Significance
The carotenoid fucoxanthin gives diatoms, haptophytes, and kelps (brown algae) their distinct brown color by extending the spectral range of photosynthetic light harvesting. Despite the global abundance and pharmaceutical potential of fucoxanthin, its biosynthetic pathway has been elusive. We have identified key enzymes and previously unknown intermediates in the fucoxanthin biosynthetic pathway, enabling us to propose the complete pathway in diatoms and haptophytes. It is much more intricate than previously speculated and evolved by duplications of genes originally involved in the formation of photoprotective carotenoids. This evolutionary trajectory is further manifested by our observation that the intermediate diadinoxanthin serves two purposes: diatoxanthin biosynthesis for protective energy dissipation under excessive light and fucoxanthin biosynthesis for light harvesting under limiting light.
Keywords: fucoxanthin, biosynthesis, xanthophyll cycle, diatoms, haptophytes
Abstract
Fucoxanthin is a major light-harvesting pigment in ecologically important algae such as diatoms, haptophytes, and brown algae (Phaeophyceae). Therefore, it is a major driver of global primary productivity. Species of these algal groups are brown colored because the high amounts of fucoxanthin bound to the proteins of their photosynthetic machineries enable efficient absorption of green light. While the structure of these fucoxanthin-chlorophyll proteins has recently been resolved, the biosynthetic pathway of fucoxanthin is still unknown. Here, we identified two enzymes central to this pathway by generating corresponding knockout mutants of the diatom Phaeodactylum tricornutum that are green due to the lack of fucoxanthin. Complementation of the mutants with the native genes or orthologs from haptophytes restored fucoxanthin biosynthesis. We propose a complete biosynthetic path to fucoxanthin in diatoms and haptophytes based on the carotenoid intermediates identified in the mutants and in vitro biochemical assays. It is substantially more complex than anticipated and reveals diadinoxanthin metabolism as the central regulatory hub connecting the photoprotective xanthophyll cycle and the formation of fucoxanthin. Moreover, our data show that the pathway evolved by repeated duplication and neofunctionalization of genes for the xanthophyll cycle enzymes violaxanthin de-epoxidase and zeaxanthin epoxidase. Brown algae lack diadinoxanthin and the genes described here and instead use an alternative pathway predicted to involve fewer enzymes. Our work represents a major step forward in elucidating the biosynthesis of fucoxanthin and understanding the evolution, biogenesis, and regulation of the photosynthetic machinery in algae.
Carotenoids are essential constituents of photosynthetic organisms, serving vital functions by protecting their photosynthetic machineries against photodamage from overexcitation and the resultant reactive oxygen species (1). Besides their protective role, carotenoids are involved in photosynthetic light harvesting (2) and have important structural roles in the assembly and stabilization of photosynthetic pigment–protein complexes (3). While the carotenoid composition in photosynthetic tissues of land plants is rather uniform and highly conserved (4), the aquatic algae use a substantially more diverse spectrum of photosynthetic carotenoids (5). Carotenoids such as peridinin and fucoxanthin extend the spectral range of photosynthetic light harvesting by enabling the efficient absorption of the blue-green light prevailing in many marine habitats (6, 7). As a result, algae containing either of these carotenoids are brown colored.
Peridinin and the various fucoxanthin species are present only in algae with so-called complex plastids that were acquired by secondary or higher-order endosymbiosis involving an early red alga (8, 9). Cryptophyte algae are viewed as the most ancient phylum with complex “red” plastids, while diatoms and other ochrophyte algae (i.e., photosynthetic stramenopiles (10)) may have acquired their plastids from a cryptophytic endosymbiont, and haptophytes from an ochrophytic endosymbiont by serial transfers (11, 12).
The fucoxanthin-containing diatoms and brown algae and the hexanoyloxy- and butanoyloxy-fucoxanthin-containing haptophytes are among the most abundant algal groups in marine environments (13–15) and are estimated to contribute more than half of the global marine primary production (15–17). Thus, fucoxanthin and its derivatives are major drivers of marine photosynthesis (18). During the last decade, fucoxanthin has also become a subject of rising interest for nutraceutical and pharmaceutical applications (19–21). While the structures of the fucoxanthin-chlorophyll proteins (FCPs) in the photosynthetic membranes of diatoms have recently been resolved (22–24), the biosynthetic pathway of their distinct chromophore is still unknown. So far, only enzymes involved in early biosynthetic steps shared by carotenoids from land plants and most algae have been identified (25–27).
Previous studies indicated that the carotenoid diadinoxanthin can be converted into fucoxanthin in diatoms (28, 29). Diadinoxanthin is part of a photoprotective xanthophyll cycle operating similar to the well-studied violaxanthin cycle known from land plants (30). The violaxanthin cycle is catalyzed by the antagonistic enzymes violaxanthin de-epoxidase (VDE) (31) and zeaxanthin epoxidase (ZEP) (32). Under high light stress, VDE converts violaxanthin via the intermediate antheraxanthin into zeaxanthin, which in turn promotes the dissipation of excessive excitation energy in the photosynthetic apparatus (33). When light becomes limiting again, ZEP catalyzes the back reaction, re-enabling efficient harvesting of light for photosynthesis (30). Many algae, such as the diatoms and haptophytes, use the diadinoxanthin cycle instead (28, 30). Here, VDE is also called diadinoxanthin de-epoxidase and converts the monoepoxide diadinoxanthin into diatoxanthin that supports energy dissipation in a fashion similar to zeaxanthin in the violaxanthin cycle (30, 34).
The genomes of diatoms and other algae with complex red plastids encode multiple paralogs for VDE and ZEP (35–37), whose physiological functions have not yet been fully resolved. One of these paralogs, the VDE-like 1 (VDL1) protein, is absent from land plants and algae with primary plastids. It was recently shown to catalyze an early step in the biosynthesis of fucoxanthin and diadinoxanthin by converting violaxanthin into neoxanthin (37), suggesting functional diversification in the VDE and ZEP families. In contrast, neoxanthin formation in land plants depends on the unrelated ABA4 protein (also referred to as NSY) (38). The homolog of this protein in diatoms appears to have a role in carotenoid biosynthesis but is not specifically linked to neoxanthin formation (39). Therefore, we aimed to explore the possible involvement of other paralogs within the VDE and ZEP families in fucoxanthin biosynthesis. By generating targeted knockout (KO) mutants in the diatom Phaeodactylum tricornutum by CRISPR-Cas9, we identified VDL2 and ZEP1 as essential for fucoxanthin formation in diatoms and haptophytes, enabling us to deduce the complete fucoxanthin pathway in these algae.
Results and Discussion
Diatom Mutants with Targeted KO of VDL2 or ZEP1 Are Devoid of Fucoxanthin.
To knock out VDL2 (locus tag: PHATRDRAFT_45846; GenBank protein ID: XP_002180051.1) or ZEP1 (locus tag: PHATRDRAFT_45845; GenBank protein ID: XP_002180238.1), we aimed for CRISPR-Cas9-mediated homology-directed insertion of a Ble marker gene into the target gene. Mutants with successful genomic integration of the Ble cassette were identified by growing the transformants on zeocin-containing agar plates. Fucoxanthin contains a carbonyl group as part of its conjugated double-bond system that is responsible for the strongly red-shifted absorbance of fucoxanthin molecules located in more polar binding environments within the FCPs for efficient capture of green light (7, 22, 40, 41). Consequently, we speculated that mutant lines of P. tricornutum with impaired fucoxanthin biosynthesis caused by the disruption of a target gene by Ble integration should appear green instead of brown. For both VDL2 and ZEP1, we readily identified several green colonies, although the majority (>100) of transformant colonies appeared brown as wild type. We used PCR and sequencing to determine whether there is an association between the green appearance and disruption of both alleles of the target gene in the diploid genome of P. tricornutum. We performed these detailed analyses on five mutants from separate transformations for each gene target (see SI Appendix for full details).
The PCR method we applied for genotyping of the mutants used two different primer combinations (see Dataset S1A for primer sequences). One primer pair (blue primers in Fig. 1A and B) enabled reliable and specific amplification of diagnostic fragments of both wild-type alleles of the target gene from genomic DNA of P. tricornutum as confirmed by sequencing of the PCR products (SI Appendix, Figs. S2 and S3 and Text S1). Target specific insertion of our Ble construct into both alleles—yielding biallelic KOs—is expected to result in a single PCR band with this primer pair, representing a fragment 1.4 kB larger than the wild-type product, which would confirm the absence of intact wild-type alleles (Fig. 1A and B and SI Appendix, Fig. S1A and B). The second primer pair (red primers in Fig. 1A and B) was designed to amplify a chimeric DNA fragment that represents a fusion of the inserted Ble construct and genomic DNA downstream of the insert. Genomic DNA from transformants yielding a band with this primer combination should contain a site-specific Ble insertion in at least one of the two alleles of the target gene.
