Abstract
Translational efficiency of an AUG, CUG, GUG, or UUG initiation codon was measured for the naturally leaderless cI mRNA from bacteriophage λ. In a cI-lacZ translational fusion, only AUG supported a high level of expression; GUG supported a low level of expression, while UUG and CUG expression was barely above background levels. Addition of an untranslated lac leader and Shine-Dalgarno sequence to cI increased expression but still showed a dependence on an AUG for maximum expression. cI-lacZ mRNA with an AUG initiation codon showed a greater in vitro ribosome binding strength and a higher level of full-length in vivo mRNA, suggesting that the initiation codon is an important determinant of ribosome binding strength and translational efficiency for mRNA with or without the 5′ untranslated leader.
Initiation is the rate-limiting step of translation and requires the placement of a start codon into the ribosomal P site. The most frequently used start codon in Escherichia coli is AUG, but GUG and UUG can also serve for initiation. The frequency of initiation codon usage varies, with AUG, GUG, and UUG having frequencies of 90, 8, and 1%, respectively (17). Besides the importance of the start codon for initiation, other translation factors and signals contribute to the binding of mRNA to ribosomes. The Shine-Dalgarno (SD) sequence is a highly characterized translation signal of mRNA (6, 7, 14, 19); the SD is located upstream of the initiation codon and is complementary to the anti-Shine-Dalgarno (ASD) sequence near the 3′ end of the 16S rRNA. The SD-ASD interaction contributes to the association of mRNA with the 30S ribosomal subunit by complementary base pairing, thereby increasing the likelihood of a start codon entering the ribosomal P site.
Even though an SD-ASD interaction is generally considered a requirement for efficient binding of mRNA to ribosomes, there are several mRNAs that lack the 5′ untranslated leader and SD sequence. Wu and Janssen (25) summarized more than 30 naturally leaderless mRNAs that are found in Archaea, Bacteria, and Eucarya. The most studied leaderless mRNA in E. coli is expressed from the bacteriophage λ cI gene during lysogeny (12). Although the mRNA features that determine cI translation levels are unknown, it has been suggested (18) that interaction between the cI downstream box (DB) and the 16S rRNA anti-DB (ADB) is important for expression; however, additional studies have not supported a DB-ADB contribution to cI translation (13, 11). It has been suggested recently that leaderless mRNAs are translated by a subset of ribosomes lacking the initiation factor IF3 (10, 21).
It has also been reported that the untranslated leader regions of mRNAs can be removed without the loss of translatability (8, 22, 25). Removal of the untranslated leader from the E. coli lacZ gene results in a sequence-specific dependence on an AUG codon for translation initiation; however, changing the start codon on lacZ mRNA with the 5′ untranslated leader (“leadered” lacZ mRNA) from AUG to GUG, UUG, or CUG resulted only in reduced expression (22). While removal of an untranslated leader results in a dependence on an AUG start codon, it is not known if this AUG dependency also occurs with a naturally leaderless mRNA. The work described here is the first examination of start codon requirements for a naturally leaderless mRNA and reveals that the start codon is an important determinant of ribosome binding strength and translational efficiency for leaderless and leadered cI mRNA.
MATERIALS AND METHODS
Bacterial strains.
E. coli DH5α [F− endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(lacIZYA-argF)] U169 deoR (φ80dlac ΔlacZ)M15] was used as the host for all plasmid DNA manipulations. E. coli RFS859 (F− thr-1 araC859 leuB6 Δlac74 tsx-274 λ− gyrA111 recA11 relA1 thi-1) (16) was used as the host for the final expression and assay of all cI-lacZ constructs.
Reagents and recombinant DNA procedures.
Radiolabeled nucleotides, [γ-32P]ATP (6,000 Ci/mmol; 150 mCi/ml) and [α-32P]dATP (3,000 Ci/mmol; 10 mCi/ml), were purchased from New England Nuclear. Isopropylthio-β-d-galactoside (IPTG) was purchased from Sigma Chemical Co. Restriction endonucleases, T4 DNA ligase, T4 polynucleotide kinase, and T7 RNA polymerase were purchased from New England BioLabs and used according to the manufacturer's specifications. RNase-free DNase I was purchased from Boehringer-Mannheim, and avian myeloblastosis virus reverse transcriptase was purchased from Life Sciences. Sequenase (Amersham) and Pfu DNA polymerase (Stratagene) were used according to the manufacturers' specifications. DNA manipulations, including miniprep plasmid isolations and preparation and transformation of competent cells, were performed according to the methods of Sambrook et al. (15). Oligonucleotides were synthesized using a Beckman 1000M oligo synthesizer. The lacZ-specific oligonucleotide 5′-GTTTTCCCAGTCACGACGTTG-3′ was used in DNA sequencing reactions, primer extensions, toeprint assays, and Northern assays and anneals to positions +92 to +73 of the lacZ coding sequences in both pNUGcI and pSD-NUGcI.
