Abstract
Studies in genetically ‘identical’ individuals indicate that as much as 50% of complex trait variation cannot be traced to genetics or to the environment. The mechanisms that generate this ‘unexplained’ phenotypic variation (UPV) remain largely unknown. Here, we identify neuronatin (NNAT) as a conserved factor that buffers against UPV. We find that Nnat deficiency in isogenic mice triggers the emergence of a bi-stable polyphenism, where littermates emerge into adulthood either ‘normal’ or ‘overgrown’. Mechanistically, this is mediated by an insulin-dependent overgrowth that arises from histone deacetylase (HDAC)-dependent β-cell hyperproliferation. A multi-dimensional analysis of monozygotic twin discordance reveals the existence of two patterns of human UPV, one of which (Type B) phenocopies the NNAT-buffered polyphenism identified in mice. Specifically, Type-B monozygotic co-twins exhibit coordinated increases in fat and lean mass across the body; decreased NNAT expression; increased HDAC-responsive gene signatures; and clinical outcomes linked to insulinemia. Critically, the Type-B UPV signature stratifies both childhood and adult cohorts into four metabolic states, including two phenotypically and molecularly distinct types of obesity.
Subject terms: Epigenetics, Obesity, Genetics research
Yang et al. show that neuronatin (NNAT) can explain part of the phenotypic variation of complex traits, independently of genetics or the environment. Such NNAT-dependent variations can stratify human cohorts into four metabolic sub-types, including two distinct types of obesity.
Main
Biomedical research is driven by a 100-year-old dogma that phenotype results from the additive effects of genes and environment1,2. Since the 1920s, a persistent and compelling body of evidence has argued for the existence of an additional dimension of phenotypic variation not explained by genetics or the environment3. Klaus Gärtner’s 30-year effort to standardize rodent models, for instance, aptly demonstrated that continuously inbred animals raised under stringent, standardized conditions continue to exhibit a remarkable degree of UPV4. Potential mediators of UPV include residual genetic variation5, mosaic genetic variation, gene–gene and gene–environment interactions (non-additive modifier effects), intergenerational and developmental programming and probabilistic mechanisms such as those underpinning organismal polyphenisms and meta-stable epiallele control6,7. For precision medicine, our limited understanding of UPV represents a massive source of untapped potential: estimates from trait concordance analyses between co-twins8 suggest that UPV is responsible for ~50% of relevant complex trait variation9–15.
Deep literature on epigenetics demonstrates the existence of highly conserved, molecular machinery that stabilize transiently plastic transcriptional units into highly stable ON or OFF transcriptional (phenotypic) outputs between isogenic cells and organisms16,17. Literature on position-effect variegation for instance, highlights the existence of hundreds of such genomic loci whose expression output is transiently probabilistic in early development and ultimately deterministic (ON or OFF) despite originating in the same tissue of the same individual in the same environment, without change in the underlying DNA sequence. These studies indicate that a fraction of UPV is likely not due to random biological ‘noise’. The existence of alternate but distinct phenotypic sub-states, as opposed to random phenotypic noise, carries profound implications for precision medicine. While not typically interpreted in this fashion, the original work that pioneered the discovery of epigenetic silencing mechanisms in yeast and Drosophila, demonstrate a complex regulatory network exists sufficient to underpin organismal UPV18–20, at least as they pertain to single reporter loci. While it is now clear that hormones and chromatin pathways can regulate UPV, we know very little about the molecular machinery that buffers against phenotypic variation and confine developmental/phenotypic outcomes to a specific range for any given gene–environment context. Notably, while conceptually related, the regulation of robustness is thought to be distinct from that of phenotypic plasticity21–23. For instance, plasticity regulators inherently mediate gene–environment interaction; robustness factors prevent phenotypic variation upon environmental perturbations22,23.
One challenge when studying UPV (and phenotypic variability in general) is the large number of experimental animals required to statistically test and validate variance heterogeneity effects (differences in distribution or variance)24,25. Experimental designs must factor in (and rule out) confounds such as paternal, maternal and litter-size effects26–29. Most biomedical experiments are not designed or powered for such analyses. Using proper design and power, we recently demonstrated that Trim28 is a robustness factor in mice; Trim28 buffers against UPV30. This work suggested Nnat and imprinted gene network 1 (IGN1) as potential mediators of Trim28-dependent UPV control.
Nnat is a paternally expressed imprinted gene that encodes for a transmembrane proteolipid of the endoplasmic reticulum (ER). It was first described as a developmentally regulated gene of the embryonic brain31,32, but is also widely expressed and associated with energy homeostasis across tissues33. Nnat expression is necessary for proper glucose-stimulated insulin secretion in differentiated pancreatic β-cells34–36, for adipogenesis and glucose disposal in adipocytes37–39, appetite in the hypothalamus40 and for energy expenditure and food intake41,42. It remains unclear whether any of these functions play a causal role in the emergence of UPV or mammalian polyphenism.
Here, we find that: (1) Nnat insufficiency triggers an overgrowth polyphenism (increased fat and lean mass) distinct and independent of Trim28-buffered UPV43; (2) Nnat- and Trim28-buffering mechanisms are distinct; (3) Nnat-buffered overgrowth is driven by cell-autonomous β-cell hyperplasia and can be abrogated by chemical intervention; and (4) that β-cell hyperplasia depends on HDAC-dependent transcriptional rewiring. Expanding our analysis to humans, we identify at least two different, recurrent patterns of human UPV among monozygotic (MZ) co-twins (Type A and Type B). Of note, Type-B UPV is associated with reduced NNAT gene expression and shares similar molecular and metabolic features with the mouse model. Critically, a Type-B UPV gene expression signature stratifies human populations into distinct molecular/metabolic sub-types and separates two types of obesity. The data reported here therefore identify NNAT as a critical regulator of mammalian UPV.
Results
Nnat buffers an overgrowth polyphenism
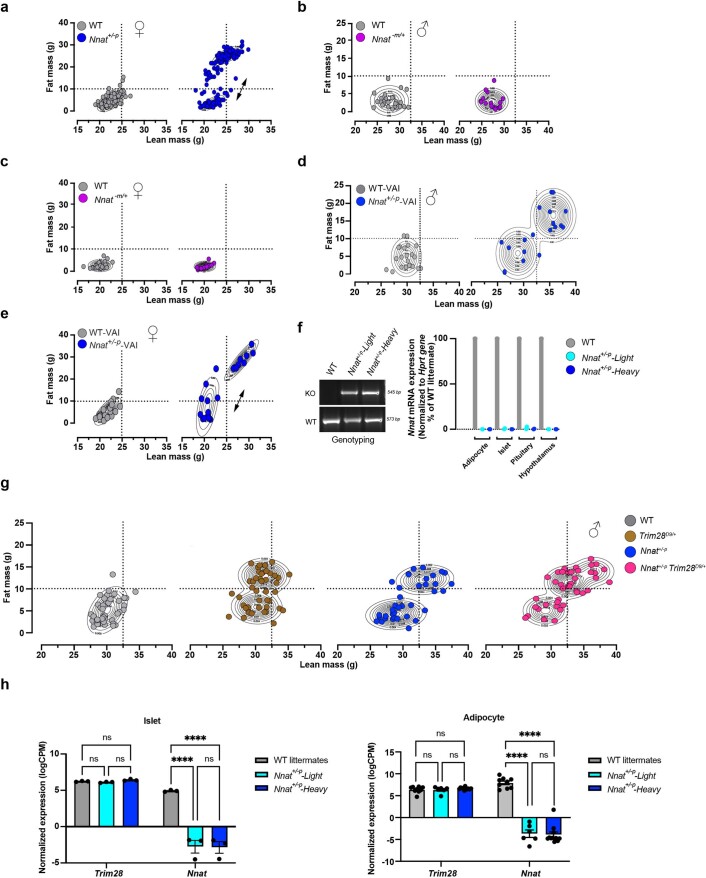
To unequivocally test Nnat’s role as a robustness factor and understand the physiological mechanisms by which it buffers against UPV, we intercrossed highly inbred B6 congenic Nnat knockout males (B6.Nnat+/-p) with wild-type (WT) FVBN/J females, generating large cohorts of Nnat-deficient (Nnat+/-p) and WT littermate matched F1 controls. This breeding scheme maximally restricted inter-individual genetic variation (through isogenicity) while maintaining substantial genome-wide heterozygosity. To minimize litter-size effects (variation attributable to differences in in utero/early-life sufficiency), we used offspring from litters of 9–12 pups and tightly controlled husbandry, environment and housing density. Nnat+/-p mice emerged into adulthood in one of two non-overlapping (bi-stable) phenotypic forms: either unremarkable (Nnat+/-p-Light) or overgrown relative to WT and Nnat+/-p-Light animals (Nnat+/-p-Heavy). Overgrowth was characterized by coordinated increases in fat and lean mass (Fig. 1a,b and Extended Data Fig. 1a), which is distinct from the previously reported Trim28-buffered polyphenism and from other reports of heterogeneity using the Nnat+/-p allele36,41–43. Nnat+/-p-Heavy animals were ~50% heavier than both their WT and Nnat+/-p-Light littermates and had increased white adipose tissue, spleen, pancreas, kidney, liver and heart mass (Fig. 1c). Notably, not all tissues were enlarged. Skeletal muscle, brown adipose tissue and brain masses were unchanged or minimally reduced in Nnat+/-p-Heavy animals (Fig. 1c, right). Skeletal morphometry confirmed a larger skeletal frame in the Nnat+/-p-Heavy morphs (Fig. 1d). Given that the Nnat+/-p-Light and Nnat+/-p-Heavy animals are isogenic, raised in highly standardized environments and reproducibly observed within litters and across multiple independent breeding pairs, these data demonstrate that Nnat acts to buffer against bi-stable overgrowth potential. Thus, Nnat deficiency triggers a polyphenism, characterized by probabilistic overgrowth and obesity.
Fig. 1. Paternal Nnat deletion triggers a bi-stable epigenetic overgrowth in mice.
a, Body composition shown for 16-week-old F1 male progeny from Nnat+/-p × FVBN/J crosses. Contour plots highlighted main clusters identified by Gaussian finite mixture modeling. b, Representative picture presented for Nnat+/p isogenic morphs and WT littermates. c, Organ masses were measured from Nnat+/p colony. Each group had at least eight animals. *Adjusted P ≤ 0.05, as assessed by one-sided Tukey’s multiple comparisons test, comparing Nnat+/p-Heavy and Nnat+/p-Light littermates. Specifically, gonadal white adipose tissue (gWAT) P < 0.0001, subcutaneous white adipose tissue (sWAT) P < 0.0001, spleen P < 0.0001, pancreas P = 0.0019, kidney P = 0.0011, liver P < 0.0001 and heart P < 0.0297. Data are presented as mean ± s.e.m. BAT, brown adipose tissue. d, The lumbar spine (L1–L5) length was measured for the Nnat+/p colony. Each group had at least five animals. In all box-plots, the lower and upper hinges represent 25th and 75th percentiles. The upper/lower whiskers represent largest/smallest observation less/greater than upper/lower hinge+1.5 × interquartile range (IQR). Central median represents 50% quantile. *Adjusted P = 0.015) as assessed by one-sided Tukey’s multiple comparisons test. e, Body composition (fat and lean mass) was measured via EchoMRI for each F1 male progeny at 4, 6, 8, 12 and 16 weeks from B6.Nnat+/-p × FVB.Trim28D9/+ crosses. Developmental trajectories according to the phenotypic groups were plotted from 4 to 16 weeks. Each trajectory had at least four animals. Data are presented as mean ± s.e.m. *P ≤ 0.05 by Student’s t-test.
Extended Data Fig. 1. Paternal but not maternal Nnat deletion causes bi-stable overgrowth.
a, The body composition was shown for 16 weeks old of F1 female wild-type and Nnat+/-p animals from Nnat+/-p x FVBN/J crosses. b-c, The body composition was shown for 16 weeks old F1 male (b) and female (c) wild-type and Nnatm/+ animals from Nnat-m/+ x FVBN/J crosses. d-e, The Nnat+/-p male (d) and female (e) body composition was plotted for the Nnat+/-p colony in the vivarium, VAI, U.S.A. f, Genotyping (DNA) was confirmed in the indicated Nnat+/-p morphs and WT littermate. Nnat mRNA expression was confirmed in metabolic tissues (adipocytes, islet, pituitary gland and hypothalamus) from male Nnat+/-p mice. Nnat mRNA expression was measured from two independent sets of littermate matched animals. The experiments were repeated independently 3 times with similar results. g, Body composition was measured by EchoMRI on 16-week-old wild-type (WT), Trim28D9/+, Nnat+/p and Nnat+/p Trim28D9/+ male progeny from F1 of B6.Nnat+/-p x FVB.Trim28D9/+ crosses. Contour plots highlight main clusters identified by Gaussian finite mixture modeling. h, Trim28 and Nnat (as control) gene expression were shown from islet (left) and adipocyte (right) transcriptomes from Nnat+/-p colony. Each group had at least 3 animals. All data were plotted as mean ± SEM, ns (not significant), **** (adjusted p < 0.0001) by one-sided Tukey’s multiple comparisons test.
We validated these findings in several ways. First, deleting the imprinted maternal Nnat allele, which generates isogenic Nnat-deletion mutants albeit with normal Nnat expression, did not trigger overgrowth in the same line (Nnat-m/+; Extended Data Fig. 1b,c). Second, we tested and observed phenotypic bi-stability in two independent mouse houses and after surviving rederivations independently from cryopreserved embryos and sperm (MPI-IE, Germany; Fig. 1a and Extended Data Fig. 1a; and VAI, USA; Extended Data Fig. 1d,e). Third, we observed bi-stability over dozens of generations, despite exclusively using Nnat+/-p-Light animals as fathers and naive WT C57BL/6J females for continuous backcrossing. Nnat deletion is clear in both Light and Heavy morphs at the DNA and messenger RNA levels (Extended Data Fig. 1f). Thus, Nnat+/-p-triggered overgrowth is robust across distinct environments and in vitro rederivation protocols. To the best of our knowledge, these data represent an unprecedented demonstration of mammalian polyphenism in a genetic context of substantial genome-wide heterozygosity (F1 hybrids as opposed to congenic lines). They rule out genome-wide homozygosity as a precondition for mammalian polyphenism.
One of the key challenges in deciphering mechanisms that regulate UPV is our limited understanding of the fidelity with which UPV effects are manifest across disparate experimental conditions. We therefore used genetic epistasis to test whether Trim28 and Nnat-buffered polyphenisms are simply context-specific forms of the same process. We crossed FVB.Trim28+/D9 (maternal) and B6.Nnat+/-p (paternal) lines to generate B6/FVB F1 hybrid offspring that were either WT at both loci, mutant only for Trim28+/D9, mutant only for Nnat+/-p or mutant for both alleles in the very same genetic background, parental and in utero contexts. Trim28+/D9 offspring (WT for Nnat) showed bi-stable growth trajectories culminating in a bi-stable obesity, whereas solely their Nnat+/-p siblings (WT for Trim28) showed distinct early bifurcating overgrowth trajectories (Fig. 1e). These data indicated the Trim28- and Nnat-induced polyphenisms are at least partially distinct. Notably and in the true test of independence, double-mutant (Nnat+/-p;Trim28+/D9) littermates showed tri-stable phenotypic trajectories (Fig. 1e and Extended Data Fig. 1g), where genetically and context-matched animals exhibited a light (WT-like), obese or overgrown phenotype. Trim28 expression was unchanged among WT, Nnat+/-p-Light and Nnat+/-p-Heavy animals (Extended Data Fig. 1h). The single-mutant analyses also indicate that Nnat-mediated buffering is agnostic to the loss of maternal Trim28; and that Trim28-dependent buffering is agnostic to the loss of paternal Nnat+/−. This data shows unprecedented genetic proof of tri-stable phenotypic potential in mammals and demonstration of independence and additivity of distinct polyphenisms. The data also indicate that the probabilistic ‘obese’ and ‘heavy’ morphs triggered by loss of Trim28 and Nnat (respectively) are distinct and they demonstrate that the mammalian genome has the capacity to canalize three reproducible and discrete developmental trajectories.