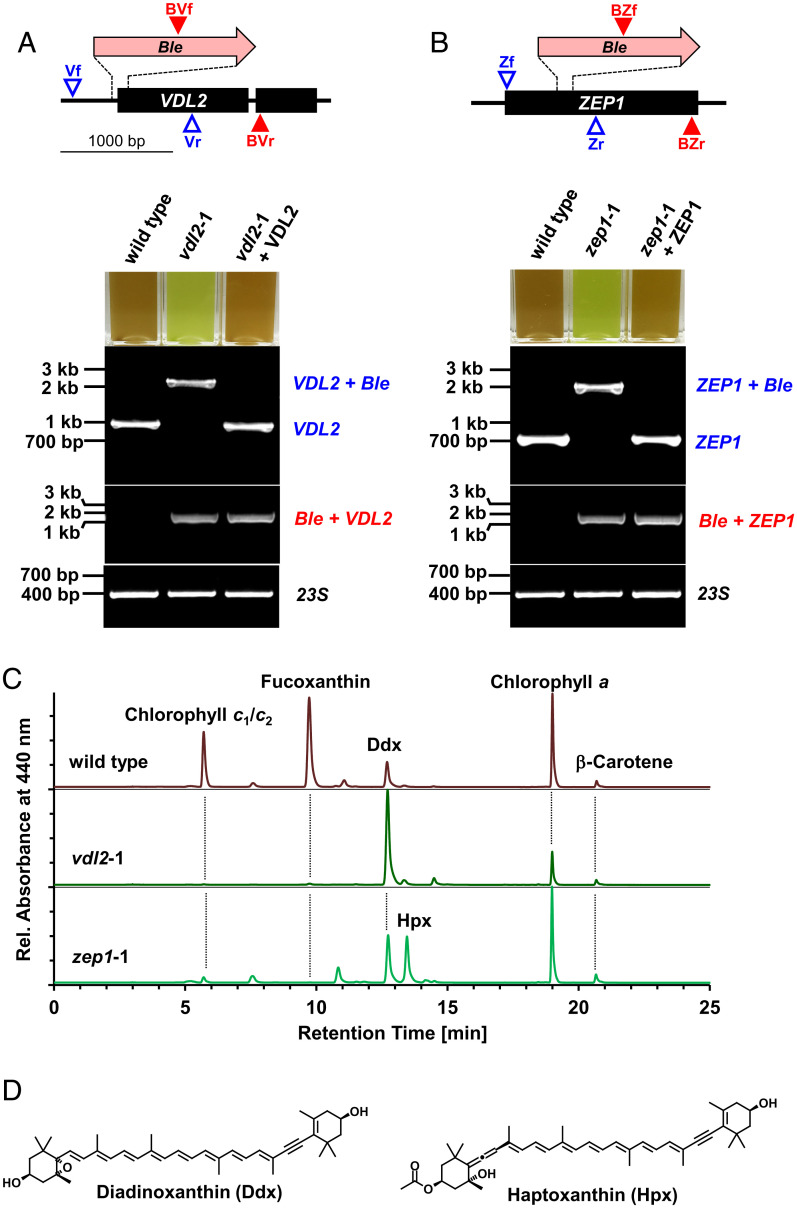
Fig. 1.
Molecular characterization and pigment phenotypes of vdl2- and zep1-knockout (KO) mutants from P. tricornutum. (A and B) Schemes showing insertion sites of the 1.4-kB zeocin resistance cassette (Ble) in the target genes; PCR primer binding sites for differentiation of wild-type (VDL2/ZEP1) and mutants (VDL2/ZEP1 + Ble) indicated by blue open triangles, for detection of the inserted Ble genes (Ble + VDL2/ZEP1) by red filled triangles; photographs show cuvettes with cultures of wild-type, mutants, and mutants complemented with randomly integrated native genes, and agarose gels of PCR products from genomic DNA confirming biallelic KOs (product colors correspond to colors of primers in the top schemes; amplification of fragments of reintroduced native genes was strongly favored over genes with Ble insertion; DNA controls amplified a fragment of the plastome-encoded 23S rRNA gene). (C) HPLC traces of pigment extracts from wild-type and the KO mutants. (D) Chemical structures of the pigments that show compensatory accumulation in the mutants.
In support of our assumption, PCR genotyping demonstrated that all green-colored mutants we investigated were biallelic KOs of the VDL2 or the ZEP1 gene (Fig. 1A and B and SI Appendix, Fig. S1A and B and Texts S1 and S3). We confirmed the predicted identity of the PCR products from DNA of all of these biallelic KO lines by sequencing (SI Appendix, Figs. S2 and S3 and Texts S1 and S3). We further validated our screening approach by also genotyping more than 10 randomly chosen brown colonies from the selective plates after transformation with the VDL2- or the ZEP1-KO constructs. None of these brown colonies were biallelic KOs (SI Appendix, Fig. S4 and Text S2). All five green mutants for each gene were devoid of fucoxanthin as confirmed by high-performance liquid chromatography (HPLC) (Fig. 1C and SI Appendix, Fig. S1C and D). Complementation of the two different mutant strains from P. tricornutum with the native genes encoding ZEP1 or VDL2, respectively, fully restored the brown appearance and the biosynthesis of fucoxanthin (Fig. 1A and B and SI Appendix, Fig. S1E). This result, together with the consistent phenotype between the multiple independent mutants, excludes the possibility that the pigment phenotypes resulted from off-target mutations caused by the CRISPR-Cas9 method or the transformation procedure.
VDL2- or ZEP1-Deficient Mutants Exhibit Defects in Photosystem II Antennae and Accumulate Other Carotenoids.
We used the mutants described above to reveal additional physiological consequences associated with fucoxanthin deficiency. Concomitant with the absence of fucoxanthin, both VDL2- and ZEP1-deficient mutants showed a severe reduction in the accessory chlorophylls c1 and c2 (Fig. 1C and SI Appendix, Fig. S1C and D). Because both fucoxanthin and chlorophyll c play a central role in capturing light in the photosynthetic antennae of diatoms (7), we investigated the loss of these pigments on light-harvesting efficiency. We observed that each mutant had a reduced functional antenna size associated with Photosystem II (σPSII) compared to wild-type controls (SI Appendix, Fig. S5). This shows the importance of fucoxanthin in facilitating photosynthesis in diatoms. The above-mentioned complemented lines were rescued in the accumulation of chlorophyll c1/c2 in addition to fucoxanthin (SI Appendix, Fig. S1E), proving that the loss of chlorophyll c1/c2 in the mutants was also correlated with the disruption of VDL2 or ZEP1. The parallel loss of fucoxanthin and chlorophylls c1 and c2 hints to a highly coordinated biosynthesis and incorporation of these pigments into the FCPs.
Mutants deficient in biosynthetic enzymes may correspondingly accumulate upstream precursor metabolites. The VDL2-deficient mutants showed a strong increase in diadinoxanthin, while the ZEP1-deficient mutants contained a carotenoid with absorbance properties similar to diadinoxanthin but with a longer retention time in our HPLC system (Fig. 1C and SI Appendix, Fig. S1 C and D). Mass spectrometry (MS) and NMR analyses identified this pigment as 7,8-didehydro-deepoxydinoxanthin (3′-ethanoyloxy-7,8,6′,7′-tetradehydro-5′,6′-dihydro-β,β-carotene-3,5′-diol) (Fig. 1D and SI Appendix, Figs. S6–S14, Table S1, and Text S4) for which we propose the trivial name haptoxanthin (after the Greek word “haptos,” meaning “touch” or “fasten,” as it was the first pigment of the pathway that we obtained and as it also is a likely intermediate of fucoxanthin biosynthesis in haptophytes).
The deduced biosynthetic pathway of fucoxanthin in diatoms.
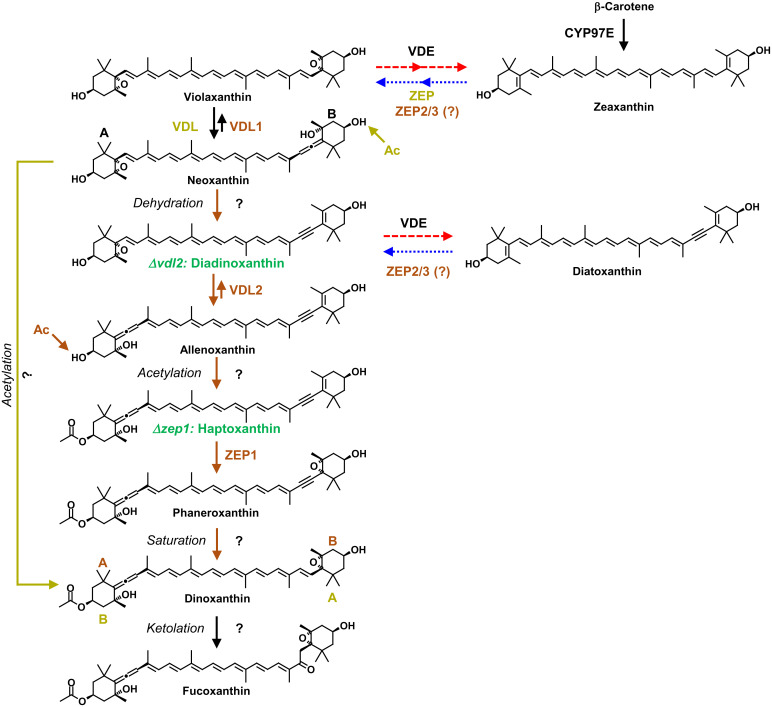
The identification of the carotenoid intermediates that specifically accumulated in the KO mutants enabled us to propose an unexpected biosynthetic pathway from violaxanthin to fucoxanthin in diatoms (Fig. 2). In the first step, VDL1 catalyzes the tautomerization of violaxanthin to neoxanthin, as shown previously (37). Elimination of water from neoxanthin by a yet unknown enzyme then yields diadinoxanthin. The strong accumulation of diadinoxanthin in the vdl2 mutant (Fig. 1C and SI Appendix, Fig. S1C) suggests this carotenoid to be the native substrate of VDL2. Based on the high sequence similarity of VDL1 and VDL2, we postulate that VDL2 catalyzes the tautomerization of diadinoxanthin to 7′,8′-didehydro-deepoxyneoxanthin (6,7,7′,8′-tetradehydro-5,6-dihydro-β,β-carotene-3,5,3′-triol) (Fig. 2), for which we propose the trivial name allenoxanthin, because it contains an allenic group and is not only a precursor of fucoxanthin but also a potential precursor of the xanthophyll alloxanthin in cryptophyte algae. The rationale is that the chemistry behind this reaction is identical to that of the conversion from violaxanthin to neoxanthin by VDL1, and allenoxanthin is similar to haptoxanthin except that it is nonacetylated.
Fig. 2.
Proposed pathways of fucoxanthin biosynthesis in algae. The fucoxanthin biosynthetic pathway and pathway-specific enzymes in algae with diadinoxanthin cycle such as diatoms and haptophytes are labeled in brown, those in algae with violaxanthin cycle such as brown algae are labeled in khaki; question marks denote the steps catalyzed by unknown enzymes. Intermediates labeled in green accumulate in the respective KO mutants of the diatom P. tricornutum (see Fig. 1C). Note that the acetylation steps (Ac) in both pathways are predicted to occur on opposite rings (A or B) of their last common precursor neoxanthin. Dashed red arrows denote high light-induced xanthophyll cycle activity, dotted blue arrows denote back reactions catalyzed in low light. VDE, violaxanthin de-epoxidase; VDL, violaxanthin de-epoxidase-like; ZEP, zeaxanthin epoxidase.
