Abstract
Fascioliasis is a highly pathogenic disease affecting humans and livestock worldwide. It is caused by the liver flukes Fasciola hepatica transmitted by Galba/Fossaria lymnaeid snails in Europe, Asia, Africa, the Americas and Oceania, and F. gigantica transmitted by Radix lymnaeids in Africa and Asia. An evident founder effect appears in genetic studies as the consequence of their spread by human-guided movements of domestic ruminants, equines and Old World camelids in the post-domestication period from the beginning of the Neolithic. Establishing the geographical origins of fasciolid expansion is multidisciplinary crucial for disease assessment. Sequencing of selected nuclear ribosomal and mitochondrial DNA markers of F. nyanzae infecting hippopotamuses (Hippopotamus amphibius) in South Africa and their comparative analyses with F. hepatica and F. gigantica, and the two Fascioloides species, Fs. jacksoni from Asian elephants and Fs. magna from Holarctic cervids, allow to draw a tuned-up evolutionary scenario during the pre-domestication period. Close sequence similarities indicate a direct derivation of F. hepatica and F. gigantica from F. nyanzae by speciation after host capture phenomena. Phylogenetic reconstruction, genetic distances and divergence estimates fully fit fossil knowledge, past interconnecting bridges between continents, present fasciolid infection in the wild fauna, and lymnaeid distribution. The paleobiogeographical analyses suggest an origin for F. gigantica by transfer from primitive hippopotamuses to grazing bovid ancestors of Reduncinae, Bovinae and Alcelaphinae, by keeping the same vector Radix natalensis in warm lowlands of southeastern Africa in the mid-Miocene, around 13.5 mya. The origin of F. hepatica should have occurred after capture from primitive, less amphibious Hexaprotodon hippopotamuses to mid-sized ovicaprines as the wild bezoar Capra aegagrus and the wild mouflon Ovis gmelini, and from R. natalensis to Galba truncatula in cooler areas and mountainous foothills of Asian Near East in the latest Miocene to Early Pliocene, around 6.0 to 4.0 mya and perhaps shortly afterwards.
Keywords: Fasciola and Fascioloides species, paleobiogeographical origins, F. nyanzae from hippopotamus, lymnaeid snail vectors, southeastern Africa, Asian Near East
Introduction
The trematode species Fasciola hepatica and F. gigantica are liver flukes which infect livestock and humans causing a worldwide parasitic disease called fascioliasis. These two helminths are transmitted by specific freshwater snails of the family Lymnaeidae, including species of the Galba/Fossaria group in the transmission of F. hepatica in Europe, Asia, Africa, the Americas and Oceania, and species of the Radix group as intermediate hosts or vectors of F. gigantica in Africa and Asia (1). The zoonotic characteristics of this disease are due to the low specificity of the adult stage, which is able to develop in many different definitive hosts, mainly herbivore mammals and also omnivores such as humans and pigs. However, these liver flukes do not produce eggs in all definitive host species and, if they do, the fasciolid eggs shed by several definitive host species show low viability rates which are insufficient to maintain the parasite life cycle in the long term and therefore they do not play the role of a reservoir host (2).
Ruminants such as sheep, goats, cattle and buffaloes are the main reservoirs. Secondarily, equids and Old World camelids also participate in the transmission and spread of this disease typical of rural areas (1). The high pathogenicity along the first invasive, migratory or acute phase of the disease and during the long biliary, obstructive or chronic phase underlies great veterinary losses and impact on livestock husbandry (3). In public health, human fascioliasis has been emerging as a wide problem along the last three decades (4), with the description of human fascioliasis endemic areas in many countries and a progressively increasing number of case reports in the five continents (5). This worrying scenario adds to its high pathogenicity (6–8), virulence (9), long-term post-treatment sequelae (10), and immunosuppression in the acute phase (11) and chronic phase (12, 13). The latter underlies usual coinfections with other pathogenic protozoans and helminths leading to high morbidity (14, 15), and even mortality, mainly in hyperendemic areas of low income countries (16). According to all this, the World Health Organization (WHO) included fascioliasis among the group of Foodborne Trematodiases in the priority list of Neglected Tropical Diseases (NTDs) in the WHO NTD Roadmap for 2030 (17), and has very recently emphasized the convenience of achieving the Roadmap targets through a cross-cutting One Health approach (18).
This disease is the only trematodiasis showing a worldwide distribution. It reached such a wide coverage by taking advantage of human-guided movements of domestic ruminants, equids and Old World camelids along the 12,000–10,000 year post-domestication period. This was possible due to the wide geographical distribution of their snail vector species belonging to Lymnaeidae, a very old family of snails with species present in all continents. Moreover, the amphibious characteristics of these snails facilitate their passive transport by domestic animals when in mud attached to their hooves. Reproduction by selfing and quick multiplication further add to the high spreading capacity of these snails. Thus, the large animal-accompanied movements of human populations throughout the Neolithic, and also subsequently in present times, were decisive in fasciolid spread. DNA marker sequencing indicated a worldwide radiation with evident founder effect from the main domestication regions, illustrating the link of these parasites to mankind history (1). Similar results have been recently observed in genome-wide comparisons of these trematodes (19).
Therefore, assessing the paleobiogeographical origins of both F. hepatica and F. gigantica in the pre-domestication period is crucial to establish the sources of their radiations. This enables to understand the spreading capacities of these parasites and offers the baseline for disease control and prevention measures. It should be considered that this human disease follows different transmission patterns and shows different epidemiological situations. This is in part due to the diversity of human infection sources linked to diet, behavior and traditions markedly differing according to human population ethnics (20). However, the aforementioned disease heterogeneity also depends on (i) which Fasciola species and the presence of only one or the two species in the same area, (ii) the number of lymnaeid vector species involved and their ecology, behavior, population dynamics and seasonality, and also (iii) the mammal reservoir species participating in the disease transmission. All these factors are partly consequences of previous introductions by human-guided animal movements when the range of environmental and climatic characteristics allows for fasciolids and lymnaeids to colonize and establish in an area according to their development requirement thresholds.
The only two other species included in the genus Fasciola are F. nyanzae infecting hippopotamuses in Africa and F. jacksoni specific of Asian elephants (1). However, the mitochondrial genome of the latter has recently demonstrated that this species should better be included in the other fasciolid genus Fascioloides (21). Fascioloides was so far only including the large American liver fluke species Fascioloides magna which infects cervids and bovids originally in North American and recently anthropogenically introduced into Europe (22, 23). The recent finding of F. nyanzae infecting hippopotamuses in three natural reserves of northeastern South Africa (Figure 1) has given the chance for the obtaining of the complete sequences of the DNA markers of the 18S gene and the spacers ITS-1 and ITS-2 of the nuclear ribosomal operon (rDNA), and the cox1 and nad1 genes of the mitochondrial genome (mtDNA).
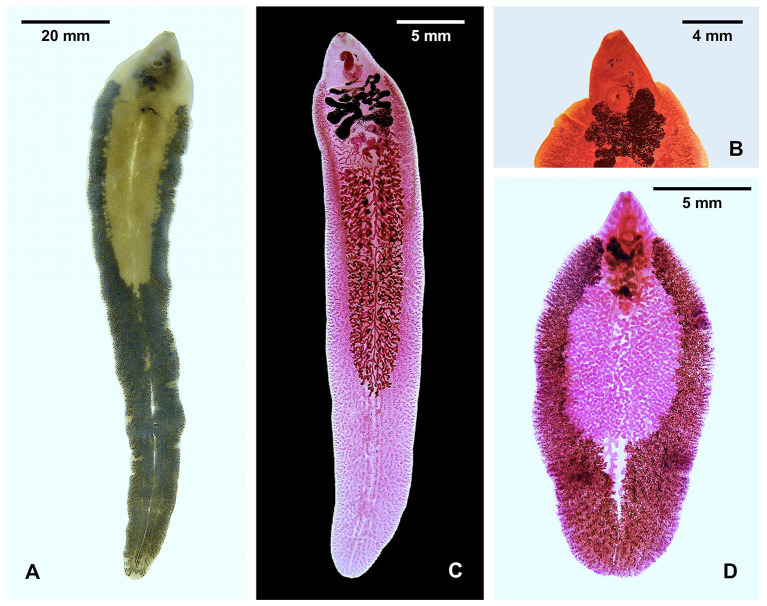
Figure 1.
