Significance
DNA is packed inside the nucleus of the cell by wrapping around histone proteins, which are synthesized in the cytoplasm and must be rapidly imported into the nucleus. IMPORTIN-4 is the major nuclear import receptor for histones H3 and H4, and ASF1 is the most abundant chaperone for H3–H4 dimers in the cytoplasm. It is not understood how these proteins assemble for nuclear import. This paper presents atomic-resolution structural information that explains how H3–H4 and ASF1 form a heterotetrameric complex with IMPORTIN-4 in the cytoplasm and how this nuclear import complex is dissociated in the nucleus.
Keywords: karyopherins, histones, nuclear import, nucleosome, importin
Abstract
IMPORTIN-4, the primary nuclear import receptor of core histones H3 and H4, binds the H3–H4 dimer and histone chaperone ASF1 prior to nuclear import. However, how H3–H3–ASF1 is recognized for transport cannot be explained by available crystal structures of IMPORTIN-4–histone tail peptide complexes. Our 3.5-Å IMPORTIN-4–H3–H4–ASF1 cryoelectron microscopy structure reveals the full nuclear import complex and shows a binding mode different from suggested by previous structures. The N-terminal half of IMPORTIN-4 clamps the globular H3–H4 domain and H3 αN helix, while its C-terminal half binds the H3 N-terminal tail weakly; tail contribution to binding energy is negligible. ASF1 binds H3–H4 without contacting IMPORTIN-4. Together, ASF1 and IMPORTIN-4 shield nucleosomal H3–H4 surfaces to chaperone and import it into the nucleus where RanGTP binds IMPORTIN-4, causing large conformational changes to release H3–H4–ASF1. This work explains how full-length H3–H4 binds IMPORTIN-4 in the cytoplasm and how it is released in the nucleus.
During S phase of the cell cycle, core histones H3, H4, H2A, and H2B are rapidly synthesized in the cytoplasm, transported into the nucleus, and incorporated into newly replicated DNA to form nucleosomes. Following translation in the cytoplasm, H3 and H4 are passed to heat shock proteins and histone chaperones to be folded into H3–H4 heterodimers, which are then acetylated at several lysine side chains (1–3). Multiple studies have shown that H3 and H4 are actively transported across the nuclear pore complex into the nucleus by karyopherin-β nuclear import receptors, which are also called IMPORTINs (4–6). The major IMPORTINs that copurify with H3 and H4 from Saccharomyces cerevisiae (Sc) and HeLa cell extracts are Sc KAP123 and human IMPORTIN-4 (IMP4), respectively, while smaller amounts of Sc KAP121 and human IMPORTIN-5 (IMP5) associate with H3 and H4 as secondary or backup IMPORTINs (1, 7–9).
Early studies showed that the basic N-terminal tail peptides of both H3 and H4 (H3tail and H4tail, Fig. 1A) can bind and be imported by at least six different IMPORTINs (5, 10, 11), but nuclear import of the full-length histones appears more complex. For example, the minor IMPORTINs for H3 and H4 (Kapβ2, IMP9, and IMP5) bind histone tails more tightly than the major/primary IMPORTIN IMP4 (10). Crystal structures of the Sc homolog of IMP4, KAP123, bound separately to the H3tail (residues 1 to 28) and H4tail (residues 1 to 34) showed interactions with only a few histone residues in each structure (12), and cellular studies of H3 and H4 without N-terminal tails suggested that tailless histones are imported into the nucleus in a manner dependent on their histone-fold domain (13, 14). Most importantly, H3–H4 heterodimers were detected in the cytoplasm of HeLa cells in complex with both IMP4 and the histone chaperone ASF1, suggesting that the cargo transported by IMP4 is the H3–H4–ASF1 complex (1). Of several IMPORTINs tested, IMP4 also binds the tightest to the H3–H4–ASF1 complex, consistent with its role as the major H3 and H4 importer (10). In the absence of an atomic-resolution structure of IMP4 bound to H3–H4–ASF1, it was unclear how the H3–H4 globular histone-fold domain, histone tails, and ASF1 contribute to the formation of the IMP4–H3–H4–ASF1 import complex.
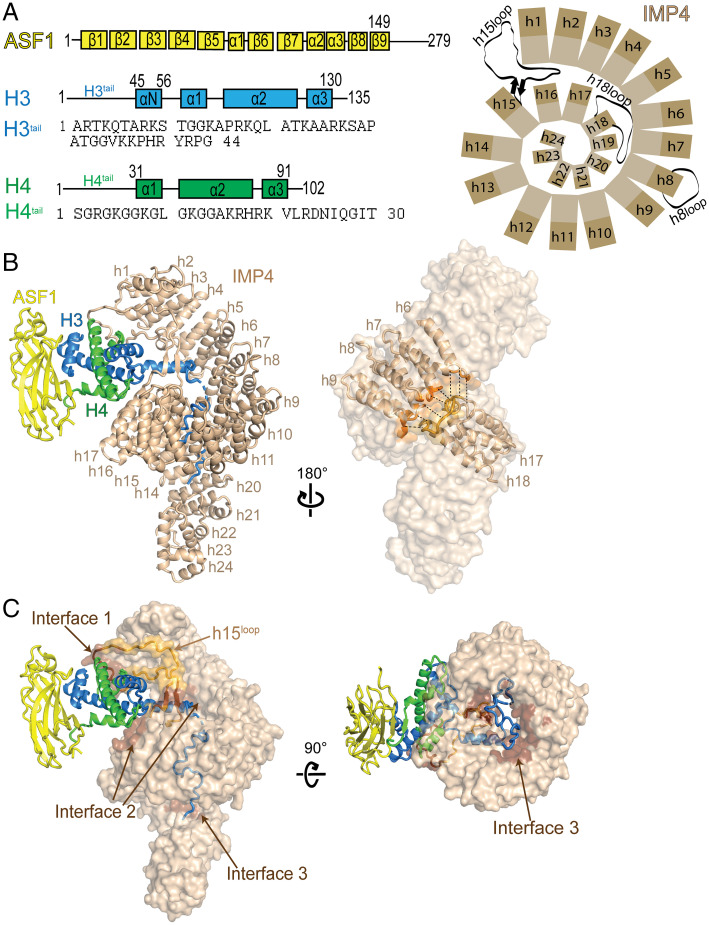
Fig. 1.
