Summary
Numerous proteins experience and respond to mechanical forces as an integral part of their cellular functions, but measuring these forces remains a practical challenge. Here, we present a compact, 11-kDa molecular tension sensor termed STReTCh (sensing tension by reactive tag characterization). Unlike existing genetically encoded tension sensors, STReTCh does not rely on experimentally demanding measurements based on Förster resonance energy transfer and is compatible with typical fix-and-stain protocols. Using a magnetic tweezers assay, we calibrate the STReTCh module and show that it responds to physiologically relevant, piconewton forces. As proof of concept, we use an extracellular STReTCh-based sensor to visualize cell-generated forces at integrin-based adhesion complexes. In addition, we incorporate STReTCh into vinculin, a cytoskeletal adaptor protein, and show that STReTCh reports on forces transmitted between the cytoskeleton and cellular adhesion complexes. These data illustrate the utility of STReTCh as a tool for visualizing molecular-scale forces in biological systems.
Keywords: mechanobiology, biosensor, cytoskeleton, cell adhesion, mechanical force
Graphical abstract

Highlights
-
•
STReTCh is a genetically encoded sensor for detecting molecular-scale forces
-
•
STReTCh visualizes forces using standard immunofluorescence protocols
-
•
STReTCh is applied to detect forces present at adhesion complexes in mammalian cells
Motivation
Visualizing the molecular-scale forces present in living cells can provide insights into numerous biological processes, but existing tools for force measurement can be challenging to use. To reduce barriers to measuring molecular forces in biological systems, we designed a molecular tension sensor, termed STReTCh, whose use follows common “fix-and-stain” workflows, making it broadly accessible to the cell biological community.
Zhong et al. describe STReTCh, a genetically encoded force-sensing module that provides a readout of molecular forces in biological systems. STReTCh relies on typical “fix-and-stain” immunofluorescence protocols, making it broadly accessible.
Introduction
The ability of living cells to generate and sense molecular-scale mechanical forces is crucial to embryonic development, cell motility, and other biological processes. Nanoscale mechanical forces such as those produced by molecular motors typically range from single to tens of piconewtons (pN) (Borghi et al., 2012; Chang et al., 2016; Finer et al., 1994; Grashoff et al., 2010; Tan et al., 2020; Yao et al., 2016). Existing methods for characterizing forces experienced by proteins in intact cells rely on Förster resonance energy transfer (FRET) or similar phenomena, where energy transfer between a pair of fluorophores flanking a molecular spring of known force constant is used to infer the extension of the spring and the force across the tension sensor (Grashoff et al., 2010; LaCroix et al., 2015; Morimatsu et al., 2013). While these sensors have yielded important biological insights (Borghi et al., 2012; Chang et al., 2016; Grashoff et al., 2010; Morimatsu et al., 2013; Tan et al., 2020), quantitative FRET measurements require specialized equipment, involved analysis techniques, and careful control measurements (Fischer et al., 2021), factors that limit the broad accessibility of such sensors. Consequently, existing molecular tension sensors are often used for qualitative assessments of relative forces. Moreover, FRET-based tension-sensing modules are relatively large at ∼60 kDa in size, increasing the likelihood that their insertion may disrupt the function of the host protein. DNA hairpin-based tension sensors (Wang et al., 2016; Zhang et al., 2014) represent another well-established modality for high-precision measurements of molecular mechanical forces, but such sensors are not currently viable for in-cell measurements.
Here, we present an 11-kDa molecular tension sensor module, which we term STReTCh (sensing tension by reactive tag characterization), that is sensitive to forces in the single-pN range and whose use follows typical fix-and-stain protocols. We characterize the biophysical behavior of STReTCh under mechanical force and demonstrate the utility of STReTCh as a sensor of biologically relevant forces, both at cell-substrate interfaces and inside cells.
Results
Design and characterization of the STReTCh sensor
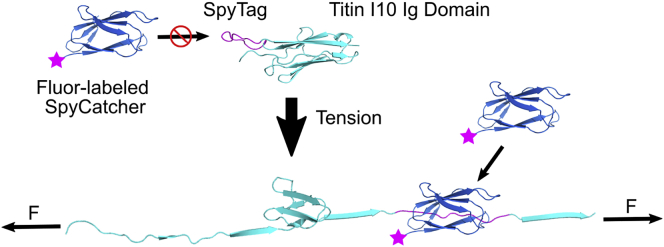
As a general approach, we sought to embed an epitope tag into a larger mechanosensitive domain, so the tag could not be recognized by its binding partner unless the domain was unfolded. We reasoned that the I10 immunoglobulin-like domain from human titin, which reversibly folds and unfolds at ∼6 pN on the seconds timescale (Rivas-Pardo et al., 2016), could serve as the basis for a sensor of the pN-scale forces across individual proteins inside of cells. Specifically, we inserted the 13-amino acid SpyTag in the C-terminal loop of titin I10 to generate the STReTCh tension sensor (Figure 1). SpyTag forms a covalent, isopeptide bond with its binding partner SpyCatcher (Li et al., 2014; Zakeri et al., 2012). Thus, a fluorescently labeled SpyCatcher can be used as a labeling agent for the force-unfolded STReTCh sensor in a similar manner as antibodies for immunocytochemistry. In designing STReTCh, we tested whether insertion of SpyTag into each of the six loops of titin I10 could yield a candidate for a force sensor. However, only insertion of SpyTag into the loop closest to the C terminus of titin I10 (see STAR Methods) yielded a protein that was well expressed in BL21 E. coli (data not shown). Consequently, we moved forward with characterization and validation of this construct as the STReTCh sensor.
Figure 1.
Design and operation of the STReTCh tension-sensing module
SpyTag is inserted into the Titin I10 domain, and is only recognized by SpyCatcher when STReTCh is unfolded under tension. Unfolded STReTCh can be visualized using fluorophore-labeled SpyCatcher. An illustrative representation of STReTCh was generated using RosettaRemodel structure prediction based on the Titin I10 crystal structure (PDB: 4QEG).
Initial in vitro characterization of the STReTCh sensor revealed a lower melting temperature than that of the unmodified titin I10 domain (Figure S1C), though this melting temperature was still higher than physiological temperatures (i.e., 37°C). Nevertheless, we anticipated this destabilization could reflect different force-dependent behavior of STReTCh compared with what has been reported for titin I10 (Rivas-Pardo et al., 2016). To better characterize the unfolding behavior of STReTCh under tension, we used magnetic tweezers to apply tension in the single-pN range to individual surface-tethered STReTCh molecules (Figure 2A). Individual STReTCh molecules exhibited reversible, steplike unfolding transitions of ∼10–15 nm (Figure 2B) between folded and unfolded states, consistent with previous characterizations of titin I10 unfolding (Rivas-Pardo et al., 2016). These transitions occurred at forces below 2 pN, consistent with the lower observed melting temperature relative to titin I10 (Figure 2B). The fractions of time spent in the folded state for STReTCh molecules at various forces are plotted in Figure 2C, where the size of each data point corresponds to the total duration of time over which the molecule was observed. We fit the folded and unfolded state lifetimes measured using magnetic tweezers to a Bell-Evans kinetic model (Bell, 1978; Evans and Ritchie, 1997) to obtain a maximum-likelihood estimate of the threshold unfolding force for STReTCh, which we found to be ∼1 pN. Extrapolation of single-molecule measurements suggests that in the absence of force, STReTCh is in a folded state ∼95% of the time, which allows for effective distinction between loaded and non-loaded STReTCh molecules. STReTCh is thus among the most sensitive molecular force sensors described to date, and it is capable of responding to the full range of biologically relevant, molecular-scale forces, which span 2 to ∼100 pN (Borghi et al., 2012; Brockman et al., 2020; Chang et al., 2016; Grashoff et al., 2010; Morimatsu et al., 2013; Tan et al., 2020; Yusko and Asbury, 2014; Zhang et al., 2014).
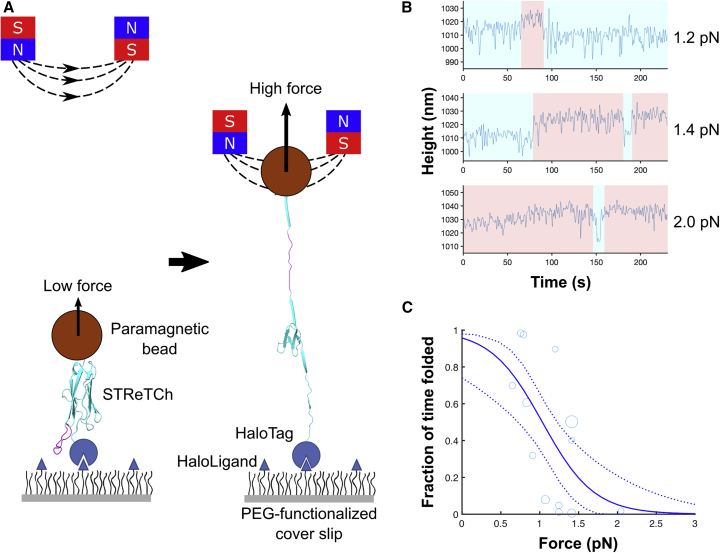
Figure 2.
The STReTCh module is largely unfolded at forces above 1 pN
(A) Single-molecule magnetic tweezers assay (not to scale). STReTCh molecules are covalently tethered to a microscope cover slip and bound at the other end to paramagnetic beads. Forces in the pN range are applied via a magnetic field gradient created by two permanent magnets, with increasing force as the magnets are brought closer to the sample (see Figures S1A and S1B).
