Abstract
A cluster of 18 open reading frames (ORFs), 15 of which are homologous to genes involved in division and cell wall synthesis, has been identified in Neisseria gonorrhoeae and Neisseria meningitidis. The three additional ORFs, internal to the dcw cluster, are not homologous to dcw-related genes present in other bacterial species. Analysis of the N. meningitidis strain MC58 genome for foreign DNA suggests that these additional ORFs have not been acquired by recent horizontal exchange, indicating that they are a long-standing, integral part of the neisserial dcw gene cluster. Reverse transcription-PCR analysis of RNA extracted from N. gonorrhoeae strain FA19 confirmed that all three ORFs are transcribed in gonococci. One of these ORFs (dca, for division cluster competence associated), located between murE and murF, was studied in detail and found to be essential for competence in the gonococcal but not in the meningococcal strains tested. Computer analysis predicts that dca encodes an inner membrane protein similar to hypothetical proteins produced by other gram-negative bacteria. In some meningococcal strains dca is prematurely terminated following a homopolymeric tract of G's, the length of which differs between isolates of N. meningitidis, suggesting that dca is phase variable in this species. A deletion and insertional mutation was made in the dca gene of N. gonorrhoeae strain FA19 and N. meningitidis strain NMB. This mutation abrogated the ability of the gonococci to be transformed with chromosomal DNA. Thus, we conclude that the dca-encoded gene product is an essential competence factor for gonococci.
The proteins encoded by bacterial division and cell wall (dcw) gene clusters are essential for viability (16). Significant progress has been made in recent years in understanding the genes involved in cell wall biosynthesis and division (5, 6, 7, 11, 32, 62). Fifteen genes located at 2 min on the Escherichia coli chromosome have been identified and are called the dcw cluster (4, 64). The genes of this cluster are tightly packed, some overlapping, and are oriented in one direction (4, 64). Although the dcw cluster is highly conserved among evolutionarily diverse bacterial species (48), some variations in the clusters have been reported. These variations include the location of some dcw genes at separate chromosomal locations, such as ftsW, murF, and ddlA of Staphylococcus aureus (48), and the addition of species-specific genes within the dcw cluster, such as spoVD and spoVE of Bacillus subtilis (12, 31). In the case of these B. subtilis sporulation genes, spoVD (12) and spoVE (31) have homology to the dcw cluster genes pbpB and ftsW, respectively. Although not essential for vegetative growth, these genes are essential for sporulation (12, 31).
Since the products of the dcw genes encode proteins involved in peptidoglycan synthesis and cellular division, it is necessary to use conditionally lethal mutants to study these genes (5, 6, 11, 16, 27). Moreover, since they are typically expressed at low levels, it has been difficult to examine their regulation and to map their promoter elements (13). However, using promoter reporter constructs, a putative promoter (Pmra) upstream of the E. coli dcw cluster has been identified. This promoter is required for transcription of at least the first nine genes of the cluster (yabB to ftsW) and may provide the majority of the transcript for the downstream genes (30, 43).
Neisseria gonorrhoeae and N. meningitidis are important, human-specific pathogens and are causative agents of gonorrhea and of meningitis and septicemia, respectively. MtrR is a transcriptional regulator, in gonococci, of both the mtr operon and the far operon, which encode efflux pump proteins (28, 39). While investigating the regulation of the mtr efflux system by MtrR in N. gonorrhoeae, we performed a search of the gonococcal genome sequence database for the recognition site of MtrR (40) in an attempt to identify other genes that might be subject to its regulation. A potential MtrR-binding sequence was found upstream of an open reading frame (ORF) with homology to yabB of E. coli (4). A homologue of yabB is the first gene of the dcw cluster in all of the bacterial species studied to date (4, 13, 20, 38, 48, 64). Since the dcw cluster of the pathogenic Neisseria spp. had not been previously described at that time, we decided to investigate this gene cluster in the pathogenic Neisseria spp. A recent report (21) has described the gonococcal dcw cluster to some extent, although two ORFs (ftsL and the homologue of NMB0417) were not identified and a hypothetical gene was defined as the most 3′ gene of the cluster, rather than the conserved ftsZ. We report that while the dcw clusters of gonococci and meningococci are broadly similar to those described in other species, they contain three additional ORFs internal to the dcw cluster that are not present in the dcw clusters of other the bacterial species studied to date. We demonstrate that these ORFs do not have features of recent horizontal acquisition and that they are all transcribed. One of these ORFs, dca, has features suggestive of phase variability associated with polymorphisms between strains and is associated with natural competence in the gonococcal strains, although not in the meningococcal strains tested.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
We used N. gonorrhoeae strains FA19 and FA1090 (kindly provided by P. F. Sparling and J. Cannon, respectively, of the University of North Carolina School of Medicine, Chapel Hill). N. meningitidis invasive disease isolate strains (NMB [capsular serogroup B], 0929 [serogroup Y], and 2633 [serogroup Y]) and six commensal Neisseria spp. (N. flavescens, N. lactamica, N. macacae, N. polysaccharea, N. sicca, and N. subflava) were kindly provided by D. Stephens (Emory University School of Medicine, Atlanta, Ga.). A transformant of strain FA19 containing an insertional inactivation and deletion in the recA gene was constructed by transformation using pC68a (kindly provided by C. Gibbs and T. Meyer), which contains recA::ermC. All strains were propagated on GCB agar or in GCB broth (Difco Laboratories, Detroit, Mich.) containing the Kellogg supplements and ferric nitrate (36) at 37°C under 3.8% (vol/vol) CO2. Growth in liquid media employed nonpiliated, transparent colony variants with growth monitored by determining the optical density at 600 nm. Spontaneous streptomycin-resistant (Strr) mutants of gonococci were selected by plating 109 CFU of strain FA19 onto GCB agar plates containing 100 μg of streptomycin per ml. Strr mutants of meningococci were made by transformation of strain NMB with Strr gonococcal chromosomal DNA. Transformants were selected on GCB agar plates containing either 100 μg of streptomycin per ml or 1 μg of erythromycin per ml. All antimicrobial agents were obtained from Sigma Chemical Company (St. Louis, Mo.).
PCR amplification, plasmid preparation, and DNA sequencing.
Chromosomal DNA extractions were performed using the method of McAllister and Stephens (41). PCR from chromosomal DNA was performed as described previously (29). Plasmids were purified using the QIAprep Spin Miniprep Kit according to the manufacturer's instructions (QIAGEN, Valencia, Calif.). DNA sequencing of PCR products and plasmids was performed using automated sequencing by the Emory DNA Sequencing Core Facility (Atlanta, Ga.) using a PE Applied Biosystems 377 automated DNA sequencers (Foster City, Calif.). Sequencing was carried out by the modified method of Sanger et al. (50) using BigDye reaction chemistry (ABI PRISM; PE Applied Biosystems, Norwalk, Conn.). PCR products for sequencing of dca were generated with combinations of oligonucleotide primers orfA#1, orfA#3, orfA#5, orfA#6, and murE#4 (Fig. 1 and Table 1). DNAStar was used for nucleotide and amino acid sequence analysis and alignments (8). Protein topology predictions were generated using the algorithm of von Heijne (60, 61), as implemented by TopPredII (10), and PSORT (45).