Construction of leadered and leaderless cI-lacZ fusions with NUG start codons.
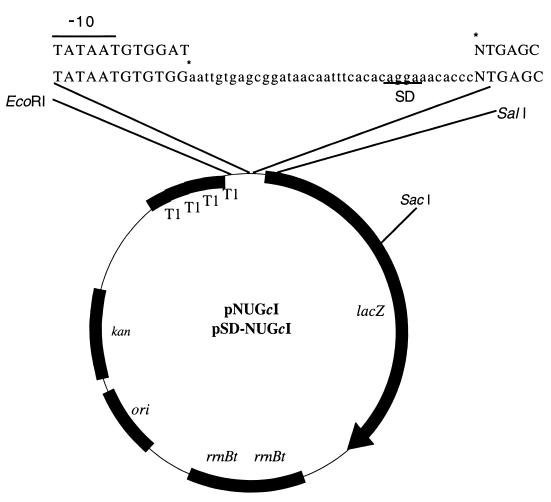
PCR amplification was used to clone cI codons 1 to 16 between the EcoRV and SalI sites of pUL-AUGcI (22). PCR amplification reactions were then used to prepare cI DNA fragments with NTG start codons (where N is A, C, G, or T), essentially as described by Van Etten and Janssen (22). The mutagenic oligonucleotide 5′-NTGAGCACAAAAAAGAAACCATTACC-3′ (where N is A, C, G, or T) anneals to positions +1 (or +2) through +26 of the cI sequence present in the cI-lacZ fusion of pUL-AUGcI (22) and was used as the upstream primer to create DNA fragments with alternate (NUG) cI start codons. The oligonucleotide 5′-ACGCTCATCGATAATTTCACCGCC-3′ anneals to positions +843 to +820 of the lacZ coding sequence and was used as a downstream primer in the PCR amplifications. After PCR-directed mutagenesis of the cI start codon, a DNA fragment containing 16 cI codons was cloned into translation fusion plasmids containing the lacUV5 promoter (25) and a portion of the lacZ gene (Fig. 1) with (pSOD1) or without (pM1108A) a modified lac untranslated leader upstream to the cI coding sequence. Transcription is predicted to initiate at the first nucleotide of a modified lac leader upstream to the cI-lacZ fusion (pSOD1 deravitives) or at the first nucleotide of the cI start codon (pM1108A derivatives). The cloned region was then sequenced to ensure the presence of the desired mutation and the absence of unwanted secondary site changes. The resulting 2.1-kb EcoRI-SacI fragment containing the lacUV5 promoter and cI codons 1 to 16 fused to lacZ, with or without the lac leader, were subcloned into the pACYC177 derivative pUL-AUGcI (22), resulting in leaderless cI-lacZ fusions (pNUGcI) or lac-leadered cI-lacZ fusions (pSD-NUGcI) with an AUG, CUG, GUG, or UUG start codon.
FIG. 1.
Plasmid map of leadered and leaderless cI-lacZ fusion vectors with alternate start codons. DNA sequences above the plasmid map report the differences between leaderless (upper), and leadered (lower) cI sequences (where N is A, G, T, or C) fused to lacZ. Abbreviations and indications: lowercase letters indicate the lac untranslated leader sequence; asterisks indicate transcriptional start sites; nucleotides identifying the lacUV5 promoter −10 region are overlined; SD sequence is underlined; kan, kanamycin resistance; ori, pACYC177 origin of replication; rrnBt, E. coli rrnB T1 and T2 transcriptional terminators; T1, E. coli rrnB T1 transcriptional terminator; lacZ, E. coli β-galactosidase gene.
β-Galactosidase activity measurements.
E. coli RFS859 cells containing pACYC177-derivative plasmids expressing leadered or leaderless cI-lacZ fusions were grown to an optical density at 600 nm of 0.3 to 0.6 in 2× YT (per liter, 16 g of Difco Bacto Tryptone, 10 g of Difco Bacto yeast extract, 10 g of NaCl, pH 7.4) supplemented with kanamycin (25 μg/ml) and 0.2 mM IPTG at 37°C. At least three β-galactosidase assays (9) were performed on each of triplicate cultures for each strain.
Radiolabeling of oligonucleotides.
Oligonucleotides were end labeled as described previously (22).
RNA isolation and Northern blots.
Total RNA was isolated as described previously (25). Northern blotting for the detection of size-fractionated mRNA was carried out with a Schleicher and Schuell Nytran membrane (0.1-μm pore size) as previously described (15). The radiolabeled lacZ-specific oligonucleotide was used at 106 cpm per ml of hybridization solution and was hybridized at 68°C. A control lane was excised from the gel and equilibrated in 0.25 M ammonium acetate with ethidium bromide (2 μg/ml) for 30 min, and the 16S and 23S rRNA bands were visualized with UV light. Size estimations of hybridization signals were extrapolated from a plot of rRNA size versus distance migrated.