Nnat loss triggers bi-stable β-cell hyperplasia
Previous work suggested that Nnat deletion causes a stochastic obesity (B6 background; several groups/vivaria42,43) and partially penetrant, early-life growth restriction that is alleviated in later life through increased food intake (129S2/Sv background; one group41). To capture early growth kinetics of the Nnat+/-p-Light and Nnat+/-p-Heavy animals, we tattooed animals at birth and tracked body composition. Nnat+/-p-Light and Nnat+/-p-Heavy animals exhibited comparable birth weights (Extended Data Fig. 2a), but Nnat+/-p-Heavy morphs initiated the overgrowth developmental trajectory just after 4 weeks of age (Fig. 1e and Extended Data Fig. 2b; top and middle). The phenotypic bifurcation is distinct from that observed in Trim28 mutant mice that bifurcate (toward lean or obese end states) in early adulthood (8–12 weeks; Fig. 1e). To better understand the origins of phenotypic bifurcation, we measured food intake and body composition changes of the Nnat+/-p animals between 4 and 7 weeks of age. Of note, increases in lean and fat mass were measurable several weeks before any detectable hyperphagia, arguing against hyperphagia as a driver of Nnat+/-p-associated overgrowth and adiposity (Extended Data Fig. 2b,c).
Extended Data Fig. 2. Nnat+/-p-Heavy morphs exhibit accelerated post-weaning growth kinetics associated with hyperinsulinemia.
a, Birth weight was measured for Nnat+/-p male newborns. Each group had at least 22 animals. Data are presented as mean ± SEM. b, Fat (top) and lean (middle) mass deposition between 4 to 7 weeks were plotted for male Nnat+/-p colony. Daily food intake (button) was measured and the average daily food intake were calculated per week between 4 to 7 weeks for male Nnat+/-p colony. All data were plotted as mean ± SEM. Adjusted p-values by one-sided Tukey’s multiple comparisons test. Top panel *** p < 0.0001; middle panel * p = 0.016, *** p < 0.0001; lower panel * p = 0.0019. c, Daily food intake normalized to body weight (top) and body weight deposition normalized to average daily food intake per week (button) between 4 to 7 weeks were plotted for male Nnat+/-p colony. All data were plotted as mean ± SEM. Adjusted p-values by one-sided Tukey’s multiple comparisons test. Lower panel * p = 0.0104, ** p = 0.002, *** p < 0.0001. d, Circulating growth factors (insulin-like growth factor 1, IGF1; growth hormone, GH; and insulin, INS) were detected in plasma of Nnat+/p males and WT littermates at 4 and 6 weeks of age. Each group had at least 3 animals. All data were plotted as mean ± SEM. Adjusted p-values by one-sided Tukey’s multiple comparisons test. ** p = 0.0001, *** p < 0.0001.
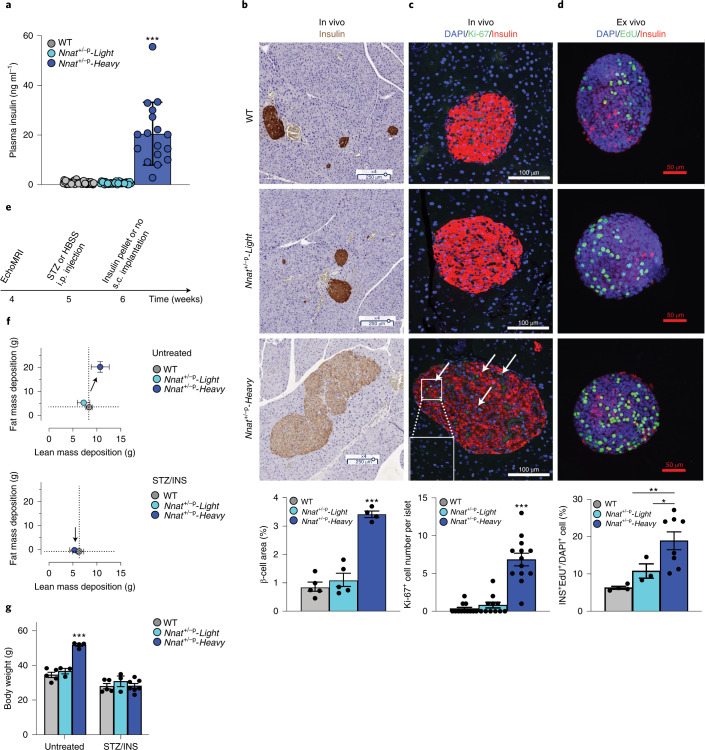
Clinically, overgrowth typically results from hyperactive growth hormone (GH)/insulin-like growth factor (IGF) signaling. We found no increase in GH, IGF1 or IGF2 (undetectable) in Nnat+/-p-Heavy mice, suggesting non-canonical overgrowth (Extended Data Fig. 2d). GH was slightly reduced in Nnat+/-p-Heavy mice at 4 weeks, but not significantly. Notably, plasma levels of insulin (a non-canonical activator of IGFR signaling) showed marked increases in Nnat+/-p-Heavy animals simultaneously with overgrowth bifurcation (Extended Data Fig. 2d), reaching an exceptional ~20-fold normal levels by 16 weeks of age (Fig. 2a).
Fig. 2. Nnat+/-p-overgrowth exhibits autonomous β-cell hyperplasia and hyperinsulinemia.
a, Plasma insulin was measured from 16-week-old male animals fasted for 6 h. Each group had at least 17 animals. ***Adjusted P ≤ 0.001, as assessed by one-sided Tukey’s multiple comparisons test. b, Insulin-positive β-cells (brown) in Nnat+/-p pancreata were detected by immunohistochemistry staining. Scale bar, 250 µm. β-cell area was quantified as percentage of the entire pancreas area. Each group had at least four animals. ***Adjusted P ≤ 0.001, as assessed by one-sided Tukey’s multiple comparisons test. c, In vivo immunofluorescence was performed for proliferating β-cells (white arrows) in primary islets from 16-week-old animals (red, insulin; blue, DAPI; green, Ki-67). Scale bar, 100 µm. Ki-67+ β-cells were quantified and each group had at least 11 islets. ***Adjusted P ≤ 0.001, as assessed by one-sided Tukey’s multiple comparisons test. DAPI, 4,6-diamidino-2-phenylindole. d, Ex vivo immunofluorescence was performed for proliferating β-cells by EdU-incorporation. Size-matched primary islets from 5–6-week-old mice were cultured for 3 days before the EdU-incorporation assay (red, insulin; blue, DAPI; green, EdU). Scale bar, 50 µm. EdU+ proliferating β-cells were quantified and each group had at least three islets. *Two-tailed P ≤ 0.05, **two-tailed P ≤ 0.01, by Welch’s t-test. e, STZ (300 mg kg−1) was administered at 5 weeks of age when the Nnat+/-p-Heavy morphs first show signs of accelerated weight gain. An equal number of subcutaneous (s.c.) insulin implants were administered after 5 days and 1 month after the STZ injection to all STZ groups such that relative euglycemia was maintained. i.p., intraperitoneal. f, Lean and fat mass gained between 4 and 12 weeks of age for untreated and STZ-treated Nnat+/-p littermates. Each group has at least three animals. INS, insulin. g, Body weight at termination highlights how Nnat+/-p-Heavy mice fail to exhibit the overgrowth phenotype on combined STZ/insulin treatment. ***Adjusted P = 0.0001, as assessed by one-sided Tukey’s multiple comparisons test. All data are presented as mean ± s.e.m.
We then analyzed morphology, function and turnover of the pancreatic islet β-cell compartment, the primary source of insulin in the body. Nnat+/-p-Heavy mice showed marked increases in β-cell mass (Fig. 2b and Extended Data Fig. 3a,b) relative to both WT and Nnat+/-p-Light groups. Nnat+/-p-Heavy animals also showed reduced insulin immunoreactivity (Fig. 2b), consistent with previous work showing that β-cell-specific loss of Nnat (in otherwise normal developmental contexts) impairs insulin secretion and storage36. Overall, Nnat+/-p-Heavy animals showed total pancreatic insulin content ~2.5-fold higher than WT and Nnat+/-p-Light groups (Extended Data Fig. 3c). Thus, Nnat deletion leads to distinct programming of β-cell mass in Nnat+/-p-Light and Nnat+/-p-Heavy mice.
Extended Data Fig. 3. Nnat+/-p-Heavy morphs exhibit normal glucose tolerance and islet functionality.
a, H&E staining was performed to locate islets (light pink areas, arrows) in Nnat+/-p pancreata. Scale bar, 500 µm. The experiment was repeated independently 3 times with similar results. b, Islet area was quantified as percent of the entire pancreas area. Each group had 3 thin sections from at least 6 animals. All data were plotted as mean ± SEM, *** (adjusted p < 0.0001) by one-sided Tukey’s multiple comparisons test. c, Total insulin content was extracted from whole pancreata, normalized to total insulin in WT littermates. Each group had at least 5 animals. All data were plotted as mean ± SEM, *** (adjusted p = 0.0008) by one-sided Tukey’s multiple comparisons test. d, Glucagon and somatostatin staining was performed on Nnat+/-p pancreata. Scale bar, 100 µm. The experiment was repeated independently 3 times with similar results. e, Cell death (apoptosis) event was examined via TUNEL assay in the pancreatic section from Nnat+/-p and WT littermates at 16 weeks old. DNase treated sample as positive control from the same animals. The black circle heights the islet area. Each group had at least 3 animals. Scale bar, 100 μm f, Basal insulin secretion was measured from size-matched β-cell spheroids. Each group had 8 spheroids. All data were plotted as mean ± SEM. g, h Glucose-stimulated insulin secretion assays were performed on primary islets (g) and spheroids (h) from 16 weeks old Nnat+/-p and WT littermates. 2.8 mM glu.: 2.8 mM glucose and 16.7 mM glu.: 16.7 mM glucose. At least 6 primary islets and 4 spheroids were in each group. All data were plotted as mean ± SEM. i, Oral glucose tolerance test (OGTT) was performed in 16 weeks old Nnat+/-p and WT mice (n = 4-5) fasted for 6 hours and showed relatively normal glucose tolerance in Nnat+/-p-Heavy morphs. All data were plotted as mean ± SEM. j, Growth trajectories for untreated and STZ-treated Nnat+/-p animals and WT littermates between 4 and 12 weeks of age. All data were plotted as mean ± SEM.
Nnat+/-p-Heavy animals showed increased numbers of Ki-67-positive β-cells in vivo relative to Nnat+/-p-Light animals (Fig. 2c). No differences were observed in islet organization and rates of cell death (TUNEL) (Extended Data Fig. 3d,e). We therefore measured proliferation in islets ex vivo to validate these findings and assess the stability and cell autonomy of the hyperproliferative program. Increased β-cell proliferation was readily measurable in Nnat+/-p-Heavy islets after 3 days of ex vivo equilibration culture and after 2 days of 5-ethynyl-2'-deoxyuridine (EdU) incubation (Fig. 2d). These findings indicated islet-autonomous hyperproliferative programming in Nnat+/-p-Heavy animals. No measurable differences were observed in insulin release in both primary islets and reconstituted islet spheroids, under steady-state or glucose-stimulated conditions (Extended Data Fig. 3f–h). In line with these data, glucose tolerance was largely normal in Nnat+/-p-Heavy animals despite the marked hyperinsulinemia (Extended Data Fig. 3i). These data suggested that the overgrowth polyphenism is driven by an alternate β-cell hyperplasia program.
Nnat+/-p triggered overgrowth is insulin-dependent
To test whether probabilistic Nnat+/-p overgrowth is driven by β-cell hyperplasia and resulting hyperinsulinemia, we artificially ‘clamped’ in vivo insulin levels at equal levels across groups by injecting animals with a single high-dose injection of streptozocin (STZ) to deplete the endogenous β-cell pool44 and implanting slow-release subcutaneous insulin pellets to restore insulin sufficiency equally across animals (Fig. 2e). Treatment was initiated at ~5 weeks of age in longitudinally tracked cohorts. Animals initiating their Nnat+/-p-Heavy trajectory (defined as a 3 g fat mass gain within 5 days) were randomly sorted into treatment or control groups. Notably and where parallel control cohorts of Nnat+/-p-Heavy morphs gained ~15 g of fat and ~3 g of lean mass beyond that of their WT and Nnat+/-p-Light siblings (Fig. 2f,g top; arrow from WT/crosshair), combination therapy completely abrogated this alternate phenotype, yielding lean and fat mass accumulations comparable to WT animals (Fig. 2f (bottom) and Extended Data Fig. 3j). All treated animals completed normal growth trajectories, reaching healthy mature body mass levels of ~30 g (Fig. 2g). Thus, Nnat+/-p-Heavy overgrowth is insulin-dependent.
The alternate β-cell hyperplasia program is HDAC-dependent
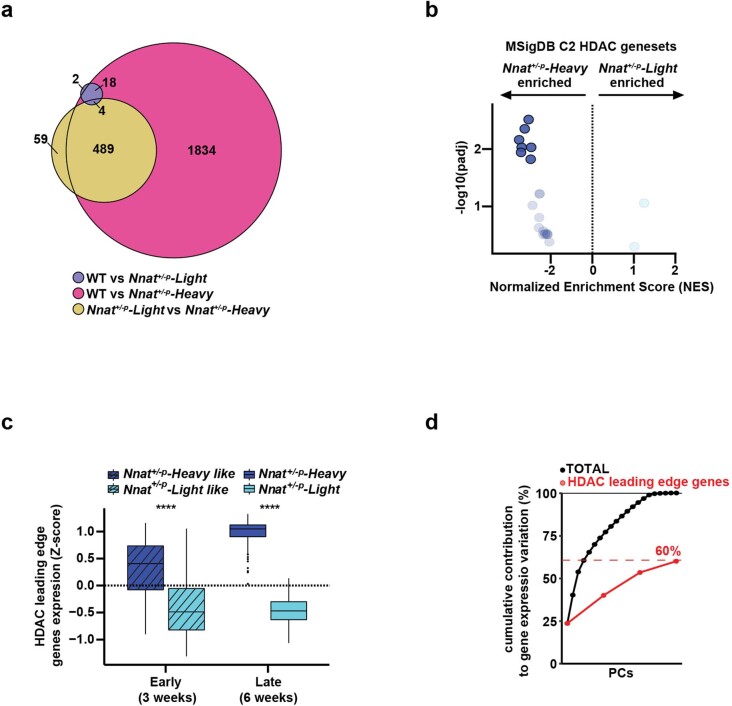
To gain insight into the developmental switch driving β-cell hyperplasia in Nnat+/-p-Heavy morphs, we performed RNA-sequencing (RNA-seq) on Nnat+/-p islets before and after the onset of detectable overgrowth bifurcation (3 and 6 weeks). Transcriptomes from WT and Nnat+/-p-Light islets showed minimal differences (Extended Data Fig. 4a), consistent with the phenotypic similarities between the two genotypes. These data demonstrate that whole-body Nnat deletion, by itself, is not sufficient to drive β-cell dysregulation and imply that Nnat’s primary function is to buffer against phenotypic variation. In contrast, Nnat+/-p-Heavy samples showed major transcriptional rewiring (Fig. 3a,b and Extended Data Fig. 4a). Consistent with the observed hyperplasia, gene set enrichment analysis (GSEA) revealed upregulation of pathways and leading-edge signatures associated with cell cycle (Cdk6, Ccnl2, Myc and Tp53), proliferation (Wnt7a/b, Mapk13, Foxj1, Fos and Smad3) and growth factor signaling (Egr1, Fgfr2 and Epn3) (Fig. 3b,c). Downregulated factors included islet endocrine lineage hormone genes (Gcg and Sst), ER-processing (Pdia3/4, Lman1/2, Rpn1/2, Hsp90b1, Dnajb9 and Ssr4) and protein export pathways (Spcs3, Srps and Sec61b/g) (Fig. 3b,c). Unexpectedly, Nnat+/-p-Heavy islets showed upregulation of a functionally disparate set of HDAC-responsive genes (Fig. 3c and Extended Data Fig. 4b). This result was particularly noteworthy because histone acetylation dynamics have been implicated in regulating insect polyphenisms45,46 and cell proliferation in cancer47.