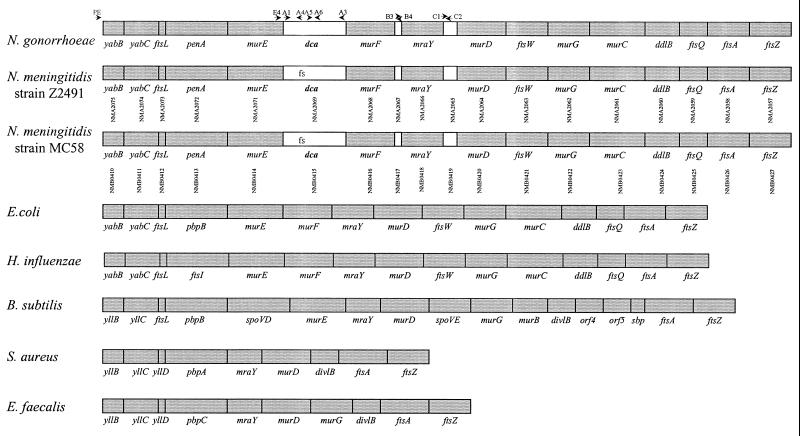
FIG. 1.
Comparison of the dcw clusters from N. gonorrhoeae, N. meningitidis strain Z2491 (46), N. meningitidis strain MC58 (59), E. coli (4, 64), H. influenzae (20), B. subtilis (13, 38), S. aureus, and E. faecalis (48). All genes are transcribed from left to right. Due to variations in nomenclature between the species, functionally conserved genes have different names; the penA, ftsI, and pbp genes are homologues, as are the yab and yll genes. The first two ORFs of the dcw clusters in this figure are named based on the consistent designations of these ORFs in GenBank. The spoVD and spoVE genes of B. subtilis are homologues of pbpB and ftsW, respectively, and the proteins they encode have a sporulation-specific function related to that of the PbpB and FtsW function in vegetative growth (12, 31). NMA and NMB annotation numbers are indicated below each ORF (46, 59). White boxes in N. gonorrhoeae and N. meningitidis represent ORFs not seen in other dcw clusters. The NMB0417 and NMB0419 homologues are of unknown function and are therefore not named in N. gonorrhoeae. The relative positioning of the frameshift in dca in N. meningitidis is indicated (fs). The relative positions of the oligonucleotides from Table 1 are shown (arrows). PE, yabBPE; E4, murE#4; A1, orfA#1; A3, orfA#3; A4, orfA#4; A5; orfA#5; A6, orfA#6; B3, dcaB#3; B4, dcaB#4; C1, dcaC#1; C2, dcaC#2. Apparent overlap is due to the limitations of the figure.
TABLE 1.
Oligonucleotides used for PCR and sequencing
| Primer | Oligonucleotide sequence (5′-3′) |
|---|---|
| orfA#1 | ATGAAACAATCCGCCCGAATA |
| orfA#3 | CGGCATTTTTTCTGTACGTA |
| orfA#4 | AATCAGTAAAACAACTGAAG |
| orfA#5 | ATCCTTATCCAAACGTGTGCA |
| orfA#6 | ATGCCGGCCTGTTGTTGAATAT |
| murE#4 | GGATAAAGTCGTCGTAACCA |
| dcaB#3 | TGAAAATGAAAAGCCGACG |
| dcaB#4 | TAACGTGCAGGGACTTTC |
| dcaC#1 | CCGCCCTGATTGCCTTGG |
| dcaC#2 | CTTCAGGCGTTGGTCATTGTC |
| yabBPE | CTGTCGATGCTTAATTCGTGTGCGCCGCCGAACAT |
DNS.
A DNA sequence can be considered to consist of a string of dinucleotides. Analysis of the proportions of dinucleotides in large sequences has revealed that species differences are consistently present in genome sequence compositions (34, 35). This approach has been used to identify regions of horizontally transferred DNA in N. meningitidis MC58 (59). We used a modification of this methodology that addresses single ORFs (44; J. Mirsky, N. J. Saunders, J. F. Peden, and S. Jarvis, unpublished data). We have used dinucleotide signature analysis (DNS) to evaluate the possibility that the ORFs within the dcw cluster were acquired by recent horizontal transfer of DNA.
Nucleotide sequence searches.
The Basic Local Alignment Search Tool (BLAST) (1) was used to search publicly available microbial genome sequences and GenBank. The sequences of yabB, yabC, ftsL, penA, murE, dca, murF, the homologue of NMB0417, mraY, the homologue of NMB0419, murD, ftsW, murG, murC, ddlB, ftsQ, ftsA, and ftsZ for gonococci were obtained from the N. gonorrhoeae Genome Sequencing Project at the University of Oklahoma (http://www.genome.ou.edu/gono.html). The corresponding meningococcal sequences from serogroup A strain Z2491 were obtained from the Sanger Centre. This sequence data was produced by the N. meningitidis Sequencing Group at the Sanger Centre and can be obtained online (http://www.sanger.ac.uk/Projects/N_meningitidis/) (46). The meningococcal sequence from serogroup B strain MC58 was produced by The Institute for Genomic Research (http://www.tigr.org/tdb/CMR/gnm/htmls/SplashPage.html) (59). The sequence of dca from N. meningitidis serogroup C strain FAM18 was produced by the Sanger Centre (http://www.sanger.ac.uk /Projects/N_meningitidis/seroC.shtml). GenBank was accessed through the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). The sequences of the Actinobacillus actinomycetemcomitans HK1651 dca homologues were obtained from the University of Oklahoma (http://www.genome.ou.edu/act.html). The sequence of the Salmonella enterica serovar Typhi strain CT18 dca homologue was obtained from the Sanger Centre (http://www.sanger.ac.uk/Projects/S_typhi/).
Primer extension analysis.
Total RNA was prepared using the hot-phenol method (3). Primer extension products were generated from the yabB-specific oligonucleotide, yabBPE (Table 1), hybridized to 20 μg of total RNA. The DNA sequence was generated using the same oligonucleotide as a primer and therefore corresponds to that of the mRNA (28).
RT-PCR.
RNA was prepared as for primer extension analysis. cDNA was generated from 1 μg of RNA reverse transcribed with SuperScript II RNase H− reverse transcriptase (Gibco-BRL, Grand Island, N.Y.) and ORF-specific oligonucleotide primers orfA#4, dcaB#4, and dcaC#2 (Table 1). The cDNA was PCR amplified with oligonucleotide primer combinations of orfA#4 and orfA#1, dcaB#4, and dcaB#3 and dcaC#2 and dcaC#1 (Fig. 1 and Table 1). In order to detect contaminating DNA in the RNA preparation, control reverse transcription-PCR (RT-PCR) reactions were conducted in the absence of reverse transcriptase.
Southern blotting.