Primer extension.
Primer extension reactions containing 80 μg of RNA and 2 pmol of an end-labeled lacZ specific oligonucleotide were performed as previously described (3). The primer extension reactions were electrophoresed against the appropriate dideoxy sequencing reactions and visualized by autoradiography.
Primer extension inhibition (toeprint) assays.
Messenger RNAs for use in the toeprint assays were generated in vitro using T7 RNA polymerase (8). In brief, DNA fragments used in the T7 transcription reactions were prepared by PCR using either pNUGcI or pSD-NUGcI (where N is A, C, G, or T) plasmids as template. For the PCRs using pNUGcI as the template, a downstream primer, 5′-TTCCCGCTAGCCACGCCCGG-3′, and an upstream primer 5′-GGAATT CTAATACGACTCACTATAGNTGAGCACAAAAAAGAAACCATTA-3′ (where N is A, C, G, or T), were used, resulting in DNA fragments encoding a T7 promoter and leaderless cI-lacZ fragments containing AUG, CUG, GUG, or UUG initiation codons. PCRs were also carried out using pSD-NUGcI as the template (where N is A, C, G, or U) with the downstream primer 5′-TTCCCGCTAGCCACGCCCGG-3′ and the upstream primer 5′-GGAATCTAATACGACTCACTATAGAATTGTGAGCGG-3′, resulting in DNA fragments encoding a T7 promoter and lac-leadered cI-lacZ fragments containing AUG, CUG, GUG, or UUG initiation codons. Transcription reactions contained 15 mM dithiothreitol, 4 mM nucleoside triphosphates, 40 mM Tris-HCl (pH 7.9), 8 mM MgCl2, 2 mM spermidine, 1 μg of template DNA, and 500 U of T7 RNA polymerase (NEB) and were carried out at 37°C for 1 h, followed by treatment with 5 U of RNase-free DNase I for 30 min at 37°C. The transcription reactions were then extracted with phenol-chloroform-isoamyl alcohol (25:24:1), and the RNA was precipitated with 0.3 M sodium acetate (pH 6) and isopropanol. The final RNA concentration was determined by absorbance at 260 nm (assuming one unit of optical density at 260 nm is 40 μg ml−1 and 330 g mol−1 nucleotide−1).
The toeprint assays were carried out according to the method of Martin-Farmer and Janssen (8).
RESULTS
Construction of leaderless and leadered cI-lacZ fusions with alternate start codons.
To investigate the contribution of the initiation codon to translation of leaderless and lac-leadered cI, six site-directed mutations were constructed from plasmids containing a lacZ reporter gene without (pNUGcI) or with (pSD-NUGcI) an untranslated leader (Fig. 1). Six mutations were constructed to change the start codon of pSD-AUGcI and pAUGcI to CUG, GUG, or UUG.
Identification of transcriptional start sites.
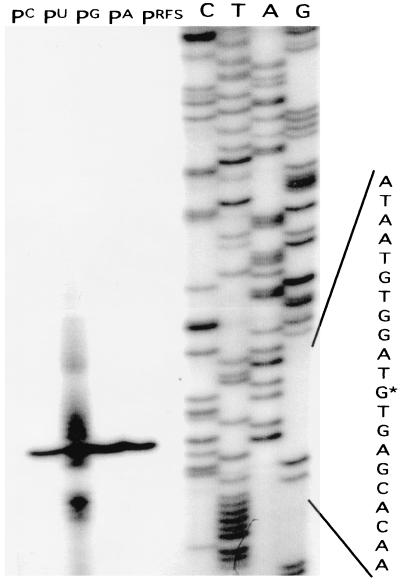
Primer extension analysis of leaderless cI-lacZ fusions revealed that transcription initiated at the first position of the cI start codon regardless of whether it was AUG, GUG, or CUG; analysis of cI-lacZ initiation containing a UUG start codon revealed several bands in addition to the expected start site (Fig. 2). The same pattern of bands was observed with RNA extracted from cells containing a second, independently constructed pUUGcI (data not shown). Dideoxy sequencing of the mutagenized region revealed that the UUG start codon was present with no additional deletions or rearrangements (data not shown). This suggests that the presence of a UUG start codon somehow destabilizes initiation from the lacUV5 promoter at the cI start site. However, a primer extension signal is observed corresponding to position +1 of the UUG start codon and indicates the presence of mRNA with an intact UUG start codon. Various amounts of the primer extension reaction products were needed to visualize the transcriptional start sites, presumably an indication of the in vivo steady-state abundance of cI-lacZ mRNA levels. Transcription of the lac-leadered cI-lacZ fusions is expected to initiate at the first nucleotide of the 38-nucleotide lac leader, as demonstrated previously by Van Etten and Janssen (22) with this promoter and leader sequence.