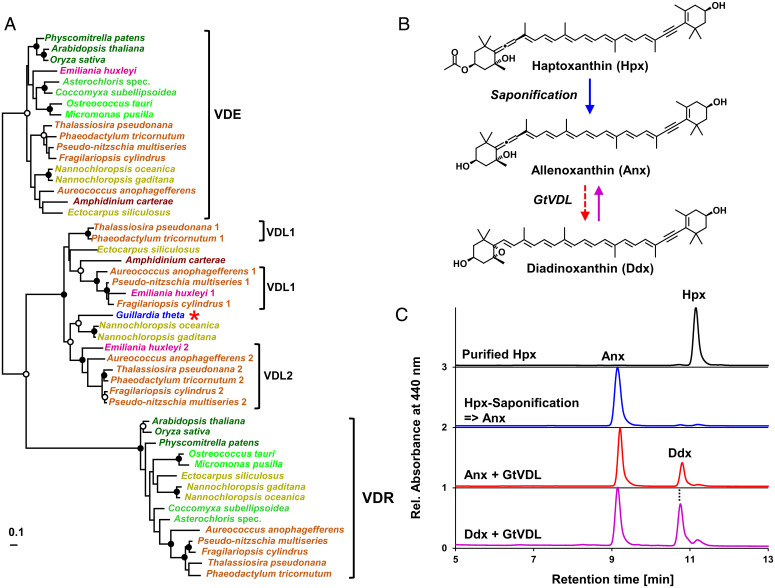
The VDL2- and ZEP1-deficient mutants enabled purification of pigment intermediates as substrates for in vitro enzyme activity assays. The proposed intermediate allenoxanthin could be prepared by saponification of haptoxanthin. In a previous study, we failed to detect the activity of recombinant VDL2 with diadinoxanthin in vitro (37). We now have found, however, that a VDL protein from the cryptophyte Guillardia theta (locus tag: GUITHDRAFT_109359; GenBank protein ID: XP_005831564.1; see also Dataset S1D), which is closely related to VDL1 and VDL2 from P. tricornutum and other diatoms (Fig. 3A and Dataset S2), was able to tautomerize diadinoxanthin to allenoxanthin (and vice versa) in vitro without the addition of any cosubstrate (Fig. 3B and C). This result demonstrates a flexibility in the substrate specificity of VDL proteins supporting the feasibility for VDL2 to catalyze the conversion of diadinoxanthin to allenoxanthin. We attribute the inactivity of VDL2 from P. tricornutum with diadinoxanthin in vitro to its tight regulation in vivo by an as-yet unknown factor that was absent from our in vitro assay. The fucoxanthin biosynthetic pathway proposed in Fig. 2 should indeed demand a tight regulation of VDL2 to enable the accumulation of a diadinoxanthin pool sufficiently large for photoprotective xanthophyll cycle activity.
Fig. 3.
The VDL protein from the cryptophyte G. theta (GtVDL) is closely related to the VDL2 protein from P. tricornutum and other diatoms and catalyzes the isomerization of allenoxanthin and diadinoxanthin in vitro. (A) Maximum likelihood tree showing the close phylogenetic relation of GtVDL (red asterisk) with VDL1 and VDL2 from diatoms and haptophytes (species names are color-coded according to their taxonomic affiliation: Viridiplantae, different shades of green; ochrophytes with Vx cycle, khaki; ochrophytes with fucoxanthin and Ddx cycle, light brown; Dinophyta, dark brown; Haptophyta, magenta; Cryptophyta, blue; bootstrap support (100 replicates) for nodes indicated by black dots if higher than 90%, by white dots if higher than 75%. (B) Reaction scheme showing saponification of haptoxanthin (Hpx) to allenoxanthin (Anx) by cleavage of the acetyl ester, followed by enzymatic isomerization of Anx to diadinoxanthin (Ddx) by GtVDL. (C) HPLC chromatograms of Hpx purified from the zep1 mutant of P. tricornutum (black trace) and of Anx resulting from saponification of Hpx (blue trace). Incubation of Anx for 2 h with recombinant GtVDL yielded a mixture of Anx and its tautomer Ddx (Anx + GtVDL, red trace); incubation of Ddx purified from wild type cells of P. tricornutum with GtVDL also resulted in a mixture of Anx and Ddx (Ddx + GtVDL, purple trace). The experiments were completed twice, with similar results.
Acetylation of allenoxanthin at position C3 by an unknown enzyme results in the formation of haptoxanthin. Previously, it was proposed that fucoxanthin formation in diatoms proceeds via the ketolation of neoxanthin to fucoxanthinol, which is then acetylated to form fucoxanthin as the final step in the pathway (SI Appendix, Fig. S15) (42). Our discovery of diadinoxanthin and the acetylated carotenoid haptoxanthin as intermediates in the biosynthesis of fucoxanthin disproves those previous speculations.
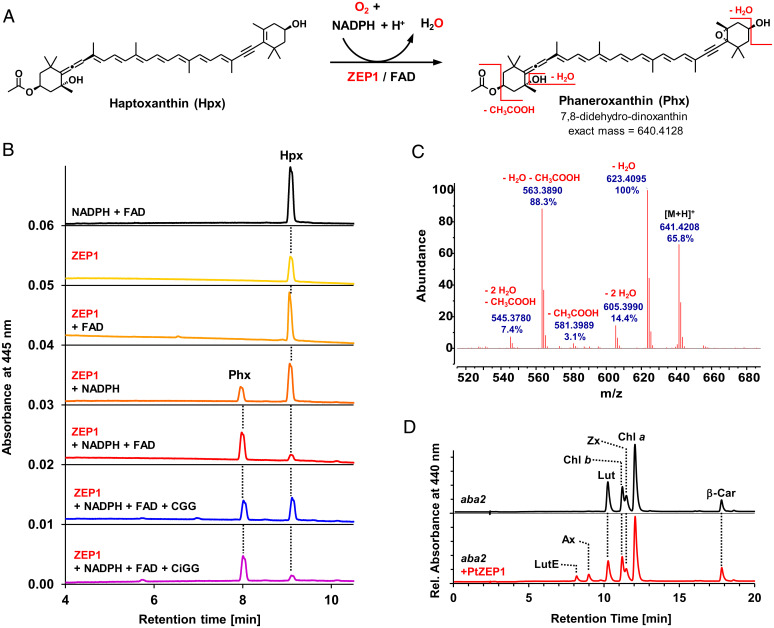
In the next step, ZEP1 performs the epoxidation of haptoxanthin to 7,8-didehydro-dinoxanthin (5,6-epoxy-3′-ethanoyloxy-7,8,6′,7′-tetradehydro-5,6,5′,6′-tetrahydro-β,β-carotene-3,5′-diol) for which we propose the trivial name phaneroxanthin (after the Greek word “phaneros,” meaning “visible” or “manifest,” because its identification manifested the proposed pathway of fucoxanthin biosynthesis). We could successfully demonstrate this reaction in vitro using recombinant ZEP1 of P. tricornutum isolated from Escherichia coli and haptoxanthin prepared from the zep1 mutant as substrate (Fig. 4). The ZEP from land plants uses molecular oxygen as a cosubstrate and nicotinamide adenine dinucleotide phosphate (NADPH) and enzyme-bound flavin adenine dinucleotide (FAD) as electron donors for C5,C6-epoxidation of zeaxanthin (43). Agrobacterium-mediated transient expression of ZEP1 from P. tricornutum in the leaves of the ZEP-deficient aba2 mutant of tobacco (Nicotiana plumbaginifolia) (32) resulted in the accumulation of low but detectable amounts of the monoepoxides antheraxanthin and lutein epoxide (Fig. 4D), demonstrating that ZEP1 also has C5,C6-epoxidase activity. In line with its conserved catalytic activity, ZEP1 activity was also dependent on the presence of NADPH and FAD (Fig. 4B). Moreover, removal of molecular oxygen from the ZEP1 assay by the addition of glucose oxidase, glucose and catalase to the reaction buffer resulted in a strong reduction in phaneroxanthin formation (Fig. 4B). This indicates that the epoxy oxygen introduced by ZEP1 also originates from molecular oxygen. The mass spectrum of the product from the ZEP1 in vitro assay was also in agreement with introduction of a C5,C6-epoxy group to haptoxanthin (Fig. 4C and SI Appendix, Fig. S16).
Fig. 4.
ZEP1 from P. tricornutum catalyzes the 5,6-epoxidation of haptoxanthin to phaneroxanthin, and requires FAD as prosthetic group and NADPH and molecular oxygen as cosubstrates. (A) Reaction scheme showing the epoxidation of Hpx to phaneroxanthin (Phx) and some characteristic MS fragments of the latter. (B) In the in vitro assays, Hpx was incubated in reaction buffer for 2 h at room temperature with different additives, followed by pigment extraction and HPLC analysis. The resulting chromatograms showed no enzymatic activity when only NADPH and FAD (black trace), only ZEP1 (yellow), or ZEP1 and FAD (light orange) were added. Addition of ZEP1 and NADPH (dark orange) resulted in partial conversion of Hpx to Phx, indicating that a fraction of the enzyme prepared from E. coli already contained FAD produced by the bacteria. Addition of ZEP1, NADPH, and FAD (red trace) led to almost complete conversion. Lowering dissolved oxygen in the assay by addition of catalase, glucose oxidase and glucose (CGG) resulted in a reduced conversion (blue trace), indicating that molecular oxygen is the donor of the epoxy group in Phx. When this experiment was repeated with heat-inactivated glucose oxidase (CiGG), the conversion was almost complete again (purple trace). (C) APCI-MS scan (positive ion mode) of Phx resulting from in vitro epoxidation of Hpx by ZEP1(48 spectra averaged; calculated mass of 641.4201 for [M+H]+); the differences between expected and measured masses were below 2 ppm (below 0.001 U) for all fragment peaks analyzed). (D) Transient expression of ZEP1 in leaves of the ZEP-deficient aba2 mutant of Nicotiana plumbaginifolia resulted in the formation of antheraxanthin (Ax) and lutein epoxide (LutE); other pigments: Car, carotene; Chl, chlorophyll; Lut, lutein; Zx, zeaxanthin.
