Abstract
Lung ultrasound (LUS) has been increasingly expanding since the 1990s, when the clinical relevance of vertical artifacts was first reported. However, the massive spread of LUS is only recent and is associated with the coronavirus disease 2019 (COVID-19) pandemic, during which semi-quantitative computer-aided techniques were proposed to automatically classify LUS data. In this review, we discuss the state of the art in LUS, from semi-quantitative image analysis approaches to quantitative techniques involving the analysis of radiofrequency data. We also discuss recent in vitro and in silico studies, as well as research on LUS safety. Finally, conclusions are drawn highlighting the potential future of LUS.
Key Words: Lung ultrasound, Artificial intelligence, Image processing, Signal processing, Quantitative lung ultrasound, In vitro, In vivo, Review
Introduction
The ability of ultrasound waves to penetrate media having similar acoustic impedances (e.g., soft tissue) makes them particularly suitable for medical imaging. Moreover, the presence of similar speeds of sound in the human body is fundamental to reconstruct the anatomy with ultrasound imaging. Specifically, these two characteristics allow clinicians to anatomically investigate the internal parts of the human body in real time without exposing patients to ionizing radiations. However, the anatomical investigation of aerated organs is not possible as the standard ultrasound imaging assumptions of similar acoustic impedances and quasi-homogeneous speed of sound in the volume of interest are unmet because of the presence of air. Indeed, the acoustic impedance of air significantly differs from that of soft tissues, causing ultrasound waves to be almost completely reflected when encountering an acoustic interface formed by these two media. This is extremely relevant in lungs, as they consist of millions of air-filled alveoli.
The first studies on the use of ultrasound imaging for lung investigation were conducted by Dunn and Fry in 1961, in which they attempted to estimate ultrasound absorption and reflection in lung tissue. Similar studies focused on the acoustical characterization of lung tissue were performed in the following years both in vitro (Bauld and Schwan 1974; Dunn 1974, 1986; Pedersen and Ozcan 1986; Towa et al. 2002; Oelze et al. 2008) and in vivo (Sagar et al. 1978). However, these studies did not spread to clinical practice because of the difficulty in achieving reproducible estimations.
On the other hand, the first clinical applications of lung ultrasound (LUS) date back to 1967, when Joyner et al. reported the possible existence of characteristic ultrasound patterns in patients with pulmonary embolism and pleural effusion (Joyner et al. 1967; Miller et al. 1967). Fifteen years later, Ziskin et al. (1982) observed the so-called “comet tail artifact” in a patient with a shotgun wound in the abdomen. These artifacts were described as “dense horizontal reverberation echoes likely caused by the strong reflection of objects having significantly different acoustic impedances with the background medium” (Ziskin et al. 1982). However, given their vertical extent, these artifacts are generally referred to as vertical artifacts or comet-tail artifacts. The “comet-tail artifact” was indeed detected by the authors also in canine liver in correspondence of lead pellets and by imaging glass and metallic rods in a water tank (Ziskin et al. 1982). The clinical relevance of these artifacts was then highlighted in 1983, when Thickman et al. (1983) observed them in several tissue–gas interfaces (e.g., diaphragm/aerated lung interface). A similar artifact, called “ring-down,” was reproduced in vitro by Avruch and Cooperberg (1985), who observed these vertical artifacts when a bubble tetrahedron formed by two layers of soapy bubbles (diameter from 1 to 7 mm) was imaged with an ultrasound transducer. However, only in the 1990s the clinical use of these vertical artifacts (first called “comet tail” artifacts) started to rapidly increase thanks to Lichtenstein et al. (1997), who observed a correlation between this artifact and the presence of alveolar–interstitial syndrome. We should also mention an important previous study that compared the sonographic appearance of pulmonary infarctions with their pathological reports (Mathis and Dirschmid 1993). However, contrary to Lichtenstein et al. (1997), who compared in vivo the sonographic signs with computed tomography (CT) anatomical images of patients, this study was performed ex vivo (Mathis and Dirschmid 1993).
Following the work of Lichtenstein et al. (1997), the use of LUS techniques based on visual interpretations of imaging artifacts spread rapidly in the clinical world. For example, LUS vertical artifacts (called “comets” or “B-lines”) were exploited to assess extravascular lung water (Jambrik et al. 2004; Picano et al. 2006) and differentiate acute cardiogenic pulmonary edema (CPE) from acute respiratory distress syndrome (ARDS) (Copetti et al. 2008).
In addition to the presence of artifacts, ultrasound can be also used to detect subpleural consolidations, which consist of anatomical findings that could be imaged at the lung surface (Lichtenstein et al. 2004). Specifically, consolidations are areas where lung tissue is substantially de-aerated, thus making the lung significantly dense. Indeed, in these cases, lung is partially characterized by acoustical properties similar to those of soft tissues, and can thus be anatomically imaged.
In clinical practice, the common approach adopted by clinicians consists of visually inspecting LUS videos to detect the above-mentioned patterns. These qualitative techniques were reviewed and included in an international consensus on LUS, which is currently the only consensus publication on LUS (Volpicelli et al. 2012).
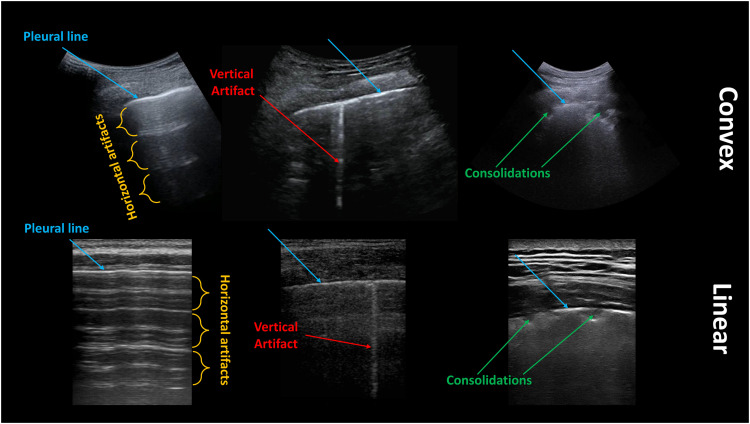
These approaches form the basis of the so-called semi-quantitative techniques, which nowadays represent the most diffused and used strategies in LUS analyses. These techniques are indeed based on the visual interpretation of LUS patterns, where a score is assigned depending on the observed patterns, which correlate with the state of the lung. As an example, the lowest score of a scoring system is generally associated with a continuous pleural line (i.e., the image representation of the acoustic interface formed by intercostal tissues and air within lungs) with associated horizontal artifacts (also known as A-lines), which generally correlate with a healthy lung (Soldati et al. 2020d). Horizontal artifacts represent equally spaced horizontal repetitions of the pleural line, and their genesis is linked to the presence of two strong reflectors (the probe and the pleural line), which causes ultrasound waves to bounce between these two interfaces (Soldati et al. 2019). As a healthy lung is characterized by a high level of aeration, with alveoli close to each other, the pleural line forms an acoustic interface having a reflection coefficient of about 1, thus generating horizontal artifacts in the image (Soldati et al. 2019). Figure 1 contains examples of horizontal and vertical artifacts, as well as consolidations.
Fig. 1.
Examples of lung ultrasound images acquired with convex (top) and linear (bottom) probes. Pleural lines, horizontal artifacts, vertical artifacts and consolidations are indicated in blue, orange, red, and green, respectively.
As the widespread use of these approaches accelerated with the recent outbreak of the COVID-19 pandemic, a significant part of the literature on semi-quantitative LUS is related to COVID-19 applications (Allinovi et al. 2020; Bonadia et al. 2020; Dargent et al. 2020; Smargiassi et al. 2020b; Soldati et al. 2020a, 2020c, 2020d, 2021a, 2021b, 2022; Sultan and Sehgal 2020; Zhao et al. 2020; Demi et al. 2021, 2022; Lerchbaumer et al. 2021; Mento et al. 2021b; Perrone et al. 2021; Russell et al. 2021). Nevertheless, these techniques are generally heterogeneous and are influenced by confounding factors, such as imaging frequency, focal depth and utilized probes, which are generally not mentioned in the publications (Demi 2020). As a consequence, findings obtained using different approaches are extremely difficult to compare (Demi 2020). Another main problem is related to the strong operator dependence in the evaluation of LUS patterns and, thus, to the assigned score.
These limitations could be mitigated by adopting a standardized imaging protocol and scoring system, including technical aspects such as imaging parameters (Soldati et al. 2020d). Moreover, artificial intelligence (AI) algorithms could be exploited to automatically classify LUS data based on scores, resulting in a more robust and reproducible semi-quantitative method (Carrer et al. 2020; Roy et al. 2020; Chen et al. 2021; Mento et al. 2021a; Xue et al. 2021; Roshankhah et al. 2021b; Frank et al. 2022).
Because the necessity to develop acquisition strategies specifically designed for lung has been highlighted not only by clinicians, but also by researchers with technical backgrounds, the development of quantitative approaches could represent the future of LUS (Demi et al. 2014). The evaluation of correlations between anatomical findings observed with standard imaging modalities (e.g., chest x-rays and CT), which are linked to histological observations, and LUS patterns (Soldati et al. 2016, 2017; Smargiassi et al. 2019, 2020a) should represent an important starting point for developing ultrasound approaches specifically dedicated to lungs. This comparison could indeed allow researchers to study the link between the air spaces' (alveoli) resizing and spatial reorganization, as caused by different pathologies, and the quantifiable features of LUS artifacts (Soldati et al. 2019, 2020b). Then, these studies could be exploited to better comprehend LUS artifact genesis, allowing the development of quantitative LUS techniques aimed at estimating physical properties of lung surface (Demi et al. 2017, 2020a; Mohanty et al. 2017; Zhang et al. 2017; Mento and Demi 2020; Mento et al. 2020; Wiley et al. 2021).
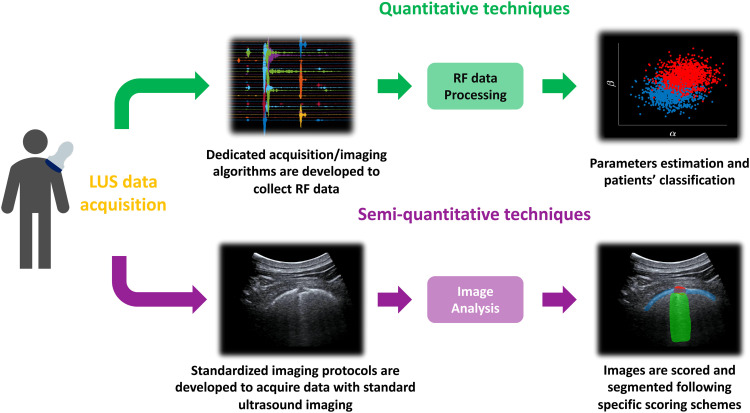
The development of quantitative approaches, however, requires the study and analysis of radiofrequency (RF) data; thus, these solutions are not yet available with clinical ultrasound imaging scanners (Demi et al. 2014). Figure 2 illustrates a simplified flowchart depicting the different applications of quantitative and semi-quantitative LUS techniques.
Fig. 2.
Simplified flowchart depicting the different applications of quantitative and semiquantitative LUS techniques. LUS = lung ultrasound; RF = radiofrequency.
In this review we discuss the main technical publications on LUS, from image analysis techniques (semi-quantitative) to quantitative studies (in vitro, in silico and in vivo). Then, we discuss the main literature investigating LUS safety (Child et al. 1990; Zachary and O'Brien 1995; O'Brien and Zachary 1996; O'Brien et al. 2000; Zachary et al. 2001; Miller et al. 2019) and, finally, draw conclusions.
In this review we cite several pre-clinical and clinical studies. The majority of the studies cited were published in journals for which a condition of publication was either Institutional Animal Care and Use Committee (IACUC) approval, if animals were studied, or approval by an ethics committee or institutional review board and/or receipt of informed consent from each study participant. For those studies for which we were unable to confirm the above we have no reason to doubt that the work was compliant with the ethics and safety guidelines of the institutes involved in the study.
Image Analysis
In this section we analyze the main contributions of LUS image analysis techniques. Specifically, model-based techniques are described in the first subsection, and deep learning approaches in the second subsection.
Model-based techniques
Contreras-Ojeda et al. (2020) proposed an approach to distinguish muscular tissues (above pleural line) from the artifactual structures below the pleural line in ultrasound images of healthy lung tissues of 13 pediatric patients. Symlet (SYM) and Daubechies (DB) wavelet-based feature extraction, principal component analysis (PCA) and recursive backward elimination (RBE) were employed for feature selection, followed by K-nearest-neighbor-based classification. This approach was able to achieve 97.5% accuracy in discriminating muscular tissues from artifactual structures. Even though the results seem promising, the computational cost of the feature extraction phase could represent a relevant drawback for this application. This limitation could indeed strongly affect the advantage of real-time imaging of LUS.