FIG. 2.
Transcriptional start sites of leaderless cI-lacZ fusions with alternate start codons by primer extension analysis. The DNA sequence presented on the right is the sense strand from +10 to −10 relative to the transcriptional start site (+1) present in pGUGcI and is the complement of the DNA sequence presented in the autoradiogram. Lanes G, A, T, and C indicate the dideoxy termination sequencing reactions. The transcriptional start site is indicated on the DNA sequence with an asterisk. Lanes PA, PG, PU, PC, and PRFS represent the primer extension reaction products resulting from RNA isolated from E. coli RFS859 (PRFS), and E. coli RFS859 containing the plasmids pAUGcI (PA), pGUGcI (PG), pUUGcI (PU), or pCUGcI (PC). Dilutions of the primer extension reactions were performed as follows: pAUGcI, 1:20; pGUGcI, 1:9; pUUGcI, 1:4; and pCUGcI, undiluted.
Both leaderless and leadered cI-lacZ fusions require an AUG start codon for efficient expression.
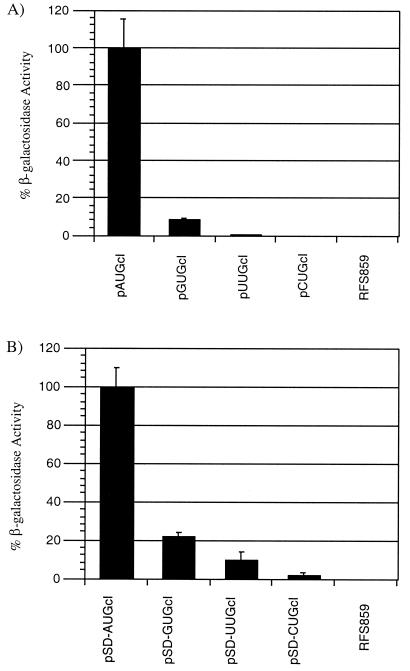
β-Galactosidase assays were performed on both leaderless and leadered cI-lacZ fusions with alternate start codons. The β-galactosidase activity of the leaderless constructs containing an AUG, GUG, UUG, or CUG start codon was 4,481 (=100%), 360 (=8.0%), 15 (=0.3%), and 3 (≤0.1%) Miller units (9), respectively (Fig. 3A). The lac-leadered cI-lacZ fusions expressed significantly more β-galactosidase than the leaderless constructs. β-Galactosidase activity from cells expressing the leadered cI-lacZ fusions with AUG, GUG, UUG, or CUG start codons was 25,244 (=100%), 6,638 (=26.3%), 2,825 (=11.2%), and 432 (=1.7%) Miller units, respectively (Fig. 3B). In both the leaderless (pNUGcI) and the leadered (pSD-NUGcI) constructs, AUG was the most efficient start codon used for initiation, followed by GUG, UUG, and very little expression from CUG (Fig. 3). This observation fits the model of start codon hierarchy proposed for E. coli leadered mRNA (14), in which AUG was most efficient followed by GUG, UUG, and CUG, respectively.
FIG. 3.
β-Galactosidase expression of leaderless (A) and leadered (B) cI-lacZ fusions with alternate start codons and the host negative control. (A) A value of 4,481 Miller units, obtained for the pAUGcI construct, is represented as 100%; (B) A value of 25,244 Miller units, obtained for the pSD-AUGcI construct, is represented as 100%.
The presence of an AUG initiation codon correlates with a greater abundance of full-length lacZ mRNA.
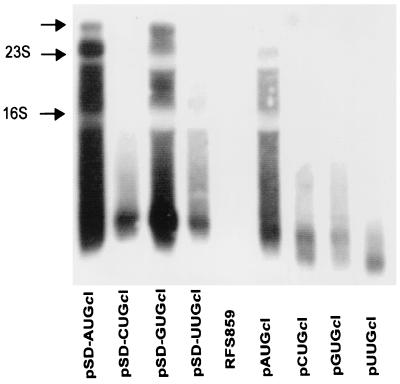
RNAs extracted from E. coli RFS859 cells containing either leaderless or leadered cI-lacZ fusions were size fractionated in an agarose gel and analyzed by Northern hybridization. RNA from cells containing pAUGcI or pSD-AUGcI revealed a greater abundance of high-molecular-weight cI-lacZ mRNA relative to the signals observed for the leaderless or leadered fusions containing GUG, UUG, or CUG initiation codons (Fig. 4). The largest hybridizing signal, approximately 4,200 nt, is the predicted size of full-length cI-lacZ mRNA, suggesting that cells containing cI-lacZ fusions with AUG start codons contain more full-length functional mRNA. The abundant low-molecular-weight hybridization signals expressed from non-AUG cI-lacZ fusions are estimated to be 300 to 800 nt, suggesting that they represent stable 5′-terminal fragments of cI-lacZ mRNA.
