Abstract
Characterization of a series of urease-negative transposon mutations of Actinobacillus pleuropneumoniae revealed that many of the mutants had insertions 2 to 4 kbp upstream of the urease gene cluster. A 5-kbp upstream region of DNA was sequenced and found to contain six open reading frames (ORFs) transcribed in the same orientation as the urease genes. As well, a partial ORF, apuR, 202 bp upstream of these six ORFs, is transcribed in the opposite orientation. The predicted product of this partial ORF shows homology with many members of the LysR family of transcriptional regulators. Five of the ORFs (cbiKLMQO) appear to form an operon encoding a putative nickel and cobalt periplasmic permease system. The cbiM and cbiQ genes encode proteins that have sequence similarity with known cobalt transport membrane proteins, and the cbiO gene encodes a cobalt transport ATP-binding protein homologue. The product of the cbiK gene is predicted to be the periplasmic-binding-protein component of the system, though it does not show any sequence similarity with CbiN, the cobalt-binding periplasmic protein. Escherichia coli clones containing this putative transport operon together with the urease genes of A. pleuropneumoniae were urease positive when grown in unsupplemented Luria-Bertani broth, whereas a clone containing only the minimal urease gene cluster required the addition of high concentrations of NiCl2 for full urease activity. This result supports the hypothesis that nickel is a substrate for this permease system. The sixth ORF, utp, appears to encode an integral membrane protein which has significant sequence identity with mammalian urea transport proteins, though its function in A. pleuropneumoniae remains to be determined.
The genes required for urease activity in various bacterial species typically include those encoding the structural subunits and those encoding the accessory proteins involved in insertion of two nickel ions within the catalytic site (24). In bacteria with urea-inducible gene clusters, the regulatory gene, ureR, is also present (24). Because of the requirement for nickel as a cofactor, genes such as Helicobacter pylori nixA and Alcaligenes eutrophus hoxN that encode nickel uptake systems have also been shown to affect urease activity (23, 41).
We recently identified the urease gene cluster of the gram-negative swine pathogen Actinobacillus pleuropneumoniae (4). The organization of the A. pleuropneumoniae gene cluster was found to be similar to that of other bacterial ureases, with the first three genes encoding the structural subunits (UreABC), and the accessory proteins (UreEFGD) encoded by four contiguous genes downstream. There was no evidence of a regulatory gene (ureR) upstream of the cluster.
In order to further characterize the urease activity of A. pleuropneumoniae, a bank of transposon mutants of strain CM5 Nalr was generated and screened for urease-negative isolates. A large number of insertions in the urease-negative mutants mapped upstream of the urease gene cluster. Therefore, the 5-kbp region upstream of the urease cluster was sequenced and analyzed.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A spontaneously nalidixic acid-resistant derivative of the virulent A. pleuropneumoniae serotype 1 strain CM5 was isolated and designated CM5 Nalr. This strain was routinely grown at 37°C with 5% CO2 on brain heart infusion (BHI; Difco Laboratories, Detroit, Mich.) agar supplemented with 0.01% NAD and 20 μg of nalidixic acid per ml (BHIV-NAL). Escherichia coli S17-1(λ-pir) was used as the conjugal donor strain for plasmid pLOF/Km and was grown at 37°C on Luria-Bertani (LB) medium supplemented with 50 μg of kanamycin (KAN) per ml and 100 μg of ampicillin (AMP) per ml. Transconjugants were grown on BHIV-NAL plus KAN at 50 μg/ml (BHIV-NAL-KAN). E. coli DH5 α containing recombinant plasmids was grown at 37°C on LB agar supplemented with AMP (100 μg/ml).
Transposon mutagenesis.
The suicide plasmid pLOF/Km containing a mini-Tn10 transposon was conjugally transferred from E. coli S17-1(λ-pir) into CM5 Nalr as previously described (39). Transconjugants were selected on BHIV-NAL-KAN, and loss of the suicide plasmid was confirmed by patching colonies onto BHIV-NAL-KAN-AMP. Individual isolates were cultured in wells of a microtiter plate containing 100 μl of BHIV-NAL-KAN broth per well and stored at −70°C with 8% dimethyl sulfoxide. Detection of Ure− mutants was done by subculturing isolates in 96-well plates containing 120 μl of BHIV-NAL-KAN broth per well. After overnight incubation at 37°C, 30 μl of 5× urea base medium (Difco) per well was added, and the wells were sealed. Urease-positive isolates turned the medium pink, whereas urease-negative isolates turned the medium yellow. Urease-negative isolates were confirmed by plating onto BHIV-NAL-KAN agar and urea agar.
Mapping of transposon insertions.