The final steps from phaneroxanthin to fucoxanthin are speculative. Labeling experiments indicated dinoxanthin as an intermediate in the biosynthesis of peridinin in dinoflagellates (44) and it has been proposed to be a precursor of fucoxanthin as well, based on chemosystematic considerations and its tentative identification in diatoms (45, 46). Accordingly, we propose that the triple bond in phaneroxanthin is reduced to a double bond yielding dinoxanthin, which then is converted into fucoxanthin by ketolation at C8. As explained in more detail below, this proposal also allows for the most parsimonious scenario of evolution of fucoxanthin biosynthesis in brown algae.
Evidence for two different pathways to fucoxanthin in algae.
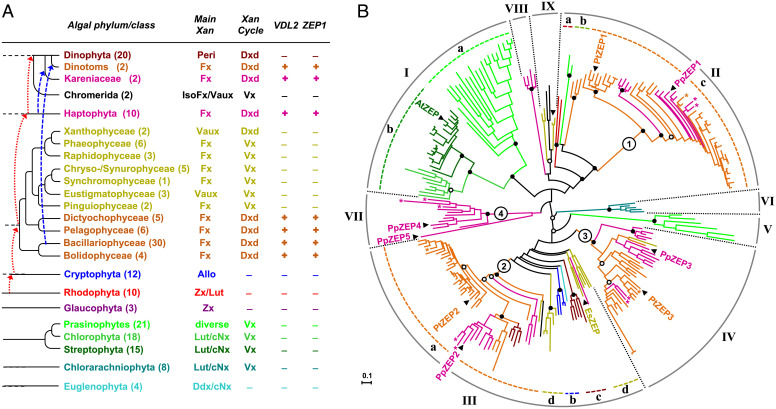
The similar carotenoid composition of diatoms and haptophytes, and particularly the shared presence of diadinoxanthin, implies that species of both phyla may use the same biosynthetic pathway to fucoxanthin. In line with this assumption, an investigation of publicly available genomic or transcriptomic data from haptophytes revealed putative orthologs of VDL2 and ZEP1 in all 10 species examined (Fig. 5A and Dataset S3). Moreover, expression of the VDL2 candidate gene from the haptophyte Emiliania huxleyi in the vdl2 mutant from P. tricornutum restored fucoxanthin biosynthesis, as did expression of the ZEP1 candidate from the haptophyte Prymnesium parvum in the zep1 mutant (SI Appendix, Fig. S17). These results strongly suggest that our findings for the diatom P. tricornutum also apply to haptophyte algae.
Fig. 5.
Comparative genomics of enzymes involved in fucoxanthin biosynthesis. (A) Occurrence of VDL2 and ZEP1 in publicly available sequence data from 194 species of photosynthetic eukaryotes (see Dataset S3 for species and sequence accessions). For each phylum/class, the number of examined species (in brackets), major light-harvesting xanthophyll(s) (Main Xan), and type of xanthophyll cycle (Xan cycle) are indicated. Branches indicate phylogenetic relations between taxa; dotted arrows in red and dashed arrows in blue indicate putative plastid acquisitions by endosymbioses. Allo, alloxanthin; cNx, cis-neoxanthin; Ddx, diadinoxanthin; IsoFx, isofucoxanthin; Lut, lutein; Peri, peridinin; Vaux, vaucheriaxanthin ester; Vx, violaxanthin; Zx, zeaxanthin. (B) Midpoint-rooted radial maximum-likelihood tree of the ZEP family from land plants and algae inferred from a protein alignment (324 amino acid positions) of 239 sequences from 121 species and using the same taxonomic color code as in (A) (sequences from Kareniaceae and dinotoms labeled with asterisks; see Dataset S4 for species and sequence accessions). Numbered circles indicate basal nodes of the main ZEP paralog clusters in ochrophytes and haptophytes. The paralogs from P. tricornutum (PtZEP1-3) and the haptophyte Prymnesium parvum (PpZEP1-5) and the single sequences from Arabidopsis thaliana (AtZEP) and the phaeophyte Ectocarpus siliculosus (EsZEP) are labeled. Bootstrap support (100 replicates) for major nodes indicated by black dots if higher than 90%, by white dots if higher than 75%.
A broader phylogenomic analysis of publicly available sequence data revealed that genes encoding ZEP1 and VDL2 are strictly confined to the group of algae that harbor the diadinoxanthin cycle and synthesize fucoxanthin (Fig. 5A and Dataset S3). Besides haptophytes, diatoms, and several other classes of ochrophyte algae, this group includes dinoflagellates that have replaced their ancestral peridinin-containing plastid with a haptophyte alga (Kareniaceae) or a diatom (so-called dinotoms) by tertiary endosymbiosis (47) (Fig. 5A and Dataset S3). In combination with the phenotypes of the KO mutants, our phylogenomic data thus demonstrate that diadinoxanthin is an essential intermediate in the formation of fucoxanthin in diatoms and all other fucoxanthin-containing algae using the diadinoxanthin cycle for photoprotection. However, orthologs of VDL2 and ZEP1 are absent from fucoxanthin-containing algae lacking diadinoxanthin, such as brown algae (Phaeophyceae) or golden-brown algae (Chrysophyceae). Consequently, these algae must synthesize fucoxanthin via an alternative route.
Based on chemosystematic considerations and the presence of minor amounts of fucoxanthinol (i.e., deacetylated fucoxanthin), it is commonly assumed that brown algae synthesize fucoxanthin by the ketolation of neoxanthin to fucoxanthinol, which is then acetylated to fucoxanthin (SI Appendix, Fig. S15) (48, 49). Diatoms also contain small amounts of fucoxanthinol (50) that, according to our findings, is not an intermediate in their pathway to fucoxanthin but only a minor side product. Therefore, fucoxanthinol in brown algae may also be a side product and fucoxanthin may instead be synthesized by the acetylation of neoxanthin to dinoxanthin followed by ketolation to fucoxanthin (SI Appendix, Fig. S15) (46). Accordingly, the pathways in diatoms and brown algae may split after neoxanthin and converge again at the level of dinoxanthin (Fig. 2), and the development of the hypothesized pathway in brown algae from that in diatoms would demand only an altered substrate specificity of the predicted acetyltransferase.
Physiological and evolutionary implications of the two fucoxanthin pathways.
The findings in this research provide biochemical and genetic evidence that diatoms and haptophytes can reuse photoprotective xanthophyll cycle pigments that accumulated during periods of high light stress for biosynthesis of their major light-harvesting xanthophylls under light-limited conditions (Fig. 2) (28, 29). The conversion of diatoxanthin or zeaxanthin to fucoxanthin consumes less energy than its de novo synthesis from intermediates of the Calvin cycle (51), thus reducing metabolic costs under energy limitation. Moreover, the observation that both violaxanthin and diadinoxanthin are obligate intermediates in the formation of fucoxanthin identifies VDL in algae with the violaxanthin cycle and VDL2 in algae with the diadinoxanthin cycle as gatekeepers controlling the flux from the photoprotective carotenoids of the respective xanthophyll cycle toward the light-harvesting xanthophyll fucoxanthin (Fig. 2).
VDL2 and ZEP1 evolved by the duplication of genes encoding for enzymes with more ancient roles in carotenoid biosynthesis, particularly in the photoprotective xanthophyll cycle. We exemplified this by a phylogenetic analysis of the ZEP family with broad taxon sampling (Fig. 5B and Dataset S4), revealing the presence of three ZEP paralogs in most diatoms and other ochrophytes with the diadinoxanthin cycle and up to five paralogs in haptophyte algae, whereas land plants contain only a single ZEP and green algae one or two ZEP candidates (Fig. 5B and Dataset S4). In the P. tricornutum genome, VDL2 and ZEP1 as well as VDE and ZEP3 are arranged next to each other, indicating that the two gene pairs evolved by duplication of a VDE/ZEP tandem (36). In support of this idea, we found the VDL2/ZEP1 cluster to be conserved in five and the VDE/ZEP3 cluster in four out of eight other diatom genomes (Dataset S5). These observations demonstrate that gene duplication and neofunctionalization have been major drivers in the evolution of carotenoid biosynthesis from photoprotection to photosynthetic light harvesting in algae.
Based on the current views on the phylogeny of ochrophyte algae (52, 53), early branching lineages such as Bacillariophyceae, Bolidophyceae, Pelagophyceae and Dictyochophyceae synthesize fucoxanthin via diadinoxanthin (Fig. 5A). Thus, the fucoxanthin biosynthetic pathway in the late-branching ochrophytes using the violaxanthin cycle may have evolved via the reduction of the probably more ancient pathway in algae with the diadinoxanthin cycle. Once the biosynthesis of fucoxanthin became independent of diadinoxanthin, several ochrophyte classes probably lost the gene necessary for diadinoxanthin formation, while xanthophyte algae may have retained it (Fig. 5A).
Materials and Methods
Algal and Plant Cultivation.
Wild-type and mutant lines of Phaeodactylum tricornutum strain CCAP 1055/1, and Guillardia theta CCMP2712 were grown in f/2 medium, Emiliania huxleyi SAG 33.90 in SWES medium, and Prymnesium parvum SAG 127.79 in 1/2SWES medium (https://www.uni-goettingen.de/de/list-of-media-and-recipes/186449.html). For semipreparative isolation of haptoxanthin for saponification to allenoxanthin and MS analyses, the zep1 mutant line of P. tricornutum was grown to high density in modified ASP-2 medium (28). Algae were grown at 18 °C and a photosynthetic photon flux density (PPFD) of 20 or (in the case of P. tricornutum) 40 to 80 µmol m−2 s−1 with a 16-h light/8-h dark cycle. For mutant isolation and pigment phenotyping, P. tricornutum was also grown on f/2 agar plates (1% w/v). The aba2 mutant of N. plumbaginifolia was grown in soil pots on irrigation trays containing Hoagland′s nutrient solution at 22 °C and a PPFD of 60 μmol m−2 s−1 with a 15-h light/9-h dark cycle.