Species of Fasciola. (A) Unstained F. nyanzae from hippopotamus in South Africa (note maximum width at the level of ovary). (B) Anterior part of stained F. nyanzae showing big apical cone and pronounced shoulders. (C) F. gigantica from cattle in Burkina Faso (note less pronounced shoulders and almost parallel lateral body walls). (D) Specimen highlighting the vitelline follicles of F. hepatica from sheep in Spain (note non-parallel lateral body walls). See decreasing distance between testis end and posterior extremity of the body.
This trematode material enables not only for the analysis of the intraspecific variability of F. nyanzae, but also the comparison of its sequences with those of F. hepatica and F. gigantica (Figure 1), as well as with those of Fs. jacksoni and Fs. magna. The purpose of the present study includes genetic distances and divergence estimates by means of the 18S molecular clock assessment, to allow for an accurate re-definition of the palaeobiogeographical origins of F. hepatica and F. gigantica, by additionally considering the present paleontological knowledge on the evolution of hippopotamuses and ruminants, present data on the fasciolid infection in the wild fauna, and the geographical distribution of the respective lymnaeid vector species.
Materials and methods
Sampling for DNA markers and potential drawback analyses
For the characterization of the fasciolid flukes, the sequences of the complete rDNA small subunit (18S rRNA gene), the complete inter-genic nuclear ribosomal DNA (rDNA) region including the spacers ITS-1 and ITS-2 and the 5.8S gene, and the complete sequences of the two protein-coding genes cox1 and nad1 of the mitochondrial DNA (mtDNA) were selected according to the purposes of this study. The usefulness of these markers for the molecular characterization of Fasciola species and strains has already been verified both at local and regional levels (24, 25) and in worldwide analyses, including the assessment of the spreading routes (1).
Only adult flukes collected from livers of the respective hosts were used for the sequences obtained in the present study. All hosts furnishing fasciolid materials were dead before the autopsy. Only fasciolid samples from selected countries (whenever possible personally collected to assure the obtaining of complete information details) and sequenced by the authors are included in this study. This allowed each marker to be sequenced in its complete length and each mutation detected to be verified in the electropherograms and by specimens re-sequencing when needed. Only in the case of Fs. magna, sequences were used from other authors available in GenBank. No sequence fragments were used to avoid information bias (26).
Countries for the obtaining of fasciolid species were selected to assure that only “genetically pure” specimens were used for sequencing. Samples of F. hepatica were only from countries of Europe and Latin America (see list of countries in next section), where hybrids do not occur because of the absence of F. gigantica and Radix lymnaeids (25). Similarly, samples of F. gigantica were only from African countries (see list of countries in next section), where hybrids do not occur because of the absence of F. hepatica and Galba/Fossaria lymnaeids (1, 27). Specimens of F. hepatica and F. gigantica were obtained from cattle and sheep. It should be considered that metacercariae of these two fasciolid species are similarly infective to cattle and sheep independently of the host isolate of origin (28).
Fasciola nyanzae poses no problem in this sense, because it is specific to hippopotamus (29). A total of 15 liver fluke specimens were obtained from naturally infected Hippopotamus amphibius (two males and five females, aged from 7 to 15 years old), from the following three nature reserves in South Africa: (i) eight liver fluke specimens from Private Nature Reserve 1, Hoedspruit, (ii) one liver fluke specimen Private Nature Reserve 3 (New camp dam), and (iii) six liver fluke specimens from Nature Reserve 2, all located in the Mpumalanga Province (Figure 2). Unfortunately, we could not extract proper DNA from the only specimen from the Private Nature Reserve 3 because of bad fixation.
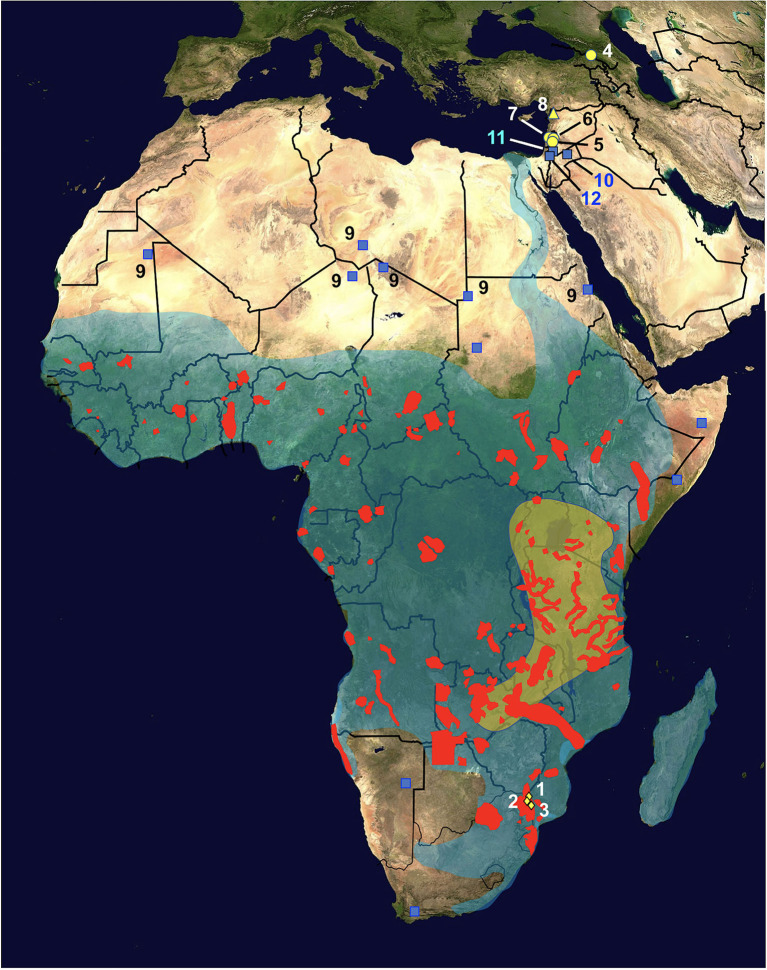
Figure 2.
Map of Africa and Asian Near East showing the distribution of hippopotamus definitive host and radicine snail vector of Fasciola nyanzae. Diamonds = natural reserves where the infected hippopotamuses were found, located in the Mpumalanga province, northeastern South Africa, including Private Nature Reserve 1, Hoedspruit (No. 1), Private Nature Reserve 3 (New camp dam) (2), and Private Nature Reserve 2 (3); red areas = present fragmented distribution of Hippopotamus amphibius; transparent green area = oldest region of the African hippopotamus according to molecular assessments; yellow circles = fossil remains of Hexaprotodon georgicus in the Caucasus mountains (4) and Hexaprotodon gorgops in the Levantine corridor in archeological sites of northern Israel in Ubeidiyah in the Jordan Rift Valley (5), Gesher Benot Yaakov (6), and Evron (7); yellow triangle = fossil remains of H. amphibius found in Latamne, Syria (8); transparent blue area = present distribution of its snail vector Radix natalensis; blue squares = isolated populations of R. natalensis outside the general area, including fossil ones in the Sahara desert (9); note populations in Jordan found in Azraq oasis (10) and the Jordan Valley (11), and in Kishda, Palestina (12). Information from various sources (see text). Geographic background from composed satellite map of Africa orthographic projection by NASA (public domain) via Wikimedia Commons. Schema S. Mas-Coma.
Fascioloides jacksoni is specific to the Asian elephant and it was collected on the island of Sri Lanka, where both F. hepatica and F. gigantica are absent. Regarding Fs. magna, this is the only species whose sequences were obtained from GenBank and a priori its potential hybridization capacity with F. hepatica or F. gigantica may be excluded, as such hybrids have never been reported.
In the case of the three lymnaeid vector species considered for the present paleobiogegraphical analysis, namely Radix natalensis, R. auricularia and Galba truncatula, only the rDNA markers of the 18S gene and the spacers ITS-1 and ITS-2 were obtained and used for the analysis of relative genetic divergences by means of molecular clock assessments.
Fasciolid and lymnaeid samples
Specimens of the fasciolid species were collected from various countries representing different regions, full-length sequences of the aforementioned rDNA and mtDNA markers obtained and deposited in GenBank (Accession Numbers noted in parentheses), and used for the assessments in this paleobiogeographical study:
18S rDNA: 21 sequences including:
- F. hepatica: six sequences from Spain (two), Peru (two) and Argentina (two) (ON661086).
