Abstract
Pseudomonas putida strain TW3 is able to metabolize 4-nitrotoluene via 4-nitrobenzoate (4NBen) and 3, 4-dihydroxybenzoic acid (protocatechuate [PCA]) to central metabolites. We have cloned, sequenced, and characterized a 6-kbp fragment of TW3 DNA which contains five genes, two of which encode the enzymes involved in the catabolism of 4NBen to PCA. In order, they encode a 4NBen reductase (PnbA) which is responsible for catalyzing the direct reduction of 4NBen to 4-hydroxylaminobenzoate with the oxidation of 2 mol of NADH per mol of 4NBen, a reductase-like enzyme (Orf1) which appears to have no function in the pathway, a regulator protein (PnbR) of the LysR family, a 4-hydroxylaminobenzoate lyase (PnbB) which catalyzes the conversion of 4-hydroxylaminobenzoate to PCA and ammonium, and a second lyase-like enzyme (Orf2) which is closely associated with pnbB but appears to have no function in the pathway. The central pnbR gene is transcribed in the opposite direction to the other four genes. These genes complete the characterization of the whole pathway of 4-nitrotoluene catabolism to the ring cleavage substrate PCA in P. putida strain TW3.
Nitroaromatic compounds are widely distributed pollutants which have been present in the environment for a relatively short period of time due to their use in the industrial syntheses of many dyes, pesticides, and explosives; for example 2- and 4-nitrotoluenes and 2, 4- and 2, 6-dinitrotoluenes are precursors in the production of 2, 4, 6-trinitrotoluene. Their presence in the environment has apparently selected microorganisms that are capable of their degradation. Such bacteria use a number of different biochemical strategies for the removal of the nitro group during the conversion to central metabolites. Some pathways proceed via an initial monooxygenase attack on the aromatic ring with subsequent release of the nitro group as nitrite, as in the degradation of 2-nitrophenol (45), 4-nitrophenol (39), and 4-chloro-2-nitrophenol (7). In other examples, the initial attack is by a dioxygenase which results in a hypothetical partially reduced and unstable diol intermediate from which nitrite is subsequently eliminated to form a catechol (1, 2-dihydroxybenzene), as has been reported for 2, 4-dinitrotoluene (40), 2, 6-dinitrotoluene (30), 2-nitrotoluene (17), nitrobenzene (31), 2, 6-dinitrophenol (12), and 3-nitrobenzoic acid (29). Alternatively, the nitro group can be partially reduced and ultimately released as ammonium. The initial reduction is to a hydroxylamino group via a nitroso intermediate. This can then undergo a mutase-mediated rearrangement to ortho-aminophenols (as in the cases of nitrobenzene [31], 3-nitrophenol [37, 38], and 4-chloronitrobenzene [21]) with the later release of ammonia; alternatively, the hydroxylamino compound can be converted directly to the corresponding catechol by a lyase-mediated reaction with direct elimination of ammonia, such as in the degradation of 4-nitrobenzoate (14, 15, 43), 4-nitrotoluene (16, 35), and 3-nitrophenol (27).
During the catabolism of 4-nitrotoluene in Pseudomonas putida strain TW3 (35), the nitro group is retained during the sequential oxidation of the methyl group to form 4-nitrobenzoate (4NBen). The genes encoding the enzymes for the initial steps in the catabolism of 4-nitrotoluene to 4NBen are very similar in sequence and organization to the TOL plasmid-encoded upper pathway genes of toluene catabolism (42), with the addition of a novel NAD(P)+-independent alcohol dehydrogenase (18, 19). 4NBen is then further converted to the ring cleavage substrate protocatechuate (PCA), with the release of the nitro group as ammonia (35). Biochemical evidence for conversion of 4NBen to PCA was first described in the 4-nitrobenzoate-degrading Comamonas acidovorans strain NBA-10 (14, 15): 4-nitrosobenzoate and 4-hydroxylaminobenzoate were shown to be intermediates, and the final reaction was a lyase-catalyzed conversion of 4-hydroxylaminobenzoate to PCA. This appears to be the general pathway for 4NBen catabolism and has subsequently been described in other strains (28, 43, 46).
Preliminary reports have described cloning the 4NBen catabolic genes of Ralstonia pickettii YH105 (43) and Pseudomonas sp. strain YH102 (46; L. M. Newman and G. J. Zylstra, Abstr. 97th Gen. Meet. Am. Soc. Microbiol., abstr. Q341, p. 512, 1997). We describe here the cloning and nucleotide sequencing of the genes and the functional analysis of those enzymes involved in the latter stages of 4-nitrotoluene catabolism, from 4NBen to PCA in P. putida TW3, complementing our earlier reports of its genes for metabolism of 4-nitrotoluene to 4NBen (18, 19).
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used
| Bacterial strain or plasmid | Genotype or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| P. putida | ||
| TW3 | Wild type, 4NT+a Pnb+ | 35 |
| PaW340 | Trp− Strr plasmid-free derivative of P. putida mt-2 (PaW1) | 20 |
| E. coli | ||
| DH5α | φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+elA1 deoR Δ(lacZYA-argF)V169 | Promega |
| DH5αPRO | deoR endA1 gyrA96 hsdR17 (rK− mK+) recA1 relA1 supE44 thi-1 Δ(lacZYA-argFV169) φ80dlacZΔM15 F− λ− PN25/tetR PLacIq/LacI Spr | Clontech |
| MHB101 | F′ thi-1 hsdS20 (rB− mB−) supE44 recA13 ara-14 leuB6 proA2 lacY1 galK2 rpsL20 (Strr) xyl-5 mtl-1 | Promega |
| Plasmids | ||
| pLAFR5 | IncP Tcr; derivative of pLAFR3 containing a double cos cassette | 22 |
| pRK2013 | IncP Tra Rk2+ ΔrepRk2 repEI+ Kmr | 13 |
| pPROLar.A122 | Plac/ara-1 Kmr p15A origin | Clontech |
| pHN150 | 1.9-kbp XbaI fragment in pUC19 containing protocatechuate 4, 5-dioxygenase genes (ligA and ligB) | 33 |
| pTW3.11 | 6.6-kbp EcoRI clone containing orf1, pnbR, pnbB, and orf2 in pUC18 | This study |
| pTW3.12 | 3.6-kbp EcoRI/SacI subclone of pTW3.11 | This study |
| pTW3.13 | 6-kbp SacI/SmaI clone containing pnbA, orf1, pnbR, pnbB, and orf2 in pLAFR5 | This study |
| pTW3.14 | 4.9-kbp SmaI subclone of pTW3.13 | This study |
| pTW3.15 | 3.4-kbp SalI/PstI subclone of pTW3.14 | This study |
| pTW3.15NX | pTW3.15 with frameshift introduced in XhoI site to create knockout of pnbA | This study |
| pTW3.15NH | Deletion of 366-bp HindIII fragment in pTW3.15 to create knockout of orf1 | This study |
| pTW3.16 | Insertion of PCR fragment carrying pnbB into EcoRI site of pUC18 | This study |
| pPROpnbA | KpnI/BamHI fragment containing pnbA in pPROLar.A122 | This study |
4NT, 4-nitrotoluene.
