Abstract
Previous work has identified the conjugative transposon Tn5397 from Clostridium difficile. This element was shown to contain a group II intron. Tn5397 can be conjugatively transferred from C. difficile to Bacillus subtilis. In this work we show that the intron is spliced in both these hosts and that nonspliced RNA is also present. We constructed a mutation in the open reading frame within the intron, and this prevented splicing but did not prevent the formation of the circular form of the conjugative transposon (the likely transposition intermediate) or decrease the frequency of intergeneric transfer of Tn5397. Therefore, the intron is spliced, but splicing is not required for conjugation of Tn5397.
Conjugative transposons are genetic elements that encode their own integration, excision, and transfer functions. They are remarkably promiscuous and are capable of being transferred across large phylogenetic distances. They are important clinically, as they are one of the major vectors involved in the spread of antibiotic resistance among bacterial pathogens. There are several recent reviews describing the properties of conjugative transposons (2, 17, 19, 20).
Tn5397 is a conjugative transposon isolated from the gram-positive anaerobic pathogen Clostridium difficile strain 630 (15, 16). Tn5397 encodes tetracycline resistance via the tet(M) gene and has been shown to be transferable by a conjugation-like process from C. difficile 630 to Bacillus subtilis CU2189 and back to C. difficile CD37 (16). It has also been shown to be able to transfer between C. difficile strains (16). Furthermore, Tn5397 has been shown to readily transfer from a B. subtilis donor to a Streptococcus acidominimus recipient in a model oral biofilm community, indicating that the element is likely to be able to transfer to a new host in the natural environment (18). Physical and genetic analysis has shown that Tn5397 is related to the extensively studied conjugative transposon Tn916 (7, 15, 16, 24). There are, however, some important differences. The ends of the two elements are completely different, with the xis and int genes of Tn916 being replaced by a gene called tndX in Tn5397. TndX is a member of the family of large resolvase/invertase proteins. This protein is responsible for the insertion and excision of Tn5397 (24).
The other major difference between Tn916 and Tn5397 is that Tn5397 contains a group II intron, inserted into a gene that is almost identical to orf14 from Tn916 (15). The Tn5397 version of this gene is termed orf14*. Group II introns are a class of genetic elements that were first discovered in the genomes of eukaryotic organelles in fungi and in plants. They are categorized by their secondary structure, which is essential for splicing (12). As well as being capable of splicing, group II introns can also transpose to allelic sites (a process called homing) at high frequency and to ectopic sites at a much lower frequency (12). Some group II introns encode a multifunctional protein, with maturase, reverse transcriptase, and endonuclease activities (4). This protein acts on both RNA and DNA and, together with the catalytic RNA of the intron, forms a ribonucleoprotein (RNP) particle which is required to promote homing, splicing, and transposition activities (3, 4, 27, 28).
Over the past few years, group II introns have been found in a number of different bacteria (5, 6, 8, 10, 13, 15, 26). In most cases, however, splicing and mobility have not been reported to occur in vivo. The main exception to this has been the demonstration that the Lactococcus lactis intron Ll.ltrB, inserted into the putative relaxase gene (ltrb) of the conjugative element pRS01, is spliced and is capable of transposition (13, 14). Furthermore, splicing of this intron was found to be required for conjugal transfer of pRS01 (13). As the group II intron within Tn5397 is inserted into a gene shown to be required for conjugative transfer of the related conjugative transposon Tn916 (21), the work described in this paper was designed to see if the intron is spliced in vivo and if splicing is required for conjugative transfer of Tn5397. We demonstrate that the intron is spliced but that splicing is not a requirement for conjugative transfer of the host conjugative transposon.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All bacterial strains and plasmids used are listed in Table 1. C. difficile and B. subtilis were routinely grown on or in brain heart infusion (BHI) agar or broth (Oxoid, Basingstoke, U.K.). C. difficile strains for RNA preparations were grown in Wilkins-Chalgren medium (Oxoid, Basingstoke). E. coli strains were grown on or in Luria-Bertani (LB) agar or broth. C. difficile strains were grown anaerobically in an anaerobic chamber (Don Whitley Scientific) (10% hydrogen, 10% carbon dioxide, and 80% nitrogen), and B. subtilis and E. coli were grown aerobically. Antibiotics were used at concentrations of 10 μg/ml for tetracycline, 25 μg/ml for kanamycin, 25 μg/ml for rifampin, and 50 μg/ml for ampicillin. All antibiotics were obtained from Sigma Aldrich, UK.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant properties | Reference or source |
|---|---|---|
| C. difficile | ||
| 630 | Wild-type strain containing Tn5397, Tetr | 7 |
| CD37 | Recipient strain, Tets | 7 |
| FM1A | CD37::Tn5397, Tetr | This work |
| B. subtilis | ||
| BS6A | B. subtilis::Tn5397, Tetr | 18 |
| BS16A | BS6A::Tn5397 IntronΔkan, Tetr Kanr | This work |
| E. coli | ||
| DH5αMCR′ | Host strain for pGEM T-easy vector | Gibco-BRL |
| Plasmids | ||
| pGEM T-easy vector | PCR product cloning vector | Promega |
| pUC18 | Cloning vector | 25 |
| pPPM70 | pUC18 containing IntronΔkan | This work |
Mating experiments.