Southern blot analysis (32) was used to determine the number of Tn10 insertions within each mutant. Briefly, DNA was extracted from each mutant strain using a genomic DNA extraction kit (Qiagen Inc., Chatsworth, Calif.) and digested with either ClaI, XhoI, or EcoRI (Pharmacia Biotech Inc., Baie D'Urfé, Québec, Canada). The digested DNA was electrophoresed on 0.7% agarose and transferred to positively charged nylon membranes (Boehringer Mannheim Canada, Laval, Québec, Canada) by capillary transfer. Blots were probed with the kanamycin resistance cassette from pLOF/Km labeled with digoxigenin (Boehringer Mannheim). Hybridization was carried out at 65°C overnight. Blots were washed twice for 5 min at room temperature with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% sodium dodecyl sulfate (SDS), followed by two 15-min washes at 65°C with 0.1× SSC and 0.1% SDS. Blots were developed using antidigoxigenin-alkaline phosphatase conjugate and CSPD (Boehringer Mannheim) according to the manufacturer's instructions.
The locations of the Tn10 insertions were mapped by sequencing specific DNA fragments generated by PCR using a combination of primers designed to span the urease gene cluster and the 5-kbp upstream region (Fig. 1A), as well as a primer (Kan) designed from a sequence within the transposon (Table 1).
FIG. 1.
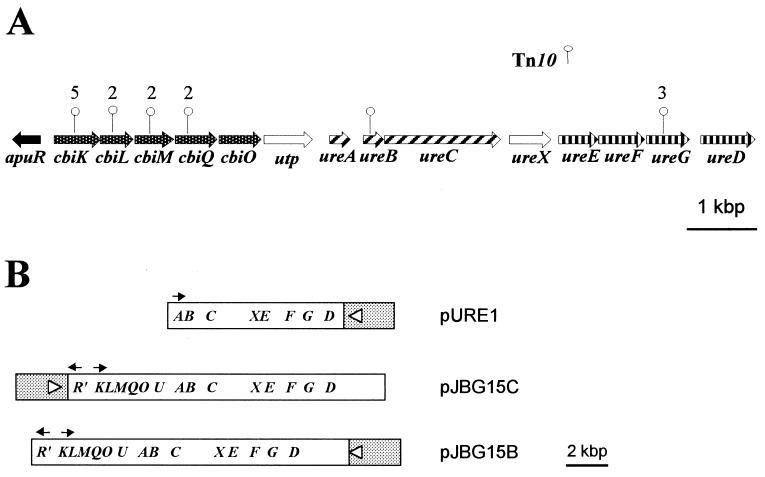
(A) Organization of urease genes and upstream sequences in A. pleuropneumoniae. The sites of the Tn10 insertions in the various urease-negative mutants are indicated with vertical lines. The numbers indicate the number of insertions at a particular location. (B) Organization of various urease plasmid constructs. The pBluescript II KS+ vector sequence is shaded and is not to scale.
TABLE 1.
Oligonucleotides used for PCR
| Oligonucleotide | Positiona | Strand | Length (bases) | Sequence, 5′−3′b |
|---|---|---|---|---|
| Kan | NA | NA | 21 | CCTCTTCCGACCATCAAGCAT |
| 59F1 | 318–340 | + | 22 | GGCTAATAACCGGCGGAGAAAC |
| 59R1 | 5632–5612 | − | 20 | CCGTAAGTCGCTACATACTG |
| 59R2c | 5223–5243 | + | 20 | TTCTGCTGCTCTCTCTCAAG |
| Urease/Xba | 11120–11100 | − | 32 | CCGGCCTCTAGAAGCGAAGCACGAATTAAGTT |
| Urease/Sac | 4527–4548 | + | 32 | GCGGCGAGCTCAGTTGGTTATTATTGGCGGTT |
| Tn10 | NA | NA | 21 | GATCATATGACAAGATGTGTA |
Position 5′ to 3′ within the compiled urease and upstream sequences (see Fig. 1). Note that the Kan and Tn10 primers were designed from sites within the Kan cassette of and near the end of the mini-Tn10 transposon sequence, respectively. NA, not applicable.
Extensions containing the restriction enzyme-cut sites (in bold) are shown.
Sequencing of the upstream region.
Sequencing of the 5.9-kbp insert of plasmid pJBG5.9 was continued as previously described (4). This plasmid contains part of the urease gene cluster as well as the 5-kbp upstream region. Both strands of the 5.9-kbp insert were completely sequenced, and the data were compiled and analyzed as previously described (4). In addition, database searches were performed using BLAST (http://www.ncbi.nlm.nih.gov/BLAST); transmembrane (TM) domains were predicted using TMPred (http://dot.imgen.bcm.tmc.edu:9331/seq-search/struc-predict.html); identification of N-terminal signal sequences and prediction of cellular localization of proteins were performed using SignalP (http://genome.cbs.dtu.dk/services/SignalP/) and PSORT (http://psort.nibb.ac.jp:8800/form.htm); potential promoter sequences were identified using NNPP (http://www-hgc.lbl.gov/projects/promoter.html); and Pfam (http://www .sanger.ac.uk/Software/Pfam/), and Prosite pattern search (http://www.expasy.ch /tools/scnpsite.html) were used to search for known protein motifs. In addition, Block Maker (http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html) was used to search for potential motifs in aligned protein sequences, and Boxshade 3.21 (http://www.ch.embnet.org/software/BOX_form.html) was used to compile consensus sequences.
