the bgl operon, one upstream of the promoter, and one within the bglG gene. However, binding to these two sites can be followed by H-NS oligomerization, possibly with the formation of a DNA loop. While DNA binding to either one or the other of its two binding sites alone can give rise to mild repression, it seems that binding alone is insufficient to cause the strong bgl operon repression that is caused by H-NS. Instead, our preliminary results, presented herein, suggest that the associative properties of wild type H-NS, lacking in the mutant form of the protein (H-NSL30P), are required for the strong repression that is responsible for silencing the expression of the bgl operon in the absence of an IS insertional event. This possibility will be the subject of a future publication, which is a work currently in progress (Lam et al., manuscript in preparation). Thus, it seems that the “on/off switch” that results from IS insertion/excision in the promoter upstream region of the bgl operon is largely due to the repressive effect of H-NS, and possibly, to a lesser extent, or under different conditions, due to StpA and/or Fis.
Using a bgl-activated strain, we showed that the deletion of fis caused a moderate increase in operon transcription, revealing Fis’s role as a weak repressor of the activated operon. Two Fis binding sites have been identified to overlap with the Crp binding site in Pbgl, and in vitro assays showed that these two proteins (one repressor and one activator) compete with each other to bind to the same DNA region within Pbgl [55]. This antagonistic relationship between Crp and Fis is supported by our in vivo assays, which demonstrated that when crp is deleted, Fis exhibits a greater repressive effect due to stronger binding. On the other hand, our data show that Fis has almost no effect on the bgl promoter. This is probably because Pbgl is already a strong promoter, and Crp successfully outcompetes Fis to activate it. In addition, Fis and Crp exert negligible effects on the non-activated bgl operon in wild type cells. This is probably because with H-NS-mediated DNA looping, these DNA-binding proteins are incapable of accessing their binding sites on Pbgl, which is embedded within the loop.
Another surprising observation concerns the bgl operon repression by StpA in the absence of H-NS. StpA can form heterodimers with a C-terminally truncated H-NS (still able to dimerize/ oligomerize but unable to bind DNA) to bind to the same DNA as for H-NS homodimers [51,66], suggesting that StpA homodimers alone may bind to the same DNA (that is, the bgl promoter and the bglG gene) especially when it is produced at high levels. Wolf et al. show that StpA has no appreciable inhibitory effects on upstream (that is, Pbgl) or downstream (that is, bglG) silencing in the wild type or the hns deletion mutant [28]. In the wild type strain, the StpA level is minimal due to the strong repression by H-HS and the self-autorepression [67,68]. Therefore, it is not surprising that the deletion of stpA has little effect on Pbgl and the operon expression (this study and [28]) since H-NS alone is already sufficient to silence the operon. To show that StpA does not repress the downstream site within bglG, Wolf et al. [28] used a reporter in which a strong constitutive promoter stimulates bglG and lacZ (only carrying the downstream regulatory element). When driven by a strong promoter, the downstream bglG repression by H-NS has been reported to be lost [43] as RNA polymerase can transcribe through the site bound with H-NS. A similar mechanism may explain why StpA does not exert an inhibitory effect at the bglG site, that is, strong transcription may help RNA polymerase pass through the site bound by StpA. In this study, we used our operon reporter, the native Pbgl driving bglG and lacZ, which carries both upstream- and downstream-regulatory elements. The repression by H-NS via these two elements has been reported to be synergistic [43]. In the absence of H-NS, it is conceivable that high levels of StpA proteins can form enough homodimers [69], resulting in an increased (synergistic) repression to the bgl operon transcription, probably by binding to Pbgl and bglG. Further experiments will be needed to show the direct binding of StpA homodimers to these sites within the bgl operon.
It is also interesting to note that even in the presence of β-glucosides in the medium during the stationary growth phase, RpoS does not appear to have an appreciable effect on bgl operon transcription. RpoS is required for the silencing of the bgl operon mediated by an H-NS mutant lacking the DNA-binding domain [57]. It has been further shown to directly repress bgl operon expression [56]. However, as a sigma factor, the activity of RpoS is positively affected by Crl [70], an RNA polymerase assembly factor [59]. In the absence of Crl, RpoS does not contribute to the silencing of the bgl operon. However, the crl gene is deleted from some commonly used lab E. coli strains including our parental strain, BW25113, which explains why RpoS did not repress bgl operon expression in our study.
Further studies will be required to establish the detailed repressive mechanism of this unusual operon, as well as that responsible for the very interesting process by which IS elements activate it. This class of mutations grants wild type E. coli cells the capacity to switch the expression of the operon using insertion sequence (IS) elements as triggers, thereby enabling the protection of the cell from toxic β-glucosides while benefiting from the presence of nutritious β-glucosides [71]. It seems likely that this is another example in which small bacterial transposons have been used to allow directed mutation to occur only under appropriate environmental stress conditions, as discussed previously [23,72,73,74,75,76,77,78,79,80].