Filter mating were carried out according to Wang et al. (24).
RNA preparation.
C. difficile RNA was isolated as described by Hundsberger et al. (9), but the C. difficile strains were grown in Wilkins-Chalgren medium and not BHI. B. subtilis RNA was isolated using the Qiagen RNeasy kit (Qiagen, Basingstoke, U.K.), according to the manufacturer's instructions.
RT-PCR and PCR.
cDNA for reverse transcriptase PCR (RT-PCR) was prepared using Superscript Rnase H reverse transcriptase according to the manufacturer's instructions (Gibco-BRL). The primers used in these experiments were I1 (5′TGCCGAAGGCAGTCATGTGT), I2 (5′AATCTATCCTCAATTGTTGGGAT), I3 (5′TTTCAGGGCGTTACTCCGTC), I4 (5′AAATTACGTGCTTTAGCTGGGA), I5 (5′ACCACGAACCGCACAACAA), and I6 (5′GCCAATCACAACCTTTTTA). PCR was carried out using a program of 94°C for 4 min, followed by 35 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 to 3 min, followed by a final extension of 4 min at 72°C. PCR from DNA templates was carried out as above. The primers used in these experiments were 5397 (5′ACGTGTATCAAGCAGAGGGAATCG) and 5397t2 (5′CCGGCCAAGCTTAATTAGTAAGCGTCTTGCTC). Products were analyzed on a 1 to 1.5% agarose gel.
Recombinant DNA techniques.
B. subtilis was transformed as described by Agnoustopoulos and Spizizen (1). Genomic DNA was prepared using the Puregene Gram-Positive DNA isolation kit (Flowgen). Plasmid preparations were carried out using the Qiagen miniprep kit according to the manufacturer's instructions. DNA sequencing was carried out using the ABI BigDye terminator mix (PE Biosystems) and analyzed on a Perkin Elmer 310 genetic analyzer. All restriction enzymes were obtained from Promega.
Construction of the intron mutant.
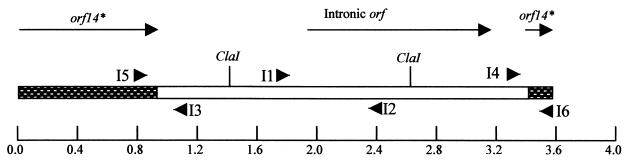
The ClaI fragment of the intron (Fig. 1) was replaced with the aphA-3 kanamycin resistance gene (23). The resulting mutant intron, IntronΔkan, was cloned into the BamHI site of pUC18, generating pPPM70. Plasmid pPPM70 was linearized by digestion with ScaI and transformed into B. subtilis::Tn5397 (BS6A; Table 1), selecting for kanamycin and tetracycline resistance. Strains in which a double crossover had occurred with resultant replacement of the intronic ClaI fragment with IntronΔkan (as demonstrated by PCR) were selected for further study.
FIG. 1.
Genetic organization of the region of Tn5397 containing the group II intron. orf14∗ is represented by the shaded boxes, and the group II intron is represented by the open box. The arrows at the top indicate the position and direction of transcription of the ORFs within this area. The positions of the ClaI sites used for constructing the mutant intron are shown. The binding sites and direction of the oligonucleotide primers used in this investigation are shown by arrows; the names of the primers are beside the arrows. The scale is shown on the bottom line (in kilobases).
RESULTS
Spliced mRNA can be detected from C. difficile containing Tn5397.