(B) Sample traces of STReTCh under fixed levels of tension. STReTCh exhibits steplike changes in length of ∼10–15 nm at forces of 1–2 pN. Periods of time when the molecule is folded and unfolded are shaded in blue and red, respectively. Data shown were acquired at 50 Hz and filtered with a 7th order Butterworth filter with a cutoff frequency of 0.5 Hz.
(C) Unfolding behavior of STReTCh under mechanical force suggests that STReTCh transitions from primarily folded to unfolded at 1 pN. Each data point represents one trace acquired at the corresponding force, with the total dataset drawn from seven distinct molecules measured across six independent experiments. The size of each marker is proportional to the total duration of the corresponding trace. The solid curve represents the maximum likelihood estimate fit of the data to the Bell-Evans model, with 95% confidence intervals represented by the dotted curves. Parameters for the Bell-Evans model and corresponding confidence intervals are listed in Table S1. See also Figure S1.
Measuring extracellular integrin-ECM forces using STReTCh
As a functional test of the STReTCh sensor, we evaluated whether STReTCh could detect forces generated at integrin-based cellular adhesions, hereafter generically termed focal adhesions (FAs), to the extracellular matrix (ECM) (Hoffman et al., 2011; Kanchanawong et al., 2010). We seeded human foreskin fibroblasts (HFFs) transfected with focal adhesion marker GFP-paxillin (GFP-Pxn) onto a substrate functionalized with the STReTCh sensor fused to a C-terminal fibronectin-derived RGD peptide that mediates integrin-based adhesion (Pierschbacher and Ruoslahti, 1984). We hypothesized that FA-localized forces exerted by HFFs would preferentially unfold STReTCh at FAs. After allowing the HFFs to adhere to the substrate, we treated the HFFs with 0.5% paraformaldehyde (PFA) prior to imaging to minimize movement of cells during imaging, and then we “stained” the sample with a truncated, minimal SpyCatcher (Li et al., 2014) labeled with Alexa Fluor 647 (647-SC) for 10 min to visualize unfolded sensors and obtain a snapshot of the force distribution. Following washout of 647-SC, the cells were imaged at 37°C using total internal reflection fluorescence (TIRF) microscopy (Figures 3A and 3B).
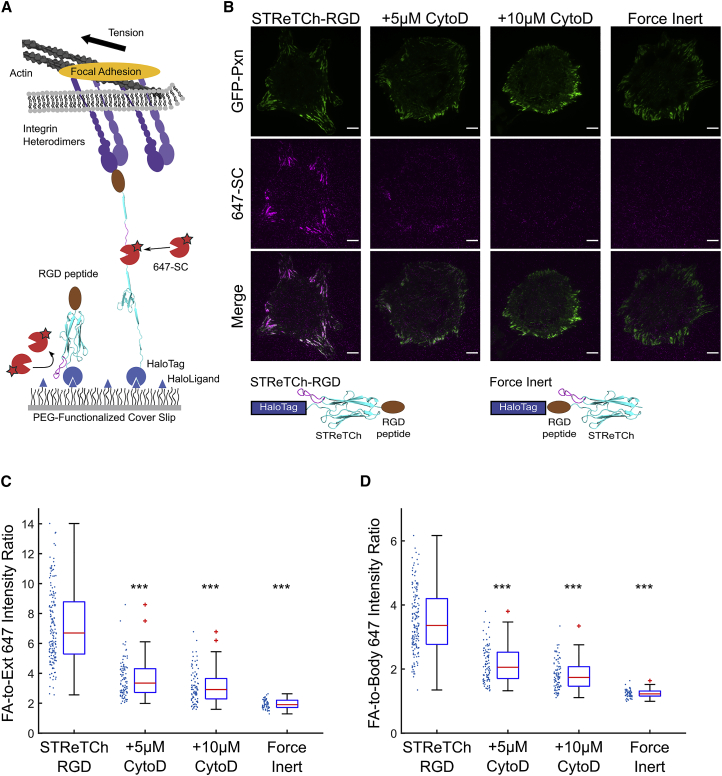
Figure 3.
STReTCh detects cell-adhesive forces across integrin-RGD bonds
(A) Experimental setup for extracellular force assay with STReTCh. Cells are seeded on a surface functionalized with STReTCh fused to an RGD ligand. Sensors under tension are visualized using SpyCatcher labeled with Alexa Fluor 647.
(B) Representative images of GFP-paxillin and 647-SC for HFFs adhering to STReTCh-RGD in the absence and presence of cytochalasin D, and for HFFs adhering to a force-inert construct. Scale bars, 10 μm.
(C) Ratio of the average 647-SC intensity within FAs compared with background signal outside of cells for conditions described in (B). ∗∗∗ denotes p < 0.001 compared with STReTCh-RGD (two-tailed Mann-Whitney). Red lines indicate medians. Top and bottom of blue boxes indicate 75th and 25th percentiles, respectively, black bars indicate range, excluding outliers, and outliers are plotted as red plus signs. All subsequent boxplots follow this convention. Values for individual cells are plotted to the left of each bar.
(D) Ratio of average 647-SC intensity within focal adhesions and average intensity outside of focal adhesions but underneath cell bodies. ∗∗∗ denotes p < 0.001 compared with STReTCh-RGD (two-tailed Mann-Whitney). N = 162 cells for STReTCh-RGD, 95 for 5 μM CytoD, 95 for 10 μM CytoD, and 59 for the force-inert sensor. Data for each condition are pooled from a minimum of three independent experiments. See also Figures S2, S3, and S7.
We observed strong colocalization of 647-SC and GFP-Pxn, indicative of 647-SC recruitment to FAs. As a control for staining in the absence of force, we seeded HFFs on a substrate coated with a force-inert variant of the fusion protein in which the STReTCh domain is C-terminal to the RGD peptide. Cells seeded on this variant can adhere to the surface through the RGD ligand, but tension is not transduced through the STReTCh domain (Morimatsu et al., 2013). As anticipated, we observed minimal staining in the force-inert control (Figures 3B–3D). Quantification of the average Alexa Fluor 647 fluorescence intensity inside of FAs compared with the intensity outside of the cell yielded a signal-to-background ratio of ∼7:1 (Figures 3C and S7A). Overall signal intensities increased with increasing concentrations of 647-SC added (Figures S2A–S2B). Staining of STReTCh-RGD for HFFs imaged live, without fixation, yielded similar results (Figure S2C). The 647-SC signal at FAs was reduced in cells treated with cytochalasin D, which disrupts actin-mediated cytoskeletal tension in a concentration-dependent manner (Figures 3C, 3D, and S7A). Thus, upon reduction of forces in a functional context, STReTCh refolds to obscure SpyTag, as expected from our calibration experiments. These observations indicate that STReTCh robustly detects extracellular forces across individual integrins.
Measuring intracellular forces across vinculin using STReTCh
After validating the functionality of STReTCh as an extracellular force sensor, we asked whether STReTCh could be genetically encoded to detect intracellular tension. In the experiments described above, 647-SC was added to cells after light fixation, but prior to permeabilization. Control experiments revealed some nonspecific sticking of 647-SC in fully fixed and permeabilized cells, including low levels of recruitment to FAs that presumably reflects the interaction of 647-SC with one or more cytoskeletal proteins (Figure S3). We took advantage of the covalent nature of the SpyTag-SpyCatcher interaction and incorporated more stringent post-staining wash steps with EDTA and guanidine-HCl (Gdn-HCl), which reduced this nonspecific staining (Figure S4).
To demonstrate the ability of STReTCh to measure intracellular tension, we inserted STReTCh into vinculin, a protein that reinforces connections between cell adhesion complexes and the actin cytoskeleton (Grashoff et al., 2010; Hu et al., 2007). Previous measurements found that vinculin on average bears ∼2.5 pN of force at FAs in mouse embryonic fibroblasts (MEFs) (Grashoff et al., 2010). In analogy to previous FRET-based tension sensors (Aird et al., 2020; Grashoff et al., 2010), we inserted the STReTCh sensor between the vinculin head and tail domains in a flexible linker region known to be mechanosensitive. We further tagged this fusion protein with EGFP to quantify the expression levels and localization of the sensor construct and denote this construct Vin-STReTCh (Figure 4A). As a force-inert control, we placed STReTCh at the C terminus of the GFP-vinculin fusion protein (Vin-STReTCh-CT), so STReTCh would not bear tension (Figure 4A). We stably transduced vinculin-null MEFs with these vinculin constructs and seeded these MEFs on fibronectin-coated coverslips. The MEFs were then fixed, permeabilized, blocked with bovine serum albumin, and incubated with 647-SC. Note that previous measurements show that measurable cytoskeletal tension persists up to 1 h or more after fixation and permeabilization (Brockman et al., 2020; Chang et al., 2016), a timescale that is compatible with labeling with 647-SC. Finally, cells were washed with 2 mM EDTA and 3 M Gdn-HCl to remove residual, nonspecifically adsorbed 647-SC. These additional wash steps were particularly useful in reducing nonspecific sticking in the context of visualizing intracellular mechanical forces (Figure S4).