Sequence Data Mining and Analyses.
Protein and nucleotide sequence data from algae available in GenBank (https://www.ncbi.nlm.nih.gov/), the Joint Genome Institute (JGI) Genome Portal (https://genome.jgi.doe.gov/portal/pages/tree-of-life.jsf), the Microbial Eukaryote Transcriptome Sequencing Project (54), and the Online Resource for Community Annotation of Eukaryotes (ORCAE; https://bioinformatics.psb.ugent.be/orcae/) were searched with the Basic Local Alignment Search Tool (BLAST) (55) using the amino acid sequences of the VDE and ZEP family members (35) from P. tricornutum as input. Protein sequences were aligned with CLUSTALW 1.83 (56) and manually refined using BIOEDIT 5.0.9 (57). Maximum likelihood (ML) trees were inferred from the protein alignments using PThreads in RAxML 8.0.14 (58) and the WAG substitution model (59) with gamma rate distribution (“PROTGAMMAWAG,” four discrete rate categories). Branch support was estimated with 100 bootstrap replicates using the rapid bootstrap algorithm (60) implemented in RAxML PThreads. The phylogenetic tree of the ZEP family was visualized using Dendroscope 3.5.8 (61), other trees were visualized in TreeView 1.6.6 (62); trees were reformatted using PowerPoint (Microsoft). Accessions of sequence data used for the in silico analyses are compiled in Datasets S2–S5.
Generation of Targeted-KO Lines from P. tricornutum.
CRISPR-Cas9-mediated homology directed repair was used to generate the KO mutants presented in Fig. 1. Guide sequences were designed by querying the exon sequence of the target gene for the motif (5′-N20NGG-3′) against the P. tricornutum genome (NCBI: txid556848) using the online design tool crispor.tefor.net (63). Oligo pairs composed of 20-bp complementary spacers and 5′ overhangs of TCGA (forward oligo) or AAAC (reverse oligo) were annealed in a 20-µL reaction mixture (5 µM each oligo, 1 × T4 ligase buffer [NEB]) by incubating at 95 °C for 2 min in a thermocycler and slowly cooling down to room temperature. The product was ligated into the BsaI-HFv2 (NEB) digested vector pKS diaCas9_sgRNA (Addgene #74923), which contains a diatom-codon optimized Cas9 expression cassette fused with the P. tricornutum LHCF2 promoter and LHCF1 terminator, and a single-guide RNA (sgRNA) cassette driven by the P. tricornutum U6 promoter (64). The homology donor plasmids were synthesized at Gene Universal on the backbone of vector pUC57. They were designed to include 1-kb upstream and downstream sequences of the guide RNA targeting site as the 5′ and 3′ homologous arms and a Ble gene (65) between the two arms controlled by the LHCF11 promoter and LHCF1 terminator to confer resistance to the antibiotic zeocin. Oligos and the homology arm sequences used for this design are listed in Dataset S1A.
The plasmids pKS diaCas9_sgRNA and the homology donor were linearized with KpnI-HF and NdeI (NEB) before the cotransformation into wild-type P. tricornutum via electroporation, as previously described (66). After 24 h low light incubation (∼30 µmol photons s−1 m−2) at 18 °C, electroporated cells were spread onto 1% agar f/2 plates with 100 µg/mL zeocin for selection of mutants successfully transformed with the Ble gene. A total of five independent transformations were performed for each target gene. Both mutants with green and brown appearances were isolated and restreaked at least three times before clones were analyzed for genotype (Fig. 1 and SI Appendix, Figs. S1–S4).
Rescue of KO Mutants with Native Genes.
For KO line complementation with the native genes, the genomic sequence, including the promoter and terminator, was PCR amplified from purified P. tricornutum genomic DNA, and together with the gene encoding blasticidin S deaminase (67) (BSD; gift of Christiaan Karreman) assembled in the pPtPuc3 expression vector flanked by the endogenous histone 4 promoter and LHCF1 terminator using gateway cloning (Thermo Fisher Scientific), resulting in pPtPuc3_pH4_BSD_LHCF1. The BSD expression cassette was PCR amplified using pPtPuc3_pH4_BSD_LHCF1 as a template. The target gene and BSD expression cassette PCR products were purified and coelectroporated into the corresponding KO mutant with a molar ratio of 1:1. Electroporations and selection were performed as described above, with one exception. Transformed suspension was spread onto f/2 agar plates supplemented with 10 µg/mL blasticidin S HCl (Thermo Fisher Scientific) for the selection of transformants that had incorporated the BSD gene. Five brown colonies each randomly picked from a separate transformation were verified in genotypes (SI Appendix, Fig. S1). Primers used for this purpose are listed in Dataset S1A.
Rescue of KO Mutants with Orthologs from Haptophytes.
The plasmid pPtPuc3_pNR_EYFP_nosT_H4_BSD_FcpA was used as the backbone of all of the rescuing plasmids. It contains the blasticidin resistance gene cassette described above and an enhanced yellow fluorescence protein (YFP) expression cassette. The plasmid was digested with AvrII and SacI-HF to remove the enhanced YPF. The insertion fragments with overlapping sequences were amplified by PCR and ligated into the digested backbone plasmid by using the GeneArt Gibson Assembly HiFi kit (Invitrogen), following the manufacturer’s instructions. Plasmids for heterologous rescue were linearized by restriction enzyme BspDI (NEB) before the electroporation transformation; subsequent methods of transformation and strain isolation were as described above.
The intron-less VDL2 gene of E. huxleyi (EhVDL2) was cloned from genomic DNA isolated using a modified cetyltrimethyl ammonium bromide DNA extraction protocol (68). The gene encoding ZEP1 from the haptophyte P. parvum (PpZEP1) was cloned from total RNA that was prepared with the InnuPREP Plant RNA kit (Analytik Jena). cDNA was synthesized from total RNA using the Transcriptor High Fidelity cDNA Synthesis kit (Roche Life Science) and an anchored-oligo(dT)18 primer. PCR-amplified genes and gene fragments for expression constructs were cloned into pGEM-T Easy (Promega GmbH) by PCR using Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific) with templates and gene-specific primers as listed in Dataset S1B.
Proper targeting of EhVDL2 and PpZEP1 into the secondary plastids of P. tricornutum was achieved by replacing the haptophyte targeting peptides with the native targeting signals of PtVDL2 or PtZEP1, respectively. The lengths and putative cleavage sites of the respective targeting sequences were predicted using SignalP 3.0 (69), SignalP 4.1 (70), and ChloroP 1.1 (71), as described previously (37). The protein sequences expressed by these constructs are also listed in Dataset S1B.
Mutant Genotyping.
Biomass from individual colonies was suspended in 20 µL lysis buffer (100 mmol/L Tris⋅HCl, pH 8.0, 1% sodium dodecyl sulfate, 100 mmol/L NaCl, 2% Triton X-100) and incubated at 85 °C for 10 min. Nuclease-free water at 80 µL was added after cooling to room temperature. A total of 3 µL of this crude genomic DNA was used as the template in a 1 × Gotaq Green Master Mix (Promega) PCR. Primers for genotyping are listed in Dataset S1C.
Expression of ZEP1 from P. tricornutum in Tobacco.
Transient expression of PtZEP1 in the ZEP-deficient aba2 mutant of N. tabacum was achieved by Agrobacterium-mediated transformation of leaves of the ZEP-deficient aba2 mutant of N. plumbaginifolia (32) using a pPZP200BAR-based (72) expression construct. A gene fragment encoding the C-terminal 522 amino acids of the mature protein was PCR amplified from cDNA using primers given in Dataset S1D and placed in-frame behind a fragment encoding the targeting sequence of the ZEP from A. thaliana for targeting of the protein to the plastid stroma as previously described (73). For transient expression, leaves of the upper part of 6- to 8-wk-old tobacco plants were infiltrated with Agrobacterium transformants harboring the expression construct, as previously described (73). Four days after infiltration, infiltrated and untreated control leaves were harvested, 1 to 3 leaf disks of 7-mm diameter taken per leaf, immediately frozen in liquid nitrogen, lyophilized, and stored at −20 °C until further analysis.
Production of Recombinant Algal Enzymes for In Vitro Assays.
Recombinant VDL from G. theta was generated by heterologous expression in E. coli Rosetta (Novagen) using pET-44a (Novagen). A 1,128-bp gene fragment of GtVDL encoding the mature protein and a short remainder of the predicted targeting sequence was cloned from cDNA generated as described for cloning of ZEP1 from haptophytes into the destination vector (see Dataset S1D). Expression in E. coli BL21(DE3) (Novagen), preparation, and renaturation of the recombinant VDL protein were performed as previously described (37, 74). For the generation of recombinant ZEP1 from P. tricornutum, the entire cDNA sequence of the gene was PCR amplified with primers listed in Dataset S1D and cloned by recombination with the seamless-cloning method (ClonExpress, Vazyme) into the pMAL-c5x vector (NEB), which encodes a maltose-binding protein fusion at the N terminus. The vector was then transformed into E. coli strain BL21(DE3) (Novagen). Induction of PtZEP1 expression was performed at 16 °C by supplementation of isopropyl thio-β-d-galactoside to a final concentration of 0.4 mmol/L. Bacterial cells were harvested 12 to 15 h later in precooled column buffer and ruptured by passage through a French press. The lysate was centrifuged at 13,000 × g for 15 min at 4 °C. The supernatant was collected for protein purification. Affinity chromatography of recombinant protein was carried out by use of an amylose resin column (NEB) according to the manufacturer’s instructions. Further purification was achieved by using SOURCE Q columns (anion and cation exchange) in an AKTA (GE Healthcare) system. Finally, the protein was enriched in a small volume (∼1 mL) by using Amicon MWCO 30KD Ultra tubes (Millipore). Protein concentration was determined using a bicinchoninic acid assay kit (Sangon Biotech) according to the manufacturer’s instructions.
In Vitro Assays with Recombinant Algal Enzymes.