Extended Data Fig. 4. Dysregulated HDAC-related transcriptome precedes the Nnat+/-p-overgrowth.
a, Venn diagram of differential gene expression analyses of islets transcriptome of 6 weeks old mice comparing between Nnat+/-p-Heavy, Nnat+/-p-Light morphs and WT littermates. b, GSEA results of HDAC-responsive gene sets between the Nnat+/-p-Light and Nnat+/-p-Heavy morphs, showing a specific enrichment in the latter. Solid and transparent colored dots, highlight either statistically significant or not significant enrichments, respectively (adjusted p-value cutoff < 0.05). c, Gene expression (Z-score) comparison was performed for HDAC gene set leading-edge genes between Nnat+/-p-Heavy-like and Nnat+/-p-Light-like morphs (early stage) and Nnat+/-p-Heavy and Nnat+/-p-Light morphs (late stage). **** (p ≤ 0.001) as assessed by two-tails t-tests. In all box-plots, the lower and upper hinges = 25th and 75th percentiles. The upper/lower whiskers = largest/smallest observation less/greater than upper/lower hinge + 1.5 * IQR. Central median = 50% quantile. d, Estimate of the contribution of the HDAC-responsive genes, to the overall transcriptional variability between WT, Nnat+/-p-Heavy and Nnat+/-p-Light mice. Following PCA, the dotplot shows either the cumulative contribution of all principal components (PCs) to gene expression variation (black dots/line), or the contribution of the top four PCs (red dots/line), mostly associated with the HDAC-leading-edge genes from (b). The cumulative contribution of these four PCs is describing 58.7% of total gene expression variation.
Fig. 3. HDAC mediates Nnat+/-p-driven β-cell hyperplasia.
a, Heat map showing the expression of 552 differentially expressed genes (DEGs), defined as Nnat+/-p-Heavy and Nnat+/-p-Light islets transcriptome, in the indicated animals. b, Volcano plot showing DEGs and highlighting the enriched or depleted biologically relevant genes in Nnat+/-p-Heavy morphs. P values as assessed by negative binomial generalized linear model. c, The cytoscape plot of the GSEA showed the enriched or depleted gene sets in Nnat+/-p-Heavy morphs. TFs, transcription factors. d, PCA demonstrated transcriptional similarity of Nnat+/-p-Heavy-like at early stages (3 weeks) and Nnat+/-p-Heavy morphs at late stages (6 weeks) apart from Nnat+/-p-Light-like at early stages (3 weeks) and Nnat+/-p-Light morphs at late stages (6 weeks). e, Heat map shows HDAC gene set leading-edge gene expression in Nnat+/-p-Heavy-like and Nnat+/-p-Light-like morphs (early stage) and Nnat+/-p-Heavy and Nnat+/-p-Light morphs (late stage). f–h, Proliferating β-cells were counted by EdU-incorporation from Nnat+/-p-Heavy and Nnat+/-p-Light morphs and are normalized to WT littermates in untreated (control, f), HDACi-treated (g) and HATi-treated (h) conditions. At least 19 islets were quantified and plotted per condition.
To test whether HDAC regulation was causally linked to the overgrowth polyphenism, we first examined whether the HDAC transcriptional signatures were already present before the phenotypic bifurcation. Notably, the 3-week RNA-seq data revealed that Nnat+/-p islet transcriptomes definitively separate into Nnat+/-p-Light-like or Nnat+/-p-Heavy-like clusters, before phenotypic distinctions are detectable (Fig. 3d). Fully 60% of the variation in the RNA-seq dataset correlated with the same HDAC-responsive gene sets (Fig. 3e and Extended Data Fig. 4c,d), indicating that the HDAC-associated genes are fundamentally responsible for the phenotypic variation in that moment of early life. The data also suggested that dysregulation of HDAC-responsive genes might cause the β-cell hyperproliferation phenotype in Nnat+/-p-Heavy animals. In either case, HDAC-associated transcriptional rewiring precedes the phenotypic bifurcation toward overgrowth.
Second, we cultured islets from the three genotype-phenotype combinations with histone acetylase inhibitors (HATi) or histone deacetylase inhibitors (HDACi) and tracked β-cell proliferation in vitro. Consistent with the data above, Nnat+/-p-Heavy islets showed islet-autonomous β-cell hyperplasia at baseline (Fig. 3f). Notably, HDACi treatment had no observable effect on WT and Nnat+/-p-Light islets, but the treatment was sufficient to abrogate Nnat+/-p-Heavy β-cell hyperplasia and return proliferation back to WT levels (Fig. 3g). These data demonstrate that HDAC-sensitive gene regulation is required for control of β-cell programming and that Nnat’s buffering effect on phenotypic variation is mediated through HDAC-responsive genes. HATi treatment, on the other hand, had no effect on Nnat+/-p-Heavy β-cells (Fig. 3h), indicating a necessary directionality to the regulatory process. Parallel treatment of Nnat+/-p-Light β-cells showed no substantial regulation by either inhibitor (Fig. 3f–h), further highlighting the specificity of the HDAC-dependence. Thus, Nnat buffers against probabilistic phenotypic variation by preventing the activation of an HDAC-dependent β-cell hyperplasia program.
Identification of phenotypic variation patterns in humans
The foregoing data identify a Nnat-buffered axis that regulates probabilistic phenotypic variation. They demonstrate islet-autonomous underpinnings, reproducible epigenome dysregulation and reversibility at in vitro and organismal scales, all of which are unprecedented findings for a mammalian polyphenism. Critically, they demonstrate epistatic independence and additivity with Trim28-buffered phenotypic variation and thus identify two independent pathways for buffering alternate but bi-stable developmental trajectories and phenotypic heterogeneity.
In our previous work43, we found bimodal body mass distributions in large epidemiological cohorts, which raised the possibility that bi-stable UPV might also exist in human populations. That Nnat also buffers against bi-stable UPV again raised the question about regulatory control and polyphenism potential in humans. We therefore analyzed monozygotic (MZ) and dizygotic (DZ) twin data8,48–52 for potential signatures of human UPV. While twin analyses do not rule out genetic and environmental trait variation5,53,54, they substantially reduce these contributions. We performed a high-dimensional analysis of 35 anthropometric traits measured across 153 MZ co-twin pairs from the TwinsUK’s Multiple Tissue Human Expression Resource (MuTHER) cohort55–57. The high-dimensional approach served two purposes. First, the analysis makes no assumptions about how UPV should manifest, but instead hypothesizes that if regulated UPV systems exist in humans, their consequences should be reproducible. Second, searching for patterns of variation (twin discordance), as opposed to single-trait discordances, reduces the impact of measurement errors in any given trait.
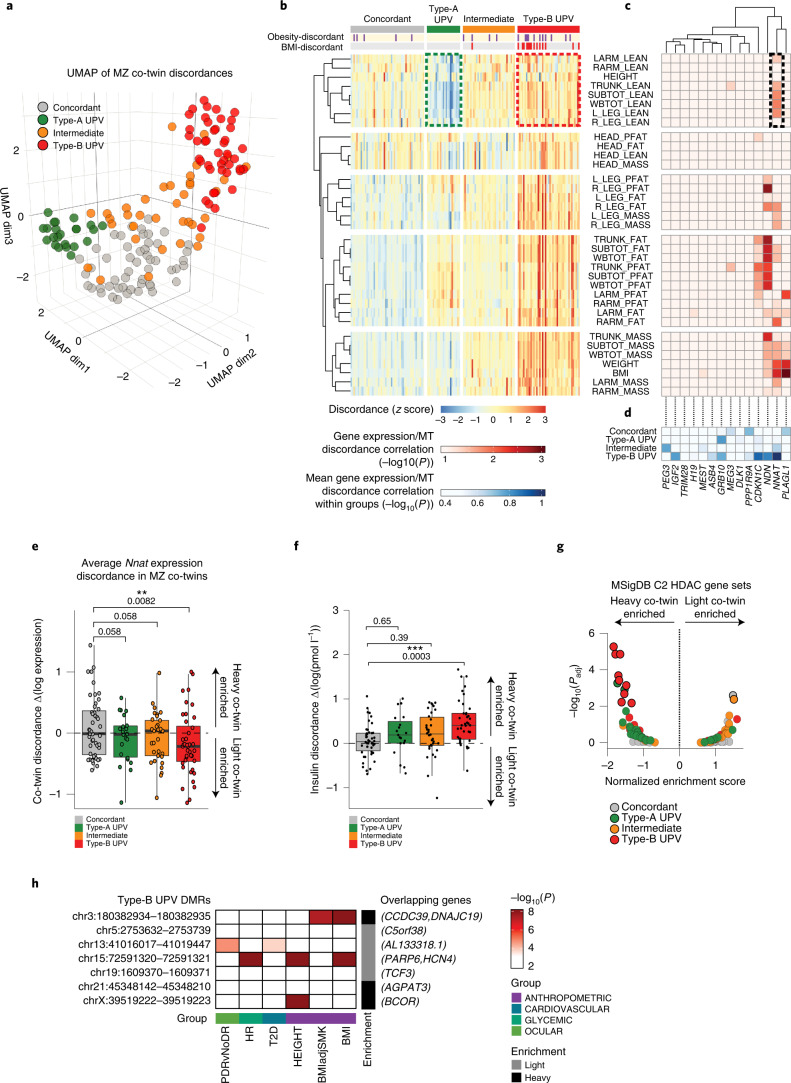
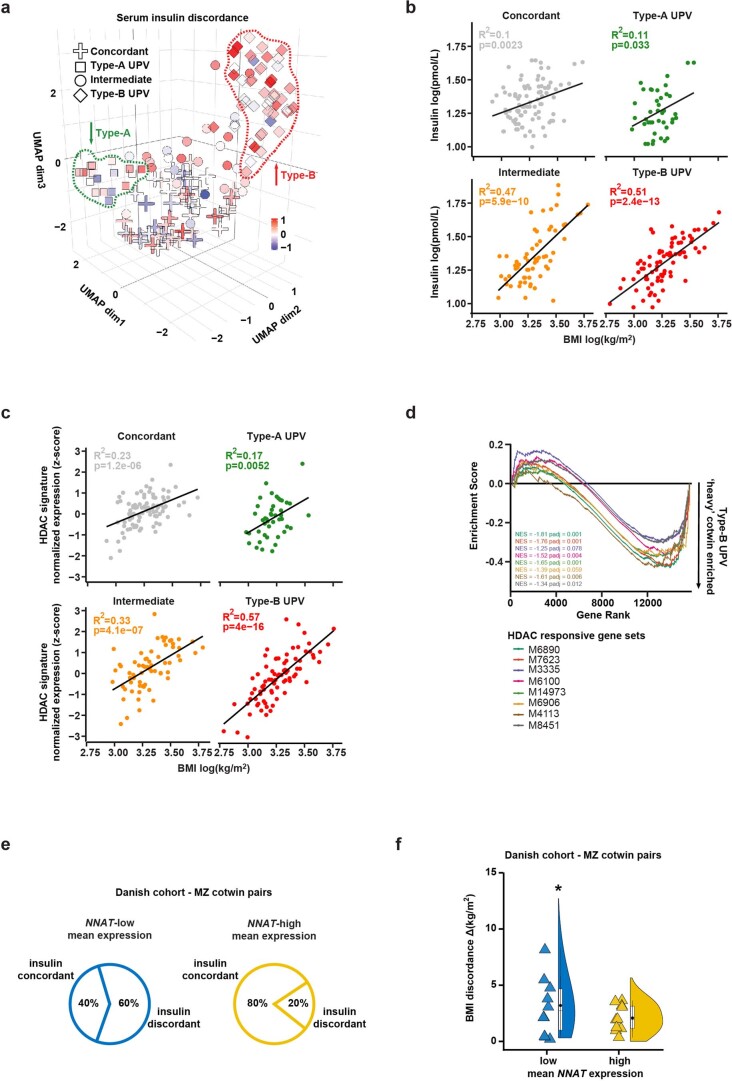
We calculated co-twin trait discordance for each trait, which included weight, height and fat and lean masses of the head, trunk and limbs. Discordance between each trait was calculated for each co-twin pair, setting the co-twin with the lower body mass index (BMI) as the reference. Discordances for all traits were uniformly lower in MZ co-twins than DZ co-twins, as expected (Extended Data Fig. 5a). We then focused on MZ twin pairs and performed a Uniform Manifold and Projection (UMAP) dimensional reduction58 on the 35 trait discordance × 153 co-twin pair matrix (Fig. 4a). Notably, the analysis revealed four clusters or ‘patterns’ of phenotypic variation in the cohort: two were distinctly discordant clusters (Type-A (green) and Type-B (red); Fig. 4a,b) and one was a central concordant cluster (gray). An ‘Intermediate’ UPV cluster (orange) was also identified tending toward Type-B UPV. Thus, we identified four candidate patterns of human UPV.
Extended Data Fig. 5. Characterization of the concordant/discordant MZ co-twin groups.
a, Boxplot showing the inter-quartile ranges (IQR) of co-twin discordance indices for all the available features in either MZ or dizygotic (DZ) co-twins. b, Boxplot showing the MZ co-twin discordance indices for the indicated morphometric features, in the concordant/discordant groups identified. A horizontal dashed line highlights zero values of the discordance index (that is, concordance). Vertical dashed lines separate fat/lean/total mass and percentage of fat from the indicated body parts. c, UMAP projection of MZ co-twin couples from TwinsUK (n = 153), according to 35 morphometric discordances. Each observation represents a twin pair, colored by the whole-body fat (above) or lean (below) mass discordance (calculated as the difference between log-transformed measurements). The different shapes of the co-twin pairs represent the identified groups, as indicated. Dotted red/green lines highlight the Type-A and Type-B UPV and show their differences with respect to lean mass discordance. d, Box-plots representing the average height, between the ‘light’ and ‘heavy’ MZ co-twins, belonging to the indicated groups. Solid horizontal lines represent medians. The p-value is calculated by ANOVA. e, Boxplot showing the MZ co-twin discordance indexes for the indicated morphometric features, after height-normalization, in the concordant/discordant groups identified. A horizontal dashed line highlights the zero value of the discordance index (that is, concordance). Box plots in (b,d,e) show the lower and upper hinges = 25th and 75th percentiles. The upper/lower whiskers = largest/smallest observation less/greater than upper/lower hinge + 1.5 * IQR. Central median = 50% quantile.
Fig. 4. Characterization of human UPV.
a, UMAP projection of MZ co-twin couples, according to 35 morphometric discordances. b, Heat map of hierarchical clustering of morphometric discordances among MZ co-twin couples. Obesity-discordant co-twins indicate that only one co-twin is affected by obesity (BMI > 30). BMI discordant co-twins, BMI difference >5 BMI points. Dashed colored boxes highlight distinct lean mass discordances between Type-A and Type-B UPV. c, Heat map showing the hierarchical clustering of Trim28/IGN1 genes based on the correlation of their expression discordance and indicated phenotypic discordances. A dashed black box highlights NNAT expression discordance correlation with phenotypic discordances of those traits that distinguish Type-A and Type-B UPV. d, Heat map showing the hierarchical clustering of Trim28/IGN1 genes based on the average correlation of their expression discordance and all phenotypic discordances, stratified by four co-twin pairs’ clusters. e, Box-plots representing discordance of NNAT expression, among MZ co-twins, belonging to the indicated clusters. **P = 0.0082, as assessed by one-tailed t-tests. f, Box-plots representing serum insulin discordance, among MZ co-twins, belonging to the indicated groups. ***P = 0.0003 as assessed by one-sided Tukey’s multiple comparisons test, following one-way analysis of variance (ANOVA). In all box-plots, lower and upper hinges indicate 25th and 75th percentiles. The upper/lower whiskers indicate largest/smallest observation less/greater than upper/lower hinge + 1.5 ×IQR. Central median indicates 50% quantile. g, GSEA results of HDAC-responsive gene sets between the ‘light’ and ‘heavy’ co-twins, belonging to the indicated MZ co-twins groups. Solid and transparent colored dots, highlight either statistically significant or not significant enrichments, respectively (adjusted P value cutoff <0.01). h, Heat map showing association of single-nucleotide polymorphisms (SNPs) and indicated phenotypic traits, within the DMRs identified between ‘light’ and ‘heavy’ Type-B UPV co-twins. White boxes indicate no significant associations (P > 1 × 10−3), dark-red boxes indicate genome-wide significant associations (P < 1 × 10−8). Nearest are reported. Gray and black boxes indicate the enrichment of DMR in either the ‘light’ or ‘heavy’ co-twin. BMIadjSMK, BMI adjusted by smoking; T2D, type 2 diabetes; HR, heart rate; PDR, proliferative diabetic retinopathy; PDRvNoDR, PDR versus no PDR.