- F. gigantica: six sequences from Burkina Faso (two), Nigeria (two) and Cameroon (two) (ON661087- ON661089).
- F. nyanzae: six sequences from (i) Private Nature Reserve 1, Hoedspruit (three) and (ii) Private Nature Reserve 2 (three), South Africa (ON661084).
- Fs. jacksoni: three sequences from Sri Lanka (ON661085).
Intergenic rDNA region (ITS1-5.8S-ITS2): 64 sequences including:
- F. hepatica: 18 sequences including 10 (one by country) from Spain, France, Poland, Mexico, Venezuela, Ecuador, Peru, Bolivia, Uruguay, and Argentina (MG569980), six (one by country) from Spain, Andorra, Mexico, Ecuador, Bolivia, and Uruguay (MG569978, MG569981), and two from Ecuador (MK212150).
- F. gigantica: 30 sequences from Burkina Faso (15), Niger (three), Nigeria (five), Senegal (three), Cameroon (two) (AJ853848), and Algeria (two) (ON661090, ON661091).
- F. nyanzae: 13 sequences from Private Nature Reserve 1, Hoedspruit (seven) and Private Nature Reserve 2 (six), South Africa (ON661092).
- Fs. jacksoni: three sequences from Sri Lanka (ON661093).
mtDNA cox1 gene: 57 sequences including:
- F. hepatica: 10 sequences from Spain, Poland, Mexico, Venezuela, Ecuador, Peru, Bolivia, Uruguay, and Argentina (MW867310- MW867317, MW867324-MW867326).
- F. gigantica: 31 sequences from Burkina Faso (15), Niger (three), Nigeria (five), Senegal (three), Cameroon (two) (MT094380-MT094390), and Algeria (three) (MN913872, MN913873).
- F. nyanzae: 13 sequences from the Private Nature Reserve 1, Hoedspruit (seven) and the Private Nature Reserve 2 (six), South Africa (ON661094-ON661100).
- Fs. jacksoni: three sequences from Sri Lanka (ON733331).
mtDNA nad1 gene: 57 sequences including:
- F. hepatica: 10 sequences from Spain, Poland, Mexico, Venezuela, Ecuador, Peru, Bolivia, Uruguay, and Argentina (MW867318-MW867323, MW867327-MW867329).
- F. gigantica: 31 sequences from Burkina Faso (15), Niger (three), Nigeria (five), Senegal (three), Cameroon (two) (MT094391-MT094405), and Algeria (three) (MN913874).
- F. nyanzae: 13 sequences from Private Nature Reserve 1, Hoedspruit (seven) and the Private Nature Reserve 2 (six), South Africa (ON661077-ON661084).
- Fs. jacksoni: three sequences from Sri Lanka (ON713419).
For comparison purposes, the following sequences of F. nyanzae and Fs. magna were obtained from GenBank. Concerning F. nyanzae, sequences retrieved included ITS-1 and ITS-2 (MT909820-21, MW046870, MW046872, MT893586-MT893588, MT893595) and partial cox1 sequences (MT909542- MT909550) from Zimbabwe (30). Concerning Fs. magna, sequences retrieved included the 18S gene (EF051080) and ITS-1 and ITS-2 (EF051080, KU232369) from USA (31, 32), and cox1 (EF534998) and nad1 (EF535001) from USA (33). Fasciolopsis buski 18S and ITSs (L06668, MN970005) (34, 35) and cox1 (MF287794) and nad1 (MF287793) from Vietnam (36) were included in the corresponding data matrices for phylogenetic reconstructions.
Additionally, specimens of the three lymnaeid snail vector species involved in the present study were collected in 14 different countries, full-length sequences of the following nuclear rDNA markers obtained and deposited in the GenBank (Accession Numbers noted in parentheses), and used for the assessments of relative genetic divergence estimates:
18S rDNA: 18 sequences including:
- R. natalensis: six sequences from Egypt (one), Burkina Faso (three), and Angola (two) (ON720979).
- R. auricularia: six sequences from Spain (two), France (one), Georgia (two) and Iran (one) (Z73980).
- G. truncatula: six sequences from Spain (two), Egypt (one), Georgia (two), and Iran (one) (Z73985).
rDNA ITS-2: 41 sequences including:
- R. natalensis: 13 sequences from Egypt (two), Burkina Faso (six), Nigeria (one) and Angola (four) (ON729287, ON729288).
- R. auricularia: 14 sequences from Spain (three), France (three), Czech Republic (two), Georgia (four) and Iran (two) (AJ319628-AJ319632, ON729289, ON729290).
- G. truncatula: 14 sequences representing Spain, France, Morocco, Egypt, Georgia, Iran Venezuela, Bolivia, and Argentina (AJ243017, AJ296271, AJ272051).
DNA sequencing
For DNA extraction, a small part of the anterior body region of fasciolids was individually processed (24, 25, 37). Materials were suspended in 400 μl of lysis buffer (10 mM Tris-HCl, pH 8.0, 100 mM EDTA, 100 mM NaCl, 1% sodium dodecyl sulfate SDS) containing 500 μg/ml Proteinase K (Promega, Madison, WI, USA). The digestion was performed during 2 h at 55°C, including alternate shaking every 15 min. Methods previously outlined were followed concerning the procedure steps (24, 38). The phenol-chloroform extraction and ethanol precipitation method was applied for total DNA isolation. Each pellet was dried and resuspended in 30 μl sterile TE buffer (pH 8.0), and subsequently this suspension was stored at −20°C until needed.
Each DNA marker was amplified by PCR in an independent way for each liver fluke individual. Each PCR product was sequenced for a bona-fide haplotype characterization. A set of eight conserved oligonucleotide primers was used for the amplification of five superimposed fragments of the 18S rRNA gene using specific primers (26, 38, 39) and a standard protocol to amplify specific 18S rDNA regions (39). Forward and reverse primers were designed in the regions flanking the rRNA genes 18S and 28S for the subsequent amplification of the complete ITS-1, 5.8S, ITS-2 region (24, 37). The complete cox1 and nad1 gene sequences were obtained using forward and reverse primers designed in regions flanking these genes (1, 24, 27).
For the PCR amplification, the Biotools DNA polymerase® (Biotools B&M Labs. S.A., Madrid, Spain) was used in a Verity-96 Well Thermal Cycler (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA USA). The programs differed according to the marker.
– For the 18S rDNA, it comprised one cycle of 3 min at 94°C, 34–39 cycles of 30 min at 94°C, 40 s at 50–53°C and 1.5 min at 72°C each, preceded by 3 min at 72°C, and followed by a final cooling at 4°C.
– For the rDNA inter-genic region, it comprised one cycle of 2 min at 94°C, 35 cycles of 1 min at 93°C, 1 min at 55°C and 1 min at 72°C each, preceded by 2 min at 72°C, and followed by a final cooling at 4°C.
– For the mtDNA cox1 and nad1 genes, it comprised one cycle of 1 min at 94°C, 40–42 cycles of 1 min at 93°C, 1 min at 52–55°C and 2–3 min at 72°C each, preceded by 5 min at 72°C and followed by a final cooling at 4°C.
For the purification of the PCR product, the Ultra Clean™ PCR Clean-up DNA Purification System (MoBio, Solana Beach, CA, USA) was used following the manufacturer's protocol and eluted in 50 μl of 10 mM TE buffer (pH 7.6). The final DNA concentration (in μg/ml) and the absorbance at 260/280 nm were determined in a Eppendorf BioPhotometer (Hamburg, Germany).
Each molecular marker was sequenced on both strands by the dideoxy chain-termination method performed with the Taq dye-terminator chemistry kit on an Applied Biosystems 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA), by using the PCR primers.
Sequence analyses
The software Sequencher v. 5.4.6 (Gene Codes Co. MI, USA) was used to edit and assemble the sequences, and ClustalW to align them by means of default parameters in MEGA X software (40). Corresponding penalties for gaps were included in pairwise and multiple alignments. Total character differences were used to measure the divergence of the sequences within and among each one of the 18S, ITS-1 and ITS-2, cox1 and nad1 markers. All changes, comprising transitions (ts), transversions (tv) and insertions/deletions (indels), were considered as character states in MEGA X.