Chemicals and growth media.
Aromatic and aliphatic substrates were obtained from Aldrich Chemical Co. 4-Hydroxylaminobenzoic acid and 4-nitrosobenzoic acid were synthesized chemically (6, 9). P. putida TW3 was grown on minimal salts medium (MM) (5) supplemented with either solid 4-nitrotoluene (0.5 g/lit), sodium 4-nitrobenzoate (5 mM), or sodium succinate (10 mM). Escherichia coli strains were grown on Luria-Bertani (LB) medium (36). Where appropriate, ampicillin was added at 100 μg/ml, kanamycin and spectinomycin were added at 50 μg/ml, and tetracycline was added at 25 μg/ml. p-Toluidine plates for detecting the accumulation of catechols were prepared as described by Parke (34).
DNA manipulations.
Unless otherwise stated, standard methods for DNA manipulation were used (36). Total DNA was prepared from P. putida TW3 by the method of Ausubel et al. (4). Plasmid DNA was prepared from E. coli strains by CONCERT rapid plasmid miniprep systems (GibcoBRL), and cosmid DNA prepared by CONCERT high-purity plasmid midiprep systems (GibcoBRL). DNA fragments were recovered from agarose gels by Qiaquick columns (Qiagen). Southern blot analyses were carried out as described by Sambrook et al. (36). Hybridizations were carried out with enhanced chemiluminescence direct labeling (Amersham) according to the manufacturer's instructions.
Preparation of P. putida TW3 cosmid library.
TW3 genomic DNA was partially digested with Sau3AI and ligated to pLAFR5 arms previously digested with ScaI and BamHI. Ligation and packaging reactions were carried out as described by Sambrook et al. (36).
Triparental matings for transfer of cosmid DNA into PaW340.
Donor, recipient, and E. coli HB101, carrying pRK2013 as helper plasmid, were grown in LB medium until they reached an optical density at 600 nm (OD600) of 0.6. Then 500 μl of each culture was mixed and centrifuged, and the pellets washed in MM. The pellets were finally resuspended in 50 μl of MM and dispensed onto a sterile nylon membrane (Bio-Rad) laid on the surface of an LB plate. Following incubation overnight at 30°C, the cells were washed off the filter into 2 ml of MM, and appropriate dilutions were spread onto selective media. Donor-only and recipient-only controls were treated in the same way.
DNA sequencing and sequence analysis methods.
Nucleotide sequences of both DNA strands were determined by MWG-Biotech Ltd. (Ebersberg, Germany). PCR primers were designed with the aid of the Lasergene software package (DNAStar, Inc., Madison, Wis.). Searches of the GenBank and Swissprot databases were carried out with BLASTN and BLASTX, respectively (1). Multiple sequence alignments were done using ClustalW.
Expression of pnbA and pnbB in E. coli.
The pnbA gene was amplified by PCR from plasmid pTW3.15 with Pfu DNA polymerase (Promega). Primers were designed to incorporate a KpnI site in the forward primer and a BamHI site in the reverse primer. Primer sequences, with restriction sites underlined and altered bases in boldface, were as follows: forward, 5′-GTGGAGGTACCTATGGCTTTGCTTACTGATG (corresponding to positions 1759 to 1789); and reverse, 5′-GCTGGATCCTCAATAGCGATGGGC (positions 2492 to 2469). PCR amplifications were carried out in a 50-μl reaction volume containing 50 to 100 ng of template DNA, 50 pmol of each primer, 200 μM each deoxynucleoside triphosphate, 1× Pfu buffer [20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 2mM MgSO4, 0.1 mg of bovine serum albumin/ml, 0.1% Triton X-100] and 1 U of Pfu polymerase. After an initial denaturation at 95°C for 1 min, the reaction mixtures were given 30 cycles of 1 min at 95°C, 30 s at 55°C, and 3 min at 74°C, followed by a final extension at 74°C for 5 min. The PCR product was cut with KpnI and BamHI and ligated into pPROLar.A122 cut with KpnI and BamHI, placing the pnbA gene downstream of the lac/ara-1 promoter (Plac/ara-1), to form plasmid pPROpnbA (Table 1). The sequence of the cloned gene was confirmed by double-strand DNA sequencing. The PnbA protein was expressed in E. coli DH5αPRO grown in LB containing kanamycin and spectinomycin to an OD600 of 0.5 and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and 0.2% arabinose for 4 h prior to harvesting.
The pnbB gene was amplified by PCR from plasmid pTW3.14 with Pfu DNA polymerase (Promega). Primers incorporating EcoRI sites into them were designed. Primer sequences, with restriction sites underlined and modified bases in boldface, were as follows: forward, 5′-GAGCGTGAATTCATATGAATAAGCGTGATG (corresponding to positions 4304 to 4333). and reverse, 5′-TAAAGAATTCGCATTCAACCTTGTGCCTGGA (positions 4863 to 4833). PCR amplifications were carried out as for pnbA. The PCR product was cut with EcoRI and ligated into EcoRI-cut pUC18, and recombinants were screened for those with the pnbB gene located downstream of the lacZ promoter; the resultant plasmid was called pTW3.16 (Table 1). The sequence of the cloned pnbB gene was confirmed by double-strand DNA sequencing. PnbB was expressed in E. coli DH5α(pTW3.16) grown in LB broth containing ampicillin to an OD600 of approximately 1.8 prior to harvesting.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out by the method of Laemmli (24) on a Mini-PROTEAN II electrophoresis cell (Bio-Rad, Hemel Hempstead, United Kingdom).