In vitro assays with recombinant VDL protein from the cryptophyte G. theta (GtVDL) were carried out in reaction buffer at pH 5.2 containing 40 mmol/L MES, 10 mmol/L KCl, and 5 mmol/L MgCl2 and a final volume of 1 mL (75, 76). 10 µL of a 40-µmol/L ethanolic stock solution of the respective carotenoid substrate and 10 µL of a 1.16-mmol/L methanolic monogalactosyldiacylglycerol (Lipid Products) stock were thoroughly mixed in a 1.5-mL reaction tube, followed by the rapid addition of 920 µL reaction buffer and vortexing for 10 s. The reaction was started by the addition of 50 µL enzyme in buffer and vortexing for 10 s. Reaction tubes were incubated in a water bath at 20 °C. Reactions were stopped by the addition of 300 µL of 1 mol/L NaOH and incubation on ice. Pigment/lipid aggregates were harvested by centrifugation (18,000 × g, 2 min), the supernatant removed, the pellets incubated at −20 °C for 10 min, and then dissolved in 200 µL HPLC extraction medium (81.1% methanol [v/v], 10.8% ethyl acetate [v/v], 8.1% water [v/v], 180 mmol/L ammonium acetate) by vortexing and brief sonication. After centrifugation (18,000 × g, 2 min), pigments in the supernatant were analyzed by injecting 100 µL into HPLC system II (see below).
The conditions for the in vitro assay using recombinant ZEP1 protein were adopted from a previously published report on the ZEP from land plants (43). Purified carotenoids were dissolved in reaction buffer (0.1% Triton X-100, 100 mmol/L Tris, 10 mmol/L MgCl2, 1 mmol/L dithiothreitol, pH 7.4) with sonication. The final concentration of haptoxanthin in the assays was 0.19 μmol/L; final concentrations of other additives were as follows: FAD, 100 μmol/L; NADPH, 1 mmol/L. In addition, some assays contained CGG (4 μg/mL catalase, 30 μg/mL glucose oxidase, 150 μg/mL glucose) to lower dissolved oxygen. Enzyme assays were started with the addition of 50 μg purified enzymes into a 200-μL reaction buffer containing appropriate substrate and incubated at room temperature in the dark for 2 h. To end the reaction, 200 μL of acetone and 200 μL of diethyl ether were added to each reaction tube (or a 200-μL aliquot at each time point for the time course experiment) followed by centrifugation at 12,000 × g for 5 min. The upper, organic phase was collected, dried under vacuum, and redissolved in 50 μL 90% methanol for analysis using HPLC system III (see below).
HPLC Systems Used for Pigment Preparation and Analysis.
HPLC system I consisted of a Waters Alliance 2795 Separation Module coupled with a Waters 2996 photodiode-array detector (Waters), equipped with a ProntoSIL 200–5 C30, 5.0-µm, 250 × 4.6-mm column and a ProntoSIL 200–5-C30, 5.0-µm, 20 × 4.0-mm guard column (Bischoff Analysentechnik), and operated with a ternary gradient (eluent A: 85% aqueous methanol buffered with 75 mmol/L ammonium acetate, eluent B: 90% aqueous acetonitrile, eluent C: ethyl acetate) and other conditions as previously described (77). HPLC systems II and IIb were run on a Waters Alliance e2695 Separation Module with Waters 2998 photodiode-array detector, an EC 250/4 Nucleosil 300–5 C18 column protected by a CC 8/4 Nucleosil 300–5 C18 guard column (Macherey-Nagel) also applying ternary gradients (same eluents as for system I) with run times of either 20 min (II) or 25 min (IIb) and other conditions, as previously described (37, 73). HPLC system III consisted of a Waters Acquity ultra-performance LC (UPLC)-MS system with PDA and QDA detector and an Acquity UPLC HSS T3 1.8 μm (3 μm, 100 Å, 2.1 × 150 mm) column. The mobile phase was designed as phase A, consisting of a mixture of acetonitrile:methanol:methyl t-butyl ether (70:20:10,v:v:v), and phase B, which was 10 mmol/L ammonium acetate. The column temperature was 45 °C and the elution flow rate was 0.3 mL/min, with the following gradient with linear changes from 0 min: 60% A and 40% B to 4 min: 75% A and 25% B to 12 min: 100% A. The injection volume was set to 3 μL. HPLC system IV comprised a Thermo Ultimate 3000 UHPLC equipped with a UV/visible light detector and a C-18 column (5 μm, 100 Å, 250 × 4.6 mm, Waters). The pigments were eluted at 20 °C at a flow rate of 1 mL min−1 with the following steps: 0 to 20 min, linear gradient of buffer A (methanol:water = 90:10) from 100 to 0; 20 to 22 min, 100% buffer B (ethyl acetate); 22 to 23 min, linear gradient of buffer B from 100 to 0; and 23 to 28 min, 100% buffer A. The injection volume was set to 10 μL.
Analysis of Pigments from Algae and Tobacco.
For analysis of pigments from liquid cultures of wild-type and KO mutant lines from P. tricornutum, cells were harvested by filtering 5 mL of algal culture through glass fiber filters (GF6, Whatman), applying mild suction. Filter samples were immediately transferred to 1.5 mL cryovials, frozen in liquid nitrogen, and lyophilized. For extraction of the filters in the cryovials, 700 µL precooled extraction medium (ExMed: 81.1% methanol [v/v], 10.8% ethyl acetate [v/v], 8.1% water [v/v], 180 mmol/L ammonium acetate) and a spatula tip of glass beads (1 part 0.25 to 0.55 mm, 3 parts 0.75 to 1 mm, w/w) were added and the filters homogenized in a Mini-Beadbeater-1 (BioSpec) for 2 × 10 s at 5,000 rpm with intermittent cooling on ice. After centrifugation with 18,000 × g for 1 min, 50 to 100 µL of the supernatant were used for pigment analysis using HPLC system IIb. Pigments of complemented mutant lines were analyzed by extraction from colonies grown on agar plates. Using a pipette tip, algal material was transferred into a 1.5-mL reaction tube containing 500 µL precooled ExMed and a spatula tip of glass beads (1 part 0.25 to 0.55 mm, 3 parts 0.75 to 1 mm, w/w). Pigment extraction and further preparation for analysis using HPLC system II was performed as described for algae on lyophilized filters. Pigment extracts of the lyophilized tobacco leaf discs using precooled ExMed were produced as previously described (73) and analyzed using HPLC system I.
Preparation of Pigments for In Vitro Assays.
For in vitro assays with GtVDL, diadinoxanthin was isolated from the wild type and haptoxanthin from the ZEP1-deficient mutant of P. tricornutum. Cells from liquid cultures were harvested and homogenized in ExMed as described above, the pigments separated and collected using HPLC system II, extracted from the eluent by consecutive addition of equal volumes of diethyl ether and 5 mol/L NaCl, and the pigment-containing ether phase split into aliquots that were evaporated to dryness with nitrogen. Allenoxanthin for the GtVDL assay was prepared by saponification of haptoxanthin that was dissolved in 1 mL diethyl ether, mixed with 1 mL ethanolic 5% KOH solution by sonification, and incubated in the dark at 25 °C for 2 h. Allenoxanthin was separated from the reaction mixture by washing 4 times with 2 mL of 5 mol/L NaCl followed by evaporation of the pigment-containing ether phase to dryness with nitrogen. Haptoxanthin for the ZEP1 assays were aliquots from the samples used for NMR analysis (see below).
Preparation and Analysis of Pigments for MS and NMR.
For MS of haptoxanthin and its saponification product, pigments were prepared by consecutive purification on HPLC systems I and II. Multiple aliquots of pigment extracts from 400-mL batch cultures of the ZEP1-deficient mutant were separated on HPLC system II, the pigments of interest collected, and, after phase separation into diethyl ether, dried by flushing with nitrogen. Pigments were redissolved in ExMed, injected to HPLC system I, and the collected pigment fractions again purified by phase separation into diethyl ether and subsequent evaporation to dryness. For saponification, an aliquot containing 10 mg of the purified haptoxanthin was dissolved in 2 mL diethyl ether, mixed with 2 mL ethanolic 5% KOH solution by sonification, and incubated in the dark at 25 °C for 8 h. The saponification products were purified by phase separation into diethyl ether and dried as described.
Haptoxanthin used for structural characterization by NMR was prepared from an additional ZEP1 mutant strain of P. tricornutum generated in the background of Xiaobo Li’s laboratory strain XLF5 by an episome-based CRISPR-Cas9 procedure. Briefly, a vector was generated to include the Cas9 gene derived from Addgene plasmid #109219 (78) and driven by the promoter of γ-tubulin (79), the Blasticidin S Deaminase gene (80) driven by the promoter of the gene encoding the fucoxanthin-chlorophyll protein FCPb (79), and the P. tricornutum U6 promoter (79) to drive the expression of guide RNAs. Design of the guide sequences, annealing of primers, and ligation of the annealed product into the CRISPR plasmid were similar to the methods described above to generate the mutants used in Fig. 1 (see Dataset S1E for oligos used). The conjugation-facilitating plasmid pTA-Mob (81) and the CRISPR plasmids containing Cas9 and guide RNA expression cassettes were sequentially transformed into the E. coli strain EPI300. Conjugation between E. coli and P. tricornutum was performed using previously described methods (82). Approximately 10 d later, transformant colonies with a green-yellow appearance were isolated and restreaked at least 3 times on the same media before clones were subjected to PCR (primers listed in Dataset S1E) and sequencing. Clones that yielded uniform sequencing results were regarded as biallelic mutants and chosen for phenotyping by HPLC system IV. The characterization of a representative ZEP1 mutant (zep1-b) is shown in SI Appendix, Fig. S18. Pigments from the mutant lines (∼1 × 107 cells/sample) were extracted with 250 μL cold 90% acetone (v/v) by treatment in an ultrasonic cleaner for 10 min in the dark. After centrifugation, the supernatant was collected and used to determine the pigment concentration using HPLC system IV. Absorbance of the elute was detected at 445 nm, as previously described, to verify their identities (83). Collected pigments were dried under a nitrogen stream and stored under −80 °C.
High-Resolution MS.
