Abstract
The sodA gene encoding the Corynebacterium melassecola manganese-cofactored superoxide dismutase (SOD) has been cloned in Escherichia coli and sequenced. The gene is transcribed monocistronically; the predicted polypeptide is 200 amino acids long and associates in a homotetrameric, manganese-dependent form, able to complement an SOD-deficient E. coli mutant. A second open reading frame, coding for a putative 217-amino-acid protein with high homology to peptide methionine sulfoxide reductases from various origins, has been identified immediately upstream of sodA in the opposite transcription orientation. The sodA gene was inactivated by insertion of an integrative vector carrying a kanamycin resistance gene. The growth rate of the SOD-deficient integrant was only slightly affected in BHI rich medium as well as in BMCG chemically defined medium, but was strongly affected by the presence of the redox-cycling agent paraquat. The SOD deficiency had, on the other hand, a deleterious effect on viability as soon as the culture entered the stationary phase of growth in BHI medium. Surprisingly, SOD deficiency was able to rescue the dramatic loss of viability observed for the wild-type strain in BMCG synthetic medium when glucose was not the limiting growth factor.
Reactive oxygen species are formed in aerobic organisms as by-products of normal metabolism, as a consequence of partial reduction of molecular oxygen to superoxide anion (O2·−), hydrogen peroxide (H2O2), and hydroxyl radical ( · OH). To protect cells from damage by these reactive species to DNA, proteins, and lipids, aerobic organisms have evolved detoxification and repair systems (69). Superoxide dismutase (SOD; EC 1.15.1.1) is considered a key enzyme in oxygen defense systems by catalyzing the dismutation of O2·− into oxygen and H2O2 (22), the latter being broken in turn to water by catalase or peroxidase.
SODs are classified in different groups depending on the type of metal cofactors: copper-and zinc-containing (Cu,Zn-SOD) (46), iron-containing (Fe-SOD) (83), manganese-containing (Mn-SOD) (39), and nickel-containing (Ni-SOD) (84) enzymes. Another type of small metalloprotein with superoxide dismutase activity, unrelated to classical SODs, has also been recently identified in the sulfate-reducing bacterium Desulfovibrio (65). Most bacteria possess an Mn-SOD or an Fe-SOD in their cytoplasm, while Cu,Zn-SODs have been identified in the periplasm or periphery of pathogenic and endosymbiotic bacteria (4, 5, 80). The Mn- and Fe-SODs have highly similar sequences and structures, usually have a strict metal selectivity, and are evolutionarily unrelated to other SODs. On the other hand, some SODs have been found to be active with either manganese or iron incorporated into the same apoprotein (47, 63, 82). These enzymes were named cambialistic. Some bacteria, like Escherichia coli and Pseudomonas aeruginosa, have both an Fe-SOD and an Mn-SOD, which differ not only in their primary sequences but also in their regulation (29, 72) and, at least for E. coli, in their intracellular localization, the Fe-SOD being located closer to the inner membrane than the Mn-SOD enzyme (32).
With the exception of E. coli, which has been investigated in detail (72), little is known about bacterial SOD expression and regulation, especially in gram-positive bacteria, although many genes have been cloned and sequenced. Most work concerns pathogenic species, mainly because of the relationship between antioxidant defenses and pathogenicity (1, 4, 85). Less is known about their nonpathogenic relatives. Although the amino acid-producing, telluric, and nonpathogenic Corynebacteria species have been well studied for decades, most of the work has focused on biochemical pathways or metabolic fluxes, and nothing is known about their antioxidant defenses. The so-called “amino acid-producing corynebacteria” group contains the closely related Corynebacterium glutamicum, Corynebacterium melassecola, Brevibacterium flavum, and Brevibacterium lactofermentum species (42), sometimes gathered together under the sole C. glutamicum name, and belongs to the actinomycetes suborder along with the Mycobacterium and Nocardia genera (66). Considering the high oxygen requirement of the aerobic C. melassecola species (15), we found it of great interest to study their antioxidant defenses. We therefore investigated and characterized the sodA gene and the corresponding protein of C. melassecola and studied the role of this gene in the response to superoxide and in survival after starvation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and vectors used in this study are listed in Table 1. C. melassecola strains were grown aerobically (250 rpm) at 34°C, either in brain heart infusion (BHI; Difco) rich medium or in BMCG chemically defined medium containing 2% glucose, as described by Guillouet and Engasser (26). Growth was followed by measuring the optical density at 570 nm (OD570) in a DU 7400 Beckman spectrophotometer. Kanamycin (25 μg/ml) and chloramphenicol (6 μg/ml) were added to the medium when needed. The E. coli DH5α strain was used for molecular biology procedures. E. coli QC1799 sodA sodB and its parental GC4468 strain were kindly provided by Danièle Touati (Institut Jacques Monod, Université Paris VII, Paris, France). E coli strains were grown aerobically at 37°C in Luria-Bertani (LB) medium (Difco) or in M63 glucose minimal medium as described by Carlioz and Touati (13). Growth was monitored at 600 nm (OD600). Ampicillin (100 μg/ml), kanamycin (25 μg/ml), and chloramphenicol (30 μg/ml) were added to the medium when needed. All chemicals were purchased from Sigma.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristics | Sourcea or reference |
|---|---|---|
| C. melassecola | ||
| ATCC 17965 | Wild-type strain | ATCC |
| CGL10016 | ATCC 17965 sodA::viMM12 (Kmr) | This study |
| E. coli | ||
| DH5α | F− Δ(lac-argF)U169 recA1 endA1 hsdR (rK− mK+) supE44 gyrA1 relA1 deoR thi-1 (Φ80dlacZΔM15) | BRL |
| GC4468 | F− Δ(lac-argF)U169 rpsL | 73 |
| QC1799 | F− Δ(lac-argF)U169 rpsL ΔsodA3, Φ(sodB-kan)Δ2 | 73 |
| Plasmids | ||
| pGEM-T | Cloning vector for PCR fragments, ColE1 origin, Apr | Promega |
| pMM7 | pGEM-T with 0.29-kb S1/S3 amplified fragment from C. melassecola, Apr | This study |
| pUN121 | Cloning vector, ColE1 origin, Apr Tcs | 53 |
| pMM8 | pUN121 with 10-kb C. melassecola insertion including sodA, Apr Tcr | This study |
| pCGL243 | Shuttle vector, pACYC184 origin for E. coli, pBL1 origin for C. melassecola, Kmr | 59 |
| pMM12 | pCGL243 with a 0.36-kb BamHI-HindIII fragment internal to sodA (S6/S7 amplification fragment), Kmr | This study |
| pKK388-1 | E. coli expression vector with trc promoter, Apr | 11 |
| pCGL482 | Shuttle vector, pACYC184 origin for E. coli, pBL1 origin for C. melassecola, Cmr | 57 |
| pMM23 | pCGL482 with the sodA gene on a 0.8-kb NcoI-SmaI fragment from pMM8, under the trc promoter as a BamHI-NcoI fragment from pKK388-1, Cmr | This study |
| viMM12 | Integrative vector derived from pMM12 by NotI digestion and religation of the 1.8-kb fragment carrying aphIII and the internal sodA fragment, Kmr | This study |
ATCC, American Type Culture Collection, Rockville, Md.; BRL, Bethesda Research Laboratory, Gaithersburg, Md.; Promega Corporation, Madison, Wis.
C. melassecola-specific microbial procedures.
Sensitivity to paraquat and hydrogen peroxide (H2O2) was monitored by growth inhibition on plates. Bacteria were grown in liquid BHI rich medium to late log phase, and 100-μl aliquots from a 1,000-fold dilution were spread on BHI agar plates. A 5-mm sterile filter paper disk was placed in the middle of each plate and impregnated with 10 μl of 10-mg/ml paraquat or 10% H2O2. After 2 days of incubation at 34°C, sensitivity was determined by measuring the diameter of the growth inhibition zone surrounding the disk. For the determination of bacterial viability, C. melassecola cultures were serially diluted at room temperature in NaCl (0.9 g/liter), and aliquots were spread on BHI plates. After 2 days of incubation at 34°C, the colonies were counted, and viability was calculated as CFU per OD570 unit (CFU/OD).
General molecular biology techniques.
Standard molecular biology procedures were used according to Sambrook et al. (62). Enzymes were obtained from Promega and Boehringer Mannheim. Plasmid DNA was prepared with the Wizard kit from Promega, with 1 h of pretreatment at 37°C for C. melassecola in Wizard resuspension solution containing lysozyme (20 mg/ml). DNA fragments were isolated from agarose gels with the Jetsorb kit (Genomed). Preparation of C. melassecola genomic DNA and Southern blotting were performed as described elsewhere (59). DNA probes were labeled with [α-32P]dCTP (Amersham) using the T7 Quick Prime random priming kit (Pharmacia).
Total C. melassecola RNA was prepared from cells grown until the late exponential phase as described by Igo and Losick (34). For Northern blot analysis, approximately 10 μg of RNA was separated on a 1% agarose gel with the RNA ladder (Promega) as size markers. Transfer to a Hybond-N (Amersham) membrane, probe labeling, and hybridization at 65°C without formamide were done as described by Sambrook et al. (62). The probe used for sodA transcript monitoring was the PCR fragment isolated from pMM7 (see below). Methylene blue staining of RNAs on blots was performed according to Wilkinson et al. (78).
Cloning of the sodA gene. (i) Amplification of an internal fragment by PCR.