Enzyme assays.
Cells were harvested by centrifugation, washed with 50 mM Na-K phosphate buffer (pH 7.4), and resuspended in the same buffer at approximately 0.2 g (wet weight)/ml. Cells were disrupted by sonication for four periods of 30 s at an amplitude of 6 to 7 μm, and particulates were removed by centrifugation at 45,000 × g and 4°C for 30 min. Dithiothreitol was added at a final concentration of 5 mM to cell extracts containing 4-hydroxylaminobenzoate lyase (PnbB). The activity of 4NBen reductase (PnbA) was determined by measuring the decrease in absorbance at 340 nm due to NADH oxidation in a 1ml assay mixture containing 50 mM Na-K phosphate buffer (pH 7.4), 400 μM NADH, and 50 μM 4NBen. The reaction was initiated by adding 100 μl of cell extract. The molar extinction coefficient for NADH at 340 nm was taken to be 6,220 M−1 cm−1. The stoichiometry of the reaction was calculated from the change in A340 resulting from the total conversion of amounts of substrate varying from 10 nmol to 100 nmol in the presence of excess NADH.
Activity of 4-hydroxylaminobenzoate lyase was observed by repeated spectral scans from 200 to 500 nm to follow the conversion of 4-hydroxylaminobenzoate to PCA. Reactions were performed in 1-ml cuvettes containing 50 mM Na-K phosphate buffer (pH 7.4), 50 μM 4-hydroxylaminobenzoate, and 50 μl of cell extract. The presence of PCA as the lyase product was confirmed by addition to the reaction of extracts of E. coli(pHN150) containing protocatechuate 4, 5-dioxygenase (Table 1) and following the spectral changes in the range 200 to 500 nm, which indicated the formation of 2-hydroxy-4-carboxymuconic semialdehyde (3). Protocatechuate 4, 5-dioxygenase was also used to determine the stoichiometry of PCA formed by the action of PnbB. Amounts of substrate (4NBen or 4-hydroxylaminobenzoate) varying from 10 to 100 nmol were subjected to complete conversion by either PnbA plus PnbB (for 4NBen) or PnbB alone (for 4-hydroxylaminobenzoate). The resulting reaction was then incubated with protocatechuate 4, 5-dioxygenase, and the final change in A410 produced was compared with that produced from standard amounts of authentic PCA under the same reaction conditions. The molar extinction coefficient of 2-hydroxy-4-carboxymuconic semialdehyde under these conditions was estimated to be 973 M−1 cm−1.
The amount of ammonium production from the lyase reaction on 4-hydroxylaminobenzoate, either authentic or produced from 4-nitrobenzoate by the action of PnbA, was determined by the complete conversion of various amounts of substrate from 100 to 500 nmol using Nessler's reagent (2).
Nucleotide sequence accession number.
The DNA sequence obtained in this study has been added to the GenBank database (accession no. AF292094).
RESULTS
Cloning of genes involved in the 4NBen degradation pathway.
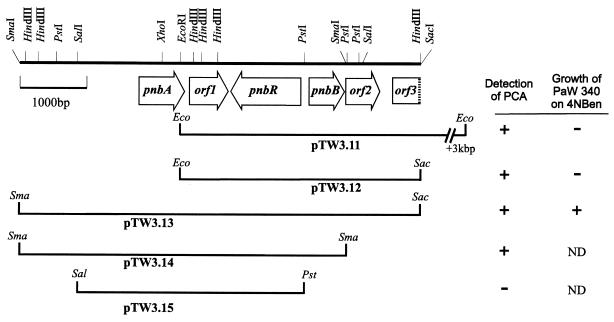
P. putida strain TW3 is able to grow on 4NBen as the sole carbon and nitrogen source (35). To identify the genes essential for 4NBen degradation, P. putida PaW340 was used as the recipient for a library of TW3 genomic DNA, inserted in the broad-host-range cosmid pLAFR5. Transconjugants were selected on MM plates with either succinate or 4NBen as carbon source and supplemented with tryptophan, streptomycin and tetracycline. The number of transconjugants able to grow on 4NBen plates (Pnb+ cells) comprised about 0.9% of those able to grow on succinate. Multiple restriction digests of cosmid DNA isolated from six of the Pnb+ transconjugants showed that all contained a common 6.6-kbp EcoRI restriction fragment together with other EcoRI fragments of different sizes.
Subcloning of catabolic genes.
Subclones of EcoRI-digested cosmids were constructed in pUC18 and transformed into E. coli, and clones were screened for the accumulation of a catechol (in this case, expecting PCA) on plates containing 4NBen and p-toluidine (34). Only with the 6.6-kbp subcloned EcoRI fragment (in plasmid pTW3.11) did PCA accumulate in the media, and subsequently we found that a 3.6-kbp EcoRI/SacI subclone of pTW3.11 (plasmid pTW3.12) produced the same effect (Fig. 1; Table 1).
FIG. 1.
Map of the pnb gene cluster of TW3. Locations of the five open reading frames are marked by open arrows, the direction of the arrowheads indicating the direction of transcription. The inserts of recombinant plasmids are represented below. pTW3.11 and pTW3.13 were subcloned from a large cosmid clone, and pTW3.12, pTW3.14, and pTW3.15 were derived from them by further subcloning. Only restriction sites relevant to the clones and constructs described in this paper are shown. The symbols at the right indicate which plasmids in E. coli accumulated PCA in media containing p-toluidine and 4NBen, and which were able to confer on PaW340 the ability to grow on 4NBen (ND, not determined).