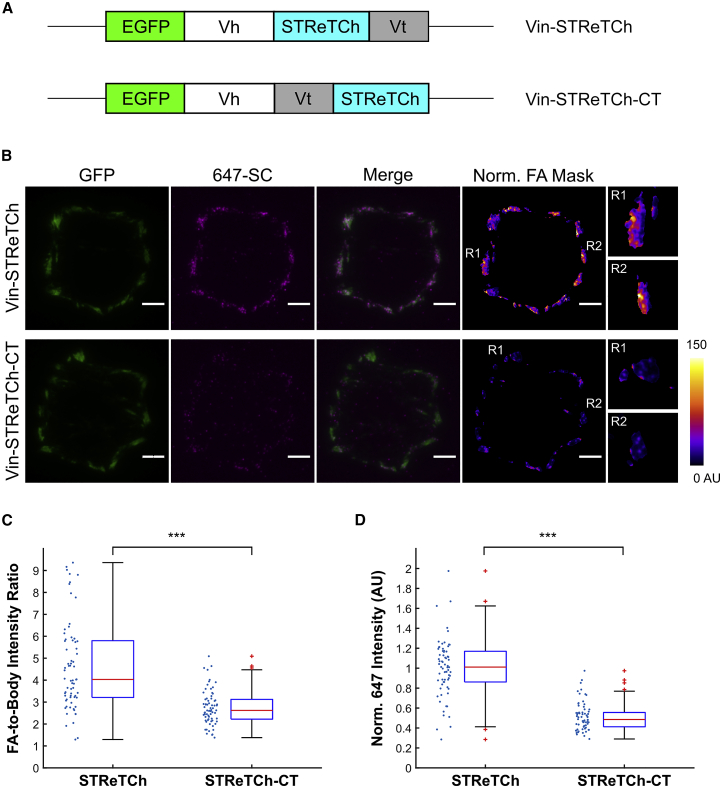
Figure 4.
STReTCh detects intracellular tension across vinculin
(A) Constructs for vinculin-STReTCh (Vin-STReTCh) and a C-terminal force-inert control (Vin-STReTCh-CT). Vh denotes vinculin head. Vt denotes vinculin tail.
(B) Representative images of GFP, 647 intensity, and 647 intensity normalized by GFP intensity within FAs for MEFs expressing Vin-STReTCh and Vin-STReTCh-CT and fixed and stained with 647-SC. Scale bars, 5 μm. R1 and R2 panels depict enlarged images of normalized 647 intensity for selected representative adhesions.
(C) Ratios of 647-SC intensity within FAs to the average intensity outside of FAs but within the cell boundary for MEFs expressing Vin-STReTCh and Vin-STReTCh-CT. ∗∗∗p < 0.001 by two-tailed Mann-Whitney.
(D) Quantification of 647 intensity normalized by GFP intensity within FAs for Vin-STReTCh versus Vin-STReTCh-CT MEFs. N = 68 cells for STReTCh and 70 for STReTCh-CT, and data for each condition are pooled from three independent experiments. ∗∗∗ denotes p < 0.001 by two-tailed Mann-Whitney. See also Figures S4–S7.
TIRF images of the fixed and stained Vin-STReTCh and Vin-STReTCh-CT MEFs are presented in Figure 4B. The 647-SC signal was increased in the FAs of Vin-STReTCh MEFs compared with Vin-STReTCh-CT MEFs, indicative of force-dependent exposure and labeling of the SpyTag sequence (Figures 4B–4D, S4, and S7B). Both the absolute and GFP-normalized 647-SC signal within FAs indicated higher degrees of 647-SC staining in Vin-STReTCh compared with Vin-STReTCh-CT (Figures 4B–4D). Expression levels of vinculin-STReTCh fusion proteins were relatively well matched in both populations of cells, and 647-SC signal was higher in Vin-STReTCh MEFs irrespective of the abundance of the vinculin fusion proteins at individual FAs (Figures S4D and S5). Additionally, STReTCh inserted in vinculin was sensitive to loss of cytoskeletal tension upon treating MEFs with the ROCK inhibitor Y-27632, but STReTCh appended C-terminally was not (Figure S6). Overall, our observations on the behavior of STReTCh in Vin-STReTCh and Vin-STReTCh-CT MEFs demonstrate that STReTCh can be inserted into proteins of interest as a genetically encoded sensor to provide a readout of intracellular tension.
Discussion
We engineered an 11-kDa tension-sensing protein module, termed STReTCh, by inserting SpyTag into the human titin I10 domain, so SpyTag is labeled when the I10 domain undergoes force-dependent unfolding. The smaller size of the STReTCh module compared with existing sensors may reduce the likelihood that STReTCh would perturb biological function when inserted into a protein of interest. We used magnetic tweezers to characterize the unfolding behavior of STReTCh, tethered at one end to a glass surface and the other to a magnetic bead, thus mimicking the pulling geometry experienced by STReTCh when inserted into a protein of interest. STReTCh undergoes reversible unfolding at 1–2 pN of applied force, making it among the most sensitive molecular tension sensors of which we are currently aware. The extension length of ∼10–15 nm that is associated with the unfolding of STReTCh is comparable to length changes of previously validated FRET-based force sensors that make use of protein conformational changes (Ringer et al., 2017), as well as length changes of endogenous mechanosensitive proteins such α-catenin (Yao et al., 2014), talin (Yao et al., 2016), and integrins (Li and Springer, 2017). Single-molecule magnetic tweezers data indicate that STReTCh is ∼95% folded in the absence of external load (Figure 2). As expected, recruitment of SpyCatcher to STReTCh required active force generation by the cell cytoskeleton (Figures 3 and 4), indicating that the STReTCh reports on cell-generated forces. These properties enable STReTCh to robustly detect physiologically relevant mechanical forces in biological systems.
We demonstrated the ability of STReTCh to detect biologically relevant forces associated with cell-ECM adhesion, both intra- and extracellularly. However, the properties of STReTCh make it potentially well suited for measuring forces at other structures, for example at cadherin-based adherens junctions. More broadly, the cryptic tag binding strategy of STReTCh represents a conceptually distinct paradigm for visualizing molecular-scale forces, in contrast to many existing molecular force measurement tools, which primarily rely on energy transfer phenomena. Thus, the experiments reported here represent a proof of concept that may stimulate further development of new force sensors that further increase the accessibility of molecular force measurements in biological systems.
Limitations of the study
Like any laboratory tool, STReTCh has limitations that should be considered when evaluating its appropriateness for a given measurement. The background signal due to nonspecific interaction of SpyCatcher with intracellular proteins currently limits signal to noise of the current STReTCh sensor. Optimization of SpyCatcher to reduce nonspecific interactions or implementation of alternative tag/binder pairs may address this limitation. From a practical standpoint, the signal-to-background ratio will depend on the concentration of SpyCatcher and duration of the staining step. Thus, we recommend that STReTCh users optimize staining concentration and time (e.g., perform serial dilution, time sweeps, etc.) to maximize the signal contrast between the force signal of the protein of interest and an unloaded negative control. Practically speaking, this corresponds to identifying labeling conditions so signal in the unloaded negative control is at the edge of detection of the user’s optical setup, as this corresponds roughly to conditions that generate maximal signal from mechanically unfolded STReTCh with minimal detectable background signal (see Methods S1). Unlike some FRET-based sensors (Chang et al., 2016; Grashoff et al., 2010; Ringer et al., 2017; Tan et al., 2020), STReTCh is not meant to yield quantitative force measurements. However, quantitative force measurements are arguably required in only a minority of situations. For this reason, we anticipate that the ability of STReTCh to report on forces above a biologically meaningful force of ∼2 pN will make it a useful addition to the available suite of tools for force measurement.