The concordant cluster was characterized by minimal co-twin trait discordance across all traits, which is what we would normally expect from stereotypical ‘identical twins’ (Fig. 4b and Extended Data Fig. 5b). Type-A UPV was characterized by increased fat masses and a modest reduction of lean masses in heavier co-twins (dashed green box; Fig. 4b and Extended Data Fig. 5b,c). By contrast, Type-B UPV was characterized by increases in both fat and lean masses across body parts in the heavier co-twin (Fig. 4b, dashed red box and Extended Data Fig. 5b,c). These two patterns were distinct and identified an inherently reproducible substructure in ‘non-genetic’ human variation. The findings provided a refined view of twin variation relative to arbitrary BMI and obesity cutoffs59,60 (Fig. 4b; for example ‘BMI’ and ‘obesity’ discordance top annotations). No differences were observed in mean height or height discordance across clusters (Extended Data Fig. 5b,d) and repeat analysis using height-adjusted trait discordances captured the same fundamental UPV patterns (Extended Data Fig. 5e). Notably, we also analyzed genotyping data across the individuals of our four identified UPV groups of MuTHER UK twins57. We found no evidence of consistent genotypic differences between MZ co-twins that extended beyond expected data missingness and that could conceivably underlie the observed patterns (Extended Data Fig. 6a–e). Thus, we identified two ‘non-genetic’ patterns of human phenotypic variation, Type-A UPV, characterized by reciprocal fat and lean mass differences (a relative adiposity) and Type-B UPV, characterized by coordinated fat and lean mass dysregulation (a relative overgrowth).
Extended Data Fig. 6. UPV groups are not determined by genetic differences between MZ cotwins.
a, Violin-plots showing the distribution of missing data (as percentage of the total) among individuals’ genotypes, stratified by UPV groups. This data show that the degree of genome wide missingness is not correlated to UPV groups. b, Table summarizing the amount of genetically identical loci among MZ cotwins and genetic differences, only due to missing data. c, Barplot summarizing the data in table b. MZ cotwins were identical on > 99.9% of the analyzed loci and differences due to missing data account on average for ~ 0.06% of the total data, among all UPV groups. d, Heatmaps showing the distribution of SNPs that resulted different between MZ cotwins, only due to missing data. On the left, the cotwin pairs are ordered by the UPV sub-types. On the right, they are ordered according to hierarchical clustering. These data show that neither the degree of missingness, nor the specific genomic positions of missing data showed any correlation to UPV sub-types. e, Same as in a, on the indicated genesets. These data show no specific enrichment of missingness in any UPV group, nor any genesets, arguing against evidence for genotypic differences underlying the detected transcriptional signatures. All p-values as assessed by ANOVA.
Type-B human UPV parallels Nnat+/-p-Heavy overgrowth
We next explored adipose tissue transcriptomic data from the same co-twin pairs and asked whether NNAT expression was associated with any of the human UPV clusters. Correlative analysis revealed substantial correlations between expression of several IGN1 genes and trait discordances, including NNAT, NDN, CDKN1C and PLAGL1 (Fig. 4c). Of note, NNAT was the only gene whose expression discordance consistently correlated with both fat and lean mass discordances, features that were also specific to Type-B UPV (Fig. 4c; dashed box). Indeed, when data were stratified by UPV sub-type, NNAT expression associated strongly with trait discordance specifically in Type-B UPV (Fig. 4d) with expression decreased in heavier Type-B UPV co-twins (Fig. 4e). These results suggest that NNAT discordance (and downregulation) is a hallmark signature of Type-B human UPV. TRIM28 expression showed no correlation with trait discordances at either the cohort or UPV sub-type levels (Fig. 4c,d). Thus, NNAT gene expression associates with Type-B human UPV.
Given this clear and specific association between NNAT expression and Type-B UPV, we tested whether other key features of the murine overgrowth polyphenism were also altered in human Type-B UPV. For insulinemia, we found that plasma insulin discordances were highest in Type-B co-twin pairs (Fig. 4f and Extended Data Fig. 7a). Indeed, the Type-B UPV group exhibited the clearest correlation between insulinemia and BMI levels (R2 = 0.51; P = 2.4 × 10−13), an association that extended across the BMI spectrum (Extended Data Fig. 7b) and was well above the concordant and Type-A UPV groups (R2 = 0.1 and R2 = 011, respectively). These data indicate that Type-B UPV represents a unique metabolic state where insulinemia and BMI are tightly coupled. Similarly, we found a substantially more robust HDAC-target gene regulation associated with BMI specifically in Type-B UPV (R2 = 0.57; P = 4 × 10−16; Extended Data Fig. 7c). Consistent with the directionality of the Nnat+/-p mouse system, GSEA analysis showed that HDAC-target genes were upregulated in heavier Type-B co-twins (Fig. 4g and Extended Data Fig. 7d). Thus, hyperinsulinemia, HDAC-target gene upregulation and lean and fat mass excesses are species-conserved features of NNAT-associated UPV. From the clinical perspective, these data suggest a model where NNAT buffers against emergence of a distinct phenotypic state (polyphenism) where BMI is exceptionally coupled to insulinemia, lean and fat mass excess.
Extended Data Fig. 7. Type-A and Type-B UPV show specific metabolic and molecular profiles.
a, UMAP projection of MZ cotwin couples form the TwinsUK (n = 153), according to 35 morphometric discordances. Each dot represents a cotwin pair, colored by their serum insulin level discordance (calculated as the difference between log-transformed measurements). The different shapes of the cotwin pairs represented the identified groups. Dotted red/green lines highlight the Type-A and Type-B UPV and show the increase insulin levels (that is, relative hyperinsulinemia) in Type-B ‘heavy’ cotwins. b,c, Scatterplots showing the correlation between BMI and insulin level (b) or the HDAC-signature expression (c), in the indicated cotwin groups. Both showing stronger correlation in the Type-B UPV cotwins (BMI/insulin: R2 = 0.51, p-value=2.4-13, BMI/insulin: R2 = 0.57, p-value=4-16). R2 and p-values as calculated by fitted linear regression models. d, Gene set enrichment analysis (GSEA) results of HDAC-related gene sets between the ‘light’ and ‘heavy’ cotwins, belonging to the Type-B UPV group. Gene sets specifically enriched in ‘heavy’ cotwins of the Type-B UPV cluster are shown. NES = normalized enrichment score; padj = adjusted p-value. e, Pie-charts, showing the distribution of insulin-concordant/discordant MZ cotwin pairs, from the Danish twin cohort, in either low- (blue chart), or high-NNAT expressing couples (yellow chart). The definition of insulin-concordant/discordant couples was obtained by Gaussian finite mixture modeling (see Methods section). f, Box- and violin-plots of BMI discordance distributions, represented as inter co-twin differences in the Danish twin cohort. Solid horizontal lines and black points represent means and medians in the box-plots, respectively. * p-value = 0.03, as assessed by the Bartlett’s test of homogeneity of variances. Blue and yellow triangles represent co-twin pairs expressing averaged low and high NNAT levels, respectively. In box-plots, the lower and upper hinges = 25th and 75th percentiles. The upper/lower whiskers = largest/smallest observation less/greater than upper/lower hinge + 1.5 * IQR. Central median = 50% quantile.
To validate these findings, we examined adipose NNAT gene expression levels and available anthropomorphic traits in a small, independent Danish twin cohort. The cohort consisted of 20 MZ same-sex co-twin pairs61. As the cohort size and available traits precluded the same clustering analysis (as in Fig. 4), we stratified the cohort into upper and lower quantiles of NNAT expression. Notably, we validated the key findings from the MuTHER cohort: we observed increased insulinemia discordance (Extended Data Fig. 7e) and increases in BMI discordance specifically in MZ co-twin pairs with low NNAT expression (Extended Data Fig. 7f). Thus, reduced NNAT expression is associated with increased phenotypic variation in BMI and insulinemia in an independent twin cohort.
Human UPV sub-types are epigenetically distinct
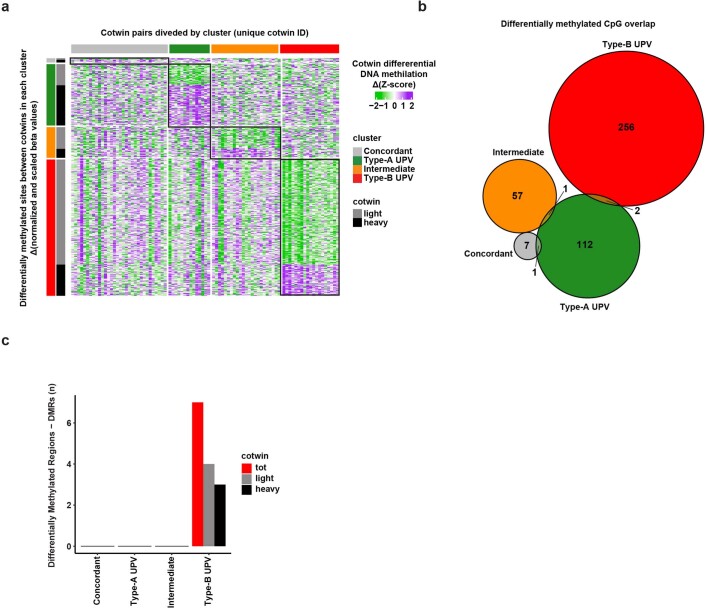
Altered epigenetic control is believed to be a key mediator of developmental programming effects62,63. We tested whether any of our human UPV types showed evidence of unexpected epigenetic alteration. We analyzed Infinium HumanMethylation450 DNA methylation profiles from the same MuTHER cohort adipose samples and called differentially methylated sites between heavy versus light co-twins within each of the four UPV groups (Extended Data Fig. 8a). Concordant co-twins had the fewest differentially methylated sites (Extended Data Fig. 8a,b). Type-A and Type-B discordant co-twins exhibited hundreds of differential sites, with Type-B UPV showing the most differently methylated sites (Extended Data Fig. 8a,b). Whereas Type-A and Type-B UPVs are both characterized by relative increases in fat mass (they are both relative adiposities), their adipose tissue DNA methylation profiles were clearly distinct. On average, Type-A UPV was characterized by DNA hypermethylation in the heavier co-twin, as opposed to DNA hypomethylation in Type-B UPV (Extended Data Fig. 8a). Further, the differentially methylated sites of Type-A and Type-B discordant co-twins showed almost no overlap, indicating that the two types of human phenotypic variation are truly and fundamentally distinct in their nature (Extended Data Fig. 8b). Consistent with this idea, a search for differentially methylated regions (DMRs), as opposed to differentially methylated sites, identified only DMRs between co-twins of the Type-B UPV group (Extended Data Fig. 8c). Seven DMRs were identified that reproducibly distinguish the heavy and light Type-B co-twins. Four of the seven Type-B DMRs were hypomethylated in the heavy co-twin. Notably, these DMRs were enriched for proximity to significant genome-wide association study (GWAS) variants for BMI, height, body fat percentage, insulin sensitivity, insulinogenic index, diabetes and diabetes-associated cardiovascular disease (Fig. 4h). These findings directly link NNAT and Type-B UPV to epigenetic changes near causal, metabolic disease loci. Of the 15 genes neighboring the DMRs, a substantial fraction is involved in energy metabolism (PARP6 and AGPAT3), transcriptional and epigenetic regulation (TCF3 and BCOR) and associated either directly or indirectly to inherited syndromes causing lipodystrophy, hypotonia and intellectual and heart disorders (C5orf58, AGPAT3, DNAJC19 and HCN4). Thus, human Type-B UPV is characterized by epigenetic regulation near human metabolic disease loci.
Extended Data Fig. 8. Type-A and Type-B UPV are associated to distinct DNA methylation patterns.
a, heat map showing the differentially DNA methylated sites among ‘heavy’ and ‘light’ cotwins belonging to the indicated groups identified in the TwinsUK cohort. SeSAMe cutoffs: adjusted p-value < 0.05; effect size > 0.05. Dark-gray and black boxes highlight DNA methylation enrichment in either the ‘light’ or ‘heavy’ cotwin, respectively. b, Venn diagram showing the overlap between differentially DNA methylated sites from the indicated cotwin groups and highlighting the specificity of these epigenetic profiles. c, Barplots showing the amount of differentially methylated regions (DMRs) among ‘heavy’ and ‘light’ cotwins belonging to the indicated groups identified in the TwinsUK cohort. Only in Type-B UPV, DMRs were detected. Dark-gray and black bars highlight DNA methylation enrichment in either the ‘light’ or ‘heavy’ cotwin, respectively.
Type-B UPV stratifies human into distinct metabolic subgroups
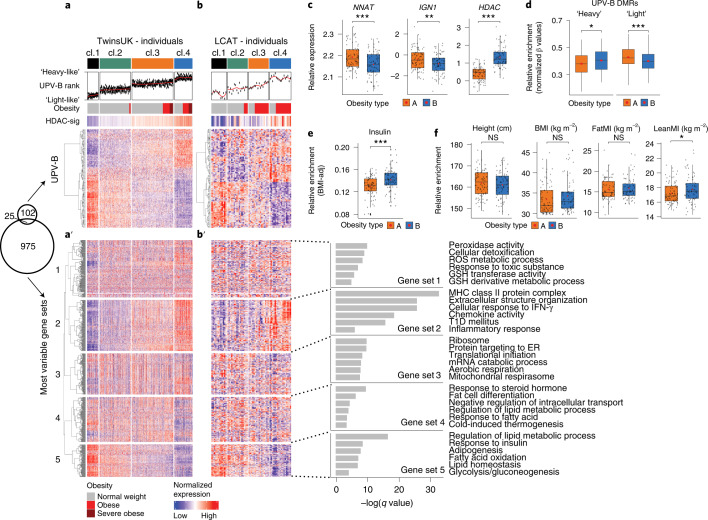
The analysis above showed that at least 30% of twin pairs in the MuTHER cohort exhibited Type-B UPV. If Type-B UPV is truly so common, then Type-B signatures should be readily detectable in the general population. We tested this idea in the greater MuTHER cohort, which includes hundreds of other individuals56,57 (Fig. 5a). First, we generated a Type-B UPV gene expression signature by identifying the 127 genes differentially regulated between heavy and light co-twins in the Type-B phenotypic variation cluster, but not differentially regulated between co-twins of the three other phenotypic variation clusters (Extended Data Fig. 9a and Methods). We then used hierarchical clustering to stratify the cohort according to expression of these 127, Type-B UPV genes. This analysis revealed four reproducible clusters of individuals in the general population (clusters 1–4 top, Fig. 5a). Individuals in clusters 2 and 3 showed little if any coordinated regulation of Type-B UPV genes (Fig. 5a). Clusters 1 and 4, however, showed strong coordinated regulation of Type-B UPV genes (Fig. 5a; heat map and rank analysis (UPV-B rank)). Cluster 4 individuals were specifically enriched in ‘heavy-like’ UPV-B transcriptome signatures, whereas cluster 1 individuals had ‘light-like’ gene expression profiles (Fig. 5a, UPV-B rank). Notably, clusters 1 and 4 also exhibited anti-correlated expression of NNAT (Extended Data Fig. 9b) and the non-overlapping HDAC-responsive genes. The latter indicated that the Type-B UVP gene signature (127 genes) captures the NNAT-buffered axis of phenotypic variation observed in the mouse and Type-B co-twins (Fig. 5a, HDAC-signature and Extended Data Fig. 9b). Thus, we find molecular evidence for Type-B human phenotypic variation (or plasticity) in the general population.