To test whether the inserts from pTW3.11 and pTW3.12 conferred the ability to grow on 4NBen, they were both inserted into pLAFR5 and mobilized back into PaW340. Surprisingly, although both appeared to be responsible for the conversion of 4NBen to PCA in E. coli, neither conferred a Pnb+ phenotype to PaW340. The EcoRI/SacI insert from pTW3.12 was used in Southern blots to probe restriction digests of one of the original cosmids which did confer a Pnb+ phenotype to PaW340. We identified a hybridizing fragment which, when reinserted into pLAFR5 to form pTW3.13 (Fig. 1; Table 1) and mobilized into PaW340, conferred on it the ability to grow on 4NBen. The 4NBen catabolic genes were further assigned to a 4.7-kb SmaI subfragment which was inserted into pUC18 to form pTW3.14. In E. coli, this was shown to cause the accumulation of PCA from 4NBen by screening on p-toluidine plates. The overlapping DNA sequences of the inserts of pTW3.12 and pTW3.14 were determined over 5,996 bp.
Analysis of nucleotide and protein sequences.
In the nucleotide sequence, five complete open reading frames were identified (Fig. 1). The first two open reading frames from the 5′ end were 684 and 579 bp, respectively, and have been designated pnbA and orf1. The putative translation products of both genes exhibit significant similarity to reductase enzymes from various other bacteria (Table 2). Immediately downstream is an open reading frame (designated pnbR) which is 1,047 bp long and is convergently transcribed. PnbR appears to be a regulatory protein in the LysR family and shows greatest similarity to other regulators associated with 4NBen catabolism from Pseudomonas sp. strain YH102 (46; Newman and Zylstra, Abstr. 97th Gen. Meet. Am. Soc. Microbiol.) and Ralstonia pickettii YH105 (43). The product of a fourth open reading frame (PnbB), transcribed divergently from pnbR, exhibits similarity only to 4-hydroxylaminobenzoate lyases from Pseudomonas sp. strain YH102 and R. pickettii YH105. Immediately downstream of pnbB is a fifth open reading frame of 549 bp (orf2), the product of which is similar to Orf2 of unknown function from Pseudomonas sp. strain YH102 and also to a lyase, UbiC, which catalyzes the conversion of chorismate to 4-hydroxybenzoate in E. coli (Table 2). Further downstream is the 5′ end of a sequence for a protein Orf3 which is homologous only to that encoded by a gene in the same relative position downstream of the pnb genes in Pseudomonas sp. strain YH102 (accession no. AF 187880).
TABLE 2.
P. putida TW3 genes and gene products
| Gene designation | Putative function of gene product | Position in sequence | Most similar gene product(s) [species] (accession no.) (% amino acid identity) |
|---|---|---|---|
| pnbA | 4NBen reductase | 1771–2454 | 4NBen reductase [R. pickettii YH105] (AF187879)(42%), putative oxidoreductase [Streptomyces coelicolor A3(2)] (AL109950)(36%) |
| orf1 | Possible reductase | 2527–3105 | Unknown [Pseudomonas sp. strain YH102] (AF187880)(84%), reductase homolog [S. cyanogenus] (AF080235)(30%) |
| pnbR | Regulator | 4191–3145 | PnbR [Pseudomonas sp. strain YH102] (AF187880)(85%), PnbR [R. pickettii YH105] (AF187879)(45%), MexT protein [P. aeruginosa] (AJ007825)(30%) |
| pnbB | 4-Hydroxylaminobenzoate lyase | 4319–4849 | 4-Hydroxylaminobenzoate lyase [Pseudomonas sp. strain YH102] (AF187880)(84%), 4-Hydroxylaminobenzoate lyase [R. pickettii YH105] (AF187879)(59%) |
| orf2 | Possible lyase | 4850–5398 | Unknown [Pseudomonas sp. strain YH102] (AF187880)(64%), chorismate-pyruvate lyase [E. coli] (X66619)(31%) |
| orf3 (incomplete) | Unknown | 5606–5996 | Unknown [Pseudomonas sp. strain YH102] (AF187880)(64%) |
Expression of pnbA in E. coli and nitroreductase assays.
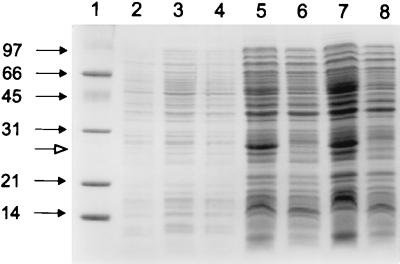
The pnbA gene was copied by PCR into the expression vector pPROLar.A122 to form pPROpnbA (Table 1) and overexpressed in E. coli DH5αPRO. Cell extracts were able to oxidize NADH to NAD+ only in the presence of 4NBen and with a specific activity of 400 mU/mg of protein. SDS-PAGE of the same extracts showed high levels of a polypeptide of ∼25 kDa (Fig. 2). No activity or enhanced 25-kDa protein band was detectable in controls where expression of the protein was not induced or where the expression vector contained no insert.
FIG. 2.
SDS-PAGE of overexpressed 4NBen reductase (PnbA) in E. coli (pPROpnbA) on a 15% gel. Lane 1, low-range marker (Bio-Rad); lane 2, uninduced cell extracts of E. coli(pPROpnbA); lane 3, cell extracts of E. coli(pPROpnbA) obtained immediately after induction with IPTG and arabinose (0 h); lanes 5 and 7, cell extracts of E. coli(pPROpnbA) obtained at 4 and 20 h, respectively, after induction; lanes 4, 6, and 8, cell extracts of E. coli(pPROLar.A122) obtained at 0, 4, and 20 h, respectively after induction. Marker sizes are indicated at the left in kilodaltons. The molecular mass of the overexpressed protein (indicated by the open arrow) is ∼25 kDa.