4. Materials and Methods
4.1. E. coli Strains and Growth Media
Except for DH5α pir, used for cloning purposes, and some CGSC-deletion mutants from the E. coli Stock Center, all other strains used in this study were derived from K12 strain BW25113 [81], and they are described in Supplementary Table S1. Bacterial strains were routinely cultured in LB media at 30 °C or 37 °C. For β-galactosidase assays, test strains were grown in M63 minimal media with either 0.5% (w/v) glycerol, 0.5% (w/v) salicin, or both at 0.5% as carbon sources [82]. M63 salt solution contains 2 g (NH4)2SO4, 13.6 g KH2PO4, and 0.5 mg FeSO4·7H2O; the solution was then brought to pH 7.5 using KOH. It was supplemented with 10−4 % thiamine, 0.05% casamino acids, and 1.7 mM MgSO4. This minimal medium was used to prepare precultures and cultures prior to β-galactosidase assay. When necessary, ampicillin, kanamycin, and tetracycline were added to the media at 100 µg/mL, 25 µg/mL, and 12 µg/mL, respectively.
4.2. Construction of Deletion Mutants
CGSC strains JW3701-2, JW5955-1, JW2205-2, JW0075-2, JW1225, JW2644-3, and JW3229-1 (E. coli Genetic Stock Center, Yale Univ.) carry the deletion mutations of bglG, bglJ, rcsB, leuO, hns, stpA, and fis, respectively. For each of these mutants, a kanamycin resistance (kmr) gene was substituted for the target gene. These mutations were individually transferred to strain BW25113 (wild type; [81]) by P1 transduction, and the kmr gene was subsequently flipped out by pCP20 [81], yielding the deletion mutant strains ∆bglG, ∆bglJ, ∆rcsB, ∆leuO, ∆hns, ∆stpA and ∆fis, respectively (Supplementary Table S1). The bglJ mutation was transferred into strain ∆rcsB, yielding the ∆bglJ∆rcsB double mutant. The stpA mutation was transferred into strain ∆hns, yielding the ∆hns∆stpA double mutant.
4.3. Construction of the bgl Promoter Transcriptional Reporter Pbgl-lacZ
To create the bglGFB promoter-lacZ transcriptional fusion used to measure the promoter activities, the promoter region (−205 to + 54 relative to the transcriptional start site, + 1) without the first terminator upstream of bglG, was amplified using oligos Pbgl-Xho-F and Pbgl-Bam-R (Supplementary Table S2), digested with XhoI and BamHI, and then cloned into the same XhoI/BamHI sites of the integration vector, pKDT [83], yielding pKDT-Pbgl. The region carrying the kmr, rrnBT and Pbgl (kmr:rrnBT:Pbgl) was PCR-amplified using oligos bgl-Z-P1 and Pbgl-Z-P2 (Supplementary Table S2) and then integrated into the chromosomal default strain EQ42 [83] to replace the lacI gene and the lacZ promoter. The resultant strain carried the kmr:rrnBT:Pbgl cassette followed by lacZ’s ribosomal binding site (RBS) and the lacZ structural gene within the lac locus. After being confirmed by PCR and sequencing, the promoter reporter, Pbgl driving lacZ expression (that is, Pbgl-lacZ) was transferred into BW25113 and various genetic backgrounds by P1 transduction. This yielded the bgl promoter reporter strains BW_Pbgl-Z, ∆bglG_Pbgl-Z, ∆bglJ_Pbgl-Z, ∆rcsB_Pbgl-Z, ∆leuO_Pbgl-Z, ∆hns_Pbgl-Z, ∆stpA_Pbgl-Z, ∆fis_Pbgl-Z, ∆bglJ∆rcsB_Pbgl-Z, and ∆hns∆stpA_Pbgl-Z, respectively. The Pbgl-Z reporter was transferred into a crp deletion mutant [73], yielding the strain ∆crp_Pbgl-Z. In addition, the same reporter was transferred into one previously isolated Bgl+ mutant (carrying a reverse-oriented IS5 element at -207.5, located upstream of the bglG translation start site), yielding the strain Bgl+_Pbgl-Z (Supplementary Table S1). To determine how an IS5 insertion affects Pbgl activities, the regulatory region carrying both IS5 (the same IS5 as for Bgl+) and Pbgl was PCR-amplified using the same oligos as for Pbgl amplification from the genomic DNA of Bgl+ cells. The resultant product, IS5Pbgl, was cloned into pKDT, yielding pKDT-IS5Pbgl. The IS5Pbgl cassette was chromosomally integrated within the lac locus as described above for the Pbgl cassette. This promoter reporter was transferred into BW25113, ∆crp, ∆hns, and ∆hns∆stpA, yielding the strains BW_IS5Pbgl-Z, ∆crp_IS5Pbgl-Z, ∆hns_IS5Pbgl-Z, and ∆hns∆stpA_IS5Pbgl-Z, respectively.