Beyond that, various pathologies can be assessed by examining the pleural line, the subpleural regions and presence or absence of horizontal or vertical artifacts. To this end various techniques have been proposed.
In this regard, work has been done to automate the detection and localization of the pleural line (Carrer et al. 2020). The authors proposed an unsupervised method based on the hidden Markov model and Viterbi algorithm. Furthermore, support vector machine (SVM)–based classification was used to evaluate the characteristics of the pleural line, and the score value for each frame of a given LUS acquisition was assessed based on severity of the pathology. The proposed method was tested on a subset of the Italian COVID-19 LUS Database (ICLUS-DB), acquired from multiple clinical centers. Results revealed high image-based accuracy for both unsupervised detection of the pleural line along with the classification performance. This study indicated the potential of real-time implementation, as the algorithms required a total of 4 s (per image) to detect the pleural line and assign a score.
Anantrasirichai et al. (2017) presented a method for restoration of lines in speckle images, followed by the automatic identification of vertical artifacts. To that purpose, deblurring was performed in the radon domain using a total variation blind deconvolution method. On the other hand, local maxima technique in the radon transform domain was used to identify the vertical artifacts. To evaluate line restoration and detection, 50 simulated images of varying size were used. It was found that, for smaller regularization norms, noise from most of the lines was removed. Detection of vertical artifacts was evaluated using bedside data on 23 children. Although the approach produced promising results, there is a high computational cost (45.75 s per image), making it unfeasible for practical application. Moshavegh et al. (2019) presented a multistep study for automatic detection and visualization of vertical artifacts in LUS scans. They identified the vertical artifacts as connected regions below the pleural line to the bottom end of the scan. Evaluation of 3200 frames from healthy subjects and patients with pulmonary edema revealed that the average number of vertical artifacts was significantly higher in the patients than the healthy participants. Similarly, another method was presented by Karakus et al. (2020). This approach was formulated as a non-convex regularization problem involving a sparsity-enforcing Cauchy-based penalty function and use of an inverse radon transform. The proposed method was validated in both the radon and image domains, over examination of six male and three female patients. Results revealed promising detection accuracy with improved identification of vertical artifacts when compared with the method of Anantrasirichai et al. (2017). However, this approach requires a processing time for a single frame of about 11-13 s, thus not allowing real-time imaging.
When discussing vertical artifacts, it is important to highlight how, given the strong dependence of vertical artifacts on imaging parameters (Kameda et al. 2019; Mento and Demi 2020, 2021), the count of vertical artifacts represents a qualitative application, which does not offer high reproducibility.
Moving from the analysis of vertical artifacts to examination of the pleural and subpleural space, a gray-level co-occurrence matrix (GLCM) with second-order statistical textural-based digital analysis of pleural and subpleural space was proposed by Brusasco et al. (2022). They investigated the discriminating characteristics of a GLCM for differentiating ARDS from acute CPE. The analysis was performed on 47 participants (16 diagnosed with CPE and 8 with ARDS, and 23 declared as healthy). Results revealed statistical significance for 9 of 11 GLCM features on comparison of ARDS and CPE patient subgroups with the healthy participants. Similar statistical significance was reported on comparison of ARDS with CPE. Conclusively, the proposed method showed potential in differentiating those with ARDS and CPE from healthy participants along with differentiating between patients with ARDS and CPE.
Conclusively, it is again important to highlight how LUS analysis is highly subjective and that several confounding factors (e.g., imaging parameters and types of scanners) influence the appearance of the relevant imaging patterns. This leads to high variability among the generated images, affecting the reproducibility of these results. Moreover, the ground truth used to train AI algorithms remains based on subjective labeling of clinicians.
Deep learning techniques
Deep learning (DL)–based analysis of LUS patterns can be applied to assess different pulmonary diseases and pathologies.
Kulhare et al. (2018) developed a convolutional neural network (CNN)–based algorithm to identify some of the lung features linked to pathological lung conditions. These features were defined as vertical artifacts, merged vertical artifacts, lack of lung sliding, consolidation and pleural effusion. Ultrasound data from swine lung pathology models were captured for both normal and abnormal lungs in the form of 100 exams with 2200 videos collected in total. The single-shot detection (SSD) framework was applicable to all LUS features, achieving at least 85% in sensitivity and specificity for all features. Lung features critical for diagnosing abnormal lung conditions were detected with greater than 85% accuracy.
To detect and localize the presence of vertical artifacts, Van Sloun and Demi (2020) presented a DL-based method for automatic detection and localization of vertical artifacts in an ultrasound scan. A fully CNN was trained to perform this task on B-mode images of dedicated ultrasound phantoms in vitro and on patients in vivo. The in vitro study included 3162 frames from 10 tissue-mimicking phantoms, while the in vivo study included 5370 frames from 10 patients. Both showed high classification performance in localizing vertical artifacts. A gradient-weighted class activation map (grad-CAM) approach was used to guarantee a minimum level of explainability. Conclusively the method enabled detection and localization of vertical artifacts in real time.
Another method proposed by Kerdegari et al. (2021) was also aimed at automatically detecting and localizing vertical artifacts in LUS videos using DL networks trained with weak labels. CNN combined with a long short-term memory (LSTM) network and a temporal attention mechanism was evaluated on LUS scans from 60 patients, totaling 298 examinations. These architectures varied in terms of convolutional networks (2-D or 3-D), presence and absence of temporal attention module along with the LSTM networks. The 2-D convolutional network with LSTM and attention module outperformed the other models, allowing the capture of features from both spatial and temporal dimensions. The model was able to achieve a classification F1 score of 0.81 with a localization accuracy of 67.1% within frames with vertical artifacts.
Baloescu et al. (2020) developed and tested a DL algorithm to assess the presence and absence of vertical artifacts in LUS. A total of 400 consecutive thoracic ultrasound clips, each from a unique patient, were used. Each of the 400 clips was split into several 2415 subclips, rated by two emergency physician point-of-care ultrasound (POCUS) experts. Rating was based on a pre-determined ordinal scale from 0 (none) to 4 (severe), representing the number of vertical artifacts. When compared with expert interpretation for the presence or absence of vertical artifacts, the model for binary classification produced promising results with a weighted κ of 0.88 with a 95% confidence interval (CI) of 0.79–0.97. Similar performance was also observed in the severity assessment using multiclass classification.
Deep learning–based techniques have also been used to count the vertical artifacts present in the LUS scan. In this regard, Wang et al. (2019) proposed a study using a CNN to count vertical artifacts on a 4864-image LUS data set labeled by clinicians. Furthermore, correlation between the automated count and the clinical parameters was examined. The clinical data set was composed of 152 LUS videos corresponding to a total of 4864 images. The available labels for each image ranged between 0 and 6, corresponding to the number of vertical artifacts in the image. The neural network (NN) agreed with the observer (true value) in 43.4% of the images, with an additional 40.8% in images with a deviation of 1. The intra-class correlation (ICC) for observer difference also revealed that the agreement between the human count and the output of the NN is high (ICC = 0.791).
Lung sliding is the respirophasic back-and-forth movement of the visceral and parietal pleural surface. Its presence indicates complete aeration of the lung at the site of probe placement on the chest wall, while its absence indicates the possibility of pneumothorax. DL-based techniques have been developed to automatically identify the presence or absence of lung sliding. In this regard, Mehanian et al. (2019) developed and compared several DL methods for identifying pneumothoraces in 3-s ultrasound videos collected with a handheld ultrasound system. The first group of methods were based on CNNs paired with time-mapping pre-processing algorithms, including reconstructed M-mode and the proposed simplified optical flow transform (SOFT). The second class of algorithms used a DL architecture that combined CNN for processing spatial information (Inception V3) with a recurrent network (LSTM) for temporal analysis. To evaluate the methods, a total of four swine models were used, forming a total of 10 collection sessions per animal. As a result, a total of 130 positive videos with absence of lung sliding caused by pneumothorax and 122 negative videos with normal lung sliding were formed. The performance of the four methods in identifying the absence of lung sliding in swine pulmonary ultrasound videos was compared. All models learned informative representations of the data, all achieving area under the curve (AUC) greater than 0.83 on unseen data.
Similarly, Jascur et al. (2021) presented a novel DL-based automated M-mode classification method to detect the absence of lung sliding motion in LUS. Automated M-mode classification leveraged semantic segmentation to select 2-D slices across the temporal dimension of the video recording. The data set used to evaluate the study contained recordings of patients after thoracic surgery and were divided by physicians into two classes based on the presence and absence of lung sliding. They generated 17,338 frames. All presented models were pre-trained on the ImageNet database and then fine-tuned for a maximum of 15 epochs. The training and validation data sets followed the patient-wise split, and the ratio between them was maintained at 1.5/1. Balanced accuracy ranged from 62% to 78%, with the best-performing model at 64 frames and the worst-performing model at 256 frames.
Pleural effusion refers to the buildup of excess fluid between the pleural layers outside the lungs. Tsai et al. (2021) aimed to develop an automated system for the interpretation of LUS images of pleural effusion. The standardized protocol followed involved scanning of six anatomical regions combined with a DL algorithm using a spatial transformer network (STN) providing the basis for automatic pathology classification on an image-based level. In this work, the DL model was trained using supervised and weakly supervised approaches, which used frame and video-based ground truth labels, respectively. In total, 623 ultrasound videos were acquired resulting in 99,209 2-D ultrasound images. To perform cross-validation, 10 folds of training and test sets were created, in which each patient appeared at least once in the test set. The video-based labeling approach reached 91.12% mean accuracy in the test set over the 10-folds, while the frame-based labeling approach reached 92.38%. In addition, a t-test on the accuracy of the two labeling approaches revealed no statistically significant difference in performance between the video-based and frame-based labeling approaches. This significantly reduced the input required from clinical experts to provide ground-truth labels.
Interstitial lung disease (ILD), appearing as fibrotic and stiffened lung parenchyma, may lead to symptoms such as dyspnea, causing respiratory failure. Lung mass is not uniformly distributed in the lung, and it increases with the degree of fibrosis. Zhou and Zhang (2018a) developed a method for analyzing lung mass density of superficial lung tissue using a deep neural network (DNN) and synthetic data of wave speed measurements with LUS surface wave elastography (LUSWE). Data were generated for fibrotic lung tissue, pulmonary congestion and edema, resulting in a total of 792,000 data measurements consisting of surface wave speed, excitation frequency, lung mass density and viscoelasticity. Analyzing the convergence of different optimizers in terms of the validation loss over the epochs, the Adam optimizer had the highest validation accuracy, 0.992. When assessing the performance of the DNN (trained with synthetic data) with a sponge phantom, the predicted density from the DNN matched well the measured density of the sponge phantom, yielding an accuracy of 92%.
Zhou et al. (2020) proposed another study to develop a method for analyzing lung mass density of superficial lung tissue of patients with ILD and healthy participants using a DNN and LUSWE. Surface wave speeds at three frequencies, predicted forced expiratory volume (FEV1% pre), ratio of forced expiratory volume to forced vital capacity (FEV1% = FVC%), age and weight of patients and healthy participants were used as features for training machine learning (ML) models. Random forest revealed that the contributions of age and weight were not as high as those of other features and, hence, these were not used in the training of the DNN model. As it was a retrospective study, 57 patients and 20 healthy participants underwent LUSWE. The evaluation of the performance of the model was based on a train/validation/test (80/10/10) scheme. For the ReLU (rectified linear unit) and ELU (exponential linear unit), training and validation loss significantly decreased as the number of training epochs increased. Accuracies came out to be 84% and 89% for ReLU and ELU, respectively. Comparison of correlation coefficients with different activation functions in the DNN and ML for the testing data set revealed that ELU in the DNN performed comparatively better.
As an extension to the previous study, Zhou et al. (2021) proposed in vivo prediction of lung mass density for patients with ILD using different gradient boosting decision tree (GBDT) algorithms based on measurements from LUSWE and pulmonary function testing (PFT) (Zhou et al. 2021). The study used data similar to those used by Zhou et al. (2020). A fivefold cross-validation was conducted to assess the performance of different algorithms. Among the XGBoost, CatBoost and LightGBM, mean square errors (MSE) and correlation coefficients of the test data set of three algorithms revealed that XGBoost obtained the best results.
Deep learning models require high computational power and resources, making them unsuitable for deployment over lightweight devices such as mobiles and the Internet of Things (IoT). Furthermore, these models require efficient tuning of the hyperparameters. In the light of these concerns, Almeida et al. (2020) explored computer-aided assessment of pneumonia semiology based on lightweight NNs (MobileNets). Multitask learning was performed from online available COVID-19 data sets, for which semiology (overall abnormality, vertical artifacts, consolidations and pleural thickening) was annotated by two radiologists. The data set consisted of a total of 12,718 images extracted from different LUS videos. A 75%/25% train/test split at the image level was used for validation and testing. MobileNet outperformed the naive approach for all semiology indicators, with 95% accuracy for all semiology cases. Furthermore, in classification accuracy of MobileNet trained with labels provided by a senior radiologist, in comparison to labels independently provided by the junior radiologist, a high level of inconsistency was detected for mild conditions, with a mean accuracy of 77%.