The enzyme activity of the overexpressed pnbA gene product was compared with that of the 4NBen reductase in extracts of wild-type P. putida TW3 and of E. coli carrying pTW3.15, or constructs in which pnbA or orf1 had been inactivated (Table 3). In TW3, 4NBen reductase activity was elevated only in 4NBen-grown cells, whereas negligible activity was detected in uninduced (succinate-grown) cells. In E. coli carrying both pnbA and orf1 (pTW3.15), cells contained high activity of reductase activity. To demonstrate that the reduction was due to PnbA alone and that the orf1 product played no part, two constructs were made from pTW3.15. In pTW3.15NH, a 366-bp fragment internal to orf1 was deleted; in pTW3.15NX, the single XhoI site within pnbA was filled in with T4 DNA polymerase enzyme to create a frameshift mutation within the gene (Table 1). Whereas pTW3.15NH conferred reductase activity similar to that of its parent plasmid, pTW3.15NX conferred negligible activity (Table 3). The stoichiometry of the reaction determined by pPROpnbA, pTW3.15, and pTW3.15NH was found to be ∼1.8 mol of NADH oxidized/mol of 4NBen utilized in all cases (Table 3).
TABLE 3.
Specific activities and stoichiometry of 4NBen reductase
| Source of cell extract | Growth or induction | Sp act (mU/mg of protein) | Stoichiometry of NADH oxidation (mol of NADH oxidized/mol of 4NBen utilized) |
|---|---|---|---|
| P. putida TW3 | 4NBen | 260 | 1.79 |
| Succinate | <10 | NDa | |
| E. colib(pTW3.15) | Uninduced | 230 | 1.85 |
| E. colib(pTW3.15NH) | Uninduced | 590 | 1.87 |
| E. colib(pTW3.15NX) | Uninduced | <10 | ND |
| E. colib(pUC18) | Induced | <10 | ND |
| E. colic(pPROpnbA) | Induced | 400 | 1.84 |
| E. colic(pPROpnbA) | Uninduced | <10 | ND |
| E. colic(pPROLar.A122) | Induced | <10 | ND |
ND, not determined.
Strain DH5α.
Strain DH5αPRO.
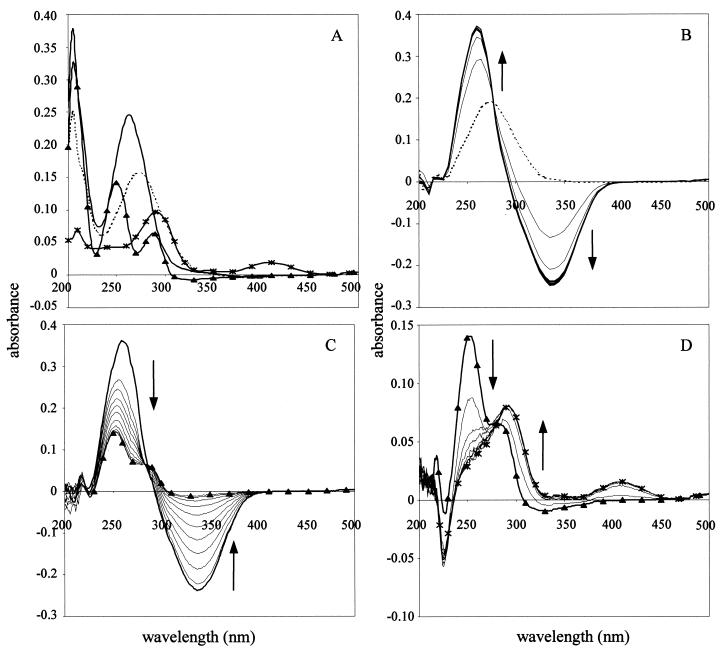
Spectroscopic scans during the incubation of PnbA with 4NBen and NADH resulted in the accumulation of a product (Fig. 3B), which was tentatively identified as 4-hydroxylaminobenzoate on the basis of the identity of its absorption spectrum to that of authentic compound with a λmax of 262 nm (Fig. 3A).
FIG. 3.
(A) Absorption spectra of authentic 4-NBen (––), 4-hydroxylaminobenzoate (——), PCA (▴), and 2-hydroxy-4-carboxymuconic semialdehyde (∗). The latter was prepared by the action of cloned protocatechuate 4, 5-dioxygenase in E. coli(pHN150) on PCA (see Material and Methods). (B) Conversion of 4NBen (––) to 4-hydroxylaminobenzoate (——) by cell extracts of E. coli containing overexpressed PnbA (4NBen reductase) in the presence of NADH. (C) Formation of PCA (▴) from 4-hydroxylaminobenzoate (——) by cell extracts containing overexpressed PnbB added to the 4-hydroxylaminobenzoate formed by PnbA action on 4NBen (see above). An identical result was obtained by the action of PnbB on authentic 4-hydroxylaminobenzoate (data not shown). (D) Authentication that PCA (▴) is formed in panel C by its conversion to 2-hydroxy-4-carboxymuconic semialdehyde (∗) by cell extracts of E. coli(pHN150) containing protocatechuate 4, 5-dioxygenase added to the PnbB assay product. An identical spectrum was obtained by the action of protocatechuate 4, 5-dioxygenase on authentic PCA (A). Scans in panels B to D were made every 60 s. over a period of 10 min.
Analysis of PnbB activity.
Cell extracts containing only PnbB expressed in E. coli(pTW3.16) converted both authentic 4-hydroxylaminobenzoate and the compound produced from 4NBen by PnbA (see above) to PCA. This was identified by the absorption spectrum with λmax of 254 and 290 nm (Fig. 3A and C) but also by incubating it with cell extracts containing cloned protocatechuate 4,5-dioxygenase (expressed from plasmid pHN150) and following its conversion to 2-hydroxy-4-carboxymuconic semialdehyde with λmax of 292 and 410 nm (Fig. 3D). Incubation of PnbA (expressed from pPROpnbA) with NADH and 4NBen followed by the addition of PnbB (from pTW3.16) resulted in a near stoichiometric conversion of 4NBen to protocatechuate (0.88 mol of PCA formed/mol of 4NBen utilized) and ammonium (0.93 mol/mol).