A dca-specific probe of 1.6 kb was generated by PCR amplification of FA19 chromosomal DNA with orfA#1 and orfA#3 (Table 1) and labeled using the Boehringer-Mannheim Non-radioactive DNA Genius 2 Labeling Kit according to the manufacturer's instructions (Indianapolis, Ind.). Three micrograms of chromosomal DNA, determined using a Beckman EU65 spectrophotomer (Fullerton, Calif.) at A260, was digested with ClaI, separated by 0.6% (wt/vol) agarose gel electrophoresis, and blotted to a 0.45-μM (pore-size) MagnaGraph nylon transfer membrane (Micron Separations, Inc., Westboro, Mass.). Hybridization, washes, and detection were performed as described in the Genius System User's Guide for Membrane Hybridization using anti-digoxigenin alkaline phosphatase conjugate and CSPD (Boehringer Mannheim).
dca-inactivation and transformation.
dca was PCR-amplified from genomic DNA of strain FA19 using gene-specific primers (orfA#1 and orfA#3) and inserted into the pCR 2.1-TOPO vector using the TOPO TA Cloning Kit according to manufacturer instructions (Invitrogen, Carlsbad, Calif.). dca was excised with EcoRI and cloned into pUC19 (New England Biolabs, Beverly, Mass.). For construction of pLS1, the nonpolar aphA-3 cassette (42) was inserted into the NsiI site 1,319 bp into dca, disrupting the gene at the 3′ end. To create pLS3, a PCR product of dca was created that would include 349 bp of DNA upstream of the BssHII site, 62 bp within dca using primers murE#4 and orfA#3. This product was then inserted into the pCR 2.1-TOPO vector, excised with EcoRI, and inserted into pUC19, as for pLS1. This construct was then cut with BssHII and NsiI, deleting 1,257 bp from dca, and the aphA-3 cassette was inserted. Restriction endonucleases were obtained from New England Biolabs (Beverly, Mass.). The pLS1 and pLS3 constructs were used as templates for PCR. Piliated gonococci (strain FA19) were transformed with PCR products as described by Gunn and Stein (26). Kanamycin-resistant (Kmr) transformants were selected using 50 μg of kanamycin (Sigma) per ml. Sequencing of the representative transformants confirmed the insertion of the aphA-3 cassette in strains LS1 and LS3 and the deletion of 1,257 bp from dca and that this transformation produced no changes in the upstream murE gene in strain LS3 (data not presented). RT-PCR studies showed no detectable polar effect of either the aphA-3 cassette or the deletion on the transcription of the downstream murF (data not presented). These transformants, along with strains FA19 and FA19 recA::ermC, were tested for natural competence by the method of Gunn and Stein (26) and by the agar overlay procedure of Sparling et al. (55) using chromosomal DNA containing the Strr marker.
Accession numbers.
The GenBank accession numbers for dca are as follows: N. gonorrhoeae strain FA19, AF195057; N. meningitidis strain NMB, AF195058; N. meningitidis strain 2633, AF195059; and N. meningitidis strain 0929, AF195060.
RESULTS
Identification and organization of the dcw gene cluster in gonococci and meningococci.
The dcw gene cluster of N. gonorrhoeae was originally identified in a search of the FA1090 genome sequence database for sites with homology to the MtrR regulator consensus binding sequence (40). One putative MtrR-binding site (18 of 31 bases identical) is located 883 bp 5′ of an ORF predicted to encode a protein with homology to the yabB-encoded protein of E. coli (32.9% amino acid similarity over the whole protein) (4). Subsequent experiments failed to demonstrate specific binding of MtrR to this site (data not presented), suggesting either that MtrR does not bind to this site or that additional cofactors or sequence determinants are required for its binding. However, the DNA sequence 3′ of the putative yabB homologue includes an additional 17 tightly packed ORFs orientated in the same direction (Fig. 1), ending with ftsZ, usually ascribed as the most 3′ gene of the dcw cluster (48). Of the 18 ORFs in this cluster, 15 encode putative proteins with homology to those encoded by dcw genes of other bacterial species (Fig. 1). These have between 28.4 and 55.3% amino acid similarity with the corresponding E. coli gene products, except for ftsL (17% amino acid similarity). This is consistent with ftsL frequently being the least-conserved dcw gene between different species. For example, although only 16% amino acid identity exists between the E. coli and B. subtilis genes, Daniel et al. (14) recently named the B. subtilis gene ftsL on the basis of its position in the dcw cluster, the ORF length, and the predicted protein features. Using this approach and the homologies to E. coli proteins, we were able to ascribe gene identifications to 15 of the 18 genes in the pathogenic Neisseria spp. Of the genes in this cluster, only penA, encoding penicillin-binding protein 2, and ftsZ, encoding cell division protein FtsZ, have been previously investigated in the Neisseria spp. (49, 56, 57). These ascribed identifications are consistent with the combined annotations of the recently published N. meningitidis genome sequences, including the annotation of NMB0412 as ftsL (46, 59). Recently, Francis et al. (21) published a description of the dcw cluster of N. gonorrhoeae, but our interpretation differs from theirs in several respects, including the definition of the 3′ end of the cluster as the conserved ftsZ rather than an undefined ORF, the number of genes in the cluster as 18 rather than 17, the identification of ftsL, and the identification of an additional novel ORF within the cluster, the homologue of NMB0417. Additionally, the present work provides a comparative analysis of the dcw cluster from the three sequenced pathogenic Neisseria spp. strains, including N. meningitidis strains MC58 and Z2491 (Fig. 1).
Using PCR with combinations of oligonucleotide primers that anneal within each of the different ORFs predicted from the FA1090 genome sequence database, we confirmed the presence of a similar dcw cluster in the same sequence location in the clonally unrelated gonococcal strain FA19. A search of the two meningococcal genome sequences (strains Z2491 and MC58) revealed that meningococci contain a similar dcw gene cluster (Fig. 1) that also includes the three additional ORFs (46, 59). The homologues of these ORFs are designated NMB0415, NMB0417, and NMB0419 in the N. meningitidis genome sequence annotation (59). Once we established (see below) that the NMB0415 homologue was involved in competence for transformation, we named it dca (division cluster competence associated). The remaining two ORFs have not been named and will be referred to on the basis of their homology with NMB0417 and NMB0419.
A promoter, Pmra, located 3′ of yabB in E. coli has been described that controls transcription of yabB to ftsZ and is essential for transcription of the first nine dcw cluster genes (30, 43). Primer extension analysis was used to seek a similarly located promoter 5′ of the putative gonococcal dcw cluster in strain FA19. An extension product ending in the region of a T residue located 68 bp 5′ of yabB was detected. Analysis of the DNA sequence in this region revealed candidate −10 (TATAGT) and −35 (TTGACC) regions, separated by 17 bp, generating an optimally spaced ς70 consensus binding site located 74 bp 5′ of yabB (data not presented). The equivalent putative promoter regions in N. meningitidis strains Z2491 and MC58 are identical. These results suggest that the neisserial dcw cluster has a promoter, similar to that seen in E. coli, 5′ of yabB. Other putative and identified promoters are likely to contribute to the transcription of genes internal to the dcw cluster (21; L. A. S. Snyder, unpublished data). Francis et al (21) identified six promoters at the 3′ end of the cluster that would contribute to ftsQAZ transcription: PQ1 within ddlB, PA1 and PA2 within ftsQ, and PZ1, PZ2, and PZ3 between ftsA and ftsZ. In order to learn whether dca and the NMB0417 and NMB0419 homologues are transcribed in gonococci, total RNA from strain FA19 was used in RT-PCR reactions that included ORF-specific primers. RT-PCR products were obtained for each ORF (data not presented), indicating that all three were transcriptionally active.