On the other hand, Hou et al. (2020) proposed the use of an interpretable subspace approximation with adjusted bias (SAAB) multilayer network to screen the LUS images. They demonstrated the advantage of using SAAB subspace network to design a low-complexity, low-cost, low-power-consumption solution for interpreting and visualizing features of LUS images to confirm the classifier recommendation. A data set of 2800 images was used for this study, consisting of 740 horizontal artifact images, 1150 vertical artifact images and 910 consolidation images. Five hundred sixty images were used for testing, and 2240 images were used for training. Greater than 96% accuracy over the testing data was obtained. In comparison to the CNN models, SaabNet needed to solve only 2800 eigenvalues to yield an embedding vector of 1183 elements and could be employed on any low-cost simple board computers.
Erfanian Ebadi et al. (2021) proposed a method for fast and reliable interpretation of LUS images by use of DL, based on the Kinetics-I3D network. The trained model could classify an entire LUS scan obtained at point of care, without requiring the use of pre-processing or a frame-by-frame analysis. The proposed video classifier was compared with ground-truth classification annotations provided by a set of expert radiologists and clinicians, which included horizontal artifacts, vertical artifacts, consolidation and pleural effusion. A total of 1530 videos were acquired corresponding to 287,549 frames. The models were trained and tested with fivefold cross-validation that creates training and testing sets with 80% (1225 videos) and 20% (305 videos), respectively, for each fold. The model was able to produce a classification of an ultrasound video with 240 frames in 220 ms with accuracy of 90%. The model learned to classify the severe disease cases (consolidation and/or pleural effusion) with a high F1 score.
Shifting from reducing the computational complexity of the network to that of the data, Khan et al. (2022) proposed a method to analyze the impact of data compression on an automated scoring system. The authors presented an automated scoring framework for reduced LUS data acquired from 20 patients with COVID-19, corresponding to 91,277 frames. LUS frames underwent spatial downsampling and reduced quantization levels by factors of 2, 3, 4 and 2, 4, 8, respectively. It was found that the prognostic agreement between expert LUS clinicians and the automatic algorithm employed ranged from 72.35% to 82.35% when reducing the data up to 32 times of its original size. This lays the foundation for efficient automated scoring in resource-constrained environments.
Roy et al. (2020) presented a novel fully annotated data set of LUS image collected from several Italian hospitals, with labels indicating the degree of disease severity at the frame, video and pixel levels (segmentation masks). To evaluate the data set they introduced several deep models that addressed relevant tasks for the automatic analysis of LUS images. In particular, they used a novel deep network, derived from STN, which simultaneously predicts the disease severity score associated with an input frame and provides localization of pathological artifacts in a weakly supervised manner. They also introduced a new method based on uninorms for effective frame score aggregation at the video level. The data were acquired from ICLUS-DB, which at the time included a total of 277 LUS videos from 35 patients (17 COVID-19–positive patients, 4 COVID-19–suspected patients and 14 healthy participants), corresponding to 58,924 frames. Data were split between test and train sets at the patient level; that is, the same patient was not included in both the train and test data sets. On the entire test set, the frame-level Fl score was 70.3%.
To further improve DL performance, Frank et al. (2022) proposed a framework for training through integration of domain knowledge into DNNs by inputting anatomical features and LUS artifacts in the form of additional channels containing pleural and vertical artifact masks along with the raw LUS frames. They used their framework to fine-tune a ResNet-18 model to classify each frame in its annotated severity score. The trained strategy and data set were the same as those of Roy et al. (2020). The performance overcame the state of the art with an Fl score of 75.2%, highlighting the potential of domain knowledge integration to improve DL performance in LUS frame classification.
La Salvia et al. (2021) developed a system based on modern DL methodologies to automatically classify patients based on a seven-level scoring system. For this purpose, they selected ResNet-18 and ResNet-50 architectures. Two thousand nine hundred and eight frames were carefully selected from a total of 5400 videos (consisting of 60,000 frames) to train the models. The data were randomly split into training (75%), validation (15%) and test (10%) sets. By considering this split strategy and the amount of data (less than 3000 frames), an accuracy above 96% was achieved.
Mento et al. (2021a) reported on the level of agreement between DL models and LUS experts when evaluating LUS videos. As a result, they evaluated an empirical threshold approach to aggregate labeled frames to obtain a video-level score. The population analyzed consisted of 82 COVID-19–positive patients corresponding to 314,879 frames. The overall video-level agreement reached its maximum value (51.61%) when a 1% frame-level threshold was applied. This means that a video was classified with the highest score appearing in at least 1% of frames in the video. A more relevant result was obtained when evaluating the percentage of agreement between experts and DL in the stratification between patients at high risk of clinical worsening and patients at low risk. In particular, the approach achieved 85.96% agreement, thus highlighting the possibility of using DL approaches to automatically stratify patients with COVID-19. This aggregation strategy was later applied to a larger cohort of patients, comprising 100 COVID-19–positive patients and 120 post–COVID-19 patients (patients not positive at the time of LUS examination) (Demi et al. 2022). Demi et al.'s study illustrated how the prognostic agreement was 80.45% for patients with COVID-19 and 72.50% for post–COVID-19 patients. The reduced performance on post–COVID-19 patients could be associated with the presence of LUS patterns not fully compatible with those obtained from healthy or acute patients (Demi et al. 2022). Therefore, it is feasible that the AI models, which were trained on LUS data from COVID-19–positive patients, were not able to correctly recognize these patterns (Demi et al. 2022).
Xue et al. (2021) proposed a novel method for severity assessment of patients with COVID-19 from LUS and clinical information. Specifically, the authors stratified the task into three different steps. In the first step, they performed an LUS pattern segmentation at frame level by using a Visual Geometry Group (VGG) encoder. Then, in the second step, they classified LUS videos based on different features, including pattern segmentations obtained from the first step. Finally, they used the LUS score obtained in the second step and the patient's clinical information to assess the overall patient condition. The reported results illustrated how classification performance at the video level increased when pattern segmentations provided in the first step were exploited as additional input features. Furthermore, performance at the patient level improved when video-level LUS scores (provided in second step) were included as additional features.
Chen et al. (2021) developed a technique to automatically classify LUS frames based on a scoring system. A total of 45 patients were used to acquire 1527 images assigned with scores and included in the study. Two different split strategies were tested. In the first, frames were randomly split between train and test sets, whereas, in the second, data were split at the patient level. In other words, the split was made to avoid having similar frames in the training and test data sets. The results illustrated how performance with the first strategy was higher than that with the second. Therefore, these results highlighted the importance of the splitting strategy when training an automatic algorithm.
Roshankhah et al. (2021b) proposed an automatic segmentation method using a CNN to automatically classify LUS images based on a scoring system. The study was evaluated by application of a randomly assigned and simple 90%/10% train set/test set split. Furthermore, the impact of splitting the training/test data was analyzed by repeating the process by performing the split between the train and test data at the patient level. The accuracy of the whole model at the frame level was 95% when data were randomly split. In contrast, it was 68.7% when data were split at the patient level. These results, consistent with the study of Chen et al. (2021), highlighted the importance of reliable splitting strategies when evaluating the performance of AI in classifying LUS data. Specifically, a random split at the frame level strongly affects the performance of AI algorithms, leading to overestimation of the capabilities of the automatic system. The studies presented in this section are summarized in Table 1 .
Table 1.
Details on image analysis publications*
| Publication | Study type | Amount of data | LUS pattern |
| Contreras-Ojeda et al. (2020) | Clinical | 22 images, NP videos, 13 patients | PL |
| Carrer et al. (2020) | Clinical | 3,315 images, 58 videos, 29 patients | PL |
| Anantrasirichai et al. (2017) | Clinical | 50 in silico, 100 clinical images, NP videos, 23 patients |
PL, HA, VA |
| Moshavegh et al. (2019) | Clinical | 3200 images, 64 videos, 8 patients | VA |
| Karakus et al. (2020) | Clinical | 100 images, NP videos, 9 patients | PL, VA |
| Brusasco et al. (2022) | Clinical | 564 images, 564 videos, 47 patients | PL |
| Kulhare et al. (2018) | Animal | NP images, 2200 videos, NP models | PL, VA, CON, LS |
| Van Sloun and Demi (2020) | In vitro, clinical | 3162 in vitro and 5770 clinical images, 10 in vitro and 27 clinical videos, 10 models and 10 patients | VA |
| Kerdegari et al. (2021) | Clinical | NP images, NP videos, 60 patients | VA |
| Baloescu et al. (2020) | Clinical | 28,980 images, 400 videos, 400 patients | VA |
| Wang et al. (2019) | Clinical | 4864 images, 152 videos, NP patients | VA |
| Mehanian et al. (2019) | Animal | NP images, 252 videos, 4 models | LS |
| Jascur et al. (2021) | Clinical | 17,338 images, 48 videos, 48 patients | LS |
| Tsai et al. (2021) | Clinical | 99,209 images, 623 videos, 70 patients | PE |
| Zhou and Zhang (2018a)‡ | In silico, in vitro | 792,000 parameters in silico, 165 parameters in vitro, NP models in silico, 1 model in vitro | None |
| Zhou et al. (2020)‡ | Clinical | NP parameters, 77 patients | None |
| Zhou et al. (2021)‡ | Clinical | NP parameters, 77 patients | None |
| Almeida et al. (2020) | Clinical | 12,718 images, 60 videos, NP patients | VA, PT, CON |
| Hou et al. (2020) | Clinical | 2800 images, NP videos, NP patients | HA, VA, CON |
| Erfanian Ebadi et al. (2021) | Clinical | 287,549 images, 1530 videos, 300 patients | HA, VA, CON, PE |
| Khan et al. (2022)† | Clinical | 91,277 images, 448 videos, 20 patients | HA, VA, CON |
| Roy et al. (2020) | Clinical | 58,924 images, 277 videos, 35 patients | HA, VA, CON |
| Frank et al. (2022) | Clinical | 58,924 images, 277 videos, 35 patients | HA, VA, CON |
| La Salvia et al. (2021) | Clinical | 2908 images, 5400 videos, 450 patients | HA, VA, CON, PL |
| Mento et al. (2021a)† | Clinical | 314,879 images, 1488 videos, 82 patients | HA, VA, CON |
| Demi et al. (2022)† | Clinical | 772,780 images, 3481 videos, 220 patients | HA, VA, CON |
| Xue et al. (2021) | Clinical | 6926 images, 1791 videos, 313 patients | HA, VA, CON, PL |
| Chen et al. (2021) | Clinical | 1527 images, NP videos, 31 patients | PL, HA, VA |
| Roshankhah et al. (2021b) | Clinical | 1863 images, 203 videos, 32 patients | HA, VA, CON, PL |
CON = consolidations; HA = horizontal artifacts; LS = lung sliding; LUS = lung ultrasound; NP = not provided (number of images, videos or patients [models for in vitro, in silico and animal studies] was not provided in the study); PE = pleural effusion; PL = pleural line; PT = pleural thickening; VA = vertical artifacts.
The first column contains references to the publications. The second column indicates the study type (clinical, in vitro, in silico, or on animals). The third column reports the amount of data used in each study. The fourth column indicates which LUS patterns are investigated.
Whether a clinical value exists in the study. For clinical value we here refer to the existence of a proven relation between the investigated LUS patterns and/or scores and the clinical state of the patient.
Study concerning the assessment of parameters rather than images.
Investigation of Artifact Genesis and Quantitative Studies
In this section, we present the main LUS publications focused on investigation of vertical artifact genesis and the main LUS quantitative studies. We first discuss in vitro and animal studies, followed by in silico studies, and, finally, the quantitative LUS approaches tested in human clinical studies.
In vitro and animal studies
One of the first experimental in vitro studies was performed by Soldati et al. (2011), where the appearance of LUS vertical artifacts was correlated to the density of wet, synthetic and partially aerated polyurethane sponges. Specifically, the authors immersed 10 sponges (phantoms) in water and scanned them with a linear probe in five temporal phases during the drying process to visually observe the different LUS patterns appearing in the image. The scans revealed a transition from the first phase (completely wet sponge), characterized by a homogeneously echogenic field of view, also called “white lung,” to phases in which vertical artifacts appeared to be more spatially separated and, thus, the field progressively less echogenic. Therefore, for these models, LUS vertical artifacts seem to be density-correlated phenomena caused by modification of phantoms' porosity, which leads to different acoustic permeabilities (Soldati et al. 2011). In particular, a lower porosity (higher quantity of water filling air spaces) seems to induce greater acoustic permeability and greater acoustic interactions with the aerated superficial structures (Soldati et al. 2011). Similar results were obtained with ex vivo rabbit lungs, which were examined during mechanical inflation (Soldati et al., 2012, Soldati et al., 2014). Specifically, ex vivo deflated lungs were imaged with an ultrasound scanner at different levels of expansion, ranging from 87% (maximum lung expansion) to 40% (naturally collapsed lungs) of air content within lungs (Soldati et al. 2012). By decreasing the air content, hence increasing acoustic permeability, the observed LUS imaging patterns pass from a continuous pleural line with horizontal artifacts (associated to healthy lung) to the progressive presence of multiple vertical artifacts and, finally, “white lung” (Soldati et al. 2012). These results seem to confirm how acoustic lung permeability is a density-related phenomenon (Soldati et al. 2012). However, the authors believe that the investigation on vertical artifacts' genesis should be focused on the porosity in terms of shape and disposition, as altered density of peripheral lung can be simply seen as an epiphenomenon of altered peripheral airspace (PAS) geometry (Soldati et al. 2014).