DISCUSSION
Previous metabolic studies on TW3 had shown that 4NBen, formed as a metabolite of 4-nitrotoluene, was converted to PCA with the elimination of the nitro group as ammonia, with subsequent intradiol cleavage by protocatechuate 3,4-dioxygenase (19, 35), but no characterization of the enzymes or genes for the steps between 4NBen and PCA was attempted. In this study we have isolated the genes and demonstrated their function by creating knockouts, expressing them in E. coli, and carrying out functional assays of the enzymes.
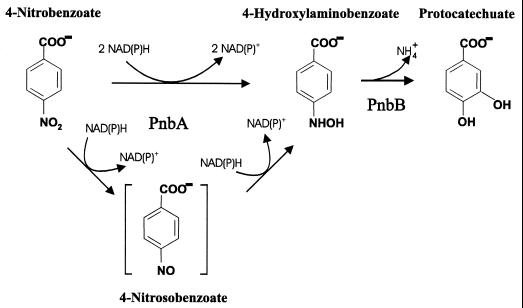
The first open reading frame, pnbA, encodes an enzyme with homologies to other reductases. Adjacent to it and transcribed in the same direction is a second gene, orf1, also encoding a reductase homolog. The pathway for conversion of 4NBen to 4-hydroxylaminobenzoate proposed by Groenewegen et al. (14, 15) involved two successive reductions with 4-nitrosobenzoate as an intermediate. Surprisingly, the two adjacent and possibly cotranscribed reductase genes (pnbA orf1) are not necessary for the two-stage reduction. PnbA is sufficient and is able solely to carry out the double reduction. Two moles of NAD+ were formed per mole of 4NBen, using plasmids carrying only pnbA or carrying both pnbA and orf1 but in which orf1 had been inactivated by an internal deletion. We were unable to detect intermediate 4-nitrosobenzoate formation, and the conversion shows an isobestic point characteristic of a straight conversion of substrate to product (Fig. 3B). We also failed to reduce a sample of authentic 4-nitrosobenzoate in vitro using extracts containing PnbA, but the compound appeared to undergo a spontaneous chemical reaction in the assay mixture and the results could not easily be interpreted (data not shown). We therefore propose that 4-nitrosobenzoate probably represents a transient intermediate (Fig. 4) and never leaves the enzyme active site. This may represent a major biochemical difference between P. putida TW3 and C. acidovorans NBA-10 (14, 15).
FIG. 4.
Reactions for the catabolism of 4NBen to PCA in P. putida TW3. The evidence presented in this paper does not distinguish whether or not 4-nitrosobenzoate is a transient intermediate in the PnbA-catalyzed reaction as proposed by Groenewegen et al. (14, 15).
The rest of the pathway for the conversion of 4NBen to PCA involves only one other gene, pnbB. Its product, PnbB, is 4-hydroxylaminobenzoate lyase, which converts 4-hydroxylaminobenzoate directly to PCA and ammonium in stoichiometric amounts. The combined action of PnbA and PnbB plus 2 mol of NADH is all that is required for the complete conversion of 4NBen to the same products.
Another open reading frame, orf2, is immediately adjacent to pnbB and might be cotranscribed with it. However, a plasmid (pTW3.14) created from pTW3.13 by deletion of orf2 was still able to encode the complete transformation of 4NBen to PCA (data not shown). This shows that, like Orf1, Orf2 is not directly involved in the conversion of 4NBen to PCA. This does not, however, exclude either or both being involved in some other, as yet uncharacterized reaction since they are so closely linked to and transcribed in the same directions as pnbA and pnbB, respectively. What is noteworthy is that the sequence comparisons of each of Orf1 and Orf2 show they are in the same functional class as their respective functional neighbors, Orf1 being a reductase homolog and Orf2 being a lyase homolog.
There is one minor experimental anomaly in our results. In the original identification of the pnb DNA, plasmids pTW3.11 and pTW3.12 both caused the accumulation of PCA from 4NBen in E. coli hosts (Fig. 1), yet further analysis of the genes has shown that pnbA, encoding the primary reduction of 4NBen, is not located on either plasmid. However, there is substantial evidence that nitro groups can be reduced nonspecifically by a variety of dehydrogenase/reductase reactions (10, 23, 26, 32), and in many situations where there are no specific degraders present, reduced derivatives often accumulate; this is particularly true in anaerobic environments where the nitro groups can be reduced completely to the corresponding amino groups (11). It is possible that nitroreductases in E. coli such as NfsA and NfsB (8, 25, 41, 44) can effect the partial reduction of 4NBen, thus complementing pnbA and allowing PnbB on plasmids pTW3.11 and pTW3.12 to produce sufficient quantities of PCA to be visualized on the p-toluidine plates.
It is probable that pnbR is the regulator of one or both of the two pnb genes, although we have no evidence to support this. However, in the case of Pseudomonas sp. strain YH102, the homologous pnbR gene has been inactivated by insertion, causing loss of both the ability to grow on 4NBen and the inducibility of 4NBen reductase (46). We are currently investigating the regulation and operon structure of the TW3 pnb genes.
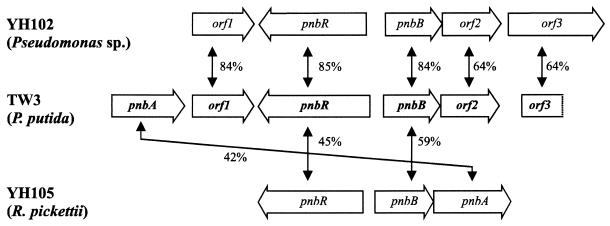
The first report of cloning genes for 4NBen degradation was from R. pickettii YH105 (43); these genes have now been sequenced, and the data have been deposited in GenBank (accession no. AF187879). Analogous genes have also been cloned from Pseudomonas sp. strain YH102 (46), and the sequence data have been deposited (accession no. AF 187880). No functional analysis of the enzyme activities in either strain has yet been published, and in the case of strain YH102, there is no available information on the location of pnbA. Comparison of the TW3 data with those of YH102 and YH105 (Fig. 5) shows that the two Pseudomonas strains (YH102 and TW3) have almost identical arrangements of genes, even including the apparently functionless orf1, orf2, and the downstream orf3. However, in YH102 pnbA is not immediately upstream of orf1, as it is in TW3, and has yet to be located (G. J. Zylstra, personal communication). In R. pickettii YH105, the nucleotide sequences of the genes are less similar to the two Pseudomonas strains and pnbA is on the opposite side of the pnbRpnbB gene pair. Given that this is a very small sample, the similarity (of sequence and gene order) matches the taxonomy of the host cells, and the fact, pointed out by Zylstra et al. (46), that the PnbB protein sequences bear very little similarity to other proteins in the database, it seems likely that the pnb genes are of fairly ancient origin and not recently recruited for nitroaromatic catabolism (46).