For PCR experiments, primers S1 [5′-GGCCACACCAACCACTCCGT(C/G)TT-3′] and S3 [5′-AGGTAGAA(A/T)GCGTGCTCCCACAT-3′] (see Fig. 3) were chosen on the basis of highly conserved regions in bacterial Fe- and Mn-SODs (30) and according to the codon preference in Corynebacterium-related species (45). PCR was carried out with 2.5 U of thermostable DNA polymerase (AmpliTaqGold; Perkin-Elmer) in a reaction mixture containing 30 ng of genomic C. melassecola DNA, 0.2 mM each deoxynucleotide triphosphates (Promega), 0.5 μM each of both primers, 2 mM MgCl2, and 1× AmpliTaqBuffer in a final volume of 50 μl. For the amplification reaction, after 10 min at 94°C, 35 identical cycles (1 min at 94°C, 1 min at 50°C, and 1 min at 72°C) were followed by a final elongation step of 5 min at 72°C. The amplified DNA fragment of the expected size was cloned into the pGEM-T vector (Promega) to give plasmid pMM7. Partial sequencing of the insertion showed high homology with known Fe- and Mn-SODs.
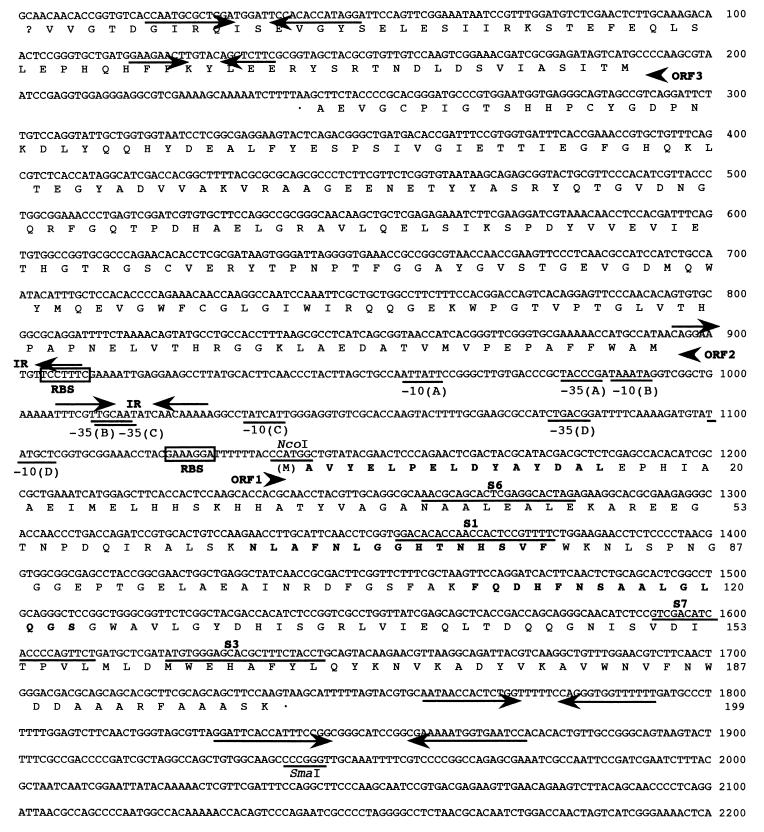
FIG. 3.
Nucleotide sequence of C. melassecola sodA region. ORF1 corresponds to the SOD and ORF2 to the PMSR encoding sequences; ORF3 is incomplete. The nucleotide sequence corresponds to the sodA coding and to the msrA-ORF3 noncoding strand. The deduced amino acid sequences are given below the nucleotide sequence. Amino acids that were determined by amino acid sequencing of the purified C. melassecola SOD are indicated in bold. Putative ribosome-binding sites (RBS) are boxed, and possible promoter regions are underlined. The two inverted sequences (IR) in the promoter region and the putative strem-loop structures downstream of the ORFs are shown with arrows. The NcoI and SmaI restriction sites used for pMM23 construction and the locations of PCR primers are underlined (5′-3′ orientation for S6 and S1, 3′-5′ orientation for S7 and S3).
(ii) Cloning of the entire sodA gene.
The amplified fragment isolated from pMM7 was radiolabeled and used as a probe for colony hybridization (62). One clone (pMM8) was selected from a previously described C. melassecola ATCC 17965 genomic library (59). The pMM8 plasmid was shown to carry a 10-kb inserted DNA fragment. From Southern experiments, this DNA fragment clearly originated from the C. melassecola chromosomal DNA, without detectable structural alteration. The DNA sequence was determined for 2.2 kb on both stands by primer walking in the region covering the sodA gene, using an ABI model 373 DNA sequencer (Applied Biosystem).
Disruption of the sodA chromosomal gene.
An internal sodA fragment (see Fig. 3) was amplified by PCR using primers S6 (5′-GCAGGATCCAACGCAGCACTCGAGGCACTA-3′), including a BamHI site (underlined), and S7 (5′-ATCAAGCTTCAGAACTGGGGTGATGTCGAT-3′), including an HindIII site (underlined). The 371-bp PCR fragment was digested with BamHI and HindIII and inserted into the E. coli-C. melassecola shuttle vector pCGL243 (59) to create pMM12. This vector was used to construct the integron, essentially as described by Reyes et al. (59). The pMM12 plasmid was introduced into C. melassecola by electroporation (9). Plasmid pMM12 was extracted from this host and digested by NotI. The 1.8-kb fragment containing the aphIII kanamycin resistance gene and the internal sodA fragment was isolated and self-ligated to create the viMM12 integron. Transformants from C. melassecola wild-type strain with the integron were the result of a single crossing-over, as verified by Southern blotting. The disrupted strain was named CGL10016, and two independant integrants were used for physiological analysis.
Construction of an expression vector for C. melassecola sodA.
The trc promoter was isolated from pKK388-1 (11) as a 0.36-kb BamHI-NcoI fragment, and sodA was isolated from pMM8 as a 0.8-kb NcoI-SmaI fragment, including the entire open reading frame (ORF) and the putative terminator (see Fig. 3). The two fragments were ligated together and inserted into a BamHI-SmaI-digested pCGL243 shuttle vector to give plasmid pMM23.
Preparation of crude extracts.
Bacterial cells were harvested and resuspended in 50 mM sodium phosphate buffer (pH 7.8) to 50 OD570 units for C. melassecola. Cells were disrupted by vortexing three times 1 ml of suspension with 1 g of 0.5-mm glass beads for 2 min in a 2-ml Eppendorf tube. The suspension was centrifuged at 10,000 × g for 15 min, and the resulting supernatant was used as the crude extract for enzymatic determinations. The protein concentration was measured by the method of Lowry et al. (44) using bovine immunoglobulin G as a standard.
Determination of SOD, catalase, aconitase, and isocitrate dehydrogenase activities.
SOD specific activity was determined by the method of Ukeda et al. (74) based on the inhibition of a tetrazolium salt reduction by superoxide generated from a xanthine-xanthine oxidase reaction. Purified E. coli Mn-SOD (Sigma) was used as a standard, and 1 U is defined as the amount of enzyme that inhibits the rate of reduction of cytochrome c by 50% in the standard xanthine oxidase/cytochrome c assay. SOD activity in nondenaturing 10% acrylamide gels was visualized by tetrazolium negative staining according to Schmidt et al. (64). Catalase activity was determined according to Goldberg and Hochman (24) by measuring the disappearance of H2O2 at 240 nm, with an initial concentration of 20 mM. Aspergillus niger catalase (Sigma) was used as a standard, and 1 U is defined as the amount of enzyme catalyzing the dismutation of 1 μmol of H2O2 per min. Aconitase and isocitrate dehydrogenase activities were determined as described by Hanson and Cox (28).
Purification of SOD from C. melassecola.
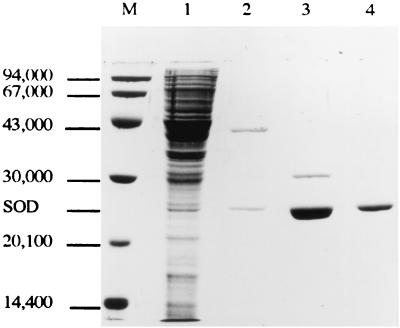
One liter of bacterial culture in BHI medium was harvested at an OD570 of 8.5, washed twice in 0.5 volume of 25 mM sodium phosphate (pH 7.5) buffer, and resuspended in 40 ml of the same buffer containing 10 mM MgCl2, RNase A (1 μg/ml), and DNase I (1 μg/ml). Cells were disrupted in a French press (SLM Aminco) at 16,000 lb/in2. The extract was centrifuged at 10,000 × g for 15 min at 4°C. The supernatant was ultracentrifuged at 130,000 × g for 2 h at 4°C, and the resulting supernatant was used as the cytosoluble fraction. This supernatant was fractionated with ammonium sulfate. Proteins precipitating in the range of 90 to 100% ammonium sulfate saturation, the fraction containing most of the SOD activity, were collected by centrifugation, dissolved in 50 mM Tris-Cl (pH 8.0) buffer, dialyzed against the same buffer, and applied to a Bioscale Q2 (Bio-Rad) ion-exchange column equilibrated with the same buffer. Proteins were eluted with a linear gradient of 0 to 0.5 M NaCl in the same buffer. The most active fractions were pooled, dialyzed against the Tris-Cl (pH 8.0) buffer, and run on a Sephacryl S300 (Pharmacia) column equilibrated with the same buffer. The fractions containing the eluted SOD were pooled and concentrated with a Centriprep-30 (Amicon). After this step, 5 μg of protein was analyzed for purity. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining according to Laemmli (40), only a single band could be observed (see Fig. 2).