The cryo-EM structure of IMP4–H3–H4–ASF1. (A) A schematic showing the organization of Homo sapiens IMP4, Sc ASF1, X. laevis H3 and H4, and the sequence of the H3 and H4 N-terminal tails. (B) (Left), a cartoon representation of IMP4 (beige) bound to H3–H4–Asf1 (H3 is blue, H4 is green, and ASF1 is yellow), with IMP4 HEAT repeats h1 to h24 labeled. (Right), IMP4 in surface representation, highlighting the two regions, h6 to h9 (dark orange) and h17 and 18 (dark yellow), that interact to close the central ring of the superhelix. (C) IMP4 (beige surface) contacts H3–H4 at interfaces 1, 2, and 3 (dark brown surface), with the long IMP4 h15loop colored light orange. A 90° rotation about the horizontal axis shows the tight central ring of IMP4 on the (Right). HEAT repeats h23 and h24 are not shown for a clear view of the ring.
We assembled full-length human IMP4, Xenopus laevis H3–H4, and Sc ASF1 (residues 1 to 160) to form a 1:1:1 stoichiometry IMP4–H3–H4–ASF1 complex and solved its structure to 3.5-Å resolution by cryoelectron microscopy (cryo-EM; Fig. 1 A–C and SI Appendix, Fig. S1 A and B and Table S1). The structure shows one IMP4 molecule bound to one H3–H4–ASF1 complex; the latter is almost identical to a previous H3–H4–ASF1 structure (Protein Data Bank ID [PDBID]: 2HUE; rmsd: 0.607 for 243 Cα atoms) (15). IMP4 makes extensive interactions with H3–H4. The globular H3–H4 histone-fold domain and the αN helix of H3 bind to the N-terminal half of IMP4, while the H3tail binds to the C-terminal half of the IMPORTIN (Fig. 1 B and C). ASF1 binds the H3–H4 dimer but makes no contact with IMP4. No density is observed for the H4tail and the C-terminal tails of the histones. The structure of IMP4 and its full-length multiprotein cargo H3–H4–ASF1 reveals another case where the three-dimensional context of the protein(s) is essential for cargo recognition by IMPORTINs (16–20).
Histone-bound IMP4 is an elongated superhelix of 24 HEAT repeats (h1 to h24; named for helical repeats first observed in the proteins Huntingtin, Elongation Factor 3, Protein Phasphatase 2A and TOR1 Kinase), each comprising a pair of A and B α-helices (Fig. 1 A and B). Like its yeast homolog Kap123 (12), most IMP4 helices are connected by short loops except for the helices in h8, h15, and h18. The h8loop is 26 residues long and partially disordered (Fig. 1 A and B). The h15loop is 50 residues long (residues 618 to 668), contains a β-strand pair at its proximal end, and has a highly acidic distal portion. This loop extends from the middle of the IMP4 superhelix toward the IMP4 N terminus where it interacts with repeats h1 to h3 and with the histone-fold domain of H3–H4 (Fig. 1 A–C and SI Appendix, Fig. S2A). Interestingly, the h15loop in the unliganded and H3tail peptide-bound Kap123 structures (PDBID: 5VE8 and 5VCH) are disordered and not modeled (12). The highly acidic h18loop (812DTDEEEEEEADDQAEYD828) of H3–H4–ASF1-bound IMP4 is also involved in intramolecular interactions, making electrostatic interactions with a series of lysine side chains in the h7 repeat (Fig. 1 A and B and SI Appendix, Fig. S2B). These h18loop–h7 interactions and additional interactions between h17 to h18 and h6 to h9 bring together the two distant sets of HEAT repeats to form a small ring (inner diameter of ∼20 Å) in the middle of the IMP4 superhelix that the H3tail threads through (Fig. 1 B and C). The H3tail-bound and the unliganded KAP123 superhelices are similar, with repeats h17 to h18 and h6 to h9 in close proximity (SI Appendix, Fig. S2C).