FIG. 4.
Analysis of lacZ mRNA stability by Northern blotting. Autoradiogram of size-fractionated RNA probed for cI-lacZ mRNA. The labels across the bottom indicate the source of the RNA. The arrows on the side indicate positions of the 23S and 16S rRNAs as indicated. The upper arrow indicates the predicted position of full-length lacZ mRNA, estimated to be approximately 4,200 nt. The abundant smaller signals present near the bottom of the blot are estimated to be 300 to 800 nt.
Efficient in vitro ribosome binding of leaderless and leadered cI-lacZ mRNA requires an AUG initiation codon.
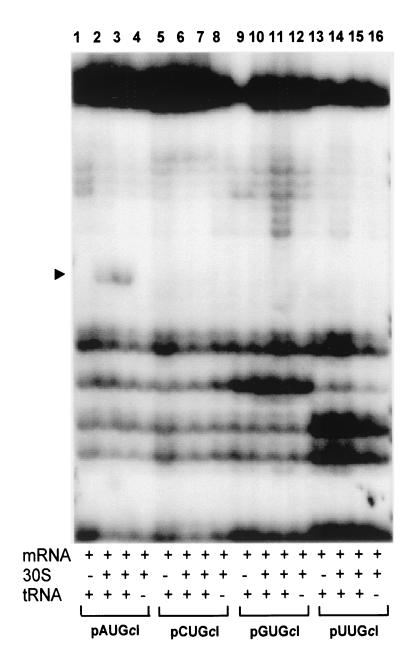
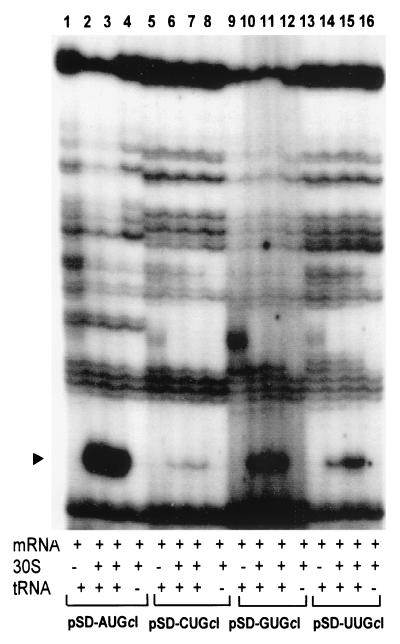
Primer extension inhibition (toeprint) assays (5) were carried out to investigate the relative ribosome binding strength of leadered and leaderless cI-lacZ mRNAs with NUG start codons. Using two different ribosome/mRNA ratios, a ternary complex (i.e., mRNA, 30S ribosomal subunit, and initiator tRNA)-dependent toeprint signal was observed at position +16 relative to the first position (+1) of the start codon for leaderless cI-lacZ mRNA containing an AUG initiation codon (Fig. 5, lanes 2, 3). Under identical reaction conditions, leaderless cI-lacZ mRNA with CUG, GUG, or UUG initiation codons did not produce a detectable ternary complex-dependent toeprint signal (Fig. 5). Leadered cI-lacZ mRNAs were also assayed under similar conditions. A ternary complex dependent toeprint signal was observed with leadered cI-lacZ mRNAs containing AUG, CUG, GUG, and UUG initiation codons (Fig. 6). The relative intensity of the toeprint signals differed significantly, with AUG being the strongest, followed by GUG, and UUG, and only a weak toeprint signal was observed with the CUG initiation codon.
FIG. 5.
Primer extension inhibition (toeprinting) assays for leaderless cI-lacZ mRNAs containing alternate start codons. Toeprinting assays were performed without 30S subunits (lanes 1, 5, 9, and 13) or with 30S subunits in a 3-fold (lanes 2, 6, 10, and 14) or 12-fold (lanes 3, 7, 11, and 15) molar excess over mRNA or a 12-fold molar excess of 30S subunits without initiator tRNA (lanes 4, 8, 12, and 16). The labels across the bottom indicate the plasmid templates used to synthesize the mRNA, while the arrowhead indicates the position of the toeprint signal (+16) relative to the first position of the NUG start codon; the migratory position of the toeprint signal was identified by simultaneous electrophoresis of a dideoxy DNA-sequencing reaction adjacent to the toeprint reactions (not shown).
FIG. 6.
