Significance
Arsenic is a centuries-old, naturally occurring toxicant, and its cellular effect is still poorly understood. The significance of this work is three-fold. First, among a battery of oxidizing and alkylating agents, arsenite exposure caused a unique reprogramming of wobble queuosine in the transfer RNA (tRNA) epitranscriptome, which depended upon the micronutrient precursor queuine and was linked to codon-biased shifts in the translation of metabolic proteins known to be linked to arsenite toxicity. Second, we showed that the tRNA epitranscriptome is dynamically and differentially regulated by exposure to a variety of toxicants. Finally, the results have implications for the role of queuine as a micronutrient that determines the human cell response to toxic stresses.
Keywords: epitranscriptome, tRNA modifications, arsenite, metabolism, codon-biased translation
Abstract
Cells respond to environmental stress by regulating gene expression at the level of both transcription and translation. The ∼50 modified ribonucleotides of the human epitranscriptome contribute to the latter, with mounting evidence that dynamic regulation of transfer RNA (tRNA) wobble modifications leads to selective translation of stress response proteins from codon-biased genes. Here we show that the response of human hepatocellular carcinoma cells to arsenite exposure is regulated by the availability of queuine, a micronutrient and essential precursor to the wobble modification queuosine (Q) on tRNAs reading GUN codons. Among oxidizing and alkylating agents at equitoxic concentrations, arsenite exposure caused an oxidant-specific increase in Q that correlated with up-regulation of proteins from codon-biased genes involved in energy metabolism. Limiting queuine increased arsenite-induced cell death, altered translation, increased reactive oxygen species levels, and caused mitochondrial dysfunction. In addition to demonstrating an epitranscriptomic facet of arsenite toxicity and response, our results highlight the links between environmental exposures, stress tolerance, RNA modifications, and micronutrients.
Human cells respond to environmental toxicant and xenobiotic exposure with highly coordinated mechanisms involving signaling pathways, altered gene expression, and protein secondary modification, with the integrated response often leading to cell death and carcinogenesis (1). Inorganic arsenic compounds are among the most abundant, naturally occurring environmental toxicants and are classified as group I carcinogens, with chronic exposure linked to cardiovascular disease, hypertension, and cancer (2). While arsenic is a major public health threat, little is known about its mechanisms of toxicity and carcinogenicity, although there is strong evidence for mitochondrial dysregulation, oxidative stress, and associated genotoxicity with arsenic exposure (3, 4).
Here we sought to determine the role of the transfer RNA (tRNA) epitranscriptome in the cell response to chemical stressors, with a focus on arsenite toxicity. The human tRNA epitranscriptome of ∼50 modified ribonucleotides is a subset of the ∼170 modifications in all organisms, with each tRNA molecule containing an average of 13 modifications critical for efficient and accurate protein synthesis, as well as tRNA maturation and structural stability (5–8). The chemical complexity of these modifications ranges from simple methylations to hypermodified ribonucleotides, such as queuosine (Q; Fig. 1) (9, 10). Q has received particular attention due to its strong conservation across eubacteria, plants, and animals and to its location at the wobble position 34 of tRNAs with a 5′-GUN-3′ anticodon (tyrosine, asparagine, aspartate, histidine; N is any canonical nucleotide), where it replaces guanine (G) and plays a direct role in messenger RNA (mRNA) codon recognition (11). As demonstrated in computational modeling, Q restricts the anticodon loop flexibility of tRNAs by establishing an extended intraresidue and intramolecular hydrogen-bonding network, enabling the recognition of both cognate codons NAC and NAU (wobble poitions underlined) without favoring one or the other (12). Interestingly, the presence of Q is not limited to cytosolic tRNAs but also occurs in the simpler and often less modified mitochondrial tRNAs, demonstrating the importance of enhanced codon recognition by Q (13). This idea is supported by a recent study showing that Q-modified tRNAs in Trypanosoma brucei are preferentially imported into mitochondria to allow optimal organellar protein synthesis (14).
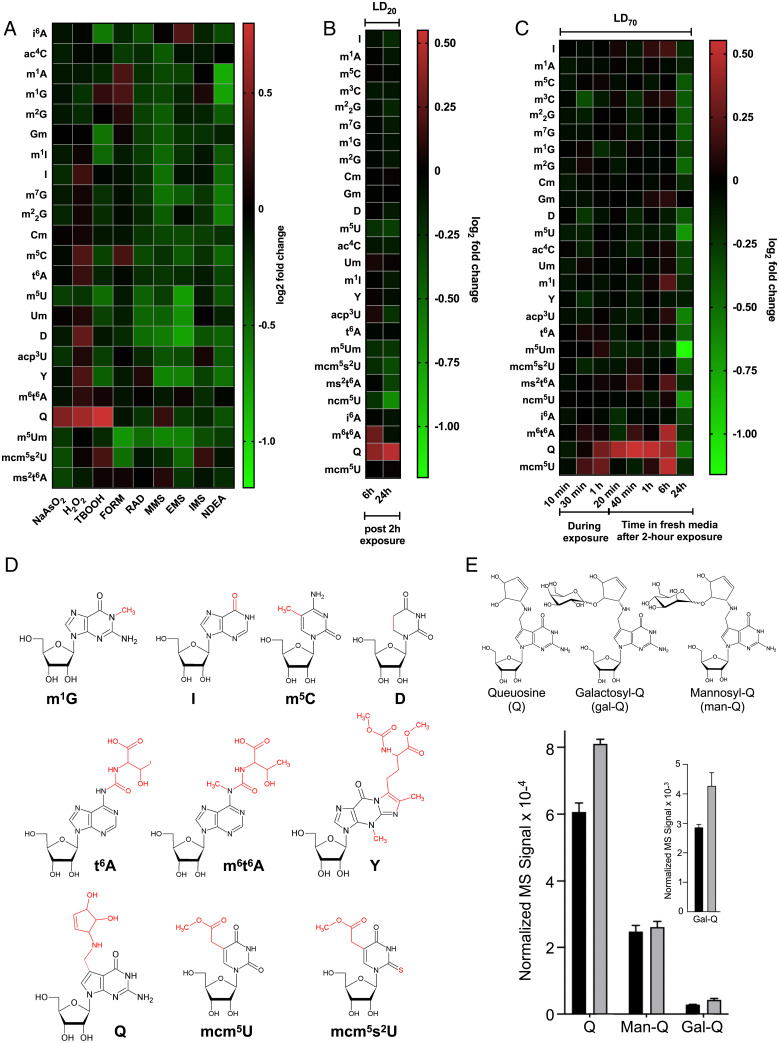
Fig. 1.
Epitranscriptomic reprogramming identifies Q and other tRNA modifications as signature markers of oxidative stress and NaAsO2 exposure. (A) tRNA modification reprogramming in HepG2 cells exposed to LD20 doses of oxidizing and alkylating agents for 2 h, with RNA harvested 24 h postexposure. (B and C) Epitranscriptomic response of HepG2 cells to (B) a 2 h exposure to an LD20 dose of NaAsO2 followed by 6 h and 24 h postexposure incubation and (C) a LD70 dose at various times postexposure. (D) Chemical structures and abbreviations of the modifications affected by oxidative stressors and NaAsO2 exposure in the present studies. (E) Q, Gal-Q, and Man-Q were quantified by LC-MS in tRNA extracted from HepG2 cells exposed to an LD20 dose of arsenite for 6 h. Black bars: unexposed; gray bars: arsenite-exposed. Inset shows a magnification of data for Gal-Q on the 10−3 scale. Data represent mean ± SD for three experimental replicates. See data in Dataset S1.
While bacteria can synthesize Q de novo in a complex biosynthetic pathway starting with GTP (15), eukaryotes lack the early synthesis pathways for Q and rely on exogenous sources of its free base precursor queuine, a micronutrient from the diet, the gut microbiota, and the Q-modified tRNA salvage pathway, with subsequent enzymatic transglycosylation into tRNA (16, 17). Q is emerging as a critical determinant of human health, as changes in its abundance have been implicated in translational dysregulation and activation of the unfolded protein response (18), cell proliferation and differentiation (19, 20), bacterial virulence (21), tyrosine biosynthesis (22), cancer (23), and neurological and psychiatric disorders (24). Queuine and Q have also been demonstrated to be involved in oxidative stress tolerance. For example, in a mouse model of transplanted lymphoma, the addition of queuine was shown to increase the activity of antioxidant enzymes such as catalase, superoxide dismutase, glutathione peroxidase, and glutathione reductase, thereby inhibiting oxidative stress and tumorigenesis (25). Furthermore, Q has also been found to protect its cognate tRNAs for histidine and asparagine against cleavage by ribonucleases when human cells were exposed to oxidative stress (26).