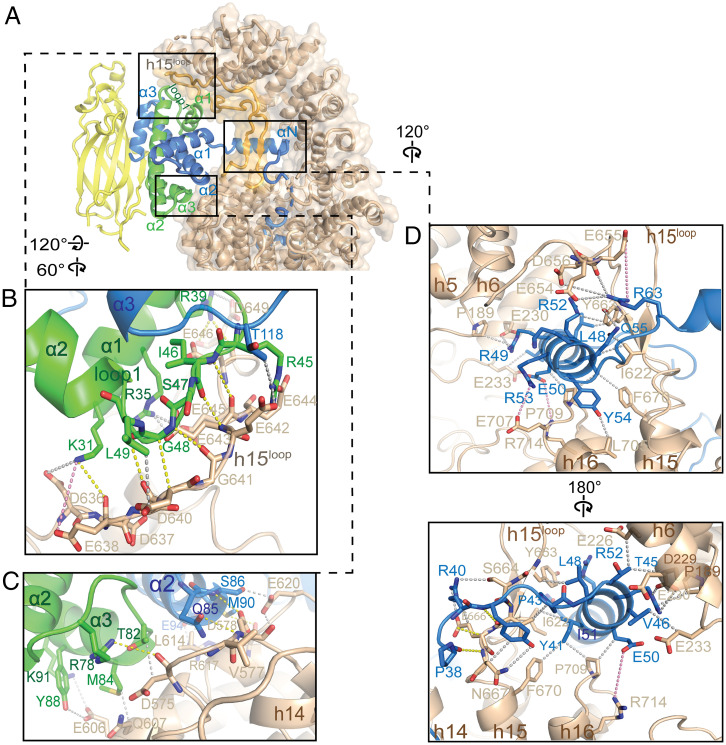
IMP4 makes extensive interactions with H3–H4 through three noncontiguous interfaces 1 to 3 that cover a total surface area of 1,969 Å2 on the histone dimer (Fig. 1C). The H3–H4 histone-fold domain is clamped by the N-terminal half of the IMP4 superhelix through interactions at interface 1 with the distal part of the IMP4 h15loop (which also interacts with IMP4 repeats h1 and h2) and with IMP4 h14 to h16 (part of interface 2; Fig. 2A). h15loop residues 636DDESDGEEEEELMDED651 interact with helix α1 of H4 and the loop that follows (loop 1, Fig. 2B). Interactions here involve many main-chain contacts, including hydrogen bonds that resemble β-sheet interactions. Many acidic h15loop side chains make electrostatic interactions with basic H4 side chains; hydrophobic interactions are also present. On the other side of the histone-fold domain, residues in the α2 helix of H3 and residues in the region that span α2 to α3 of H4 interact with IMP4 repeats h14 and h15 through a myriad of interaction types to form part of interface 2 (Fig. 2C). The other part of interface 2 involves interactions of the H3 αN helix (αNH3, residues 45 to 58). αNH3 adopts different orientations relative to the histone-fold domain depending on the binding partners. αNH3 in the nucleosome interacts with and is part of the histone-fold domain as it also makes multiple contacts with DNA, whereas αNH3 in the H3–H4–ASF1 crystal structure is likely highly dynamic and hence not modeled (15, 21). However, the αNH3 helix in our IMP4–H3–H4–ASF1 structure protrudes away from the histone-fold domain and is surrounded by several different IMP4 elements—B helices of IMP4 repeats h4 to h7, the IMP4 h16A helix, and the β-strand pair of the IMP4 h15loop—that make many electrostatic, polar, and hydrophobic interactions with the αNH3 helix (Fig. 2D).
Fig. 2.
The histone-fold domain of H3–H4 and the αNH3 helix binds the N-terminal half of IMP4. (A) Cartoon and surface representations of the N-terminal half of IMP4 (beige) clamping the histone-fold domain of the ASF1-bound H3–H4 (ASF1 is yellow, H3 is blue, and H4 is green), interacting at interfaces 1 and 2. The IMP4 h15loop is colored light orange. (B–D) Details of interfaces 1 and 2. Select interactions are shown with yellow dashed lines (interactions that involve main chains), gray dashes (interactions between side chains), and pink dashes (long-range electrostatic interactions). (B) Details of interface 1 between IMP4 h15loop residues and the region of H4 that spans α1 to α2. (C) Details of interface 2 between residues of IMP4 HEAT repeats h14 and h15 and the other end of the H3–H4 domain from B, involving residues from α2 to α3 of H4 and from the α2 helix of H3. (D) Details of the remaining portion of interface 2, where the proximal part of the h15loop, HEAT repeats h14 to h16, and h4 to h6 of IMP4 surround the αNH3 helix.
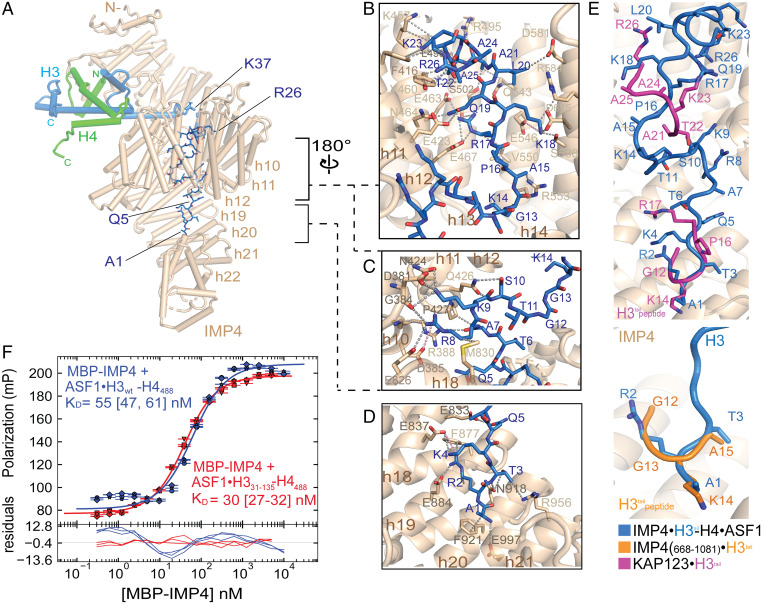
The αNH3 helix is positioned at the opening of the IMP4 central ring. Immediately N-terminal, residues 37 to 43 of αNH3 approach the ring, threading the H3tail into and through the ring. No density is observed for H3 residues 27 to 36. Residues 1 to 26 bind to B helices in the C-terminal half of IMP4 through a variety of interactions—electrostatic, polar, and hydrophobic (Fig. 3 A–D and SI Appendix, Fig. S3 A and B). H3tail segment 1ARTKQ5 binds C-terminal HEAT repeats h18 to h22, while H3tail residues 7 to 26 line the inner surface of the IMP4 central ring (interface 3, Fig. 3 A–D). Within the IMP4 ring, the H3tail segment 7ARKS10 contacts h17 and h18 and h10 and h11 (Fig. 3C). Density for 10STGG13 is weak and noncontiguous, consistent with few to no contacts with IMP4, and H3tail 14KAPRKQLATKAAR26 makes extensive interactions with h11 to h15 (Fig. 3B).
Fig. 3.