The Tn5397 group II intron is contained within orf14*, which is almost identical to orf14 from Tn916 (15, 24). The genetic organization of this region is shown in Fig. 1. In order to determine if the intron is spliced, the primers I5 and I6 were used in an RT-PCR on RNA isolated from C. difficile 630. A product of 330 bp was obtained from cDNA generated with random primers (Fig. 2A) and with cDNA template generated with a specific primer from the coding strand. However, no PCR product was observed when cDNA generated with a specific primer from the noncoding strand was used as the template, indicating, as expected, that splicing only occurs in RNA identical to the coding strand (results not shown). The 330-bp PCR product was sequenced, and it consisted of an in-frame ligation of the two flanking exons, the splice site being located exactly as predicted from the secondary structure of the intron (15). That the intronic open reading frame (ORF) is transcribed was confirmed by RT-PCR using primers I1 and I2 (results not shown). The same results were obtained when the cells were harvested at early, mid-, or late exponential phase (results not shown).
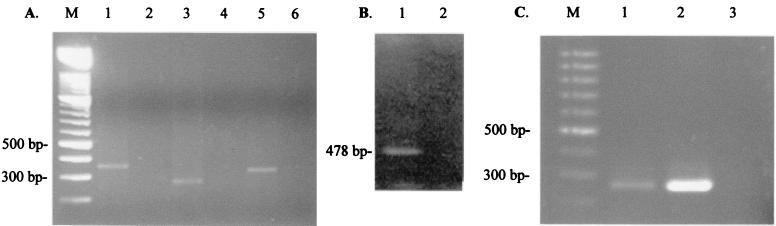
FIG. 2.
RT-PCR and PCR showing the presence of spliced and nonspliced RNA from C. difficile and B. subtilis and the presence of the circular molecule in wild-type and mutant B. subtilis. (A) Template is RNA from C. difficile 630 containing wild-type Tn5397. Lane M, molecular size markers; other lanes contain amplification products of primer combinations as follows: 1, I5 and I6; 2, I5 and I6, no RT; 3, I5 and I3; 4, I5 and I3, no RT; 5, I4 and I6; 6, I4 and I6, no RT. (B) Template is RNA from strain B. subtilis BS6A, which contains the intron mutant IntronΔkan. Lane 1, amplification products with primers I6 and I5; lane 2, primers I6 and I5, no RT. (C) PCR with primers 5397 and 5397t2 to amplify the junction of the circular form. Lane M, molecular size markers. Lanes: 1, BS6A genomic DNA template; 2, BS16A genomic DNA template; 3, B. subtilis CU2189 genomic DNA template.
Nonspliced intron-exon borders can be detected from C. difficile containing Tn5397.
To determine if unspliced intron was also present, RT-PCR with primer pairs I5 and I3 was used to amplify the 5′ intron-exon junction, and primers I4 and I6 were used to amplify the 3′ intron-exon junction. The results are shown in Fig. 2A and show that both the 5′ and 3′ intron-exon junctions are present in the C. difficile RNA. These results were confirmed by DNA sequence analysis.
Spliced and nonspliced RNA can also be detected in B. subtilis.
RNA was prepared from the B. subtilis strain BS6A (18), which contains Tn5397, to see if the intron is spliced in this host. Essentially the same experiments as those described for C. difficile 630 were carried out, and the RT-PCR gave products of identical size as those shown in Fig. 2A for the intron in C. difficile 630. Again, DNA sequencing of the PCR products confirmed that they represented ligated exons and intron-exon borders, indicating that both spliced and nonspliced RNA is present in B. subtilis (results not shown).
Determination of relative amounts of spliced and nonspliced intron.
A dilution series of cDNA template prepared from C. difficile strains 630 and FM1A (Table 1) were used in RT-PCRs to determine the relative amounts of 5′ intron-exon junction, 3′ intron-exon junction, and ligated exons. The results of this analysis are shown in Table 2. The results show that the 5′ intron-exon junction was apparently present at about a 106-fold-lower amount than the 3′ intron-exon junction. There also seems to be as much 3′ intron-exon junction as spliced intron. This could mean that the splicing of the 5′ intron occurs 106 times faster than that of the 3′ intron. Alternatively, some of the intron intermediates may get degraded before splicing is complete. Lariat is more stable than linear RNA, so if splicing were aborted before finishing, the 3′ half (with the lariat) would be expected to persist longer than the 5′ half.