For high-resolution MS of haptoxanthin and its saponification product allenoxanthin, 10 to 15 µg dried pigment was dissolved in 100 μL acetonitrile, diluted by 1:200 with acetonitrile and 10 μL injected via the autosampler of a 1260 Infinity II HPLC system (Agilent Technologies) and using an eluent composed of 20% A (2% acetonitrile, 98% H2O) and 80% B (98% acetonitrile, 2% H2O + 0.1% formic acid) into a 6545 QTOF mass spectrometer (Agilent Technologies) equipped with an Agilent atmospheric pressure chemical ionization (APCI) source. The drying gas temperature was set to 300 °C with a flow rate of 8 mL/min, the vaporizer temperature was 350 °C, and the nebulizer pressure was 20 psig. High-purity nitrogen was used as the nebulizer, drying, and collision gas. The corona current was 4 μA in positive ion mode, the capillary voltage was set to 3,000 V, and the fragmentor voltage was 50 V. For MS/MS, the fragmentor voltage was increased to 65 V and the collision energy set to 15 V. The scan range for MS was m/z 100 to 2,100, with a scan rate of 1 spectrum/s. For MS/MS, the range was m/z 50 to 1,000, with a scan rate of 4 spectra/s. For MS/MS analysis of haptoxanthin, precursors were selected with an isolation width of 1.5 m/z, while for its saponification product, the isolation width was 4 m/z. MS data were recorded using the Agilent MassHunter LC/MS Data Acquisition B.08.00 software, and the mass spectra for publication prepared with MestReNova version 14.0.1 (Mestrelab Research).
High-resolution MS analysis of phaneroxanthin synthesized from haptoxanthin by ZEP1 in vitro was performed on an UPLC-time-of-flight mass spectrometer (Synapt-G2-Si, Waters) equipped with APCI. Chromatographic separation was carried out on an Acquity UPLC BEH C18 column (1.7 μm, 2.1 × 50 mm) using HPLC system III at a flow rate of 0.4 mL/min. The corona current was 1.0 μA, and the voltages applied to sampling cone and source offset were 20 and 60 V, respectively. The probe and source temperature were 400 and 150 °C, respectively. High nitrogen flow of 900 L/h was used for desolvation. All of the samples were analyzed in positive ion mode, with the collision energy fixed at 28 eV for MS/MS analysis.
NMR Analyses of Haptoxanthin.
Samples were dissolved in 420 μL CDCl3 and transferred into 5 mm NMR tubes for NMR analysis. The amount of haptoxanthin used was ∼1.59 mg. All of the NMR experiments were performed at 298 K (25 °C) on a Bruker Avance NEO 600 MHz NMR spectrometer equipped with an QCI-F Cryoprobe with helium gas cooling.
Determination of the PSII Functional Absorption Cross-section (σPSII).
Wild-type and KO mutants and complementary lines of P. tricornutum CCAP1055/1 were grown at 18 °C under constant light of 60 ± 5 µmol photons m−2 s−1 on a shaker. Cultures (200 µL) at exponential phase were collected and added into 1.8 mL f/2 medium. Chlorophyll fluorescence was immediately measured by a fluorescence induction and relaxation fluorometer (Satlantic) with blue light source (450 nm) for excitation, and the fluorescence signal was measured at 678 nm. σPSII was calculated using the MATLAB program Fireworx (https://sourceforge.net/projects/fireworx/).
Supplementary Material
Acknowledgments
We thank Christopher J. Kampf (Department of Chemistry, Analytical Core Facility, Johannes Gutenberg-University Mainz) for recording and discussing the mass spectra of haptoxanthin and its saponification product; Christiaan Karreman (In-Vitro Toxicology and Biomedicine, University Konstanz, Germany) for supplying the gene encoding BSD; and Yangmin Gong (Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences), Hanhua Hu (Institute of Hydrobiology, Chinese Academy of Sciences), Run-Zhou Liu (Westlake University), and Jing Huang (Westlake University) for discussion. We also acknowledge the support from the Instrumentation and Service Center for Molecular Sciences and the Biomedical Research Core Facilities at Westlake University. Parts of the project were funded by a grant from the National Key R&D Program of China (2019YFA0906300) to X.L., a grant from the US Department of Energy, Office of Science, Biological, and Environmental Research (DE-SC0018344) to G.P., a grant from the Westlake Education Foundation to X.L., and a grant from the China Postdoctoral Science Foundation (2020M681939) to T.C.
Footnotes
Competing interest statement: T.C. and X.L. have filed a patent application related to the work on the ZEP1 gene described here.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2203708119/-/DCSupplemental.
Data, Materials, and Software Availability
All of the study data are included in the article and/or supporting information. Previously published data were used for this work (protein and nucleotide sequence data from algae available in GenBank (https://www.ncbi.nlm.nih.gov/), the JGI Genome Portal (https://genome.jgi.doe.gov/portal/pages/tree-of-life.jsf), the Microbial Eukaryote Transcriptome Sequencing Project, and the ORCAE (https://bioinformatics.psb.ugent.be/orcae/) were searched with the BLAST tool using the amino acid sequences of the VDE and ZEP family members from P. tricornutum as input.
References
- 1.Bassi R., Dall’Osto L., Dissipation of light energy absorbed in excess: The molecular mechanisms. Annu. Rev. Plant Biol. 72, 47–76 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Polívka T., Frank H. A., Molecular factors controlling photosynthetic light harvesting by carotenoids. Acc. Chem. Res. 43, 1125–1134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paulsen H., “Carotenoids and the assembly of light-harvesting complexes” in The Photochemistry of Carotenoids, Frank H. A., Young A. J., Britton G., Cogdell R. J., Eds. (Springer Netherlands, Dordrecht, the Netherlands, 1999), pp. 123–135. [Google Scholar]
- 4.Matsubara S., et al. , Sun-shade patterns of leaf carotenoid composition in 86 species of neotropical forest plants. Funct. Plant Biol. 36, 20–36 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Jeffrey S. W., Wright S. W., Zapata M., “Micoralgal classes and their signature pigments” in Phytoplankton Pigments. Characterization, Chemotaxonomy and Applications in Oceanography, Roy S., Llewellyn C. A., Egeland E. S., Johnsen G., Eds. (Cambridge University Press, Cambridge, UK, 2011), pp. 3–77. [Google Scholar]
- 6.Larkum A. W. D., Barrett J., Light-harvesting processes in algae. Adv. Bot. Res. 10, 1–219 (1983). [Google Scholar]
- 7.Büchel C., Light harvesting complexes in chlorophyll c-containing algae. Biochim. Biophys. Acta Bioenerg. 1861, 148027 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Sibbald S. J., Archibald J. M., Genomic insights into plastid evolution. Genome Biol. Evol. 12, 978–990 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gould S. B., Waller R. F., McFadden G. I., Plastid evolution. Annu. Rev. Plant Biol. 59, 491–517 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Adl S. M., et al. , Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 66, 4–119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stiller J. W., et al. , The evolution of photosynthesis in chromist algae through serial endosymbioses. Nat. Commun. 5, 5764 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petersen J., et al. , Chromera velia, endosymbioses and the rhodoplex hypothesis–plastid evolution in cryptophytes, alveolates, stramenopiles, and haptophytes (CASH lineages). Genome Biol. Evol. 6, 666–684 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pierella Karlusich J. J., Ibarbalz F. M., Bowler C., Phytoplankton in the Tara Ocean. Annu. Rev. Mar. Sci. 12, 233–265 (2020). [DOI] [PubMed] [Google Scholar]
- 14.de Vargas C., et al. ; Tara Oceans Coordinators, Ocean plankton. Eukaryotic plankton diversity in the sunlit ocean. Science 348, 1261605 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Malviya S., et al. , Insights into global diatom distribution and diversity in the world’s ocean. Proc. Natl. Acad. Sci. U.S.A. 113, E1516–E1525 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Field C. B., Behrenfeld M. J., Randerson J. T., Falkowski P., Primary production of the biosphere: Integrating terrestrial and oceanic components. Science 281, 237–240 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Liu H., et al. , Extreme diversity in noncalcifying haptophytes explains a major pigment paradox in open oceans. Proc. Natl. Acad. Sci. U.S.A. 106, 12803–12808 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peloquin J., et al. , The MAREDAT global database of high performance liquid chromatography marine pigment measurements. Earth Syst. Sci. Data 5, 109–123 (2013). [Google Scholar]
- 19.Galasso C., Corinaldesi C., Sansone C., Carotenoids from marine organisms: Biological functions and industrial applications. Antioxidants 6, 33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ávila-Román J., García-Gil S., Rodríguez-Luna A., Motilva V., Talero E., Anti-inflammatory and anticancer effects of microalgal carotenoids. Mar. Drugs 19, 531 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christaki E., Bonos E., Giannenas I., Florou-Paneri P., Functional properties of carotenoids originating from algae. J. Sci. Food Agric. 93, 5–11 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Wang W., et al. , Structural basis for blue-green light harvesting and energy dissipation in diatoms. Science 363, eaav0365 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Pi X., et al. , The pigment-protein network of a diatom photosystem II-light-harvesting antenna supercomplex. Science 365, eaax4406 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Nagao R., et al. , Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex. Nat. Plants 5, 890–901 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Dambek M., et al. , Biosynthesis of fucoxanthin and diadinoxanthin and function of initial pathway genes in Phaeodactylum tricornutum. J. Exp. Bot. 63, 5607–5612 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eilers U., Dietzel L., Breitenbach J., Büchel C., Sandmann G., Identification of genes coding for functional zeaxanthin epoxidases in the diatom Phaeodactylum tricornutum. J. Plant Physiol. 192, 64–70 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Cui H., et al. , Cloning, identification and functional characterization of two cytochrome P450 carotenoids hydroxylases from the diatom Phaeodactylum tricornutum. J. Biosci. Bioeng. 128, 755–765 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Lohr M., Wilhelm C., Algae displaying the diadinoxanthin cycle also possess the violaxanthin cycle. Proc. Natl. Acad. Sci. U.S.A. 96, 8784–8789 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohr M., Wilhelm C., Xanthophyll synthesis in diatoms: Quantification of putative intermediates and comparison of pigment conversion kinetics with rate constants derived from a model. Planta 212, 382–391 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Goss R., Jakob T., Regulation and function of xanthophyll cycle-dependent photoprotection in algae. Photosynth. Res. 106, 103–122 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Bugos R. C., Yamamoto H. Y., Molecular cloning of violaxanthin de-epoxidase from romaine lettuce and expression in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 93, 6320–6325 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marin E., et al. , Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 15, 2331–2342 (1996). [PMC free article] [PubMed] [Google Scholar]