Fig. 5. Type-B UPV signature separates adults and children into distinct phenotypic and metabolic sub-types.
a, Heat map of k-means clustering of TwinsUK individuals. Four clusters were generated according to expression of the UPV-B signature. The UPV-B ranks annotation show the median rank of everyone according to their level of expression of UPV-B signature genes, discriminating Type-B ‘heavy-like’ and ‘light-like’ individuals. The obesity annotation is based on arbitrary cutoffs of BMI (obesity, >30 BMI; severe obesity, >35 BMI). The average expression of HDAC-signature (HDAC-sig) genes (leading-edge genes from Extended Data Fig. 6d) is reported. a′, Heat map (bottom), the expression profile of the most variable genes (top 1,000) across all samples is reported, after k-means clustering into five gene sets. Venn plot (left) shows the overlap between the most variable genes and the UPV-B. b–b′, Same as in a–a′, but on the LCAT cohort. The obesity annotation is based on standardized BMI arbitrary cutoffs (BMI standard score (SDS), obesity >1.88). On the right, representative results from Gene Ontology (GO) and pathway enrichment analysis for the five gene sets from the heat map of the TwinsUK individuals (a′). GO, KEGG and Molecular Signatures Database (MSigDB) databases were assessed. Related to the extended analysis in Extended Data Fig. 8g. c–f, Box-plots showing the distributions of indicated gene expression profiles (c), normalized DNA methylation on UPV-B DMRs (d), metabolic traits (e) and morphometric measurements (f), between Type-A and Type-B obesities (TwinsUK individuals affected by obesity and belonging to clusters 3 and 4 (cl.3 and 4) from the heat map in a). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, NS, not significant, as assessed by two-tailed Student’s t-tests. NNAT P = 0.00036; IGN1 P value = 0.0067; HDAC P = 2.2 × 10–16; UPV-B DMRs ‘heavy’ P = 0.028; UPV-B DMRs ‘light’ P = 0.00026; insulin P = 0.001; height P = 0.4; BMI P = 0.21; FatMI P = 0.46; LeanMI P = 0.047. In all box-plots, the lower and upper hinges indicate 25th and 75th percentiles. The upper/lower whiskers indicate largest/smallest observation less/greater than upper/lower hinge + 1.5 × IQR. Central median indicates 50% quantile. GSH, glutathione; MHC, major histocompatibility; IFN, interferon; T1D, type 1 diabetes.
Extended Data Fig. 9. Characterization of adults’ and childrens’ clusters.
a, Heat map of hierarchical clustering of DEGs between cotwins belonging to indicated groups. b, Boxplot showing the mean expression of HDAC-signature (above) or NNAT (below), among indicated clusters. p-values (2.2e-16; 1e-10) from ANOVA. c, Barplot showing obesity distribution among clusters. d, Boxplot showing normalized beta values for differentially methylated sites (above, p = 2.2e-16 and 2.2e-16) or regions (below, p = 0.0005 and 2.6e-8), among cotwins. ‘Heavy’- and ‘light’ enriched DNA methylated sites are reported. *** (p ≤ 0.001) from two-tails t-tests. e, UPV-B genes estimate of contribution to overall transcriptional variability in the TwinsUK cohort. Cumulative contribution of all principal components (black dots/line) and contribution of the most UPV-B-associated PCs (red dots/line) are reported. f, Boxplot showing mean expression of genes belonging to five genesets form Fig. 5b, among clusters. *** (p = 2.2e-16) from one-sided Tukey’s multiple comparisons test, following significant ANOVA. g, Complete gene ontology and pathway enrichment analysis for the 5 genesets from the heat map of TwinsUK individuals. h, Boxplot showing mean expression of HDAC-signature (above) or NNAT (below), among the clusters of the LCAT cohort. p-values from ANOVA. i, Barplot showing obesity distribution, among cluster in the LCAT cohort. j, Boxplot showing mean expression of genes belonging to the five genesets form Fig. 5b, among clusters in the LCAT cohort. * (p = 0.004), *** (p = 2.2e-16) from one-sided Tukey’s multiple comparisons test, following significant ANOVA. k, Box-plots showing distributions of serum insulin levels normalized on BMI-SDS, among children with obesity belonging to indicated clusters. ** (p = 0.0016) from two-tails t-tests. l, Barplots showing average cell-type compositions among transcriptional profiles of individuals belonging to indicated clusters. ASPC = adipose stem and progenitor cells, LEC = lymphatic endothelial cells, SMC = smooth muscle cells. m-n, Box-plots showing CDKN1C expression between ‘light’ and ‘heavy’ Type-B UPV cotwins (m) and Type-A/-B obesities (n). In all box-plots, lower and upper hinges = 25th and 75th percentiles. The upper/lower whiskers = largest/smallest observation less/greater than upper/lower hinge + 1.5 * IQR. Central median = 50% quantile. p-values from one-tail (m) or two-tails (n) t-tests.
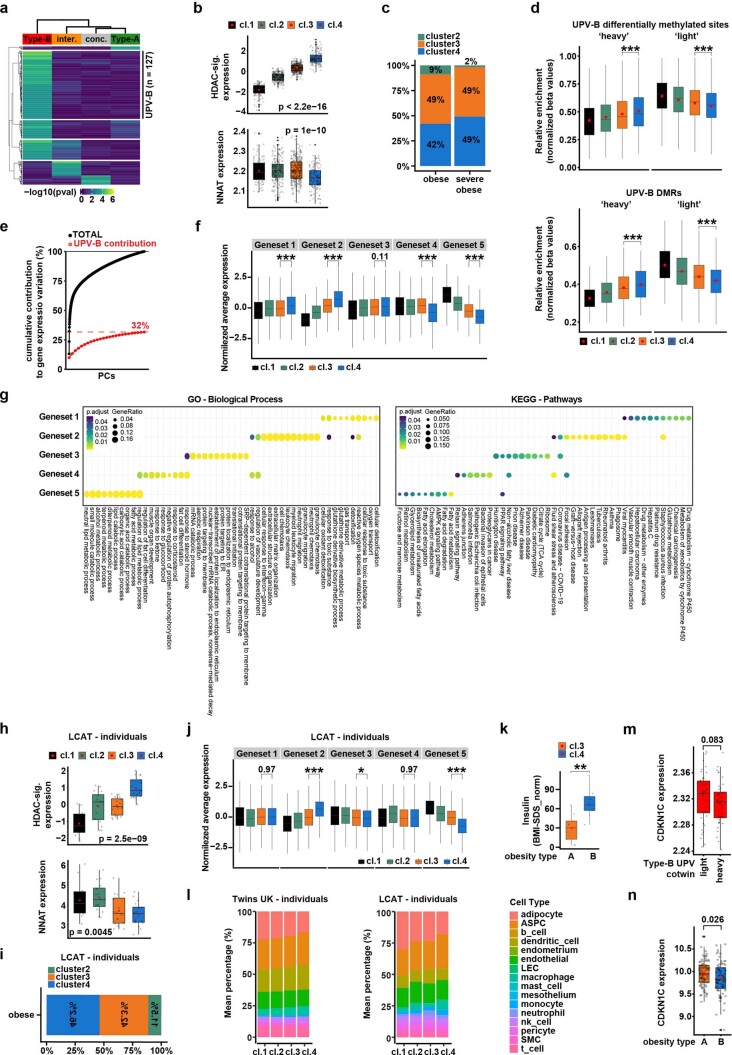
Consistent with their ‘light-like’ gene expression signature, cluster 1 individuals were all not affected by obesity (Fig. 5a, obesity annotation (top) and Extended Data Fig. 9c). In contrast, cluster 4 was enriched for individuals with obesity (Fig. 5a, obesity annotation (top) and Extended Data Fig. 9c). What is even more notable, is that cluster 4 still included many individuals without obesity (~50% of the cluster) despite strong Type-B UPV and NNAT/HDAC gene expression signatures (Fig. 5a and Extended Data Fig. 9c). These data are consistent with the fact that approximately 62% of Type-B ‘heavy’ co-twins were also not affected by obesity (BMI < 30). Along similar lines, there was no correlation between Type-B gene expression and BMI within population clusters (Fig. 5a; data within clusters are ordered from low to high BMI). Examination of Type-B differentially methylated sites and DMRs showed a clear enrichment of heavy-like methylation patterns in cluster 4 individuals (Extended Data Fig. 9d). Thus, Type-B UPV captures a dimension of population-level variation that is distinct from obesity per se (Fig. 4b).
Type-B UPV shows an inflammatory transcriptional program
Principal-component analysis (PCA) revealed that approximately one-third of all variation in the transcriptional dataset correlates tightly with the Type-B UPV gene expression signature (Extended Data Fig. 9e). This indicates that a major dimension of population-level phenotypic variation can be attributed to the Type-B ‘state’. To better understand the potential health implications of this finding, we analyzed the 1,000 most variable genes in the same transcriptional dataset. Of note, these 1,000 genes were distinct from the 127-gene Type-B UPV signature itself (Fig. 5a; Venn diagram). Consistent with the cumulative PCA analysis, two of the five major groups of the 1,000 most variable genes correlated strongly with the Type-B UPV gene signature (Fig. 5a′; gene sets 2 and 5). Cluster 4 (UPV-B heavy-like) individuals expressed high levels of inflammatory, immune and reactive oxygen species (ROS)-associated genes (Fig. 5a′, gene set 2, and Extended Data Fig. 9f,g). Conversely, the same individuals showed downregulation of genes involved in adipogenesis, lipid and glucose homeostasis (Fig. 5a′, gene set 5, and Extended Data Fig. 9f,g). Of note and as with the Type-B UVP gene signature itself, gene sets 2 and 5 were not correlated with obesity within clusters. These data again argue that the phenotypic outcomes of NNAT downregulation and Type-B UPV are not obesity per se. Rather, Type-B UPV consists of an altered metabolic state characterized by enhanced adipose tissue inflammatory signatures and reduced adipogenesis, nutrient uptake and metabolism pathway expression. Notably, cluster 4 included both individuals with and without obesity. Type-B UPV therefore also captures what seems to be a clear group of ‘normal weight obesity’ (individuals with normal BMI but with hyperinsulinemia and a highly consistent inflammatory gene expression pattern in adipose tissue).
Type-B UPV is present in childhood
Guided by the fact that we identified Type-B UVP in controlled genetic contexts (isogenic Nnat+/-p mice and MZ twins) and that the Type-B gene signature exhibited the most striking and distinct epigenetic characteristics (HDAC-signatures and DNA methylation signatures), we reasoned that Type-B phenotypic variation represents a state of altered epigenetic programming initiated in early life64. We therefore repeated our analysis using the Leipzig Childhood Adipose Tissue (LCAT) cohort65, a childhood cohort that had equivalent (and relevant) phenotypic and adipose tissue transcriptomic data (Fig. 5b). Notably, this analysis recapitulated all key findings above. The Type-B UPV gene expression signature stratified the childhood cohort into four corresponding clusters: cluster 4 children showed the strongest Type-B, heavy-like gene expression patterns (Fig. 5b) and cluster 1 showed the least. Cluster 4 children showed the strongest enrichment of HDAC-responsive genes (Fig. 5b (top) and Extended Data Fig. 9h) and reduced NNAT expression (Extended Data Fig. 9h). As with the adult cohort, all clusters included lean individuals; clusters 3 and 4 were enriched for individuals with obesity (Extended Data Fig. 9i); and while cluster 4 was the mostly enriched for obesity, it again included a substantial number of lean individuals. Also, in line with the adult analysis, cluster 4 children showed increased expression of inflammatory genes (Fig. 5b′, gene set 2, and Extended Data Fig. 9j) and downregulation of the lipid metabolism and adipocyte-specific genes (Fig. 5b′, gene set 5, and Extended Data Fig. 9j). Thus, Type-B UPV is readily observed in childhood and stratifies children into comparable metabolic ‘states’.
Type-B UVP gene expression identifies two major obesity types
One of the most relevant implications of these data is that they suggest the existence of fundamentally distinct metabolic disease sub-types and in particular, two major sub-types of obesity. To test this idea, we performed a focused comparison of the individuals with obesity in clusters 3 and 4 (Type-B UPV gene signature stratified) (Fig. 5c–f). Relative to cluster 3 obesity, cluster 4 individuals with obesity showed enrichment of the Type-B UPV gene expression signature (Fig. 5a, heat map and rank analysis (UPV-B rank)); reduced NNAT/IGN1 expression (Fig. 5c); increased HDAC-responsive gene expression (Fig. 5c); increased inflammatory and decreased adipogenesis gene expression (Fig. 5a′, gene set 2 and 5); and marked dysregulation of Type-B-specific DMRs (Fig. 5d). Phenotypically, cluster 4 individuals with obesity also exhibited consistently higher serum insulin levels (Fig. 5e); and while not different in height, BMI or relative fat mass (fat mass index; FatMI), cluster 4 individuals with obesity showed increased relative lean mass (lean mass index; LeanMI) (Fig. 5f). The distinctions between cluster 3 and cluster 4 obesities were validated in the LCAT childhood cohort for all available parameters (Fig. 5b,b′, gene sets and Extended Data Fig. 9k). Notably, this finding suggests that the pathophysiological consequences of Type-B UPV can already arise in early life. The data show that cluster 3 and cluster 4 obesities are distinct from phenotypic, transcriptional and epigenetic points of view. Given the parallels to co-twin-based Type-B UPV, we refer to these as Type-A obesity (cluster 3) and Type-B obesity (cluster 4).
Finally, to gain deeper insight into the transcriptional signatures themselves and their clinically relevance, we performed cell-type deconvolution using CibersortX66 and a recently published human adipose tissue single-cell atlas67. The analysis, which derives estimates of relative cell-type content in the MuTHER and LCAT cohort adipose biopsies, revealed high consistency across groups and cohorts (Extended Data Fig. 9l). Cluster 4, notably, showed a relative increase in pre-adipocyte-to-adipocyte ratio relative to cluster 3 and increased estimates for macrophage and monocyte content. These data are in line with the observed transcriptional signatures and indicated that Type-B (obesity and ‘normal weight obesity’) are characterized by increased adipose tissue inflammation and an adipocyte compartment skew toward adipose tissue progenitors.
Thus, we identify two major forms of obesity over and above the distinct classes of human phenotypic variation.
Discussion
UPV is pervasive in complex traits and disease, even among highly inbred animals and MZ twins. Yet we have very limited understanding of the regulatory mechanisms underpinning UPV. In precision medicine and according to canonical models, UPV is typically attributed to uncharacterized early (developmental programing) or later-life environmental effects or dismissed as random biological ‘noise’. Here, we demonstrate the existence of a conserved, NNAT-regulated axis that buffers against phenotypic variation (as a trait) and control an overgrowth polyphenism. We show that the same axis stratifies human populations into unique metabolic classes and two common obesity sub-types that are distinct in their clinical presentation, transcriptional signatures and epigenetic control. The data provide support for probabilistic, multi-stable phenotypic canalization events being a major driver of metabolic outcomes.
One key translational question raised by these data is whether NNAT-associated phenotypic variation and bi-stable effects are influenced by genetic or environmental interactions. Since our original description43, several groups41,42 generated anecdotal evidence for the existence of genetic and environmental modifiers of NNAT-dependent buffering of UPV. We have directly tested and validated the idea. We observe bi-stable adiposity and bi-stable overgrowth in parallel crosses to C57BL/6J and FVB/N dams, respectively42,43. The finding indicates a substantial background genetic interaction with the NNAT-buffered UPV axis. These data highlight the challenge of examining and dissecting UPV effects even in mice where genetics and environment can be controlled. So how might plasticity axes be investigated in very large human cohorts? Aside from building dedicated cohorts whose design accounts for both documented and undocumented genetic, environmental and parental modifier effects, possibilities include stratification by, or GWAS of, meta-traits (for example, all tangential Type-A versus Type-B covarying phenotypes); and evaluation of phenotypic associations using ‘surrogate’ polygenic risk scores, for instance composed of variants at or near UPV-type-specific differentially expressed genes.
Based on knockout mouse data, we hypothesize that the cluster 4 metabolic state and the Type-B obesities therein, are developmentally programmed states. Incorporating probabilistic developmental effects into models of phenotypic variation and clinical practice is still very challenging, because we have very little understanding of the regulatory axes or mechanisms that control phenotypic variation (as a trait), how these axes interact to generate complex traits, or how the probabilistic regulatory mechanisms ‘switch’ and canalize development toward altered phenotypic states and/or disease. The finding that Nnat+/-p and Trim28+/D9 triggered effects are independent within the same in utero context is profound. It demonstrates that multiple and distinct regulatory axes exist that act in parallel to canalize UPV outcomes. The observation also provides proof for the existence of multiple independent variation-buffering mechanisms and represents an example of a tri-stable mammalian polyphenism. They set the precedent that mammals have the genomic architecture and physiological networks to canalize at least three distinct and reproducible developmental trajectories within the same gene–environment context. The findings indicate that without assays such as epigenome profiling, major precision medicine efforts will be blind to an entire dimension of phenotypic regulation.