4.4. Construction of the bgl Operon Transcriptional Reporter Strains
Recently, we reported the construction of a bgl operon transcriptional reporter Pbgl-bglG-lacZ (referred to as G-Z) [23]. Located within the lac locus, this operon reporter construct carries the bglGFB promoter and the first gene, bglG, including the 2nd terminator downstream of the bglG translational stop codon (the 205th nucleotide to the 1127th nucleotide relative to the transcriptional start site) followed by a stop codon, lacZ’s RBS, and the lacZ structural gene. In addition to three operon reporter strains (BW_Z, Ptet-G_Z, and Iq-G_Z), this operon reporter was transferred by P1 transduction to other genetic backgrounds. This yielded Bgl+_G-Z, ∆crp_G-Z, ∆bglG_G-Z, ∆bglJ_G-Z, ∆rcsB_G-Z, ∆bglJ∆rcsB_G-Z, ∆leuO_G-Z, ∆hns_G-Z, ∆stpA_G-Z, ∆fis_G-Z, ∆hns∆stpA_G-Z. To examine the effect of the IS insertion on expression of the bgl operon, a new bgl operon transcriptional reporter, IS5Pbgl-bglG-lacZ (referred as IS5G-Z), was constructed, in which a reverse-oriented IS5 element was inserted upstream of the original operon reporter Pbgl-bglG-lacZ (the same IS5 as for Bgl+). To achieve this, the regulatory region containing IS5 and Pbgl-bglG was amplified from the Bgl+ cells (containing the reverse-oriented IS5 at the same location as for the operon reporter) using the same oligos as for Pbgl-bglG. The resultant IS5Pbgl-bglG product was cloned into pKDT, yielding pKDT_IS5Pbgl-bglG. The IS5Pbgl-bglG cassette was chromosomally integrated within the lac locus as recently reported in [23]. This operon reporter was transferred to BW25113, Bgl+, ∆crp, ∆hns, Bgl+∆fis, Bgl+∆crp, and Bgl+∆crp∆fis, yielding operon reporter strains BW25113_IS5G-Z, Bgl+_IS5G-Z, ∆crp_IS5G-Z, ∆hns_IS5G-Z, Bgl+∆fis_IS5G-Z, Bgl+∆crp_IS5G-Z and Bgl+∆crp∆fis_IS5G-Z, respectively. To make a truncated operon transcriptional reporter (tPbgl-bglG-lacZ or referred to as tG-Z), a smaller DNA region (−93 to + 1127 relative to the transcriptional start site, + 1), supposedly not carrying the H-NS binding site on Pbgl, was cloned into pKDT, yielding pKDT-tPbgl-bglG. This DNA fragment, “kmr:rrnBT:tPbgl-bglG”, was chromosomally integrated within the same lac locus as for Pbgl-bglG-lacZ [23], yielding tPbgl-bglG, which drives lacZ expression. This truncated operon reporter was transferred into BW25113 and Bgl+, yielding the strains BW25113_tG-Z and Bgl+_tG-Z, respectively.
4.5. Construction of the hnsL30P Strain Using Positive/Negative Selection
H-NS is the primary silencer of the bgl operon. The H-NS protein usually exists in oligomeric forms, and these contribute to its biological activity [84], promoting the formation of structures such as DNA loops and bridges [40,85]. The N-terminal domain is responsible for H-NS oligomerization. The leucine residue at position 30 is essential for H-NS:H-NS binding [84]. To test the possible looping mechanism by which H-NS silences the bgl operon, we used a two-step recombineering protocol based on TetA-SacB positive-selection and counter-selection [86] to change the leucine codon CTG (88 to 90 relative to the first hns codon ATG) to a proline codon CCT in the hns gene. “TG” in the leucine codon was first replaced by the tetA-sacB cassette that was amplified from the chromosomal DNA of strain T-SACK [86] using chimeric oligos hns-AB-F and hns-AB-R (Supplementary Table S2). These long oligos carry the appropriate homologous arms flanking the “TG” nucleotides in the hns gene. The replacement of “TG” by tetA-sacB in some tetracycline (Tc)-resistant mutants was confirmed by colony PCR and sequencing. A 100-bp single strand DNA fragment, which covers the region of the hns gene (38 to 138 relative to the ATG) with CT replacing “TG” in the middle, was synthesized. This fragment was amplified using oligos hns-F and hns-R (Supplementary Table S2), and PCR products were electroporated into the cells of a Tc resistant mutant expressing Lambda-Red proteins encoded by pKD46 [81]. After one-hour incubation, the electroporated cells were applied onto TetA/SacB counter-selection agar (plus 6% sucrose and 24 mg fusaric acid per liter). After incubation at 42 °C for about two days, 10 colonies resistant to sucrose and fusaric acid were purified on LB agar plates and tested for both sensitivity to Tc and resistance to sucrose. Several Tc-sensitive/sucrose-resistant colonies were confirmed for the replacement of the tetA-sacB cassette with “CT” by PCR and sequencing. The resultant altered strain was named hnsL30P, in which the 30th codon was changed from leucine to proline.