Following this hypothesis, Demi et al. (2017) designed an in vitro study to evaluate the possibility of characterizing the lung structure (alveolar disposition and shape) by analyzing frequency spectra of vertical artifacts. Lung phantoms were produced by trapping a layer of monodisperse microbubbles in tissue-mimicking gel. Two different populations of phantoms were produced, one with 80-μm-diameter microbubbles and the other with 170-μm-diameter microbubbles (Demi et al. 2017). These sizes were selected to mimic the alveolar size reduction (normal alveolar size = 280 μm), which is typical of various pathologies (Demi et al. 2017). The phantoms were scanned with a research platform using a multifrequency approach in which images were sequentially generated using orthogonal subbands centered at different frequencies (3, 4, 5 and 6 MHz with 1-MHz bandwidth) (Demi et al. 2017). Vertical artifacts appeared with significantly stronger amplitude in specific portions of the frequency spectrum, highlighting the strong frequency dependence of these artifacts. This was also confirmed by an analysis on raw RF data (Demi et al. 2017). These results suggest exploitation of the native frequency (i.e., the frequency at which the vertical artifact appears with stronger intensity) to characterize the state of the lung surface, thus opening to the possibility of developing a quantitative technique based on analysis of the artifact frequency spectrum (Demi et al. 2017). To design such a quantitative technique, the dependence of vertical artifacts on different transmission parameters should be quantitatively evaluated. This was the aim of two recent quantitative studies, in which the dependence of these artifacts' intensity on different parameters was assessed in microbubble (Mento and Demi 2020) and thorax (Mento and Demi 2021) phantoms. The study on microbubble phantoms revealed no correlation between the intensity of vertical artifacts and beam size, thus highlighting how changes in the lateral resolution do not affect the intensity of vertical artifacts (Mento and Demi 2020). Moreover, the center frequency was found to be the most impactful parameter in vertical artifact characterization, followed by focal point position and number of transmitting elements (Mento and Demi 2020). These results were confirmed by the second study, which also highlighted the importance of considering the impact of bandwidth and ultrasound beam angle of incidence when evaluating artifact intensity (Mento and Demi 2021). These parameters indeed represent a significant source of variability in artifact evaluation and, thus, should be carefully considered when implementing a quantitative approach (Mento and Demi 2020, 2021). On one hand, center frequency could be used to characterize the state of lung surface, whereas, on the other hand, the other imaging parameters should be carefully set to prevent them from causing strong variations in artifact intensity (Mento and Demi 2020, 2021).
A completely different quantitative approach was proposed by Mohanty et al. (2017)—a technique based on ultrasound multiple scattering that exploits the complex propagation of sound waves in the lung structure. This approach is a near-field method based on assessment of the growth of a diffusive halo (Mohanty et al. 2017; Demi et al. 2020b). Specifically, it aims at estimating diffusion constant and transport mean free path (L*) of lung parenchyma by using an acquisition approach in which the elements of an array were fired one by one (Mohanty et al. 2017). Then, for each transmitted pulse, the received signals were collected on all the elements and these two parameters estimated (Mohanty et al. 2017). This technique was tested in vitro with a sponge phantom with varying air volume fractions and both in vivo and ex vivo in rat lungs with induced pulmonary edema (Mohanty et al. 2017). The results highlighted how a change of 10% of the air volume fraction corresponded to significant variation of L*. Indeed, when the quantity of fluid increases, the mean distance between scatterers increases, thus increasing L* (Mohanty et al. 2017). In the following years, similar techniques were experimentally tested to differentiate rat lungs with pulmonary fibrosis (PF) from healthy lungs (Mohanty et al. 2020), detect pulmonary nodules (regions inside lungs containing no scatterers) (Roshankhah et al. 2021a) and differentiate rat lungs affected by PF (or edema) from healthy lungs of control rats (Lye et al. 2021).
In 2017 another group developed a quantitative approach based on LUSWE (Zhang et al. 2017), which was then tested in vitro with a cellulose sponge filled with water in specific locations (Zhou and Zhang 2018b). Specifically, water was injected at three locations of the phantom, and a shaker was placed in contact with its surface to generate harmonic vibrations at different frequencies (100, 150 and 200 Hz) (Zhou and Zhang 2018b). An ultrasound probe was then placed 0.5 cm from the shaker to detect and measure the waves propagating on the phantom surface (Zhou and Zhang 2018b). The measurements were taken at six different stages: dry phantom and phantom injected with volumes of water from 3 to 15 mL (in 3-mL steps) (Zhou and Zhang 2018b). The results indicated how surface wave speeds were generally higher when higher shaker frequencies were used, resulting in behavior similar to that observed in LUSWE measurements on human lung (Zhou and Zhang 2018b). Nevertheless, by considering the six stages, the surface wave speeds appeared to be similar, highlighting the difficulty of assessing the quantity of water by means of the LUSWE technique (Zhou and Zhang 2018b).
As discussed in the preceding paragraphs, the design of lung phantoms represents an important step in the development of new LUS quantitative techniques and the investigation of vertical artifact genesis in a controlled environment. This was the main aim of a recent work in which different devices (phantoms) were fabricated by using agar cylinders (or disks), computer numerical control (CNC) milling machines and polyvinyl chloride (PVC) containers (Demi 2021). If imaged with an ultrasound probe, these phantoms were able to in vitro reproduce vertical artifacts, which changed their visual appearance when selected imaging parameters were differently set (e.g., pulse amplitude and center frequency) (Demi 2021). In a similar study, Kameda et al. (2019, 2022) produced simple experimental models able to generate vertical artifacts. Two different models were fabricated: the first consisted of different materials (i.e., a drop of ultrasound gel, a spindle-shaped juice sac of a mandarin orange and a string shaped glucomannan gel) placed on a polypropylene sheet (simulating pleural line), and the second was made by placing glass beads and plates of different sizes on the sheet (Kameda et al. 2019, 2022). The authors then imaged these phantoms by evaluating the visual effect of spatial compound imaging, focal point, center frequency and probe type (convex vs linear) on the appearance of vertical artifacts (Kameda et al. 2019, 2022). They qualitatively illustrated how the impact of these ultrasound machine settings on vertical artifact morphology cannot be considered negligible (Kameda et al. 2019, 2022).
As mentioned above, the reproduction of LUS patterns in experimental phantoms has been proven to be an effective strategy in investigating their genesis. However, these artificial models could significantly differ from human lungs. Therefore, the use of a large animal model to reproduce LUS imaging patterns could represent an important tool to mimic the behavior of human lungs. This was the scope of a recent study that assessed the usability of a pig model to reproduce LUS patterns of viral pneumonia (Wolfram et al. 2020). Specifically, after anesthetization and intubation, six pigs were mechanically ventilated, and a saline liquid was progressively instilled within their lungs using the one-lung flooding technique (Wolfram et al. 2020). Then, the lungs were re-ventilated and imaged with an ultrasound scanner, and the acquired videos were classified based on a scoring system (Wolfram et al. 2020). The LUS score increases (higher score corresponds to a worse state of lung) as the instilled saline fraction increases. Moreover, different LUS patterns associated with ARDS-related pneumonia (e.g., COVID-19 pneumonia) were observed (Wolfram et al. 2020). Given the results obtained and the easy implementation of the model, it could represent an important instrument for LUS research, as it provides, in a more controlled environment, a model similar to humans.
In silico studies
Only recently researchers have started to perform studies on LUS in silico. The first study investigated the dependence of vertical artifacts on alveolar diameter and spacing (i.e., distance between alveoli) (Peschiera et al. 2021). The numerical simulations were performed using the k-Wave MATLAB toolbox (Treeby and Cox 2010), which was used to replicate a simplified lung structure. Specifically, the first 2 cm of depth was composed of muscle tissue, whereas air inclusions (alveoli) were introduced beyond 2 cm (Peschiera et al. 2021). These alveoli were periodically arranged to maintain the same distance between each other, in both the axial and lateral directions. The spacing and diameter of these air inclusions were then varied, and the volume of interest was imaged at different center frequencies (from 1 to 5 MHz with a 1-MHz step size and 1-MHz pulse bandwidth) (Peschiera et al. 2021). In particular, the spacing was set to 198, 263 and 395 μm, which correspond to half of the wavelength in muscle tissue at 4, 3 and 2 MHz. The quantitative results illustrated how the relation between artifact intensity and imaging frequency depends on the complex interaction between wavelength and alveolar geometries. By evaluation of the intensity when spacing was set to half of the wavelength, a possible correlation between artifact strength and the ratio between wavelength and spacing was found (Peschiera et al. 2021). Moreover, as also proven by studies on rabbit lungs (Soldati et al., 2012, Soldati et al., 2014), a higher density of lung (percentage of muscle in the air inclusion area) correlated with more intense vertical artifacts (Peschiera et al. 2021).
The second study aimed at reproducing in silico primary LUS patterns, such as horizontal and vertical artifacts (Ostras et al. 2021). The authors used a custom Fullwave numerical simulator to model different acoustic traps, which were able to simulate different fluid portions in an affected lung region (Ostras et al. 2021). The area above the simulated lung consisted of simulated intercostal tissues, extracted by an optical human data set (Ostras et al. 2021). The volume of interest was imaged with a simulated clinical phased array transducer. The results illustrated how these simulations were able to reproduce horizontal and vertical artifacts in silico (Ostras et al. 2021). Furthermore, consistent with other ex vivo (Soldati et al., 2012, Soldati et al., 2014) and in silico (Peschiera et al. 2021) studies, the authors found a correlation between the density of lung (in terms of fluid portions in the alveoli) and vertical artifact appearance. Specifically, a 55% fluid portion is needed to detect vertical artifacts in these simulations (Ostras et al. 2021).
Quantitative LUS imaging, human clinical studies
There have been few LUS clinical studies in humans aimed at testing or developing quantitative techniques. Indeed, only few researchers have recently moved in this direction. The first quantitative studies in humans were performed by Zhang et al. (2017), who developed and applied LUSWE to estimate superficial lung tissue elastic properties. Specifically, they aimed at differentiating patients with interstitial lung disease (ILD) and healthy participants by using LUSWE (Zhang et al. 2017). The implemented technique was the same at that applied by Zhou and Zhang (2018b) for the in vitro study, in which a handheld shaker was used to generate a 0.1-s harmonic vibration on the phantom surface. During the clinical study, the measurements were taken on both lungs of 10 ILD patients and 10 healthy participants, who were examined through six intercostal spaces (Zhang et al. 2017). The lung surface wave speed was measured three times for each location, that is, once for each frequency (100, 150 and 200 Hz). The results revealed significant differences in the surface wave speed between healthy participants and patients with ILD (Zhang et al. 2017). A similar study was conducted in 2019, in which the authors tried to use LUSWE to assess ILD patients and systemic sclerosis (SSC) (Zhang et al. 2019b). The technique was tested on 91 ILD patients (41 with SSC and 50 without SSC) and 30 healthy participants. As in the previous study, the surface wave speeds of patient lungs were significantly higher than those of healthy participants. Nevertheless, no significant differences were reported between ILD patients with SSC and ILD patients without SSC (Zhang et al. 2019b). The same group applied the LUSWE technique to perform two follow-up studies, aimed at assessing disease progression in patients with ILD (Zhang et al. 2019a) and pulmonary edema (Wiley et al. 2021). In both studies, correlations between changes in lung surface speed and clinical assessments were found, highlighting the possibility of using LUSWE for quantitative assessment of ILD and edema progression (Zhang et al. 2019a; Wiley et al. 2021).