Minimal urease subclone.
A 6.5-kbp fragment of DNA containing the urease gene cluster from strain CM5 Nalr was generated by PCR using the Expand High Fidelity PCR system (Boehringer Mannheim) and the primers Urease/Sac and Urease/Xba (Table 1). These primers introduced SacI and XbaI sites at either end of the urease cluster, which were subsequently used to subclone the fragment into the plasmid vector pBluescript II KS+ (Stratagene, La Jolla, Calif.). The resultant plasmid, pURE1 (Fig. 1B), was electroporated into E. coli DH5α, and transformants were selected on LB-AMP plates. Urease-positive clones were identified by patching colonies onto urea agar.
Urease assay.
The urease activity of the minimal subclone (DH5α/pURE1) was compared to that of clones (DH5α/pJBG15B and DH5α/pJBG15C) containing the entire urease cluster plus the 5-kbp upstream region (Fig. 1B). Cultures were grown overnight in LB-AMP broth, and NiCl2 was added at various concentrations (0, 2, 20, and 200 μM). Cultures were standardized to an optical density at 600 nm of 1.0, and urease activity in whole cells in the presence of excess urea (100 mM) was quantitatively measured using a urea nitrogen assay kit (Sigma Chemical Co., St. Louis, Mo.) as previously described (4).
Production of urease apoenzyme by Ure− mutants.
In order to determine if the urease mutants were producing inactive apoenzyme, immunoblot analysis was performed. Cytosolic extracts were prepared from representative mutants 8G12 (cbiK::Tn10), 15E10 (cbiL::Tn10), 16D4 (cbiM::Tn10), 17E10 (cbiQ::Tn10), 30A10 (ureB::Tn10), and 32D6 (ureG::Tn10) and from the parental strain (CM5 Nalr) as described previously (4). The protein content of each lysate was determined using a protein assay kit (Bio-Rad Laboratories Ltd., Mississauga, Ontario, Canada). All samples were standardized to 4 mg of protein per ml and quantitatively assayed for urease activity.
Nondenaturing polyacrylamide gel electrophoresis was performed using a modification of the method of Senior et al. (36). Briefly, samples containing 20 μg of protein were mixed in equal volumes with tracking dye (50% [wt/vol] sucrose, 0.1% [wt/vol] bromophenyl blue) and loaded onto a 6% resolving gel (30:0.8, acrylamide-bisacrylamide) in 0.375 M Tris-HCl (pH 8.9) with a 3.75% stacking gel in 0.0625 M Tris HCl (pH 6.7) in a mini-Protein II apparatus (Bio-Rad). Gels were electrophoresed in electrode buffer containing 0.38 M glycine and 0.005 M Tris at 75 V for 2.5 h. Gels were then either stained for urease activity (36) and subsequently with Coomassie brilliant blue R250 or electrophoretically transferred to TransBlot nitrocellulose membranes (Bio-Rad) at 30 V overnight at 4°C, using a Trans Blot apparatus (Bio-Rad). Immunoblots were reacted either with rabbit serum raised against UreB, the large urease subunit of H. pylori (a generous gift from H. Mobley), or with normal rabbit serum. Protein A conjugated to alkaline phosphatase (Sigma Chemical Co.) was used to detect bound antibodies, and the blots were developed using 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium (Sigma Chemical Co.) according to the manufacturer's instructions.
Nucleotide sequence accession number.
The nucleotide sequence reported here has been submitted to GenBank and assigned no. AF167577.
RESULTS
Analysis of mutants.
A bank of over 6,000 individual transposon mutations was screened, and 19 Ure− mutants were identified. Four of 19 Tn10 insertions were located within known urease genes, while insertions in 11 of 15 of the remaining mutants mapped within four open reading frames (ORFs) located 2 to 4 kbp upstream of the urease genes (Fig. 1A). The remaining four mutants have either a spontaneous mutation within these regions or Tn10 insertions within as yet unidentified urease-related genes. Of the four mutants with insertions in either the ureB or ureG gene, all had additional inserts within their chromosome, as did one of the cbiK::Tn10 mutants. Also, the insertions in the three ureG::Tn10 mutants were in the same location in a sequence known to be a hotspot (5′-NGCTNAGCN-3′) for Tn10 insertion (2).
Analysis of upstream gene sequence.