4.6. β-Galactosidase Assays
E. coli reporter strains were cultured in 4 mL of LB contained in glass test tubes (1.5 cm in diameter × 15 cm in length) with shaking at 37 °C for 8 h. An amount of 30 µL of LB cultures were used to inoculate 3 mL of M63 minimal media in smaller glass tubes (1.2 cm × 12 cm), and the tubes were shaken at 37 °C overnight. The carbon sources were 0.5% glycerol, 0.5% salicin, or both. To improve the growth of the hns-deletion mutant and its derivatives, casamino acids (CAA) were added to all minimal M63 media to 0.05%. The overnight M63 cultures (precultures) were inoculated into 5 mL of the same media in larger test tubes (1.8 cm × 15 cm) with an initial OD600 of 0.03. The tubes were rotated at 250 rpm and 37 °C, and cell densities (OD600) were measured with a Bio-Rad spectrophotometer. During the exponential growth phase, four samples were collected in the range of OD600 from 0.1 to 1. The samples (roughly 0.3 mL for promoter reporter strains, and 0.6 mL for operon reporter strains) were immediately frozen at −20 °C prior to β-galactosidase assays. To test RpoS effects, samples were collected in the range of OD600 from 1 to 4 when the cultures entered the early and late stationary phases.
To measure β-galactosidase activities in bgl promoter reporter strains, 0.8 mL of Z-buffer containing β-mercaptoethanol (2.7 μL/mL) and sodium dodecyl sulfate (SDS) (0.005%) was mixed with 0.2 mL of sample and 25 μL of CHCl3 in test tubes. Alternatively, for bgl operon reporter strains, 0.5 mL of Z-buffer was mixed with 0.5 mL of the sample. The tubes were vortexed twice (each time for 10 s at a constant speed) and incubated in a 37 °C water bath until equilibration. A 0.2 mL aliquot of O-nitrophenyl galactoside (ONPG) substrate (4 mg/mL) was then added to each test tube. When a yellow color developed, the reaction was stopped by adding 0.5 mL of 1 M Na2CO3 followed by vortexing. Reaction mixtures were centrifuged (15,000 rpm, 3 min), and the absorbance values of the supernatants were measured at 420 nm and 550 nm. A control tube was run in parallel using M63 salts instead of the test sample. β-galactosidase activity (Miller units) = [(OD420−1.75 × OD550)/(sample volume in mL × time in min)] × 1000 [87]. For a given test strain, the slope of OD600 values versus β-galactosidase activities was referred to as the promoter activity or the operon activity.
Acknowledgments
We thank Peter Kopkowski for expert assistance in the preparation of this manuscript.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms231810343/s1.
Author Contributions
Conceptualization, Z.Z. and M.H.S.J.; methodology, Z.Z. and D.T.; investigation, D.T. and K.J.K.L.; data curation, D.T. and Z.Z; writing—original draft preparation, M.H.S.J. and Z.Z.; writing—review and editing, M.H.S.J. and Z.Z.; supervision, M.H.S.J. and Z.Z.; funding acquisition, M.H.S.J. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Funding Statement
This research was funded by NIH grant GM077402 to Milton H. Saier.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Prasad I., Schaefler S. Regulation of the beta-glucoside system in Escherchia coli K-12. J. Bacteriol. 1974;120:638–650. doi: 10.1128/jb.120.2.638-650.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schnetz K., Toloczyki C., Rak B. Beta-glucoside (bgl) operon of Escherichia coli K-12: Nucleotide sequence, genetic organization, and possible evolutionary relationship to regulatory components of two Bacillus subtilis genes. J. Bacteriol. 1987;169:2579–2590. doi: 10.1128/jb.169.6.2579-2590.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahadevan S., Reynolds A.E., Wright A. Positive and negative regulation of the bgl operon in Escherichia coli. J. Bacteriol. 1987;169:2570–2578. doi: 10.1128/jb.169.6.2570-2578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gulati A., Mahadevan S. The Escherichia coli antiterminator protein BglG stabilizes the 5'region of the bgl mRNA. J. Biosci. 2001;26:193–203. doi: 10.1007/BF02703643. [DOI] [PubMed] [Google Scholar]
- 5.Houman F., Diaz-Torres M.R., Wright A. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell. 1990;62:1153–1163. doi: 10.1016/0092-8674(90)90392-R. [DOI] [PubMed] [Google Scholar]
- 6.Amster-Choder O., Houman F., Wright A. Protein phosphorylation regulates transcription of the beta-glucoside utilization operon in E. coli. Cell. 1989;58:847–855. doi: 10.1016/0092-8674(89)90937-9. [DOI] [PubMed] [Google Scholar]