Sequence composition of the additional neisserial ORFs within the dcw gene cluster.
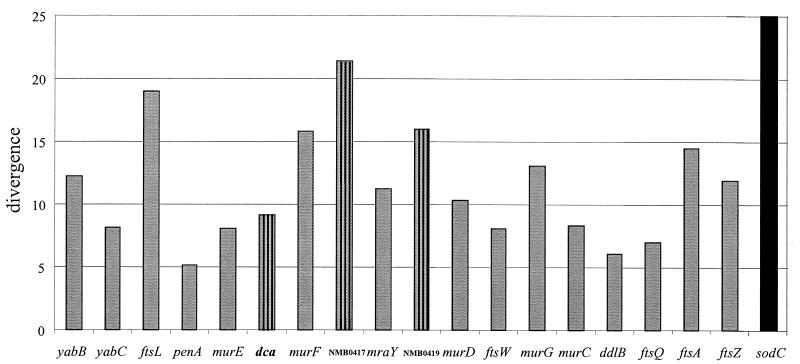
The pathogenic Neisseria spp. are naturally transformable, and this can lead to the acquisition of genes from both related and unrelated bacterial species (51). Given that the pathogenic Neisseria spp. examined contain ORFs within the dcw cluster that are not present in other bacterial species, we sought to investigate the evolutionary history of these genes by determining whether they have features of recent horizontal acquisition. The dcw cluster of N. meningitidis strain MC58 was analyzed using a modification of DNS that can assess single ORFs (44; Mirsky et al. unpublished) (Fig. 2). This analysis failed to detect evidence of recent interspecies horizontal acquisition of any of the genes in the dcw cluster, including the additional ORFs. The most atypical of the ORFs are murF and ftsA, but the degree of divergence is less than that normally associated with horizontally transferred genes in this genome, such as sodC (37) (Fig. 2). The short nature of ftsL (264 bp) and NMB0417 (177 bp) is sufficient to account for their elevated values in this analysis due to a sampling effect, and the moderate elevation of the NMB0419 homologue (381 bp) can be accounted for on a similar basis.
FIG. 2.
Dinucleotide signature of the genes within the dcw cluster. The figure shows the divergence of the dinucleotide signature for each gene within the dcw cluster in N. meningitidis strain MC58 from the mean value determined for the complete genome sequence (59) using the method of Mirsky (44). Genes which have been recently horizontally transferred tend to have a divergence of greater than 16% using this type of analysis in this genome (N. J. Saunders, unpublished data), once elevations due to sampling in short ORFs has been accounted for. For example, the sodC gene, which is believed to have been horizontally acquired from or via H. influenzae (37), has a dinucleotide signature divergence from the mean of 25% (black bar). Striped bars indicate the three additional dcw genes: dca, NMB0417, and NMB0419.
dcw cluster-associated ORF dca.
We selected dca, which is located between murE and murF in both gonococci and meningococci, for further investigation on the basis of its homology to hypothetical proteins present in other gram-negative species and the presence of a homopolymeric tract suggestive of phase variation (see below). The sequence of dca (1,647 bp) from gonococcal strain FA19 was determined. dca encodes a predicted protein of 548 amino acids with five transmembrane domains located within the N-terminal region between residues 16 and 174 and an N-terminal signal sequence of 35 amino acids. Similar hypothetical proteins are present in other microbial genome sequences, but they appear to be restricted to gram-negative bacteria. Alignment of the gonococcal sequence with related proteins of A. actinomycetemcomitans, E. coli (ybiP), H. influenzae (HI1005), and serovar Typhi are shown in Fig. 3. Protein structural predictions, obtained by using TopPredII, suggest that these homologues have a structure and conformation similar to that of the predicted dca product.
FIG. 3.
dca homologues. Alignment of the predicted amino acid sequence of dca from N. gonorrhoeae FA19 (N.g.) with hypothetical proteins from A. actinomycetemcometans (A.a. 1 and A.a. 2), H. influenzae HI1005 (H.i.) (20), E. coli ybiP (E.c.) (4), and serovar Typhi (S.t.). Identical residues are indicated by black boxes.
Perrin et al. (47) recently identified regions of the chromosome of strains Z2491 and FA1090 that are specific to the pathogenic Neisseria spp. and are not present in N. lactamica, a nonpathogenic commensal organism, which includes a region with homology to HI1005 of H. influenzae. As shown in Fig. 3, Dca is homologous to the H. influenzae ORF HI1005. In order to extend this observation from the single commensal species to more nonpathogenic species and other strains of the pathogenic species, a Southern blot of ClaI-digested DNA from six gonococcal strains, three meningococcal strains, and six commensal Neisseria strains was probed with a PCR product containing only dca. While the pathogenic Neisseria spp. produced hybridizing bands (12 kb for all gonococcal strains, 6.6 kb for meningococcal strains NMB and 2633, and 3.2 kb for meningococcal strain 0929) for the dca-containing ClaI fragment, no hybridization bands were observed for the nonpathogenic commensal strains (data not presented). These results, which include N. lactamica, are consistent with those of Perrin et al. and expand the repertoire of Neisseria spp. without a detectable dca to include N. flavescens, N. macacae, N. polysaccharea, N. sicca, and N. subflava.
Analysis of the dca sequence from the meningococcal strains Z2491 and MC58 predicted that they encode a protein of only 145 amino acids in length, compared to the predicted 548-amino-acid gonococcal protein. These shorter ORFs are a consequence of premature termination following a homopolymeric tract (HPT) of nine G's located 420 bp through the ORF. The sequence in the equivalent location, including polymorphisms, in N. gonorrhoeae is eight bases, which is associated with the presence of the full length ORF (Fig. 4). This suggests the potential for alteration in the expression of dca mediated by variation in the length of this HPT (52). In N. gonorrhoeae this sequence includes polymorphisms (changes of the first and third bases to T's) which reduce the length of the poly(G) repeat to five bases (Fig. 4). When repeats of this shorter length are present in the phase-variable lgt LPS biosynthesis genes in N. meningitidis they are relatively stable (33). The presence of the shorter (G)5 repeat element in strains FA19 and FA1090 suggests that this gene would also be stable in these strains. Following identification of the HPT, we investigated this sequence in clinical isolates of N. meningitidis. We sequenced the dca region from an additional capsular serogroup B isolate strain (NMB) and two serogroup Y invasive disease isolate strains (2633 and 0929). NMB contained a (G)9 HPT, and the serogroup Y strains each contained a (G)8 HPT. Preliminary reads from the sequencing project on the serogroup C meningococcus strain FAM18 indicate that this strain also contains a (G)8 HPT. This suggests that the sequenced dca genes from these capsular group A and B strains encode a frame-shifted Dca protein, while the serogroup Y and C strains and N. gonorrhoeae strains encode a full-length protein (Fig. 4). These observations are consistent with Dca being a phase-variable protein in which the homopolymeric tract acts as a translational switch.