FIG. 2.
Purification of SOD from C. melassecola. SDS-PAGE of cytosoluble extract from C. melassecola (lane 1), extract after ammonium sulfate fractionation (lane 2), and SOD-containing fractions after Bioscale Q2 chromatography (lane 3), and Sephacryl S300 chromatography (lane 4). Lane M, molecular weight standards as described in Materials and Methods.
The molecular mass of the purified native SOD was estimated by gel filtration on Sephacryl S300 using chymotrypsinogen (25 kDa), ovalbumin (45 kDa), bovine serum albumin (68 kDa), and aldolase (158 kDa) as molecular mass markers. The molecular mass of the monomeric protein was estimated by SDS-PAGE using LMW markers (Pharmacia) as molecular mass markers.
The N-terminal sequence of the purified enzyme and sequences from internal fragments generated by trypsin digestion were determined with an ABI model 473A protein sequencer.
Atomic absorption for the identification of the metal cofactor present in the purified SOD was carried out on a Perkin Elmer 2280 atomic absorption spectrometer.
Nucleotide sequence accession number.
The assigned accession number for the nucleotide and deduced amino acid sequences of sodA and msrA is AF236111 in the GenBank nucleotide sequence database.
RESULTS
Heterologous expression of the cloned gene.
The C. melassecola sodA gene was cloned, through a PCR-based method, as a pMM8-borne DNA fragment as described in Materials and Methods. Since many C. melassecola genes have been proven to be expressed in E. coli (56), the cloned DNA fragment was tested for its ability to complement the QC1799 sodA sodB mutant of E. coli. This mutant cannot grow aerobically in minimal medium without addition of a subset of amino acids and exhibits enhanced sensitivity to the superoxide generator paraquat (13). The QC1799 E. coli mutant was transformed with plasmid pMM8. The resulting transformed strain showed a growth pattern in minimal medium similar to that of the parental strain. To ensure that E. coli complementation depended only on C. melassecola sodA gene expression, and for further expression studies in C. melassecola, the sodA coding region, identified after DNA sequencing (see below), was placed under the control of the E. coli hybrid trc promoter, which was previously shown to promote efficient expression of genes in both E. coli (11) and C. glutamicum (17). The resulting pMM23 plasmid was used to transform QC1799, and the recombinant strain showed a growth pattern in minimal medium (Fig. 1a) and in LB medium containing 50 μM paraquat (Fig. 1b) similar to that of the parental SOD-proficient strain and grew even faster than the nontransformed E. coli SOD-deficient strain in LB without paraquat (Fig. 1b).
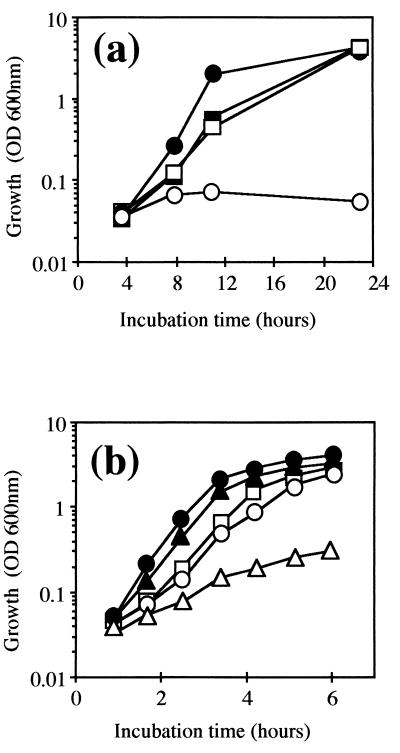
FIG. 1.
Complementation of SOD defect in E. coli by the C. melassecola sodA gene. (a) Growth in M63 minimal medium was measured by OD600 for the QC1799 sodA sodB mutant transformed by the control vector pCGL482 (○), for QC1799 transformed by pMM23 (□), for the nontransformed parental strain GC4468 (●), and for GC4468 transformed by pMM23 (■). Transformed strains were grown in the presence of chloramphenicol (30 μg/ml). (b) Growth of GC4468 (●, ▴), nontransformed QC1799 (○, ▵), and QC1799 transformed by pMM23 (□) was monitored by OD600 in LB medium in the presence (▴, ▵, □) or absence (●, ○) of 30 μM paraquat.
SOD specific activity was determined in cell extracts from transformed and nontransformed strains. Activities from GC4468, QC1799, QC1799(pMM8), and QC1799(pMM23) were 34.8 ± 4.3, not detected, 12.3 ± 2.4, and 20.4 ± 0.7 U/mg of protein, respectively.
It can therefore be concluded that the SOD deficiency in E. coli was complemented by the sodA gene from C. melassecola.
Purification and biochemical characterization of SOD.
To obtain information on the characteristics of the C. melassecola SOD, it was purified as described in Materials and Methods from the wild-type strain. A representative scheme for purification is given in Table 2. The isolated protein was estimated to be pure from SDS-PAGE analysis (Fig. 2). The SOD specific activity of the purified protein was estimated to 3,900 U/mg of protein. The amino acid sequences of the N terminus and two internal peptides isolated from the purified protein were determined (see below). These sequences unambiguously confirmed that the purified protein corresponded to an SOD. The molecular weight of native SOD was estimated to approximately 80,000 from gel filtration chromatography. Its monomeric molecular weight, determined by SDS-PAGE, was 24,000 (Fig. 2). Atomic absorption spectrometry for identification of the metal cofactor in the purified protein was carried out. Manganese was clearly identified, and the calculated stoichiometry was 0.43 atom/subunit. This unbalanced stoichiometry could be explained by either release of manganese from the enzyme during purification or manganese limitation in the growth medium. No signal corresponding to the presence of iron was detected.
TABLE 2.
Purification of SOD from cell extracts of C. melassecola
| Fraction | Vol (ml) | Total amt (mg) of protein | Total activity (U) | Sp act (U mg−1) | Recovery of activity (%) | Purification (fold) |
|---|---|---|---|---|---|---|
| Cytosoluble | 28 | 227 | 10,200 | 45 | 100 | 1 |
| (NH4)2SO4 | 15.8 | 4.9 | 5,900 | 1,200 | 58 | 27 |
| Bioscale Q2 | 0.5 | 0.31 | 1,300 | 4,100 | 13 | 91 |
| Sephacryl S300 | 0.54 | 0.09 | 380 | 3,900 | 4 | 87 |
Nucleotide sequence analysis of the cloned DNA fragment.
The nucleotide sequence of a 2,200-bp fragment from pMM8 was determined by a primer walking strategy from both strands by dideoxy chain termination. Computer analysis revealed two complete and one incomplete ORF, as shown in Fig. 3, extending from ATG (bp 1139) to TAA (bp 1741) for ORF1, from ATG (bp 892) to TAA (bp 239) for ORF2, and from ATG (bp 188) to bp 1 for the incomplete ORF3. ORF2 and ORF3 are in the reverse orientation with respect to ORF1. Database searches with the deduced polypeptides of these three ORFs revealed that the amino acid sequence deduced from ORF1 shows significant homology to known Fe- and Mn-SODs (see below), that deduced from ORF2 shows homology to peptide methionine sulfoxide reductases (PMSR; EC 1.8.4.5) from various organisms (see below), and that deduced from the incomplete ORF3 shows homology to N-acyl-l-aminoacid hydrolases from the Campylobacter jejuni hippuricase family (27). These results indicated that ORF1 may correspond to the sodA gene of C. melassecola and ORF2 may correspond to the msrA gene. ORF3 has not been named.
Analysis of the sodA gene.
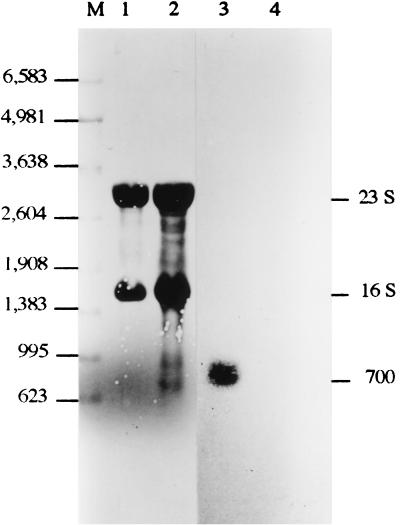
The predicted sodA gene product consists of 200 amino acids (Fig. 3), with a molecular weight of 22,104, which is in agreement with the size of the subunit of the purified C. melassecola SOD protein (Mr 24,000), as estimated from SDS-PAGE. The G+C content of 57% in the coding region corresponds to a clear preference for C or G at the wobble position, with a codon bias index of 0.79 as calculated according to Malumbres et al. (45). This is consistent with the relative abundance (about 1% of cytosoluble proteins) calculated by the ratio between specific SOD activity in cytosoluble extract (45 U/mg of protein) and for the purified enzyme (3,900 U/mg). In order to confirm the predicted translation initiation site, the N-terminal amino acid sequence of the purified SOD from C. melassecola was determined. This sequence was identical to the predicted one with the exception of the initial formyl-methionine, which was missing in the purified enzyme, as described for SODs from various origins (35, 70) and in some C. glutamicum proteins (31). The sequences of two internal peptides obtained by trypsin digestion of the purified enzyme were also identical to the deduced sequence. A ribosome-binding site (5′-GAAAGGA-3′), complementary to the 3′ end of the Brevibacterium lactofermentum 16S rRNA (3′-AAUCUUUCCUCC-5′) (3), was identified 10 bp upstream from the ATG initiation codon (Fig. 3). Two putative promoter regions with nucleotide sequences close to the consensus sequence for C. glutamicum promoters (56) are located upstream from the putative ribosome-binding site. An almost perfect palindromic sequence covers the −35 region from the distal putative promoter. Such a structure could be a binding site for a regulatory factor. Two inverted repeats with putative stable RNA stem-loop structures (ΔG, −61.6 and −87.6 kJ mol−1), located 18 and 88 bp downstream of the stop codon, may represent transcription termination regions. Northern hybridization experiments were performed in order to analyze the size of the sodA transcript. Hybridization of C. melassecola RNA to an internal sodA probe resulted in a single signal at approximately 700 nucleotides (Fig. 4). This size is consistent with the translation initiation and termination regions described above and indicates that the sodA gene is monocistronic.