We now provide insights into the role of queuine in the oxidative stress response by describing how arsenic pathobiology is regulated by the environmental availability of queuine and by Q-specific tRNA reprogramming. This mechanism involves our previously identified systems-level cell stress response network in bacteria (27), parasites (28), yeast (29–32), and human cells (33), which centers around tRNA modification-based translational regulation of gene expression. In this system, stress-specific reprogramming of an organism’s ∼50 modified ribonucleotides in tRNAs leads to proteome changes linked to codon-biased mRNAs representing families of genes involved in the response to the specific stress (27, 29–33). Here we demonstrate that queuine and Q in tRNA are needed to prevent arsenite-induced cell death and mitochondrial dysfunction, with these endpoints linked to increased reactive oxygen species (ROS), metabolic shift toward glycolysis, and altered translation. Notably, we have used proteomic studies and codon analytics to show that key proteins regulating glycolysis and the pentose phosphate pathways are up-regulated in response to arsenic, with their corresponding mRNAs biased for specific Q-decoded codons as well as other codons. These studies highlight the important role of Q in regulating the human cellular response to arsenite exposure.
Results
Q is selectively up-regulated in tRNA by oxidative stressors.
We previously showed that the exposure of yeast to oxidizing and alkylating agents caused highly predictive agent-, class-, and dose-specific reprogramming of 23 tRNA modifications, with the agent being the chemical name of the stressor and the class indicating a group of agents that cause the same type of stress to the cells (e.g., oxidants versus alkylators) (30, 31). To determine whether this behavior was recapitulated in human cells, we used chromatography-coupled tandem mass spectrometry (LC-MS/MS) to profile 23 modified ribonucleosides in the tRNA epitranscriptome in human hepatocellular carcinoma (HepG2) cells exposed to equitoxic concentrations of three chemical oxidizing agents (sodium meta-arsenite [NaAsO2], hydrogen peroxide [H2O2], tert-butyl hydroperoxide [TBOOH]), five alkylating agents (methyl methanesulfonate [MMS], ethyl methanesulfonate [EMS], isopropyl methanesulfonate [IMS], formaldehyde [FORM], N-nitrosodiethylamine [NDEA]), and ionizing radiation (RAD) (SI Appendix, Fig. S1). The first set of experiments involved exposing HepG2 cells to - concentrations of toxicants at a lethal dose for 20% of the cells (LD20) for 2 h followed by a 24 h incubation in toxicant-free medium. LC-MS/MS analysis revealed agent- and class-specific changes in the levels of tRNA modifications as shown by the fold-change values relative to untreated control cells in Fig. 1A (data detailed in Dataset S1). The general trend observed for simple alkylating agents was that exposure caused decreased levels of most of the analyzed tRNA modifications, with the exception of agent-specific increases in ms6t6A* and Q for MMS, i6A for EMS, and mcm5s2U for IMS. While FORM can be considered an alkylating agent, the resulting chemical transfer is distinct and we observed increased levels of m5C, m1G, and m1A after exposure. A general decrease in modification levels was also observed for all 23 analyzed tRNA modifications in response to RAD, similar to previous observations in yeast (30, 31), and to NDEA exposure. NDEA is a complex toxicant that is metabolized by cytochrome P450 to yield a methylating agent and FORM. In contrast to the RNA modification decreases observed for simple alkylating agents and RAD, the chemical oxidants all induced significantly increased levels of Q. As a group, the chemical oxidants caused increases in eight tRNA modifications: m1G, I, m5C, t6A, D, Y, Q, and mcm5s2U (Fig. 1). While Q was the only modification increased by NaAsO2 exposure, six modifications increased in response to H2O2 (Q, Y, D, t6A, m5C, I) and three modifications increased after TBOOH exposure (m1G, Q, mcm5s2U). These results point to more complicated rules of stress-induced epitranscriptomic reprogramming in human cells compared to yeast (30, 31), with Q increases as a signature marker for oxidative stress.
NaAsO2 exposure up-regulates mcm5U, m6t6A, Q, and Gal-Q in tRNA.
Among the oxidizing agents, NaAsO2 exposure caused increases in only Q at 24 h after a 2-h exposure to an LD20 concentration (Fig. 1A). To better understand the potential role of Q as an early oxidative stress and arsenite exposure marker, two sets of studies assessed epitranscriptomic reprogramming at earlier times following NaAsO2 treatment. First, HepG2 cells were exposed to an LD20 concentration of NaAsO2 for 2 h and 26 tRNA modifications were then quantified at 6 h and 24 h after a change to fresh, toxicant-free medium (Fig. 1B). Both m6t6A and Q were up-regulated at the 6-h postexposure time point, which places m6t6A and Q as early responses to NaAsO2 exposure. We observed that while Q levels further increased to 24 h postexposure, m6t6A levels returned to control levels and there was a continued down-regulation of five RNA modifications (m5U, m5Um, mcm5s2U, ncm5U, and ms2t6A) (Fig. 1B).
For a more granular view of the arsenic response time course, we exposed cells to an LD70 concentration of NaAsO2 and assessed tRNA modification changes 10, 30, and 60 min after NaAsO2 addition. We also assessed tRNA modification changes after a 2-h NaAsO2 exposure at 20 min, 40 min, 1 h, 6 h, and 24 h after changing to toxicant-free medium (Fig. 1C). At a higher concentration and earlier time points, mcm5U increased as soon as 30 min after exposure but returned to control levels as soon as 20 min after the media was replaced at 2 h of exposure. On the other hand, Q started increasing later at 1 h and remained high up to 6 h postexposure. The 6-h time point reveals that there is up-regulation of multiple RNA modifications, highlighted by mcm5U, Q, and m6t6A (Fig. 1C), which supports the idea that there is coordinated epitranscriptomic reprogramming occurring after exposure. Together, the heatmaps (Fig. 1) support the idea that multiple tRNA modifications are regulated in response to NaAsO2 (Fig. 1D). However, the 24-h time point data should be viewed with caution, as many cells are dead at this point in the time course.
Finally, given the evidence for the glycosylated forms of Q, galactosyl- and mannosyl-Q (Gal-Q and Man-Q; Fig. 1E), we quantified arsenite-induced changes in the levels of the three wobble modifications at 6 h postexposure to an LD20 dose of NaAsO2. Here we identified Gal-Q and Man-Q by high-resolution mass spectrometric fragmentation analysis of total tRNA hydrolysates resolved by high-performance liquid chromatography (HPLC), with the reversed-phase HPLC retention times and relative quantities of Gal-Q and Man-Q similar to those observed by Thumbs and colleagues (SI Appendix, Fig. S2) (34). Using these parameters, we observed a 40% increase in the level of Gal-Q (P < 0.03) and a 25% increase in Q (P < 0.002) but no change in Man-Q levels (P < 0.40), following 6 h of an LD20 exposure to arsenite (Fig. 1E). Current understanding suggests that Q is located on tRNA-Tyr(QUA), tRNA-Asp(QUC), tRNA-His(QUG), and tRNA-ASN(QUU), but only tRNA-Tyr(gal-QUA) and tRNA-Asp(QUC) are further glycosylated to form Gal-Q and Man-Q, respectively (34). However, it is not known how the hypermodified Q wobble modifications alter codon reading or affect tRNA levels.
Queuine deprivation reduces Q in tRNA and sensitizes cells to arsenite-induced mitochondrial toxicity.
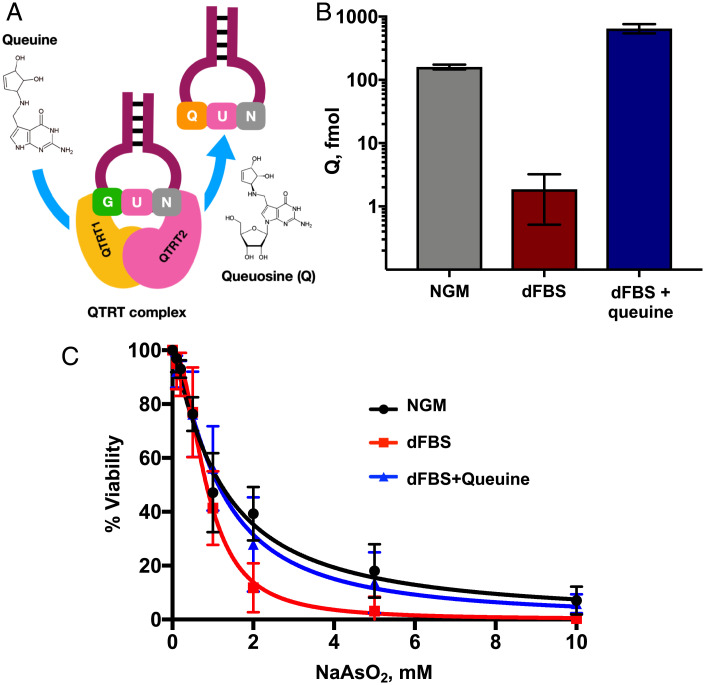
Building on the observation that NaAsO2 exposure increased Q in tRNA (Fig. 1 A–C) and the mammalian auxotrophy for queuine (Fig. 2A), we explored the role of environmental queuine in the NaAsO2 stress response. We first quantified the effect of queuine deprivation on Q levels in tRNA by growing HepG2 cells in culture medium containing 10% dialyzed fetal bovine serum (dFBS) for 10 passages (25 d). LC-MS/MS analysis revealed a ∼100-fold reduction in Q in tRNA (Fig. 2B) compared to cells grown in growth medium with normal FBS (normal growth medium [NGM]). A complete Q depletion was not expected due to the ability of human cells to salvage queuine from full-length tRNAs. Supplementation of dFBS culture medium with queuine (1 μM; dFBS+queuine) restored Q in tRNA to three-fold higher levels than in normal growth medium (Fig. 2B). These results reveal significant regulation of Q levels in tRNA by queuine availability in HepG2 cells. To quantify the role of queuine in the cell response to arsenite exposure, HepG2 cells were grown in each of the three media (NGM, dFBS, dFBS+queuine) and then treated with varying concentrations (2–10 mM) of NaAsO2 for 2 h, with cell survival assayed with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) 24 h after treatment. As shown in Fig. 2C, HepG2 cells grown in queuine-depleted medium (dFBS) were significantly (P < 0.03) more sensitive to NaAsO2, while supplementation of dFBS with queuine restored Q in tRNA and reduced the sensitivity of cells to NaAsO2 to control levels.