The other LUS quantitative approach that was tested in human clinical studies is based on LUS spectroscopy (Demi et al. 2020a; Mento et al. 2020), and was developed following the findings of a previous in vitro study (Demi et al. 2017). Specifically, the theory underlying these studies consists of hypothesizing that vertical artifacts originate from the interaction between ultrasound waves and acoustic traps (or channels) formed at the lung surface when the lung becomes pathological (Demi et al. 2020a). The genesis of vertical artifacts seems indeed to be related to the formation of channels consisting of media that can be penetrated by ultrasound, such as blood, water and tissue (Soldati et al. 2019, 2020b; Demi et al. 2020c). When a sufficient quantity of energy enters these channels, it can be trapped and progressively irradiated toward the probe with a periodicity depending on the channel's size and the ultrasound pulse's frequency (Demi et al. 2020a, 2020c). Estimation of the sizes of these traps could be relevant in discriminating different pathologies that are characterized by completely different alveolar dispositions and, thus, trap geometries. Specifically, the native frequency and bandwidth of vertical artifacts could carry information on, respectively, the size and geometry of the acoustic traps (Demi et al. 2020a, 2020c). By exploiting these concepts, Demi et al. (2020a) presented an image formation protocol able to capture the frequency dependence of LUS vertical artifacts and visualize it in real time. The final scope consisted of providing a quantitative evaluation of signals received from lung. Ten patients with various lung diseases (e.g., PF, pneumonia and adenocarcinoma) were enrolled, and radiofrequency data were acquired by means of a research platform. The acquisition strategy was based on a multifrequency approach in which four ultrasound images were formed with pulses having different center frequencies (3, 4, 5 and 6 MHz) (Demi et al. 2020a). The pulse repetition frequency (PRF) was fixed at 4 kHz to guarantee that the volume of interest did not significantly change when considering four images acquired with different center frequencies. Moreover, to evaluate more precisely the change in vertical artifacts along the frequency spectrum, a narrow bandwidth (1 MHz) was employed for each center frequency (Demi et al. 2020a). The results illustrated how the appearance of vertical artifacts significantly varied from patient to patient, and different native frequencies and bandwidths of these artifacts were estimated (Demi et al. 2020a). The multifrequency approach of Demi et al. (2020a) was later proposed by Mento et al. (2020) to differentiate PF from other lung pathologies. Specifically, 26 patients were enrolled; half had idiopathic PF (IPF) and the other half were affected by different lung diseases (e.g., emphysema, pulmonary hypertension and asthma). The patients were examined by following the same procedure described by Demi et al. (2020a), and RF data were acquired and analyzed (Mento et al. 2020). One thousand twenty-nine vertical artifacts were detected and their main features (native frequency, bandwidth and total intensity) were analyzed (Mento et al. 2020). Results revealed how these three parameters could be exploited to discriminate patients with PF from patients with other lung diseases. In particular, when all three parameters were considered, an empirically defined binary classifier was able to achieve 92% specificity and sensitivity. Moreover, statistical analysis revealed that native frequency and total intensity were significant in discriminating PF from other lung pathologies, whereas bandwidth was not (Mento et al. 2020). The results obtained in statistical analysis were consistent with the results achieved when standard classifiers receiving these three features as input were employed to differentiate patients (Mento et al. 2020). In conclusion, this study illustrated the potential to discriminate fibrotic patients by exploiting a quantitative approach based on the frequency and intensity analysis of vertical artifacts (Mento et al. 2020).
Lung ultrasound safety
The use of ultrasound as a diagnostic tool is considered safe when imaging soft tissues, as long as the mechanical index (MI) is kept below 1.9. However, the safety of LUS should be carefully analyzed further, as the presence of air in the volume of interest could give rise to completely different phenomena.
One of the first studies in this direction was performed by Child et al. (1990), who were aimed at verifying the effect of exposing mouse lung to pulsed ultrasound. This work was based on the hypothesis linking LUS damage to cavitation phenomena. A cavitation-like phenomenon is linked to the presence of air bubbles that are subjected to high peak pressure variations, which can cause the bubbles to implode. Therefore, these kind of phenomena should be associated with a temporal peak pressure exceeding a given threshold rather than a continuous ultrasound exposure with high temporal average intensities that do not exceed the threshold. In this study, the authors found a non-thermal effect with characteristics that could be associated with cavitation (Child et al. 1990). However, no clear proof of this hypothesis was reported. Similar results were obtained a few years later by Penney et al. (1993). In the same year, Raeman et al. (1993) performed a study investigating the effect of ultrasound on mouse lungs exposed to an approximately constant total “on-time" but different total exposure periods. Specifically, the three temporal conditions were as follows: (i) PRF = 17 Hz for 3-min continuous exposure, (ii) PRF = 1000 Hz for 1-s exposure repeated at 1-min intervals three times, (iii) PRF = 1000 Hz for 3-s continuous exposure (Raeman et al. 1993). The results indicated that the threshold at which lung damage appeared was lower when a continuous exposure was employed (Raeman et al. 1993). Therefore, this result seems not to be consistent with the cavitation hypothesis, which should occur on overcoming a peak positive pressure (PPP) threshold, regardless of the exposure duration.
Following studies strengthened the hypothesis that LUS-induced damage is probably not linked to inertial cavitation (O'Brien et al. 2000; Frizzell et al. 2003). Specifically, O'Brien et al. (2000) directly contradicted the cavitation hypothesis by performing an experimental study in which an elevated hydrostatic pressure was used to suppress the involvement of inertial cavitation. In particular, if cavitation were responsible for lung damage, elevated hydrostatic pressures should cause less lung hemorrhage at each ultrasound pressure level. However, the results illustrated how the impact of ultrasound pressure on lung lesion severity was enhanced by higher hydrostatic pressures, suggesting cavitation was not the cause of lung hemorrhage (O'Brien et al. 2000). Similar results were achieved 3 y later by Frizzell et al. (2003).
A potential problem in the aforementioned studies is the animal model used. Indeed, the animal model used (mouse or rat) differs significantly from humans. Therefore, evaluation of the different effects caused by exposure of the lungs to ultrasound in various animal species could be relevant to reliable extrapolation of the findings to humans. This was the aim of two studies that compared the effects of LUS exposure on mice, rabbits and pigs (Zachary and O'Brien 1995; O'Brien and Zachary 1996). When the same exposure conditions were applied to each animal species, mouse lung was the most sensitive to ultrasound-induced damage, followed by rabbit and, then, pig (O'Brien and Zachary 1996). Specifically, on assessment of ultrasound-induced damage according to the same criteria (i.e., scoring system ranging from 0 to 5), the ratios of lung damage scores were 3.9, 3.7 and 14.4 for mouse/rabbit, rabbit/pig and mouse/pig, respectively (O'Brien and Zachary 1996). These results could be associated with the different anatomies of these animals. As an example, the total lung volumes of rabbits and pigs are 100 and 5000 times larger than that of mice, respectively (O'Brien and Zachary 1996). Given the anatomical characteristics of pigs, they should be chosen as a more reliable animal model in which to investigate LUS-induced damage.
Nevertheless, because of the difficulty involved in managing large animal models, most of the following studies on LUS-induced damage were conducted on small animals. The most recent studies were performed by Miller et al., who analyzed the effect of diagnostic ultrasound (Miller 2012), fixed-beam focused ultrasound (Miller et al. 2015) and shear wave elastography (Miller et al. 2019) on rat lungs. The findings on diagnostic ultrasound illustrated how an MI of 0.44 was sufficient to cause hemorrhage in rat lungs, highlighting a greater sensitivity to LUS than was expected from previous results (Miller 2012). This result on MI was obtained when the rat lung was exposed to ultrasound for 5 min (Miller 2012). Comparable results were obtained by using a shear wave elastography system, although with a significantly reduced exposure time (Miller et al. 2019). In contrast, fixed-beam pulsed ultrasound exposures generated lower PPP thresholds than diagnostic ultrasound (Miller et al. 2015).
Discussion
Lung ultrasound represents a relatively novel application of ultrasound technology, which has been increasingly expanding since the 1990s (Lichtenstein et al. 1997). However, contrary to standard ultrasound imaging, which was developed primarily for imaging non-invasively the anatomy of internal body parts, LUS is based mainly on the visual interpretation of imaging artifacts. Among these, the so-called vertical artifacts are particularly important as they correlate with various pathologies (Jambrik et al. 2004; Picano et al. 2006; Copetti et al. 2008). The main limitations associated with this type of pattern analysis remain its subjectivity and limited reproducibility. Moreover, the understanding and exploitation of the mechanisms underlying the genesis of vertical artifacts are just beginning.
To overcome LUS limits and study vertical artifacts as a mean to characterize the lung surface, various experimental studies have been recently performed (Kameda et al. 2019; Mento and Demi 2020, 2021) and mathematical models have been proposed (Demi et al. 2020a, 2020c). Specifically, the acoustic trap theory suggests that vertical artifacts originate from multiple reflections of ultrasound waves trapped within channels that can form between alveoli when lung tissue becomes pathological (Demi et al. 2020a, 2020c). By exploiting this concept and the dependence of vertical artifacts on imaging frequency (Demi et al. 2017; Mento and Demi 2020), Demi et al. (2020a, 2020c) proposed a frequency characterization of these artifacts to indirectly estimate the dimensions of acoustic channels (or traps). In particular, a lower native frequency should indicate a greater trap size. This theory associates the genesis of vertical artifacts with specific resonance phenomena, as also reported in recent in vitro studies (Mento and Demi 2020, 2021).
The dependence of vertical artifacts on different imaging parameters (center frequency, bandwidth, focal point position and the beam's angle of incidence) was further investigated in vitro, both qualitatively (Kameda et al. 2019) and quantitatively (Mento and Demi 2020, 2021). These studies recommended positioning the focal point at the pleural line depth, as the structures that can be analyzed are indeed those along the lung surface (Kameda et al. 2019; Mento and Demi 2020, 2021). The findings of these in vitro studies are relevant for the development of quantitative LUS clinical approaches as they quantify the impact of potentially confounding factors. Moreover, the results described in these studies highlighted how a visual interpretation of these artifacts leads to subjective and qualitative analysis, as their appearance strongly depends on several imaging parameters that are seldom considered in the design of clinical studies. As a consequence, the technique often used in clinical practice, that is, the count of vertical artifacts in the image, should be considered qualitative and poorly reproducible.
In contrast, the main advantage of quantitative techniques consists of providing a physical measure able to estimate the state of the lung surface, avoiding a subjective evaluation based only on visual interpretation of LUS artifacts. However, even though different quantitative techniques have been proposed since 2017 (Demi et al. 2017; Mohanty et al. 2017; Zhang et al. 2017), they have not been fully validated in large clinical trials, and specific limitations exist for each technique. The technique proposed by Mohanty et al. (2017) showed promise in differentiating healthy participants from patients with lung diseases, but represents a practical limitation related to the impact of the intercostal layer thickness. It has so far been tested only with the probe directly in contact with the lung. Moreover, it was applied only to rat lungs, which have different characteristics compared with humans (Mohanty et al. 2017). On the other hand, Zhang et al. tested LUSWE in human studies and obtained encouraging results in terms of differentiation between healthy participants and patients affected by different diseases (Zhang et al. 2017, Zhang et al., 2019a, Zhang et al., 2019b; Wiley et al. 2021). However, as suggested by an in vitro study (Zhou and Zhang 2018b), the positioning of the shaker could lead to strong variability in the findings and, thus, to less reliable results. The multifrequency technique proposed by Demi et al. (2017) yielded promising results in differentiating patients with PF from patients with different lung diseases (Mento et al. 2020). Nevertheless, only results on a relatively small cohort of patients have been obtained so far.
At present, semi-quantitative techniques still represent the main available strategy to exploit LUS to assess the state of the lung surface. Nevertheless, the use of these techniques should be guided by proper definitions and standardization of acquisition protocols. Specifically, given the dependence of vertical artifacts on imaging parameters (Kameda et al. 2019; Mento and Demi, 2020, Mento and Demi, 2021), standardization of the imaging protocols represents a key methodology to reduce confounding factors. Unfortunately, many protocols are heterogeneously defined, often lacking details on the imaging settings (Dargent et al. 2020; Demi 2020; Zhao et al. 2020). Also the scoring systems, as well as the amount and location of the scanning areas, are often arbitrarily defined (Allinovi et al. 2020). Differently, to develop and validate proper imaging protocols and scoring systems to be used depending on the disease being investigated, it is of fundamental importance to compare the performance of different protocols following an evidence-based approach (Mento et al. 2021b; Demi et al. 2022). As an example, during the COVID-19 pandemic, an LUS standardized imaging protocol was proposed (Soldati et al. 2020d), tested (Mento et al. 2021b; Smargiassi et al. 2020b; Demi et al. 2022), clinically validated (Perrone et al. 2021) and adopted (Bonadia et al. 2020).
An additional problem related to semi-quantitative approaches is the subjectivity of the analysis. Indeed, even though scoring systems are used, they are based only on the coding of visually interpreted LUS patterns into scores. Hence, the operator dependence is impossible to overcome. To this end, the use of AI to automatically score LUS data could be instrumental in reducing subjectivity in the evaluation of LUS patterns. Nevertheless, given the strong subjectivity of the task (Lerchbaumer et al. 2021), it is not reliable to expect levels of agreement between AI and human operators at the video or frame level around 90%–100% (Mento et al. 2021a; Demi et al. 2022). Indeed, the use of AI algorithms could lead to more reproducible analyses but cannot completely avoid subjectivity, as AI training remains based on the subjective labeling performed by clinicians.