The sequence upstream of the urease cluster in A. pleuropneumoniae (GenBank accession number AF167577) revealed six closely linked ORFs, all transcribed in the same direction as the urease cluster, and a partial ORF transcribed in the opposite orientation, 202 bases upstream of the six ORFs (Fig. 1A). The alternate start codon, GUG, is used in the third ORF, while the remaining ORFs all have the predicted AUG start codons. All but the second and fourth ORFs are preceded by sites similar to the E. coli ribosome-binding site consensus sequence (6). Putative ς70 promoter sequences (13) are present upstream of the first and sixth ORFs.
The first five ORFs encode polypeptides that show from 42 to 63% sequence identity with a gene cluster of unknown function in Haemophilus influenzae (10). The polypeptide sequences predicted from the third, fourth, and fifth ORFs also show sequence similarity with bacterial cobalt transport membrane proteins CbiM (19 to 32%) and CbiQ (16 to 56%) and cobalt transport ATP-binding protein CbiO (28 to 40%). Accordingly, we have designated these ORFs cbiM, cbiQ, and cbiO, respectively. Neither of the first two ORFs has any significant similarity with the cbiN gene, which encodes the periplasmic binding protein component of known cobalt permeases. The close proximity of these ORFs with the cbiMQO genes, however, suggests that they belong to this operon, and therefore we designated these two ORFs cbiK and cbiL. The sixth ORF encodes a polypeptide which has considerable sequence identity (20 to 30% over the entire sequence) with eukaryotic urea transport proteins and has been designated utp. The truncated upstream ORF, which we designated apuR, encodes 141 amino acid residues that show extensive homology with members of the LysR family of transcriptional regulators.
Computer analyses of the predicted polypeptide sequences revealed that CbiK is likely a periplasmic protein. A potential N-terminal signal sequence was detected, with a likely cleavage site at position 21. The CbiL sequence was predicted to be a cytoplasmic membrane protein. It contains two strong TM helices, the first of which corresponds to a predicted N-terminal signal sequence, with a cleavage site at position 22. Both the CbiM and CbiQ polypeptides appear to be integral cytoplasmic membrane proteins, with five and four predicted TM helices, respectively. The CbiM and CbiQ proteins do not contain consensus sequences related to any of the known hydrophobic membrane protein families of periplasmic permeases (33). However, the A. pleuropneumoniae CbiM protein was found to have a region of high homology with a number of CbiM proteins. This sequence, located approximately 100 residues from the C terminus, contains several conserved glycine residues. The CbiQ protein also contains a region (amino acids 117 to 159) with homology to a consensus sequence common to all known CbiQ proteins. The CbiO protein was identified as a member of the ATP-binding cassette (ABC) transport family, with the conserved ATP/GTP binding motif, and was predicted to be localized in the cytoplasm. The Utp protein was predicted to be an integral cytoplasmic membrane protein, with seven predicted TM helices.
Urease activity in the presence and absence of NiCl2.
Like the other constructs, the minimal urease subclone DH5 α/pURE1 was urease positive when patched from LB-AMP plates onto urea agar. However, quantitative urease assays using cultures grown in the presence of various concentrations of NiCl2 showed that the level of urease activity of this clone was reduced compared to that of DH5α/pJBG15B or DH5α/pJBG15C, clones containing the upstream genes in addition to the urease genes (Table 2 and Fig. 1B). Maximal expression of urease activity by the minimal subclone required the addition of ≥200 μM NiCl2, whereas addition of more than 2 μM NiCl2 did not greatly enhance the urease activities of either DH5α/pJBG15B or DH5α/pJBG15C (Table 2). The level of urease expression also differed depending on the orientation of the 15-kbp insert (Table 2 and Fig. 1B). Urease activity was higher in DH5α/pJBG15C, where the truncated apuR gene is immediately downstream of and in opposite orientation from the lac promoter of the vector. In contrast, E. coli DH5α/pJBG15B produced levels of urease activity similar to that of the pURE1 clone in the absence of added nickel. However, unlike DH5α/pURE1, the addition of only 2 μM NiCl2 resulted in production of nearly maximal urease activity by DH5α/pJBG15B, although this level was still less than that of DH5α/pJBG15C (Table 2).
TABLE 2.
Urease activity in whole-cell samples of E. coli clones containing the plasmids pURE1, pJBG15B, and pJBG15C and grown in the presence or absence of added NiCl2
| NiCl2 concn (μM) | Mean urease activity (μM NH3/min/ml) ± SD in E. coli carrying:
|
||
|---|---|---|---|
| pURE1 | pJBG15B | pJBG15C | |
| 0 | 0.96 ± 0.01 | 1.69 ± 0.15 | 5.57 ± 0.20 |
| 2 | 1.18 ± 0.02 | 4.42 ± 0.18 | 8.72 ± 0.06 |
| 20 | 1.37 ± 0.08 | 4.79 ± 0.03 | 10.03 ± 0.21 |
| 200 | 3.66 ± 0.08 | 5.14 ± 0.05 | 9.63 ± 0.23 |
Production of urease apoenzyme.