FIG. 4.
Northern analysis of the C. melassecola sodA gene. RNA was extracted from wild-type C. melassecola (lanes 1 and 3) and from the SOD-deficient C. melassecola strain CGL10016 (lanes 2 and 4). RNAs were monitored either with a radioactively labeled sodA internal probe (lanes 3 and 4) or by methylene blue staining (lanes 1 and 2) as described in Materials and Methods. The positions of RNA standards (Promega) are given (lane M) (in nucleotides).
Analysis of the deduced SOD amino acid sequence from C. melassecola.
Comparison of the deduced SOD amino acid sequence with protein sequences from databases using the National Center for Biotechnology Information Blast program (2) revealed high similarity to Fe- and Mn-SODs of prokaryotic and eukaryotic origin. The highest scores were obtained with deduced partial sequences of SODs from the pathogenic Corynebacterium diphtheriae and Corynebacterium pseudodiphtheriticum species (81% identity) (86) and from the closely related Mycobacterium species (61% identity with Mycobacterium leprae Mn-SOD). Homology was higher with mitochondrial Mn-SODs (54% identity with human Mn-SOD) than with other bacterial species like Bacillus stearothermophilus (51% identity) and E. coli Mn-SOD (42%) and Fe-SOD (40%). The 12 strictly conserved residues in Fe/Mn-SODs (33), including the four metal-binding residues His27, His76, Asp160, and His164 (C. melassecola SOD numbering), could be identified. Among the residues involved in metal selectivity, Met24, Leu26, Gly71, Gly72, His73, Phe79, Gln145, and Gln169 are predicted to be discriminant for an Mn-dependent enzyme (37, 55). Manganese as a cofactor was clearly identified in the purified C. melassecola enzyme by atomic adsorption spectrometry, as described previously.
The predicted folding profile of the C. melassecola SOD subunit with α-helices in the N-terminal region and β-sheets together with α-helices in the C-terminal region, as predicted by the PredictProtein program (60, 61), is similar to the typical structure described by crystallographic studies of different Fe/Mn-SODs (10, 16, 67, 68). The molecular mass of the native protein was approximately 80 kDa, as estimated by gel filtration analysis of the purified enzyme, with a monomer molecular weight of approximately 24,000, as seen before (Fig. 2). The active enzyme is thus a homotetramer, as is the case for Thermus thermophilus Mn-SOD (67), mitochondrial human Mn-SOD (10), and Mycobacterium tuberculosis Fe-SOD (16).
Analysis of the msrA and adjacent ORF3 genes and products.
The 654-bp-long msrA ORF is located only 246 bp upstream from the sodA ORF, in the opposite transcriptional orientation (Fig. 3). The predicted msrA gene product consists of 217 amino acids, with a calculated molecular weight of 23,892, in agreement with the size of known bacterial PMSRs. The predicted msrA gene product showed similarities along its entire length to PMSRs from mammals (50% identity with bovine PMSR [52]) and from bacteria (48% with E. coli [58] and 36% with Streptococcus pneumoniae [79]). A codon bias index of 0.48 was calculated for msrA as described by Malumbres et al. (45), which may indicate a rather low expression level for this gene. A possible ribosome-binding site with the sequence 5′-GAAAGGA-3′ is located 12 bp upstream from the initiation codon, and two possible promoter regions with reasonable homologies to consensus sequences are located further upstream. Two short inverted repeats, one covering the ribosome-binding site and the other located at the distal −35, corresponding also to a potential −35 for the sodA gene (see above), could be regions involved in transcriptional regulation.
No clear transcription termination region for msrA could be identified, the free energies of the two putative stem-loop structures located downstream of the stop codon being relatively low (ΔG, −28.9 and −27.2 kJ mol−1). Moreover, these stem-loop structures are located within the coding region of ORF3. The end of the msrA coding region and the putative ATG for ORF3 are separated by a short stretch of only 51 nucleotides. ORF3 is not preceded by a typical ribosome-binding site. A codon bias index of 0.39 was calculated as described by Malumbres et al. (45) for the truncated ORF3, which may indicate an expression level for this gene as low as that for msrA. The predicted product for the incomplete ORF3 shows significant homologies with N-acyl-l-amino acid amidohydrolases from the C. jejuni hippuricase family (27), an enzyme of unclear physiological function. These observations raise the question of a possible operon organization. Such a hypothesis was not confirmed, since no transcript was identified by Northern blotting experiments with an msrA probe under our experimental conditions.
Disruption of the chromosomal sodA gene.
To study the physiological effects of sodA deficiency, the chromosomal copy of this gene was disrupted in C. melassecola. For this purpose, the integrative vector viMM12, carrying an internal fragment of the sodA coding region and the aphIII gene, which confers resistance to kanamycin, was constructed as described in Materials and Methods. Integration of viMM12 by a single crossing-over event in the sodA chromosomal locus led to sodA gene disruption, as seen by Southern blotting on several transformants (data not shown). Lack of sodA transcription and expression in the disrupted CGL10016 strain was confirmed as follows. In Northern blotting experiments with CGL10016 RNAs, no sodA transcript was seen (Fig. 4). Activity staining on native polyacrylamide gels did not reveal the presence of any active SOD (not shown). Furthermore, SOD activity in crude extracts from the disrupted strain was 0.24 ± 0.18, versus 17.4 ± 1 U/mg of protein in crude extracts from the wild-type strain. The residual SOD activity in cell extracts from the disrupted strain probably resulted from the presence of about 0.5% genotypic revertants at the stage of cell harvesting (see below).
Effect of sodA disruption on growth.
Various growth characteristics of the SOD-proficient wild-type C. melassecola strain ATCC 17965 and the corresponding SOD-deficient strain CGL10016 were compared.
The wild-type strain showed pink pigmentation after growth on BHI plates, whereas the CGL10016 mutant remained white. This indicated an SOD-dependent alteration in the biosynthesis or reduction level of a pigment in the mutant strain.
Exponential growth of the sodA mutant was only slightly impaired in BHI rich medium as well as in BMCG chemically defined medium without amino acid. In the two media, the mutant strain had a generation time only 10 min longer than that of the wild type (60 versus 50 min) (Fig. 5a). The mutant strain reached the wild-type growth rate after transformation with pMM23 (SOD specific activity of 32 ± 2 U/mg) or addition of the 20 amino acids to the medium.
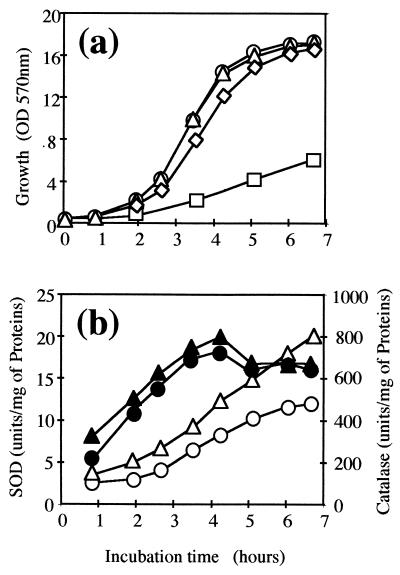
FIG. 5.
Effect of paraquat on growth and SOD and catalase activities in C. melassecola. (a) Growth of ATCC 17965 (○, ▵) and SOD-deficient CGL10016 (◊, □) was monitored in BHI or BMCG medium and in BHI with 100 μM paraquat (ATCC 17965, ▵; CGL10016, □), by measuring OD570. CGL10016 was cultivated in the presence of kanamycin (25 μg/ml). (b) SOD (▴, ●) and catalase (▵, ○) activities in ATCC 17965 crude extracts were measured as described in Materials and Methods in the presence (▴, ▵) and absence (●, ○) of 100 μM paraquat in BHI medium. Activity values are the means of at least three experiments that differed by less than 20%.