- 7.Schnetz K., Rak B. Regulation of the bgl operon of Escherichia coli by transcriptional antitermination. EMBO J. 1988;7:3271–3277. doi: 10.1002/j.1460-2075.1988.tb03194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aymerich S., Steinmetz M. Specificity determinants and structural features in the RNA target of the bacterial antiterminator proteins of the BglG/SacY family. Proc. Natl. Acad. Sci. USA. 1992;89:10410–10414. doi: 10.1073/pnas.89.21.10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prasad I., Young B., Schaefler S. Genetic determination of the constitutive biosynthesis of phospho-glucosidase A in Escherichia coli K-12. J. Bacteriol. 1973;114:909–915. doi: 10.1128/jb.114.3.909-915.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan M.A., Isaacson R.E. In vivo expression of the beta-glucoside (bgl) operon of Escherichia coli occurs in mouse liver. J. Bacteriol. 1998;180:4746–4749. doi: 10.1128/JB.180.17.4746-4749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnetz K. Silencing of Escherichia coli bgl promoter by flanking sequence elements. EMBO J. 1995;14:2545–2550. doi: 10.1002/j.1460-2075.1995.tb07252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Defez R., De Felice M. Cryptic operon for beta-glucoside metabolism in Escherichia coli K12: Genetic evidence for a regulatory protein. Genetics. 1981;97:11–25. doi: 10.1093/genetics/97.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dole S., Nagarajavel V., Schnetz K. The histone-like nucleoid structuring protein H-NS represses the Escherichia coli bgl operon downstream of the promoter. Mol. Microbiol. 2004;52:589–600. doi: 10.1111/j.1365-2958.2004.04001.x. [DOI] [PubMed] [Google Scholar]
- 14.Mukerji M., Mahadevan S. Characterization of the negative elements involved in silencing the bgl operon of Escherichia coli: Possible roles for DNA gyrase, H-NS, and CRP-cAMP in regulation. Mol. Microbiol. 1997;24:617–627. doi: 10.1046/j.1365-2958.1997.3621725.x. [DOI] [PubMed] [Google Scholar]
- 15.Reynolds A.E., Mahadevan S., LeGrice S.F., Wright A. Enhancement of bacterial gene expression by insertion elements or by mutation in a CAP-cAMP binding site. J. Mol. Biol. 1986;191:85–95. doi: 10.1016/0022-2836(86)90424-9. [DOI] [PubMed] [Google Scholar]
- 16.Ueguchi C., Ohta T., Seto C., Suzuki T., Mizuno T. The leuO gene product has a latent ability to relieve bgl silencing in Escherichia coli. J. Bacteriol. 1998;180:190–193. doi: 10.1128/JB.180.1.190-193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giel M., Desnoyer M., Lopilato J. A mutation in a new gene, bglJ, activates the bgl operon in Escherichia coli K-12. Genetics. 1996;143:627–635. doi: 10.1093/genetics/143.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiNardo S., Voelkel K.A., Sternglanz R., Reynolds A.E., Wright A. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell. 1982;31:43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- 19.Dole S., Klingen Y., Nagarajavel V., Schnetz K. The protease Lon and the RNA-binding protein Hfq reduce silencing of the Escherichia coli bgl operon by H-NS. J. Bacteriol. 2004;186:2708–2716. doi: 10.1128/JB.186.9.2708-2716.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madhusudan S., Paukner A., Klingen Y., Schnetz K. Independent regulation of H-NS-mediated silencing of the bgl operon at two levels: Upstream by BglJ and LeuO and downstream by DnaKJ. Microbiology. 2005;151:3349–3359. doi: 10.1099/mic.0.28080-0. [DOI] [PubMed] [Google Scholar]
- 21.Reynolds A.E., Felton J., Wright A. Insertion of DNA activates the cryptic bgl operon in E. coli K12. Nature. 1981;293:625–629. doi: 10.1038/293625a0. [DOI] [PubMed] [Google Scholar]
- 22.Humayun M.Z., Zhang Z., Butcher A.M., Moshayedi A., Saier M.H., Jr. Hopping into a hot seat: Role of DNA structural features on IS5-mediated gene activation and inactivation under stress. PLoS ONE. 2017;12:e0180156. doi: 10.1371/journal.pone.0180156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z., Zhou K., Tran D., Saier M. Insertion Sequence (IS) Element-Mediated Activating Mutations of the Cryptic Aromatic beta-Glucoside Utilization (BglGFB) Operon Are Promoted by the Anti-Terminator Protein (BglG) in Escherichia coli. Int. J. Mol. Sci. 2022;23:1505. doi: 10.3390/ijms23031505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amster-Choder O. The bgl sensory system: A transmembrane signaling pathway controlling transcriptional antitermination. Curr. Opin. Microbiol. 2005;8:127–134. doi: 10.1016/j.mib.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 25.Frendorf P.O., Lauritsen I., Sekowska A., Danchin A., Norholm M.H.H. Mutations in the Global Transcription Factor CRP/CAP: Insights from Experimental Evolution and Deep Sequencing. Comput. Struct. Biotechnol. J. 2019;17:730–736. doi: 10.1016/j.csbj.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai M.J., Wang J.R., Yang C.D., Kao K.C., Huang W.L., Huang H.Y., Tseng C.P., Huang H.D., Ho S.Y. PredCRP: Predicting and analysing the regulatory roles of CRP from its binding sites in Escherichia coli. Sci. Rep. 2018;8:951. doi: 10.1038/s41598-017-18648-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gulati A., Mahadevan S. Mechanism of catabolite repression in the bgl operon of Escherichia coli: Involvement of the anti-terminator BglG, CRP-cAMP and EIIAGlc in mediating glucose effect downstream of transcription initiation. Genes Cells. 2000;5:239–250. doi: 10.1046/j.1365-2443.2000.00322.x. [DOI] [PubMed] [Google Scholar]