Another problem that emerged during the COVID-19 pandemic was the extensive development of AI solutions regardless of domain knowledge on LUS. As an example, AI systems have been proposed for the diagnosis of COVID-19 based on the simple evaluation of LUS videos or frames (Horry et al. 2020; Awasthi et al. 2021; Diaz-Escobar et al. 2021). This application of AI is fallacious as it is not possible to diagnose COVID-19 based on LUS. Indeed, positive patients may still not present any alterations along the lung surface. Moreover, LUS imaging patterns are strongly unspecific (Demi 2020). As an example, different LUS artifacts (e.g., vertical artifacts and “white lung") or anatomical findings (e.g., consolidations) that were observed in patients with COVID-19 could be observed in many other diseases (e.g., CPE, PF and ARDS).
Conclusions
Quantitative approaches represent the future of LUS, as they could provide a physical metric able to characterize the lung surface by applying an acquisition technique specifically designed for the lung. Nevertheless, to develop these techniques, the genesis of vertical artifacts needs to be more deeply investigated and understood by means of controlled in vitro and in silico studies. In the meantime, semi-quantitative approaches based on image analysis techniques should be exploited to estimate the state of lungs by detecting and recognizing specific LUS patterns that do signal different levels of aeration. However, to reduce the impact of confounding factors, standardization of the imaging protocols and scoring systems is essential. In this context, AI algorithms could be used to guide the acquisition and analysis of LUS data.
Finally, the potential risks associated with the application of unnecessary high-pressure fields to lung examinations should be carefully investigated. Indeed, the presence of air within lungs seems to suggest the use of a MI significantly below the limit of 1.9 imposed for standard ultrasound imaging in soft tissues. This is moreover not conflicting with the possibility to acquire high-quality LUS data. Indeed, the pleural line lies only a few centimeters from the skin, and lower MIs are thus sufficient for imaging. In particular, it is suggested that the MI be set below 0.7 for LUS applications (Soldati et al. 2020d).
Declaration of Competing Interest
All authors declare no conflicts of interest.
References
- Allinovi M, Parise A, Giacalone M, Amerio A, Delsante M, Odone A, Franci A, Gigliotti F, Amadasi S, Delmonte D, Parri N, Mangia A. Lung ultrasound may support diagnosis and monitoring of COVID-19 pneumonia. Ultrasound Med Biol. 2020;46:2908–2917. doi: 10.1016/j.ultrasmedbio.2020.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida A, Bilbao A, Ruby L, Rominger MB, Lopez-De-Ipina D, Dahl J, Elkaffas A, Sanabria SJ. Lung ultrasound for point-of-care COVID-19 pneumonia stratification: Computer-aided diagnostics in a smartphone. First experiences classifying semiology from public datasets. Proc IEEE Int Ultrason Symp. 2020:1–4. [Google Scholar]
- Anantrasirichai N, Hayes W, Allinovi M, Bull D, Achim A. Line detection as an inverse problem: Application to lung ultrasound imaging. IEEE Trans Medical Imaging. 2017;36:2045–2056. doi: 10.1109/TMI.2017.2715880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avruch L, Cooperberg PL. The ring-down artifact. J Ultrasound Med. 1985;4:21–28. doi: 10.7863/jum.1985.4.1.21. [DOI] [PubMed] [Google Scholar]
- Awasthi N, Dayal A, Cenkeramaddi LR, Yalavarthy PK. Mini-COVIDNet: Efficient lightweight deep neural network for ultrasound based point-of-care detection of COVID-19. IEEE Trans Ultrason Ferroelectr Freq Control. 2021;68:2023–2037. doi: 10.1109/TUFFC.2021.3068190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloescu C, Toporek G, Kim S, McNamara K, Liu R, Shaw MM, McNamara RL, Raju BI, Moore CL. Automated lung ultrasound B-line assessment using a deep learning algorithm. IEEE Trans Ultrason Ferroelectr Freq Control. 2020;67:2312–2320. doi: 10.1109/TUFFC.2020.3002249. [DOI] [PubMed] [Google Scholar]
- Bauld TJ, Schwan HP. Attenuation and reflection of ultrasound in canine lung tissue. J Acoust Soc Am. 1974;56:1630–1637. doi: 10.1121/1.1903488. [DOI] [PubMed] [Google Scholar]
- Bonadia N, Carnicelli A, Piano A, Buonsenso D, Gilardi E, Kadhim C, Torelli E, Petrucci M, Di Maurizio L, Biasucci DG, Fuorlo M, Forte E, Zaccaria R, Franceschi F. Lung ultrasound findings are associated with mortality and need for intensive care admission in COVID-19 patients evaluated in the emergency department. Ultrasound Med Biol. 2020;46:2927–2937. doi: 10.1016/j.ultrasmedbio.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusasco C, Santori G, Tavazzi G, Via G, Robba C, Gargani L, Mojoli F, Mongodi S, Bruzzo E, Tro R, Boccacci P, Isirdi A, Forfori F, Corradi F. UCARE (Ultrasound in Critical Care and Anesthesia Research Group). Second-order grey-scale texture analysis of pleural ultrasound images to differentiate acute respiratory distress syndrome and cardiogenic pulmonary edema. J Clin Monit Comput. 2022;36:131–140. doi: 10.1007/s10877-020-00629-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrer L, Donini E, Marinelli D, Zanetti M, Mento F, Torri E, Smargiassi A, Inchingolo R, Soldati G, Demi L, Bovolo F, Bruzzone L. Automatic pleural line extraction and COVID-19 scoring from lung ultrasound data. IEEE Trans Ultrason Ferroelectr Freq Control. 2020;67:2207–2217. doi: 10.1109/TUFFC.2020.3005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, He C, Yin J, Li J, Duan X, Cao Y, Sun L, Hu M, Li W, Li Q. Quantitative analysis and automated lung ultrasound scoring for evaluating COVID-19 pneumonia with neural networks. IEEE Trans Ultrason Ferroelectr Freq Control. 2021;68:2507–2515. doi: 10.1109/TUFFC.2021.3070696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child SZ, Hartman CL, Schery LA, Carstensen EL. Lung damage from exposure to pulsed ultrasound. Ultrasound Med Biol. 1990;16:817–825. doi: 10.1016/0301-5629(90)90046-f. [DOI] [PubMed] [Google Scholar]
- Contreras-Ojeda SL, Dominguez-Jimenez JA, Contreras-Ortiz SH. Proc SPIE 11583, 16th International Symposium on Medical Information Processing and Analysis, 115830F (3 November 2020) The International Society for Optical Engineering; 2020. Analysis and classification of lung and muscular tissues in ultrasound images using 2D wavelet transform and machine learning. [Google Scholar]
- Copetti R, Soldati G, Copetti P. Chest sonography: A useful tool to differentiate acute cardiogenic pulmonary edema from acute respiratory distress syndrome. Cardiovasc Ultrasound. 2008;6:16. doi: 10.1186/1476-7120-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dargent A, Chatelain E, Kreitmann L, Quenot JP, Cour M, Argaud L, Antoine M, Baudry T, Bertrand PJ, Bougnaud J, Charbonnieras F, Damien M, Delsol C, Gimet P, Moutou A, Nourry E, Pinede A, Lougnon Z, Roccia H, Romain M, Simon M, Srage K, Venet M. Lung ultrasound score to monitor COVID-19 pneumonia progression in patients with ARDS. PLoS One. 2020;15 doi: 10.1371/journal.pone.0236312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demi L. Lung ultrasound: The future ahead and the lessons learned from COVID-19. J Acoust Soc Am. 2020;148:2146–2150. doi: 10.1121/10.0002183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demi M. The impact of multiple concurrent factors on the length of the ultrasound pulmonary vertical artifacts as illustrated through the experimental and numerical analysis of simple models. J Acoust Soc Am. 2021;150:2106–2115. doi: 10.1121/10.0006413. [DOI] [PubMed] [Google Scholar]
- Demi L, Demi M, Smargiassi A, Inchingolo R, Faita F, Soldati G. Ultrasonography in lung pathologies: New perspectives. Multidiscip Respir Med. 2014;9:27. doi: 10.1186/2049-6958-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demi L, van Hoeve W, van Sloun RJG, Soldati G, Demi M. Determination of a potential quantitative measure of the state of the lung using lung ultrasound spectroscopy. Sci Rep. 2017;7:12746. doi: 10.1038/s41598-017-13078-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demi L, Demi M, Prediletto R, Soldati G. Real-time multi-frequency ultrasound imaging for quantitative lung ultrasound first clinical results. J Acoust Soc Am. 2020;148:998–1006. doi: 10.1121/10.0001723. [DOI] [PubMed] [Google Scholar]
- Demi L, Egan T, Muller M. Lung ultrasound imaging, a technical review. Appl Sci. 2020;10:462. [Google Scholar]
- Demi M, Prediletto R, Soldati G, Demi L. Physical mechanisms providing clinical information from ultrasound lung images: Hypotheses and early confirmations. IEEE Trans Ultrason Ferroelectr Freq Control. 2020;67:612–623. doi: 10.1109/TUFFC.2019.2949597. [DOI] [PubMed] [Google Scholar]
- Demi L, Mento F, Perrone T, Fiengo A, Smargiassi A, Inchingolo R, Soldati G. Agreement between expert sonographers and artificial intelligence in the evaluation of lung ultrasound data acquired from COVID-19 patients. ERJ Open Res. 2021;7:61. [Google Scholar]
- Demi L, Mento F, Di Sabatino A, Fiengo A, Sabatini U, Macioce VN, Robol M, Tursi F, Sofia C, Di Cienzo C, Smargiassi A, Inchingolo R, Perrone T. Lung ultrasound in COVID-19 and post-COVID-19 patients, an evidence-based approach. J Ultrasound Med. 2022;41:2203–2215. doi: 10.1002/jum.15902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Escobar J, Ordonez-Guillen NE, Villarreal-Reyes S, Galaviz-Mosqueda A, Kober V, Rivera-Rodriguez R, Lozano Rizk JE. Deep-learning based detection of COVID-19 using lung ultrasound imagery. PLoS One. 2021;16 doi: 10.1371/journal.pone.0255886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn F. Attenuation and speed of ultrasound in lung. J Acoust Soc Am. 1974;56:1638–1639. doi: 10.1121/1.1903489. [DOI] [PubMed] [Google Scholar]
- Dunn F. Attenuation and speed of ultrasound in lung: Dependence upon frequency and inflation. J Acoust Soc Am. 1986;80:1248–1250. doi: 10.1121/1.393818. [DOI] [PubMed] [Google Scholar]
- Dunn F, Fry WJ. Ultrasonic absorption and reflection by lung tissue. Phys Med Biol. 1961;5:401–410. doi: 10.1088/0031-9155/5/4/302. [DOI] [PubMed] [Google Scholar]
- Erfanian Ebadi S, Krishnaswamy D, Bolouri SES, Zonoobi D, Greiner R, Meuser-Herr N, Jaremko JL, Kapur J, Noga M, Punithakumar K. Automated detection of pneumonia in lung ultrasound using deep video classification for COVID-19. Inform Med Unlocked. 2021;25 doi: 10.1016/j.imu.2021.100687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank O, Schipper N, Vaturi M, Soldati G, Smargiassi A, Inchingolo R, Torri E, Perrone T, Mento F, Demi L, Galun M, Eldar YC, Bagon S. Integrating domain knowledge into deep networks for lung ultrasound with applications to COVID-19. IEEE Trans Med Imaging. 2022;41:571–581. doi: 10.1109/TMI.2021.3117246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frizzell LA, Zachary JF, O'Brien WD., Jr. Effect of pulse polarity and energy on ultrasound-induced lung hemorrhage in adult rats. J Acoust Soc Am. 2003;113:2912–2918. doi: 10.1121/1.1559176. [DOI] [PubMed] [Google Scholar]