TABLE 2.
Template dilution PCRsa
| C. difficile strain | Primers used | Lowest dilution giving positive result |
|---|---|---|
| 630 | I5 + I3 | 10−2 |
| I5 + I6 | 10−7–10−10 | |
| I4 + I6 | 10−8–10−9 | |
| FM1A | I5 + I3 | 10−2 |
| I5 + I6 | 10−7–10−10 | |
| I4 + I6 | 10−8–10−10 |
Only the 330-bp product corresponding to ligated exons was observed using primer pair I5 and I6.
Deletion of part of the intronic ORF prevents splicing but does not prevent conjugative transposition of Tn5397.
An intron mutant was constructed in B. subtilis strain BS6A (see Materials and Methods). The mutant was designated BS16A. When RT-PCR using primers I5 and I6 was performed, a PCR product of 478 bp was detected (lane 1, Fig. 2B). This product did not correspond to the size expected of spliced intron. DNA sequencing of the product showed that it resulted from mispriming with primer I5. Therefore, we could not detect splicing in the mutant intron.
Previous work in our laboratory has shown that Tn5397 produces a circular form and that this molecule is likely to be the conjugative transposition intermediate (24). In order to determine if the B. subtilis strain containing the intron mutant was still capable of excision and producing the circular form of the transposon, primers reading out from the ends of the element were used for PCR, as in Wang et al. (24). Using these primers, a PCR product would only be produced if the element circularizes and the left and right ends are ligated together. A PCR product of the expected size (272 bp) corresponding to the ligated ends of the transposon was produced in B. subtilis BS6A (B. subtilis strain containing wild-type Tn5397) and BS16A (BS6A::Tn5397 IntronΔkan) (Fig. 2C). DNA sequencing of the PCR product showed that the two ends of Tn5397 had been ligated together with a GA dinucleotide at the joint between the ends of the element, as is the case in the wild-type Tn5397 (24). These results show that splicing is not required for production of the circular form of the element.
The B. subtilis strains BS6A and BS16A were used as donors in a filter mating experiment with C. difficile CD37 as the recipient. Transconjugants arose at a frequency of 2 × 10−7 per donor, a frequency similar to that previously reported for the transfer of Tn5397 from B. subtilis to C. difficile (16, 24). All transconjugants still contained the IntronΔkan allele (confirmed by PCR and DNA sequencing; not shown). Therefore, preventing splicing does not prevent conjugal transfer.
DISCUSSION
In this work, we have demonstrated that the intron from Tn5397 splices in vivo in both C. difficile and B. subtilis. This is an important observation as, although group II introns have recently been found in a number of different bacteria (5, 6, 8, 10, 13, 15, 26), only the Lactococcus lactis group II intron Ll.ltrB has previously been showed to be capable of in vivo splicing (13). The same group II intron has also been found independently in the putative relaxase gene of a conjugative element inserted in the chromosome of L. lactis 712 (22). Furthermore, the Ll.ltrB intron is more closely related to the mitochondrial group II introns than to the other introns found in bacteria (13). The Tn5397 intron falls within a group composed of only bacterial introns. A secondary-structure comparison of the introns also showed that the lactococcal intron belongs to subgroup IIA and the Tn5397 intron to subgroup IIB (S. Zimmerly, personal communication). Therefore, this is the first demonstration of in vivo splicing in a member of this family of group II introns.
Most of the group II introns found in bacteria (including the mitochondrion-like Ll.ltrB intron) have been found associated with genes that are proven or likely to be involved in DNA mobility or contained within mobile genetic elements (5, 6, 8, 10, 13, 15, 26). Splicing of the lactococcal intron Ll.ltrB was required for conjugative transfer of the host element (13). In contrast, however, our results show that splicing of the intron is not required for conjugative transfer of Tn5397, even though the gene interrupted by the intron in Tn5397 has been shown to be required for conjugative transfer of the related conjugative transposon Tn916 (2). As the regions concerned with conjugation in Tn916 and Tn5397 are very closely related, we expected that the orf14* gene product would also be required for transfer of Tn5397. However, as the intron is located near the 3′ end of the gene, lack of splicing would only result in the translated protein's losing the last 23 amino acids, which may not be required for full function. Furthermore, we cannot rule out the possibility that splicing is required in C. difficile but not in B. subtilis due to different host factors which may substitute for the function provided by Orf14*.