The prototrophy of the sodA-disrupted strain raised the question of the possible presence of another SOD-encoding gene in C. melassecola, for example, sodB. This seems unlikely for the following reasons. Gardner and Fridovich (23) demonstrated that in a sodA-deficient strain of E. coli, the residual SOD activity, corresponding to the Fe-SOD encoded by sodB, is 30% of the total SOD activity in the wild-type cells, whereas SOD activity is not detected in a sodA sodB double mutant. They also demonstrated that the activity of aconitase, known as a superoxide-sensitive enzyme, is lowered to 78% of that of the wild type in a sodA mutant and to 30% in a sodA sodB double mutant (23). In C. melassecola, the SOD activity in the disrupted strain is only 1.4% of that of the wild-type strain and probably results from reversion events, as described below. The specific activity for aconitase in the CGL10016 strain is lowered to 20% of that of the wild-type strain (16 ± 7 and 79 ± 8 nmol min−1 mg−1, respectively) when the control isocitrate dehydrogenase activity is not very different (2.47 ± 0.27 and 3.14 ± 0.13 μmol min−1 mg−1, respectively).
Effect of sodA disruption on oxidative stress response.
Responses of the mutant, the pMM23-transformed mutant, and wild-type strains to oxidative stresses generated by paraquat and H2O2 were investigated by growth inhibition tests on plates (Table 3) as described in Materials and Methods. Growth of the wild-type strain was not inhibited in the presence of 0.1 mg of paraquat, but the disrupted strain showed a bright inhibition zone. This growth inhibition of the SOD-deficient strain was completely abolished after transformation by pMM23. On the other hand, all strains exhibited the same sensitivity to H2O2.
TABLE 3.
Sensitivity of C. melassecola strains to oxidizing agentsa
| Inhibitor | Strain | Diam of growth inhibition zone (mm) |
|---|---|---|
| Paraquat | ATCC 17965 | 7 |
| CGL10016 | 29 | |
| CGL10016(pMM23) | 6 | |
| H2O2 | ATCC 17965 | 25 |
| CGL10016 | 25 |
Cells were grown on BHI agar plates. Growth inhibition by paraquat or H2O2 was tested as described in Materials and Methods. Strains and plasmid pMM23 are described in Table 1.
When the wild-type strain was grown in BHI liquid medium in the presence of 100 μM paraquat, growth was not altered and no significant modification in SOD specific activity was observed (Fig. 5). This held true when the paraquat concentation was increased up to 400 μM. On the other hand, catalase was clearly induced when cells were grown in the presence of paraquat (Fig. 5b). This indicates that paraquat efficiently enters cells but does not induce sodA expression. At a paraquat concentration of 100 μM, the growth of the disrupted strain was dramatically impaired, indicating that redox cycling was effectively induced and not rescued in the SOD-deficient strain (Fig. 5a).
Effect of sodA disruption on viability during stationary phase of growth.
To test the effect of SOD deficiency on cell viability after extensive stationary-phase cultivation, cultures of wild-type ATCC 17965 and SOD-deficient CGL10016 C. melassecola strains in BHI rich medium and BMCG chemically defined medium were maintained under aeration at 34°C for 6 days. All reached the stationary growth phase in less than 1 day, the OD570 then remaining stable at approximatly 20 U (Fig. 6). Viability was estimated at various cultivation times by the CFU/OD ratio. Microscopic observations at each cultivation time did not reveal cellular aggregation.
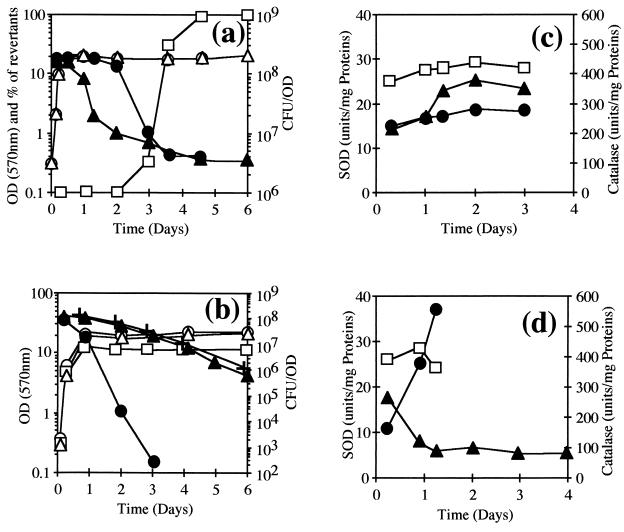
FIG. 6.
Viability and SOD and catalase activities of C. melassecola during the stationary phase of growth. (a) Growth (○, ▵), viability (●, ▴), and proportion of phenotypic revertants (□) for SOD-deficient CGL10016 (▵, ▴, □), and parental ATCC 17965 (○, ●) strains cultivated in BHI, in the presence or absence of kanamycin (25 μg/ml) for CGL10016. Growth was measured by OD570, and cellular viability was determined by plating on BHI and calculated as CFU/OD. The experiment was done at least three times without significant differences. (b) Growth (○, ▵, □) and viability (●, ▴, +) determined as described above for ATCC 17965 (○, ●) and CGL10016 (▵, ▴) in BMCG (2% glucose) and for ATCC 17965 in BMCG containing only 1% glucose (□, +). Kanamycin (25 μg/ml) was added for CGL10016. (c and d) SOD (□) and catalase (●, ▴) activities measured as described in Materials and Methods, from crude extracts of ATCC 17965 (□, ●) and CGL10016 (▴) strains cultivated in BHI (c) or BMCG with 2% glucose (d) in the presence of kanamycin (25 μg/ml) for CGL10016. Values are the means of at least two experiments that differed by less than 20%.
In BHI rich medium, viability was stable at 1.5 × 108 CFU/OD during logarithmic and early stationary phase for the two strains (Fig. 6a). Viability of the wild-type strain was stable during the first 2 days, then reduced to 2% of the initial value, and remained constant until the end of the experiment. On the other hand, viability of the SOD-deficient strain decreased soon after entry into stationary phase. Cell viability dropped to 5% within the first 2 days and then remained relatively stable.
Loss of viability during these experiments could not be associated with a clear loss of antioxidant activities (Fig. 6c), since catalase specific activity remained relatively constant in the wild-type strain (250 to 280 U/mg) and even increased in the disrupted one (340 to 380 U/mg), and SOD activity was stable in the wild-type strain (26 to 29 U/mg).
Genotypic revertants, phenotypically characterized as pink, kanamycin-sensitive colonies, appeared during the first 2 days of culture of CGL10016. The frequency of reversion increased rapidly to give nearly no SOD-deficient colonies after 4 days. Reversion occurred the same way whether the culture was grown in the presence or absence of kanamycin. Reversion probably resulted from excision of the integrative vector viMM12. As a control, a C. melassecola malE mutant (25) and a dcp mutant (unpublished data), obtained by a similar disruption strategy, were analyzed in the same conditions, and their insertions remained stable even after 7 days of cultivation.
The same analysis was carried out in BMCG chemically defined medium for both strains (Fig. 6b). After 3 days of cultivation, the viability of the disrupted strain was close to 10% of the initial 1.5 × 108 CFU/OD value and 0.1% after 6 days. The level of genotypic revertants remained below 1% even after 6 days of cultivation. More surprisingly, the viability of the wild-type strain dropped in a more dramatic way: 13% of the initial viability remained after 1 day of cultivation, and only 0.02% remained after 2 days. The strain became nonculturable after 4 days, with more than a 108-fold drop in viability. When the glucose concentration in BMCG medium was lowered from 2 to 1%, the wild-type strain viability profile was the same as for the SOD-deficient strain in BMCG containing 2% glucose.
As observed during experiments in BHI, loss of viability during these experiments could not be correlated to the loss of antioxidant activities (Fig. 6d).
DISCUSSION
We identified the sodA gene encoding SOD in C. melassecola ATCC 17965. As estimated from activity-staining experiments, Northern experiments, SOD and aconitase activity measurements, and response of the wild-type and corresponding sodA-disrupted mutant to paraquat, the cloned gene seems to be the unique SOD-encoding gene in this organism. From biochemical experiments on the purified enzyme, SOD from C. melassecola was shown to be a homotetramer. From the deduced amino acid sequence, the enzyme belongs to the Mn-dependent SOD family, as experimentally confirmed by atomic adsorption spectrometry. Since SOD from Mycobacterium tuberculosis contains iron even though its sequence is closer to that of Mn-SODs than Fe-SODs (85), and since the Mycobacterium smegmatis cambialistic enzyme contains both metals (82), it would be interesting to determine if only manganese is present in the SOD of C. melassecola after growth in different media or if an iron-substituted enzyme would retain activity.
Putative promoter sequences for the vegetative sigma 70 factor were proposed upstream of the initiation codon, with respect to the promoter consensus sequences from C. glutamicum (56). The initiation site for transcription should be determined to clearly identify the actual promoter. The gene was shown to be monocistronic by Northern experiments, and the transcript size is fully compatible with the putative initiation and termination signals of transcription. We were unable to demonstrate regulation of C. melassecola sodA gene expression under various growth conditions. No standard binding site for the transcription factors SoxS (41) and Fur (20), involved in the regulation of sod genes in other species (29, 72), were found. The SOD specific activity was not significantly different in the wild-type strain grown in the presence or in the absence of paraquat, even if we assumed from experimental data that paraquat-mediated redox cycling readily occurred in this strain. The absence of a change in activity profile upon paraquat addition could be explained either by considering an immediate removal of superoxide radicals by the existing SOD enzyme or by the failure of superoxide radicals to induce C. melassecola sodA expression. Nevertheless, the expression of sodA could be somehow regulated, as in standard laboratory growth conditions in BHI medium the SOD specific activity was shown to vary. Activity increased during the logarithmic phase of growth and reached a plateau as soon as the culture entered stationary phase, with a maximum at transition phase. It would therefore be interesting to follow the expression of sodA in different conditions, in the presence of other stress inducers, for instance, H2O2, or when the strain is less likely to produce superoxide radicals, as should be the case under low aeration. It would thus be important for further study of sodA expression to put a reporter gene under the control of the upstream sequences of sodA.