- 33.Ruban A. V., Johnson M. P., Duffy C. D. P., The photoprotective molecular switch in the photosystem II antenna. Biochim. Biophys. Acta 1817, 167–181 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Buck J. M., et al. , Lhcx proteins provide photoprotection via thermal dissipation of absorbed light in the diatom Phaeodactylum tricornutum. Nat. Commun. 10, 4167 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coesel S., Oborník M., Varela J., Falciatore A., Bowler C., Evolutionary origins and functions of the carotenoid biosynthetic pathway in marine diatoms. PLoS One 3, e2896 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frommolt R., et al. , Ancient recruitment by chromists of green algal genes encoding enzymes for carotenoid biosynthesis. Mol. Biol. Evol. 25, 2653–2667 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Dautermann O., et al. , An algal enzyme required for biosynthesis of the most abundant marine carotenoids. Sci. Adv. 6, eaaw9183 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.North H. M., et al. , The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J. 50, 810–824 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Shen X., et al. , A DUF4281 domain-containing protein (homologue of ABA4) of Phaeodactylum tricornutum regulates the biosynthesis of fucoxanthin. Algal Res. 65, 102728 (2022). [Google Scholar]
- 40.Zigmantas D., et al. , Effect of a conjugated carbonyl group on the photophysical properties of carotenoids. Phys. Chem. Chem. Phys. 6, 3009–3016 (2004). [Google Scholar]
- 41.Premvardhan L., Bordes L., Beer A., Büchel C., Robert B., Carotenoid structures and environments in trimeric and oligomeric fucoxanthin chlorophyll a/c2 proteins from resonance Raman spectroscopy. J. Phys. Chem. B 113, 12565–12574 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Kuczynska P., Jemiola-Rzeminska M., Strzalka K., Photosynthetic pigments in diatoms. Mar. Drugs 13, 5847–5881 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Büch K., Stransky H., Hager A., FAD is a further essential cofactor of the NAD(P)H and O2-dependent zeaxanthin-epoxidase. FEBS Lett. 376, 45–48 (1995). [DOI] [PubMed] [Google Scholar]
- 44.Swift I. E., Milborrow B. V., Jeffrey S. W., Formation of neoxanthin, diadinoxanthin and peridinin from [14C]zeaxanthin by a cell-free system from Amphidinium carterae. Phytochemistry 21, 2859–2864 (1982). [Google Scholar]
- 45.Lohr M., “Beziehungen zwischen den Xanthophyllzyklen und der Biosynthese von Lichtsammelxanthophyllen in Chlorophyll a/c-haltigen Algen” in Fachbereich Biologie (Johannes Gutenberg-Universität, Mainz, Germany, 2001), 186 pp. [Google Scholar]
- 46.Lohr M., “Carotenoid metabolism in phytoplankton” inPhytoplankton Pigments. Characterization, Chemotaxonomy and Applications in Oceanography, Roy S., Llewellyn C. A., Egeland E. S., Johnsen G., Eds. (Cambridge University Press, Cambridge, UK, 2011), pp. 113–161. [Google Scholar]
- 47.Dorrell R. G., Howe C. J., Integration of plastids with their hosts: Lessons learned from dinoflagellates. Proc. Natl. Acad. Sci. U.S.A. 112, 10247–10254 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haugan J. A., Liaaen-Jensen S., Algal carotenoids 54. Carotenoids of brown algae (Phaeophyceae). Biochem. Syst. Ecol. 22, 31–41 (1994). [Google Scholar]
- 49.Nègre D., et al. , Genome-scale metabolic networks shed light on the carotenoid biosynthesis pathway in the brown algae Saccharina japonica and Cladosiphon okamuranus. Antioxidants 8, 564 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pennington F., Guillard R. R. L., Liaaen-Jensen S., Carotenoid distribution patterns in bacillariophyceae (diatoms). Biochem. Syst. Ecol. 16, 589–592 (1988). [Google Scholar]
- 51.Lichtenthaler H. K., The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 47–65 (1999). [DOI] [PubMed] [Google Scholar]
- 52.Brown J. W., Sorhannus U., A molecular genetic timescale for the diversification of autotrophic stramenopiles (Ochrophyta): Substantive underestimation of putative fossil ages. PLoS One 5, e12759 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang E. C., et al. , Supermatrix data highlight the phylogenetic relationships of photosynthetic stramenopiles. Protist 163, 217–231 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Keeling P. J., et al. , The marine microbial eukaryote transcriptome sequencing project (MMETSP): Illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12, e1001889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Altschul S. F., et al. , Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chenna R., et al. , Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hall T. A., BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 (1999). [Google Scholar]
- 58.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whelan S., Goldman N., A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699 (2001). [DOI] [PubMed] [Google Scholar]
- 60.Stamatakis A., Hoover P., Rougemont J., A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57, 758–771 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Huson D. H., Scornavacca C., Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61, 1061–1067 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Page R. D. M., TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358 (1996). [DOI] [PubMed] [Google Scholar]
- 63.Concordet J. P., Haeussler M., CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46 (W1), W242–W245 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nymark M., Sharma A. K., Sparstad T., Bones A. M., Winge P., A CRISPR/Cas9 system adapted for gene editing in marine algae. Sci. Rep. 6, 24951 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Apt K. E., Kroth-Pancic P. G., Grossman A. R., Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol. Gen. Genet. 252, 572–579 (1996). [DOI] [PubMed] [Google Scholar]
- 66.Zhang C., Hu H., High-efficiency nuclear transformation of the diatom Phaeodactylum tricornutum by electroporation. Mar. Genomics 16, 63–66 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Karreman C., New positive/negative selectable markers for mammalian cells on the basis of Blasticidin deaminase-thymidine kinase fusions. Nucleic Acids Res. 26, 2508–2510 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petersen R., Djozgic H., Rieger B., Rapp S., Schmidt E. R., Columnar apple primary roots share some features of the columnar-specific gene expression profile of aerial plant parts as evidenced by RNA-seq analysis. BMC Plant Biol. 15, 34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bendtsen J. D., Nielsen H., von Heijne G., Brunak S., Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 (2004). [DOI] [PubMed] [Google Scholar]
- 70.Petersen T. N., Brunak S., von Heijne G., Nielsen H., Signal P., SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 (2011). [DOI] [PubMed] [Google Scholar]
- 71.Emanuelsson O., Nielsen H., von Heijne G., Chloro P., ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hajdukiewicz P., Svab Z., Maliga P., The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994 (1994). [DOI] [PubMed] [Google Scholar]
- 73.Dautermann O., Lohr M., A functional zeaxanthin epoxidase from red algae shedding light on the evolution of light-harvesting carotenoids and the xanthophyll cycle in photosynthetic eukaryotes. Plant J. 92, 879–891 (2017). [DOI] [PubMed] [Google Scholar]
- 74.Morosinotto T., Baronio R., Bassi R., Dynamics of chromophore binding to Lhc proteins in vivo and in vitro during operation of the xanthophyll cycle. J. Biol. Chem. 277, 36913–36920 (2002). [DOI] [PubMed] [Google Scholar]
- 75.Yamamoto H. Y., Higashi R. M., Violaxanthin de-epoxidase. Lipid composition and substrate specificity. Arch. Biochem. Biophys. 190, 514–522 (1978). [DOI] [PubMed] [Google Scholar]
- 76.Goss R., et al. , Role of hexagonal structure-forming lipids in diadinoxanthin and violaxanthin solubilization and de-epoxidation. Biochemistry 44, 4028–4036 (2005). [DOI] [PubMed] [Google Scholar]
- 77.Blatt A., Bauch M. E., Pörschke Y., Lohr M., A lycopene β-cyclase/lycopene ε-cyclase/light-harvesting complex-fusion protein from the green alga Ostreococcus lucimarinus can be modified to produce α-carotene and β-carotene at different ratios. Plant J. 82, 582–595 (2015). [DOI] [PubMed] [Google Scholar]
- 78.Sharma A. K., Nymark M., Sparstad T., Bones A. M., Winge P., Transgene-free genome editing in marine algae by bacterial conjugation - comparison with biolistic CRISPR/Cas9 transformation. Sci. Rep. 8, 14401 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Slattery S. S., et al. , An expanded plasmid-based genetic toolbox enables Cas9 genome editing and stable maintenance of synthetic pathways in Phaeodactylum tricornutum. ACS Synth. Biol. 7, 328–338 (2018). [DOI] [PubMed] [Google Scholar]
- 80.Buck J. M., Río Bártulos C., Gruber A., Kroth P. G., Blasticidin-S deaminase, a new selection marker for genetic transformation of the diatom Phaeodactylum tricornutum. PeerJ 6, e5884 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Strand T. A., Lale R., Degnes K. F., Lando M., Valla S., A new and improved host-independent plasmid system for RK2-based conjugal transfer. PLoS One 9, e90372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Karas B. J., et al. , Designer diatom episomes delivered by bacterial conjugation. Nat. Commun. 6, 6925 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu C., et al. , Structural basis for energy transfer in a huge diatom PSI-FCPI supercomplex. Nat. Commun. 11, 5081 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the study data are included in the article and/or supporting information. Previously published data were used for this work (protein and nucleotide sequence data from algae available in GenBank (https://www.ncbi.nlm.nih.gov/), the JGI Genome Portal (https://genome.jgi.doe.gov/portal/pages/tree-of-life.jsf), the Microbial Eukaryote Transcriptome Sequencing Project, and the ORCAE (https://bioinformatics.psb.ugent.be/orcae/) were searched with the BLAST tool using the amino acid sequences of the VDE and ZEP family members from P. tricornutum as input.