While we expect that the small size of STReTCh will reduce the likelihood of perturbing biological function, users should validate that protein function is not disrupted when inserting STReTCh into new proteins of interest and identify optimal sites of insertion. The current version of STReTCh is not compatible with live-cell measurements using intracellular probes, though it is sufficiently sensitive to measure residual intracellular forces in fixed cells, which persist approximately 1 h after fixation (Brockman et al., 2020; Chang et al., 2016). Live-cell force measurements are possible for extracellular probes, though we note that short incubation times with SpyCatcher are required to capture a close-to-instantaneous measurement of cell-generated forces (Figure S2C). Live, intracellular force measurements may potentially be addressed by future versions of STReTCh in which the interaction of SpyCatcher and SpyTag (or other tag/binder pair) is reversible rather than covalent.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| BL21(DE3) chemically competent E. coli | New England Biolabs | C2527H |
| Chemicals, peptides, and recombinant proteins | ||
| DNase I, recombinant | Roche | Cat# 04536282001 |
| EcoRI-HF | New England Biolabs | Cat# R3101 |
| BamHI-HF | New England Biolabs | Cat# R3136 |
| Biotin-PEG3-CoenzymeA | SiChem | Cat# SC-8618 |
| SYPRO Orange | ThermoFisher | Cat# S6650 |
| Lambda phage DNA | New England Biolabs | Cat# N3011 |
| HaloTag Succinimidyl Ester (O4) Ligand | Promega | Cat# P6751 |
| Sulfosuccinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate | ThermoFisher | Cat# A39268 |
| 5% casein in water | Sigma-Aldrich | Cat# C4765 |
| 16% paraformaldehyde | Electron Microscopy Sciences | Cat# 15710 |
| Cytochalasin D | Enzo Life Sciences | Cat# BMLT1090001 |
| Bovine serum albumin | Sigma Aldrich | Cat# A9418 |
| Fibronectin | Corning | Cat# 356008 |
| Y-27632 | STEMCELL Technologies | Cat# 72304 |
| Critical commercial assays | ||
| HisPur Ni-NTA Resin | Thermo Scientific | Cat# 88221 |
| Experimental models: Cell lines | ||
| GFP-paxillin human foreskin fibroblasts | Morimatsu et al., 2015 | N/A |
| Human foreskin fibroblasts, CCD-1070Sk | ATCC | Cat# CRL-2091 |
| Vinculin-null mouse embryonic fibroblasts | Rothenberg et al., 2018 | N/A |
| EGFP-Vinculin-STReTCh mouse embryonic fibroblasts | This study | N/A |
| EGFP-Vinculin-STReTCh-CT mouse embryonic fibroblasts | This study | N/A |
| Oligonucleotides | ||
| Primer: STReTCh Gibson Insert Fwd: GTTGGAGATCAGCGGCGGTGCTG GCGAGTTCATGGAAACTCTGCATAT TACCAAAACC |
Elim Biopharmaceuticals | N/A |
| Primer: STReTCh Gibson Insert Rev: TACCTTGCTCGAAGTACAGATTCTCG GAAGTCACGGTCAGGGTCGC |
Elim Biopharmaceuticals | N/A |
| Primer: SpyCatcher Cys SDM Fwd: CATCACCATCACCATCACGATTGCG ACATCCCAAC |
Elim Biopharmaceuticals | N/A |
| Primer: SpyCatcher Cys SDM Rev: GTTGGGATGTCGCAATCGTGATG GTGATGGTGATG |
Elim Biopharmaceuticals | N/A |
| Primer: pDEST Vector amplify Fwd: TAAATGGTTGATGCTTGAGGATC | Elim Biopharmaceuticals | N/A |
| Primer: pDEST Vector amplify Rev: TTTGAGAATTTAATATGGGTAGCAC TATCGCCCTGAAAATACAGGTTTTC |
Elim Biopharmaceuticals | N/A |
| Primer: SCMin fragment amplify Fwd: GATAGTGCTACCCATATTAAATTC | Elim Biopharmaceuticals | N/A |
| Primer: SCMin fragment amplify Rev: GATCCTCAAGCATCAACCATTTAGCC ATTTACAGTAACCTGAC |
Elim Biopharmaceuticals | N/A |
| Primer: Titin I10 Q5 Fwd: AACGATCAG GTGAGCGCG |
Elim Biopharmaceuticals | N/A |
| Primer: Titin I10 Q5 Rev: GCCGCACAC AAAGGTATATTC |
Elim Biopharmaceuticals | N/A |
| Recombinant DNA | ||
| pJ414-STReTCh-6xHis | This study | N/A |
| pJ414-HaloTag-STReTCh-YbbR-EGFP-6xHis | This study | N/A |
| pET28a-GGG-ELP(120nm)-Cys | Addgene | #91572 |
| Sfp pet29b C-terminal His Tag | Addgene | #75015 |
| pJ414-HaloTag-(GPGGA)8-RGD-6xHis | Morimatsu et al., 2015 | N/A |
| pJ414-HaloTag-STReTCh-RGD-6xHis | This study | N/A |
| pJ414-HaloTag-RGD-STReTCh-6xHis | This study | N/A |
| pDEST14-SpyCatcher | Addgene | #35044 |
| pDEST14-Cys-SCMin | This study and Li et al. (2014) | N/A |
| pJ509-02, PiggyBac vector | Price et al. (2018) | N/A |
| PiggyBac-EGFP-Vin-STReTCh | This study | N/A |
| PiggyBac-EGFP-Vin-STReTCh-CT | This study | N/A |
| Other | ||
| Glass coverslips | Fisher Scientific | Cat# 12-544-14 |
| 3-well coverwell perfusion chambers | Grace Biolabs | Cat# 622103 |
| 4-well coverwell perfusion chambers | Grace Biolabs | Cat# 622104 |
| Polybeads sampler kit | Polysciences | Cat# 19822-1 |
| Dynbeads M-270 Streptavidin | ThermoFisher | Cat# 65305 |
| DMEM, high glucose, phenol free | Gibco | Cat# 21063-029 |
| Fetal bovine serum, heat inactivated | Corning | Cat# 35011CV |
| Non essential amino acids | Gibco | Cat# 11140050 |
| Penicillin-streptomycin | Gibco | Cat# 15140122 |
| Lonza P4 Primary Cell Nucleofector Kit | Lonza | Cat# V4XP-4032 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Alex Dunn (alex.dunn@stanford.edu).
Materials availability
Plasmids generated in this study will be shared upon request to the lead contact, until made publicly available on Addgene.
Experimental model and subject details
Cell lines and transfection
GFP-paxillin expressing HFFs were prepared from CCD-1070Sk HFFs (ATCC CRL-2091) as previously described (Chang et al., 2016; Morimatsu et al., 2013; Tan et al., 2020). Both HFFs and MEFs were cultured in DMEM high-glucose medium (Gibco, 21063–029) in the absence of phenol red and supplemented with 10% fetal bovine serum (FBS, Corning, 35011CV), sodium pyruvate (1 mM, Gibco), MEM nonessential amino acids (1×, Gibco), and penicillin/streptomycin (100 U/ml and 100 μg/mL, Gibco). Cells were cultured on tissue culture plastic and grown at 37°C with 5% CO2.
Vin-STReTCh(-CT) MEFs were prepared by transfecting vin−/− MEFs, a gift from K. Rothenberg and B. Hoffman (Duke University). Vin−/− MEFs were stably transduced with EGFP-Vinculin-STReTCh or EGFP-Vinculin-STReTCh-CT using a Lonza P4 kit. Cells were trypsinized, pelleted, resuspended in medium without FBS and penicillin/streptomycin, then counted. 5⋅105 cells were repelleted, then resuspended in a solution of 82 μL P4 nucleofector solution and 18 μL P4 supplement. 2.5 μg of DNA was added to the cells and gently flicked before transferring to a Lonza nucleofection cuvette. Cuvettes were placed in a Lonza 4D-Nucleofector system and program C2167 (for MEFs) was used. 500 μL of warm medium was added to the cuvette to transfer the cells to a six-well plate with medium equilibrated at 37°C using a pipette bulb without pipetting up and down. 24 h after transfection, MEFs with the stably integrated constructs were selected for using puromycin (Gibco) at increasing concentrations (1–2 μg/mL) over the course of 5 days, then sorted into populations using fluorescence-assisted cell sorting based on GFP to enrich for construct-expressing cells and isolate populations of Vin-STReTCh and Vin-STReTCh-CT MEFs with similar levels of protein expression.
Method details
DNA constructs
The STReTCh tension sensing module was designed by taking the titin I10 domain (residues 2880 to 2967 of H. sapiens titin, Q8WZ42 on UniProt) and inserting the 13-amino acid SpyTag (AHIVMVDAYKPTK) between residues 2956 and 2957. The amino acid sequence for the STReTCH module linked to a C-terminal 6xHis tag through a triglycine linker is:
METLHITKTMKNIEVPETKTASFECEVSHFNVPSMWLKNGVEIEMSEKFKIVVQGKLHQLIIMNTSTEDSAEYTFVCGAHIVMVDAYKPTKNDQVSATLTVTGGGHHHHHH
For melting temperature characterization, the DNA encoding this construct was cloned into the pJ414 expression vector (DNA 2.0) by Epoch Life Sciences Inc. (Missouri City, TX). DNA encoding the wild type H. sapiens titin I10 domain was cloned by removing the SpyTag using Q5 site-directed mutagenesis (New England Biolabs) following manufacturers’ instructions with the primers 5′-AACGATCAGGTGAGCGCG-3′ and 5′-GCCGCACACAAAGGTATATTC-3′.
For magnetic tweezers force spectroscopy measurements, DNA encoding a protein composed of STReTCh fused to HaloTag at the N-terminus and a YbbR tag (Yin et al., 2005), TEV cleavage sequence, EGFP, and 6xHis tag at the C-terminus was cloned into pJ414 using custom cloning from Epoch Life Sciences Inc. Elastin-like polypeptide for functionalization of coverslips was expressed from pET28a-GGG-ELP(120nm)-Cys, a kind gift from Dr. Hermann Gaub (Addgene plasmid #91572 ; http://n2t.net/addgene:91572 ; RRID: Addgene_91572). A plasmid encoding the phosphopantetheinyl transferase Sfp from B. subtilis (Sfp synthase) was a kind gift from Dr. Michael Burkart (Addgene plasmid # 75015 ; http://n2t.net/addgene:75015 ; RRID: Addgene_7015).
For measurement of extracellular forces, the DNA encoding STReTCh-RGD was constructed from a previously described RGD force sensor construct encoding an N-terminal HaloTag, followed by a flagelliform-based tension sensing domain and a C-terminal RGD ligand (Chang et al., 2016; Morimatsu et al., 2013; Tan et al., 2020). STReTCh was inserted in place of the flagelliform tension sensing domain using Gibson Assembly (New England Biolabs). The previous force sensor construct was digested at EcoRI (New England Biolabs) and BamHI (New England Biolabs) restriction sites to excise the flagelliform tension sensing domain from the backbone. STReTCh flanked by the appropriate overlaps for insertion into the backbone by Gibson Assembly was amplified using primers 5′-GTTGGAGATCAGCGGCGGTGCTGGCGAGTTCATGGAAACTCTGCATATTACCAAAACC-3′ and 5′-TACCTTGCTCGAAGTACAGATTCTCGGAAGTCACGGTCAGGGTCGC-3′. The amino acid sequence of STReTCh-RGD (with STReTCh in bold) is:
MGSEIGTGFPFDPHYVEVLGERMHYVDVGPRDGTPVLFLHGNPTSSYVWRNIIPHVAPTHRSIAPDLIGMGKSDKPDLGYFFDDHVRFMDAFIEALGLEEVVLVIHDWGSALGFHWAKRNPERVKGIAFMEFIRPIPTWDEWPEFARETFQAFRTTDVGRKLIIDQNVFIEGTLPMGVVRPLTEVEMDHYREPFLNPVDREPLWRFPNELPIAGEPANIVALVEEYMDWLHQSPVPKLLFWGTPGVLIPPAEAARLAKSLPNAKAVDIGPGLNLLQEDNPDLIGSEIARWLSTLEISGGAGEFMETLHITKTMKNIEVPETKTASFECEVSHFNVPSMWLKNGVEIEMSEKFKIVVQGKLHQLIIMNTSTEDSAEYTFVCGAHIVMVDAYKPTKNDQVSATLTVTSENLYFEQGTVYAVTGRGDSPASSAAHHHHHH.