By means of the ALTER web server (41), the sequences aligned were collapsed to haplotypes, counting gaps as differences. Closely related sequences were searched by utilizing the BLASTN programme from the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST). The sequences were analyzed by comparing the rDNA and mtDNA sequences of F. hepatica, F. gigantica and F. nyanzae in the genus Fasciola, and those of Fs. jacksoni together with GenBank sequences of Fs. magna in the genus Fascioloides.
DNA haplotype nomenclature
The terminology to identify the haplotype (H) of the aforementioned DNA markers follows the previously proposed combined haplotyping (CH) nomenclature (1, 42). According to this nomenclature, 18S haplotypes are defined by numbers, ITS-2 haplotypes are defined by numbers, and ITS-1 haplotypes by capital letters. Numbers are also utilized for the nucleotide and protein haplotypes of the mtDNA cox1 and nad1 genes. Worth mentioning is that haplotype codes are only definitive when the sequences are complete, i.e., full length sequences. When dealing with fragments or incomplete sequences, haplotype codes are considered only provisional.
Phylogenetic analyses
Due to different limitations recently shown by mtDNA markers for interspecific sequence analyses in invertebrates (26, 43, 44), phylogenetic reconstruction by combined sequences data sets was made from ribosomal and mitochondrial markers separately. Three different phylogenetic analyses were performed, including only complete sequences of the respective markers.
A first phylogenetic tree was obtained by using the concatenated rDNA sequences of the 18S gene, the ITS-1, the 5.8S gene and the ITS-2. The data matrix included 12 sequences and 3,092 characters. In this analysis, the trematode species Paramphistomum cervi (KJ459937) was included as outgroup. Sequences of the Giant Asian fasciolid species Fasciolopsis buski infecting the pig and humans in the Far East and southern Asia (45) were also included (L06668 and MN970005). It should be here considered that sequences of nuclear rDNA and mtDNA markers have recently suggested that Fps. buski populations from China and Vietnam are similar whereas those of India show sufficient nucleotide differences as to consider that they represent distinct taxa (46).
The second phylogenetic tree was obtained by using the mtDNA cox1 sequences, with the data matrix including 33 sequences and 1,578 characters and using Fps. buski from Vietnam (MF287794) as outgroup. A third phylogenetic tree was obtained by means of the mtDNA nad1 sequences, with the corresponding data matrix including 33 sequences and 903 characters and using Fps. buski from Vietnam (MF287793) as outgroup. In both analyses, haplotypes of F. hepatica, F. gigantica F. nyanzae, Fs. jacksoni and Fs. magna were included.
For the phylogenetic reconstruction in each of the aforementioned trees, all the alignments were visually inspected for anomalies and trimmed to the largest possible consensus. The best substitution model selection analysis was run in MEGA X, considering the BIC scores (Bayesian Information Criterion), the AICc value (Akaike Information Criterion, corrected), the Maximum Likelihood value (lnL) and the number of parameters (including branch lengths) for each model. The evolutionary history was inferred by using the Maximum Likelihood (ML) and the Neighbor-Joining (NJ) methods. Initial tree(s) for the heuristic search were obtained automatically with the Nearest-Neighbor-Interchange (NNI) method by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach. To assess the reliability of the nodes in the ML and NJ trees a bootstrap analysis using 1,000 replicates was made using Bootstrap method in MEGA X.
Results
Nuclear rDNA sequences
The small subunit or 18S gene of F. nyanzae has a length of 1,980 bp and a GC content of 50.91%. The sequence of this gene proved to be identical in specimens of F. nyanzae collected in hippopotamuses inhabiting the Klaserie Private Nature Reserve, Hoedspruit and the Sabi Sand Nature Reserve, located in the Mpumalanga Province (Figure 2). For the analysis of this sequence, it was aligned with those of the same gene we obtained in the close fasciolid species F. gigantica, F. hepatica and Fs. jacksoni from the elephant in Sri Lanka, plus that of Fs. magna infecting cervids and bovids obtained in the USA. The resulting alignment was 1,980-bp long, including 1,966 conserved and 14 variable positions, of which eight were parsimony informative positions and six were singleton sites (Table 1).
Table 1.
Polymorphic sites in the sequence comparison of the complete 18S rRNA gene sequence of Fasciola hepatica, F. gigantica and F. nyanzae together with Fascioloides jacksoni and Fs. magna.
| Fasciolid species | 18S haplotypes | Country | GenBank Acc. No. | 18S rRNA gene variable positions | Length (bp) | GC (%) | |||
|---|---|---|---|---|---|---|---|---|---|
| 11 | 11 | ||||||||
| 1 | 3377777846 | 89 | |||||||
| 1 | 1111111178 | 1501999299 | 70 | ||||||
| 1234567890 | 1234567818 | 2152034917 | 71 | ||||||
| F. hepatica | Fh-18S-1 | Spain | ON661086 | CTGGTTGATC | CTGCCAGTTG | TCGTTCTTTT | TT | 1,980 | 50.91 |
| F. hepatica | Fh-18S-1 | Peru | ON661086 | . . . . . . . . . . | . . . . . . . . . G | . . . . . . . . . . | . . | 1,980 | 50.91 |
| F. hepatica | Fh-18S-1 | Argentina | ON661086 | . . . . . . . . . . | . . . . . . . . . G | . . . . . . . . . . | . . | 1,980 | 50.91 |
| F. gigantica | Fg-18S-1 | Burkina Faso | ON661089 | . . . . . . . . . . | . . . . . . . . GG | . . . . . . C . . . | C. | 1,980 | 51.06 |
| F. gigantica | Fg-18S-1 | Nigeria | ON661088 | . . . . . . . . . . | . . . . . . . . GG | . . . . . . C . . . | C. | 1,980 | 51.06 |
| F. gigantica | Fg-18S-1 | Cameroon | ON661087 | . . . . . . . . . . | . . . . . . . . GG | . . . . . . C . . . | C. | 1,980 | 51.06 |
| F. nyanzae | Fn-18S-1 | South Africa-K | ON661084 | . . . . . . . . . . | . . . . . . . . . G | . . . . . . . . . . | . . | 1,980 | 50.91 |
| F. nyanzae | Fn-18S-1 | South Africa-S | ON661084 | . . . . . . . . . . | . . . . . . . . . G | . . . . . . . . . . | . . | 1,980 | 50.91 |
| Fs. jacksoni | Fsj-18S-1 | Sri Lanka | ON661085 | . . . . . . . . . . | . . . . . . . . . A | . . . CCT . ACC | . C | 1,980 | 51.06 |
| Fs. magna | Fsm-18S-1 | Oregon, USA | EF051080 | - - - - - - - - - - | - - - - - - - -. A | CTAC . TCA . . | . C | 1,962 | 50.87 |
Positions, numbers (to be read in vertical) refer to variable positions obtained in the alignment made with MEGA X; . = identical; shaded positions with a gap = not sequenced; K, Klaserie Private Nature Reserve; S, Sabi Sand Nature Reserve.
This sequence is identical to that of the 18S gene in F. hepatica, but shows three mutations with regard to F. gigantica, despite identical length, in positions 71 (T in F. nyanzae but G in F. gigantica), 794 (T vs. C), and 1,878 (T vs. C) (Table 1). This gene is also 1,980-bp long in Fs. jacksoni, but has been reported to only have 1,962 bp (lacking 18 nucleotides in the 5' extremity) in Fs. magna. When comparing in the corresponding alignment (Table 1), eight mutations appear between F. nyanzae and Fs. jacksoni, all of them in positions differing from the aforementioned three ones distinaguishing F. nyanzae from F. gigantica. Nine mutations appear between F. nyanzae and Fs. magna, of which seven are different from the eight in Fs. jacksoni.
The whole intergenic region of the rDNA operon of F. nyanzae, including ITS-1, 5.8S gene and ITS-2, proved to be 951-bp long, with a 50.47% GC content. No intraspecific variability was found in the specimens from the two reserves. The lengths of these three markers were 432, 154, and 365 bp, respectively. These lengths agree with the 951-bp-long intergenic region of F. hepatica and differ from the 950-bp-long sequence of F. gigantica because of one deletion in the ITS-2 of this species. The alignment of the intergenic sequences of these three Fasciola species allows for the comparison of the two spacer markers (Table 2). Interestingly, nucleotides differing between the ITS sequences of F. nyanzae and those of F. hepatica and F. gigantica are very few and only appear in the same positions differing between the two latter. Moreover, the differing nucleotides in F. nyanzae coincide with those of one or the other of the two fasciolids, which indicates that the ITSs of F. nyanzae are of intermediate characteristics. Without counting the two positions of the ITS-2 in which intraspecific variability is known in F. hepatica and F. gigantica, F. nyanzae only showed two differences in the ITS-1 and other two in the ITS-2 when compared to F. hepatica, whereas three and two, respectively appeared when compared to F. gigantica.