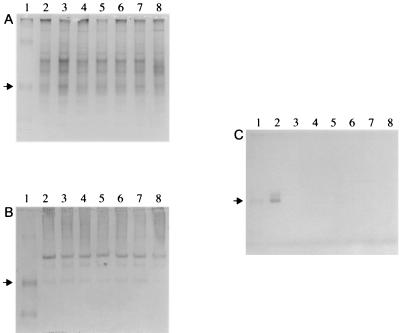
None of the A. pleuropneumoniae Tn10 mutants tested showed detectable levels of urease activity in cell-free cytosolic fractions, whereas the parental strain (CM5 Nalr) exhibited a urease activity of 21 ± 2 U/mg of protein. This was confirmed in a nondenaturing polyacrylamide gel stained for urease activity (Fig. 2C), where only the CM5 Nalr strain produced a bright pink band. In contrast, a band at the same level as the active urease was detected in all samples except the ureB::Tn10 mutant using rabbit polyclonal antiserum against the UreB subunit of H. pylori in immunoblot analysis (Fig. 2B). Other nonspecific bands were also detected by the polyclonal rabbit antiserum in all of the samples. The Coomassie blue-stained gel (Fig. 2A) verified that equal amounts of all samples were loaded.
FIG. 2.
Nondenaturing polyacrylamide gels of ultracentifuged cell lysates from A. pleuropneumoniae strains CM5 Nalr (lane 2), cbiK::Tn10 (lane 3), cbiL::Tn10 (lane 4), cbiM::Tn10 (lane 5), cbiQ::Tn10 (lane 6), ureG::Tn10 (lane 7), and ureB::Tn10 (lane 8). Prestained broad-range molecular size markers (lane 1) were used for reference only, not to indicate the sizes of polypeptides. The arrow indicates the position of the urease band. (A) Coomassie blue-stained gel. (B) Immunoblot reacted with antiserum against H. pylori UreB (diluted 1:250). (C) Urease activity gel.
DISCUSSION
Analysis of urease-negative transposon mutants has led to the identification of a putative ABC transport operon (cbiKLMQO) upstream of the urease cluster that is required for urease activity in A. pleuropneumoniae (Fig. 1A). The production of inactive urease apoenzyme by these mutants suggests that the Ure− phenotype is not due to a polar effect on the downstream urease genes (Fig. 2). Although the upstream genes are not absolutely required for urease activity in E. coli, they do enhance it. E. coli DH5α/pJBG15C, containing the upstream genes along with the urease cluster, expressed urease activity when grown in unsupplemented LB-AMP broth (which likely contains trace quantities of nickel). Urease activity of this clone was not greatly improved by addition of more than 2 μM NiCl2. In contrast, urease activity was barely detectable with the minimal subclone (DH5α/pURE1) grown in unsupplemented LB-AMP broth. This activity was greatly enhanced, however, in the presence of high concentrations (200 μM) of NiCl2, suggesting that the upstream genes may be needed for nickel transport.
The orientation of the 15-kbp insert in pJBG15B and pJBG15C also affected the level of urease activity (Table 2). This may have been the result of increased expression of the cbi genes by readthrough from the lac promoter (Fig. 1B). Alternatively, production of the truncated ApuR protein could have affected expression of the nickel uptake operon. Urease genes in Klebsiella aerogenes and Bordetella bronchiseptica have been reported to be regulated by LysR-like proteins, NAC and BbuR, respectively (3, 20), and it was recently reported that the 100 amino-terminal residues of NAC are sufficient for all of the known activities of this protein (25). Since 141 amino-terminal residues of the truncated ORF are present in pJBG15B and pJBG15C, and since there are sequences similar to typical LysR binding sites (interrupted dyad with T-N11-A [34]) upstream of both cbiK (TCTT-N12-AAGA) and ureA (ATT-N11-AAT), it is possible that ApuR′ may be involved in the regulation of expression of these genes. Given the orientation of apuR′ with respect to the lac promoter, it could be predicted that ApuR′ expression would be higher in DH5α/pJBG15B than in DH5α/pJBG15C, and if ApuR was acting as a repressor, urease expression would be accordingly lower. Urease expression was indeed two to three times higher in DH5α/pJBG15C (Table 2), but further experiments are required to conclusively demonstrate the role of ApuR in the transcriptional regulation of these genes.
At high concentrations, nickel transport can be mediated through magnesium uptake system such as the Salmonella enterica serovar Typhimurium CorA, MgtA, and MgtB (12, 19) which have Kms for Ni(II) of 200, 30, and 13, respectively. Since nickel availability in serum is less than 0.5 ± 0.3 μM, the affinity of these systems for Ni(II) is too low to be physiologically relevant (27, 30). At lower concentrations, specific high-affinity transport of nickel may be mediated either via a single integral membrane protein, as in the case of NixA (23), HoxN (9, 41), HupN (11), and UreH (18), or via an ABC transport system, such as the Nik operon of E. coli (26). The cbiKLMQO genes of A. pleuropneumoniae have extensive sequence identity with and show similar organization to a gene cluster in H. influenzae (10), another ureolytic member of the family Pasteurellaceae. Both of these gene clusters have the appearance of periplasmic binding protein-dependent permease operons (1), although the function of the H. influenzae gene cluster is unknown.