The H3tail binds the C-terminal half of IMP4. (A) A view (IMP4 is in beige, H3 is in blue, H4 is in green, and ASF1 is in yellow) showing H3tail residues 1 to 25 (blue sticks) binding to the C-terminal half of IMP4. (B–D) Details of IMP4–H3tail interactions, which mainly involve side chains (gray dashed lines) and include a few long-range electrostatic contacts (pink dashes). (B) H3tail residues 14 to 25 contact residues from HEAT repeats h11 to h15 of IMP4. (C) H3tail residues 7 to 10 bind to HEAT repeats h8 to h11 and h18. (D) H3tail residues 1 to 5 bind to repeats h18 to h22. (E) Superposition of H3tail residues of three different structures: IMP4–H3–H4–ASF1 (blue), IMP4668 to 1081–H31 to 18 (orange; PDBID: 5XBK) (22), and KAP123–H31 to 28 (magenta; PDBID: 5VE8) (12). Backbone residues of the H3tail are shown in ribbon representations, and the side chains are shown in sticks. H3tail IMP4 is beige and in cartoon representation. (F) Fluorescence polarization binding assays with MBP–IMP4 and ASF1–H3WT–H4AF488 (green line) or ASF1–H331 to 135–H4AF488 (blue line). H4 is labeled with the XFD488 fluorophore conjugated to residue 63, which is mutated to cysteine. Fitted binding curves are overlaid onto data points, with error bars representing the mean and SD of triplicate titrations. KD values are reported in the graphs with CIs of 68% in brackets.
No density is observed for the N-terminal tail of H4 or the C-terminal tails of H3 and H4, suggesting that they are flexible and disordered. Locations of the N-terminal most modeled H4 residue and the last modeled residues of H3 and H4 place these tails far from IMP4, further suggesting that the N-terminal H4tail and the C-terminal tails of H3 and H4 do not contribute to the binding of IMP4 (Fig. 3A). This observation also suggests that the two well-established acetylation sites of H4 in the cytoplasm (K5 and K15) are accessible in the IMP4–H3–H4–ASF1 complex, and acetylation at these sites is unlikely to affect IMP4 binding (1, 23, 24). However, IMP4 shields most of the posttranslational modification sites in the H3tail, consistent with the modifications occurring only after the release of H3–H4 in the nucleus (1).
Previous biochemical studies had suggested a role for the H3tail in IMP4 binding (10, 13, 25). However, H3tail density in our cryo-EM map is not contiguous and is weaker than that of the H3–H4 histone-fold domain (SI Appendix, Fig. S3 A and B). We deleted the N-terminal tail from H3 (H3Δtail), assembled the wild-type (wt) and mutant ASF1–H3–H4 complexes, and measured their affinities for IMP4 by fluorescence polarization (Fig. 3F and SI Appendix, Fig. S4). The wt and H3Δtail ASF1–H3–H4 complexes bind IMP4 with similar high affinities (dissociation constants [KD values] of 55 nM and 30 nM, respectively), suggesting minor energetic contributions of the H3tail to formation of the IMP4–H3–H4–ASF1 complex.
The H3tail in our structure binds somewhat differently to IMP4 than in previous crystal structures of a H3tail peptide bound to an IMP4 fragment and to the homologous KAP123, respectively. In the IMP4668 to 1081–H3tail structure (PDBID: 5XBK), only four H3tail residues (12GGKA15) of the 28-residue tail peptide were modeled into a site occupied by H3 residues 1ART3 in our structure but in a different orientation (Fig. 3E, Bottom) (12, 22, 26). A slightly longer 12GGKAPR17 segment was modeled into the same site of the KAP123–H3tail structure (5VE8) along with another short H3tail segment 22SKAAR26 binding to KAP123 h11 to h13 (12, 22, 26), close to the binding site for H3tail 14KAPR17 in our structure (Fig. 3E, Top). Our IMP–H3–H4–ASF1 cryo-EM structure is also different from an integrative model that was built based on cross-linking and negative-stained EM data (22). In the integrative model, IMP4 adopts a more open conformation and binds mostly only to the H3tail. The placement of the histone-fold domain and ASF1 close to the IMP4 C terminus is quite different from how the ASF1-bound histone-fold domain is clamped in the N-terminal half of IMP4 in our cryo-EM structure.
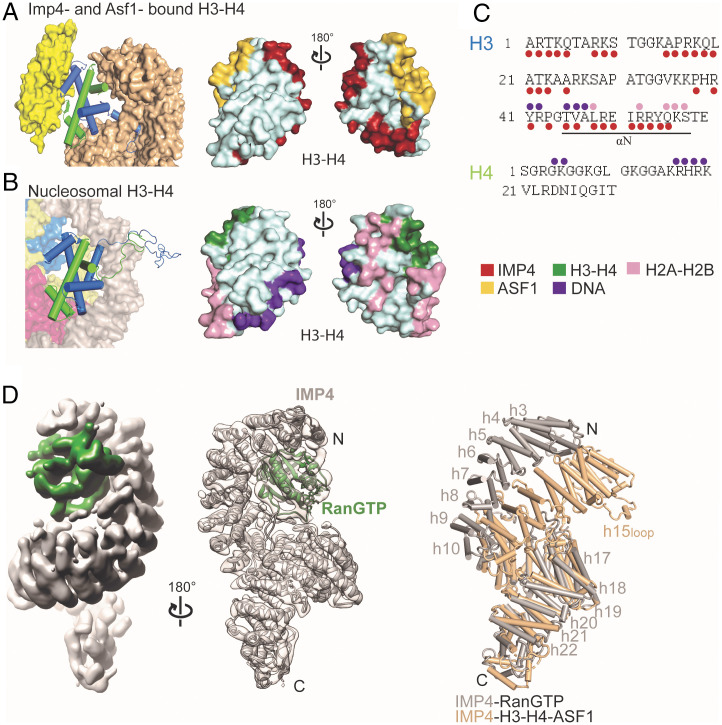
ASF1 binds the globular domain of H3–H4 but makes no direct contact with IMP4. The histone chaperone shields the site for H3–H4 tetramerization and likely also stabilizes the otherwise unstable globular fold of the H3–H4 dimer, allowing it to bind IMP4 (15, 27). IMP4 further shields the H2A–H2B and DNA sites of H3–H4 found in the nucleosome (Fig. 4 A–C). IMP4 and ASF1 appear to act in concert to shield much of the nucleosome interaction sites of H3–H4 (Fig. 4 A–C). This is yet another case of the relatively novel concept or example of histone cochaperone complexes, where multiple chaperones function together to optimize histone delivery from synthesis in the cytoplasm to nucleosome deposition (28). IMP4 is also another example of IMPORTINs acting as histone chaperones while transporting the very abundant and highly charged histones that are prone to aggregation; IMPORTIN-9 is an effective chaperone of its cargo H2A–H2B, and the IMPORTIN-7/IMPORTIN-β heterodimer effectively shields the linker histone H1 (17, 18).