Table 2.
Polymorphic sites in the sequence comparison of the nuclear rDNA complete intergenic region and in the ITS-1 and ITS-2 between haplotypes of “genetically pure” Fasciola hepatica from livestock in Europe and the Americas, haplotypes of “genetically pure” F. gigantica from livestock in African countries, and the haplotype of F. nyanzae from hippopotamuses in the nature reserves in South Africa.
| Fasciola spp. and combined haplotypes |
Polymorphic sites
Intergenic region (ITS-1, 5.8S, ITS-2) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positions | 24 | 114 | 208 | 286 | 306 | 821 | 834 | 860 | 866 | 874 | 917 | 924 |
| Polymorphic sites ITS-1 | Polymorphic sites ITS-2 | |||||||||||
| Positions | 24 | 114 | 208 | 286 | 306 | 234 | 248 | 273 | 279 | 288 | 330 | 337 |
| F. hepatica 1A | C | A | C | T | C | T | A | C | C | C | T | G |
| F. hepatica 2A | C | A | C | T | C | T | A | C | C | T | T | G |
| F. hepatica 1/2A* | C | A | C | T | C | T | A | C | C | C/T | T | G |
| F. gigantica 1A | T | T | T | A | T | C | A | T | T | C | - | A |
| F. gigantica 2A | T | T | T | A | T | C | C | T | T | C | - | A |
| F. gigantica 1/2A** | T | T | T | A | T | C | C/A | T | T | C | - | A |
| F. nyanzae 1A | T | A | T | T | C | T | A | T | C | C | T | A |
Heterozygotic in position 874/288 not differentiating between F. hepatica and F. gigantica.
Heterozygotic in position 834/248 not differentiating between F. hepatica and F. gigantica- also designed as H3A in Chougar et al. (27).
Opposite to this, nucleotide differences were numerous in the two spacers when compared to Fs. jacksoni and Fs. magna (Tables 3, 4) and underlie the great genetic distances between the two genera Fasciola and Fascioloides (Tables 5A–C), which agree with the marked morpho-anatomical differences in the adult stages of their species.
Table 3.
Polymorphic sites in the sequence comparison of the nuclear rDNA ITS-1 between haplotypes of “genetically pure” Fasciola hepatica from livestock in Europe and the Americas, “genetically pure” F. gigantica from livestock in African countries, F. nyanzae from hippopotamuses in the nature reserves in South Africa and from snails and one hippopotamus in Zimbabwe, Fascioloides jacksoni from elephants in Sri Lanka, and Fs. magna from cervids in USA.
| Fasciolid species | haplotype or isolate (I) | Country | Acc. No. | ITS-1 variable positions | Length (bp) | GC (%) | |||
|---|---|---|---|---|---|---|---|---|---|
| 111111 | 1111111222 | 2222333333 | |||||||
| 1244577777 | 7799111246 | 6677789056 | 7789001227 | ||||||
| 6568902345 | 6737356275 | 6826735977 | 2878571591 | ||||||
| F. hepatica | Fh-ITS1-A | Spain + a | MG569980 | -CACGTGCGG | ACAAGACTCT | CCGTTAGCTT | T A T A T C A T C C | 365 | 48.49 |
| F. gigantica | Fg-ITS1-A | Burkina Faso + b | AJ853848 | -T . . . . . . . . | . . . . . T . . . . | . . . . . . . T . . | . . A . . T . . . . | 364 | 48.08 |
| F. nyanzae | Fn-ITS1-A | South Africa-K | ON661092 | -T . . . . . . . . | . . . . . . . . . . | . . . . . . . T . . | . . . . . . . . . . | 365 | 47.95 |
| F. nyanzae | Fn-ITS1-A | South Africa-S | ON661092 | -T . . . . . . . . | . . . . . . . . . . | . . . . . . . T . . | . . . . . . . . . . | 365 | 47.95 |
| F. nyanzae | I S8bLN | Zimbabwe | MT893595 | -T . . . . . . . . | . . . . . . . . . . | . . . . . . . T . . | . . . . . . . . . . | 365 | 47.95 |
| F. nyanzae | I 483 | Zimbabwe | MT893588 | -T . . . . . . . . | . . . . . . . . . . | . . . . . . . T . . | . . . . . . . . . . | 365 | 47.95 |
| F. nyanzae | I 481 | Zimbabwe | MT893587 | -T . . . . . . . . | . . . . . . . . . . | . . . . . . . T . . | . . . . . . . . . . | 365 | 47.95 |
| F. nyanzae | I 480 | Zimbawbe | MT893586 | -T . . . . . . . . | . . . . . . . . . . | . . . . . . . T . . | . . . . . . . . . . | 365 | 47.95 |
| F. nyanzae | I 394 | Zimbabwe | MW046872 | -T . . . . . . . . | . . . . . . . . . . | . . . . . . . T . . | . . . . . . . . . . | 365 | 47.95 |
| F. nyanzae | I 393 | Zimbabwe | MW046870 | -T . . . . . . . . | . . . . . . . . . . | . . . . . . . T . . | . . . . . . . . . . | 365 | 47.95 |
| F. nyanzae | I 2 | Zimbabwe | MT909820 | -T . . . . . . . . | . . . . . . . . . . | . . . . . . . T . . | . . . . . . . . . . | 365 | 47.95 |
| F. nyanzae | I 1 | Zimbabwe | MT909821 | -T . . . . . . . . | . . . . . . . . . . | . . . . . . . T . . | . . . . . . . . . . | 365 | 47.95 |
| Fs. jacksoni | Fsj-ITS1-A | Sri Lanka | ON661093 | A ... A . . . . . | . . CG. . ACTC | TT-AG . A . . . | . . . . . TG. . T | 364 | 48.90 |
| Fs. magna | n.d. | Oregon, USA | KU232369 | A . GT. GTT- - | - T . . CGTCTC | T. - GGG. . . C | C G . CCT. CT. | 364 | 50.00 |
| Fs. magna | n.d. | MS, USA | EF051080 | A . GT. GTT- - | - T . . CGTCTC | T. - GGG. . CC | C G . CCT. CT. | 364 | 50.00 |
Differences observed in a 433-bp-long alignment including 398 conserved and 35 variable positions plus 5 gapped positions, of which 34 were parsimony informative positions and 1 was a singleton site. Positions, numbers (to be read in vertical) refer to variable positions obtained in the alignment made with MEGA X; . = identical; - = insertion/deletion; n.d., not determined.
a, Andorra, France, Poland, Mexico, Venezuela, Ecuador, Peru, Bolivia, Uruguay, Argentina.
b, Niger, Nigeria, Senegal, southern Algeria, Cameroon.
K, Klaserie Private Nature Reserve; S, Sabi Sand Nature Reserve.
Table 4.
Polymorphic sites in the sequence comparison of the nuclear rDNA ITS-2 between haplotypes of “genetically pure” Fasciola hepatica from livestock in Europe and the Americas, “genetically pure” F. gigantica from livestock in African countries, F. nyanzae from hippopotamuses in the nature reserves in South Africa, and from snails and one hippopotamus in Zimbabwe, Fascioloides jacksoni from elephants in Sri Lanka, and Fs. magna from cervids in USA.