Fig. 2.
Queuine promotes viability after exposure of HepG2 cells to NaAsO2. (A) Mechanism by which the QTRT enzyme incorporates Q into the wobble position of tRNA isoacceptors for four amino acids. (B) Q in tRNA is decreased in HepG2 cells grown in dFBS, relative to NGM or dFBS supplemented with queuine (dFBS+queuine). LC-MS/MS analysis of the Q ribonucleoside was performed on tRNA isolated from cells grown in each medium condition. (C) NaAsO2 dose–response curves of HepG2 cells grown in medium containing NGM, dFBS, or dFBS+queuine. Cells were exposed to NaAsO2 and viability was assessed using the MTT assay 24 h after exposure with significant differences between NGM and dFBS at 2 (P < 0.03) and 5 mM (P < 0.01), as determined using a Student t test in GraphPad Prism 6.
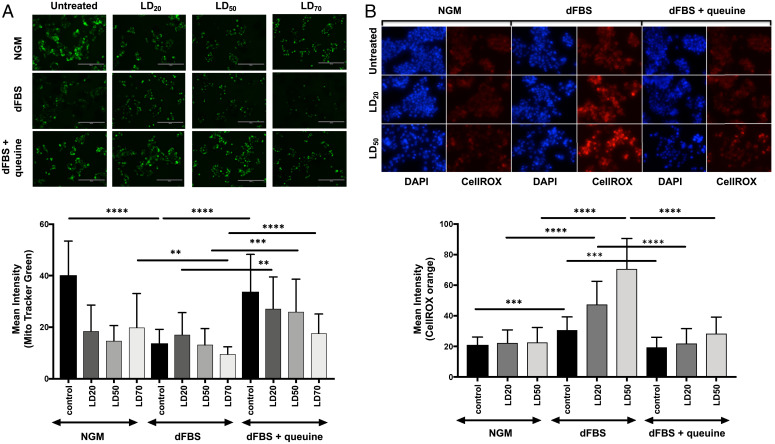
Queuine-dependent resistance to arsenite-induced mitochondrial mass reduction was also apparent in cells treated with LD20 and LD50 concentrations of NaAsO2 for 2 h. MitoTracker Green staining after 24 h revealed that cells grown in dFBS had a 2.5-fold reduction in mitochondrial mass compared to cells grown in NGM, with no significant changes caused by NaAsO2 exposure at either concentration (Fig. 3A). The mitochondrial mass returned to levels observed with NGM when queuine was added back to the growth medium (dFBS+queuine), for both untreated and NaAsO2 conditions (Fig. 3A). These results demonstrate that queuine, likely as Q in tRNA, plays an important role in preventing mitochondrial sensitivity to arsenite toxicity. The queuine dependency of mitochondrial mass changes following arsenite exposure (Fig. 3B) raised the possibility of the queuine-dependent disruption of electron transport by arsenite. In support of this idea, previous studies have proposed that arsenite uncouples the respiratory chain by inhibiting complexes I, II, and IV, with a subsequent increase in ROS (35). Here we tested this model in HepG2 cells using CellROX Orange to quantify superoxide and other ROS in cells exposed to arsenite under varying conditions of queuine availability. As shown in Fig. 3B, we did not observe an increase in ROS levels in arsenite-exposed cells grown in NGM. However, cells grown in dFBS experienced a significant increase in ROS levels upon arsenite exposure, with ROS reduced to control levels by supplementation with queuine (Fig. 3B). These results demonstrate that queuine, possibly through Q in tRNA, regulates ROS production or management and protects cells from arsenite-induced ROS increases.
Fig. 3.
Queuine prevents mitochondrial and ROS changes after exposure of HepG2 cells to NaAsO2. (A) MitoTracker Green (green) staining of HepG2 cells grown under different media and NaAsO2 exposure conditions (Top), with quantitation of MitoTracker Green (Bottom). (B) CellRox Orange (orange) and DAPI staining was performed on HepG2 cells grown under different media and NaAsO2 exposure conditions (Top), with quantitation of CellROX Orange (Bottom). Data in all panels represent mean ± SD for three experimental replicates. For (A) and (B): Black bars represent untreated cells, while gray bars shaded represent different NaAsO2 exposure conditions (LD20, LD50, LD70). Data were compared using Student t test, with ****P < 0.0001, ***P < 0.002, and **P < 0.005 in GraphPad Prism 6.0. The concentrations represent the LD20, LD50, and LD70 doses used in NGM.
We next tested the translational effects of arsenite exposure to analyze for queuine-dependent changes in global translation. To quantify queuine effects on global translation, we measured polysome profiles for cells grown in NGM, dFBS, and dFBS+queuine. As shown in SI Appendix, Fig. S3 A and B, after scaling the polysome data to the 60S peak, we found that cells grown in NGM and dFBS had similar polysome levels, while cells grown in dFBS+queuine had reduced but still robust polysome levels. However, after an LD20 concentration of NaAsO2, polysome levels were significantly reduced in cells grown in dFBS compared to cells growing NGM and dFBS+queuine (SI Appendix, Fig. S3C). Surprisingly, NaAsO2-treated cells grown in dFBS+queuine trended toward increased polysome levels relative to NGM. These data support the idea that there is a global change in translation in queuine-depleted cells and a dramatic decrease in response to NaAsO2, with both phenomena linked to decreased Q levels in tRNA. In addition, the observed increased ROS and changes to mitochondrial physiology are likely further promoting global changes to translation.
NaAsO2 exposure alters mitochondrial function and shifts metabolism toward glycolysis.
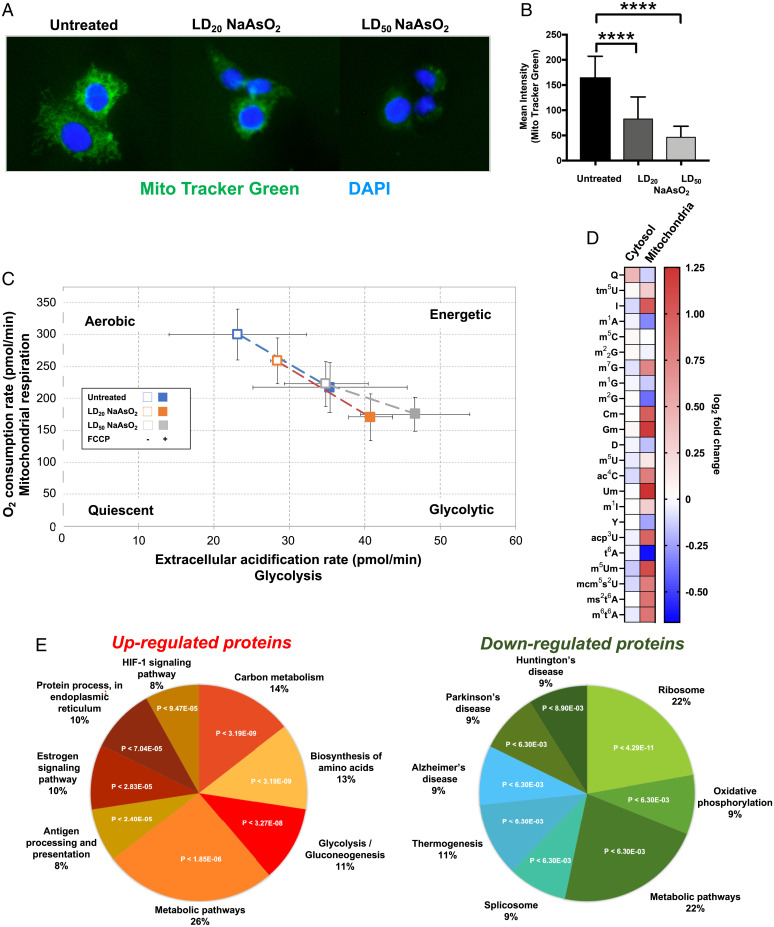
The dramatic changes in translation we identified using depleted media highlight the importance of Q and translation in the response to arsenite, but the global changes make identifying the exact mechanism challenging. Next, we used a series of experiments in NGM to quantitatively define pathways of cellular response to NaAsO2, to reveal the connections between Q levels in tRNA and NaAsO2 toxicity. We first looked at mitochondrial pathology given the evidence that mitochondria are an important cellular target of arsenic toxicity, including damage to mitochondrial DNA; uncoupling the respiratory chain by inhibiting complexes I, II, and IV; and increased production of ROS (35–38). The first set of studies addressed the effect of NaAsO2 on HepG2 mitochondria as a gross measure of pathology, using MitoTracker Green dye, which accumulates in mitochondria, to quantify mitochondrial mass as a function of NaAsO2 exposure (Fig. 4 A and B). We observed concentration-dependent decreases in MitoTracker Green intensity and therefore reduced mitochondrial mass. We next defined the effect of arsenite exposure on HepG2 cell metabolism using the Seahorse XF24 Analyzer to measure real-time effects of NaAsO2 exposure on the oxygen consumption rate (OCR) as an index of mitochondrial respiration and extracellular acidification (ECAR) as an index of glycolytic activity in HepG2 cells. We used cells that were left untreated or exposed to either LD20 or LD50 doses of NaAsO2. These cells were then treated with carbonyl cyanide-4-trifluoromethoxyphenylhydrazone (FCCP) as a positive control reagent to drive oxygen consumption rates to their highest levels. FCCP uncouples oxidative phosphorylation by depolarizing the mitochondrial membrane and disrupting adenosine triphosphate synthesis. As shown in Fig. 4C, NaAsO2 caused a concentration-dependent reduction in basal OCR and an increase in basal ECAR, which represents a shift toward glycolysis and away from mitochondrial respiration. These results are consistent with previous observations (1, 2) and support the idea that NaAsO2 exposure shifts cellular metabolism to glycolysis in HepG2 cells. We next focused on epitranscriptomic changes in the mitochondria. We observed that the levels of 17 RNA modifications found in the mitochondrial fraction were altered by NaAsO2 exposure (12 increased and five decreased), with notable examples being m6t6A and mcm5U (Fig. 4D). The dramatic change in the mitochondrial epitranscriptome further supports that there are NaAsO2-induced changes in mitochondrial physiology.