We identified a second gene, msrA, immediately upstream from sodA, coding for a putative PMSR (52, 58, 79), whose catalytic activity is the reduction of oxidized methionine residues, a modification leading to functional inactivation of numerous proteins (75). A role for PMSR in protecting against oxidative damage has been described in bacteria (51) and yeasts (50). Thus, the location of the msrA gene upstream of sodA in C. melassecola could be more than just a coincidence. The genes are in opposite transcription orientations, and the two initiation codons are separated by only 246 bp. This kind of divergent organization, with two genes involved in the same physiological process and with coordinate regulation, has often been characterized. For example, it is the case for the E. coli soxR and soxS genes, involved in superoxide response (81), and for the Fur-regulated fepA-fes genes, involved in siderophore production (21). In M. tuberculosis and M. leprae, the alkyl hydroperoxide reductase and peroxide stress regulator homolog-encoding genes ahpC and oxyR are divergently transcribed (18). Furthermore, a sod gene has been localized upstream from a peroxidase gene in Helicobacter pylori (76). Nevertheless, this is the first time that divergent sod and msrA genes have been reported. Although a sodA transcript has been clearly identified in the C. melassecola wild-type strain, no msrA transcript could be detected, even in the sodA-disrupted strain, in which the superoxide level is assumed to be increased. This may be due either to a low transcription level of this gene under our conditions or to poor technology for the extraction and/or detection of large RNA of C. melassecola in the case of an operon organization, since msrA is immediately followed by ORF3, the physiological role of which is unknown. We now need to look for conditions and methods to detect msrA gene expression in C. melassecola and to determine if the expression of the divergent sodA and msrA genes is connected. Northern identification of the msrA transcript will also highlight the possible operon organization suggested by the presence of ORF3 immediately downstream of msrA.
A sodA null mutant was constructed by disruption of the chromosomal gene by an integrative vector carrying a kanamycin resistance gene. Unlike the wild-type strain, the disrupted strain showed an increased sensitivity to superoxide radicals generated by the redox cycling agent paraquat. This indicated that the sodA mutation could not be rescued by another activity in C. melassecola cells. The growth rate of the disrupted strain was only slightly impaired in BHI rich medium, with a generation time of 60 min, compared to 50 min for the wild-type strain. This is consistant with the behavior of other organisms (13, 36). Much more surprising was the growth of the disrupted strain in BMCG chemically defined medium. Even without supplementary amino acids, its growth rate was the same as in BHI rich medium, whereas the growth of other species of sod mutants was dramatically impaired in minimal medium without amino acids (13, 36). Oxidative stress has been proven to exist in the C. melassecola mutant strain, as revealed by the decrease in aconitase activity. The prototrophy of the C. melassecola SOD-deficient strain could then be explained by a lower sensitivity of amino acid biosynthesis pathways to superoxide radicals generated by aerobic metabolism. Another explanation could be partial compensation for SOD deficiency by either an unidentified enzymatic activity or the presence of an intracellular metabolite or metal ion able to detoxify superoxide radicals. Since the BMCG medium contains 70 μM FeSO4, 10 times the concentration in E. coli M63 minimum medium, and since it has been reported that growth in iron-enriched medium can partially compensate for SOD deficiency in E. coli (7), iron-mediated compensation of SOD deficiency could be arguable in our case. Note that, as even the wild-type C. melassecola ATCC 17965 strain cannot grow in M63, the control experiment was not possible. Carotenoid pigments can play a role in singlet oxygen and also superoxide scavenging, (8, 12, 49). Carotenoids have been identified in some Corynebacterium species (54, 77). Even if it has not been demonstrated that the C. melassecola ATCC 17965 pink pigment belongs to this family, its role in superoxide scavenging could be arguable.
While only a small difference in growth rate on BHI medium was observed between wild-type and SOD-deficient strains, they behave in a different way in late stationnary phase with regard to their viability. The viability of the SOD-deficient strain decreased dramatically soon after entry into stationary phase, while the wild-type strain kept a high viability for at least 2 days. Viability of the disrupted strain dropped to 5% of the initial value within 2 days of culture, and almost no sodA mutant cells could be recovered after 4 days, when close to 100% of the population consisted of genotypic revertants. Viability loss in SOD-deficient C. melassecola is not surprising, since the same type of behavior has been described for other organisms (6, 14, 43, 48), but it still remains unclear why the viability of the wild-type strain decreased after 2 days of culture to reach only 2% of the initial value after 4 days.
When viability was followed in BMCG chemically defined medium, the behavior of the two strains was much more surprising. Viability of the SOD-deficient strain decreased less rapidly than in BHI medium, but continued to decrease for 6 days, while no real increase in reversion was observed, contrary to the behavior in BHI. The most important difference concerns the evolution of viability of the wild-type strain. Viability loss was so dramatic that no viable cells could be recovered after 4 days of cultivation. As the viability profile of the wild-type strain was similar to that of the disrupted strain when the initial glucose concentration was lowered from 2 to 1%, we can conclude that this dramatic loss of viability was the result of an excess of glucose, by a physiological mechanism that is still to be defined, and which does not take place, or takes place to a lesser extent, in the SOD-deficient strain.
Even if wild-type and SOD-deficient strains do not behave in the same way in BHI rich medium and in BMCG chemically defined medium concerning viability, it appeared that loss of viability during the culture cannot be attributed to a decrease in SOD or catalase antioxidant activities. Moreover, catalase activity increased in the SOD-deficient strain in BHI and SOD-proficient strain in BMCG, but only after viability started to decrease.
We have to consider that nonviable cells, as determined by our viability test, could in fact be viable but noncultivable cells, or in a so-called dormant state (19, 38). Nevertheless, this is the first time that a SOD-deficient strain has shown a selective advantage over a wild-type strain. Efforts will now be engaged to understand this at the metabolic level.
ACKNOWLEDGMENTS
This work was supported by grant BIO4-CT96-0145 within the 4th Framework Program of the European Community. M. Merkamm was supported in part by a grant from Eurolysine.
We are grateful to D. Touati for the kind gift of SOD-deficient E. coli strains, to A. Boussac for atomic adsorbtion spectrometry experiments, and to M. Blight for critical reading of the manuscript.
REFERENCES
- 1.Alcendor D J, Chapman G D, Beaman B L. Isolation, sequencing and expression of the superoxide dismutase-encoding gene (sod) of Nocardia asteroides strain GUH-2. Gene. 1995;164:143–147. doi: 10.1016/0378-1119(95)00476-m. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amador E, Castro J M, Correia A, Martin J F. Structure and organization of the rrnD operon of ‘Brevibacterium lactofermentum’: analysis of the 16S rRNA gene. Microbiology. 1999;145:915–924. doi: 10.1099/13500872-145-4-915. [DOI] [PubMed] [Google Scholar]
- 4.Battistoni A, Pacello F, Folcarelli S, Ajello M, Donnarumma G, Greco R, Ammendolia M G, Touati D, Rotilio G, Valenti P. Increased expression of periplasmic Cu,Zn superoxide dismutase enhances survival of Escherichia coli invasive strains within nonphagocytic cells. Infect Immun. 2000;68:30–37. doi: 10.1128/iai.68.1.30-37.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck B L, Tabatabai L B, Mayfield J E. A protein isolated from Brucella abortus is a Cu-Zn superoxide dismutase. Biochemistry. 1990;29:372–376. doi: 10.1021/bi00454a010. [DOI] [PubMed] [Google Scholar]
- 6.Benov L, Fridovich I. A superoxide dismutase mimic protects sodA sodB Escherichia coli against aerobic heating and stationary-phase death. Arch Biochem Biophys. 1995;322:291–294. doi: 10.1006/abbi.1995.1465. [DOI] [PubMed] [Google Scholar]