Primer extension inhibition (toeprinting) assays for leadered cI-lacZ mRNAs containing alternate start codons. Toeprinting assays were performed without 30S subunits (lanes 1, 5, 9, and 13) or with 30S subunits in a 3-fold (lanes 2, 6, 10, and 14) or 12-fold (lanes 3, 7, 11, and 15) molar excess over mRNA or a 12-fold molar excess of 30S subunits without initiator tRNA (lanes 4, 8, 12, and 16). The labels across the bottom indicate the plasmid templates used to synthesize the mRNA, while the arrowhead indicates the position of the toeprint signal (+16) relative to the first position of the NUG start codon; the migratory position of the toeprint signal was identified by simultaneous electrophoresis of a dideoxy DNA-sequencing reaction adjacent to the toeprint reactions (not shown). The autoradiogram was overexposed in order to visualize the toeprint signal with mRNA transcribed from pSD-CUGcI.
The toeprinting results indicate that an AUG initiation codon was required to produce a toeprint signal with leaderless cI-lacZ mRNA and 30S subunits. The inability to toeprint non-AUG cI-lacZ mRNAs suggests that the low in vivo expression levels (Fig. 3A) relate to the inefficient binding of ribosomes to leaderless mRNA with a non-AUG initiation codon. The start codon effects on in vitro ribosome binding strength, as measured by toeprinting assays for both leaderless and leadered cI-lacZ mRNAs, can be correlated with the relative abundance of full-length in vivo mRNA (Fig. 4) and in vivo expression (Fig. 3), suggesting that ribosome binding contributes to mRNA functional stability, possibly by protecting it from degradation.
DISCUSSION
This report is the first analysis of start codon efficiency for translation of a naturally leaderless mRNA. Changing the start codon of the leaderless cI-lacZ fusion from AUG to GUG resulted in a 12-fold reduction in expression; alterations from AUG to UUG or CUG reduced expression to background levels. Northern blots suggested that the start codon contributed to mRNA stability, resulting in a greater amount of functional mRNA with an AUG start codon relative to mRNA containing GUG, UUG, or CUG start codons. Toeprinting analysis further suggested that an AUG initiation codon was needed for efficient binding of 30S ribosomal subunits to leaderless cI-lacZ mRNA in vitro. The strength of the ribosome binding site has been correlated with the amount of full-length mRNA present (27); a strong ribosome binding site, supporting frequent translation, can provide ribosome-mediated protection of mRNA from ribonuclease degradation. As evident from the toeprinting assays and Northern analysis, the increased amount of cI-lacZ expression from mRNA with an AUG initiation codon correlates well with a stronger ribosome binding site and more full-length mRNA. The dramatic reduction in in vitro ribosome binding strength, in vivo expression levels, and full-length message abundance when a non-AUG start codon was used suggests that an AUG start codon is an essential feature for efficient translation of leaderless cI mRNA. The AUG requirement for efficient cI expression is similar to recent observations with lacZ and gusA genes from which the untranslated leader region had been deleted (22); alterations of the AUG start codon to GUG greatly reduced expression, while UUG and CUG failed to support measurable levels of expression. The similarity of these results suggests that both mRNAs initiate translation by a similar mechanism, even though one occurs naturally as a leaderless mRNA and the other is genetically engineered to be leaderless.
Addition of the lac leader to the cI coding sequence allowed translation with non-AUG initiation codons (Fig. 3B); relative expression from the leadered cI-lacZ mRNAs was much reduced from that observed with the leadered lacZ and gusA genes containing non-AUG start codons (22), suggesting that an AUG start codon might be a stronger translational signal for cI mRNA. In vitro ribosome binding strength, as estimated from toeprint assays, revealed that 30S subunits bound leadered mRNA containing an AUG initiation codon most efficiently, followed by GUG, UUG, and only slight binding to mRNA containing CUG. Northern analysis revealed that the amount of leadered cI-lacZ full-length mRNA containing alternate start codons could be correlated with the strength of in vitro ribosome binding and in vivo expression levels. These results suggest that the start codon affects the mRNA's ribosome binding strength; the strength of ribosome binding would then influence the frequency of translation, in vivo expression levels, and mRNA stability.
Our results differ from those reported by Wagner et al. using lacZ mRNAs with NUG start codons and alternate SD sequences (24). Although ribosome binding was not directly measured, Wagner et al. concluded that mRNA stability was maintained by ribosomal binding via the SD sequence and was not affected by changing the start codon from AUG to CUG, GUG, or UUG. We show here, using leadered cI-lacZ messages with a constant SD sequence, the in vitro ribosome binding strength was directly related to the start codon sequence. In addition, the in vivo expression levels and abundance of full-length mRNA correlate with the toeprint signal intensities, suggesting that the start codon contributes importantly to in vivo ribosome binding and translation efficiency. The difference between our findings and those of Wagner et al. (24) might relate to the cI coding sequence present in our cI-lacZ fusions, the alterations introduced to their untranslated leader sequence, or stability of their mRNA fragment containing the NUG start codons.