The force-inert, inverted RGD sensor was cloned into pJ414 by Epoch Life Sciences Inc. and has the following amino acid sequence:
MGSEIGTGFPFDPHYVEVLGERMHYVDVGPRDGTPVLFLHGNPTSSYVWRNIIPHVAPTHRSIAPDLIGMGKSDKPDLGYFFDDHVRFMDAFIEALGLEEVVLVIHDWGSALGFHWAKRNPERVKGIAFMEFIRPIPTWDEWPEFARETFQAFRTTDVGRKLIIDQNVFIEGTLPMGVVRPLTEVEMDHYREPFLNPVDREPLWRFPNELPIAGEPANIVALVEEYMDWLHQSPVPKLLFWGTPGVLIPPAEAARLAKSLPNAKAVDIGPGLNLLQEDNPDLIGSEIARWLSTLEISGGAGEFTVYAVTGRGDSPASSAAGGGMETLHITKTMKNIEVPETKTASFECEVSHFNVPSMWLKNGVEIEMSEKFKIVVQGKLHQLIIMNTSTEDSAEYTFVCGAHIVMVDAYKPTKNDQVSATLTVTSENLYFEQGHHHHHH.
DNA encoding a truncated, minimal SpyCatcher (SCMin) with a cysteine for thiol-maleimide labeling was cloned from pDEST14-SpyCatcher (a gift from Mark Howarth, Addgene plasmid # 35044; http://n2t.net/addgene:35044; RRID:Addgene_35044). A cysteine was introduced through a Y8C mutation downstream of the N-terminal 6xHis Tag using QuikChange II XL site-directed mutagenesis (Agilent) with primers 5′-CATCACCATCACCATCACGATTGCGACATCCCAAC-3′ and 5′-GTTGGGATGTCGCAATCGTGATGGTGATGGTGATG-3′. The full-length SpyCatcher was then truncated using Gibson Assembly to remove amino acids 21–43 and the C-terminal 9 amino acids. The vector backbone was amplified using primers 5′-TAAATGGTTGATGCTTGAGGATC-3′ and 5′-TTTGAGAATTTAATATGGGTAGCACTATCGCCCTGAAAATACAGGTTTTC-3′. The minimal SpyCatcher insert with appropriate overlaps for Gibson cloning was amplified using primers 5′-GATAGTGCTACCCATATTAAATTC-3′ and 5′-GATCCTCAAGCATCAACCATTTAGCCATTTACAGTAACCTGAC-3′. The final amino acid sequence for the minimal SpyCatcher (SCMin) is:
HHHHHHDCDIPTTENLYFQGDSATHIKFSKRDEDGKELAGATMELRDSSGKTISTWISDGQVKDFYLYPGKYTFVETAAPDGYEVATAITFTVNEQGQVTVNG
DNA constructs for Vin-STReTCh and Vin-STReTCh-CT were cloned from a PiggyBac plasmid containing G. gallus vinculin downstream of the hEF1α promoter (pJ509-02, DNA 2.0) (Price et al., 2018). EGFP linked to a GS linker (SGLGGSGGGGSGGGGSGG) was fused at the N-terminus of vinculin. STReTCh was inserted either after amino acid 883 of vinculin (in a linker region between the head and tail domains) or at the C-terminus as illustrated in Figure 4. This insertion into the linker region has previously been shown not to perturb biological function (Grashoff et al., 2010). These fusion proteins were cloned into the PiggyBac vector downstream of the hEF1α promoter by Epoch Life Sciences Inc. to give EGFP-Vinculin-STReTCh(-CT). All constructs were verified by sequencing.
Protein expression, purification, and labeling
All proteins were expressed in BL21(DE3) chemically competent Escherichia coli (New England Biolabs) and purified using Ni-NTA chromatography, except for the ELP. Cultures were induced at an optical density of 0.6 with 1mM isopropyl-β-D-thiogalactopyranoside (VWR Scientific) and grown overnight at 18°C. The cultures were then spun down at 6000×g for 15 min, and bacterial pellets were resuspended in lysis buffer (50 mM sodium phosphate, 300 mM NaCl, and 10 mM imidazole, pH 8) with a cOmplete EDTA-free protease inhibitor cocktail (11873580001, Roche), 70 μg/mL lysozyme (90082, ThermoFisher), and 7 U/mL DNase I (04536282001, Roche). The resuspended cells were rotated end-over-end for 45 min at 4°C, lysed with a tip sonicator, and spun at 12,000×g for 30 min. The supernatant was incubated with 1.5 mL of Ni-NTA HisPur Resin (ThermoFisher) for every 500 mL of culture and rotated end-over-end at 4°C for 2 h. The solution was then packed into a gravity column, washed three times with 5 mL of wash buffer (50 mM sodium phosphate, 300 mM NaCl, and 20 mM imidazole, pH 7.4 with 2 mM β-mercaptoethanol; BME), and protein was eluted by incubating the resin bed with 1 mL of elution buffer (50 mM sodium phosphate, 300 mM sodium chloride, and 250 mM imidazole, pH 7.4 and 2 mM BME) for 5 min, four times. The eluate was collected, concentrated, and buffer-exchanged into storage buffer (1× phosphate-buffered saline, 2 mM BME) using Amicon centrifugal filter units (MilliporeSigma) of the appropriate molecular-weight cutoff, then flash-frozen and stored at −80°C. For magnetic tweezers experiments, the HaloTag-STReTCh-YbbR-GFP fusion protein was further purified using anion exchange chromatography with a 1 mL HiTrap Q column (Cytiva Life Sciences) in 50mM Tris, pH 8 supplemented with 2 mM BME using an AKTA Pure FPLC (GE Healthcare). FPLC purified protein was then buffer-exchanged into PBS+2 mM BME before freezing. Eluates were characterized by SDS–polyacrylamide gel electrophoresis (SDS-PAGE), and the concentration was determined by ultraviolet–visible (UV-Vis) spectroscopy.
For magnetic tweezer experiments, ELP was expressed in BL21(DE3) E. coli and purified using a thermoprecipitation and redissolution strategy described previously (Ott et al., 2017), then buffer exchanged into 50 mM HEPES prior to flash freezing. Biotinylated STReTCh was generated by labeling HaloTag-STReTCh-YbbR-GFP with biotin-PEG3-CoenzymeA (SiChem GmbH, Bremen, Germany). 5 μM HaloTag-STReTCh-YbbR-GFP was incubated with 0.5 μM Sfp synthase, 10 μM biotin-PEG3-CoenzymeA, and 10 mM MgCl2 in PBS overnight rotating at 4°C to give HaloTag-STReTCh-Biotin-GFP. The reaction was then buffer exchanged into PBS+2 mM BME using a 7 kDa Zeba desalting column to remove excess biotin-PEG3-CoenzymeA, flash-frozen, and stored at −80°C for later use.
For in-cell experiments, Alexa Fluor 647 labeled SpyCatcher (647-SC) was generated by first further purifying SCMin using size exclusion chromatography on an AKTA Pure FPLC with a Sephadex 75 Increase 10/300 GL (Cytiva Life Sciences) column in PBS+2 mM BME. Purified SCMin was buffer-exchanged into buffer containing 100 mM phosphate, 150 mM NaCl and 1 mM EDTA using three 7 kDa Zeba desalting columns (ThermoFisher) in series to remove excess BME. 100 μL of 116 μM desalted SpyCatcher was then incubated with excess tris(2-carboxyethyl)phosphine (TCEP) for 1 h at room temperature, followed by 2 equivalents of Alexa Fluor 647 C2-maleimide (ThermoFisher, A20347), rotating at 4°C overnight. The reaction was then quenched by adding 20 μL of PBS +2 mM BME. Excess free dye was removed using 2 Sephadex G-25 PD MiniTrap desalting columns (Cytiva Life Sciences) in series, using gravity-driven flow. Labeling efficiency was 72% as measured by UV-Vis. The labeled 647-SC was flash-frozen at −80°C and stored in PBS until later use.
Melting temperature characterization
STReTCh or titin I10 were mixed with SYPRO Orange dye (ThermoFisher) at a final concentration of 5 μM protein and 5× SYPRO Orange. 3 replicates of 35 μL of the protein-dye mixture were loaded into a 96-well PCR plate (ThermoFisher) which was spun down in a swinging bucket centrifuge prior to measurement. SYPRO Orange fluorescent signal was measured at temperatures from 25°C to 95°C at 0.5°C increments using a StepOnePlus Real-Time PCR instrument (ThermoFisher).