- 7.Benov L, Fridovich I. Growth in iron-enriched medium partially compensates Escherichia coli for the lack of manganese and iron superoxide dismutase. J Biol Chem. 1998;273:10313–10316. doi: 10.1074/jbc.273.17.10313. [DOI] [PubMed] [Google Scholar]
- 8.Black H S. Radical interception by carotenoids and effects on UV carcinogenesis. Nutr Cancer. 1998;31:212–217. doi: 10.1080/01635589809514705. [DOI] [PubMed] [Google Scholar]
- 9.Bonamy C, Guyonvarch A, Reyes O, David F, Leblon G. Interspecies electro-transformation in corynebacteria. FEMS Microbiol Lett. 1990;66:263–269. doi: 10.1016/0378-1097(90)90294-z. [DOI] [PubMed] [Google Scholar]
- 10.Borgstahl G E, Parge H E, Hickey M J, Beyer W F J, Hallewell R A, Tainer J A. The structure of human mitochondrial manganese superoxide dismutase reveals a novel tetrameric interface of two 4-helix bundles. Cell. 1992;71:107–118. doi: 10.1016/0092-8674(92)90270-m. . (Erratum, 72:476, 1993.) [DOI] [PubMed] [Google Scholar]
- 11.Brosius J. Expression vectors employing lambda-, trp-, lac- and lpp-derived promoters. In: Rodriguez R L, Denhardt D T, editors. Vectors: a survey of molecular cloning vectors and their uses. Boston, Mass: Butterworth; 1988. pp. 205–225. [DOI] [PubMed] [Google Scholar]
- 12.Carbonneau M A, Melin A M, Perromat A, Clerc M. The action of free radicals on Deinococcus radiodurans carotenoids. Arch Biochem Biophys. 1989;275:244–251. doi: 10.1016/0003-9861(89)90370-6. [DOI] [PubMed] [Google Scholar]
- 13.Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clements M O, Watson S P, Foster S J. Characterization of the major superoxide dismutase of Staphylococcus aureus and its role in starvation survival, stress resistance, and pathogenicity. J Bacteriol. 1999;181:3898–3903. doi: 10.1128/jb.181.13.3898-3903.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cocaign-Bousquet M, Guyonvarch A, Lindley N D. Growth rate-dependent modulation of carbon flux through central metabolism and the kinetic consequences for glucose-limited chemostat cultures of Corynebacterium glutamicum. Appl Environ Microbiol. 1996;62:429–436. doi: 10.1128/aem.62.2.429-436.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper J B, McIntyre K, Badasso M O, Wood S P, Zhang Y, Garbe T R, Young D. X-ray structure analysis of the iron-dependent superoxide dismutase from Mycobacterium tuberculosis at 2.0 angstrom resolution reveals novel dimer-dimer interactions. J Mol Biol. 1995;246:531–544. doi: 10.1006/jmbi.1994.0105. [DOI] [PubMed] [Google Scholar]
- 17.Delaunay S, Uy D, Baucher M F, Engasser J M, Guyonvarch A, Goergen J L. Importance of phosphoenolpyruvate carboxylase of Corynebacterium glutamicum during temperature triggered glutamic acid fermentation. Metabolic Eng. 1999;1:334–343. doi: 10.1006/mben.1999.0131. [DOI] [PubMed] [Google Scholar]
- 18.Dhandayuthapani S, Mudd M, Deretic V. Interactions of OxyR with the promoter region of the oxyR and ahpC genes from Mycobacterium leprae and Mycobacterium tuberculosis. J Bacteriol. 1997;179:2401–2409. doi: 10.1128/jb.179.7.2401-2409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dick T, Lee B H, Murugasu-Oei B. Oxygen depletion induced dormancy in Mycobacterium smegmatis. FEMS Microbiol Lett. 1998;163:159–164. doi: 10.1111/j.1574-6968.1998.tb13040.x. [DOI] [PubMed] [Google Scholar]
- 20.Escolar L, Perez-Martin J, de Lorenzo V. Binding of the fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J Mol Biol. 1998;283:537–547. doi: 10.1006/jmbi.1998.2119. [DOI] [PubMed] [Google Scholar]
- 21.Escolar L, Perez-Martin J, de Lorenzo V. Coordinated repression in vitro of the divergent fepA-fes promoters of Escherichia coli by the iron uptake regulation (Fur) protein. J Bacteriol. 1998;180:2579–2582. doi: 10.1128/jb.180.9.2579-2582.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fridovich I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 23.Gardner P R, Fridovitch I. Superoxide sensitivity of the Escherichia coli aconitase. J Biol Chem. 1991;266:19328–19333. [PubMed] [Google Scholar]
- 24.Goldberg I, Hochman A. Three different types of catalases in Klebsiella pneumoniae. Arch Biochem Biophys. 1989;268:124–128. doi: 10.1016/0003-9861(89)90572-9. [DOI] [PubMed] [Google Scholar]
- 25.Gourdon P, Baucher M-F, Lindley N D, Guyonvarch A. Cloning of the malic enzyme gene from Corynebacterium glutamicum and role of the enzyme in lactate metabolism. Appl Environ Microbiol. 2000;66:2981–2987. doi: 10.1128/aem.66.7.2981-2987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guillouet S, Engasser J M. Sodium and proline accumulation in Corynebacterium glutamicum as a response to an osmotic saline upshock. Appl Environ Microbiol. 1995;43:315–320. [Google Scholar]
- 27.Hani E K, Chan V L. Expression and characterization of Campylobacter jejuni benzoylglycine amidohydrolase (hippuricase) gene in Escherichia coli. J Bacteriol. 1995;177:2396–2402. doi: 10.1128/jb.177.9.2396-2402.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanson R S, Cox D P. Effect of different nutritional conditions on the synthesis of tricarboxylic acid cycle enzymes. J Bacteriol. 1967;93:1777–1787. doi: 10.1128/jb.93.6.1777-1787.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hassett D J, Schweizer H P, Ohman D E. Pseudomonas aeruginosa sodA and sodB mutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. J Bacteriol. 1995;177:6330–6337. doi: 10.1128/jb.177.22.6330-6337.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinzen R A, Frazier M E, Mallavia L P. Coxiella burnetii superoxide dismutase gene: cloning, sequencing, and expression in Escherichia coli. Infect Immun. 1992;60:3814–3823. doi: 10.1128/iai.60.9.3814-3823.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hermann T, Wersch G, Uhlemann E M, Schmid R, Burkovski A. Mapping and identification of Corynebacterium glutamicum proteins by two-dimensional gel electrophoresis and microsequencing. Electrophoresis. 1998;19:3217–3221. doi: 10.1002/elps.1150191827. [DOI] [PubMed] [Google Scholar]
- 32.Hopkin K A, Papazian M A, Steinman H M. Functional differences between manganese and iron superoxide dismutases in Escherichia coli K-12. J Biol Chem. 1992;267:24253–24258. [PubMed] [Google Scholar]
- 33.Hunter T, Ikebukuro K, Bannister W H, Bannister J V, Hunter G J. The conserved residue tyrosine 34 is essential for maximal activity of iron-superoxide dismutase from Escherichia coli. Biochemistry. 1997;36:4925–4933. doi: 10.1021/bi9629541. [DOI] [PubMed] [Google Scholar]
- 34.Igo M M, Losick R. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis. J Mol Biol. 1986;191:615–624. doi: 10.1016/0022-2836(86)90449-3. [DOI] [PubMed] [Google Scholar]
- 35.Inaoka T, Matsumura Y, Tsuchido T. Molecular cloning and nucleotide sequence of the superoxide dismutase gene and characterization of its product from Bacillus subtilis. J Bacteriol. 1998;180:3697–3703. doi: 10.1128/jb.180.14.3697-3703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inaoka T, Matsumura Y, Tsuchido T. SodA and manganese are essential for resistance to oxidative stress in growing and sporulating cells of Bacillus subtilis. J Bacteriol. 1999;181:1939–1943. doi: 10.1128/jb.181.6.1939-1943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackson S M, Cooper J B. An analysis of structural similarity in the iron and manganese superoxide dismutases based on known structures and sequences. Biometals. 1998;11:159–173. doi: 10.1023/a:1009238214394. [DOI] [PubMed] [Google Scholar]
- 38.Kaprelyants A S, Gottschal J C, Kell D B. Dormancy in nonsporulating bacteria. FEMS Microbiol Rev. 1993;10:271–285. doi: 10.1111/j.1574-6968.1993.tb05871.x. [DOI] [PubMed] [Google Scholar]
- 39.Keele B B J, McCord J M, Fridovich I. Superoxide dismutase from Escherichia coli B. A new manganese-containing enzyme. J Biol Chem. 1970;245:6176–6181. [PubMed] [Google Scholar]
- 40.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 41.Li Z, Demple B. Sequence specificity for DNA binding by Escherichia coli SoxS and Rob proteins. Mol Microbiol. 1996;20:937–945. doi: 10.1111/j.1365-2958.1996.tb02535.x. [DOI] [PubMed] [Google Scholar]
- 42.Liebl W, Ehrmann M, Ludwig W, Schleifer K H. Transfer of Brevibacterium divaricatum DSM 20297T, “Brevibacterium flavum” DSM 20411, “Brevibacterium lactofermentum” DSM 20412 and DSM 1412, and Corynebacterium glutamicum and their distinction by rRNA gene restriction patterns. Int J Syst Bacteriol. 1991;41:255–260. doi: 10.1099/00207713-41-2-255. [DOI] [PubMed] [Google Scholar]
- 43.Longo V D, Gralla E B, Valentine J S. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae: mitochondrial production of toxic oxygen species in vivo. J Biol Chem. 1996;271:12275–12280. doi: 10.1074/jbc.271.21.12275. [DOI] [PubMed] [Google Scholar]
- 44.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 45.Malumbres M, Gil J A, Martin J F. Codon preference in corynebacteria. Gene. 1993;134:15–24. doi: 10.1016/0378-1119(93)90169-4. [DOI] [PubMed] [Google Scholar]