How does the ribosome discriminate between alternate start codons on an mRNA? IF3 inspects the P site codon-anticodon interaction and destabilizes unfavorable pairings. It has been shown by Hartz et al. (5a) that IF3 does not discriminate against GUG or UUG but does discriminate against an AUU start codon, suggesting that the start codon third position is important for IF3 inspection. Therefore, it seems unlikely that IF3 selects against GUG, UUG, and CUG start codons on leadered mRNA. The presence of a suppressor initiator tRNA was sufficient to restore expression to a leadered cI-lacZ mRNA with a UAG start codon (22), suggesting that codon-anticodon complementarity is an important, perhaps sufficient, feature for initiation on leadered mRNA. The reduced efficiency of GUG, UUG, and CUG start codons on leadered mRNA might relate to reduced stability of noncomplementary pairings and/or stereospecific constraints of the P site anticodon U pairing with a start codon's first position purine (A or G) or pyrimidine (C or U).
The leaderless cI-lacZ mRNA requires an AUG start codon for efficient expression. This requirement is unlikely to reflect a need for codon-anticodon complementarity because leaderless cI-lacZ with a noncognate UAG start codon was not expressed in the presence of a UAG-suppressing initiator tRNA (22), suggesting that some component other than IF3 discriminates against non-AUG start codons of leaderless mRNA. Also, it has been reported recently (21) that IF3 discriminates against a 5′ AUG on leaderless mRNA, suggesting that IF3 might antagonize expression of all leaderless mRNAs in vivo. How is the 5′-terminal start codon of a leaderless mRNA selected if IF3 discriminates against all 5′-terminal start codons? One possibility might involve the selection of leaderless mRNA and start codon inspection by an IF2-formylmethionyl-tRNA complex already present on a subset of 30S subunits, an initiation pathway reported for translation of some mRNAs in E. coli (26). Additional evidence to support IF2-enhanced translation of leaderless mRNA has been reported recently (4). Alternatively, leaderless mRNAs might bind 70S monosomes in vivo, as has been demonstrated in vitro (1), and start codon discrimination might occur in the absence of initiation factors during the initial interaction of the mRNA 5′ end with assembled ribosomes.
Interestingly, two actinomycete genes encoding leaderless mRNAs initiate with GUG (2, 23). The results presented here suggest that the GUG start codons might function to down-regulate expression from these genes. Alternatively, actinomycete translation systems might differ from E. coli in their ability to initiate efficiently from GUG start codons on leaderless mRNA. Additional analysis of leaderless mRNA translation signals will contribute to our understanding of translation initiation and ribosome interactions with mRNA.
ACKNOWLEDGMENT
This work was supported by grant GM45923 from the National Institutes of Health.
REFERENCES
- 1.Balakin A G, Skripkin E A, Shatsky I N, Bogdanov A A. Unusual ribosome binding properties of mRNA encoding bacteriophage λ repressor. Nucleic Acids Res. 1992;20:563–571. doi: 10.1093/nar/20.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibb M J, White J, Ward J M, Janssen G R. The mRNA for the 23S RNA methylase encoded by the ermE gene of Saccharopolyspora erythraea is translated in the absence of a conventional ribosome-binding site. Mol Microbiol. 1994;14:533–545. doi: 10.1111/j.1365-2958.1994.tb02187.x. [DOI] [PubMed] [Google Scholar]
- 3.Brown J W, Thomm M, Beckler G S, Frey G, Stetter K O, Reeve J N. An archaebacterial RNA polymerase binding site and transcription initiation of the hisA gene in Methanococcus vanniellii. Nucleic Acids Res. 1988;16:135–150. doi: 10.1093/nar/16.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grill S, Gualerzi C O, Londel P, Blasi U. Selective stimulation of translation of leaderless mRNA by initiation factor 2: evolutionary implications for translation. EMBO J. 2000;19:1–10. doi: 10.1093/emboj/19.15.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartz D, McPheeters D S, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 5a.Hartz D, Binkley J, Hollingsworth T, Gold L. Domains of initiator tRNA and initiation codon crucial for initiator tRNA selection by Escherichia coli IF3. Genes Dev. 1990;4:1790–1800. doi: 10.1101/gad.4.10.1790. [DOI] [PubMed] [Google Scholar]