Magnetic tweezer construction and calibration
A magnetic tweezer was constructed on a Nikon Ti-E microscope equipped with a motorized 3-axis piezo stage (Mad City Labs). Two 6 mm-diameter cylindrical N52 magnets (K&J Magnets), arranged as in Figure S1A, were affixed to a custom-fabricated aluminum bracket, which was attached to a 0.5″-travel XYZ translation stage equipped with a motorized Z-axis actuator (Thorlabs, Newton, New Jersey, USA). The stage position was homed before measurement by stepping the Z-actuator until the magnet bracket just touched the top of the sample. Measurements were performed while the Nikon Perfect Focus feedback system was turned on.
Samples were illuminated from the objective side via a mint green LED (Thorlabs, MINTL5) reflected into the back focal plane of a 100× Plan-Apo TIRF objective using a polarizing beamsplitter (Edmund Optics). Reflected, cross-polarized light was transmitted through the beamsplitter and collected by a sCMOS camera (Hamamatsu Orca Flash v4). Bead images were collected using a custom-written LabView script or MicroManager 1.4 software (Edelstein et al., 2014).
Magnetic tweezers force calibration was performed as described previously (Löf et al., 2019) using transverse fluctuations of a magnetic bead tethered to a glass coverslip via lambda-phage DNA (New England Biolabs).
Magnetic tweezers force spectroscopy
Coverslips functionalized with HaloLigand-ELP were prepared similarly to a previously described protocol (Chang et al., 2016; Morimatsu et al., 2013; Tan et al., 2020). ELPs were first functionalized at the N-terminus with HaloLigand by reacting 300 μM ELP with 3 mM HaloTag Succinimidyl Ester (O4) Ligand (Promega, P6751) in 50 mM HEPES, pH 7.5 for 4 h at room temperature. During this reaction, glass coverslips (Fisher Scientific, No. 12-544-14) were cleaned and silanized as described previously (Chang et al., 2016; Morimatsu et al., 2013; Tan et al., 2020) to generate a nucleophilic, amine-functionalized substrate. Sulfosuccinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate (ThermoFisher, A39268) (Sulfo-SMCC) was diluted to 10 mM in 50 mM HEPES, pH 7.5, and a coverslip “sandwich” was formed by adding 90 μL of the Sulfo-SMCC solution between a pair of silanized coverslips. These coverslip sandwiches were incubated at room temperature in a humid environment for 1 h to generate maleimide-functionalized coverslips. Coverslips were washed by dipping in a beaker of MilliQ water to remove excess Sulfo-SMCC, then gently dried under a stream of nitrogen.
After the 4 h incubation was complete, the HaloLigand-ELP reaction was quenched by adding Tris buffer (pH 7) to a final concentration of 5 mM for 15 min. In preparation for functionalizing the ELP to the coverslip using cysteine-maleimide chemistry, excess TCEP was added to the reaction mixture and incubated for 30 min at room temperature. The reaction was then buffer exchanged into ELP conjugation buffer (50 mM phosphate, 50 mM NaCl, 10 mM EDTA, pH 7.2) using two 7 kDa Zeba desalting columns in series. Coverslip sandwiches using the maleimide-functionalized coverslips were incubated with 90 μL of the ELP solution for 2 h at room temperature in a humid environment. Coverslips were then washed by dipping in MilliQ water as described above, and were sufficiently dry upon slow removal of the coverslips from the water due to the relatively hydrophobic surface properties of the ELP-functionalized surface. Finally, ELP-functionalized coverslip sandwiches were incubated with 100 μL of 10 mM L-cysteine for 1 h at room temperature in a humid environment to quench remaining free maleimides. These coverslips were then washed by dipping in water and were sufficiently dry for storage without additional drying steps. Coverslips were stored in vacuum-sealed bags at room temperature for up to several weeks, or at −20°C for longer term storage.
Non-magnetic 2.0 μm polystyrene beads (Polybeads, Polysciences) were non-specifically attached to the HaloLigand-ELP coverslips to use as reference beads to account for drift during magnetic tweezer measurements. 1 drop (∼50 μL) of polystyrene bead stock suspension was diluted in 1.5 mL ethanol, and 100 μL was spin-coated onto the coverslip at 3500 rpm for 40 s. The coverslip was then baked at 75°C for 10 min, then washed by dipping in MilliQ water to remove non-adhered polystyrene beads.
3-well coverwells (Grace Biolabs) of ∼100 μL well volumes were attached to HaloLigand-ELP functionalized coverslips. Wells were surface passivated by treatment for 1 h at room temperature with 110 μL of a 1% (w/v) solution of casein, diluted from a 5% stock (Sigma-Aldrich, C4765). Wells were then washed twice with 200 μL PBS. 110 μL of HaloTag-STReTCh-Biotin-GFP was added to the wells at concentrations ranging from 100 pM to 1 nM and incubated at room temperature for 30 min to allow for covalent attachment to the coverslip. Wells were washed twice with 200 μL PBS to remove unbound protein.
Streptavidin-coated superparamagnetic beads (Dynabeads M−270 Streptavidin, ThermoFisher, 65305) were washed by diluting the stock 30× in PBS +0.1% Tween 20 and vortexing the diluted bead suspension for 20 s. Beads were pulled down using a magnet, and washed twice more with PBS +0.1% Tween 20. Beads were then resuspended in PBS +0.1% Tween 20 + 1% casein at a 30× dilution from the stock concentration and equilibrated with the buffer for 15 min. 110 μL of this bead suspension was added to the HaloTag-STReTCh-Biotin-GFP functionalized wells and incubated at room temperature for 25 min. After bead incubation, magnetic tweezers were brought above the wells to apply sub-pN forces to the samples to remove a majority of the beads not tethered to the biotinylated STReTCh construct, and the wells were washed twice with 200 μL PBS. This detachment and wash process was repeated twice before making magnetic tweezers measurements. Minutes-long movies of streptavidin beads tethered to HaloTag-STReTCh-Biotin-GFP surface were then acquired at frequencies ranging from 2.5 Hz to 50 Hz, with a 20 ms exposure time, at forces ranging from sub-pN to ∼8 pN. Measurements were carried out at room temperature rather than 37°C. However, calculations on the temperature dependence of folding behavior of STReTCh using the Bell-Evans model and melting temperature data suggest that the force dependence of unfolding is not substantially altered at 37°C (see Methods S1).
Magnetic tweezers data analysis
As described previously (Löf et al., 2019), in separate calibration experiments, a nonspecifically adhered bead was imaged while the piezo stage was stepped in increments of 5 nm to generate a reference image lookup table (LUT). These LUTs were generated for both magnetic and non-magnetic beads and were used to estimate the z-position of query bead images. The analysis pipeline is schematized in Figure S1A. Briefly, the two-dimensional Fourier transform of the query bead image was calculated, and the radial profile extracted. The bead position was then estimated in one of two ways. In one method, the Pearson correlation coefficient r between the radial profile of the query image and each image in the LUT was calculated, and the LUT position that maximized r was computed by quadratic interpolation. In the second method, the one-dimensional Fourier transforms of the LUT radial profiles were computed. The phase at a high-amplitude frequency was then plotted against the bead height corresponding to the LUT slice, and a third- to fifth-order polynomial was fit to this curve. This polynomial fit was then used to directly estimate height from the Fourier transform of the query image. Images were analyzed with both methods and found to produce similar results, reducing the chance that measured bead heights were analysis artifacts.
To correct for drift during measurement, the z-position trace of either a non-magnetic polystyrene bead or occasionally an immobile, non-specifically adhered magnetic bead was subtracted from the trace of the bead of interest. To characterize the unfolding behavior of the STReTCh domain, we identified traces in which the bead of interest exhibited steplike changes in length of ∼10–15 nm. We further filtered these traces based on the height extension behavior to validate the presence of ELP extension, which has a contour length of ∼120 nm (Löf et al., 2019; Ott et al., 2017), and by confirming that the bead exhibited xy-fluctuations consistent with the expected Brownian motion under pN forces based on the Equipartition Theorem (Sarkar and Rybenkov, 2016). Steps in traces that met the quality control criteria were manually identified, and datapoints for each trace were designated as either folded or unfolded based on the localized, reference-subtracted z-position to calculate the fraction of the trace that the molecule spent in each of these two states. The folded fraction was then plotted in Figure 2C.
Magnetic tweezers data for the folding and unfolding of STReTCh was fit to the Bell-Evans kinetic model (Bell, 1978; Evans and Ritchie, 1997), which gives a sigmoidal fit for the folded fraction as illustrated in Figure 2C. The equilibrium between the folded species Pf and unfolded species Pu is:
where ku and kf are the first-order rate constants for folding and unfolding, respectively, and are dependent on the applied force F:
Here, ku0 and kf0 are force-independent rates, Δxu and Δxf represent distances to the transition state from the folded and unfolded states, kB is Boltzmann’s constant, and T is the temperature. At equilibrium, the folding and unfolding rates are equal, so concentrations of the folded and unfolded species satisfy
and the folded fraction is given by
which varies sigmoidally with the applied force F.