- 46.McCord J M, Fridovich I. Superoxide dismutase: an enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 47.Meier B, Barra D, Bossa F, Calabrese L, Rotilio G. Synthesis of either Fe- or Mn-superoxide dismutase with an apparently identical protein moiety by an anaerobic bacterium dependent on the metal supplied. J Biol Chem. 1982;257:13977–13980. [PubMed] [Google Scholar]
- 48.Melov S, Coskun P E, Wallace D C. Mouse models of mitochondrial disease, oxidative stress, and senescence. Mutat Res. 1999;434:233–242. doi: 10.1016/s0921-8777(99)00031-2. [DOI] [PubMed] [Google Scholar]
- 49.Mortensen A, Skibsted L H, Sampson J, Rice-Evans C, Everett S A. Comparative mechanisms and rates of free radical scavenging by carotenoid antioxidants. FEBS Lett. 1997;418:91–97. doi: 10.1016/s0014-5793(97)01355-0. [DOI] [PubMed] [Google Scholar]
- 50.Moskovitz J, Berlett B S, Poston J M, Stadtman E R. The yeast peptide-methionine sulfoxide reductase functions as an antioxidant in vivo. Proc Natl Acad Sci USA. 1997;94:9585–9589. doi: 10.1073/pnas.94.18.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moskovitz J, Rahman M A, Strassman J, Yancey S O, Kushner S R, Brot N, Weissbach H. Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J Bacteriol. 1995;177:502–507. doi: 10.1128/jb.177.3.502-507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moskovitz J, Weissbach H, Brot N. Cloning and expression of a mammalian gene involved in the reduction of methionine sulfoxide residues in proteins. Proc Natl Acad Sci USA. 1996;93:2095–2099. doi: 10.1073/pnas.93.5.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nilsson B, Uhlen M, Josephson S, Gatenbeck S, Philipson L. An improved positive selection plasmid vector constructed by oligonucleotide mediated mutagenesis. Nucleic Acids Res. 1983;11:8019–8030. doi: 10.1093/nar/11.22.8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ostrovsky D, Diomina G, Lysak E, Matveeva E, Ogrel O, Trutko S. Effect of oxidative stress on the biosynthesis of 2-C-methyl-d-erythritol-2, 4-cyclopyrophosphate and isoprenoids by several bacterial strains. Arch Microbiol. 1998;171:69–72. doi: 10.1007/s002030050680. [DOI] [PubMed] [Google Scholar]
- 55.Parker M W, Blake C C. Iron- and manganese-containing superoxide dismutases can be distinguished by analysis of their primary structures. FEBS Lett. 1988;229:377–382. doi: 10.1016/0014-5793(88)81160-8. [DOI] [PubMed] [Google Scholar]
- 56.Patek M, Eikmanns B J, Patek J, Sahm H. Promoters from Corynebacterium glutamicum: cloning, molecular analysis and search for a consensus motif. Microbiology. 1996;142:1297–1309. doi: 10.1099/13500872-142-5-1297. [DOI] [PubMed] [Google Scholar]
- 57.Peyret J L, Bayan N, Joliff G, Gulik-Krzywicki T, Mathieu L, Schechter E, Leblon G. Characterization of the cspB gene encoding PS2, an ordered surface-layer protein in Corynebacterium glutamicum. Mol Microbiol. 1993;9:97–109. doi: 10.1111/j.1365-2958.1993.tb01672.x. [DOI] [PubMed] [Google Scholar]
- 58.Rahman M A, Nelson H, Weissbach H, Brot N. Cloning, sequencing, and expression of the Escherichia coli peptide methionine sulfoxide reductase gene. J Biol Chem. 1992;267:15549–15551. [PubMed] [Google Scholar]
- 59.Reyes O, Guyonvarch A, Bonamy C, Salti V, David F, Leblon G. ‘Integron’-bearing vectors: a method suitable for stable chromosomal integration in highly restrictive corynebacteria. Gene. 1991;107:61–68. doi: 10.1016/0378-1119(91)90297-o. [DOI] [PubMed] [Google Scholar]
- 60.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 61.Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 62.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 63.Santos R, Bocquet S, Puppo A, Touati D. Characterization of an atypical superoxide dismutase from Sinorhizobium meliloti. J Bacteriol. 1999;181:4509–4516. doi: 10.1128/jb.181.15.4509-4516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmidt H L, Stocklein W, Danzer J, Kirch P, Limbach B. Isolation and properties of an H2O-forming NADH oxidase from Streptococcus faecalis. Eur J Biochem. 1986;156:149–155. doi: 10.1111/j.1432-1033.1986.tb09560.x. [DOI] [PubMed] [Google Scholar]
- 65.Silva G, Oliveira S, Gomes C M, Pacheco I, Liu M Y, Xavier A V, Teixeira M, Legall J, Rodrigues-Pousada C. Desulfovibrio gigas neelaredoxin: a novel superoxide dismutase integrated in a putative oxygen sensory operon of an anaerobe. Eur J Biochem. 1999;259:235–243. doi: 10.1046/j.1432-1327.1999.00025.x. [DOI] [PubMed] [Google Scholar]
- 66.Stackebrandt E, Rainey F A, Ward-Rainey N L. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol. 1997;47:479–491. [Google Scholar]
- 67.Stallings W C, Pattridge K A, Strong R K, Ludwig M L. The structure of manganese superoxide dismutase from Thermus thermophilus HB8 at 2.4-Å resolution. J Biol Chem. 1985;260:16424–16432. [PubMed] [Google Scholar]
- 68.Stallings W C, Powers T B, Pattridge K A, Fee J A, Ludwig M L. Iron superoxide dismutase from Escherichia coli at 3.1-Å resolution: a structure unlike that of copper/zinc protein at both monomer and dimer levels. Proc Natl Acad Sci USA. 1983;80:3884–3888. doi: 10.1073/pnas.80.13.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Storz G, Imlay J A. Oxidative stress. Curr Opin Microbiol. 1999;2:188–194. doi: 10.1016/s1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 70.Takeda Y, Avila H. Structure and gene expression of the E. coli Mn-superoxide dismutase gene. Nucleic Acids Res. 1986;14:4577–4589. doi: 10.1093/nar/14.11.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Touati D. Superoxide dismutases in bacteria and pathogen protists. In: Scandalios J C, editor. Oxidative stress and molecular biology of antioxidant defenses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1997. pp. 447–493. [Google Scholar]
- 73.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in Δfur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ukeda H, Maeda S, Ishii T, Sawamura M. Spectrophotometric assay for superoxide dismutase based on tetrazolium salt 3′-1-[(phenylamino)-carbonyl]-3, 4-tetrazolium-bis(4-methoxy-6-nitro)benzenesulfonic acid hydrate reduction by xanthine-xanthine oxidase. Anal Biochem. 1997;251:206–209. doi: 10.1006/abio.1997.2273. [DOI] [PubMed] [Google Scholar]
- 75.Vogt W. Oxidation of methionyl residues in proteins: tools, targets, and reversal. Free Radic Biol Med. 1995;18:93–105. doi: 10.1016/0891-5849(94)00158-g. [DOI] [PubMed] [Google Scholar]
- 76.Wan X Y, Zhou Y, Yan Z Y, Wang H L, Hou Y D, Jin D Y. Scavengase p20: a novel family of bacterial antioxidant enzymes. FEBS Lett. 1997;407:32–36. doi: 10.1016/s0014-5793(97)00302-5. [DOI] [PubMed] [Google Scholar]
- 77.Weeks O B, Andrewes A G. Structure of the glycosidic carotenoid corynexanthin. Arch Biochem Biophys. 1970;137:284–286. doi: 10.1016/0003-9861(70)90435-2. [DOI] [PubMed] [Google Scholar]
- 78.Wilkinson M, Doskow J, Lindsey S. RNA blots: staining procedures and optimization of conditions. Nucleic Acids Res. 1990;19:679. doi: 10.1093/nar/19.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wizemann T M, Moskovitz J, Pearce B J, Cundell D, Arvidson C G, So M, Weissbach H, Brot N, Masure H R. Peptide methionine sulfoxide reductase contributes to the maintenance of adhesins in three major pathogens. Proc Natl Acad Sci USA. 1996;93:7985–7990. doi: 10.1073/pnas.93.15.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu C H, Tsai-Wu J J, Huang Y T, Lin C Y, Lioua G G, Lee F J. Identification and subcellular localization of a novel Cu,Zn superoxide dismutase of Mycobacterium tuberculosis. FEBS Lett. 1998;439:192–196. doi: 10.1016/s0014-5793(98)01373-8. [DOI] [PubMed] [Google Scholar]
- 81.Wu J, Weiss B. Two divergently transcribed genes, soxR and soxS, control a superoxide response regulon of Escherichia coli. J Bacteriol. 1991;173:2864–2871. doi: 10.1128/jb.173.9.2864-2871.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yamakura F, Kobayashi K, Tagawa S, Morita A, Imai T, Ohmori D, Matsumoto T. pH-dependent activity change of superoxide dismutase from Mycobacterium smegmatis. Biochem Mol Biol Int. 1995;36:233–240. [PubMed] [Google Scholar]
- 83.Yost F J J, Fridovich I. An iron-containing superoxide dismutase from Escherichia coli. J Biol Chem. 1973;248:4905–4908. [PubMed] [Google Scholar]
- 84.Youn H D, Kim E J, Roe J H, Hah Y C, Kang S O. A novel nickel-containing superoxide dismutase from Streptomyces spp. Biochem J. 1996;318:889–896. doi: 10.1042/bj3180889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang Y, Lathigra R, Garbe T, Catty D, Young D. Genetic analysis of superoxide dismutase, the 23 kilodalton antigen of Mycobacterium tuberculosis. Mol Microbiol. 1991;5:381–391. doi: 10.1111/j.1365-2958.1991.tb02120.x. [DOI] [PubMed] [Google Scholar]
- 86.Zolg J W, Philippi-Schulz S. The superoxide dismutase gene, a target for detection and identification of mycobacteria by PCR. J Clin Microbiol. 1994;32:2801–2812. doi: 10.1128/jcm.32.11.2801-2812.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]