The first gene in bacterial periplasmic permease operons is typically the periplasmic binding protein (1). The product of the first gene in the A. pleuropneumoniae cbi operon, cbiK, shows no homology with either CbiN, the cobalt-specific binding protein of Salmonella (31), or with NikA, the nickel-binding protein of E. coli (26). It should be noted that neither CbiK nor NikA contains a typical metal-binding motif (Cys-X-X-Cys) (26). The CbiK sequence does, however, contain six histidine residues as well as eight aspartate and 20 glutamate residues which could play a role in nickel binding (42). CbiL, which is predicted to be a cytoplasmic membrane protein, also contains 5 His, 10 Asp, and 10 Glu residues. A distinct role for CbiL is not clear; however, it is possible that CbiL may work in conjunction with CbiK.
The cbiM and cbiQ genes both encode highly hydrophobic proteins with five and four predicted TM helices, respectively, which may combine to form the integral membrane portion of the transport system. The hydrophobic membrane proteins (HMPs) of ABC transport systems may show little overall sequence similarity, but most contain a conserved hydrophilic segment with an invariant glycine approximately 100 residues from the C terminus (7). The HMPs have been divided into distinct families, each with its own consensus sequence for this region (33). The A. pleuropneumoniae CbiM and CbiQ proteins did not fit into any of the known HMP families, but both proteins contain consensus sequences found in other CbiM and CbiQ proteins. These proteins may represent a new family of HMPs. The CbiO protein has strong sequence identity with a large number of ABC transport proteins, most notably the cobalt transport ATP-binding proteins from various bacteria.
Taken together, these data suggest that the CbiKLMQO proteins form a periplasmic binding protein-dependent permease for cobalt and/or related molecules such as nickel. Both cobalt and nickel are divalent cations, and there is precedent for their common transport. For example, Nh1F of Rhodococcus rhodochrous (8) and the Cnr and Ncc efflux systems in Alcaligenes eutrophus and Alealigenes xylosoxidans, respectively, have been shown to transport both nickel and cobalt (17, 35). The fact that the presence of the cbiKLMQO genes negated the need for high concentrations of nickel for urease activity in the DH5α/pJBG15C clone supports the hypothesis that nickel is a substrate for this permease system.
The utp gene also appears to encode an integral membrane protein with seven potential TM domains. It is transcribed in the same direction as the other five genes, but there is no homologous gene in H. influenzae, and it is not clear whether it is separate from the preceding gene cluster. It is interesting to note that the predicted Utp polypeptide has considerable sequence identity with mammalian urea transport proteins, which are believed to transport urea via facilitated diffusion, although there is also evidence for sodium-dependent active transport (28, 29, 37, 43). There is evidence for carrier-mediated energy-dependent urea uptake systems in various bacteria (14–16, 38), and genes (fmdABCDE) encoding an outer membrane porin and a binding protein-dependent permease system for the uptake of short-chain amides and urea in Methylophilus methylotrophus have been identified (21, 22). Also, it has recently been shown that the UreI protein of H. pylori, which is an integral membrane protein with six TM helices, is a hydrogen-gated urea channel (40). The Utp protein of A. pleuropneumoniae shows no homology with either UreI or the Fmd proteins, and it remains to be determined whether the utp gene encodes a novel bacterial urea transport protein.
The fact that A. pleuropneumoniae requires the putative nickel transport genes (cbiKLMQO) for urease activity indicates that a large number of genes are involved in maintaining the urease-positive phenotype. This finding further supports the hypothesis that urease activity is important in the pathogenesis of pleuropneumonia (5).
ACKNOWLEDGMENTS
This work was supported by a Natural Science and Engineering Research Council of Canada (NSERC) grant to J.M. J.B. was the recipient of an NSERC PGS3 scholarship and an Ontario Graduate Scholarship.
We thank Kati Anton for help in screening some of the urease mutants.