- 6.Hui A, de Boer H A. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc Natl Acad Sci USA. 1987;84:4762–4766. doi: 10.1073/pnas.84.14.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacob W F, Santer M, Dahlberg A E. A single base change in the Shine-Dalgarno region of 16S RNA of Escherichia coli affects translation of many proteins. Proc Natl Acad Sci USA. 1987;84:4757–4761. doi: 10.1073/pnas.84.14.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin-Farmer J, Janssen G R. A downstream CA repeat sequence increases translation from leadered and unleadered mRNA in Escherichia coli. Mol Microbiol. 1999;31:1025–1038. doi: 10.1046/j.1365-2958.1999.01228.x. [DOI] [PubMed] [Google Scholar]
- 9.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 10.Moll I, Resch A, Blasi U. Discrimination of 5′-terminal start codons by translation initiation factor 3 is mediated by ribosomal protein S1. FEBS Lett. 1998;436:213–217. doi: 10.1016/s0014-5793(98)01131-4. [DOI] [PubMed] [Google Scholar]
- 11.O'Connor M, Asai T, Squires C L, Dahlberg A E. Enhancement of translation by the downstream box does not involve base pairing of mRNA with the penultimate stem sequence of 16S rRNA. Proc Natl Acad Sci USA. 1999;96:8973–8978. doi: 10.1073/pnas.96.16.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ptashne M, Backman K, Humayum M Z, Jeffrey A, Maurer R, Meyer B, Sauer R T. Autoregulation and function of a repressor in bacteriophage lambda. Science. 1976;194:156–161. doi: 10.1126/science.959843. [DOI] [PubMed] [Google Scholar]
- 13.Resch A, Tedin K, Grundling A, Mundlein A, Blasi U. Downstream box-anti-downstream box interactions are dispensible for translation initiation of leaderless mRNAs. EMBO J. 1996;15:4740–4748. [PMC free article] [PubMed] [Google Scholar]
- 14.Ringquist S, Shinedling S, Barrick D, Green L, Binkley J, Stormo G D, Gold L. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol Microbiol. 1992;6:1219–1229. doi: 10.1111/j.1365-2958.1992.tb01561.x. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 16.Schleif R. Fine-structure deletion map of the Escherichia colil-arabinose operon. Proc Natl Acad Sci USA. 1972;69:3479–3484. doi: 10.1073/pnas.69.11.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider T D, Stormo G D, Gold L. Information content of binding sites on nucleotide sequences. J Mol Biol. 1986;188:415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- 18.Shean C S, Gottesman M E. Translation of the prophage lambda cI transcript. Cell. 1992;70:513–522. doi: 10.1016/0092-8674(92)90175-c. [DOI] [PubMed] [Google Scholar]
- 19.Shine J, Dalgarno L. The 3′ terminal of Escherichia coli 16S ribosomal RNA: complementarity to non-sense triplets and ribosomal binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sprengart M L, Fatscher H P, Fuchs E. The initiation of translation in Escherichia coli: apparent base pairing between the 16S RNA and downstream sequences of mRNA. Nucleic Acids Res. 1990;18:1719–1723. doi: 10.1093/nar/18.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tedin K, Moll I, Grill S, Resch A, Graschopf A, Gualerzi C O, Blasi U. Translation initiation factor 3 antagonizes authentic start codon selection on leaderless mRNAs. Mol Microbiol. 1999;31:67–77. doi: 10.1046/j.1365-2958.1999.01147.x. [DOI] [PubMed] [Google Scholar]
- 22.Van Etten W J, Janssen G R. An AUG initiation codon, not codon-anticodon complementarity, is required for the translation of unleadered mRNA in Escherichia coli. Mol Microbiol. 1998;27:987–1001. doi: 10.1046/j.1365-2958.1998.00744.x. [DOI] [PubMed] [Google Scholar]
- 23.Van Wezel G P, White J, Young P, Postma P W, Bibb M J. Substrate induction and glucose repression of maltose utilization by Streptomyces coelicolor (A2) is controlled by malR, a member of the lacI-galR family of regulatory genes. Mol Microbiol. 1997;23:537–549. doi: 10.1046/j.1365-2958.1997.d01-1878.x. [DOI] [PubMed] [Google Scholar]
- 24.Wagner L A, Gesteland R F, Dayhuff T J, Weiss R B. An efficient Shine-Dalgarno sequence but not translation is necessary for lacZ mRNA stability in Escherichia coli. J Bacteriol. 1994;176:1683–1688. doi: 10.1128/jb.176.6.1683-1688.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu C-J, Janssen G R. Translation of vph mRNA in Streptomyces lividans and Escherichia coli after removal of the 5′ untranslated leader. Mol Microbiol. 1996;22:339–355. doi: 10.1046/j.1365-2958.1996.00119.x. [DOI] [PubMed] [Google Scholar]
- 26.Wu X-Q, RajBhandary U L. Effect of the amino acid attached to Escherichia coli initiator tRNA on its affinity for the initiation factor IF2 and on IF2 dependence of its binding to the ribosome. J Biol Chem. 1997;272:1891–1895. doi: 10.1074/jbc.272.3.1891. [DOI] [PubMed] [Google Scholar]
- 27.Yarchuk O, Jacques N, Guillerez J, Dreyfus M. Interdependence of translation and mRNA degradation in the lacZ gene. Biochimie. 1992;73:1533–1541. doi: 10.1016/0022-2836(92)90617-s. [DOI] [PubMed] [Google Scholar]