Fig. 4.
NaAsO2 promotes decreased mitochondrial mass and a shift toward glycolysis. (A) HepG2 cells were exposed to various doses of NaAsO2 and stained with MitoTracker Green (green) and DAPI (blue) 24 h after exposure. (B) The intensity of MitoTracker Green in 22 cells for each condition was determined using ImageJ and plotted as a function of NaAsO2 exposure. In (A) and (B), data were compared using the Student t test, with ****P < 0.0001. (C) After 24 h exposure, Seahorse XFp Cell Energy Phenotype Analysis was performed on HepG2 cells to measure the OCR and ECAR. The open and filled boxes represent the cells in the absence or presence of FCCP for each of the three conditions. Data represent mean ± SD for three experimental replicates. (D) Epitranscriptomic changes in cytosolic and mitochondrial tRNAs of HepG2 cells 2 h after exposure to NaAsO2. (E) Significant (P < 0.05) gene ontology enrichments for proteins that were up-regulated (P < 0.05) or down-regulated (P < 0.05) in HepG2 cells in response to NaAsO2 as determined by proteomics (see SI Appendix, Tables S1 and S2).
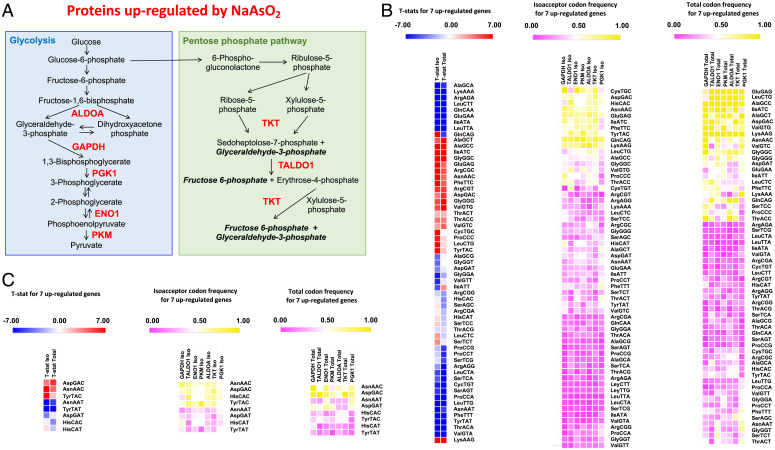
To further test arsenite-dependent changes in metabolism, we quantified changes in the steady-state proteome using isobaric tagging and unfractionated chromatography-coupled mass spectrometry with soluble protein extracts from HepG2 cells grown in NGM exposed to an LD20 concentration of NaAsO2 (Dataset S2). A total of 286 proteins were reliably quantified in all three replicates for unexposed and exposed cells. Functional analysis of significantly (P < 0.05) up- or down-regulated proteins using STRING (Fig. 4E and SI Appendix, Tables S1 and S2) revealed significant enrichment in the metabolic pathways linked to carbon metabolism, with up-regulated proteins overrepresented in the category of glycolysis and down-regulated proteins overrepresented in oxidative phosphorylation. The proteomic data matched our metabolic findings (Fig. 4 A–C) and, together with our mitochondrial data, highlight that there is a shift from oxidative phosphorylation to glycolysis. We determined that seven key proteins involved in glycolysis and the pentose phosphate pathways (ALDOA, P < 8.8E-04; PGK1, P < 2.1E-03; PKM, P < 3.4E-03; GAPDH, P < 4.6E-03; TALDO, P < 6.4E-03; ENO1, P < 1.7E-02; TKT, P < 3.7E-02) 1) were highly prominent as a STRING category among the 43 proteins significantly increased in response to NaAsO2, 2) were all located in mitochondria, and 3) all possessed biased use of Q-dependent codons. Our observation of arsenite-induced increases in glycolysis proteins is analogous to the observation by Jang and colleagues that H2O2 increased the levels of PGK1 in a dose-dependent manner (39). We used codon analytics to determine whether there was codon bias in the gene sequences corresponding to this group of glycolytic proteins. Gene-specific isoacceptor and total codon frequencies for 19,710 unique human genes were calculated, and genome averages and SDs were determined (Datasets S3 and S4). Isoacceptor codon frequencies reveal whether a gene is overusing a synonymous codon for a specific amino acid, with the number of synonymous codons ranging from two to six for each amino acid. Total codon frequencies note codon usage relative to the 64 other potential codons in a gene sequence and, in addition to documenting codon bias, can represent amino acid bias in the corresponding protein. We determined that the group of seven up-regulated glycolytic proteins was significantly biased and overrepresented with 24 codons (Fig. 5 A and B, Left and Dataset S5), as determined by the t-statistic (P < 0.05). The t-statistic (t), also known as the Test statistic, uses the sample mean (x), the population mean (μ0), the sample SD, and the sample size (), with t = (x−μ0)/(s/. We used heat maps and hierarchical clustering (Euclidean distances) of codon frequency to demonstrate that relative to other human genes, the ALDOA, GAPDH, PGK1, ENO1, PKM, TKT, and TALDO genes are enriched for many C- and G-ending codons, for both isoacceptor (Fig. 5B, Middle) and total codon frequency measures (Fig. 5B, Right). We specifically focused on the four sets of codons decoded by Q (Fig. 5C and SI Appendix, Table S3) and, as a group, ALDOA, GAPDH, PGK1, ENO1, PKM, TKT, and TALDO significantly overuse the C-ending codons for Asp (P < 0.001), Asn (P < 1.5 E-08), and Tyr (P < 1.9 E-08) based on isoacceptor codon frequency and the C-ending codons for Asn (P < 0.0003) and Asp (P < 0.000003) based on total codon usage. We observed that the His codon CAC did not show a statistically significant t-statistic trend in the group of seven glycolytic genes but was represented at high isoacceptor frequency levels in most targets.
Fig. 5.
Glycolytic proteins increased in response to NaAsO2 are codon biased. (A) Seven proteins involved in glycolysis and the pentose–phosphate pathway were significantly up-regulated after an LD20 arsenite exposure in HepG2 cells as measured by proteomics (Dataset S2). (B) Both isoacceptor and total codon frequencies for the group of seven corresponding gene sequences were analyzed to identify those codons deviating from genome averages (SI Appendix, Table S3 and Dataset S5), with t-statistics (Left; T-stat), isoacceptor codon frequencies (Middle; iso), and total codon frequencies (Right; total) hierarchically clustered and shown as heat maps. (C) Q-decoded codons are highlighted, with t-statistics (Left; T-stat), isoacceptor codon frequencies (Middle; iso), and total codon frequencies (Right); total hierarchically clustered and shown as heat maps.