FIG. 4.
Alignment of the repeat region predicted to mediate phase variation within dca from the gonococcal strains FA1090 and FA19 and the meningococcal strains Z2491, MC58, NMB, 2633, and 0929. The frameshift caused by the presence of nine guanines, as in Z2491, MC58, and NMB, brings the underlined termination codon in frame, resulting in a premature termination of the ORF. aa, amino acids.
Insertional-inactivation of dca.
Based on its location within the dcw gene cluster, we hypothesized that the dca-encoded protein might participate in the processes of cell division and/or cell wall biosynthesis. In order to investigate this possibility, we constructed two separate nonpolar mutations within dca using the Kmr-containing aphA-3 cassette and then transformed them into their original chromosomal locations in strain FA19. Transformants of strain FA19 were readily obtained with the DNA sequence generated from pLS1 and pLS3, indicating that dca is not an essential gene. The transformant generated with the pLS1 sequence contains the aphA-3 cassette inserted into the NsiI site 1,319 bp into dca, disrupting the gene at the 3′ end. The transformant generated with the sequence from pLS3 has 1,257 bp deleted from dca (between the BssHII and NsiI sites), leaving only 62 bp of dca before the insertion of the aphA-3 cassette. PCR amplification and sequencing (data not presented) was used to confirm that the aphA-3 cassette had been inserted into dca at the predicted sites within the transformants, which were termed LS1 and LS3, respectively. In transformant LS3, 1,257 bp were deleted from dca. This deletion and replacement with the nonpolar aphA-3 cassette had no detectable effect on transcription of the downstream murF gene as determined by RT-PCR (data not presented).
Strains LS1 and LS3 were compared to the parental strain FA19 in a number of experiments to identify the functional consequence(s) of dca inactivation. These mutations did not alter gonococcal susceptibility to hydrophobic or hydrophilic antibiotics, or nonionic detergents, as tested by using penicillin, erythromycin, and Triton X-100 (data not presented). Repeated experiments also failed to show any reproducible differences in growth rates between strains FA19, LS1, and LS3 in liquid media as judged by CFU and optical-density analyses (data not presented). We also monitored the rate and extent of [3-H]glucosamine uptake into peptidoglycan in growing cultures of FA19 and LS3 by the method of Dougherty (17) but found no reproducible differences (data not presented). Moreover, the rate and extent of peptidoglycan turnover, as determined by the method of Dillard and Seifert (15) using prelabeled cultures, appeared to be similar in the parent and mutant strains in repeated experiments (data not presented).
Natural competence for transformation of dca mutants.
In the course of investigating the phenotypes of the mutants, an apparent deficiency in transformation was noted in strain LS3. To determine whether strains LS1 and LS3 were deficient in transformation, we selected piliated variants (on the basis of colony morphology with the presence of pili confirmed by electron microscopy) and examined their capacity to be transformed with chromosomal DNA, using piliated variants of parental strain FA19 and FA19 recA::ermC as controls. As shown in Table 2, we found that strains FA19 and LS1 were readily transformable (frequencies of 10−3) using a chromosomal DNA marker (streptomycin resistance), while LS3 was reproducibly nontransformable (frequency of <10−8). In parallel experiments, a recA::ermC insertional mutant of strain FA19 was also nontransformable (frequency of <10−8) (Table 2). The association between the transformation defect in strain LS3 and the dca::aphA-3 mutation was confirmed by analyzing three independent dca::aphA-3 transformants of strain FA19 obtained during the original construction of LS3. All three transformants displayed the same transformation defect exhibited by the first LS3 strain (frequency of <10−8). We also examined the capacity of these piliated strains to be transformed with donor DNA using the agar overlay transformation procedure of Sparling et al. (55). However, regardless of the liquid or plate transformation conditions, transformants of strains LS3 or FA19 recA::ermC could not be recovered (frequencies of <10−8).
TABLE 2.
Dca is required for transformation of gonococci but not meningococci
| Strain | Strr CFU ml−1/total CFU ml−1a |
|---|---|
| N. gonorrhoeae | |
| FA19 | 8.3 × 10−3 |
| LS1 | 3.5 × 10−3 |
| LS3 | <7.8 × 10−8 |
| FA19 recA::ermC | <2.6 × 10−8 |
| N. meningitidis | |
| NMB | 1.8 × 10−3 |
| NMLS3 | 1.8 × 10−3 |
Three independent experiments, each done with 0.5 μg of DNA, produced similar results.
We next examined the consequence of dca inactivation on the capacity of meningococci to be transformed with meningococcal chromosomal DNA harboring the Str marker. For this purpose we used strain NMB, which encodes a truncated (145-amino-acid) Dca protein due to the presence of a phase-off HPT tract of nine G residues (Fig. 4). A dca::aphA-3 PCR product from gonococcal strain LS3 was used to transform NMB for Kmr, and the intended mutation in dca was confirmed by PCR and sequencing. A representative transformant (NMLS3), along with the parental strain, was tested for its ability to be naturally transformed by meningococcal (strain NMB) chromosomal DNA marked with Strr using the transformation method of Gunn and Stein (26). Both N. meningitidis strain NMB and the mutant strain NMLS3 were readily transformed to Strr (frequencies of 10−3; Table 2).
DISCUSSION
Horizontal gene transfer is thought to be an important component of bacterial adaptation and evolution. The Neisseria spp. are naturally competent for transformation (9, 54), meaning that they are capable of taking up external DNA and incorporating it into their chromosome by homologous recombination. Natural transformation predominantly occurs within populations of cells that are of the same or closely related species, but it is probably also involved in the acquisition of new genes from unrelated species. In Neisseria spp., species-specific targeting is largely mediated by the presence of a 10-bp uptake signal sequence (GCCGTCTGAA) which is frequently located 3′ of ORFs, (18, 25). Transformation facilitates the generation of mosaic genes through recombination between existing alleles, new gene acquisition, and possibly repair of genes whose function has been lost by mutation. Recognized examples of this process in Neisseria spp. include (i) the argF gene of N. meningitidis, which includes sections derived from a species similar to N. cinerea and from an additional unidentified source (65); (ii) the generation of Opa variants within a population (2); (iii) the acquisition of penicillin resistance through the incorporation of sequences from N. flavescens and N. cinerea (56, 57, 58); and (iv) the acquisition of foreign genes from other species (37). This is also likely to be the primary route by which silent pilus cassettes are provided for antigenic variation by recombination with the expressed pilE gene (53).