- 28.Wolf T., Janzen W., Blum C., Schnetz K. Differential dependence of StpA on H-NS in autoregulation of stpA and in regulation of bgl. J. Bacteriol. 2006;188:6728–6738. doi: 10.1128/JB.00586-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venkatesh G.R., Kembou Koungni F.C., Paukner A., Stratmann T., Blissenbach B., Schnetz K. BglJ-RcsB heterodimers relieve repression of the Escherichia coli bgl operon by H-NS. J. Bacteriol. 2010;192:6456–6464. doi: 10.1128/JB.00807-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stratmann T., Madhusudan S., Schnetz K. Regulation of the yjjQ-bglJ operon, encoding LuxR-type transcription factors, and the divergent yjjP gene by H-NS and LeuO. J. Bacteriol. 2008;190:926–935. doi: 10.1128/JB.01447-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majdalani N., Gottesman S. The Rcs phosphorelay: A complex signal transduction system. Annu. Rev. Microbiol. 2005;59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- 32.Gottesman S., Stout V. Regulation of capsular polysaccharide synthesis in Escherichia coli K12. Mol. Microbiol. 1991;5:1599–1606. doi: 10.1111/j.1365-2958.1991.tb01906.x. [DOI] [PubMed] [Google Scholar]
- 33.Salscheider S.L., Jahn A., Schnetz K. Transcriptional regulation by BglJ-RcsB, a pleiotropic heteromeric activator in Escherichia coli. Nucleic Acids Res. 2014;42:2999–3008. doi: 10.1093/nar/gkt1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breddermann H., Schnetz K. Correlation of Antagonistic Regulation of leuO Transcription with the Cellular Levels of BglJ-RcsB and LeuO in Escherichia coli. Front. Cell. Infect. Microbiol. 2016;6:106. doi: 10.3389/fcimb.2016.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitic D., Radovcic M., Markulin D., Ivancic-Bace I. StpA represses CRISPR-Cas immunity in H-NS deficient Escherichia coli. Biochimie. 2020;174:136–143. doi: 10.1016/j.biochi.2020.04.020. [DOI] [PubMed] [Google Scholar]
- 36.Oshima T., Ishikawa S., Kurokawa K., Aiba H., Ogasawara N. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 2006;13:141–153. doi: 10.1093/dnares/dsl009. [DOI] [PubMed] [Google Scholar]
- 37.Jiao Y., Wang J., Wu P., Zhao L., He C., Zhang J., Duan C. Cerium-based M4L4 tetrahedra as molecular flasks for selective reaction prompting and luminescent reaction tracing. Chemistry. 2014;20:2224–2231. doi: 10.1002/chem.201303560. [DOI] [PubMed] [Google Scholar]
- 38.Dillon S.C., Dorman C.J. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 39.Spurio R., Falconi M., Brandi A., Pon C.L., Gualerzi C.O. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 1997;16:1795–1805. doi: 10.1093/emboj/16.7.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dame R.T., Wyman C., Wurm R., Wagner R., Goosen N. Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J. Biol. Chem. 2002;277:2146–2150. doi: 10.1074/jbc.C100603200. [DOI] [PubMed] [Google Scholar]
- 41.Dorman C.J. H-NS: A universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 42.Dame R.T. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol. Microbiol. 2005;56:858–870. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- 43.Nagarajavel V., Madhusudan S., Dole S., Rahmouni A.R., Schnetz K. Repression by binding of H-NS within the transcription unit. J. Biol. Chem. 2007;282:23622–23630. doi: 10.1074/jbc.M702753200. [DOI] [PubMed] [Google Scholar]
- 44.Shin M., Song M., Rhee J.H., Hong Y., Kim Y.J., Seok Y.J., Ha K.S., Jung S.H., Choy H.E. DNA looping-mediated repression by histone-like protein H-NS: Specific requirement of Esigma70 as a cofactor for looping. Genes Dev. 2005;19:2388–2398. doi: 10.1101/gad.1316305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noom M.C., Navarre W.W., Oshima T., Wuite G.J., Dame R.T. H-NS promotes looped domain formation in the bacterial chromosome. Curr Biol. 2007;17:R913–R914. doi: 10.1016/j.cub.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 46.Lim C.J., Lee S.Y., Kenney L.J., Yan J. Nucleoprotein filament formation is the structural basis for bacterial protein H-NS gene silencing. Sci. Rep. 2012;2:509. doi: 10.1038/srep00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chib S., Mahadevan S. Involvement of the global regulator H-NS in the survival of Escherichia coli in stationary phase. J. Bacteriol. 2012;194:5285–5293. doi: 10.1128/JB.00840-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johansson J., Eriksson S., Sonden B., Wai S.N., Uhlin B.E. Heteromeric interactions among nucleoid-associated bacterial proteins: Localization of StpA-stabilizing regions in H-NS of Escherichia coli. J. Bacteriol. 