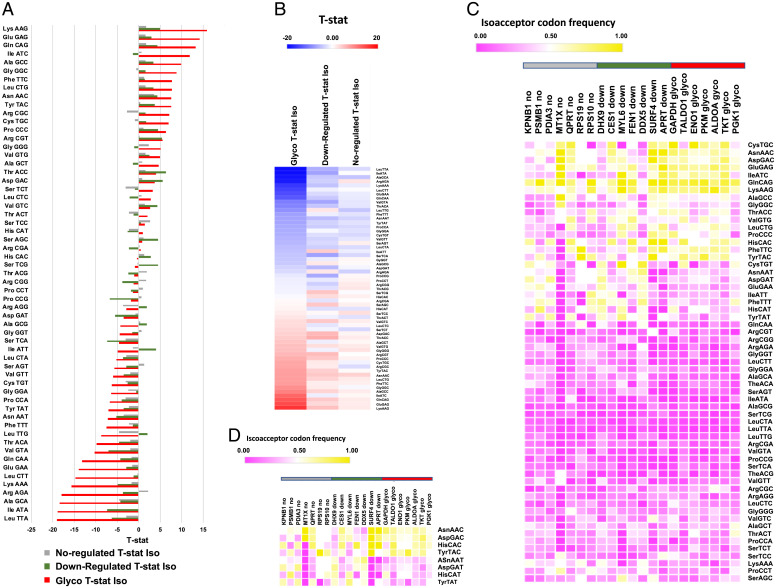
We also compared the codon usage of our seven genes encoding glycolytic enzymes to other gene sequences representing proteins identified in our proteomics data. We performed codon analysis on seven proteins whose levels did not change in response to NaAsO2 (i.e., no change) and seven proteins that decreased significantly in response to arsenite in our proteomic data set. Our findings revealed that the seven glycolytic enzymes have distinct codon usage patterns that are represented by an extreme overusage of 13 C- or G-ending codons (t-statistic > 5, maximum of 16, average of nine), with the down-regulated and not-regulated groups failing to have t-statistic values > 5.0 for any of the corresponding codons (t-statistic average for 13 corresponding codons of 2.7 and 0.9, respectively). There is an extreme underusage of 19 T- or A-ending codons for the seven glycolytic genes (t-statistic values < −5, average of −11) relative to the down-regulated (average of −3.9) and not-regulated groups (average of −1.8). Taken together, these results support the idea that the codon usage of the genes for the group of seven glycolytic enzymes has a signature of extreme overuse of C- or G-ending codons and extreme underuse of T- or A-ending codons, while the other two groups have much milder codon–usage patterns (Fig. 6 A and B). These findings are similarly supported at the gene level as the glycolytic group has distinct patterning, relative to genes in the down-regulated and not-regulated groups, with the decreased use of T- or A-ending codons notable for most codons and Q-decoded codons (Fig. 6 C and D). Our findings suggest a model in which Q increases the translation of transcripts with C-ending codons for Asp, Asn, and Tyr, with the corresponding transcripts having limited T-ending codons. Together, these codon bias data and increased levels of glycolytic enzymes support the idea that the arsenite-induced up-regulation of Q in tRNA promotes a shift to glycolysis.
Fig. 6.
Among genes for proteins altered by arsenite exposure, glycolytic genes have distinct codon usage patterns relative to genes for other proteins identified by proteomics. Codon usage comparison of the genes specific to seven up-regulated glycolytic (glyco) proteins, seven random down-regulated (down) proteins, and seven random unchanged proteins (not-regulated; no) identified in our proteomics study. Isoacceptor T-stat values for codons specific to the three groups of gene sequences in (A) bar graph and (B) heat map view. Heat maps describing isoacceptor codon frequencies for each of seven gene sequences defining the three groups of proteins for (C) all possible codons and (D) the eight Q-decoded codons. For (C) and (D), the horizontal gray, green, and red bars highlight the no, down, and glyco genes, respectively, matching the color coding used in (A).
Discussion
Our study demonstrates that the human tRNA-specific epitranscriptome is dynamically regulated in response to exposures from a broad range of oxidizing agents, alkylators, and ionizing radiation. Moreover, we have shown that the reprogramming of epitranscriptomic marks is not only agent-specific but also dose- and time-dependent. We have already shown in yeast that such tRNA modification reprogramming upon toxicant exposure regulates the selective translation of codon-biased mRNAs for critical proteins used in response to different cellular stresses (30–32). Our current studies in HepG2 cells suggest that this translational control mechanism also occurs in the human stress response. The conservation of the translational machinery from yeast to humans and the highly biased use of synonymous codons across the human genome further support our findings. However, while in yeast and bacteria the pattern of up- and down-regulation of these modifications was unique to each stressor and predictive of exposure (27, 32), the toxicant-specific reprogramming was more subtle in HepG2 cells. While we did observe some general trends in our human data sets from the 24-h time point, the epitranscriptomic reprogramming did not uniquely distinguish each toxicant on visual observation but could distinguish the chemical oxidants from the other stressors. Increased and distinct epitranscriptomic regulation may occur at earlier time points in human cells, as suggested by the observation of nearly double the number of increased tRNA modifications at 6 h after NaAsO2 exposure compared to 24 h (Fig. 1B). While we have identified changes in the levels of some tRNA modifications, whether this is due to changes in levels or activity of writers or the levels of specific tRNAs is still in question. The toxicant-induced epitranscriptomic reprogramming observed hours after exposure to each agent does support the idea that there are robust changes to protein synthesis and tRNA stability, which should be regulating gene expression outside of transcriptional changes (40).
While the exact mechanism of action of arsenite is still unknown, our work supports one emerging model in which arsenite and/or its monomethylarsonous acid metabolite react with targets in mitochondria, perhaps by bonding to protein thiols, to cause increases in superoxide (O2−) that trigger an oxidative stress response (3, 4). Our observation of mitochondrial pathology and shift to glycolysis at sublethal arsenite concentrations (Figs. 3 and 4) is consistent with a mitochondrial target in this model, while the grouping of arsenite-induced changes in tRNA modifications with H2O2 and TBOOH (Fig. 1) supports the idea that arsenite, which is itself not an oxidant, is inducing oxidative stress. Similar to studies in bacteria, yeast, and mice (27, 32, 33, 41, 42), we have shown that in human cells, toxicant treatment induced changes in the levels of wobble base modifications, with the wobble U modifications mcm5U and mcm5s2U increased by NaAsO2 and TBOOH, respectively. Specific roles in gene expression for the toxicant-regulated modifications mcm5U and mcm5s2U have been described in yeast (43, 44), as these wobble U epitranscriptomic marks are key to the translation of specific codon-biased stress response transcripts enriched for AGA and GAA codons (29, 42, 45). In addition, the mcm5U modification is found on tRNA-Sec, which is required for the specialized translation of selenoproteins, many of which are ROS-managing glutathione peroxidases and thioredoxin reductases (32). Our study also highlights that the H2O2-induced increase in m5C is conserved from yeast to humans (31, 46). Studies in yeast showed that Trm4-catalyzed m5C at the wobble position of tRNA-Leu(CAA) is needed to survive H2O2 exposure, with the translation of UUG biased mRNA in ribosomal proteins facilitated by this epitranscriptomic mark (32). It is indeed possible that some of these changes in tRNA modification levels are caused by arsenic activation of specific writer enzymes or inhibition of specific tRNA-degrading enzymes, with detailed molecular studies required to establish exact mechanisms of action, which are likely to be many. Indeed, arsenite did not alter the levels of mRNA or protein for the Q synthetic enzyme QTRT1 or mRNA for QTRT2 (SI Appendix, Fig. S4 and Table S5), so we cannot rule out posttranslational changes caused by arsenite. Our observation that Q levels in tRNA are sensitive to queuine levels raises the possibility of arsenic-induced changes in queuine uptake or metabolism.
One finding here is the induction of the RNA modification m6t6A at the 6 h time point in response to NaAsO2. m6t6A is found adjacent to the anticodon on position 37 of tRNA for Ser (10), and it does not interact with the codon triplet directly. The presence of a bulky modification like m6t6A has the potential to stabilize codon–anticodon interactions by modulating the base stacking interaction in the decoding center of the ribosome (7). m6t6A is modified from a pre-existing t6A residue by tRNA methyltransferase O (TRMO) in the presence of methyl donor SAM (47). The m6t6A modification is known to occur on the tRNASer, which has the potential to decode AGC and AGU codons (48). While we did not observe overusage of AGC or AGU codons in genes for the up-regulated glycolytic proteins, we did observe up-regulation of TRMO mRNA in response to NaAsO2 (SI Appendix, Table S5). It is indeed possible that a deeper proteome coverage would reveal arsenite-altered proteins from genes with biased use of AGC and AGU. There are limited studies detailing the roles of human TRMO, although it has recently been detailed as one of eight differentiated thyroid carcinoma–associated genes (49). Our study demonstrates that detectable levels of m6t6A increase in response to NaAsO2, which could be due to increased TRMO activity, increased tRNASer levels, or changes in the pool of tRNAs, with the role of m6t6A in the response not yet determined. The increased levels of both m6t6A and Q in tRNA early in the response NaAsO2 suggests that the translation of Ser could be affected during this time as we observed with Tyr, Asn, and Asp.