FIG. 5.
Alignment of the pnb gene cluster of TW3 with genes from Pseudomonas sp. strain YH102 (accession no. AF 187879) and R. pickettii YH105 (accession no. AF187880). The open reading frames and direction of transcription are indicated by the open arrows. Percentage identity at the amino acid level is indicated adjacent to the double-headed arrows.
ACKNOWLEDGMENTS
We thank Tomonori Sonoki, Tokyo University of Agriculture and Technology, for the gift of plasmid pHN150 and Jumáa R. Al-Dulayymi, Chemistry Department, University of Wales, Bangor, for synthesizing the 4-hydroxylaminobenzoic and 4-nitrosobenzoic acids.
This research was funded under the auspices of the Biotechnology Research Programme of the European Commission.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.American Public Health Association, American Water Works Association, and Water Pollution Control Federation. Standard methods for the examination of water and wastewater. 14th ed. Washington, D.C.: American Public Health Association, American Water Works Association, and Water Pollution Control Federation; 1976. [Google Scholar]
- 3.Arciero D M, Orville A M, Lipscomb J D. Protocatechuate 4, 5-dioxygenase from Pseudomonas testesteroni. Methods Enzymol. 1990;188:89–95. doi: 10.1016/0076-6879(90)88017-5. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D E, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 5.Bauchop T, Elsden S R. The growth of microorganisms in relation to their energy supply. J Gen Microbiol. 1960;23:457–469. doi: 10.1099/00221287-23-3-457. [DOI] [PubMed] [Google Scholar]
- 6.Bauer H, Rosenthal S M. 4-Hydroxylaminobenzenesulfonamide, its acetyl derivatives and diazotation reaction. J Am Chem Soc. 1944;66:611–614. [Google Scholar]
- 7.Bruhn C, Bayly R C, Knackmuss H J. The in vivo construction of 4-chloro-2-nitrophenol assimilatory bacteria. Arch Microbiol. 1988;150:171–177. [Google Scholar]
- 8.Bryant D W, McCalla D R, Leeksma M, Laneuville P. Type I nitroreductases of Escherichia coli. Can J Microbiol. 1981;27:81–86. doi: 10.1139/m81-013. [DOI] [PubMed] [Google Scholar]
- 9.Cartwright N J, Cain R B. Bacterial degradation of the nitrobenzoic acids. Biochem J. 1959a;71:248–261. doi: 10.1042/bj0710248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cartwright N J, Cain R B. Bacterial degradation of the nitrobenzoic acids. 2. Reduction of the nitro group. Biochem J. 1959b;73:305–314. doi: 10.1042/bj0730305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerniglia C E, Somerville C C. Reductive metabolism of nitroaromatic and nitropolycyclic aromatic hydrocarbons. In: Spain J C, editor. Biodegradation of nitroaromatic compounds. New York, N.Y: Plenum Publishing Corporation; 1995. pp. 99–115. [Google Scholar]
- 12.Ecker S, Widmann T, Lenke H, Dickel O, Fischer P, Bruhn C, Knackmuss H J. Catabolism of 2, 6-dinitrophenol by Alcaligenes eutrophus JMP134 and JMP222. Arch Microbiol. 1992;158:149–154. [Google Scholar]
- 13.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent upon a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groenewegen P E J, Breeuwer P, Vanhelvoort J, Langenhoff A A M, Devries F P, Debont J A M. Novel degradative pathway of 4-nitrobenzoate in Comamonas acidovorans NBA-10. J Gen Microbiol. 1992;138:1599–1605. doi: 10.1099/00221287-138-8-1599. [DOI] [PubMed] [Google Scholar]
- 15.Groenewegen P E J, deBont J A M. Degradation of 4-nitrobenzoate via 4-hydroxylaminobenzoate and 3, 4-dihydroxybenzoate in Comamonas acidovorans NBA-10. Arch Microbiol. 1992;158:381–386. [Google Scholar]
- 16.Haigler B E, Spain J C. Biodegradation of 4-nitrotoluene by Pseudomonas sp. strain 4NT. Appl Environ Microbiol. 1993;59:2239–2243. doi: 10.1128/aem.59.7.2239-2243.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haigler B E, Wallace W H, Spain J C. Biodegradation of 2-nitrotoluene by Pseudomonas sp. strain JS42. Appl Environ Microbiol. 1994;60:3466–3469. doi: 10.1128/aem.60.9.3466-3469.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James K D, Hughes M A, Williams P A. Cloning and expression of ntnD, encoding a novel NAD(P)+-independent 4-nitrobenzyl alcohol dehydrogenase from Pseudomonas sp. strain TW3. J Bacteriol. 2000;182:3136–3141. doi: 10.1128/jb.182.11.3136-3141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.James K D, Williams P A. ntn genes determining the early steps in the divergent catabolism of 4-nitrotoluene and toluene in Pseudomonas sp. strain TW3. J Bacteriol. 1998;180:2043–2049. doi: 10.1128/jb.180.8.2043-2049.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeenes D J, Williams P A. Excision and integration of degradative pathway genes from TOL plasmid pWW0. J Bacteriol. 1982;150:188–194. doi: 10.1128/jb.150.1.188-194.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katsivela E, Wray V, Pieper D H, Wittich R M. Initial reactions in the biodegradation of 1-chloro-4-nitrobenzene by a newly isolated bacterium, strain LW1. Appl Environ Microbiol. 1999;65:1405–1412. doi: 10.1128/aem.65.4.1405-1412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 23.Kitts C L, Green C E, Otley R A, Alvarez M A, Unkefer P J. Type I nitroreductases in soil enterobacteria reduce TNT (2, 4, 6, -trinitrotoluene) and RDX (hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine) Can J Microbiol. 2000;46:278–282. doi: 10.1139/w99-134. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Lightfoot R T, Shuman D, Ischiropoulos H. Oxygen-insensitive nitroreductases of Escherichia coli do not reduce 3-nitrotyrosine. Free Radic Biol Med. 2000;28:1132–1136. doi: 10.1016/s0891-5849(00)00208-2. [DOI] [PubMed] [Google Scholar]