Fig. 4.
IMP4 and ASF1 act together to shield H3–H4 surfaces, and large IMP4 conformational changes occur after RanGTP binding. (A) (Left), H3–H4 dimer (blue-green cartoon) bound to IMP4 (beige surface) and ASF1 (yellow surface). (Right), H3–H4 surface with IMP4 interface (red) and ASF1 interface (yellow). (B) (Left), one H3–H4 dimer (cartoon) in the nucleosome bound to DNA (gray surface), H2A–H2B (pink-yellow surfaces), and another H3–H4 dimer (blue-green surfaces). (Right), H3–H4 surface with the nucleosomal DNA interface (purple) and H2A–H2B interface (pink) and the H3–H4 interface (green). (C) H3tail and H4tail sequences; residues that bind IMP4 in the IMP4–H3–H4–ASF1 structure are marked with red circles, and residues that contact DNA and H2A–H2B in the nucleosome are marked with purple and pink circles, respectively. (D) (Left), the 7.1-Å cryo-EM map of the IMP4–RanGTP complex with IMP4 in gray and RanGTP in dark green. (Middle), cartoon model of the IMP4–RanGTP complex showing good fit in the transparent cryo-EM map. (Right), superposition of repeats h9 to h18 of IMP4–H3–H4–ASF1 (IMP4 is beige) and Imp4–RanGTP (IMP4 is gray) showing a large conformational change of the N-terminal half of IMP4 relative to its C-terminal half to accommodate RanGTP.
In the nucleus, RanGTP binds IMPORTINs with high affinity and dissociates H3–H4–ASF1 from IMP4, ending the nuclear import process (10, 29). We obtained a 7.1-Å-resolution cryo-EM map of the IMP4–RanGTP complex; the HEAT repeat helices of IMP4 can be easily modeled, and RanGTP could be reliably docked into the map by aligning the N-terminal HEAT repeats of IMP4 with the Kap121–RanGTP structure (PDBID: 3W3Z; Fig. 4D). Superposition of H3–H4–ASF1- and RanGTP-bound IMP4 proteins (Fig. 4D) and a PyMOL morph (Video S1) suggest large conformational differences in the IMP4 superhelix. HEAT repeats h2 to h8 (rmsd: 1.7 Å) and h10 to h18 (rmsd: 1.4 Å) act as two rigid bodies that move relative to each other about a hinge at h8 to h10.
The h15loop also undergoes a large conformational change in histone- versus Ran-bound IMP4. In the IMP4–H3–H4–ASF1 structure, h15loop is ordered and bound to both the N-terminal HEAT repeats and the H3–H4 globular domain (Figs. 1C and 2A). However, in the IMP4–RanGTP structure, the GTPase binds to the same IMP4 N-terminal HEAT repeats and displaces the h15loop; no density is observed for the loop (Fig. 4D). The different arrangements of the superhelix and the h15loop result in 1) a smaller N-terminal IMP4 arch that clamps the smaller H3–H4 domain along with a closed and tight central ring that the H3tail threads through in the histone-bound IMP4 (Fig. 1C) versus 2) a larger N-terminal IMP4 arch that binds RanGTP in the GTPase-bound IMP4 (Fig. 4D). Rearrangement of the Ran-bound superhelix also separates the central HEAT repeats and opens the central ring. Binding sites for H3–H4 are lost in this major rearrangement of the RanGTP-bound IMP4 superhelix.
In conclusion, the cryo-EM structure of IMP4–H3–H4–ASF1 shows the entire length of the IMP4 making extensive interactions with the H3–H4 dimer, which also binds to and is stabilized by ASF1. The N-terminal half of IMP4 binds the H3–H4 histone-fold domain and the αNH3 helix, while the C-terminal half of IMP4 binds the H3tail. We showed that despite interactions with IMP4, the H3tail is dispensable for formation of the IMP4–H3–H4–ASF1 complex. Finally, large conformational changes of the IMP4 superhelix and the long h15loop allow RanGTP to bind IMP4 and release H3–H4–ASF1 into the nucleus.
Methods
Protein Constructs.
The gene encoding the full-length human IMP4 was cloned into modified pGEX4t3 and pMALHis6 vectors that contain TEV protease cleavage sites (30, 31). The cDNA fragment corresponding to yeast ASF1 (1 to 160) was cloned into a pMALHis6 vector. Mutant and wt X. laevis H3 and H4 were cloned into pET3a, and the H4E63C construct was obtained from The Histone Source (Colorado State University). Truncated yeast Ran (residues 1 to 179) with the Q71L mutation to prevent GTP hydrolysis was cloned in pET21d (32). Sequence alignments of ASF1 and histones H3 and H4 (SI Appendix, Fig. S6) were performed in Clustal Omega (33), and the image of the alignments was generated by ENDscript (34).
Protein Expression and Purification.
IMP4.
IMP4 was expressed as maltose binding protein (MBP)–IMP4 or glutathione S-transferase (GST)–IMP4 in Escherichia coli BL21 cells grown in lysogeny broth (LB) medium and induced by adding 0.5 mM of isopropyl β-d-1-thiogalactopyranoside (IPTG) for 12 h at 25 °C. Cells were harvested, resuspended in buffer containing 50 mM Tris-HCl (pH 7.5), 500 mM NaCl, 2 mM ethylenediamine tetraacetic acid (EDTA), 20% glycerol, and protease inhibitors and lysed by high-pressure cell disruption. After centrifugation of the lysate, the supernatant was collected for purification. The supernatant was incubated with amylose resins (New England BioLabs) or glutathione resins (Cytiva), which were later washed several times with buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 20% glycerol. GST–IMP4 was then cleaved with tobacco etch virus (TEV) protease (4 °C for 12 h) to remove the GST tag, and IMP4 was eluted in the same buffer. MBP–IMP4 was eluted from the amylose resin by adding 30 mM d-maltose (Sigma) to the elution buffer, and the eluted sample was further purified by ion exchange chromatography using an HP-Q column followed by size-exclusion chromatography using a Superdex 200 Increase 10/300 GL column (Cytiva) that was preequilibrated with buffer containing 50 mM Tris-HCl (pH 7.5), 500 mM NaCl, and 20% glycerol. Peaks corresponding to IMP4 or MBP–IMP4 were collected and concentrated for the experiments.