Natural transformation is an integral part of neisserial biology and involves several genes. In addition to the requirement for the expressions of pili (22, 54), other proteins have been shown to have a role in natural transformation in the Neisseria spp. These include Tpc, mutants of which form tetracocci deficient in epithelial cell invasion and competence (23); ComA, an inner membrane protein which may be involved in translocation of DNA into the cytoplasm (19); ComL, a peptidoglycan-bound lipoprotein, mutants of which are significantly decreased in transformation (24); and PilT, which is involved in twitching motility and DNA uptake (63). Our results indicate that dca is also essential for transformation in gonococci. The fact that LS1 retained its competence indicates that the 3′ end of dca is not involved in this function. In separate experiments we also evaluated the capacity of strain FA1090 and FA1090 dca::aphA-3 (obtained using donor DNA from LS3) to be transformed with gonococcal DNA. Using an erythromycin resistance marker from strain KH15 (as FA19 but mtrR-171 [28]), we found that dca was also required for transformation in strain FA1090 (frequency of <10−8).
The ability of the meningococcal strain NMB to retain its competence suggests that N. meningitidis transformation occurs through an alternate and/or additional pathway. Alternatively, there may be functional redundancy with the dca-encoded product or the dca-associated system. Although no homologues to dca were found in either the N. meningitidis strain Z2491 or MC58 genomes in addition to dca itself and only one band appears on Southern blots probed for dca, this does not rule out the possibility of more diverse proteins with similar functions. Similar inactivation of dca in serogroup Y meningococcal strain 0929, which has a (G)8 HPT (Fig. 4), and thus a full-length dca ORF, also did not affect competence (frequency of 10−3). The fact that this ORF is retained in the chromosome and is putatively phase variable suggests that this protein has an additional function. Due to the proximity of the termination codon to the HPT (Fig. 4) and the presence of strains with both (G)8 and (G)9 sequences, this gene is a strong candidate for phase variation (52). The phase variation of this gene is currently being further investigated.
The insertion-deletion mutation in the dca mutants created for the competence experiments was generated using an aphA-3 cassette designed by Ménard et al. (42) to be nonpolar. It is demonstrated here that the mutation in dca did not affect transcription of the 3′ gene, murF, and, given that the conserved dcw genes are essential, the effects on their transcription and translation should negatively impact the organism. Due to the fact that the functions of the homologues of NMB0417 and NMB0419 are not known, the possibility still exists that this mutation is having an effect on the expression or function of those gene products, effecting the competence phenotype. Classical complementation studies, which could rule out this possibility, cannot be done due to the lack of genetic tools for complementation in the pathogenic Neisseria spp.
In summary, the dcw cluster of the pathogenic Neisseria spp. is highly homologous to those seen in other diverse organisms (Fig. 1). Similar to E. coli, there is a ς70 promoter located 5′ of yabB. It is not known if this one promoter drives transcription for the entire cluster, regardless of additional transcription from internal promoters, as Pmra is thought to do in E. coli. The three unique ORFs found within the highly conserved dcw cluster do not have features of recent horizontal acquisition (Fig. 2) but rather are integral parts of the genome. Unlike B. subtilis, which also has additional genes in its dcw cluster (12, 31), these are not homologues of other dcw genes. dca, one of these additional genes, encodes a putative inner membrane protein that has similarity to hypothetical inner membrane proteins present in several gram-negative bacteria that are located outside of their dcw clusters (Fig. 3). DNA sequencing of this ORF from several gonococcal and meningococcal strains revealed the presence of a homopolymeric tract of G's of variable length in this ORF, which suggests that this gene is phase variable (Fig. 4). We observed that while loss of the dcw cluster-associated ORF dca had no significant effect on peptidoglycan synthesis and growth rate, it had a profound effect on transformation, which could be localized to the 5′ region of the reading frame (Table 2). Thus, its loss rendered gonococci incapable of being transformed by chromosomal DNA, although meningococcal transformation was unchanged by similar mutagenesis of dca. We propose that Dca is a necessary component of the transformation machinery of gonococci, perhaps functioning to facilitate movement of DNA across the cytoplasmic membrane. The lack of necessity of Dca in the meningococci may suggest the presence of an alternate pathway in this species and that Dca may have additional, as yet undefined, functions.
ACKNOWLEDGMENTS
We thank L. Pucko for help with manuscript preparation and L. Melsen for his help with electron microscopy.
This work was supported by NIH grant AI-21150 (W.M.S.). W.M.S. is the recipient of a Senior Career Scientist Award from the VA Medical Research Service. N.J.S. was supported by a Wellcome Trust Research Fellowship in Medical Microbiology. The N. gonorrhoeae sequence was obtained from the University of Oklahoma, the Gonococcal Genome Sequencing Project (B. A. Roe, S. P. Lin, L. Song, X. Yuan, S. Clifton, T. Ducey, L. Lewis, and D. W. Dyer), which is supported by USPHS-NIH grant AI-38399. The N. meningitidis serogroup A strain Z2491 sequence and the S. typhi strain CT18 sequence were obtained from the Sanger Centre; these projects are supported by The Wellcome Trust. The N. meningitidis serogroup B strain MC58 sequence was obtained from The Institute for Genomic Research. The N. meningitidis serogroup C strain FAM18 sequence was obtained from the Sanger Centre; these projects are supported by Beowulf Genomics. The A. actinomycetemcomitans strain HK1651 sequence was obtained from the University of Oklahoma, the Actinobacillus Genome Sequencing Project (B. A. Roe, F. Z. Najar, S. Clifton, T. Ducey, L. Lewis, and D. W. Dyer), which is supported by a USPHS-NIH grant from the National Institute of Dental Research.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bhat K S, Gibbs C P, Barrera O, Morrison S G, Jahnig F, Stern A, Kupsch E-M, Meyer T F, Swanson J. The opacity proteins of Neisseria gonorrhoeae strain MS11 are encoded by a family of 11 complete genes. Mol Microbiol. 1991;5:1889–1901. doi: 10.1111/j.1365-2958.1991.tb00813.x. [DOI] [PubMed] [Google Scholar]