2001;183:2343–2347. doi: 10.1128/JB.183.7.2343-2347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams R.M., Rimsky S., Buc H. Probing the structure, function, and interactions of the Escherichia coli H-NS and StpA proteins by using dominant negative derivatives. J. Bacteriol. 1996;178:4335–4343. doi: 10.1128/jb.178.15.4335-4343.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johansson J., Dagberg B., Richet E., Uhlin B.E. H-NS and StpA proteins stimulate expression of the maltose regulon in Escherichia coli. J. Bacteriol. 1998;180:6117–6125. doi: 10.1128/JB.180.23.6117-6125.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Free A., Williams R.M., Dorman C.J. The StpA protein functions as a molecular adapter to mediate repression of the bgl operon by truncated H-NS in Escherichia coli. J. Bacteriol. 1998;180:994–997. doi: 10.1128/JB.180.4.994-997.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finkel S.E., Johnson R.C. The Fis protein: It's not just for DNA inversion anymore. Mol. Microbiol. 1992;6:3257–3265. doi: 10.1111/j.1365-2958.1992.tb02193.x. [DOI] [PubMed] [Google Scholar]
- 53.Ball C.A., Osuna R., Ferguson K.C., Johnson R.C. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J. Bacteriol. 1992;174:8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Browning D.F., Grainger D.C., Busby S.J. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr. Opin. Microbiol. 2010;13:773–780. doi: 10.1016/j.mib.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 55.Caramel A., Schnetz K. Antagonistic control of the Escherichia coli bgl promoter by FIS and CAP in vitro. Mol. Microbiol. 2000;36:85–92. doi: 10.1046/j.1365-2958.2000.01827.x. [DOI] [PubMed] [Google Scholar]
- 56.Dole S., Kuhn S., Schnetz K. Post-transcriptional enhancement of Escherichia coli bgl operon silencing by limitation of BglG-mediated antitermination at low transcription rates. Mol. Microbiol. 2002;43:217–226. doi: 10.1046/j.1365-2958.2002.02734.x. [DOI] [PubMed] [Google Scholar]
- 57.Ohta T., Ueguchi C., Mizuno T. rpoS function is essential for bgl silencing caused by C-terminally truncated H-NS in Escherichia coli. J. Bacteriol. 1999;181:6278–6283. doi: 10.1128/JB.181.20.6278-6283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schnetz K. Silencing of the Escherichia coli bgl operon by RpoS requires Crl. Microbiology. 2002;148:2573–2578. doi: 10.1099/00221287-148-8-2573. [DOI] [PubMed] [Google Scholar]
- 59.Banta A.B., Chumanov R.S., Yuan A.H., Lin H., Campbell E.A., Burgess R.R., Gourse R.L. Key features of sigmaS required for specific recognition by Crl, a transcription factor promoting assembly of RNA polymerase holoenzyme. Proc. Natl. Acad. Sci. USA. 2013;110:15955–15960. doi: 10.1073/pnas.1311642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harwani D., Zangoui P., Mahadevan S. The beta-glucoside (bgl) operon of Escherichia coli is involved in the regulation of oppA, encoding an oligopeptide transporter. J. Bacteriol. 2012;194:90–99. doi: 10.1128/JB.05837-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harwani D. Regulation of gene expression: Cryptic beta-glucoside (bgl) operon of Escherichia coli as a paradigm. Braz. J. Microbiol. 2014;45:1139–1144. doi: 10.1590/S1517-83822014000400003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jishage M., Ishihama A. Transcriptional organization and in vivo role of the Escherichia coli rsd gene, encoding the regulator of RNA polymerase sigma D. J. Bacteriol. 1999;181:3768–3776. doi: 10.1128/JB.181.12.3768-3776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jishage M., Ishihama A. A stationary phase protein in Escherichia coli with binding activity to the major sigma subunit of RNA polymerase. Proc. Natl Acad. Sci. USA. 1998;95:4953–4958. doi: 10.1073/pnas.95.9.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang W., Li G.W., Chen C., Xie X.S., Zhuang X. Chromosome organization by a nucleoid-associated protein in live bacteria. Science. 2011;333:1445–1449. doi: 10.1126/science.1204697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ueguchi C., Seto C., Suzuki T., Mizuno T. Clarification of the dimerization domain and its functional significance for the Escherichia coli nucleoid protein H-NS. J. Mol. Biol. 1997;274:145–151. doi: 10.1006/jmbi.1997.1381. [DOI] [PubMed] [Google Scholar]
- 66.Free A., Porter M.E., Deighan P., Dorman C.J. Requirement for the molecular adapter function of StpA at the Escherichia coli bgl promoter depends upon the level of truncated H-NS protein. Mol. Microbiol. 2001;42:903–917. doi: 10.1046/j.1365-2958.2001.02678.x. [DOI] [PubMed] [Google Scholar]