ASF1.
Supernatant containing ASF1 (1 to 160) was incubated with amylose resin (New England BioLabs), washed with buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 20% glycerol, and eluted by cleavage with TEV protease. The eluted ASF1 sample was further purified by anion exchange using an HiTrap Q HP column (Cytiva) and concentrated for injection onto a size-exclusion column that was preequilibrated with buffer containing 30 mM Tris (pH 8.0), 150 mM NaCl, and 10% glycerol.
H3–H4.
Wt and mutant X. laevis H3 and H4 were expressed in E. coli BL21 (DE3) pLysS cells by inducing expression with the addition of 0.5 mM IPTG to the 2XYT (yeast extract and tryptone) medium for 4 h at 25 °C. After lysis by sonication in a buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2 mM EDTA, and protease inhibitors, followed by centrifugation, the pellet was washed several times and resuspended with denaturing buffer (7 M guanidine HCl, 50 mM Tris-HCl [pH 7.5], and 10 mM dithiothreitol [DTT]) and dialyzed overnight in buffer containing 7 M urea, 20 mM sodium acetate [pH 5.2], 200 mM NaCl, 1 mM EDTA, and 5 mM β-mercaptoethanol. Unfolded H3 and H4 proteins were purified separately by cation exchange chromatography using a cation exchange column (HiTrap SP HP, Cytiva) in SAU buffer with a gradient of 200 to 600 mM NaCl. (H3–H4)2 tetramers were formed by mixing H3 and H4 in an equimolar ratio, followed by dialysis overnight in a buffer without guanidine HCl (50 mM Tris-HCl [pH 7.5], 2 M NaCl, and 5 mM β-mercaptoethanol). H3–H4 tetramers were purified by size-exclusion chromatography in a Superdex 200 column. The ASF1–H3–H4 complex was formed by mixing a 1:1 molar ratio of ASF1 (1 to 160) and (H3–H4)2 tetramers. The ASF1–H3–H4 complex was purified by size-exclusion chromatography, which removed excess ASF1. The complex IMP4–H3–H4–ASF1 was formed by incubating IMP4 and ASF1–H3–H4 in a 1:1 molar ratio for 30 min and purified by size-exclusion chromatography. Fractions were concentrated to 5 to 10 mg/mL for cryo-EM sample grid preparation.
GSP1.
Sc Ran (1 to 179, Q71L) was expressed in E. coli BL21 (DE3) by induction with 0.5 mM IPTG for 12 h at 20 °C. Cell were harvested and lysed by high-pressure cell disruption, and the supernatant was incubated with nickel nitrilotriacetate agarose resins (Qiagen) and eluted in buffer containing 50 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4), 100 mM NaCl, 10% glycerol, 2 mM MgOAc2, 2 mM DTT, and 500 mM imidazole. To load the GTPase with GTP, RAN was first incubated with 7 mM EDTA for 30 min, followed by a 30-min incubation with 20 mM MgOAc2 and 7 mM GTP. RANGTP was further purified by cation exchange chromatography using an SP column equilibrated with 20 mM HEPES (pH 7.4), 4 mM MgOAc, 1 mM DTT, and 10% glycerol and eluted with a gradient of 0 to 1 M NaCl. To form the IMP4–RANGTP complex, proteins were incubated in a 1:1 molar ratio for 30 min and purified by size-exclusion chromatography.
Cryo-EM Grid Preparation, Data Collection, and Data Processing.
Samples of IMP4–H3–H4–ASF1 and IMP4–RANGTP were buffer exchanged into a final buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 0.1% Nonidet P-40 at a final protein concentration of 3 mg/mL; 3.5 µL of the IMP4–H3–H4–ASF1 or IMP4–RANGTP samples were applied on holey carbon grids (Quantifoil R1.2/1.3, 300 mesh copper) that were previously glow discharged using a PELCO easiGlow unit (Ted Pella) for 60 s at 30 mA and frozen using the Vitrobot Mark IV system (Thermo Fisher). The grids with the best particle distribution were submitted to 24-h data collection. Datasets of both of IMP4–H3–H4–ASF1 and IMP4–RANGTP complexes used for final data processing were collected at the Cryo Electron Microscopy Facility at University of Texas Southwestern on a Titan Krios microscope (Thermo Fisher) operating at 300 kV with a postcolumn energy filter (Gatan) and a K3 direct detection camera (Gatan) using SerialEM (35). For the IMP4–H3–H4–ASF1 complex, 4,126 movies were acquired at a pixel size of 0.415 Å in superresolution counting mode, with an accumulated total dose of 52 e−/Å2 over 40 frames. The defocus range of the images was set to be −1.0 to −2.5 μm. For the IMP4–RANGTP complex, 2,933 movies were acquired at a pixel size of 0.545 Å in superresolution counting mode, with an accumulated total dose of 50 e−/Å2 over 50 frames. The defocus range of the images was set to be −1.2 to −2.7 μm.