- 3.Biran D, Brot N, Weissbach H, Ron E Z. Heat shock-dependent transcriptional activation of the metA gene of Escherichia coli. J Bacteriol. 1995;177:1374–1379. doi: 10.1128/jb.177.5.1374-1379.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Boyle D S, Khatter M M, Addinall S G, Lutkenhaus J, Donachie W D. ftsW is an essential cell-division gene in Escherichia coli. Mol Microbiol. 1997;24:1263–1273. doi: 10.1046/j.1365-2958.1997.4091773.x. [DOI] [PubMed] [Google Scholar]
- 6.Boyle D S, Donachie W D. mraY is an essential gene for cell growth in Escherichia coli. J Bacteriol. 1998;180:6429–6432. doi: 10.1128/jb.180.23.6429-6432.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buddelmeijer N, Aarsman M E G, Kolk A H J, Vicente M, Nanninga N. Localization of cell division protein FtsQ by immunoflourescence microscopy in dividing and nondividing cells of Escherichia coli. J Bacteriol. 1998;180:6107–6116. doi: 10.1128/jb.180.23.6107-6116.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cannon G C. Sequence analysis on microcomputers. Science. 1987;238:97–103. doi: 10.1126/science.3659902. [DOI] [PubMed] [Google Scholar]
- 9.Catlin B W, Cunningham L S. Transforming activities and base contents of deoxyribonulceate preparations from various neisseriae. J Gen Microbiol. 1961;26:303–306. doi: 10.1099/00221287-26-2-303. [DOI] [PubMed] [Google Scholar]
- 10.Claros M G, von Heijne G. TopPredII: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 11.Dai K, Lutkenhaus J. ftsZ is an essential cell division gene in Escherichia coli. J Bacteriol. 1991;173:3500–3506. doi: 10.1128/jb.173.11.3500-3506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniel R A, Drake S, Buchanan C E, Scholle R, Errington J. The Bacillus subtilis spoVD gene encodes a mother-cell-specific penicillin-binding protein required for spore morphogenesis. J Mol Biol. 1994;235:209–220. doi: 10.1016/s0022-2836(05)80027-0. [DOI] [PubMed] [Google Scholar]
- 13.Daniel R A, Williams A M, Errington J. A complex four-gene operon containing essential cell division gene pbpB in Bacillus subtilis. J Bacteriol. 1996;178:2343–2350. doi: 10.1128/jb.178.8.2343-2350.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniel R A, Harry E J, Katis V L, Wake R G, Errington J. Characterization of the essential cell division gene ftsL (yIID) of Bacillus subtilis and its role in the assembly of the division apparatus. Mol Microbiol. 1998;29:593–604. doi: 10.1046/j.1365-2958.1998.00954.x. [DOI] [PubMed] [Google Scholar]
- 15.Dillard J P, Seifert H S. A peptidoglycan hydrolase similar to bacteriophage endolysins acts as an autolysin in Neisseria gonorrhoeae. Mol Microbiol. 1997;25:893–901. doi: 10.1111/j.1365-2958.1997.mmi522.x. [DOI] [PubMed] [Google Scholar]
- 16.Donachie W D. The cell cycle of Escherichia coli. Annu Rev Microbiol. 1993;47:199–230. doi: 10.1146/annurev.mi.47.100193.001215. [DOI] [PubMed] [Google Scholar]
- 17.Dougherty T J. Peptidoglycan biosynthesis in Neisseria gonorrhoeae strains sensitive and intrinsically resistant to beta-lactam antibiotics. J Bacteriol. 1983;153:429–435. doi: 10.1128/jb.153.1.429-435.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elkins C, Thomas C E, Seifert H S, Sparling P F. Species specific uptake of DNA by gonococci is mediated by a 10-base-pair sequence. J Bacteriol. 1991;173:3911–3913. doi: 10.1128/jb.173.12.3911-3913.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Facius D, Meyer T F. A novel determinant (comA) essential for natural transformation competence in Neisseria gonorrhoeae and the effect of a comA defect on pilin variation. Mol Microbiol. 1993;10:699–712. doi: 10.1111/j.1365-2958.1993.tb00942.x. [DOI] [PubMed] [Google Scholar]
- 20.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, et al. Whole genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;29:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 21.Francis F, Ramirez-Arcos S, Salimnia H, Victor C, Dillon J R. Organization and transcription of the division and cell wall (dcw) cluster in Neisseria gonorrhoeae. Gene. 2000;251:141–151. doi: 10.1016/s0378-1119(00)00200-6. [DOI] [PubMed] [Google Scholar]
- 22.Fröholm L O, Jyssum K, Bovre K. Electron microscopical and cultural features of Neisseria meningitidis competence variants. Acta Pathol Microbiol Scand B Microbiol Immunol. 1973;81:525–537. doi: 10.1111/j.1699-0463.1973.tb02238.x. [DOI] [PubMed] [Google Scholar]
- 23.Fussenegger M, Kahrs A F, Facius D, Meyer T F. Tetrapac (tpc), a novel genotype of Neisseria gonorrhoeae affecting epithelial cell invasion, natural transformation competence and cell separation. Mol Microbiol. 1996;19:1357–1372. doi: 10.1111/j.1365-2958.1996.tb02479.x. [DOI] [PubMed] [Google Scholar]
- 24.Fussenegger M, Facius D, Meier J, Meyer T F. A novel peptidoglycan-linked lipoprotein (ComL) that functions in natural transformation competence of Neisseria gonorrhoeae. Mol Microbiol. 1996;19:1095–1105. doi: 10.1046/j.1365-2958.1996.457984.x. [DOI] [PubMed] [Google Scholar]
- 25.Goodman S D, Scocca J J. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1988;85:6982–6986. doi: 10.1073/pnas.85.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunn J S, Stein D C. Use of a non-selective transformation technique to construct a multiply restriction/modification-deficient mutant of Neisseria gonorrhoeae. Mol Gen Genet. 1996;251:509–517. doi: 10.1007/BF02173639. [DOI] [PubMed] [Google Scholar]
- 27.Guzman L, Barondess J J, Beckwith J. FtsL, an essential cytoplasmic membrane protein involved in cell division in Escherichia coli. J Bacteriol. 1992;174:7716–7728. [PMC free article] [PubMed] [Google Scholar]
- 28.Hagman K E, Shafer W M. Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J Bacteriol. 1995;177:4162–4165. doi: 10.1128/jb.177.14.4162-4165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hagman K E, Pan W, Spratt B G, Balthazar J T, Judd R C, Shafer W M. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology. 1995;141:611–622. doi: 10.1099/13500872-141-3-611. [DOI] [PubMed] [Google Scholar]
- 30.Hara H, Yasuda S, Horiucji K, Park J T. A promoter for the first nine genes of the Escherichia coli mra cluster of cell division and cell envelope biosynthesis genes, including ftsI and ftsW. J Bacteriol. 1997;179:5802–5811. doi: 10.1128/jb.179.18.5802-5811.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henriques A O, de Lencastre H, Piggot P J. A Bacillus subtilis morphogene cluster that includes spoVE is homologous to the mra region of Escherichia coli. Biochimie. 1992;74:735–748. doi: 10.1016/0300-9084(92)90146-6. [DOI] [PubMed] [Google Scholar]
- 32.Ikeda M, Sato T, Wachi M, Jung H K, Ishino F, Kobayashi Y, Matsuhashi M. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J Bacteriol. 1989;171:6375–6378. doi: 10.1128/jb.171.11.6375-6378.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jennings M P, Srikhanta Y N, Moxon E R, Kramer M, Poolman J T, Kuipers B, van der Lay P. The genetic basis of the phase variation repertoire of lipopolysaccharide immunotypes in Neisseria meningitidis. Microbiology. 1999;145:3013–3021. doi: 10.1099/00221287-145-11-3013. [DOI] [PubMed] [Google Scholar]