REFERENCES
- 1.Ames G F, Mimura C S, Shyamala V. Bacterial periplasmic permeases belong to a family of transport proteins operating from Escherichia coli to human: traffic ATPases. FEMS Microbiol Rev. 1990;6:429–446. doi: 10.1111/j.1574-6968.1990.tb04110.x. [DOI] [PubMed] [Google Scholar]
- 2.Bender J, Kleckner N. IS10 transposase mutations that specifically alter target site recognition. EMBO J. 1992;11:741–750. doi: 10.1002/j.1460-2075.1992.tb05107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender R A. The role of the NAC protein in the nitrogen regulation of Klebsiella aerogenes. Mol Microbiol. 1991;5:2575–2580. doi: 10.1111/j.1365-2958.1991.tb01965.x. [DOI] [PubMed] [Google Scholar]
- 4.Bossé J T, MacInnes J I. Genetic and biochemical analyses of Actinobacillus pleuropneumoniae urease. Infect Immun. 1997;65:4389–4394. doi: 10.1128/iai.65.11.4389-4394.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bossé J T, MacInnes J I. Urease activity may contribute to the ability of Actinobacillus pleuropneumoniae to establish infection. Can J Vet Res. 2000;64:145–150. [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H, Bjerknes M, Kumar R, Jay E. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 1994;22:4953–4957. doi: 10.1093/nar/22.23.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dassa E, Hofnung M. Sequence of gene malG in E. coli K12: homologies between integral membrane components from binding protein-dependent transport systems. EMBO J. 1985;4:2287–2293. doi: 10.1002/j.1460-2075.1985.tb03928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Degan O, Kobayashi M, Shimizu S, Eitinger T. Selective transport of divalent cations by transition metal permeases: the Alcaligenes eutrophus HoxN and the Rhodococcus rhodochrous Nh1F. Arch Microbiol. 1999;171:139–145. doi: 10.1007/s002030050691. [DOI] [PubMed] [Google Scholar]
- 9.Eitinger T, Friedrich B. Cloning, nucleotide sequence, and heterologous expression of a high-affinity nickel transport gene from Alcaligenes eutrophus. J Biol Chem. 1991;266:3222–3227. [PubMed] [Google Scholar]
- 10.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 11.Fu C, Javedan S, Moshiri F, Maier R J. Bacterial genes involved in incorporation of nickel into a hydrogenase enzyme. Proc Natl Acad Sci USA. 1994;91:5099–5103. doi: 10.1073/pnas.91.11.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson M M, Bagga D A, Miller C G, Maguire M E. Magnesium transport in Salmonella typhimurium: the influence of new mutations conferring Co2+ resistance on the CorA Mg2+ transport system. Mol Microbiol. 1991;5:2753–2762. doi: 10.1111/j.1365-2958.1991.tb01984.x. [DOI] [PubMed] [Google Scholar]
- 13.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jahns T. Regulation of urea uptake in Pseudomonas aeruginosa. Antonie Van Leeuwenhoek. 1992;62:173–179. doi: 10.1007/BF00582577. [DOI] [PubMed] [Google Scholar]
- 15.Jahns T, Zobel A, Kliener D, Kaltwasser H. Evidence for carrier-mediated, energy-dependent uptake of urea in some bacteria. Arch Microbiol. 1988;149:377–383. [Google Scholar]
- 16.Jahns T, Kaltwasser H. Energy-dependent uptake of urea by Bacillus megaterium. FEMS Microbiol Lett. 1989;48:13–17. doi: 10.1016/0378-1097(89)90138-9. [DOI] [PubMed] [Google Scholar]
- 17.Liesegang H, Lemke K, Siddiqui R A, Schlegel H G. Characterization of the inducible nickel and cobalt resistance determinant cnr from pMOL28 of Alcaligenes eutrophus CH34. J Bacteriol. 1993;175:767–778. doi: 10.1128/jb.175.3.767-778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maeda M, Hidaka M, Nakamura A, Masaki H, Uozumi T. Cloning, sequencing, and expression of thermophilic Bacillus sp. strain TB-90 urease gene complex in Escherichia coli. J Bacteriol. 1994;176:432–442. doi: 10.1128/jb.176.2.432-442.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maguire M E. MgtA and MgtB: prokaryotic P-type ATPases that mediate Mg2+ influx. J Bioenerg Biomembr. 1992;24:319–28. doi: 10.1007/BF00768852. [DOI] [PubMed] [Google Scholar]
- 20.McMillan D J, Mau M, Walker M J. Characterisation of the urease gene cluster in Bordetella bronchiseptica. Gene. 1998;208:243–251. doi: 10.1016/s0378-1119(97)00651-3. [DOI] [PubMed] [Google Scholar]
- 21.Mills J, Wyborn N R, Greenwood J A, Williams S G, Jones C W. An outer-membrane porin inducible by short-chain amides and urea in the methylotrophic bacterium Methylophilus methylotrophus. Microbiology. 1997;143:2373–2379. doi: 10.1099/00221287-143-7-2373. [DOI] [PubMed] [Google Scholar]