We also analyzed epitranscriptomic reprogramming over time in response to an LD70 NaAsO2 exposure. The earliest epitranscriptomic increase was observed 30 min into the stress, with the wobble uridine modification mcm5U transiently increasing until 1 h after exposure. This represents a relatively fast change in the activity of tRNA-modifying enzymes and possibly tRNA levels. Deficiencies in mcm5U and the corresponding writer enzymes yeast Trm9 or human/mouse Alkbh8 have been shown to sensitize cells to cellular stress (29, 33, 50). ALKBH8 deficiencies corrupt selenocysteine incorporation, with corresponding selenoproteins regulating ROS detoxification and linked to mitochondrial health (33, 51). The translation of these selenoproteins is facilitated by the ALKBH8-catalyzed modification (mcm5U) occurring on tRNASec (33). Specifically, a deficiency in mouse Alkbh8 has been shown to sensitize mouse embryonic fibroblasts to H2O2 and the mitochondrial toxicant rotenone and to promote increased senescence (33, 51). Mice deficient in Alkbh8 are also susceptible to killing by the toxicant naphthalene and fail to develop tolerance to this polycyclic aromatic hydrocarbon most notably found in mothballs (52). In addition to tRNAsec, the mcm5U modification is also found on tRNAs for Arg and Gly. ALKBH8 and mcm5U have known roles in regulating the translation of codon-biased transcripts for selenoproteins, and it is likely that they also protect the cellular environment in response to NaAsO2 via the regulation of selenocysteine incorporation and perhaps the translation of codons for Arg and Gly. The regulation of mcm5U early during the exposure to NaAsO2 and then again 6 h later highlights the dynamic nature of the wobble U and a late epitranscriptomic program that includes m6t6A, mcm5U, and Q.
A specific epitranscriptomic reprogramming trend we observed was that the chemical oxidants all induced increased levels of Q in tRNA, which is found at the wobble position of tRNAs that read codons for Tyr (Gal-Q), His (Q), Asn (Q), and Asp (Man-Q). We also showed that queuine, likely as Q in tRNA, is part of an epitranscriptomic program responding to and preventing mitochondrial stress and increased ROS. Our studies emphasize the importance of environmental queuine and Q in the wobble position of four tRNAs to properly respond to toxicant insult. Further, we demonstrate an epitranscriptomic determinant of cellular responses from NaAsO2 and have implicated tRNA modification and translational regulation as an important response mechanism. We sought to place translational regulation in the context of the stressor, with the shift to aerobic glycolysis in response to NaAsO2, a feature that has been previously reported to drive a Warburg-like effect in Caenorhabditis elegans. It has also been shown that defects in glycolysis or ETC genes sensitize nematodes to arsenite (53). Arsenite-mediated glycolysis is reliant on HIF-1A, a key transcription factor involved in regulating glycolytic and TCA activity (54). The reduction of Q in tRNA further enhances cellular ROS levels and mitochondrial damage in response to NaAsO2, both of which are negated by queuine repletion. Q-modified tRNAs are specific for four amino acids with Gua in the first anticodon position and include Tyr, His, Asp, and Asn. Luz et al. (53) analyzed the levels of 15 free amino acids including Asp, His, and Tyr following arsenic exposure and observed statistically significant increases in these Q-dependent amino acids. The findings that Q is the most highly regulated epitranscriptomic modification in response to NaAsO2 and that queuine deprivation increases ROS and mitochondrial damage implicate Tyr, His, Asp, and possibly Asn as critical in preserving mitochondrial integrity in response to the common environmental toxicant arsenic. Coupled with our observation that m6t6A and mcm5U are coregulated with Q, the list of critical amino acids preserving mitochondrial integrity could be expanded to include Tyr, His, Asp, Asn, Ser, Arg, Gly, and Sec.
Changes in translation are hallmarks of cancer cells, and our studies highlight that toxicant exposures that damage cells also affect translation. It has previously been shown in breast cancer cells that the enzymes ELP3 and CTU1/2, which modify the wobble U34 to mcm5s2 in tRNA Asp and tRNA Glu, are required for metastasis and invasion by translationally regulating the oncoprotein DEK1 (55). The increased DEK1 promotes the internal ribosome entry site (IRES)-dependent translation of the transcription factor LEF1, which can drive a proinvasion program. Further, in melanoma cells, it has been demonstrated that wobble U modifications promote glycolysis through the codon-dependent and translational regulation of HIF-1A (55, 56). Codon-engineered versions of DEK1 or HIF-1A can uncouple their translational regulation from the levels of specific tRNA modifications (55, 56). Our study highlights that Q in tRNA is also linked to the regulation of carbon metabolism and glycolysis, with codon-biased networks found in enzymes that regulate glycolysis and the pentose–phosphate pathway. That arsenite exposure causes Q-dependent up-regulation of glycolytic enzymes from genes enriched with C-ending GUN codons supports this model. The C-ending codons for Asn, Asp, and Tyr are significantly overused in the group of seven genes that correspond to our up-regulated proteins controlling glycolysis, supporting a model in which the translation of the corresponding mRNAs is enhanced. A recent study by Watkins and colleagues supports our findings, as they used their developed multiplex small RNA-sequencing library preparation method to assess the translational efficiency of mRNAs during stress responses (57). Their polysome-based approaches highlighted that in response to NaAsO2, translation is dependent on codon usage, with increased translational efficiency observed for transcripts enriched for codons ending in C/G and decreased in those enriched for A/T-ending codons. Further, similar to our findings, Watkins and colleagues did not observe a significant change in CAC translational efficiency either (57). Translational regulation by Q is complicated, and studies by Zaborske and colleagues in fruit flies supported a model in which Q increased the accuracy for cognate codons while reducing accuracy for near-cognate codons (58). Translational infidelity for near-cognate codons may also play a role in Q-dependent translational regulation. While this general model can provide insight into the effect of Q on translation, future studies are needed to clarify isoacceptor-specific changes for Gal-Q and Man-Q.
The observation that queuine from the environment is an important driver of translation and mitochondrial function further highlights the complex relationship between the microbiome, exposures, and humans. Our study demonstrates that queuine and Q are major determinants of the response to environmental toxicants. Q has also recently been shown to directly protect the cognate tRNAs against stress-induced ribonuclease cleavage and therefore also has the potential to influence the abundance and variety of tRNA-derived small RNA fragments in human cells (26). The response to environmental toxicants clearly includes transcriptional, posttranscriptional, and translational response. Our study provides further evidence that in addition to traditional stress response pathways, metabolic responses and epitranscriptomic reprogramming are key determinants of cellular outcomes. It was recently shown that epitranscriptomic systems are needed to respond to the polyaromatic hydrocarbon naphthalene (59). Clearly, other tRNA modifications and epitranscriptomic marks will play important roles in the response to environmental exposures, as therapeutic responses, and in cancer biology.
Materials and Methods
Cell culture conditions.
HepG2 cells were cultured in an NGM consisting of DMEM, low glucose with glutamine and pyruvate, supplemented with 10% fetal bovine serum (FBS), 1× MEM nonessential amino acids, and 1× penicillin–streptomycin solution at 37 °C in 5% CO2 atmosphere. Q-depleted HepG2 cells were generated by culturing the cells in medium in which the 10% FBS component was replaced with dialyzed fetal bovine serum (dFBS) at 10%, with growth for ≥10 passages while maintaining the cells at 70 to 80% confluency. Cells were also grown in dFBS supplemented with 1 μM queuine hydrochloride (dFBS+queuine) for 24 h prior to harvesting the cells. During treatment and replacement of growth media, the cells remained in their designated cell culture medium (NGM, dFBS, or dFBS+queuine). Mitochondria from HepG2 cells were purified using the Qproteome Mitochondria Isolation kit according to the manufacturer’s instructions and resuspended in TRI-Reagent for total RNA isolation. See SI Appendix, Extended Methods for details.
Proliferation/MTT assay and dose–response curves.
To determine the dose–response curves, HepG2 cells were treated with various concentrations of toxicants for 2 h in cell culture medium (NGM, dFBS, or dFBS + queuine). Ionizing radiation (γ-radiation; Rad) was delivered by an X-ray generator at a dose rate of 1.48 Gy per min. After treatments, the medium was replaced with fresh growth medium and, at specified times after treatment, cells were analyzed using the MTT. Dose–response curves were plotted using Prism software and a nonlinear curve fit (SI Appendix, Fig. S1). Data for LD20 and LD50 doses are noted in SI Appendix, Fig. S1. Student t tests were performed using GraphPad Prism 6 software. See SI Appendix, Extended Methods for details.
RNA extraction and purification.
Total RNA was extracted from HepG2 cells using TRIzol reagent (ThermoFisher Scientific, Waltham, MA) according to the manufacturer’s instructions. Purified total RNA was air-dried and then resuspended in an appropriate amount of RNase-free water. Samples were flash-frozen in liquid nitrogen and stored at −80 °C. See SI Appendix, Extended Methods for details.
tRNA purification and sample preparation.
Purification of tRNA from total RNA was carried out by fractional precipitation using the Quick-RNA MiniPrep kit according to the manufacturer’s instructions. tRNA hydrolysis to ribonucleosides was achieved by mixing tRNA (6 μg in 30 μL H2O) with a digestion enzyme master mix (20 μL; SI Appendix, Table S4) and incubating the mixture at 37 °C for 6 h. Enzymes were subsequently removed by filtration through 10 kDa MWCO spin columns before concentrating the ribonucleoside mixture to dryness. See SI Appendix, Extended Methods for details and SI Appendix, Fig. S5.