We assume that the system is ergodic, such that the folded fraction of an ensemble molecules can be represented by the average fraction of time that individual molecules spend in the folded state (i.e. the data presented in Figure 2C). Consequently, we used the magnetic tweezers data to perform maximum likelihood estimation (MLE) fitting for 4 parameters: ku0, kf0, Δxu/kBT and Δxf/kBT. For each trace, we define a probability of observing steps of the measured lengths by assuming that the waiting times for folding and unfolding are Poisson-distributed and that all folding and unfolding events are independent. Then, the probability density for observing a folded time step of duration tf or unfolded time step of duration tu is given by the expression
Additionally, because determining the waiting time of the last step in each trace is limited by when we stop observing the specific bead, we assign probabilities for these steps equal to the likelihood of a waiting time of at least as long as the duration that was observed. In other words, the probabilities associated with observing a folded time step tf or unfolded time step tu for the last steps in the trace are given by
Thus, for an observed trace with n observed steps (i.e. n-1 transitions between folded and unfolded states) where each step i has a duration ti, the probability of observing such a trace can be computed as
Then the probability of observing a dataset of m traces given a set of Bell-Evans parameters is
where each rate constant takes on the expression for ku or kf according to the type of step observed. The MLE fit for the 4 parameters are the values that maximize this probability expression. To calculate these estimates, we used the built-in MATLAB function fminsearch to minimize an objective function corresponding to the negative value of the natural logarithm of the probability expression above. These MLE fit parameters are tabulated in Table S1.
We also used the logarithm of the probability expression as the log-likelihood function to calculate confidence intervals for MLE parameters using a profile likelihood approach (Cole et al., 2014). Briefly, the likelihood function was computed over a grid of values for the four parameters . For a given function of these parameters , we can define a profile likelihood for the function:
where denotes a specific value of the function and denotes the Dirac delta function. For example, choosing gives the profile likelihood for that parameter, while choosing to be the folded fraction at a specific force gives the profile likelihood for that value.
We numerically integrated the above integral to find the upper and lower bounds for the 95% confidence intervals for each parameter, which are also tabulated in Table S1, and for the fraction of folded STReTCh at forces from 0 to 3 pN, which is depicted in Figure 2C.
TIRF microscopy
Total internal reflection fluorescence (TIRF) microscopy images were collected using an Apo TIRF 100× oil objective lens, numerical aperture 1.49 (Nikon) as described previously (Chang et al., 2016; Morimatsu et al., 2013; Tan et al., 2020) and controlled using Micromanager (Edelstein et al., 2014). Samples were excited with a 473-nm OBIS laser (Coherent) or 635-nm (Blue Sky Research) laser for GFP and Alexa Fluor 647, respectively. Emitted light passed through a quad-edge laser-flat dichroic with center/bandwidths of 405/60 nm, 488/100 nm, 532/100 nm, and 635/100 nm from Semrock Inc. (Di01-R405/488/532/635–25×36) and corresponding quad-pass filter with center/bandwidths of 446/37 nm, 510/20 nm, 581/70 nm, 703/88 nm band-pass filter (FF01-446/510/581/703–25). GFP and Alexa Fluor 647 images were taken through separate additional cubes stacked into the light path (GFP: 470/40 nm, 495 nm long-pass, and 525/50 nm; Alexa Fluor 647: 679/41 nm and 700/75 nm) and recorded on a Hamamatsu Orca Flash 4.0 camera.
Extracellular force sensor measurements
HaloLigand-PEG coverslips were prepared as described in the literature (Chang et al., 2016; Morimatsu et al., 2013; Tan et al., 2020). 3- and 4-well coverwells (Grace Biolabs) of ∼100 μL well volumes were attached to the functionalized coverslip. 100 μL of 200 nM Halo-STReTCh-RGD or force inert control in PBS was flowed into each channel and incubated at room temperature for 45 min. Channels were washed with 200 μL PBS to wash out unbound sensor. GFP-Pxn HFFs were seeded onto the force sensor cover slips and incubated at 37°C for 75–90 min to allow for cell spreading and adhesion onto the cover slip.
For live cell measurements, non-adhered cells were washed off with 200 μL media at 37°C after spreading. Cells were then treated with 110 μL of 20 nM 647-SC in media and incubated at 37°C for 20 min. After staining, cells were washed with 200 μL media at 37°C and imaged using TIRF microscopy with the objective maintained at 37°C. As illustrated in Figure S2C, we observed some movement of HFFs during live cell imaging. To better localize potential 647-SC signal in relation to focal adhesions, we treated cells with 110 μL 0.5% (v/v) paraformaldehyde, diluted in warm PBS from a 16% stock (Electron Microscopy Sciences) for 5 min at 37°C to lightly fix cells and reduce cell motion during imaging. Force independent of actin flow at focal adhesions in fibroblasts has shown to be preserved even when cells are treated with dilute PFA (Driscoll et al., 2020), and these forces are detectable using STReTCh. After light fixation, cells were washed with 200 μL warm PBS, followed by 200 μL warm media and subsequently treated with 110 μL 647-SC at the desired concentration (10–40 nM) for 10 min at 37°C, washed with 200 μL warm media, then imaged using TIRF microscopy at 37°C to generate the data in Figure 3. For conditions in which Cytochalasin D (CytoD, Enzo Life Sciences) was added, 110 μL of CytoD diluted at the desired concentration in warm media was added 5 min prior to the light fixation procedure.
For fixed cell measurements (Figure S3), HFFs were seeded on coverslips functionalized with Halo-STReTCh-RGD for 1 hr, after which nonadherent cells were washed off with 200 μL warm media followed by 200 μL PBS at 37°C. Cells were then fixed with 100 μL 4% PFA in PBS for 15 min and permeabilized with 100 μL 0.1% Triton X- in PBS for 10 min at room temperature. Cells were washed with 200 μL PBS after each step. Cells were then blocked with 100 μL 1% bovine serum albumin (BSA, Sigma-Aldrich) in PBS for 20 min, then stained with 20 nM 647-SC in 1% BSA/PBS for 20 min. After staining, cells were washed 3 times with 200 μL PBS. Fixed and stained cells were then imaged with TIRF microscopy at room temperature.
Intracellular force sensor measurements
3- and 4-well coverwells (Grace Biolabs) were attached to cleaned coverslips. 100 μL of fibronectin (10 μg/mL, Corning) diluted in PBS was added to the well and incubated for 45 min at room temperature. Wells were washed with 200 μL PBS, then Vin-STReTCh(-CT) MEFs were seeded by adding 100 μL of a 300,000 cells/mL suspension in warm medium. Cells were allowed to adhere to the surface for 3 h, after which, they were washed with warm media followed by warm PBS. Cells were then fixed with 100 μL 4% PFA in PBS for 15 min and permeabilized with 100 μL 0.1% Triton-X in PBS for 10 min at room temperature. Cells were washed with 200 μL PBS after each step. Cells were then blocked with 100 μL 1% BSA in PBS for 20 min, then stained with 10 nM SC-647 in 1% BSA/PBS for 10 min. After staining, cells were either washed once with 200 μL PBS, then either twice with 200 μL PBS, or washed once with 2 mM EDTA and once with 3 M Gdn-HCl. Cells were incubated for 3 min with each wash solution, washed with 200 μL PBS between washes, and washed with 2 × 200 μL PBS after the final Gdn-HCl or PBS wash, for a total of 6 washes after staining. Fixed and stained cells were then imaged with TIRF microscopy at room temperature.
For ROCK inhibitor experiments in Figure S6, MEFs were seeded at 150,000 cells/mL and allowed to adhere to the fibronectin surface overnight. 200 μL warm media either with or without 10 μM Y-27632 (STEMCELL Technologies) was added to the channels, which were placed back in the incubator for 2 h. After 2 h, cells were fixed, permeabilized, and stained as described above with 5–10 nM 647-SC for 10 min, then washed with EDTA and Gdn-HCl as described above and imaged with TIRF microscopy.
Quantification and statistical analysis
Image analysis and quantification
Image preparation and quantification of fluorescence intensities in specific regions of stained GFP-Pxn HFFs and Vin-STReTCh(-CT) MEFs was carried out in Fiji (Schindelin et al., 2012). Images were background subtracted, using either rolling ball background subtraction for GFP signal or subtracting the average intensity of an empty, unstained sample for 647 signal. Focal adhesions were segmented by generating a mask from thresholding GFP signal using Fiji’s built-in RenyiEntropy algorithm. The cell body was segmented by generating a mask from thresholding GFP signal using Fiji’s built-in Triangle algorithm applied to a Gaussian blurred GFP image. The region of the image denoted as “Body” (Figures 3 and S7) is thus defined as the region outside the FA mask but within the cell body mask. The region of the image denoted as “Ext” is defined as the region outside the cell body mask.
To compute the 647 signal normalized to the expression of Vin-STReTCh(-CT) for intracellular force measurements, GFP images were background subtracted using rolling ball background subtraction, and the 647 signal within the cell body was background subtracted using the mean cell exterior value for each image. The background subtracted 647 image was divided by the background subtracted GFP image, and this resultant image was multiplied by a FA mask generated from the GFP signal using the method described above. The mean value within this mask was measured for each cell and reported as the normalized 647 intensity in Figure 4D.
Metrics are normalized to the mean values within FAs for the force-bearing STReTCh condition when applicable to account for potential variations in imaging conditions (e.g. laser power, TIRF angle, etc.) between different experiments.
Statistical analysis
Sample sizes for each experiment and can be found in figure legends. Box plot formatting information can be found in the figure legends for Figures 3 and S2 in the supplemental information and are applicable for subsequent box plots. Significance between experimental conditions was establishing using two-tailed Mann-Whitney tests as described in figure legends.