| Fasciolid species | haplotype (H) or isolate (I) | Country | Acc. No. | ITS-2 variable positions | Length (bp) | GC (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 11111 | 1111112222 | 2222222222 | 2222223333 | 3333333333 | 333 | ||||||
| 257822234 | 4558990001 | 3333455777 | 8888890001 | 2233333334 | 555 | ||||||
| 6779001232 | 6182340341 | 3579803234 | 0168913457 | 5801235897 | 169 | ||||||
| F. hepatica | Fh-ITS2-1 | Spain + a | MG569980 | T A T T G A T G T A | G G A T C T G A G T | C T T A A A T G T C | C A G C A T G A A G | C T T T C A A G C C | TGT | 365 | 48.49 |
| F. hepatica | Fh-ITS2-2 | Spain + b | MG569981 | . . . . . . . . . . | . . . . . . . . . . | . . . . . . . . . . | . . . T . . . . . . | . . . . . . . . . . | . . . | 365 | 48.22 |
| F. hepatica | Fh-ITS2-Htz* | Ecuador | MK212150 | . . . . . . . . . . | . . . . . . . . . . | . . . . . . . . . . | . . . Y . . . . . . | . . . . . . . . . . | . . . | 365 | 48.35 |
| F. gigantica | Fg-ITS2-1 | Burkina Faso + c | AJ853848 | . . . . . . . . . . | . . . . . . . . . . | . C . . . . . . . T | T . . . . . . . . . | . . . - . . . A . . | . . . | 364 | 48.08 |
| F. gigantica | Fg-ITS2-2 | southern Algeria | ON661090 | . . . . . . . . . . | . . . . . . . . . . | . C . . C . . . T | T . . . . . . . . . | . . . - . . . A . . | . . . | 364 | 48.35 |
| F. gigantica | Fg-ITS2-Htz* | southern Algeria | ON661091 | . . . . . . . . . . | . . . . . . . . . . | . C . . M . . . T | T . . . . . . . . . | . . . - . . . A . . | . . . | 364 | 48.21 |
| F. nyanzae | Fn-ITS2-1 | South Africa-K | ON661092 | . . . . . . . . . . | . . . . . . . . . . | . . . . . . . . . T | . . . . . . . . . . | . . . . . . . A . . | . . . | 365 | 47.95 |
| F. nyanzae | Fn-ITS2-1 | South Africa-S | ON661092 | . . . . . . . . . . | . . . . . . . . . . | . . . . . . . . . T | . . . . . . . . . . | . . . . . . . A . . | . . . | 365 | 47.95 |
| F. nyanzae | I S8bLNA1 | Zimbabwe | MT893595 | . . . . . . . . . . | . . . . . . . . . . | . . . . . . . . . T | . . . . . . . . . . | . . . . . . . A . . | . . . | 365 | 47.95 |
| F. nyanzae | I 483 | Zimbabwe | MT893588 | . . . . . . . . . . | . . . . . . . . . . | . . . . . . . . . T | . . . . . . . . . . | . . . . . . . A . . | . . . | 365 | 47.95 |
| F. nyanzae | I 481 | Zimbabwe | MT893587 | . . . . . . . . . . | . . . . . . . . . . | . . . . . . . . . T | . . . . . . . . . . | . . . . . . . A . . | . . . | 365 | 47.95 |
| F. nyanzae | I 480 | Zimbabwe | MT893586 | . . . . . . . . . . | . . . . . . . . . . | . . . . . . . . . T | . . . . . . . . . . | . . . . . . . A . . | . . . | 365 | 47.95 |
| F. nyanzae | I 394 | Zimbabwe | MW046872 | . . . . . . . . . . | . . . . . . . . . . | . . . . . . . . . T | . . . . . . . . . . | . . . . . . . A . . | . . . | 365 | 47.95 |
| F. nyanzae | I 393 | Zimbabwe | MW046870 | . . . . . . . . . . | . . . . . . . . . . | . . . . . . . . . T | . . . . . . . . . . | . . . . . . . A . . | . . . | 365 | 47.95 |
| F. nyanzae | I 1 | Zimbabwe | MT909820 | . . . . . . . . . . | . . . . . . . . . . | . . . . . . . . . T | . . . . . . . . . . | . . . . . . . A . . | . . . | 365 | 47.95 |
| F. nyanzae | I 2 | Zimbawbe | MT909821 | . . . . . . . . . . | . . . . . . . . . . | . . . . . . . . . T | . . . . . . . . . . | . . . . . . . A . . | . . . | 365 | 47.95 |
| Fs. jacksoni | Fsj-ITS2-1 | Sri Lanka | ON661093 | . T. - . GCA. G | TAGA . CA . TC | T . . GG . C . . T | TGA . GC . GG . | T . A . A . GAAT | GA. | 364 | 48.90 |
| Fs. magna | n.d. | Oregon, USA | KU232369 | CTG- A. . . CG | . . GAACAGTG | T . C . . GCACT | . GA . GCAGGA | . CG . TGTAAT | . AA | 364 | 50.00 |
| Fs. magna | n.d. | MS, USA | EF051080 | CTG- A. . . CG | . . GAACAGTG | T . C . . GCACT | . GA . GCAGGA | . CG . TGTAAT | . AA | 364 | 50.00 |
Differences observed in a 365-bp-long alignment including 314 conserved and 51 variable positions plus 3 gapped positions, of which 50 were parsimony informative positions and 1 was a singleton site. Positions, numbers (to be read in vertical) refer to variable positions obtained in the alignment made with MEGA X; . = identical; - = insertion/deletion; n.d., not determined.
*Htz = pure species with heterozygotic position not differentiating between F. hepatica and F. gigantica (represented with corresponding symbol of IUPAC code for incomplete nucleic acid specification).
a, France, Poland, Mexico, Venezuela, Peru, Bolivia, Uruguay, Argentina.
b, Andorra, Mexico, Ecuador, Bolivia, Uruguay.
c, Niger, Nigeria, Senegal, Cameroon.
K, Klaserie Private Nature Reserve; S, Sabi Sand Nature Reserve.
Table 5.
Estimates of evolutionary divergences over sequence pairs between species groups in the complete sequences of the nuclear rDNA and mtDNA markers.
| (A) rDNA ITS-1 | ||||
| F. nyanzae | ||||
| F. gigantica | 3.0 | |||
| F. hepatica | 2.0 | 4.4 | ||
| Fs. jacksoni | 17.0 | 17.4 | 15.0 | |
| Fs. magna | 26.5 | 25.9 | 24.5 | 25.5 |
| (B) rDNA ITS-2 | ||||
| F. nyanzae | ||||
| F. gigantica | 2.3 | |||
| F. hepatica | 2.3 | 4.4 | ||
| Fs. jacksoni | 32.0 | 30.3 | 34.3 | |
| Fs. magna | 37.0 | 37.3 | 39.3 | 29.0 |
| (C) Complete intergenic region | ||||
| F. nyanzae | ||||
| F. gigantica | 6.4 | |||
| F. hepatica | 5.3 | 8.9 | ||
| Fs. Jacksoni | 50.0 | 47.8 | 49.3 | |
| Fs. magna | 63.5 | 64.3 | 64.8 | 55.5 |
| (D) mtDNA cox1 gene | ||||
| F. nyanzae | ||||
| F. gigantica | 119.9 | |||
| F. hepatica | 140.9 | 130.6 | ||
| Fs. Jacksoni | 215.6 | 199.8 | 212.0 | |
| Fs. magna | 198.8 | 192.9 | 205.4 | 187.7 |
| (E) mtDNA nad1 gene | ||||
| F. nyanzae | ||||
| F. gigantica | 80.8 | |||
| F. hepatica | 77.2 | 75.6 | ||
| Fs. jacksoni | 140.9 | 132.0 | 133.6 | |
| Fs. magna | 136.4 | 133.4 | 136.6 | 106.0 |
The number of base differences (ts + tv) per sequence from averaging over all sequence pairs between groups are shown. All ambiguous positions were removed for each sequence pair. Evolutionary analyses conducted in MEGA X.
Mitochondrial DNA gene sequences
The complete mtDNA cox1 gene provided seven different sequences with the same length of 1,533 bp and an average of AT content of 60.59%. Their alignment showed seven variable positions, including four parsimony-informative and three singleton, five of which close to the 3' end. The COX1 protein was 511 aa long, with start/stop codons of ATG/TAG, and provided three haplotypes. The differences between these three protein haplotypes were restricted to only one or two amino acid changes in the protein alignment (Table 6).
Table 6.