Identifying factors that buffer against phenotypic variation is challenging enough; it is equally difficult to determine how probabilistic (molecular) effects mechanistically alter organism-level physiology (such as obesity). Here, we demonstrate that the Nnat+/-p-buffered overgrowth UPV is hyperinsulinemia dependent, associated with islet-autonomous β-cell hyperproliferation and that alterations in food intake only occur after overgrowth and obesity (C57BL/6J–FVB F1s) (Fig. 2 and Extended Data Fig. 2). Notably, this type of phenotypic variation was phenocopied in human Type-B UPV. Together, these findings indicate that Type-B UPV may constitute an insulin-dependent form of adiposity. While this conclusion differs from the dogmatic view of obesity-driven hyperinsulinemia, it is consistent with reports that hyperinsulinemia may also cause or exacerbate obesity and the suggestion that its early treatment may prevent complications42. In support of this notion, ‘heavy’ Type-B discordant twins and Type-B individuals with obesity show downregulation of CDKN1C (Extended Data Fig. 9m,n), a molecular effect possibly linked to congenital hyperinsulinism68.
Our data do not rule out additional physiological mechanisms as contributors to the bimodal UPV phenotype in Nnat+/-p animals. In association with the Coll group, we previously showed that Nnat+/-p-triggered obesity (on C57BL/6J) is correlated with food intake42, suggesting a potential role of the hypothalamus; however, causality was not tested in those instances, nor was the requirement for hyperinsulinemia. Deep physiological phenotyping in these probabilistic models is warranted though choice of assay should not be decided lightly. Assays such as indirect calorimetry, feeding behavior and activity monitoring, in are experience are very challenging due to both the unpredictability of each individual’s end point and therefore very high numbers required, as well as to notable stress sensitivity that leads to numerous dropout animals such that the experimenter can no longer rule out a biologically biased sub-sampling. Based on experience, we would caution against descriptive or correlative experiments that do establish causality. Nnat+/-p animals for instance have proven challenging even for descriptive indirect calorimetry. A considerable number of animals lose weight even under home-cage and acclimatized conditions.
Our work shows that the islet-autonomous hyperproliferative program (in Nnat+/-p animals) is HDAC-dependent. This result demonstrates that HDAC modulation has a direct impact on pancreatic β-cell proliferation, which was not previously described. HDAC activity has been previously implicated in the regulation of adipogenesis, pancreas development, β-cell functionality and liver metabolism69–71 and HDAC modulation has been proposed as a potential treatment target for metabolic disease72–74. From the precision medicine perspective, our data indicate that HDAC modulation may constitute a preventative therapeutic paradigm for patients stratified by Type-B UPV gene expression.
Altogether, this work demonstrates that in addition to genetic and environmental factors, phenotypic outcomes in mammals are defined by probabilistic factors with the potential to canalize multiple distinct, stable and reproducible outcomes. The data indicate that a substantial fraction of human metabolic disease variation (and potentially associated processes such as cancer and inflammation) are defined by such processes.
Methods
All animal experiments were approved by Institutional Animal Care and Use Committee protocol no. 18-10-028 at VAI, USA and protocol no. MPI-ZH 2016-2019 at MPI, Germany. TwinsUK received ethical approval associated with TwinsUK Biobank (19/NW/0187), TwinsUK (EC04/015) or Healthy Ageing Twin Study (HATS) (07/H0802/84) studies from NHS Research Ethics Committees at the Department of Twin Research and Genetic Epidemiology, King’s College London. All samples and information were collected with written and signed informed consent. The Danish Twins study was approved by the Central Scientific-Ethical Committee of Denmark and was conducted according to the principles of the Helsinki Declaration. Furthermore, approval was obtained from the Danish Data Protection Agency. Informed consent was obtained from all participants. The LCAT study was approved by the University of Leipzig Ethics Committee (265–08, 265–08-ff) and registered in the National Clinical Trials database (NCT02208141). Written informed consent was obtained from all parents.
Trim28 and Nnat heterozygous mice
The generation of Trim28+/D9 and Nnat heterozygous mice are described elsewhere30,43,75. All mice were maintained with four to five siblings under controlled temperature (22 ± 1 °C) and humidity (50 ± 10%) and a 12 h light, 12 h dark schedule. Food and water were available ad libitum unless otherwise stated. All mice were weaned at 3 weeks of age onto a standard chow (V1185-300 MZ-Ereich, ssniff; autoclavable mouse breeder diet 5021, cat. no. 0006540, LabDiet,). Body composition was determined using an EchoMRI 4-in-1 (SYS-ID EF-036).
Genotyping
Ear biopsies were collected, boiled in 180 μl digestion buffer (50 mM NaOH and 0.1 mM EDTA) at 60 °C overnight. Twenty microliters of neutralization buffer (1 M Tris-HCl, pH 8.0) were added to stop the digestion. Two pairs of primers, one for WT and one for mutant, were mixed separately in two reactions with 1 μl biopsy lysate in 20 μl total volume including DreamTaq DNA polymerase (EP0701, Thermo Fisher) and amplified by PCR for 94 °C for 1.5 min (94 °C for 30 s, 55 °C for 30 s and 68 °C for 1 min) × 31 and 68 °C for 5 min. The primers are listed in Supplementary Table 1. The amplification products of WT (573 bp) and mutant Nnat (545 bp) bands, were confirmed on 2% agarose gels. Methods for genotyping the Trim28+/D9 mouse line are described elsewhere30,43.
Skeleton staining
Mice were killed, the skin and internal organs removed and the eviscerated mice fixed in 95% ethanol overnight and stained with Alcian blue (0.3% in 70% ethanol) for 48 h. The skeletons were incubated with 2% KOH for 48 h and stained with Alizarin red S (0.1% in 95% ethanol) overnight. The stained skeletons were cleared in 1% KOH/20% glycerol solution for up to 1 week and stored in ethanol/glycerol solution (1:1) before imaging.
Plasma growth factors measurement
Plasma insulin, IGF1/IGF2 and GH were determined by ELISA according to manufacturers’ instructions (10-1247-01, Mercodia; EMIGF1 and EMIGF2, Thermo Fisher; EZRMGH-45K, Millipore Sigma).
Glucose tolerance test and glucose-stimulated insulin secretion
Following a 6-h fast, mice were administered glucose (1 g kg−1) by oral gavage and blood samples for glucose measurement were collected from the tail vein at the indicated times. Glucose levels were measured using a OneTouch Vita glucometer. Blood samples were collected from the cheek at the indicated times into EDTA-coated tubes to prevent coagulation. Blood samples were centrifuged at 850g for 20 min at 4 °C to collect plasma. Plasma insulin was measured by ELISA (10-1247-01 or 10-1249-01, Mercodia).
Mouse islet isolation and spheroid formation
Mice were killed with CO2 and dissected according to standard procedures. The common bile duct was tied off with a thread, so perfusion buffer would flow to the pancreas rather than the liver. The pancreas was perfused with 5 ml collagenase digestion buffer (HBSS, 10 mM HEPES and 2 mg ml−1 collagenase 4 from Worthington, cat. no. LS004189) through the sphincter of Oddi. The perfused pancreas was placed in 2 ml collagenase digestion buffer and incubated for 30 min at 37 °C. After incubation, the tube was mixed by inversion 30 times. Then, 40 ml of quenching buffer (HBSS, 24 mM HEPES and 5 mg ml−1 BSA) was added to stop the collagenase activity. The tube was centrifuged at 200g for 3 min at 4 °C to form a cell/tissue pellet. The pellet was resuspended with 25 ml quenching buffer and run through a 420-µm strainer to remove undigested material. The clarified supernatant was then passed through a 70-µm strainer to collect the islets from the bulk solution. The islets were transferred into a cell culture dish containing RPMI medium (Gibco, 11875093) by inverting the strainer and dipping it in medium. The dish was placed in a 5% CO2 incubator at 37 °C overnight. The following day, islets were removed from the culture dish with a P10 pipet and transferred into a fresh RPMI dish. Recovered islets were incubated in a 5% CO2 incubator at 37 °C for another 2 days to allow islet metabolism to normalize. Then, 100 µl of islet suspension were mixed with 500 µl warm accutase (A6964-100ML, Sigma) and incubated for 2 min at 37 °C to dissociate islets into single cells. Cells were stained with DAPI and sorted by FACS to recover (DAPI-negative) live cells. Approximately 2,000 live cells were seeded per well in a 96-well spheroid plate (Corning, CLS4520-10EA) and the plate incubated in a 5% CO2 incubator for 3 days at 37 °C until spheroids formed.
Glucose-stimulated insulin secretion ex vivo
Krebs Ringer buffer (140 mM NaCl, 3.6 mM KCl, 0.5 mM NaH2PO4, 0.2 mM MgSO4, 1.5 mM CaCl2, 10 mM HEPES and 2 mM NaHCO3) and BSA (0.5 %) was prepared freshly on the treatment day and pH adjusted to 7.4. The 2.8 mM glucose and 16.7 mM glucose solutions were prepared with fresh Krebs Ringer buffer. Isolated islets or formed spheroids were transferred into 100 µl of the 2.8 mM glucose solution and incubated at 37 °C for 30 min. Then, the islets or spheroids were transferred into another well with 100 µl 2.8 mM glucose solution and incubated at 37 °C for 1 h for the basal release. Finally, the islets or spheroids were transferred into another well with 100 µl 16.7 mM glucose solution and incubated at 37 °C for 1 h for the glucose-stimulated release.
Total insulin content
Intact pancreata were weighed, cut into small pieces and incubated with 0.18 M HCl in 70% ethanol overnight at 4 °C. The resulting mixture was transferred to 1.5-ml microfuge tubes and centrifuged at 850g for 5 min at room temperature. The clarified supernatant was transferred to a new tube and stored at −20 °C until use. Insulin content was determined via ELISA according to the manufacturer’s instructions (10-1247-01, Mercodia).
Streptozocin administration and insulin pellet restoration
Five-week-old mice received a one-time i.p. injection of STZ (300 mg kg−1, Sigma, S0130-1G). Four days and again 1 month after STZ treatment, insulin pellets (Linplant, Linshin) were s.c. implanted on the back of the animals (one pellet per 20 g body weight) as per the manufacturer’s instructions.
EdU proliferation
Primary islets in RPMI medium (10% FBS, 50 IU ml−1 penicillin, 50 μg ml−1 streptomycin, 0.25 μg ml−1 amphotericin B and 50 mg ml−1 gentamicin) were stained with 10 μM EdU using a fluorescence microscopy protocol kit following the manufacturer’s instructions (iFluor 488, ab219801, Abcam). We used an A1 Plus-RSi laser scanning confocal microscope (Nikon) and z-stack function to capture sequential images of the islets and reconstruct their three-dimensional volume. The total volume of EdU-incorporated cells was then calculated with an ImageJ macro (https://visikol.com/wp-content/uploads/2019/02/Visikol-Measure-Volume-Macro.ijm).
Islet RNA extraction, library preparation and sequencing
Total RNA was extracted from isolated islets using the QIAGEN RNeasy Micro kit (cat. no. 74004). Libraries were prepared from 10 ng of total RNA. Total RNA material was converted to dsDNA using the SMART-Seq v.4 Ultra Low Input RNA kit for sequencing, v.091817 (Takara Bio). Illumina Nextera DNA Flex kit (Illumina) was used to convert the complementary DNA into Illumina-compatible sequencing libraries. The quality and quantity of the finished libraries were assessed using a combination of Agilent DNA High Sensitivity chip (Agilent Technologies), QuantiFluor dsDNA System (Promega) and Kapa Illumina Library Quantification qPCR assays (Kapa Biosystems). Individually indexed libraries were pooled and 50-bp, paired-end sequencing was performed on an Illumina NovaSeq6000 sequencer using an S1, 100 cycle sequencing kit (Illumina). Each library was sequenced to an average raw depth of 30 M reads. Base calling was conducted by Illumina RTA3 and the output of NextSeq Control Software was demultiplexed and converted to FastQ format with Illumina Bcl2fastq v.1.9.0.
Mouse transcriptomic analysis
We performed bulk messenger RNA-seq on primary islets and adipocytes taken from Nnat+/-p-Light, Nnat+/-p-Heavy mice and WT littermates. Raw sequences were aligned to mouse reference genome GRCm38.p6 with the snakePipes2 RNA-seq pipeline76. Differential expression of the raw counts was performed using DESeq2 (ref. 77). GSEA of DEG results was performed with fgsea78. Enrichment maps were generated in Cytoscape79 from results of 6-week-old Nnat+/-p-Light versus Nnat+/-p-Heavy MSigDB C2 CGP GSEA. We also performed bulk mRNA-seq on primary islets from early-stage (3-week-old) Nnat+/-p-Light and Nnat+/-p-Heavy-like mice and WT littermates. Differential gene expression analysis was performed as described above. Samples were batch corrected using ComBat and normalized count matrices were inspected using PCA. Nnat+/-p-Heavy enriched leading-edge genes from HDAC-related gene sets were assessed at early and late-stage expression profiles to be represented on the heat map. The contribution of the HDAC-signature to the overall gene expression variation was evaluated based on the PCA. The principal components (PCs) from the PCA were ordered for their association to the HDAC-signature (mean of contributions to PCs for the genes belonging to the signature) and the top four correlated were subset (the inflection point of the ordered PCs). The cumulative contribution of these PCs to gene expression variation in the dataset was visualized and compared to the overall contribution from all PCs.
HDACi and HATi treatment
Islets were isolated from 7–8-week-old animals and cultured ex vivo in RPMI medium (added 10% FBS, 50 IU ml−1 penicillin, 50 μg ml−1 streptomycin, 0.25 μg ml−1 amphotericin B and 50 mg ml−1 gentamicin) for 2 days to reach steady state. Islets were then treated with 500 nM HDACi (TSA) or 500 mM HATi (C646) for 1 day, followed by a 2-d EdU incubation (iFluor 488, Abcam) to track DNA replication.
Immunochemistry and H&E staining
Insulin (A0564, DAKO, 1× ready to use) and H&E staining were performed on sequential, 5-μm paraffin thin sections fixed in 4% phosphate-buffered formalin. Insulin immunohistochemistry was performed on an Agilent Autostainer Link 48 instrument as per the manufacturer’s instructions. Stained thin sections were digitized with an Aperio ScanScope slide scanner fitted with a ×20 objective (Leica). For each animal, we imaged three insulin and three H&E thin sections. Positively stained β-cells in each thin section were quantified with Genie (Leica) software. Reported β-cell-positive areas are the average from four animals per group and three thin sections per animal (n = 12 thin sections). We quantified apoptosis events by using the TUNEL assay kit, HRP-DAB (ab206386, Abcam) on pancreatic sections from 16-week-old animals.
Immunofluorescence
Paraffinized sections were heated, deparaffinized with xylene and rinsed in water. Antigen retrieval was performed by heating the slides at 95 °C for 20 min in Tris-EDTA, pH 9.0. Specimens were blocked in 5% goat serum PBST (0.05% Tween 20) and incubated overnight with insulin primary antibody (A0564, DAKO; 1:50 dilution) and Ki-67 primary antibody (ab15580, Abcam; 1:200 dilution); glucagon primary antibody (G2654, Sigma; 1:500 dilution); and somatostatin primary antibody (ab30788, Abcam; 1:200 dilution). Fluorochrome-conjugated secondary antibodies (Alexa Fluor 488, anti-rabbit; Alexa Fluor 555, anti-guinea pig; Alexa Fluor 647, anti-rat; Alexa Fluor 488, anti-mouse; 1:500 dilution, Invitrogen) were then added to each slide and incubated for 2 h at room temperature. Slides were rinsed three times for 10 min each in PBST buffer and air dried. A drop of VectaShield mounting medium (containing DAPI; H-1200, Vector Laboratories) and coverslip were applied to each slide and slides cured overnight at 4 °C in the dark before image acquisition. Images were acquired using an A1 Plus-RSi laser scanning confocal microscope (Nikon).
qRT–PCR
Total RNA was extracted using TRI Reagent (Sigma) and reverse transcribed into cDNA using a commercially available kit (43-688-14, Applied Biosystems). Nnat and Hprt transcripts were quantified using TaqMan gene expression assays with validated probes (Life Technologies). All probes are listed in Supplementary Table 1. All qPCR reactions were performed on a 7900HT Fast Real-Time PCR System (Applied Biosystems). Thermal cycling conditions for all genes included 2 min at 50 °C, 20 s at 95 °C and 40 cycles of 95 °C for 1 s, 60 °C for 20 s. Post-amplification melting curve analysis was performed to check for nonspecific products and probe-only controls were included as negative controls. Threshold cycles (Ct values) were normalized to Hprt within each sample to obtain sample-specific ΔCt values (Ct gene of interest − Ct housekeeping gene). The 2-ΔΔCt values were calculated to obtain fold expression levels, where ΔΔCt = (ΔCt treatment − ΔCt control).