Acknowledgments
B.L.Z. is supported by the Stanford ChEM-H Chemistry/Biology Interface Predoctoral Training Program (NIH 5T32GM120007) and the NSF Graduate Research Fellowship Program (DGE-1656518). V.T.V. is supported by the Stanford Medical Scientist Training Program (NIH T32GM007365) and the NIH NIDDK (1F30DK124985). A.R.D. acknowledges the HHMI (Faculty Scholar Award) and the NIH (R35GM130332). We are grateful to current and former members of the Dunn lab—particularly Dr. Magnus Bauer, Dr. Steven Tan, and Dr. Cayla Miller—for discussion and comments on the manuscript.
Author contributions
B.L.Z. and A.R.D. conceived the study and designed experiments. V.T.V. and B.L.Z. designed and built magnetic tweezers and implemented methods for magnetic tweezers data analysis. B.L.Z. performed all experiments. B.L.Z., V.T.V., and A.R.D. wrote and prepared the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: September 1, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2022.100278.
Supporting citation
The following reference appears in the supplemental information: Bogomolovas et al. (2016).
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Aird E., Tompkins K.J., Ramirez M.P., Gordon W.R. Enhanced molecular tension sensor based on bioluminescence resonance energy transfer (BRET) ACS Sens. 2020;5:34–39. doi: 10.1021/acssensors.9b00796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G.I. Models for specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- Bogomolovas J., Fleming J.R., Anderson B.R., Williams R., Lange S., Simon B., Khan M.M., Rudolf R., Franke B., Bullard B., et al. Exploration of pathomechanisms triggered by a single-nucleotide polymorphism in titin's I-band: the cardiomyopathy-linked mutation T2580I. Open Biol. 2016;6:160114. doi: 10.1098/rsob.160114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi N., Sorokina M., Shcherbakova O.G., Weis W.I., Pruitt B.L., Nelson W.J., Dunn A.R. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc. Natl. Acad. Sci. USA. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman J.M., Su H., Blanchard A.T., Duan Y., Meyer T., Quach M.E., Glazier R., Bazrafshan A., Bender R.L., Kellner A.V., et al. Live-cell super-resolved PAINT imaging of piconewton cellular traction forces. Nat. Methods. 2020;17:1018–1024. doi: 10.1038/s41592-020-0929-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A.C., Mekhdjian A.H., Morimatsu M., Denisin A.K., Pruitt B.L., Dunn A.R. Single molecule force measurements in living cells reveal a minimally tensioned integrin state. ACS Nano. 2016;10:10745–10752. doi: 10.1021/acsnano.6b03314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S., Chu H., Greenland S. Maximum likelihood, profile likelihood, and penalized likelihood: a primer. Am. J. Epidemiol. 2014;179:252–260. doi: 10.1093/aje/kwt245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll T., Ahn S.J., Huang B., Kumar A., Schwartz M.A. Actin flow-dependent and -independent force transmission through integrins. Proc. Natl. Acad. Sci. USA. 2020;117:32413–32422. doi: 10.1073/pnas.2010292117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A.D., Tsuchida M.A., Amodaj N., Pinkard H., Vale R.D., Stuurman N. Methods of microscope control using MicroManager software. J. Biol. Methods. 2014;1:e10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E., Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finer J.T., Simmons R.M., Spudich J.A. Single myosin molecule mechanics - piconewton forces and nanometer steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- Fischer L., Rangarajan S., Sadhanasatish T., Grashoff C., Dill K. Molecular force measurement with tension sensors. Annu. Rev. Biophys. 2021;50:595–616. doi: 10.1146/annurev-biophys-101920-064756. [DOI] [PubMed] [Google Scholar]
- Grashoff C., Hoffman B.D., Brenner M.D., Zhou R., Parsons M., Yang M.T., McLean M.A., Sligar S.G., Chen C.S., Ha T., Schwartz M.A. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman B., Grashoff C., Schwartz M.A. Dynamic molecular processes mediate cellular mechanotransduction. Nature. 2011;475:316–323. doi: 10.1038/nature10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K., Ji L., Applegate K.T., Danuser G., Waterman-Storer C.M. Differential transmission of actin motion within focal adhesions. Science. 2007;315:111–115. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]
- Kanchanawong P., Shtengel G., Pasapera A.M., Ramko E.B., Davidson M.W., Hess H.F., Waterman C.M. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCroix A.S., Rothenberg K.E., Berginski M.E., Urs A.N., Hoffman B.D., Paluch E. Construction, imaging, and analysis of FRET-based tension sensors in living cells. Methods Cell Biol. 2015;125:161–186. doi: 10.1016/bs.mcb.2014.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Springer T.A. Integrin extension enables ultrasensitive regulation by cytoskeletal force. Proc. Natl. Acad. Sci. USA. 2017;114:4685–4690. doi: 10.1073/pnas.1704171114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Fierer J.O., Rapoport T.A., Howarth M. Structural analysis and optimization of the covalent association between SpyCatcher and a peptide tag. J. Mol. Biol. 2014;426:309–317. doi: 10.1016/j.jmb.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löf A., Walker P.U., Sedlak S.M., Gruber S., Obser T., Brehm M.A., Benoit M., Lipfert J. Multiplexed protein force spectroscopy reveals equilibrium protein folding dynamics and the low-force response of von Willebrand factor. Proc. Natl. Acad. Sci. USA. 2019;116:18798–18807. doi: 10.1073/pnas.1901794116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimatsu M., Mekhdjian A.H., Adhikari A.S., Dunn A.R. Molecular tension sensors report forces generated by single integrin molecules in living cells. Nano Lett. 2013;13:3985–3989. doi: 10.1021/nl4005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimatsu M., Mekhdjian A.H., Chang A.C., Tan S.J., Dunn A.R. Visualizing the interior architecture of focal adhesions with high-resolution traction maps. Nano Lett. 2015;15:2220–2228. doi: 10.1021/nl5047335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott W., Jobst M.A., Bauer M.S., Durner E., Milles L.F., Nash M.A., Gaub H.E. Elastin-like polypeptide linkers for single-molecule force spectroscopy. ACS Nano. 2017;11:6346–6354. doi: 10.1021/acsnano.7b02694. [DOI] [PubMed] [Google Scholar]
- Pierschbacher M.D., Ruoslahti E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature. 1984;309:30–33. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- Price A.J., Cost A.L., Ungewiß H., Waschke J., Dunn A.R., Grashoff C. Mechanical loading of desmosomes depends on the magnitude and orientation of external stress. Nat. Commun. 2018;9:5284. doi: 10.1038/s41467-018-07523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringer P., Weißl A., Cost A.L., Freikamp A., Sabass B., Mehlich A., Tramier M., Rief M., Grashoff C. Multiplexing molecular tension sensors reveals piconewton force gradient across talin-1. Nat. Methods. 2017;14:1090–1096. doi: 10.1038/NMETH.4431. [DOI] [PubMed] [Google Scholar]
- Rivas-Pardo J.A., Eckels E.C., Popa I., Kosuri P., Linke W.A., Fernández J.M. Work done by titin protein folding assists muscle contraction. Cell Rep. 2016;14:1339–1347. doi: 10.1016/j.celrep.2016.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg K.E., Scott D.W., Christoforou N., Hoffman B.D. Vinculin force-sensitive dynamics at focal adhesions enable effective directed cell migration. Biophys. J. 2018;114:1680–1694. doi: 10.1016/j.bpj.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar R., Rybenkov V.V. A guide to magnetic tweezers and their applications. Front. Phys. 2016;4:48. doi: 10.3389/fphy.2016.00048. [DOI] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/NMETH.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S.J., Chang A.C., Anderson S.M., Miller C.M., Prahl L.S., Odde D.J., Dunn A.R. Regulation and dynamics of force transmission at individual cell-matrix adhesion bonds. Sci. Adv. 2020;6:eaax0317. doi: 10.1126/sciadv.aax0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Rahil Z., Li I.T.S., Chowdhury F., Leckband D.E., Chemla Y.R., Ha T. Constructing modular and universal single molecule tension sensor using protein G to study mechano-sensitive receptors. Sci. Rep. 2016;6:21584. doi: 10.1038/srep21584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M., Goult B.T., Klapholz B., Hu X., Toseland C.P., Guo Y., Cong P., Sheetz M.P., Yan J. The mechanical response of talin. Nat. Commun. 2016;7:11966. doi: 10.1038/ncomms11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M., Qiu W., Liu R., Efremov A.K., Cong P., Seddiki R., Payre M., Lim C.T., Ladoux B., Mège R.M., Yan J. Force-dependent conformational switch of alpha-catenin controls vinculin binding. Nat. Commun. 2014;5:4525. doi: 10.1038/ncomms5525. [DOI] [PubMed] [Google Scholar]
- Yin J., Straight P.D., McLoughlin S.M., Zhou Z., Lin A.J., Golan D.E., Kelleher N.L., Kolter R., Walsh C.T. Genetically encoded short peptide tag for versatile protein labeling by Sfp phosphopantetheinyl transferase. Proc. Natl. Acad. Sci. USA. 2005;102:15815–15820. doi: 10.1073/pnas.0507705102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusko E.C., Asbury C.L. Force is a signal that cells cannot ignore. Mol. Biol. Cell. 2014;25:3717–3725. doi: 10.1091/mbc.E13-12-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakeri B., Fierer J.O., Celik E., Chittock E.C., Schwarz-Linek U., Moy V.T., Howarth M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA. 2012;109:E690–E697. doi: 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ge C., Zhu C., Salaita K. DNA-based digital tension probes reveal integrin forces during early cell adhesion. Nat. Commun. 2014;5:5167. doi: 10.1038/ncomms6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.