All data processing was performed using the software cryoSPARC v3.3.1 (36). A 1/2 F-crop factor was applied during motion correction, followed by patch contrast transfer function (CTF) estimation. A small subset of ∼20 frames was used to generate the initial template for particle picking. For the IMP4–H3–H4–ASF1 complex, initially, 2,636,349 particles were extracted from all the micrographs; 217,815 particles were selected after the initial round of two-dimensional (2D) classification, and 146,050 particles were included after an additional four rounds of 2D classification for ab initio modeling, followed by heterogeneous refinement. Nonuniform refinement was carried out to generate the final 3.45-Å-resolution map. For the IMP4–RANGTP complex, 1,226,438 particles were extracted from all the micrographs. The first rounds of 2D classification showed high heterogeneity of the particles due to aggregation issues during sample preparation. Therefore, only 16,089 particles were selected after nine rounds of 2D classification and used to generate ab initio models, followed by heterogeneous refinement. Homogeneous refinement led to the final 7.1-Å-resolution map.
Cryo-EM Model Building, Refinement, and Analysis.
IMP4–H3–H4–ASF1.
The ASF1–H3–H4 (PDBID: 2HUE) (15) and KAP121 (PDBID: 3W3T) (37) structures were used to build the model of ASF1–H3–H4–IMP4. SWISS-MODEL was used to generate a three-dimensional (3D) model of IMP4 using KAP121 as template. The models were first docked into the cryo-EM map using UCSF Chimera (38), followed by multiples cycles of manual model building and refinement using Coot (39). The final model was subjected to real-space refinement in Phenix (40). The binding interfaces were then analyzed using CONTACT/ACT (41) with a contacts cutoff of 4.0 Å. These interactions were curated and analyzed in PyMOL, where the final figures were generated (42).
IMP4–RANGTP.
IMP4 was modeled into the cryo-EM map using IMP4 from our IMP4–H3–H4–ASF1 structure. RANGTP was docked into the cryo-EM map, using the PDB coordinates of KAP121–RANGTP as a template, by aligning the HEAT repeats h1 to h4 of KAP121 and IMP4. The models were submitted to cycles of model building and refinement using Coot and Phenix (39, 40).
Fluorescence Polarization Assays.
KD values of IMP4 binding to H3–H4–ASF1 complexes were obtained by fluorescence polarization assays, as previously described (32). For these assays, the H4E63C mutant protein (The Histone Source) was labeled with the XFD488 fluorophore (AAT Bioquest) by mixing H4E63C and the dye at a 1:4 molar ratio in the histone unfolding buffer (50 mM Tris-HCl [pH 7.5] and 5 M guanidine-HCl) without DTT for 4 h at 25 °C in the dark. Labeling efficiency was further confirmed by intact mass spectrometry. H3–H4E63C tetramers, ASF1–H3wt–H4E63C, and ASF1–H331 to 135–H4E63C were formed as for the wt complexes. MBP–IMP4 and the preassembled ASF1–H3–H4 complexes were separately dialyzed in buffer containing 50 mM HEPES (pH 7.5), 300 mM NaCl, and 2 mM MgCl2. MBP–IMP4 was serially diluted from 10 μM to 0.3 nM in the presence of 40 nM ASF1–H3wt–H4E63C or ASF1–H331 to 135–H4E63C in a 384-well microplate (Corning). Fluorescence signal for each titration was measured, and data were processed in, PALMIST (43) using averages of triplicate experiments. Final figures were generated with GUSSI (44). Data were processed in PALMIST (43) using averages of triplicate experiments, and the final figures were generated with GUSSI (44).
Supplementary Material
Acknowledgments
We thank Michael Soniat for the important biochemical studies that led to this work and Sheena D'Arcy for discussions. We thank the Structural Biology Laboratory and the Cryo-EM Facility at University of Texas Southwestern, which are partially supported by grant RP170644 from the Cancer Prevention & Research Institute of Texas, for cryo-EM studies and their assistance with cryo-EM data collection. We also thank the Erzenberger lab for the use of their equipment and the plate reader. This work was funded by the National Institute of General Medical Sciences of the NIH under awards R35GM141461 (Y.M.C.) and R01GM069909 (Y.M.C.), the Welch Foundation Grants I-1532 (Y.M.C.), support from the Alfred and Mabel Gilman Chair in Molecular Pharmacology, Eugene McDermott Scholar in Biomedical Research (Y.M.C.), Mary Kay International Postdoctoral Fellowship (N.E.B.), and the Gilman Special Opportunities Award (H.Y.J.F.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2207177119/-/DCSupplemental.
Data, Materials, and Software Availability
Data have been deposited in PDB (7UNK and 8DYO) (45, 46) and Electron Microscopy Data Bank (EMD-26625 and EMD-27780) (47, 48). All other data are included in the manuscript and/or supporting information.