- 26.Marvin-Sikkema F D, de Bont J A M. Degradation of nitroaromatic compounds by microorganisms. Appl Microbiol Biotechnol. 1994;42:499–507. doi: 10.1007/BF00173912. [DOI] [PubMed] [Google Scholar]
- 27.Meulenberg R, Pepi M, de Bont J A M. Degradation of 3-nitrophenol by Pseudomonas putida B2 occurs via 1,2,4-benzenetriol. Biodegradation. 1996;7:303–311. doi: 10.1007/BF00115744. [DOI] [PubMed] [Google Scholar]
- 28.Michan C, Delgado A, Haidour A, Lucchesi G, Ramos J L. In vivo construction of a hybrid pathway for metabolism of 4-nitrotoluene in Pseudomonas fluorescens. J Bacteriol. 1997;179:3036–3038. doi: 10.1128/jb.179.9.3036-3038.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nadeau L J, Spain J C. Bacterial degradation of m-nitrobenzoic acid. Appl Environ Microbiol. 1995;61:840–843. doi: 10.1128/aem.61.2.840-843.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishino S F, Paoli G C, Spain J C. Aerobic degradation of dinitrotoluenes and pathway for bacterial degradation of 2, 6-dinitrotoluene. Appl Environ Microbiol. 2000;66:2139–2147. doi: 10.1128/aem.66.5.2139-2147.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishino S F, Spain J C. Oxidative pathway for the biodegradation of nitrobenzene by Comamonas sp. strain JS765. Appl Environ Microbiol. 1995;61:2308–2313. doi: 10.1128/aem.61.6.2308-2313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishino S F, Spain J C, He Z. Strategies for aerobic degradation of nitroaromatic compounds by bacteria: process discovery to field application. In: Spain J C, Hughes J B, Knackmuss H-J, editors. Biodegradation of nitroaromatic compounds and explosives. Boca Raton, Fla: Lewis Publishers; 2000. pp. 7–61. [Google Scholar]
- 33.Noda Y, Nishikawa S, Shiozuka K I, Kadokura H, Nakajima H, Yoda K, Katayama Y, Morohoshi N, Haraguchi T, Yamasaki M. Molecular cloning of the protocatechuate 4, 5-dioxygenase genes of Pseudomonas paucimobilis. J Bacteriol. 1990;172:2704–2709. doi: 10.1128/jb.172.5.2704-2709.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parke D. Application of para-toluidine in chromogenic detection of catechol and protocatechuate, diphenolic intermediates in catabolism of aromatic compounds. Appl Environ Microbiol. 1992;58:2694–2697. doi: 10.1128/aem.58.8.2694-2697.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhys-Williams W, Taylor S C, Williams P A. A novel pathway for the catabolism of 4-nitrotoluene by Pseudomonas. J Gen Microbiol. 1993;139:1967–1972. doi: 10.1099/00221287-139-9-1967. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Schenzle A, Lenke H, Fischer P, Williams P A, Knackmuss H J. Catabolism of 3-nitrophenol by Ralstonia eutropha JMP134. Appl Environ Microbiol. 1997;63:1421–1427. doi: 10.1128/aem.63.4.1421-1427.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schenzle A, Lenke H, Spain J C, Knackmuss H J. 3-Hydroxylaminophenol mutase from Ralstonia eutropha JMP134 catalyzes a Bamberger rearrangement. J Bacteriol. 1999;181:1444–1450. doi: 10.1128/jb.181.5.1444-1450.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spain J C, Gibson D T. Pathway for biodegradation of para-nitrophenol in a Moraxella sp. Appl Environ Microbiol. 1991;57:812–819. doi: 10.1128/aem.57.3.812-819.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spanggord R J, Spain J C, Nishino S F, Mortelmans K E. Biodegradation of 2, 4-dinitrotoluene by a Pseudomonas sp. Appl Environ Microbiol. 1991;57:3200–3205. doi: 10.1128/aem.57.11.3200-3205.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whiteway J, Koziarz P, Veall J, Sandhu N, Kumar P, Hoecher B, Lambert I B. Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J Bacteriol. 1998;180:5529–5539. doi: 10.1128/jb.180.21.5529-5539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Worsey M J, Williams P A. Metabolism of toluene and xylenes by Pseudomonas putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975;124:7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yabannavar A V, Zylstra G J. Cloning and characterization of the genes for p-nitrobenzoate degradation from Pseudomonas pickettii YH105. Appl Environ Microbiol. 1995;61:4284–4290. doi: 10.1128/aem.61.12.4284-4290.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zenno S, Koike H, Tanokura M, Saigo K. Gene cloning, purification, and characterization of NfsB, a minor oxygen-insensitive nitroreductase from Escherichia coli, similar in biochemical properties to FRase I, the major flavin reductase in Vibrio fischeri. J Biochem. 1996;120:736–744. doi: 10.1093/oxfordjournals.jbchem.a021473. [DOI] [PubMed] [Google Scholar]
- 45.Zeyer J, Kearney P C. Degradation of ortho-nitrophenol and meta-nitrophenol by a Pseudomonas putida. J Agric Food Chem. 1984;32:238–242. [Google Scholar]
- 46.Zylstra G J, Bang S-W, Newman L M, Perry L L. Microbial degradation of mononitrophenols and mononitrobenzoates. In: Spain J C, Hughes J B, Knackmuss H-J, editors. Biodegradation of nitroaromatic compounds and explosives. Boca Raton, Fla: Lewis Publishers; 2000. pp. 145–160. [Google Scholar]