- 22.Mills J, Wyborn N R, Greenwood J A, Williams S G, Jones C W. Characterisation of a binding-protein-dependent, active transport system for short-chain amides and urea in the methylotrophic bacterium Methylophilus methylotrophus. Eur J Biochem. 1998;251:45–53. doi: 10.1046/j.1432-1327.1998.2510045.x. [DOI] [PubMed] [Google Scholar]
- 23.Mobley H L, Garner R M, Bauerfeind P. Helicobacter pylori nickel-transport gene nixA: synthesis of catalytically active urease in Escherichia coli independent of growth conditions. Mol Microbiol. 1995b;16:97–109. doi: 10.1111/j.1365-2958.1995.tb02395.x. [DOI] [PubMed] [Google Scholar]
- 24.Mobley H L, Island M D, Hausinger R P. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muse W B, Bender R A. The amino-terminal 100 residues of the nitrogen assimilation control protein (NAC) encode all known properties of NAC from Klebsiella aerogenes and Escherichia coli. J Bacteriol. 1999;181:934–940. doi: 10.1128/jb.181.3.934-940.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Navarro C, Wu L-F, Mandrand-Berthelot M-A. The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol Microbiol. 1993;9:1181–1191. doi: 10.1111/j.1365-2958.1993.tb01247.x. [DOI] [PubMed] [Google Scholar]
- 27.Nieboer E, Sanford W E, Stace B C. Absorption, distribution, and excretion of nickel. In: Niebor E, Nriagu J, editors. Nickel and human health. New York, N.Y: John Wiley and Sons; 1992. pp. 49–51. [Google Scholar]
- 28.Olives B, Neau P, Bailly P, Hediger M A, Rousselet G, Cartron J P, Ripoche P. Cloning and functional expression of a urea transporter from human bone marrow cells. J Biol Chem. 1994;269:31649–3152. [PubMed] [Google Scholar]
- 29.Olives B, Martial S, Mattei M G, Matassi G, Rousselet G, Ripoche P, Cartron J P, Bailly P. Molecular characterization of a new urea transporter in the human kidney. FEBS Lett. 1996;386:156–160. doi: 10.1016/0014-5793(96)00425-5. [DOI] [PubMed] [Google Scholar]
- 30.Ragsdale S W. Nickel biochemistry. Curr Opin Chem Biol. 1998;2:208–215. doi: 10.1016/s1367-5931(98)80062-8. [DOI] [PubMed] [Google Scholar]
- 31.Roth J R, Lawrence J G, Rubenfield M, Kieffer-Higgins S, Church G M. Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J Bacteriol. 1993;175:3303–3316. doi: 10.1128/jb.175.11.3303-3316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Frisch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Saurin W, Koster W, Dassa E. Bacterial binding protein-dependent permeases: characterization of distinctive signatures for functionally related integral cytoplasmic membrane proteins. Mol Microbiol. 1994;12:993–1004. doi: 10.1111/j.1365-2958.1994.tb01087.x. [DOI] [PubMed] [Google Scholar]
- 34.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt T, Schlegel H G. Combined nickel-cobalt-cadmium resistance encoded by the ncc locus of Alcaligenes xylosoxidans 31A. J Bacteriol. 1994;176:7045–7054. doi: 10.1128/jb.176.22.7045-7054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Senior B W, Bradford N C, Simpson D S. The ureases of Proteus strains in relation to virulence for the urinary tract. J Med Microbiol. 1980;13:507–512. doi: 10.1099/00222615-13-4-507. [DOI] [PubMed] [Google Scholar]
- 37.Shayakul C, Steel A, Hediger M A. Molecular cloning and characterization of the vasopressin-regulated urea transporter of rat kidney collecting ducts. J Clin Investig. 1996;98:2580–2587. doi: 10.1172/JCI119077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siewe R M, Weil B, Burkovski A, Eggeling L, Kramer R, Jahns T. Urea uptake and urease activity in Corynebacterium glutamicum. Arch Microbiol. 1998;169:411–416. doi: 10.1007/s002030050591. [DOI] [PubMed] [Google Scholar]
- 39.Tascon R I, Rodriguez-Ferri E F, Gutierrez-Martin C B, Rodriguez-Barbosa I, Berche P, Vazquez-Boland J A. Transposon mutagenesis in Actinobacillus pleuropneumoniae with a Tn10 derivative. J Bacteriol. 1993;175:5717–5722. doi: 10.1128/jb.175.17.5717-5722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weeks D L, Eskandari S, Scott D R, Sachs G. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science. 2000;287:482–485. doi: 10.1126/science.287.5452.482. [DOI] [PubMed] [Google Scholar]
- 41.Wolfram L, Friedrich B, Eitinger T. The Alcaligenes eutrophus protein HoxN mediates nickel transport in Escherichia coli. J Bacteriol. 1995;177:1840–1843. doi: 10.1128/jb.177.7.1840-1843.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu L-F. Putative nickel-binding sites of microbial proteins. Res Microbiol. 1992;143:347–351. doi: 10.1016/0923-2508(92)90027-l. [DOI] [PubMed] [Google Scholar]
- 43.You G, Smith C P, Kanal Y, Lee W-S, Stelzner M, Heidiger M A. Cloning and characterization of the vasopressin-regulated urea transporter. Nature. 1993;365:844–847. doi: 10.1038/365844a0. [DOI] [PubMed] [Google Scholar]