References
- 1.Campos E. I., et al. , The program for processing newly synthesized histones H3.1 and H4. Nat. Struct. Mol. Biol. 17, 1343–1351 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y., Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Benson L. J., et al. , Modifications of H3 and H4 during chromatin replication, nucleosome assembly, and histone exchange. J. Biol. Chem. 281, 9287–9296 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Baake M., Doenecke D., Albig W., Characterisation of nuclear localisation signals of the four human core histones. J. Cell. Biochem. 81, 333–346 (2001). [PubMed] [Google Scholar]
- 5.Mühlhäusser P., Müller E. C., Otto A., Kutay U., Multiple pathways contribute to nuclear import of core histones. EMBO Rep. 2, 690–696 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwamborn K., Albig W., Doenecke D., The histone H1° contains multiple sequence elements for nuclear targeting. Exp. Cell Res. 244, 206–217 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Pardal A. J., Fernandes-Duarte F., Bowman A. J., The histone chaperoning pathway: From ribosome to nucleosome. Essays Biochem. 63, 29–43 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alvarez F., et al. , Sequential establishment of marks on soluble histones H3 and H4. J. Biol. Chem. 286, 17714–17721 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jasencakova Z., et al. , Replication stress interferes with histone recycling and predeposition marking of new histones. Mol. Cell 37, 736–743 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Soniat M., Cağatay T., Chook Y. M., Recognition elements in the histone H3 and H4 tails for seven different importins. J. Biol. Chem. 291, 21171–21183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mosammaparast N., Guo Y., Shabanowitz J., Hunt D. F., Pemberton L. F., Pathways mediating the nuclear import of histones H3 and H4 in yeast. J. Biol. Chem. 277, 862–868 (2002). [DOI] [PubMed] [Google Scholar]
- 12.An S., Yoon J., Kim H., Song J. J., Cho U. S., Structure-based nuclear import mechanism of histones H3 and H4 mediated by Kap123. eLife 6, e30244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blackwell J. S. Jr., Wilkinson S. T., Mosammaparast N., Pemberton L. F., Mutational analysis of H3 and H4 N termini reveals distinct roles in nuclear import. J. Biol. Chem. 282, 20142–20150 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Apta-Smith M. J., Hernandez-Fernaud J. R., Bowman A. J., Evidence for the nuclear import of histones H3.1 and H4 as monomers. EMBO J. 37, e98714 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.English C. M., Adkins M. W., Carson J. J., Churchill M. E., Tyler J. K., Structural basis for the histone chaperone activity of Asf1. Cell 127, 495–508 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sankhala R. S., et al. , Three-dimensional context rather than NLS amino acid sequence determines importin α subtype specificity for RCC1. Nat. Commun. 8, 979 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Padavannil A., et al. , Importin-9 wraps around the H2A-H2B core to act as nuclear importer and histone chaperone. eLife 8, e43630 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivic N., et al. , Fuzzy interactions form and shape the histone transport complex. Mol. Cell 73, 1191–1203 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu W., et al. , Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1. Cell Host Microbe 16, 187–200 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee S. J., et al. , The structure of importin-beta bound to SREBP-2: Nuclear import of a transcription factor. Science 302, 1571–1575 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J., Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Yoon J., et al. , Integrative structural investigation on the architecture of human Importin4_histone H3/H4_Asf1a complex and its histone H3 tail binding. J. Mol. Biol. 430, 822–841 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Zhang H., Han J., Kang B., Burgess R., Zhang Z., Human histone acetyltransferase 1 protein preferentially acetylates H4 histone molecules in H3.1-H4 over H3.3-H4. J. Biol. Chem. 287, 6573–6581 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sobel R. E., Cook R. G., Perry C. A., Annunziato A. T., Allis C. D., Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl. Acad. Sci. U.S.A. 92, 1237–1241 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ejlassi A., Menil-Philippot V., Galvani A., Thiriet C., Histones H3 and H4 require their relevant amino-tails for efficient nuclear import and replication-coupled chromatin assembly in vivo. Sci. Rep. 7, 3050 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soniat M., Chook Y. M., Karyopherin-β2 recognition of a PY-NLS variant that lacks the proline-tyrosine motif. Structure 24, 1802–1809 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banks D. D., Gloss L. M., Equilibrium folding of the core histones: The H3-H4 tetramer is less stable than the H2A-H2B dimer. Biochemistry 42, 6827–6839 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Hammond C. M., Strømme C. B., Huang H., Patel D. J., Groth A., Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 18, 141–158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wing C. E., Fung H. Y. J., Chook Y. M., Karyopherin-mediated nucleocytoplasmic transport. Nat. Rev. Mol. Cell Biol. 23, 307–328 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chook Y. M., Blobel G., Structure of the nuclear transport complex karyopherin-β2-Ran • GppNHp. Nature 399, 230–237 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Chook Y. M., Jung A., Rosen M. K., Blobel G., Uncoupling Kapβ2 substrate dissociation and Ran binding. Biochemistry 41, 6955–6966 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Fung H. Y., Fu S. C., Brautigam C. A., Chook Y. M., Structural determinants of nuclear export signal orientation in binding to exportin CRM1. eLife 4, e10034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sievers F., et al. , Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robert X., Gouet P., Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi J., Matsuura Y., Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p. J. Mol. Biol. 425, 1852–1868 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Emsley P., New tools for ligand refinement and validation in coot and CCP4. Acta Crystallogr. A 74, A390 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeLano W. L., “PyMOL molecular viewer: Updates and refinements” in Abstracts of Papers of the American Chemical Society (American Chemical Society, 2009), vol. 238. [Google Scholar]
- 43.Scheuermann T. H., Padrick S. B., Gardner K. H., Brautigam C. A., On the acquisition and analysis of microscale thermophoresis data. Anal. Biochem. 496, 79–93 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brautigam C. A., Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 562, 109–133 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Bernades N. E., Fung H. Y. J., Li Y., Chen Z., Chook Y. M., Data from “Structure of IMPORTIN-4 bound to the H3–H4–ASF1 histone–histone chaperone complex.” Protein Data Bank. https://www.rcsb.org/pdb/explore/explore.do?structureId=7UNK. Deposited 11 April 2022. [DOI] [PMC free article] [PubMed]
- 46.Bernades N. E., Fung H. Y. J., Li Y., Chen Z., Chook Y. M., Cryo-EM structure of Importin-4 bound to RanGTP. Protein Data Bank. https://www.rcsb.org/pdb/explore/explore.do?structureId=8DYO. Deposited 4 August 2022. [Google Scholar]
- 47.Bernades N. E., Fung H. Y. J., Li Y., Chen Z., Chook Y. M., Data from “Structure of IMPORTIN-4 bound to the H3–H4–ASF1 histone–histone chaperone complex.” Electron Microscopy Data Bank. https://www.ebi.ac.uk/pdbe-srv/emsearch/atlas/26625_summary.html. Deposited 4 August 2022. [DOI] [PMC free article] [PubMed]
- 48.Bernades N. E., Fung H. Y. J., Li Y., Chen Z., Chook Y. M., Data from “Structure of IMPORTIN-4 bound to the H3–H4–ASF1 histone–histone chaperone complex.” Electron Microscopy Data Bank. http://www.ebi.ac.uk/pdbe-srv/emsearch/atlas/27780_summary.html. Deposited 4 August 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data have been deposited in PDB (7UNK and 8DYO) (45, 46) and Electron Microscopy Data Bank (EMD-26625 and EMD-27780) (47, 48). All other data are included in the manuscript and/or supporting information.