Nucleotide and amino acid differences found in the complete mtDNA cox1 sequence of Fasciola nyanzae from South Africa and other F. nyanzae isolate fragments from Zimbabwe.
| F. nyanzae haplotype (H) or isolate fragment (I) | Locality* | Country | Host** | GenBank Acc. No. | cox 1 nucleotide sequence | COXI amino acid sequence | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable positions | Long (bp) | AT (%) | Variable positions | Length (aa) | ||||||||
| 11 | 1111111111 | 113443 | ||||||||||
| 1222233344 | 4445666666 | 8999999900 | 0000013445 | 6373499 | ||||||||
| 9001300400 | 4492014569 | 0256669900 | 0111475992 | 8547089 | ||||||||
| 5139116538 | 1450785199 | 7783690325 | 9148130364 | |||||||||
| H cox1-a | K | South Africa | H.amph | ON661094 | CGTCTTGATA | CTCCCTTGAT | CGCCGACCGG | AAGATGCATA | 1,533 | 60.60 | MWHMIHI | 510 |
| H cox1-b | K | South Africa | H.amph | ON661095 | . . . . . . . . . . | . . . . . . . . . . | . . . . . . . . . . | . . . . . . . C . . | 1,533 | 60.53 | . . . . . P . | 510 |
| H cox1-c | S | South Africa | H.amph | ON661096 | . . . . . . . . . . | . . . . . . . . . . | . . . . . . . . . . | . . . . . AT . AG | 1,533 | 60.67 | . . . . . . N | 510 |
| H cox1-d | S | South Africa | H.amph | ON661097 | . . . . C . . . . . | . . . . . . . . . . | . . . . . . . . . . | . . . . . . . . AG | 1,533 | 60.47 | . . . . . . N | 510 |
| H cox1-e | S | South Africa | H.amph | ON661098 | . . . . . . . . . . | . . . . . . . . . . | . . . . . . . . . . | . . . . . . . . AG | 1,533 | 60.53 | . . . . . . N | 510 |
| H cox1-f | S | South Africa | H.amph | ON661099 | . . . . . . . . . . | . . . . . . . . . . | . . . . . . . . . . | . . . . . A . . AG | 1,533 | 60.60 | . . . . . . N | 510 |
| H cox1-g | K | South Africa | H.amph | ON661100 | . . . . . . . . . . | . . . T . . . . . . | . . . . . . . . . . | . . . . . AT . AG | 1,533 | 60.73 | . . Y . . . N | 510 |
| I S8b LNA 1 | Kariba | Zimbabwe | R.nat | MT909550 | TA . . . . . . C . | . . . . TA- - - - | - - - - - - - - - - | - - - - - - - - - - | 428 | 59.35 | . R . - - - - | 142 |
| I KAR S3 RAD 10 | Kariba | Zimbabwe | R.pli | MT909549 | TAA . . . . C . . | . . . . T . CCG- | - - - - - - - - - - | - - - - - - - - - - | 482 | 58.92 | K . . - - - - | 160 |
| I 483 | Kariba | Zimbabwe | R.nat | MT909548 | TA . . . . . . . . | TC . . T- - - - - | - - - - - - - - - - | - - - - - - - - - - | 425 | 59.29 | . . . - - - - | 141 |
| I 480 | Kariba | Zimbabwe | R.nat | MT909546 | TA . . . . AG . G | T . T . . - - - - - | - - - - - - - - - - | - - - - - - - - - - | 424 | 59.43 | . . . - - - - | 141 |
| I 32 | Kariba | Zimbabwe | P.col | MT909545 | TA . T . . . . . . | . . . . T - - - - - | - - - - - - - - - - | - - - - - - - - - - | 425 | 59.53 | . . . - - - - | 141 |
| I 2 | Kariba | Zimbabwe | H.amph | MT909543 | - - - T . . . . . . | . . . . T . . TG | TA TGTTTTAA | GTTGG- - - - - | 862 | 60.90 | ? . . VV - | 287 |
| I 1 | Kariba | Zimbabwe | H.amph | MT909542 | - - . . . C . . . . | . . . . . . . . GC | . . . T . . . . . . | . . . - - - - - - - | 815 | 59.39 | ? . . . - - - | 271 |
Positions, numbers (to be read in vertical) refer to variable positions obtained in the 1,533-bp-long alignment made with MEGA X; . = identical; shaded positions with a gap = not sequenced.
K, = Klaserie Private Nature Reserve; S, = Sabi Sand Nature Reserve.
H.amph, Hippopotamus amphibius; R.nat, Radix natalensis; R.pli, Radix plicatula; P.col, Pseudosuccinea columella.
When including the five 424–482-bp-long fragments of F. nyanzae cox1 obtained from lymnaeid snails and the two 815–862-bp-long fragments of the same gene obtained in only one hippopotamus, all from an artificial lake in Zimbabwe, a high number of 40 nucleotide differences appear in a 1,533-bp-long alignment (Table 6). None of the variable positions in the Zimbabwe sequences coincide with the only two observed in the corresponding fragment, and the sequence extreme where the five variable positions were detected in South African reserves was unfortunately not included in the Zimbabwe fragments.
Another alignment was performed including sequences from GenBank where appropriate comparison purposes, including the seven complete cox1 haplotypes of F. nyanzae from the South African reserves plus the complete sequences of cox1 of (i) 10 haplotypes of “genetically pure” F. hepatica from Europe and Latin America where F. gigantica is absent, (ii) 14 haplotypes of “genetically pure” F. gigantica from six African countries where F. hepatica is absent, (iii) eight isolates of F. jacksoni from Sri Lanka, and (iv) three isolates of F. magna from USA. The resulting alignment included a total of 42 sequences and was 1,546 pb long, of which 1,157 were conserved and 388 variable (359 p-info +29 singleton) (table not shown). This number of variable positions defines a genetic divergence of 25.1% for this cox1 gene in these fasciolids.
To assess the evolutionary speed of the cox1 gene inside the genus Fasciola, a genetic divergence of 14.36% was deduced from the 1,546-bp-long alignment with 222 variable positions including only the aforementioned 31 sequences of the three species of Fasciola. Additionally, a slightly smaller genetic divergence of 12.95% was deduced for Fascioloides from the 1,545-bp-long alignment with 200 variable positions including only the aforementioned 11 sequences of the two species of this genus. The mean number of base differences in cox1 between the species of Fasciola and Fascioloides are noted in Table 5D.
For its part, the mtDNA nad1 gene provided seven different sequence haplotypes with the same length of 903 bp and an average AT content of 61.50%. Their alignment showed 12 variable positions (9 p-info and three singleton). The NAD1 protein showed only one 301-aa-long haplotype with start/stop codons of GTG/TAG in all specimens analyzed, and provided three different haplotypes (Table 7).
Table 7.
Nucleotide and amino acid differences found in the complete mtDNA nad1 sequence of Fasciola nyanzae from South Africa.
| F. nyanzae haplotype (H) | Locality* | Host** | GenBank Acc. No. | nad 1 nucleotide sequence | NAD1 amino acid sequence | ||||
|---|---|---|---|---|---|---|---|---|---|
| Variable positions | Length (bp) | AT (%) | Variable positions | Length (aa) | |||||
| 1111111245 | 88 | 1 | |||||||
| 5555666280 | 16 | 555557 | |||||||
| 1489013839 | 94 | 123450 | |||||||
| H nad1-a | S | H.amph | ON661077 | GAATGGGTTC | CC | 903 | 61.35 | G S Y G E A | 300 |
| H nad1-b | S | H.amph | ON661078 | . . . . . . . C . T | . . | 903 | 61.35 | . . . . . V | 300 |
| H nad1-c | S | H.amph | ON661079 | . . . . . . . . CT | T . | 903 | 61.46 | . . . . . V | 300 |
| H nad1-d | K and S | H.amph | ON661080 | . . . . . . . . . T | . . | 903 | 61.46 | . . . . . V | 300 |
| H nad1-e | K | H.amph | ON661081 | TGTGTTC . . T | T . | 903 | 61.68 | WGLLQV | 300 |
| H nad1-f | K | H.amph | ON661082 | TGTGTTCC . T | . . | 903 | 61.46 | WGLLQV | 300 |
| H nad1-g | K | H.amph | ON661083 | TGTGTTC . . T | . . | 903 | 61.57 | WGLLQV | 300 |
Positions = numbers (to be read in vertical) refer to variable positions obtained in the 903-bp-long alignment made with MEGA X; . = identical.
K, Klaserie Private Nature Reserve; S, Sabi Sand Nature Reserve.
H.amph, Hippopotamus amphibius.
Comparative nad1 alignment analyses were performed including our seven haplotypes of F. nyanzae with other complete sequences of nad1 gene, including (i) seven haplotypes of “genetically pure” F. hepatica from Europe and Latin America, (ii) 21 haplotypes of “genetically pure” F. gigantica from six African countries where F. hepatica is absent, (iii) eight isolates of Fs. jacksoni from Sri Lanka, and (iv) three isolates of Fs. magna from the USA. The resulting 903-pb-long alignment including these 46 sequences showed a length of 903 pb, of which 675 were conserved and 228 variable (224 p-info + 4 singleton) (table not shown).