Human population studies and analyses
MuTHER TwinsUK cohort
The MuTHER cohort consists of 855 female white twins and 193 MZ co-twin pairs, aged between 40 and 87 years56,57 and is a subset of the larger TwinsUK study55 (referred to as TwinsUK in the figures and text). Subcutaneous adipose tissue samples were obtained from skin punch biopsies. Gene expression profiles were generated using Illumina’s whole genome expression array (HumanHT-12 v.3) and are available from the ArrayExpress archive under the repository no. E-MTAB-1140. For expression arrays, the original authors57 used multiple technical replicates for each sample, which were all randomized before hybridization and replicates run on different BeadChips. Expression signals were normalized separately per tissue, with quantile normalization of the replicates of an individual followed by quantile normalization across all individuals. The authors acknowledged that their approach does not adjust for shared covariance due to technical factors that may influence subsequent analysis, but previous efforts indicate that the impact on the result seemed to be minor80. Expression data were corrected for technical batch effect using ComBat81 and distributions of identified UPVs and individuals’ clusters among batches did not show specific enrichment. Differential expression analysis and GSEA were performed using limma82 and fgsea78, respectively. The differential expression analysis was performed using age as a covariate in the model. Cell-type deconvolution was performed using CibersortX66 and a recently published single-cell atlas of human white adipose tissue67 was used as a signatures reference.
Monozygotic co-twin analysis
To maximize the number of co-twin couples in the analyses (n = 153), we excluded the waist, hip and waist-to-hip ratio measurements, which were not available for ~one-third of the cohort. Discordance indices were calculated as the difference of the log-transformed values between co-twins for each measurement, after ordering the co-twins according to their BMI. Likewise, we calculated gene expression discordance from normalized expression array counts. We used a graph-based clustering approach from Seurat58 for unbiased clustering of co-twin pairs according to their morphometric discordances. Heat maps of morphometric discordances were generated with ‘pheatmap’ (https://cran.r-project.org/web/packages/pheatmap/index.html) by clustering discordances based on Euclidean distances, a complete agglomeration method and rows scaling. Correlations between gene expression and phenotypic discordances were determined by Spearman correlation and reported as the −log10(P). When performing the same analysis on the identified co-twin clusters, the average of P values from all Spearman correlations for a single gene was reported.
Danish Twins cohort
The Danish Twins cohort used in this study consists of 160 elderly individuals (88 females and 72 males), aged between 63 and 83 years. The cohort includes 20 MZ and 21 DZ same-sex co-twin pairs and is part of a larger study61. RNA samples were obtained from subcutaneous adipose tissue biopsies and NNAT expression was measured by qRT–PCR analysis, as previously reported43. The MZ co-twin pairs were divided into halves according to their average NNAT expression level. Like the TwinsUK cohort, we calculated serum insulin and BMI discordance among MZ co-twins (as log2(fold change) and difference, respectively). Gaussian finite mixture modeling from the Mclust tool83 was used to separate insulin-concordant and -discordant co-twin pairs. The proportions of insulin-concordant and -discordant co-twins among NNAT-low- and NNAT-high-expressing couples were visualized. The distributions of BMI discordances among NNAT-low- and NNAT-high-expressing couples were compared for homogeneity of variances using the Bartlett’s test.
DNA methylation analysis in the MuTHER TwinsUK cohort
The MuTHER cohort contains Infinium HumanMethylation450 BeadChips array (Illumina WG-314-1002) data from the subcutaneous fat derived of 648 TwinsUK participants. For DNA methylation arrays, the original authors reported84 that tissue samples were randomized before DNA extraction. Signal intensities were quantile normalized. Beadchip, bisulfite-sequencing (BS) conversion efficiency (assessed with the built-in BS conversion efficiency controls) and BS-treated DNA inputs were shown to contribute significantly to the variation in β levels and were included as covariates in subsequent analysis. The processed and normalized β values were previously published57,84,85 and are available from the ArrayExpress archive under the repository no. E-MTAB-1866. We analyzed the data using the ‘SeSAMe’ pipeline86,87. Normalized β values were analyzed by linear modeling to identify DMRs between ‘light’ and ‘heavy’ MZ co-twins, controlling for age as a covariate in the model. The cutoff used to define differential methylation was FP < 0.05 and effect size threshold >0.05 (DNA methylation differences under 5% were not considered biologically meaningful). The heat map of the differentially methylated CpGs between co-twin pairs belonging to the four different phenotypic variation clusters was generated with ‘ComplexHeatmap’88. The genomic regions of DMRs from the Type-B UPV co-twins were used to search for genome-wide relevant associations between SNPs and phenotypes in the T2D Knowledge Portal (https://t2d.hugeamp.org). When DMRs were defined by just a single nucleotide, we searched in ±50-kb regions. All genome-wide significant associations were reported (P = 10−8). We also visualized all the GWAS associations within our DMRs with a P < 10−3.
Genotyping data analysis in the MuTHER TwinsUK cohort
The MuTHER cohort contains genotyping data generated by Illumina 317 K, 610 k and 1 M chip arrays, from the subcutaneous fat derived of 807 TwinsUK participants. These data are available upon request at the TwinsUK consortium (https://twinsuk.ac.uk/resources-for-researchers/our-data/). The genotype annotation files were generated using IMUTE2 with 1000 Genomes Project phase 1 (interim) as a reference panel. This dataset is based on a sequence data freeze from 23 Nov 2010; the phased haplotypes were released Jun 2011. GWAS data were ‘pre-phased’ using IMPUTE2 without a reference panel. The resulting haplotypes were used to perform fast imputation from the 1000 Genomes Project phase 1 dataset. The imputation of TwinsUK1 (317 K chip) and TwinsUK23 (610 k and 1 M chips) were conducted separately and merged with GTOOL. Genotyping data were analyzed using PLINK software (v.1.9). Samples with a missing call rate exceeding 0.02 (–mind 0.02) were excluded from genetic analysis. One-way ANOVA was conducted to determine whether overall missingness was significantly different across UPV groups at the genome-wide level, without finding statistical relevant differences. After removal of overly missing samples, each co-twin pair was screened for discrepancies in genotypes to assess the extent of co-twin genetic similarity, which accounted for >99.9% of the data. These analyses were conducted in R (v.4.1.1) using the ‘stats’ package.
Whole MuTHER TwinsUK cohort analysis
To generalize the findings from MZ co-twins, we included all individuals from the MuTHER cohort and analyzed 824 gene expression profiles from s.c. adipose tissues. The Type-B UPV gene expression signature was identified by performing differential gene expression analysis between ‘heavy’ and ‘light’ co-twins from the four concordant/discordant clusters. We selected all the genes with a P value <0.001 and clustered them by k-means clustering to identify the Type-B specific signature (127 total genes). We then used the signature to stratify the TwinsUK individuals. The number of clusters for this analysis was determined by visualizing the dispersion within each cluster for k = 1–10 and selecting the number of clusters that represented the ‘saturation point’ of dispersion. Cluster stability was assessed by the Jaccard’s similarity score. These analyses were performed using the RaceID package89. The heat map of the four k-means individuals’ clusters was generated with ‘ComplexHeatmap’88. Gene clustering was based on Euclidean distances and Ward’s agglomeration method of log-transformed and scaled, normalized data. The individuals were further ordered by BMI within each group. We then ranked Type-B-specific signatures (127 genes) for each individual. High ranks were associated with a ‘heavy-like’ Type-B transcriptional profile. The median of ranks from all UPV-B genes was plotted for each individual. The contribution of the Type-B-specific gene signature to overall gene expression variation in the cohort was evaluated as follows. PCA was performed on the gene expression profiles of all individuals. The 824 PCs were ordered for their association to the Type-B gene expression signature (mean of contributions to PCs of the genes belonging to the 127-gene signature) and the top 25 correlated components were subset (inflection point of the ordered PCs). The cumulative contribution of the top 25 PCs to gene expression variation in the cohort was visualized and compared to the overall contribution from all 824 PCs. The HDAC-signature annotation reported in the heat map was derived as follows: first, we performed GSEA of HDAC-related gene sets between the ‘heavy’ and ‘light’ co-twins belonging to the Type-B phenotypic cluster. Next, we retrieved the leading-edge genes (genes driving the gene sets’ enrichment), from the ‘heavy’-enriched HDAC-related gene sets, which then defined the HDAC-signature. Annotations in the heat map show the average expression of the HDAC-signature genes for each individual. The unbiased transcriptional analysis among the four individuals’ clusters was based on the top 1,000 most variable genes among all the samples (heat map shows only the genes for which we can detect expression in the LCAT cohort). GO and pathway analysis were performed with ‘clusterProfiler’90 against the GO, KEGG and MSigDB databases. For the metabolic/morphologic characterization of the individuals’ and obesity clusters, the serum insulin levels were adjusted on BMI and the fat and lean mass normalized on the squared height, generating FatMI and LeanMI, respectively.
The LCAT cohort
The LCAT cohort consists of female and male white children aged 0–18 years who underwent elective orthopedic surgery, herniotomy/orchidopexy or other surgeries65. Exclusion criteria were severe diseases and medication that might affect adipose tissue biology, such as diabetes, generalized inflammation, malignant diseases, genetic syndromes or permanent immobilization. BMI data were standardized to age- and sex-specific centiles by applying German reference data and are represented as BMI SDS91. Overweight and obesity are defined by a cutoff of 1.28 and 1.88 SDS (90th or 97th centile), respectively. Subcutaneous adipose tissue samples were excised during surgery, washed three times in PBS and immediately frozen in liquid nitrogen for RNA isolation. For RNA-seq, gene expression profiles were generated as previously described43. Differential expression and GSEA were performed using DESeq2 (ref. 77) and fgsea78, respectively. Normalized counts were corrected for both age and sex confounders with ComBat81. For global gene expression analysis on individuals, 61 profiles from s.c. adipose tissues were analyzed (34 males and 27 females). The number of clusters selection and the heat map generation were performed as in the adult cohort. The heat map in Fig. 5 shows the Type-B specific gene clustering (top) and the same variable genes as in the TwinsUK cohort (bottom). For the metabolic/morphometric characterization of the individuals’ and obesity clusters, standardized (SDS) measurements and fasting serum insulin levels normalized on BMI SDS were visualized.
Statistics
In both human and mice analyses, equality of variances and means were assessed by Levene’s test and Student’s t-tests, respectively (unless otherwise specified). We used one-way ANOVA followed by Tukey’s honestly significant difference test (where appropriate and as indicated) for multiple comparison testing. In the mice body composition data analysis, the separation into discrete clusters was tested by Gaussian finite mixture modeling using the Mclust tool83. A supervised analysis was performed to identify the best model describing the WT as a single reference cluster. Next, the same model was applied on data from other genotypes. All data are expressed as mean ± s.e.m., unless otherwise specified. Correlations were tested by linear regression, unless otherwise specified. All reported P values are two-tailed, unless stated otherwise, where P ≤ 0.05 was considered to indicate statistical significance. Calculations to evaluate the power to detect an effect given the sample size in mouse studies, were performed with bifurcatoR (https://github.com/VanAndelInstitute/bifurcatoR). Mouse studies were designed to reach 95% power to detect effect size.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Supplementary Tables 1 and 2
Acknowledgements
We thank A. Drougard, A. Ferguson-Smith, S. Millership, D. Schones, T. Jenuwein, R. Grosschedl, R. Sawarkar, and R. Teperino for critical reading of the manuscript and theoretical discussions. We thank A. Bergsma for technical support. The original Nnat+/-p colony was a generous gift from A. P. Coll (Wellcome Trust-MRC Institute of Metabolic Science, Addenbrooke’s Hospital). We are indebted to the MPI-IE facilities, the VAI Vivarium and the VAI Pathology and Biorepository, Optical Imaging and Genomics Cores. We thank Z. Madaj, E. Wolfrum and the VAI Bioinformatics and Biostatistics Core for supporting the statistical analyses. This work was supported by funding from the MPG, the VAI, the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no. 675610, the Novo Nordisk Foundation, European Foundation for the Study of Diabetes and the Danish Council for Independent Research and National Institutes of Health awards R21HG011964 and 1R01HG012444. The LCAT cohort was supported by grants from AK German Research Foundation CRC1052 (no. 209933838), project C05 and KO3512/3-1, the German Diabetes Association and the Federal Ministry of Education and Research, Germany, FKZ, 01EO1001 (Integrated Research and Treatment Center Adiposity Diseases). TwinsUK is funded by the Wellcome Trust, Medical Research Council, European Union, Chronic Disease Research Foundation, Zoe Global Ltd. and the National Institute for Health Research-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London.
Extended data
Source data
Statistical Source Data.
Statistical Source Data.
Unprocessed and uncropped images.
Statistical Source Data.
Statistical Source Data.
Statistical Source Data.
Statistical Source Data.
Unprocessed and uncropped gel.
Statistical Source Data.
Statistical Source Data.
Unprocessed and uncropped images.
Statistical Source Data.
Statistical Source Data.
Statistical Source Data.
Statistical Source Data.
Statistical Source Data.
Author contributions
C.H.Y. and J.A.P. conceived the experiments. C.H.Y. conducted the majority of the experiments and analyzed the results. L.F. analyzed the human datasets and mice phenotypic data. C.H.Y., L.F., S.A. and S.C. analyzed mice transcriptome datasets. T.W. and V.W. performed mouse experiments and islets isolation. I.P. and E.D. supervised the in vitro and islet experiments, respectively. S.H. and A.C. supervised bioinformatics work. T.Y. and D.L. supervised the mouse skeletal analysis. J.B. and R.G. supervised and E.G. performed the genetic data analyses. M.T., L.G.G., A.A.V., L.G. and E.G. analyzed the clinical data of human twin cohorts. K.L. and A.K. analyzed and provided data of the Leipzig Childhood AT cohort. PERMUTE shaped many of the ideas and approaches used in the manuscript. C.H.Y., L.F., D.P.C. and J.A.P. wrote the manuscript.
Peer review
Peer review information
Nature Metabolism thanks Alan Attie and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Ashley Castellanos-Jankiewicz, in collaboration with the Nature Metabolism team.
Data availability
RNA-seq data from both mouse primary islets and subcutaneous adipose tissue of the LCAT cohort, have been deposited to Gene Expression Omnibus and are publicly available under the accession codes GSE205740 and GSE205668, respectively. They are collected under the GSE205741 super-series. Gene expression and DNA methylation profiles by whole genome arrays from subcutaneous adipose tissue of the MuTHER TwinsUK cohort have been deposited to Array Express and are publicly available under the accession codes E-TABM-1140 and E-TABM-1866, respectively. Morphometric and genotypic data of the MuTHER TwinsUK cohort are available upon request at https://twinsuk.ac.uk/resources-for-researchers/access-our-data/. The MSigDB is available at http://www.gsea-msigdb.org/gsea/msigdb. Source data are provided with this paper.
Code availability
No custom code or mathematical algorithm were generated in this report. All publicly available codes and tools used to analyze the data are reported and referenced in the Methods and the Reporting Summary.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chih-Hsiang Yang, Luca Fagnocchi.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
J. Andrew Pospisilik, Email: andrew.pospisilik@vai.org.
PERMUTE:
Timothy Triche, Jr, Adelheid Lempradl, Zachary J. DeBruine, Emily Wolfrum, Zachary Madaj, Tim Gruber, Brooke Grimaldi, Andrea Parham, Mitchell J. McDonald, Joseph H. Nadeau, Ildiko Polyak, Carmen Khoo, Christine Lary, Peter D. Gluckman, Neerja Karnani, David Carey, Ruth J. F. Loos, and Gabriel Seifert
Extended data
is available for this paper at 10.1038/s42255-022-00629-2.
Supplementary information
The online version contains supplementary material available at 10.1038/s42255-022-00629-2.