As well as being located within genes, at least one group II intron has been shown to be inserted in an intergenic region within the class II transposon TnMerI1 (8). Therefore, there appears to be no actual requirement for all group II introns to be spliced. However, retention of this ability increases the number of sites into which group II introns can transpose without having an adverse effect on the viability of the host cell. Therefore, it is not unexpected that these introns have retained their splicing ability.
Demonstration that the Tn5397 intron splices in both B. subtilis and C. difficile, and the results of Matsuura et al. (11) showing that the L. lactis intron is capable of in vivo splicing in both L. lactis and E. coli, indicates that group II introns are functional in a wide range of distantly related hosts. Zimmerly and coworkers (personal communication) have shown that there has been extensive horizontal gene transfer of group II introns in the past. The frequent association of these introns with broad-host-range conjugal elements, such as Tn5397 and pRS01, provides a means by which some of these introns are dispersed to distantly related hosts, and the minimal requirement for specific host factors allows the introns to be retained in their new hosts.
In conclusion, we have shown that the group II intron within Tn5397 is capable of splicing in both B. subtilis and C. difficile but that splicing is not required for conjugal transfer of Tn5397.
ACKNOWLEDGMENTS
A.P.R. was the recipient of a BBSRC studentship. The work undertaken in one of the laboratories (P.M.) was funded by the Wellcome Trust. The collaboration between the two groups (P.M. and C.v.E.S.) was generously funded by the British Council.
We thank Steven Zimmerly for helpful discussions.
REFERENCES
- 1.Anagnostopoulos G, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clewell D, Flannagan S A, Jaworski D D. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 1995;229:229–236. doi: 10.1016/s0966-842x(00)88930-1. [DOI] [PubMed] [Google Scholar]
- 3.Cousineau B, Lawrence S, Smith D, Belfort M. Retrotransposition of a bacterial group II intron. Nature. 2000;404:1081–1021. doi: 10.1038/35010029. [DOI] [PubMed] [Google Scholar]
- 4.Cousineau B, Smith D, Lawrence-Cavanagh S, Mueller J E, Yang J, Mills D, Manias D, Dunny G, Lambowitz A M, Belfort M. Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous DNA recombination. Cell. 1998;94:451–462. doi: 10.1016/s0092-8674(00)81586-x. [DOI] [PubMed] [Google Scholar]
- 5.Ferat J-L, Le Gouar M, Michel F. Multiple group II self-splicing introns in mobile DNA from Escherichia coli. CR Acad Sci Paris Sci Vie. 1994;317:141–148. [PubMed] [Google Scholar]
- 6.Ferat J-L, Michel F. Group II self-splicing introns in bacteria. Nature. 1993;364:358–361. doi: 10.1038/364358a0. [DOI] [PubMed] [Google Scholar]
- 7.Hachler H, Kayser F H, Berger-Bachi B. Homology of a transferable tetracycline resistance determinant of Clostridium difficile with Streptococcus (Enterococcus) faecalis transposon Tn916. Antimicrob Agents Chemother. 1987;31:1033–1038. doi: 10.1128/aac.31.7.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C-C, Narita M, Yamagata T, Itoh Y, Endo G. Structure analysis of a class II transposon encoding mercury resistance of the Gram-positive bacterium Bacillus megaterium MB1, a strain isolated from Minamata Bay, Japan. Gene. 1999;234:361–369. doi: 10.1016/s0378-1119(99)00184-5. [DOI] [PubMed] [Google Scholar]
- 9.Hundsberger T, Braun V, Weidmann M, Leukel P, Sauerborn M, von Eichel-Streiber C. Transcriptional analysis of the genes tcdA-E of the pathogenicity locus of Clostridium difficile. Eur J Biochem. 1997;244:735–742. doi: 10.1111/j.1432-1033.1997.t01-1-00735.x. [DOI] [PubMed] [Google Scholar]