LC-MS/MS analysis of modified ribonucleosides.
Ribonucleoside mixtures were redissolved in 5 mM ammonium acetate and resolved on a Synergi Fusion reversed-phase HPLC column coupled to an Agilent 6490 Triple Quadrupole LC/MS spectrometer with an electrospray ionization source. Modified ribonucleosides were identified using synthetic standards or by HPLC retention time and collision-induced dissociation fragmentation patterns. Biological triplicates of both exposed and unexposed control cells were used for each toxicant. The mass spectrometric peak area for each modification was corrected for variation in the quantity of input tRNA by dividing the peak area by the total ultraviolet-visible light (UV–Vis) absorbance peak area for the four canonical ribonucleosides. Fold-change values for ribonucleosides were calculated for each dose and time point and significant changes were determined by Student t test (P ≤ 0.05). See SI Appendix, Extended Methods for details and Dataset S1 for data.
Labeling mitochondria with MitoTracker Green.
To label the mitochondria, HepG2 cells were grown in NGM, dFBS, and dFBS+queuine and seeded in 12-well dishes and incubated in a humidified CO2 incubator overnight. Cells were treated with an LD20 and LD50 of NaAsO2 in culture medium for 2 h, followed by replacement with fresh growth medium and continued incubation for 6 h. Growth medium was removed and staining solution (media without FBS) containing MitoTracker green FM was added to the cells and incubated for a total of 30 min at 37 °C. After 20 min, Nuc Blue Live was added for 10 min to stain the nuclei. Cells were washed with PBS to remove excess stain and kept in PBS during microscopy. Cells were immediately imaged using the EVOS fluorescent imaging system. See SI Appendix, Extended Methods for details.
Analysis of mitochondrial function and energy metabolism.
To assess the mitochondrial function of live HepG2 cells and possible metabolic switching, we used the Seahorse XF24 extracellular flux analyzer kit, similar to our previously reported study (51). Briefly, HepG2 cells were seeded in an XF 24-well microplate overnight, allowing the cells to adhere. An XF sensor cartridge was hydrated with XF Calibrant overnight in a non-CO2 incubator at 37 °C. After seeding, the cells were treated with LD20 and LD50 doses of NaAsO2, after which growth medium was replaced with fresh growth media and samples were incubated. The assay medium was prepared according to manufacturer’s instructions. Cells were washed once with the assay medium and the XF culture plate was equilibrated in a non-CO2 incubator at 37 °C for 1 h prior to the start of the assay. During this equilibration time, the assay compounds were prepared and added to the appropriate ports of the hydrated sensor cartridge: oligomycin, FCCP, and rotenone. Using OCR and ECAR data points from the analysis, the cell energy phenotype was automatically generated using Wave software, which yielded an energy phenotype of the untreated versus treated HepG2 cells. See SI Appendix, Extended Methods for details.
Measurement of intracellular ROS levels.
To assess intracellular ROS levels of HepG2 cells grown in NGM, dFBS, and dFBS+queuine in response to NaAsO2 treatment, cells were seeded in 6-well dishes and incubated in a humidified CO2 incubator overnight. Cells were treated with LD20 and LD50 doses of NaAsO2 for 2 h, followed by replacement with fresh growth media and incubation for 24 h. CellROX Orange Reagent was then added directly to the cells and incubated in a humidified CO2 incubator at 37 °C, followed by the addition of Nuc Blue Live Stain and incubation to stain the nuclei. Cells were washed with PBS to remove excess stain and kept in PBS during microscopy. Cells were imaged using the EVOS fluorescent imaging system. CellROX Orange was detected with the RFP LED light cube and Nuc Blue Live Stain with the DAPI LED light cube. See SI Appendix, Extended Methods for details.
mRNA analysis.
Cells were grown in NGM and were seeded in a 100 mm dish and incubated overnight at 37 °C in 5% CO2. After overnight incubation, cells were treated with an LD20 dose of NaAsO2 for 2 h. Following media exchange and incubation, RNA was isolated. The sequencing library was prepared and sequencing was performed on the Illumina HiSeq platform. Data quality was analyzed using the Phred quality score with the baseline set at Q30. The indexed human genome was acquired from Biostars (60). The reads were aligned using STAR (61). The differential gene expression (DGE) analysis was performed using DESeq2 with default parameters (62). The DGE analysis was visualized using R studio (https://www.rstudio.com/). See SI Appendix, Extended Methods for details and SI Appendix, Table S5.
Polysome analysis.
The cells were exposed to their experimental conditions and cycloheximide was added with incubation for 3 min and then immediately put on ice. The plates were washed 3 times with 0.1 mg/mL cycloheximide in PBS. After washing, cell lysis buffer was added and samples were incubated for 10 min. The cells were removed from the plate using a cell scraper. The lysate solution was then incubated on ice for 10 min and then centrifuged at 4 °C for 10 min at 7,056 relative centrifugal force (RCF). Cell lysate was then laid on top of a sucrose gradient in an ultracentrifuge tube and samples were centrifuged at 104,456 x g at 4 °C for 2.75 h. After centrifugation, samples were fractionated and analyzed by UV-Vis spectroscopy. Polysome profiles were collected using an ISCO UA-6 UV-vis detector and Windaq software and scaled to the 80S peak and zeroed to the smallest detected value from the polysome. See SI Appendix, Extended Methods for details and SI Appendix, Fig. S3.
Proteomics.
Biological triplicates of NaAsO2-exposed and control HepG2 cells were lysed using cold urea lysis buffer and protein disulfides were reduced using DTT, alkylated using iodoacetamide, and finally digested using trypsin at an enzyme-to-protein ratio of 1:50. Samples were then desalted using Pierce Peptide desalting spin columns and divided into 400 µg aliquots, concentrated to ∼1 to 5 µL, and frozen at −80 °C. Peptide digests were labeled with ThermoFisher Tandem Mass Tag (TMT)-6plex reagents, combined, and dried under vacuum. Dried peptides were resuspended in 0.1% formic acid, loaded onto a precolumn, and resolved by reverse-phase HPLC. The eluent was directly injected into an Orbitrap QExactive mass spectrometer with electrospray ionization and operated in a data-dependent mode. Raw mass spectral data files (.raw) were searched using Proteome Discoverer 2.1 and Mascot version 2.4.1 against the human UniProt database. TMT quantification was obtained using Proteome Discoverer and isotopically corrected according to manufacturer instructions. The TMT values were normalized to the mean relative protein quantification ratios obtained from a total protein analysis. Without fractionation, we were able to identify 544 proteins, 286 of which satisfied the filtering criteria. Proteomics data were submitted to the PRIDE repository under submission number 1–20211230-6555 on December 30, 2021. See SI Appendix, SI Extended Methods for details.
Codon analytics.
Human cDNAs, which in some cases included multiple splice variants, for all known proteins were identified in GenBank and parsed to contain the complete coding sequence from start to stop codon. The resulting sequences were analyzed for codon isoacceptor frequency using the previously described Gene-Specific Codon Usage (GCSU) algorithm (29–32). The total codon frequency for each codon in a gene was also analyzed using GCSU. Total codon frequency measurers were determined by dividing the number of times a specific codon was found in a gene by the total number of codons in the gene, with the sum of all total codon frequencies in a gene adding to 1.0. Statistical analysis of specific gene lists to identify codon bias was determined using the t-statistic. See SI Appendix, Extended Methods for details.
Supplementary Material
Acknowledgments
Research was funded in part by grants from the NIH (R01 ES026856 and R01 ES024615 to T.J.B.) and the Swiss NSF (P2SKP3_174681 to S.M.H.). Proteomics analyses were performed in the Proteomics Facility of the MIT Koch Integrative Cancer Institute.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
*The complete chemical names of all RNA modifications discussed here can be found at the Modomics website (10).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2123529119/-/DCSupplemental.
Data, Materials, and Software Availability
Proteomics data have been deposited in the PRIDE repository [https://www.ebi.ac.uk/pride/, PXD030726 (63)].