- 34.Karlin S, Burge C. Dinucleotide relative abundance extremes: a genomic signature. Trends Genet. 1995;11:283–290. doi: 10.1016/s0168-9525(00)89076-9. [DOI] [PubMed] [Google Scholar]
- 35.Karlin S, Campbell A M, Mrazek J. Comparative DNA analysis across diverse genomes. Annu Rev Genet. 1998;32:185–225. doi: 10.1146/annurev.genet.32.1.185. [DOI] [PubMed] [Google Scholar]
- 36.Kellogg D S, Jr, Peacock W L, Jr, Deacon W E, Brown L, Pirkle C I. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J Bacteriol. 1963;85:1274–1279. doi: 10.1128/jb.85.6.1274-1279.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kroll J S, Wilks K E, Farrant J L, Langford P R. Natural genetic exchange between Haemophilus and Neisseria: intergeneric transfer of chromosomal genes between major human pathogens. Proc Natl Acad Sci USA. 1998;95:12381–12385. doi: 10.1073/pnas.95.21.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 39.Lee E, Shafer W M. The farAB-encoded efflux pump mediates resistance of gonococci to long-chain antibacterial fatty acids. Mol Microbiol. 1999;33:839–845. doi: 10.1046/j.1365-2958.1999.01530.x. [DOI] [PubMed] [Google Scholar]
- 40.Lucas C E, Balthazar J T, Hagman K E, Shafer W M. The MtrR repressor binds the DNA sequence between the mtrR and mtrC genes of Neisseria gonorrhoeae. J Bacteriol. 1997;179:4123–4128. doi: 10.1128/jb.179.13.4123-4128.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McAllister C F, Stephens D S. Analysis in Neisseria meningitidis and other Neisseria species of genes homologous to the FKBP immunophilin family. Mol Microbiol. 1993;10:13–24. doi: 10.1111/j.1365-2958.1993.tb00899.x. [DOI] [PubMed] [Google Scholar]
- 42.Ménard R, Sansonetti P J, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mengin-Lecreulx D, Ayala J, Bouhss A, vanHeijenart J, Parquet C, Hara H. Contribution of the Pmra promoter to expression of genes in the Escherichia coli mra cluster of cell envelope biosynthesis and cell division genes. J Bacteriol. 1998;180:4406–4412. doi: 10.1128/jb.180.17.4406-4412.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mirsky J. Genome analysis methodologies for the identification of horizontally acquired DNA. M.Sc. thesis. Oxford, United Kingdom: University of Oxford; 1999. [Google Scholar]
- 45.Nakai K, Kanehisa M. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 46.Parkhill J, Achtman M, James K D, Bentley S D, Churcher C, Klee S R, Morelli G, Basham D, Brown D, Chillingworth T, et al. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 47.Perrin A, Nassif X, Tinsley C. Identification of regions of the chromosome of Neisseria meningitidis and Neisseria gonorrhoeae which are specific to the pathogenic Neisseria species. Infect Immun. 1999;67:6119–6129. doi: 10.1128/iai.67.11.6119-6129.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pucci M, Thanassi J, Discotto L, Kessler R, Dougherty T. Identification and characterization of cell wall cell division gene clusters in pathogenic gram-positive cocci. J Bacteriol. 1997;179:5632–5635. doi: 10.1128/jb.179.17.5632-5635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salimnia H, Radia A, Bernatchez S, Beveridge T J, Dillon J R. Characterization of the ftsZ cell division gene of Neisseria gonorrhoeae: expression in Escherichia coli and N. gonorrhoeae. Arch Microbiol. 2000;173:10–20. doi: 10.1007/s002030050002. [DOI] [PubMed] [Google Scholar]
- 50.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saunders N J, Hood D W, Moxon E R. Bacterial evolution: bacteria play pass the gene. Curr Biol. 1999;9:R180–R183. doi: 10.1016/s0960-9822(99)80108-0. [DOI] [PubMed] [Google Scholar]
- 52.Saunders N J, Jeffries A C, Peden J F, Hood D W, Tettelin H, Pappuoli R, Moxon E R. Repeat-associated phase variable genes in the complete genome sequence of N. meningitidis strain MC58. Mol Microbiol. 2000;37:207–215. doi: 10.1046/j.1365-2958.2000.02000.x. [DOI] [PubMed] [Google Scholar]
- 53.Seifert H S, Ajioka R S, Marchal C, Sparling P F, So M. DNA transformation leads to pilin antigenic variation in Neisseria gonorrhoeae. Nature. 1988;24:392–395. doi: 10.1038/336392a0. [DOI] [PubMed] [Google Scholar]
- 54.Sparling P F. Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. J Bacteriol. 1966;192:1364–1371. doi: 10.1128/jb.92.5.1364-1371.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sparling P F, Sarubbi F A, Jr, Blackman E. Inheritance of low-level resistance to penicillin, tetracycline, and chloramphenicol in Neisseria gonorrhoeae. J Bacteriol. 1975;124:740–749. doi: 10.1128/jb.124.2.740-749.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spratt B G. Hybrid penicillin-binding proteins in penicillin-resistant strains of Neisseria gonorrhoeae. Nature. 1988;332:173–176. doi: 10.1038/332173a0. [DOI] [PubMed] [Google Scholar]
- 57.Spratt B G, Zhang Q Y, Jones D M, Hutchison A, Brannigan J A, Dowson C G. Recruitment of a penicillin-binding protein gene from Neisseria flavescens during the emergence of penicillin resistance in Neisseria meningitidis. Proc Natl Acad Sci USA. 1989;86:8988–8992. doi: 10.1073/pnas.86.22.8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spratt B G, Bowler L D, Zhang Q Y, Zhou J, Smith J M. Role of interspecies transfer of chromosomal genes in the evolution of penicillin resistance in pathogenic and commensal Neisseria species. J Mol Evol. 1992;34:115–125. doi: 10.1007/BF00182388. [DOI] [PubMed] [Google Scholar]
- 59.Tettelin H, Saunders N J, Heidelberg J, Jeffries A C, Nelson K E, Eisen J A, Ketchum K A, Hood D W, Peden J F, Dodson R J, et al. Complete genome sequence of Neisseria meningitidis serotype B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 60.von Heijne G. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature. 1989;341:456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- 61.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 62.Wang L, Khattar M K, Donachie W D, Lutkenhaus J. FtsI and FtsW are localized to the septum in Escherichia coli. J Bacteriol. 1998;180:2810–2816. doi: 10.1128/jb.180.11.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolfgang M, Lauer P, Park H S, Brossay L, Hébert J, Koomey M. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol. 1998;29:321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
- 64.Yura T, Mori H, Nagai H, Nagata T, Ishihama A, Fujita N, Isono K, Mizobuchi K, Nakata A. Systematic sequencing of the Escherichia coli genome: analysis of the 0–2.4 min region. Nucleic Acids Res. 1992;20:3305–3308. doi: 10.1093/nar/20.13.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou J, Spratt B G. Sequence diversity within the argF, fbp, and recA genes of natural isolates of Neisseria meningitidis: interspecies recombination within the argF gene. Mol Microbiol. 1992;6:2135–2146. doi: 10.1111/j.1365-2958.1992.tb01387.x. [DOI] [PubMed] [Google Scholar]