- 10.Knoop V, Brennicke A. Evidence for a group II intron in Escherichia coli inserted into a highly conserved reading frame associated with mobile DNA sequences. Nucleic Acids Res. 1994;22:1167–1171. doi: 10.1093/nar/22.7.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuura M, Saldanha R, Hongwen M, Wank H, Yang J, Mohr G, Cavanagh S, Dunny G M, Belfort M, Lambowitz A M. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev. 1997;11:2910–3555. doi: 10.1101/gad.11.21.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michel F, Ferat J-L. Structure and activities of group II introns. Annu Rev Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 13.Mills D A, Mckay L L, Dunny G. Splicing of a group II intron involved in the conjugative transfer of pRS01 in lactococci. J Bacteriol. 1996;178:3531–3538. doi: 10.1128/jb.178.12.3531-3538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mills D A, Manias D, Mckay L L, Dunny G. Homing of group II intron from Lactococcus lactis subsp. lactis ML3. J Bacteriol. 1997;179:6107–6111. doi: 10.1128/jb.179.19.6107-6111.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mullany P, Pallen M, Wilks M, Tabaqchali S. A group II intron in a conjugative transposon from the Gram-positive bacterium, Clostridium difficile. Gene. 1996;174:145–150. doi: 10.1016/0378-1119(96)00511-2. [DOI] [PubMed] [Google Scholar]
- 16.Mullany P, Wilks M, Lamb I, Clayton C, Wren B, Tabaqchali S. Genetic analysis of a tetracycline resistance determinant from Clostridium difficile and its conjugal transfer to and from Bacillus subtilis. J Gen Microbiol. 1990;136:1343–1349. doi: 10.1099/00221287-136-7-1343. [DOI] [PubMed] [Google Scholar]
- 17.Rice L B. Tn916 family of conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob Agents Chemother. 1998;42:1871–1877. doi: 10.1128/aac.42.8.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts A P, Pratten J, Wilson M, Mullany P. Transfer of a conjugative transposon, Tn5397, in a model oral biofilm. FEMS Microbiol Lett. 1999;177:63–66. doi: 10.1111/j.1574-6968.1999.tb13714.x. [DOI] [PubMed] [Google Scholar]
- 19.Salyers A A, Shoemaker N B, Stevens A M, Hing-Yew L L. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol Rev. 1995;59:579–590. doi: 10.1128/mr.59.4.579-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott J R, Churchward G G. Conjugative transposition. Annu Rev Microbiol. 1995;49:367–397. doi: 10.1146/annurev.mi.49.100195.002055. [DOI] [PubMed] [Google Scholar]
- 21.Senghas E, Jones J M, Yamamoto M, Gawron-Burke C, Clewell D B. Genetic organization of the bacterial conjugative transposon Tn916. J Bacteriol. 1988;170:245–249. doi: 10.1128/jb.170.1.245-249.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shearman C H, Gordon J J, Gasson G. Splicing of a group II intron in a functional transfer gene of Lactococcus lactis. Mol Microbiol. 1996;21:45–53. doi: 10.1046/j.1365-2958.1996.00610.x. [DOI] [PubMed] [Google Scholar]
- 23.Trieu-Cuot P, Gerbaud G, Lambert T, Courvalin P. In vivo transfer of genetic information between Gram positive and Gram negative bacteria. EMBO J. 1985;4:3583–3587. doi: 10.1002/j.1460-2075.1985.tb04120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Roberts A P, Lyras D, Rood J I, Wilks M, Mullany P. Characterization of the ends and target sites of the novel Tn916-like conjugative transposon Tn5397 from Clostridium difficile: demonstration that excision and circularization are mediated by TndX, a member of the large resolvase family. J Bacteriol. 2000;182:3775–3783. doi: 10.1128/jb.182.13.3775-3783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanisch-Perron G, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13, mp18, and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 26.Yeo C C, Tham J M, Yap M W C, Poh C L. Group II intron from Pseudomonas alcaligenes NCIB 9867 (P25X): entrapment in plasmid RP4 and sequence analysis. Microbiology. 1997;143:2833–2840. doi: 10.1099/00221287-143-8-2833. [DOI] [PubMed] [Google Scholar]
- 27.Zimmerly S, Guo H, Perlman P S, Lambowitz A M. Group II intron mobility occurs by target DNA-primed reverse transcription. Cell. 1995;82:545–554. doi: 10.1016/0092-8674(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 28.Zimmerly S, Guo H, Eskes R, Yang J, Perlman P S, Lambowitz A M. A group II intron is a catalytic component of a DNA endonuclease involved in intron mobility. Cell. 1995;83:529–538. doi: 10.1016/0092-8674(95)90092-6. [DOI] [PubMed] [Google Scholar]