References
- 1.Guéguen Y., Bontemps A., Ebrahimian T. G., Adaptive responses to low doses of radiation or chemicals: Their cellular and molecular mechanisms. Cell. Mol. Life Sci. 76, 1255–1273 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa M., Review of arsenic toxicity, speciation and polyadenylation of canonical histones. Toxicol. Appl. Pharmacol. 375, 1–4 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Hei T. K., Filipic M., Role of oxidative damage in the genotoxicity of arsenic. Free Radic. Biol. Med. 37, 574–581 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Ellinsworth D. C., Arsenic, reactive oxygen, and endothelial dysfunction. J. Pharmacol. Exp. Ther. 353, 458–464 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Motorin Y., Helm M., tRNA stabilization by modified nucleotides. Biochemistry 49, 4934–4944 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Alexandrov A., et al. , Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 21, 87–96 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Agris P. F., Vendeix F. A. P., Graham W. D., tRNA’s wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 366, 1–13 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Urbonavičius J., Qian Q., Durand J. M. B., Hagervall T. G., Björk G. R., Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 20, 4863–4873 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantara W. A., et al. , The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 39, D195–D201 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boccaletto P., et al. , MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuorto F., Lyko F., Genome recoding by tRNA modifications. Open Biol. 6, 160287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris R. C., Brown K. G., Elliott M. S., The effect of queuosine on tRNA structure and function. J. Biomol. Struct. Dyn. 16, 757–774 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Suzuki T., Suzuki T., A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 42, 7346–7357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulkarni S., et al. , Preferential import of queuosine-modified tRNAs into Trypanosoma brucei mitochondrion is critical for organellar protein synthesis. Nucleic Acids Res. 49, 8247–8260 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El Yacoubi B., Bailly M., de Crécy-Lagard V., Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 46, 69–95 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Reyniers J. P., Pleasants J. R., Wostmann B. S., Katze J. R., Farkas W. R., Administration of exogenous queuine is essential for the biosynthesis of the queuosine-containing transfer RNAs in the mouse. J. Biol. Chem. 256, 11591–11594 (1981). [PubMed] [Google Scholar]
- 17.Katze J. R., Gündüz U., Smith D. L., Cheng C. S., McCloskey J. A., Evidence that the nucleic acid base queuine is incorporated intact into tRNA by animal cells. Biochemistry 23, 1171–1176 (1984). [DOI] [PubMed] [Google Scholar]
- 18.Tuorto F., et al. , Queuosine-modified tRNAs confer nutritional control of protein translation. EMBO J. 37, e99777 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Owenby R. K., Stulberg M. P., Jacobson K. B., Alteration of the Q family of transfer RNAs in adult Drosophila melanogaster as a function of age, nutrition, and genotype. Mech. Ageing Dev. 11, 91–103 (1979). [DOI] [PubMed] [Google Scholar]
- 20.Singhal R. P., Kopper R. A., Nishimura S., Shindo-Okada N., Modification of guanine to queuine in transfer RNAs during development and aging. Biochem. Biophys. Res. Commun. 99, 120–126 (1981). [DOI] [PubMed] [Google Scholar]
- 21.Durand J. M., et al. , vacC, a virulence-associated chromosomal locus of Shigella flexneri, is homologous to tgt, a gene encoding tRNA-guanine transglycosylase (Tgt) of Escherichia coli K-12. J. Bacteriol. 176, 4627–4634 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marks T., Farkas W. R., Effects of a diet deficient in tyrosine and queuine on germfree mice. Biochem. Biophys. Res. Commun. 230, 233–237 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Gündüz U., et al. , Absence of tRNA-guanine transglycosylase in a human colon adenocarcinoma cell line. Biochim. Biophys. Acta 1139, 229–238 (1992). [DOI] [PubMed] [Google Scholar]
- 24.Skolnick S. D., Greig N. H., Microbes and monoamines: Potential neuropsychiatric consequences of dysbiosis. Trends Neurosci. 42, 151–163 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Pathak C., Jaiswal Y. K., Vinayak M., Queuine promotes antioxidant defence system by activating cellular antioxidant enzyme activities in cancer. Biosci. Rep. 28, 73–81 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Wang X., et al. , Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA 24, 1305–1313 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chionh Y. H., et al. , tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat. Commun. 7, 13302 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng C. S., et al. , tRNA epitranscriptomics and biased codon are linked to proteome expression in Plasmodium falciparum. Mol. Syst. Biol. 14, e8009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Begley U., et al. , Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell 28, 860–870 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan C. T., et al. , Highly predictive reprogramming of tRNA modifications is linked to selective expression of codon-biased genes. Chem. Res. Toxicol. 28, 978–988 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan C. T. Y., et al. , A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 6, e1001247 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan C. T. Y., et al. , Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 3, 937 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Endres L., et al. , Alkbh8 regulates selenocysteine-protein expression to protect against reactive oxygen species damage. PLoS One 10, e0131335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thumbs P., et al. , Synthesis of galactosyl-queuosine and distribution of hypermodified Q-nucleosides in mouse tissues. Angew. Chem. Int. Ed. Engl. 59, 12352–12356 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kharroubi W., et al. , Mitochondrial dysfunction, oxidative stress and apoptotic induction in microglial BV-2 cells treated with sodium arsenate. J. Environ. Sci. (China) 51, 44–51 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Partridge M. A., Huang S. X., Hernandez-Rosa E., Davidson M. M., Hei T. K., Arsenic induced mitochondrial DNA damage and altered mitochondrial oxidative function: Implications for genotoxic mechanisms in mammalian cells. Cancer Res. 67, 5239–5247 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Prakash C., Soni M., Kumar V., Biochemical and molecular alterations following arsenic-induced oxidative stress and mitochondrial dysfunction in rat brain. Biol. Trace Elem. Res. 167, 121–129 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Prakash C., Soni M., Kumar V., Mitochondrial oxidative stress and dysfunction in arsenic neurotoxicity: A review. J. Appl. Toxicol. 36, 179–188 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Jang C. H., et al. , PGK1 induction by a hydrogen peroxide treatment is suppressed by antioxidants in human colon carcinoma cells. Biosci. Biotechnol. Biochem. 72, 1799–1808 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Bramson J., O’Connor T., Panasci L., Effect of alkyl-N-purine DNA glycosylase overexpression on cellular resistance to bifunctional alkylating agents. Biochem. Pharmacol. 50, 39–44 (1995). [DOI] [PubMed] [Google Scholar]
- 41.Begley U., et al. , A human tRNA methyltransferase 9-like protein prevents tumour growth by regulating LIN9 and HIF1-α. EMBO Mol. Med. 5, 366–383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patil A., et al. , Increased tRNA modification and gene-specific codon usage regulate cell cycle progression during the DNA damage response. Cell Cycle 11, 3656–3665 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zinshteyn B., Gilbert W. V., Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet. 9, e1003675 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nedialkova D. D., Leidel S. A., Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 161, 1606–1618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng W., et al. , Trm9-catalyzed tRNA modifications regulate global protein expression by codon-biased translation. PLoS Genet. 11, e1005706 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Driscoll D. M., Copeland P. R., Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 23, 17–40 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Kimura S., Dedon P. C., Waldor M. K., Comparative tRNA sequencing and RNA mass spectrometry for surveying tRNA modifications. Nat. Chem. Biol. 16, 964–972 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kimura S., et al. , Discovery of the β-barrel-type RNA methyltransferase responsible for N6-methylation of N6-threonylcarbamoyladenosine in tRNAs. Nucleic Acids Res. 42, 9350–9365 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kulkarni O., et al. , Gene network and biological pathways associated with susceptibility to differentiated thyroid carcinoma. Sci. Rep. 11, 8932 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patil A., et al. , Translational infidelity-induced protein stress results from a deficiency in Trm9-catalyzed tRNA modifications. RNA Biol. 9, 990–1001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee M. Y., Leonardi A., Begley T. J., Melendez J. A., Loss of epitranscriptomic control of selenocysteine utilization engages senescence and mitochondrial reprogramming. Redox Biol. 28, 101375 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leonardi A., Evke S., Lee M., Melendez J. A., Begley T. J., Epitranscriptomic systems regulate the translation of reactive oxygen species detoxifying and disease linked selenoproteins. Free Radic. Biol. Med. 143, 573–593 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luz A. L., et al. , From the cover: Arsenite uncouples mitochondrial respiration and induces a Warburg-like effect in Caenorhabditis elegans. Toxicol. Sci. 152, 349–362 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao F., Severson P., Pacheco S., Futscher B. W., Klimecki W. T., Arsenic exposure induces the Warburg effect in cultured human cells. Toxicol. Appl. Pharmacol. 271, 72–77 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delaunay S., et al. , Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J. Exp. Med. 213, 2503–2523 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rapino F., et al. , Codon-specific translation reprogramming promotes resistance to targeted therapy. Nature 558, 605–609 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Watkins C. P., Zhang W., Wylder A. C., Katanski C. D., Pan T., A multiplex platform for small RNA sequencing elucidates multifaceted tRNA stress response and translational regulation. Nat. Commun. 13, 2491 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zaborske J. M., et al. , A nutrient-driven tRNA modification alters translational fidelity and genome-wide protein coding across an animal genus. PLoS Biol. 12, e1002015 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leonardi A., et al. , The epitranscriptomic writer ALKBH8 drives tolerance and protects mouse lungs from the environmental pollutant naphthalene. Epigenetics 15, 1121–1138 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parnell L. D., et al. , BioStar: An online question & answer resource for the bioinformatics community. PLOS Comput. Biol. 7, e1002216 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dedon P., Arsenic toxicity is regulated by queuine availability and oxidation-induced reprogramming of the human tRNA epitranscriptome. Proteomics. https://www.ebi.ac.uk/pride/archive?keyword=PXD030726. Deposited 27 August 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomics data have been deposited in the PRIDE repository [https://www.ebi.ac.uk/pride/, PXD030726 (63)].