- 67.Sonden B., Uhlin B.E. Coordinated and differential expression of histone-like proteins in Escherichia coli: Regulation and function of the H-NS analog StpA. EMBO J. 1996;15:4970–4980. doi: 10.1002/j.1460-2075.1996.tb00877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang A., Rimsky S., Reaban M.E., Buc H., Belfort M. Escherichia coli protein analogs StpA and H-NS: Regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 1996;15:1340–1349. doi: 10.1002/j.1460-2075.1996.tb00476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sonnenfield J.M., Burns C.M., Higgins C.F., Hinton J.C. The nucleoid-associated protein StpA binds curved DNA, has a greater DNA-binding affinity than H-NS and is present in significant levels in hns mutants. Biochimie. 2001;83:243–249. doi: 10.1016/S0300-9084(01)01232-9. [DOI] [PubMed] [Google Scholar]
- 70.Pratt L.A., Silhavy T.J. Crl stimulates RpoS activity during stationary phase. Mol. Microbiol. 1998;29:1225–1236. doi: 10.1046/j.1365-2958.1998.01007.x. [DOI] [PubMed] [Google Scholar]
- 71.Sonowal R., Nandimath K., Kulkarni S.S., Koushika S.P., Nanjundiah V., Mahadevan S. Hydrolysis of aromatic beta-glucosides by non-pathogenic bacteria confers a chemical weapon against predators. Proc. Biol. Sci. 2013;280:20130721. doi: 10.1098/rspb.2013.0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Z., Saier M.H., Jr. A mechanism of transposon-mediated directed mutation. Mol. Microbiol. 2009;74:29–43. doi: 10.1111/j.1365-2958.2009.06831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Z., Saier M.H., Jr. A novel mechanism of transposon-mediated gene activation. PLoS Genet. 2009;5:e1000689. doi: 10.1371/journal.pgen.1000689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Z., Yen M.R., Saier M.H., Jr. Precise excision of IS5 from the intergenic region between the fucPIK and the fucAO operons and mutational control of fucPIK operon expression in Escherichia coli. J. Bacteriol. 2010;192:2013–2019. doi: 10.1128/JB.01085-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Z., Saier M.H., Jr. Transposon-mediated adaptive and directed mutations and their potential evolutionary benefits. J. Mol. Microbiol. Biotechnol. 2011;21:59–70. doi: 10.1159/000333108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saier M.H., Jr., Zhang Z. Transposon-mediated directed mutation controlled by DNA binding proteins in Escherichia coli. Front. Microbiol. 2014;5:390. doi: 10.3389/fmicb.2014.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saier M.H., Jr., Zhang Z. Control of Transposon-Mediated Directed Mutation by the Escherichia coli Phosphoenolpyruvate:Sugar Phosphotransferase System. J. Mol. Microbiol. Biotechnol. 2015;25:226–233. doi: 10.1159/000375375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Z., Saier M.H., Jr. Transposon-mediated activation of the Escherichia coli glpFK operon is inhibited by specific DNA-binding proteins: Implications for stress-induced transposition events. Mutat. Res. 2016;793-794:22–31. doi: 10.1016/j.mrfmmm.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Z., Kukita C., Humayun M.Z., Saier M.H. Environment-directed activation of the Escherichia coli flhDC operon by transposons. Microbiology. 2017;163:554–569. doi: 10.1099/mic.0.000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saier M.H., Jr., Kukita C., Zhang Z. Transposon-mediated directed mutation in bacteria and eukaryotes. Front. Biosci. 2017;22:1458–1468. doi: 10.2741/4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pardee A.B., Prestidge L.S. On the nature of the repressor of beta-galactosidase synthesis in Escherichia coli. Biochim. Biophys. Acta. 1959;36:545–547. doi: 10.1016/0006-3002(59)90202-1. [DOI] [PubMed] [Google Scholar]
- 83.Klumpp S., Zhang Z., Hwa T. Growth rate-dependent global effects on gene expression in bacteria. Cell. 2009;139:1366–1375. doi: 10.1016/j.cell.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smyth C.P., Lundback T., Renzoni D., Siligardi G., Beavil R., Layton M., Sidebotham J.M., Hinton J.C., Driscoll P.C., Higgins C.F., et al. Oligomerization of the chromatin-structuring protein H-NS. Mol. Microbiol. 2000;36:962–972. doi: 10.1046/j.1365-2958.2000.01917.x. [DOI] [PubMed] [Google Scholar]
- 85.Rimsky S. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr. Opin. Microbiol. 2004;7:109–114. doi: 10.1016/j.mib.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 86.Li X.T., Thomason L.C., Sawitzke J.A., Costantino N., Court D.L. Positive and negative selection using the tetA-sacB cassette: Recombineering and P1 transduction in Escherichia coli. Nucleic Acids Res. 2013;41:e204. doi: 10.1093/nar/gkt1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Miller F. Glycopeptides of human immunoglobulins. 3. The use and preparation of specific glycosidases. Immunochemistry. 1972;9:217–228. doi: 10.1016/0019-2791(72)90087-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.