References
- 1.Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Trans. R. Soc. Edin. 1918;52:399–433. doi: 10.1017/S0080456800012163. [DOI] [Google Scholar]
- 2.Fisher RA. The causes of human variability. Eugen. Rev. 1919;10:213–220. [PMC free article] [PubMed] [Google Scholar]
- 3.Wright S. The relative importance of heredity and environment in determining the piebald pattern of guinea-pigs. Proc. Natl Acad. Sci. USA. 1920;6:320. doi: 10.1073/pnas.6.6.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gärtner K. A third component causing random variability beside environment and genotype. A reason for the limited success of a 30 year long effort to standardize laboratory animals? Lab. Anim. 1990;24:71–77. doi: 10.1258/002367790780890347. [DOI] [PubMed] [Google Scholar]
- 5.Jonsson H, et al. Differences between germline genomes of monozygotic twins. Nat. Genet. 2021;53:27–34. doi: 10.1038/s41588-020-00755-1. [DOI] [PubMed] [Google Scholar]
- 6.Panzeri I, Pospisilik JA. Epigenetic control of variation and stochasticity in metabolic disease. Mol. Metab. 2018;14:26–38. doi: 10.1016/j.molmet.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang C-H, Pospisilik JA. Polyphenism: a window into gene–environment interactions and phenotypic plasticity. Front. Genet. 2019;10:132–132. doi: 10.3389/fgene.2019.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elder SJ, et al. Genetic and environmental influences on factors associated with cardiovascular disease and the metabolic syndrome. J. Lipid Res. 2009;50:1917–1926. doi: 10.1194/jlr.P900033-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yokoi N. Epigenetic dysregulation in pancreatic islets and pathogenesis of type 2 diabetes. J. Diabetes Invest. 2018;9:475–477. doi: 10.1111/jdi.12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia Q, Grant SFA. The genetics of human obesity. Ann. NY Acad. Sci. 2013;1281:178–190. doi: 10.1111/nyas.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sayols-Baixeras S, et al. DNA methylation and obesity traits: An epigenome-wide association study. The REGICOR study. Epigenetics. 2017;12:909–916. doi: 10.1080/15592294.2017.1363951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gluckman PD. Epigenetics, the life-course and metabolic disease. Nat. Rev. Endocrinol. 2012;8:74–76. doi: 10.1038/nrendo.2011.226. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein D, Golson ML, Kaestner KH. Epigenetic control of 6ourse and metabolic diseas. Diabetes Res. Clin. Pract. 2017;123:24–36. doi: 10.1016/j.diabres.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loh M, Zhou L, Ng HK, Chambers JC. Epigenetic disturbances in obesity and diabetes: epidemiological and functional insights. Mol. Metab. 2019;27:S33–S41. doi: 10.1016/j.molmet.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elgin SCR, Reuter G. Cold Spring Harb. Perspect. Biol. 2013;5:a017780. doi: 10.1101/cshperspect.a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Timms RT, Tchasovnikarova IA, Lehner PJ. Position-effect variegation revisited: HUSHing up heterochromatin in human cells. Bioessays. 2016;38:333–343. doi: 10.1002/bies.201500184. [DOI] [PubMed] [Google Scholar]
- 18.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Ehrlich M, Lacey M. DNA methylation and differentiation: silencing, upregulation and modulation of gene expression. Epigenomics. 2013;5:553–568. doi: 10.2217/epi.13.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattacharjee S, Roche B, Martienssen RA. RNA-induced initiation of transcriptional silencing (RITS) complex structure and function. RNA Biol. 2019;16:1133–1146. doi: 10.1080/15476286.2019.1621624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sieriebriennikov B, Sommer RJ. Developmental plasticity and robustness of a nematode mouth-form polyphenism. Front. Genet. 2018;9:382. doi: 10.3389/fgene.2018.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 23.Salathia N, Queitsch C. Molecular mechanisms of canalization: Hsp90 and beyond. J. Biosci. 2007;32:457–463. doi: 10.1007/s12038-007-0045-9. [DOI] [PubMed] [Google Scholar]
- 24.Tuttle AH, Philip VM, Chesler EJ, Mogil JS. Comparing phenotypic variation between inbred and outbred mice. Nat. Methods. 2018;15:994–996. doi: 10.1038/s41592-018-0224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kentner, A. C., Speno, A. V., Doucette, J. & Roderick, R. C. The contribution of environmental enrichment to phenotypic variation in mice and rats. eNeuro10.1523/ENEURO.0539-20.2021 (2021). [DOI] [PMC free article] [PubMed]
- 26.Eisen EJ, Nagai J, Bakker H, Hayes JF. Effect of litter size at birth on lactation in mice. J. Anim. Sci. 1980;50:680–688. doi: 10.2527/jas1980.504680x. [DOI] [PubMed] [Google Scholar]
- 27.Li L, Lu X, Dean J. The maternal to zygotic transition in mammals. Mol. Asp. Med. 2013;34:919–938. doi: 10.1016/j.mam.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soubry A. Epigenetic inheritance and evolution: a paternal perspective on dietary influences. Prog. Biophys. Mol. Biol. 2015;118:79–85. doi: 10.1016/j.pbiomolbio.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Stäubli A, Peters AHFM. Mechanisms of maternal intergenerational epigenetic inheritance. Curr. Opin. Genet. Dev. 2021;67:151–162. doi: 10.1016/j.gde.2021.01.008. [DOI] [PubMed] [Google Scholar]
- 30.Whitelaw NC, et al. Reduced levels of two modifiers of epigenetic gene silencing, Dnmt3a and Trim28, cause increased phenotypic noise. Genome Biol. 2010;11:R111. doi: 10.1186/gb-2010-11-11-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joseph R, Dou D, Tsang W. Molecular cloning of a novel mRNA (neuronatin) that is highly expressed in neonatal mammalian brain. Biochem. Biophys. Res. Commun. 1994;201:1227–1234. doi: 10.1006/bbrc.1994.1836. [DOI] [PubMed] [Google Scholar]
- 32.Joseph R, Dou D, Tsang W. Neuronatin mRNA: alternatively spliced forms of a novel brain-specific mammalian developmental gene. Brain Res. 1995;690:92–98. doi: 10.1016/0006-8993(95)00621-V. [DOI] [PubMed] [Google Scholar]
- 33.Braun JL, Geromella MS, Hamstra SI, Fajardo VA. Neuronatin regulates whole-body metabolism: is thermogenesis involved? FASEB BioAdvances. 2020;2:579–586. doi: 10.1096/fba.2020-00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joe MK, et al. Crucial roles of neuronatin in insulin secretion and high glucose-induced apoptosis in pancreatic e-cells. Cell. Signal. 2008;20:907–915. doi: 10.1016/j.cellsig.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Rodríguez-Comas J, et al. Stress-Induced MicroRNA-708 Impairs 08.01.005ion and high gluc. Diabetes. 2017;66:3029–3040. doi: 10.2337/db16-1569. [DOI] [PubMed] [Google Scholar]
- 36.Millership SJ, et al. Neuronatin regulates pancreatic β cell insulin content and secretion. J. Clin. Investig. 2018;128:3369–3381. doi: 10.1172/JCI120115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suh YH, et al. Ectopic expression of neuronatin potentiates adipogenesis through enhanced phosphorylation of cAMP-response element-binding protein in 3T3-L1 cells. Biochem. Biophys. Res. Commun. 2005;337:481–489. doi: 10.1016/j.bbrc.2005.09.078. [DOI] [PubMed] [Google Scholar]
- 38.Gburcik V, Cleasby ME, Timmons JA. Loss of neuronatin promotes “browning” of primary mouse adipocytes while reducing Glut1-mediated glucose disposal. Am. J. Physiol. Endocrinol. Metab. 2013;304:E885–E894. doi: 10.1152/ajpendo.00463.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ka HI, et al. Neuronatin is associated with an anti-inflammatory role in the white adipose tissue. J. Microbiol. Biotechnol. 2017;27:1180–1188. doi: 10.4014/jmb.1702.02049. [DOI] [PubMed] [Google Scholar]
- 40.Vrang N, et al. The imprinted gene neuronatin is regulated by metabolic status and associated with obesity. Obesity. 2010;18:1289–1296. doi: 10.1038/oby.2009.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Millership SJ, et al. Neuronatin deletion causes postnatal growth restriction and adult obesity in 129S2/Sv mice. Mol. Metab. 2018;18:97–106. doi: 10.1016/j.molmet.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cimino I, et al. Murine neuronatin deficiency is associated with a hypervariable food intake and bimodal obesity. Sci. Rep. 2021;11:17571. doi: 10.1038/s41598-021-96278-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dalgaard K, et al. Trim28 haploinsufficiency triggers bi-stable epigenetic obesity. Cell. 2016;164:353–364. doi: 10.1016/j.cell.2015.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lenzen S. The mechanisms of alloxan- and streptozotocin-induced diabetes. Diabetologia. 2008;51:216–226. doi: 10.1007/s00125-007-0886-7. [DOI] [PubMed] [Google Scholar]
- 45.Simola Daniel F, et al. Epigenetic (re)programming of caste-specific behavior in the ant Camponotus floridanus. Science. 2016;351:aac6633. doi: 10.1126/science.aac6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glastad KM, et al. Epigenetic regulator CoREST controls social behavior in ants. Mol. Cell. 2020;77:338–351. doi: 10.1016/j.molcel.2019.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li, Y. & Seto, E. HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harbor Persp. Med.10.1101/cshperspect.a026831 (2016). [DOI] [PMC free article] [PubMed]
- 48.Tellegen A, et al. Personality similarity in twins reared apart and together. J. Personal. Soc. Psychol. 1988;54:1031–1039. doi: 10.1037/0022-3514.54.6.1031. [DOI] [PubMed] [Google Scholar]
- 49.Bouchard TJ, Jr., Lykken DT, McGue M, Segal NL, Tellegen A. Sources of human psychological differences: the Minnesota Study of twins reared apart. Science. 1990;250:223–228. doi: 10.1126/science.2218526. [DOI] [PubMed] [Google Scholar]
- 50.Stunkard AJ, Harris JR, Pedersen NL, McClearn GE. The body mass index of twins who have been reared apart. N. Engl. J. Med. 1990;322:1483–1487. doi: 10.1056/NEJM199005243222102. [DOI] [PubMed] [Google Scholar]
- 51.Hochberg Z, et al. Child health, developmental plasticity, and epigenetic programming. Endocr. Rev. 2011;32:159–224. doi: 10.1210/er.2009-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patti M-E. Intergenerational programming of metabolic disease: evidence from human populations and experimental animal models. Cell. Mol. Life Sci. 2013;70:1597–1608. doi: 10.1007/s00018-013-1298-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaminsky ZA, et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat. Genet. 2009;41:240–245. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- 54.Gordon L, et al. Neonatal DNA methylation profile in human twins is specified by a complex interplay between intrauterine environmental and genetic factors, subject to tissue-specific influence. Genome Res. 2012;22:1395–1406. doi: 10.1101/gr.136598.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spector TD, Williams FM. The UK Adult Twin Registry (TwinsUK) Twin Res. Hum. Genet. 2006;9:899–906. doi: 10.1375/twin.9.6.899. [DOI] [PubMed] [Google Scholar]
- 56.Nica AC, et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER Study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grundberg E, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berntzen BJ, et al. Eating behaviors in healthy young adult twin pairs discordant for body mass index. Twin Res. Hum. Genet. 2019;22:220–228. doi: 10.1017/thg.2019.43. [DOI] [PubMed] [Google Scholar]
- 60.Pietiläinen KH, et al. Global transcript profiles of fat in monozygotic twins discordant for BMI: pathways behind acquired obesity. PLoS Med. 2008;5:e51. doi: 10.1371/journal.pmed.0050051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poulsen P, et al. Increased risk of type 2 diabetes in elderly twins. Diabetes. 2009;58:1350. doi: 10.2337/db08-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bell JT, Saffery R. The value of twins in epigenetic epidemiology. Int. J. Epidemiol. 2012;41:140–150. doi: 10.1093/ije/dyr179. [DOI] [PubMed] [Google Scholar]
- 63.Castillo-Fernandez JE, Spector TD, Bell JT. Epigenetics of discordant monozygotic twins: implications for disease. Genome Med. 2014;6:60. doi: 10.1186/s13073-014-0060-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Geserick M, et al. Acceleration of BMI in early childhood and risk of sustained obesity. N. Engl. J. Med. 2018;379:1303–1312. doi: 10.1056/NEJMoa1803527. [DOI] [PubMed] [Google Scholar]
- 65.Landgraf K, et al. Evidence of early alterations in adipose tissue biology and function and its association with obesity-related inflammation and insulin resistance in children. Diabetes. 2015;64:1249. doi: 10.2337/db14-0744. [DOI] [PubMed] [Google Scholar]
- 66.Newman AM, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emont MP, et al. A single-cell atlas of human and mouse white adipose tissue. Nature. 2022;603:926–933. doi: 10.1038/s41586-022-04518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Galcheva, S., Demirbilek, H., Al-Khawaga, S. & Hussain, K. The genetic and molecular mechanisms of congenital hyperinsulinism. Front. Endocrinol.10.3389/fendo.2019.00111 (2019). [DOI] [PMC free article] [PubMed]
- 69.Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mihaylova MM, Shaw RJ. Metabolic reprogramming by class I and II histone deacetylases. Trends Endocrinol. Metab. 2013;24:48–57. doi: 10.1016/j.tem.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Emmett MJ, Lazar MA. Integrative regulation of physiology by histone deacetylase 3. Nat. Rev. Mol. Cell Biol. 2019;20:102–115. doi: 10.1038/s41580-018-0076-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Christensen DP, et al. Histone deacetylase (HDAC) Inhibition as a novel treatment for diabetes mellitus. Mol. Med. 2011;17:378–390. doi: 10.2119/molmed.2011.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ling C, Rönn T. Epigenetics in human obesity and type 2 diabetes. Cell Metab. 2019;29:1028–1044. doi: 10.1016/j.cmet.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vetere A, Choudhary A, Burns SM, Wagner BK. Targeting the pancreatic β-cell to treat diabetes. Nat. Rev. Drug Discov. 2014;13:278–289. doi: 10.1038/nrd4231. [DOI] [PubMed] [Google Scholar]
- 75.Blewitt ME, et al. An N-ethyl-N-nitrosourea screen for genes involved in variegation in the mouse. PNAS. 2005;102:7629–7634. doi: 10.1073/pnas.0409375102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bhardwaj V, et al. snakePipes: facilitating flexible, scalable and integrative epigenomic analysis. Bioinformatics. 2019;35:4757–4759. doi: 10.1093/bioinformatics/btz436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sergushichev, A. A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. Preprint at bioRxiv10.1101/060012 (2016).
- 79.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stranger BE, et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 82.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scrucca L, Fop M, Murphy TB, Raftery AE. mclust 5: clustering, classification and density estimation using Gaussian finite mixture models. R J. 2016;8:289–317. doi: 10.32614/RJ-2016-021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grundberg E, et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am. J. Hum. Genet. 2013;93:876–890. doi: 10.1016/j.ajhg.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Busche S, et al. Population whole-genome bisulfite sequencing across two tissues highlights the environment as the principal source of human methylome variation. Genome Biol. 2015;16:290. doi: 10.1186/s13059-015-0856-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Triche TJ, Jr., Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res. 2013;41:e90. doi: 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou W, Triche TJ, Jr., Laird PW, Shen H. SeSAMe: reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucleic Acids Res. 2018;46:e123. doi: 10.1093/nar/gky691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 89.Herman JS, Sagar, Grün D. FateID infers cell fate bias in multipotent progenitors from single-cell RNA-seq data. Nat. Methods. 2018;15:379–386. doi: 10.1038/nmeth.4662. [DOI] [PubMed] [Google Scholar]
- 90.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kromeyer-Hauschild K, et al. Perzentile für den body mass index für das kindes und jugendalter unter heranziehung verschiedener Deutscher stichproben. Monatsschrift Kinderheilkd. 2001;149:807–818. doi: 10.1007/s001120170107. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables 1 and 2
Data Availability Statement
RNA-seq data from both mouse primary islets and subcutaneous adipose tissue of the LCAT cohort, have been deposited to Gene Expression Omnibus and are publicly available under the accession codes GSE205740 and GSE205668, respectively. They are collected under the GSE205741 super-series. Gene expression and DNA methylation profiles by whole genome arrays from subcutaneous adipose tissue of the MuTHER TwinsUK cohort have been deposited to Array Express and are publicly available under the accession codes E-TABM-1140 and E-TABM-1866, respectively. Morphometric and genotypic data of the MuTHER TwinsUK cohort are available upon request at https://twinsuk.ac.uk/resources-for-researchers/access-our-data/. The MSigDB is available at http://www.gsea-msigdb.org/gsea/msigdb. Source data are provided with this paper.
No custom code or mathematical algorithm were generated in this report. All publicly available codes and tools used to analyze the data are reported and referenced in the Methods and the Reporting Summary.