- Horry MJ, Chakraborty S, Paul M, Ulhaq A, Pradhan B, Saha M, Shukla N. COVID-19 detection through transfer learning using multimodal imaging data. IEEE Access. 2020;8:149808–149824. doi: 10.1109/ACCESS.2020.3016780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou D, Hou R, Hou J. 2020 11th IEEE Annual Ubiquitous Computing, Electronics and Mobile Communication Conference—UEM-CON 2020. IEEE; New York: 2020. Interpretable Saab Subspace Network for COVID-19 lung ultrasound screening; p. 393. [Google Scholar]
- Jambrik Z, Monti S, Coppola V, Agricola E, Mottola G, Miniati M, Picano E. Usefulness of ultrasound lung comets as a nonradiologic sign of extravascular lung water. Am J Cardiol. 2004;93:1265–1270. doi: 10.1016/j.amjcard.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Jascur M, Bundzel M, Malik M, Dzian A, Ferencik N, Babic F. Detecting the absence of lung sliding in lung ultrasounds using deep learning. Appl Sci. 2021;11:6976. [Google Scholar]
- Joyner CR, Jr, Miller LD, Dudrick SJ, Eskin DJ, Bloom P. Reflected ultrasound in the study of diseases of the chest. Trans Am Clin Climatol Assoc. 1967;78:28–37. [PMC free article] [PubMed] [Google Scholar]
- Kameda T, Kamiyama N, Kobayashi H, Kanayama Y, Taniguchi N. Ultrasonic B-line-like artifacts generated with simple experimental models provide clues to solve key issues in B-lines. Ultrasound Med Biol. 2019;45:1617–1626. doi: 10.1016/j.ultrasmedbio.2019.03.003. [DOI] [PubMed] [Google Scholar]
- Kameda T, Kamiyama N, Taniguchi N. The mechanisms underlying vertical artifacts in lung ultrasound and their proper utilization for the evaluation of cardiogenic pulmonary edema. Diagnostics (Basel) 2022;12:252. doi: 10.3390/diagnostics12020252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakus O, Anantrasirichai N, Aguersif A, Silva S, Basarab A, Achim A. Detection of line artifacts in lung ultrasound images of COVID-19 patients via nonconvex regularization. IEEE Trans Ultrason Ferroelectr Freq Control. 2020;67:2218–2229. doi: 10.1109/TUFFC.2020.3016092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerdegari H, Nhat PTH, McBride A, Razavi R, Hao NV, Thwaites L, Yacoub S, Gomez A. Proceedings, IEEE 18th International Symposium on Biomedical Imaging. Vol. 2021. IEEE; New York: 2021. Automatic detection of B-lines in lung ultrasound videos from severe dengue patients; pp. 989–993. [Google Scholar]
- Khan U, Mento F, Giacomaz LN, Trevisan R, Smargiassi A, Inchingolo R, Perrone T, Demi L. Deep learning-based classification of reduced lung ultrasound data from COVID-19 patients. IEEE Trans Ultrason Ferroelectr Freq Control. 2022;69:1661–1669. doi: 10.1109/TUFFC.2022.3161716. [DOI] [PubMed] [Google Scholar]
- Kulhare S, Zheng X, Mehanian C, Gregory C, Zhu M, Gregory K, Xie H, McAndrew Jones J, Wilson B. Lecture Notes in Computer Science. Springer; Cham: 2018. Ultrasound-based detection of lung abnormalities using single shot detection convolutional neural networks. In: Simulation, Image Processing, and Ultrasound Systems for Assisted Diagnosis and Navigation; pp. 65–73. 11042LNCS. [Google Scholar]
- La Salvia M, Secco G, Torti E, Florimbi G, Guido L, Lago P, Salinaro F, Perlini S, Leporati F. Deep learning and lung ultrasound for Covid-19 pneumonia detection and severity classification. Comput Biol Med. 2021;136 doi: 10.1016/j.compbiomed.2021.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerchbaumer MH, Lauryn JH, Bachmann U, Enghard P, Fischer T, Grune J, Hegemann N, Khadzhynov D, Kruse JM, Lehner LJ, Lindner T, Oezkan T, Zickler D, Kuebler WM, Hamm B, Eckardt KU, Muench F. Point-of-care lung ultrasound in COVID-19 patients: Inter- and intra-observer agreement in a prospective observational study. Sci Rep. 2021;11:10678. doi: 10.1038/s41598-021-90153-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein D, Meziere G, Biderman P, Gepner A, Barre O. The comet-tail artifact: An ultrasound sign of alveolar-interstitial syndrome. Am J Resp Crit Care Med. 1997;156:1640–1646. doi: 10.1164/ajrccm.156.5.96-07096. [DOI] [PubMed] [Google Scholar]
- Lichtenstein DA, Lascols N, Meziere G, Gepner A. Ultrasound diagnosis of alveolar consolidation in the critically ill. Intensive Care Med. 2004;30:276–281. doi: 10.1007/s00134-003-2075-6. [DOI] [PubMed] [Google Scholar]
- Lye TH, Roshankhah R, Karbalaeisadegh Y, Montgomery SA, Egan TM, Muller M, Mamou J. In vivo assessment of pulmonary fibrosis and edema in rodents using the backscatter coefficient and envelope statistics. J Acoust Soc Am. 2021;150:183–192. doi: 10.1121/10.0005481. [DOI] [PubMed] [Google Scholar]
- Mathis G, Dirschmid K. Pulmonary infarction: Sonographic appearance with pathologic correlation. Eur J Radiol. 1993;17:170–174. doi: 10.1016/0720-048x(93)90098-8. [DOI] [PubMed] [Google Scholar]
- Mehanian C, Kulhare S, Millin R, Zheng X, Gregory C, Zhu M, Xie H, Jones J, Lazar J, Halse A, Graham T, Stone M, Gregory K, Wilson B. Lecture Notes Computer Science. Springer; Cham: 2019. Deep learning-based pneumothorax detection in ultrasound videos; pp. 74–82. 11798LNCS. [Google Scholar]
- Mento F, Demi L. On the influence of imaging parameters on lung ultrasound B-line artifacts, in vitro study. J Acoust Soc Am. 2020;148:975–983. doi: 10.1121/10.0001797. [DOI] [PubMed] [Google Scholar]
- Mento F, Demi L. Dependence of lung ultrasound vertical artifacts on frequency, bandwidth, focus and angle of incidence: An in vitro study. J Acoust Soc Am. 2021;150:4075–4082. doi: 10.1121/10.0007482. [DOI] [PubMed] [Google Scholar]
- Mento F, Soldati G, Prediletto R, Demi M, Demi L. Quantitative lung ultrasound spectroscopy applied to the diagnosis of pulmonary fibrosis: The first clinical study. IEEE Trans Ultrason Ferroelectr Freq Control. 2020;67:2265–2273. doi: 10.1109/TUFFC.2020.3012289. [DOI] [PubMed] [Google Scholar]
- Mento F, Perrone T, Fiengo A, Smargiassi A, Inchingolo R, Soldati G, Demi L. Deep learning applied to lung ultrasound videos for scoring COVID-19 patients: A multicenter study. J Acoust Soc Am. 2021;149:3626–3634. doi: 10.1121/10.0004855. [DOI] [PubMed] [Google Scholar]
- Mento F, Perrone T, Macioce VN, Tursi F, Buonsenso D, Torri E, Smargiassi A, Inchingolo R, Soldati G, Demi L. On the impact of different lung ultrasound imaging protocols in the evaluation of patients affected by Coronavirus Disease 2019. J Ultrasound Med. 2021;40:2235–2238. doi: 10.1002/jum.15580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DL. Induction of pulmonary hemorrhage in rats during diagnostic ultrasound. Ultrasound Med Biol. 2012;38:1476–1482. doi: 10.1016/j.ultrasmedbio.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Miller LD, Joyner CR, Jr., Dudrick SJ, Eskin DJ. Clinical use of ultrasound in the early diagnosis of pulmonary embolism. Ann Surg. 1967;166:381–393. doi: 10.1097/00000658-196709000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DL, Dou C, Raghavendran K. Pulmonary capillary hemorrhage induced by fixed-beam pulsed ultrasound. Ultrasound Med Biol. 2015;41:2212–2219. doi: 10.1016/j.ultrasmedbio.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DL, Dong Z, Dou C, Patterson B, Raghavendran K. Pulmonary capillary hemorrhage induced by supersonic shear wave elastography in rats. Ultrasound Med Biol. 2019;45:2993–3004. doi: 10.1016/j.ultrasmedbio.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty K, Blackwell J, Egan T, Muller M. Characterization of the lung parenchyma using ultrasound multiple scattering. Ultrasound Med Biol. 2017;43:993–1003. doi: 10.1016/j.ultrasmedbio.2017.01.011. [DOI] [PubMed] [Google Scholar]
- Mohanty K, Karbalaeisadegh Y, Blackwell J, Ali M, Masuodi B, Egan T, Muller M. In vivo assessment of pulmonary fibrosis and pulmonary edema in rodents using ultrasound multiple scattering. IEEE Trans Ultrason Ferroelectr Freq Control. 2020;67:2274–2280. doi: 10.1109/TUFFC.2020.3023611. [DOI] [PubMed] [Google Scholar]
- Moshavegh R, Hansen KL, Moller-Sorensen H, Nielsen MB, Jensen JA. Automatic detection of B-lines in in vivo lung ultrasound. IEEE Trans Ultrason Ferroelectr Freq Control. 2019;66:309–317. doi: 10.1109/TUFFC.2018.2885955. [DOI] [PubMed] [Google Scholar]
- O'Brien WD, Jr, Zachary JF. Rabbit and pig lung damage comparison from exposure to continuous wave 30-kHz ultrasound. Ultrasound Med Biol. 1996;22:345–353. doi: 10.1016/0301-5629(95)02055-1. [DOI] [PubMed] [Google Scholar]
- O'Brien WD, Frizzell LA, Weigel RM, Zachary JF. Ultrasound-induced lung hemorrhage is not caused by inertial cavitation. J Acoust Soc Am. 2000;108:1290–1297. doi: 10.1121/1.1287706. [DOI] [PubMed] [Google Scholar]
- Oelze ML, Miller RJ, Blue JP, Jr, Zachary JF, O'Brien WD., Jr. Estimation of the acoustic impedance of lung versus level of inflation for different species and ages of animals. J Acoust Soc Am. 2008;124:2340–2352. doi: 10.1121/1.2973186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostras O, Soulioti DE, Pinton G. Diagnostic ultrasound imaging of the lung: A simulation approach based on propagation and reverberation in the human body. J Acoust Soc Am. 2021;150:3904–3913. doi: 10.1121/10.0007273. [DOI] [PubMed] [Google Scholar]
- Pedersen PC, Ozcan HS. Ultrasound properties of lung tissue and their measurements. Ultrasound Med Biol. 1986;12:483–499. doi: 10.1016/0301-5629(86)90220-6. [DOI] [PubMed] [Google Scholar]
- Penney DP, Schenk EA, Maltby K, Hartman-Raeman C, Child SZ, Carstensen EL. Morphological effects of pulsed ultrasound in the lung. Ultrasound Med Biol. 1993;19:127–135. doi: 10.1016/0301-5629(93)90005-9. [DOI] [PubMed] [Google Scholar]
- Perrone T, Soldati G, Padovini L, Fiengo A, Lettieri G, Sabatini U, Gori G, Lepore F, Garolfi M, Palumbo I, Inchingolo R, Smargiassi A, Demi L, Mossolani EE, Tursi F, Klersy C, Di Sabatino A. A new lung ultrasound protocol able to predict worsening in patients affected by severe acute respiratory syndrome coronavirus 2 pneumonia. J Ultrasound Med. 2021;40:1627–1635. doi: 10.1002/jum.15548. [DOI] [PubMed] [Google Scholar]
- Peschiera E, Mento F, Demi L. Numerical study on lung ultrasound B-line formation as a function of imaging frequency and alveolar geometries. J Acoust Soc Am. 2021;149:2304–2311. doi: 10.1121/10.0003930. [DOI] [PubMed] [Google Scholar]
- Picano E, Frassi F, Agricola E, Gligorova S, Gargani L, Mottola G. Ultrasound lung comets: A clinically useful sign of extravascular lung water. J Am Soc Echocardiogr. 2006;19:356–363. doi: 10.1016/j.echo.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Raeman CH, Child SZ, Carstensen EL. Timing of exposures in ultrasonic hemorrhage of murine lung. Ultrasound Med Biol. 1993;19:507–512. doi: 10.1016/0301-5629(93)90126-9. [DOI] [PubMed] [Google Scholar]
- Roshankhah R, Blackwell J, Ali MH, Masuodi B, Egan T, Muller M. Detecting pulmonary nodules by using ultrasound multiple scattering. J Acoust Soc Am. 2021;150:4095–4102. doi: 10.1121/10.0006666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshankhah R, Karbalaeisadegh Y, Hastings G, Mento F, Soldati G, Smargiassi A, Inchingolo R, Torri E, Perrone T, Aylward S, Demi L, Muller M. Investigating training-test data splitting strategies for automated segmentation and scoring of COVID-19 lung ultrasound images. J Acoust Soc Am. 2021;150:4118–4127. doi: 10.1121/10.0007272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Menapace W, Oei S, Luijten B, Fini E, Saltori C, Huijben I, Chennakeshava N, Mento F, Sentelli A, Peschiera E, Trevisan R, Maschietto G, Torri E, Inchingolo R, Smargiassi A, Soldati G, Rota P, Passerini A, Sloun RJGV, Ricci E, Demi L. Deep learning for classification and localization of COVID-19 markers in point-of-care lung ultrasound. IEEE Trans Med Imaging. 2020;39:2676–2687. doi: 10.1109/TMI.2020.2994459. [DOI] [PubMed] [Google Scholar]
- Russell FM, Ehrman RR, Barton A, Sarmiento E, Ottenhoff JE, Nti BK. B-Line quantification: Comparing learners novice to lung ultrasound assisted by machine artificial intelligence technology to expert review. Ultrasound J. 2021;13:33. doi: 10.1186/s13089-021-00234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar KB, Rhyne TL, Myers GS, Lees RS. Characterization of normal and abnormal pulmonary surface by reflected ultrasound. Chest. 1978;74:29–33. doi: 10.1378/chest.74.1.29. [DOI] [PubMed] [Google Scholar]
- Smargiassi A, Inchingolo R, Chiappetta M, Ciavarella LP, Lopatriello S, Corbo GM, Margaritora S, Richeldi L. Agreement between chest ultrasonography and chest X-ray in patients who have undergone thoracic surgery: Preliminary results. Multidiscip Resp Med. 2019;14:9. doi: 10.1186/s40248-019-0171-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smargiassi A, Inchingolo R, Calandriello L, Lombardi F, Calabrese A, Siciliano M, Larici A, Demi L, Richeldi L, Soldati G. Possible role of chest ultrasonography for the evaluation of peripheral fibrotic pulmonary changes in patients affected by idiopathic pulmonary fibrosis—Pilot case series. Appl Sci. 2020;10:1617. [Google Scholar]
- Smargiassi A, Soldati G, Torri E, Mento F, Milardi D, Giacomo PD, De Matteis G, Burzo ML, Larici AR, Pompili M, Demi L, Inchingolo R. Lung ultrasound for COVID-19 patchy pneumonia: Extended or limited evaluations? J Ultrasound Med. 2020;40:521–528. doi: 10.1002/jum.15428. [DOI] [PubMed] [Google Scholar]
- Soldati G, Giunta V, Sher S, Melosi F, Dini C. Synthetic" comets: A new look at lung sonography. Ultrasound Med Biol. 2011;37:1762–1770. doi: 10.1016/j.ultrasmedbio.2011.05.024. [DOI] [PubMed] [Google Scholar]
- Soldati G, Inchingolo R, Smargiassi A, Sher S, Nenna R, Inchingolo CD, Valente S. Ex vivo lung sonography: Morphologic–ultrasound relationship. Ultrasound Med Biol. 2012;38:1169–1179. doi: 10.1016/j.ultrasmedbio.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Soldati G, Smargiassi A, Inchingolo R, Sher S, Nenna R, Valente S, Inchingolo CD, Corbo GM. Lung ultrasonography may provide an indirect estimation of lung porosity and airspace geometry. Respiration. 2014;88:458–468. doi: 10.1159/000368086. [DOI] [PubMed] [Google Scholar]
- Soldati G, Demi M, Inchingolo R, Smargiassi A, Demi L. On the physical basis of pulmonary sonographic interstitial syndrome. J Ultrasound Med. 2016;35:2075–2086. doi: 10.7863/ultra.15.08023. [DOI] [PubMed] [Google Scholar]
- Soldati G, Smargiassi A, Mariani AA, Inchingolo R. Novel aspects in diagnostic approach to respiratory patients: Is it the time for a new semiotics? Multidiscip Resp Med. 2017;12:15. doi: 10.1186/s40248-017-0098-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati G, Demi M, Smargiassi A, Inchingolo R, Demi L. The role of ultrasound lung artifacts in the diagnosis of respiratory diseases. Expert Rev Resp Med. 2019;13:163–172. doi: 10.1080/17476348.2019.1565997. [DOI] [PubMed] [Google Scholar]
- Soldati G, Giannasi G, Smargiassi A, Inchingolo R, Demi L. Contrast-enhanced ultrasound in patients with COVID-19. J Ultrasound Med. 2020;39:2483–2489. doi: 10.1002/jum.15338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati G, Smargiassi A, Demi L, Inchingolo R. Artifactual lung ultrasonography: It is a matter of traps, order, and disorder. Appl Sci. 2020;10:1570. [Google Scholar]
- Soldati G, Smargiassi A, Inchingolo R, Buonsenso D, Perrone T, Briganti DF, Perlini S, Torri E, Mariani A, Mossolani EE, Tursi F, Mento F, Demi L. Is there a role for lung ultrasound during the COVID-19 pandemic? J Ultrasound Med. 2020;39:1459–1462. doi: 10.1002/jum.15284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati G, Smargiassi A, Inchingolo R, Buonsenso D, Perrone T, Briganti DF, Perlini S, Torri E, Mariani A, Mossolani EE, Tursi F, Mento F, Demi L. Proposal for international standardization of the use of lung ultrasound for patients with COVID-19. J Ultrasound Med. 2020;39:1413–1419. doi: 10.1002/jum.15285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati G, Smargiassi A, Inchingolo R, Buonsenso D, Perrone T, Briganti DF, Perlini S, Torri E, Mariani A, Mossolani EE, Tursi F, Mento F, Demi L. Time for a new international evidence-based recommendations for point-of-care lung ultrasound. J Ultrasound Med. 2021;40:433–434. doi: 10.1002/jum.15412. [DOI] [PubMed] [Google Scholar]
- Soldati G, Smargiassi A, Perrone T, Torri E, Mento F, Demi L, Inchingolo R. There is a validated acquisition protocol for lung ultrasonography in COVID-19 pneumonia. J Ultrasound Med. 2021;40:2783. doi: 10.1002/jum.15649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati G, Smargiassi A, Perrone T, Torri E, Mento F, Demi L, Inchingolo R. LUS for COVID-19 pneumonia: Flexible or reproducible approach? J Ultrasound Med. 2022;41:525–526. doi: 10.1002/jum.15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan LR, Sehgal CM. A review of early experience in lung ultrasound in the diagnosis and management of COVID-19. Ultrasound Med Biol. 2020;46:2530–2545. doi: 10.1016/j.ultrasmedbio.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thickman DI, Ziskin MC, Jacobs Goldenberg N, Linder BE. Clinical manifestations of the comet tail artifact. J Ultrasound Med. 1983;2:225–230. doi: 10.7863/jum.1983.2.5.225. [DOI] [PubMed] [Google Scholar]
- Towa RT, Miller RJ, Frizzell LA, Zachary JF, O'Brien WD., Jr. Attenuation coefficient and propagation speed estimates of rat and pig intercostal tissue as a function of temperature. IEEE Trans Ultrason Ferroelectr Freq Control. 2002;49:1411–1420. doi: 10.1109/tuffc.2002.1041082. [DOI] [PubMed] [Google Scholar]
- Treeby BE, Cox BT. k-Wave: MATLAB toolbox for the simulation and reconstruction of photoacoustic wave fields. J Biomed Opt. 2010;15:1–12. doi: 10.1117/1.3360308. [DOI] [PubMed] [Google Scholar]
- Tsai CH, van der Burgt J, Vukovic D, Kaur N, Demi L, Canty D, Wang A, Royse A, Royse C, Haji K, Dowling J, Chetty G, Fontanarosa D. Automatic deep learning-based pleural effusion classification in lung ultrasound images for respiratory pathology diagnosis. Phys Med. 2021;83:38–45. doi: 10.1016/j.ejmp.2021.02.023. [DOI] [PubMed] [Google Scholar]
- Van Sloun RJG, Demi L. Localizing B-lines in lung ultrasonography by weakly supervised deep learning, in-vivo results. IEEE J Biomed Health Inform. 2020;24:957–964. doi: 10.1109/JBHI.2019.2936151. [DOI] [PubMed] [Google Scholar]
- Volpicelli G, Elbarbary M, Blaivas M, Lichtenstein DA, Mathis G, Kirkpatrick AW, Melniker L, Gargani L, Noble VE, Via G, Dean A, Tsung JW, Soldati G, Copetti R, Bouhemad B, Reissig A, Agricola E, Rouby JJ, Arbelot C, Liteplo A, Sargsyan A, Silva F, Hoppmann R, Breitkreutz R, Seibel A, Neri L, Storti E, Petrovic T. International evidence-based recommendations for point-of-care lung ultrasound. Intensive Care Med. 2012;38:577–591. doi: 10.1007/s00134-012-2513-4. [DOI] [PubMed] [Google Scholar]
- Wang X, Burzynski JS, Hamilton J, Rao PS, Weitzel WF, Bull JL. Quantifying lung ultrasound comets with a convolutional neural network: Initial clinical results. Comput Biol Med. 2019;107:39–46. doi: 10.1016/j.compbiomed.2019.02.002. [DOI] [PubMed] [Google Scholar]
- Wiley BM, Zhou B, Pandompatam G, Zhou J, Kucuk HO, Zhang X. Lung ultrasound surface wave elastography for assessing patients with pulmonary edema. IEEE Trans Biomed Eng. 2021;68:3417–3423. doi: 10.1109/TBME.2021.3072891. [DOI] [PubMed] [Google Scholar]
- Wolfram F, Braun C, Gutsche H, Lesser TG. In vivo assessment of lung ultrasound features mimicking viral pneumonia using a large animal model. IEEE Trans Ultrason Ferroelectr Freq Control. 2020;67:2258–2264. doi: 10.1109/TUFFC.2020.3010299. [DOI] [PubMed] [Google Scholar]
- Xue W, Cao C, Liu J, Duan Y, Cao H, Wang J, Tao X, Chen Z, Wu M, Zhang J, Sun H, Jin Y, Yang X, Huang R, Xiang F, Song Y, You M, Zhang W, Jiang L, Zhang Z, Kong S, Tian Y, Zhang L, Ni D, Xie M. Modality alignment contrastive learning for severity assessment of COVID-19 from lung ultrasound and clinical information. Med Image Anal. 2021;69 doi: 10.1016/j.media.2021.101975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachary JF, O'Brien WD., Jr. Lung lesions induced by continuous- and pulsed-wave (diagnostic) ultrasound in mice, rabbits, and pigs. Vet Pathol. 1995;32:43–54. doi: 10.1177/030098589503200106. [DOI] [PubMed] [Google Scholar]
- Zachary JF, Sempsrott JM, Frizzell LA, Simpson DG, O'Brien WD., Jr. Superthreshold behavior and threshold estimation of ultrasound-induced lung hemorrhage in adult mice and rats. IEEE Trans Ultrason Ferroelectr Freq Control. 2001;48:581–592. doi: 10.1109/58.911741. [DOI] [PubMed] [Google Scholar]
- Zhang X, Osborn T, Zhou B, Meixner D, Kinnick RR, Bartholmai B, Greenleaf JF, Kalra S. Lung Ultrasound Surface Wave Elastography: A Pilot Clinical Study. IEEE Trans Ultrason Ferroelectr Freq Control. 2017;64:1298–1304. doi: 10.1109/TUFFC.2017.2707981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhou B, Bartholmai B, Kalra S, Osborn T. A quantitative method for measuring the changes of lung surface wave speed for assessing disease progression of interstitial lung disease. Ultrasound Med Biol. 2019;45:741–748. doi: 10.1016/j.ultrasmedbio.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhou B, Osborn T, Bartholmai B, Kalra S. Lung ultrasound surface wave elastography for assessing interstitial lung disease. IEEE Trans Biomed Eng. 2019;66:1346–1352. doi: 10.1109/TBME.2018.2872907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Yu K, Zhao Q, Tian R, Xie H, Xie L, Deng P, Xie G, Bao A, Du J. Lung Ultrasound score in evaluating the severity of coronavirus disease 2019 (COVID-19) pneumonia. Ultrasound Med Biol. 2020;46:2938–2944. doi: 10.1016/j.ultrasmedbio.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Zhang X. Lung mass density analysis using deep neural network and lung ultrasound surface wave elastography. Ultrasonics. 2018;89:173–177. doi: 10.1016/j.ultras.2018.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Zhang X. A Lung phantom model to study pulmonary edema using lung ultrasound surface wave elastography. Ultrasound Med Biol. 2018;44:2400–2405. doi: 10.1016/j.ultrasmedbio.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Bartholmai BJ, Kalra S, Zhang X. Predicting lung mass density of patients with interstitial lung disease and healthy subjects using deep neural network and lung ultrasound surface wave elastography. J Mech Behav Biomed Mater. 2020;104 doi: 10.1016/j.jmbbm.2020.103682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Bartholmai BJ, Kalra S, Osborn T, Zhang X. Lung mass density prediction using machine learning based on ultrasound surface wave elastography and pulmonary function testing. J Acoust Soc Am. 2021;149:1318–1323. doi: 10.1121/10.0003575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziskin MC, Thickman DI, Goldenberg NJ, Lapayowker MS, Becker JM. The comet tail artifact. J Ultrasound Med. 1982;1:1–7. doi: 10.7863/jum.1982.1.1.1. [DOI] [PubMed] [Google Scholar]