Finally, to assess the evolutionary speed of the nad1 gene inside the genus Fasciola, a genetic divergence of 14.06% was deduced from the 903-bp-long alignment with 127 variable positions including only the aforementioned 35 sequences of the three species of Fasciola. Additionally, a similar genetic divergence of 14.16% was deduced for Fascioloides from the 903-bp-long alignment with 112 variable positions including only the aforementioned 11 sequences of the two species of this genus. The mean number of base differences in nad1 between the species of Fasciola and Fascioloides are noted in Table 5E.
Phylogenetic analyses
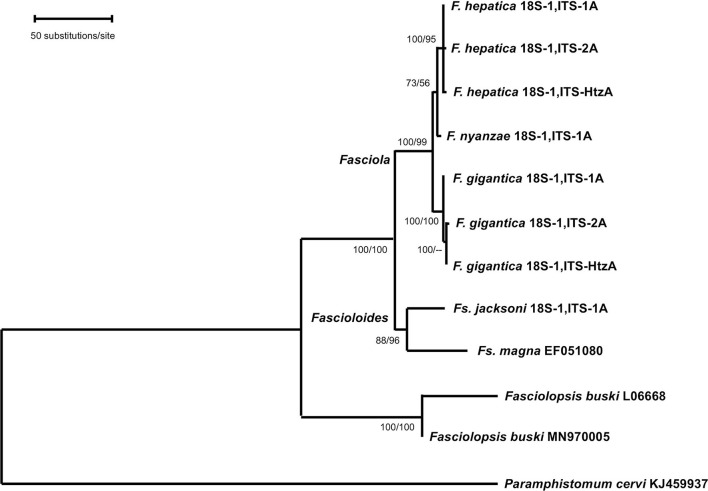
Phylogenetic analysis carried out with the combination of the 18S rRNA gene and the complete intergenic region of the ribosomal operon, including ITS-1, 5.8S and ITS-2 in a single data-set, generated a robust tree, indicating phylogenetic accordance between the ribosomal markers. The ML model best fitting this data-set was Hasegawa-Kishino-Yano model with discrete Gamma distribution (HKY + G), using a ts/tv ratio of 2,183 (LnL = −7832.871), and a +G, parameter = 0.243. In the ML tree obtained (Figure 3), the three Fasciola species cluster together with high support values (100/99 in ML/NJ), appearing F. nyanzae as a sister species to the clade of F. hepatica. The two Fascioloides species (Fs. jacksoni and Fs. magna), also cluster together, and the big clade, including both genera, Fasciola and Fascioloides, show maximum support (100/100 in ML/NJ). Although clustering in the same clade, a clear phylogenetic distance is observed between the two Fasciolopsis buski sequences appearing basal to the two groupings of Fasciola and Fascioloides. The topology obtained with the NJ algorithm using the number of differences method (figure not shown) was identical to that shown by the ML tree.
Figure 3.
Phylogenetic tree of species of Fasciola, Fascioloides, and Fasciolopsis based on maximum-likelihood (ML) estimates and reconstructed on the concatenated sequences of the nuclear rDNA 18S gene, ITS-1, 5.8S gene and ITS-2 (lnL = −7,832.871). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The tree was rooted using the sequence of Paramphistomum cervi (KJ459937) as outgroup. Supports for nodes a/b using MEGA X: a, bootstrap (1,000 replicates) with ML parameters (HKY + G); b, bootstrap (1,000 replicates) with NJ and number of differences method for distances.
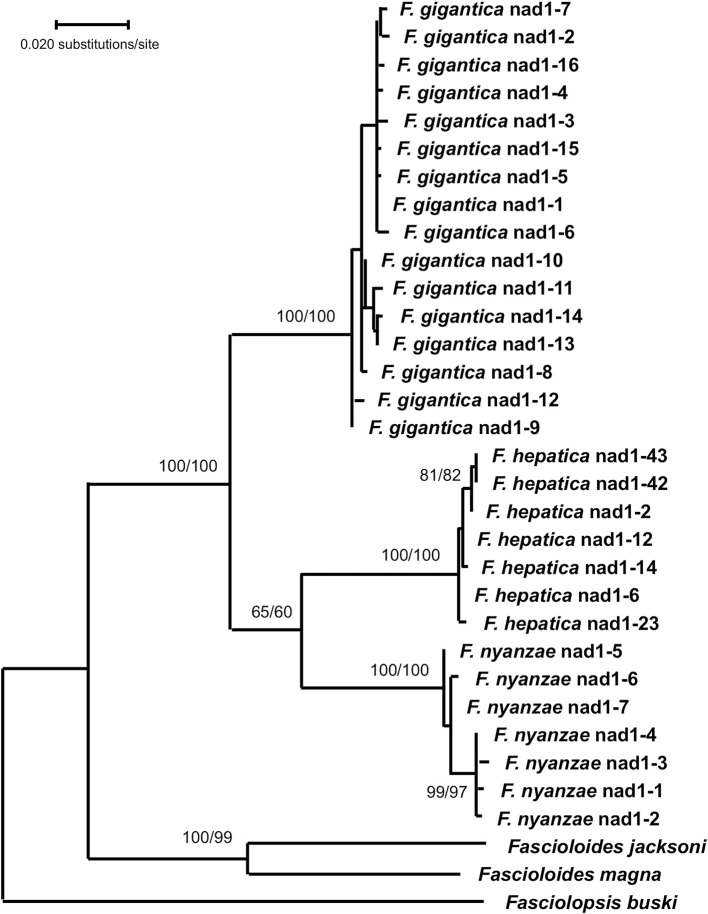
For the mtDNA cox1 data matrix, the best fitting model was Tamura 3-parameter (T92 + G) with discrete Gamma distribution (T92 + G). The resulting ML tree (log likelihood = −5,733.3311) was inferred with +G, parameter = 0.4351 and the rate variation model allowed for some sites to be evolutionarily invariable (Figure 4). In the ML tree, the seven haplotypes of F. nyanzae cluster together in a monophyletic clade, closely related and well-supported (96/96 in ML/NJ) with F. gigantica clade. Thus, all haplotypes from different African countries grouped together in a monophyletic clade. The haplotypes of F. hepatica from Europe and Latin American countries appear in a basal position regarding F. nyanzae and F. hepatica, all included in a single clade of the genus Fasciola with a high support (99/83 in ML/NJ). The two Fascioloides species (Fs. jacksoni and Fs. magna) clustered together in an independent clade also with a very high support (93/96 in ML/NJ). With the NJ algorithm, and using the Tamura 3-parameter method, identical topology of the ML tree was obtained (tree not shown).
Figure 4.
Phylogenetic tree of species of Fasciola and Fascioloides based on maximum-likelihood (ML) estimates and reconstructed on mtDNA cox1 sequences (LnL = −5,733.3311). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The tree was rooted using the sequence of Fasciolopsis buski (MF287794) as outgroup. Supports for nodes a/b using MEGA X: a, bootstrap (1,000 replicates) with ML parameters (T92 + G); b, bootstrap (1,000 replicates) with NJ and Tamura 3-p method.
In the phylogenetic analysis carried out with the mtDNA nad1 data matrix, the ML model best fitting this data-set was T92 + G. The tree with the highest log likelihood (−3,559.05) was constructed with the rate variation among sites modeled with a gamma distribution (+G, parameter = 0.3091) (Figure 5). The ML topology obtained shows a monophyletic clade for the three Fasciola species with the highest supports (100/100 in ML/NJ). The two species of Fascioloides clustered together in another clade also with a very high support (100/99 in ML/NJ), thus manifesting its clear independence from that of the Fasciola clade. With the NJ algorithm, and using the Tamura 3-p method, identical topology of the ML tree was obtained (tree not shown).
Figure 5.
Phylogenetic tree of species of Fasciola and Fascioloides based on maximum-likelihood (ML) estimates and reconstructed on mtDNA nad1 sequences (LnL = −3559.05). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The tree was rooted using the sequence of Fasciolopsis buski (MF287793) as outgroup. Supports for nodes a/b using MEGA X: a, bootstrap (1,000 replicates) with ML parameters (T92 + G); b, bootstrap (1,000 replicates) with NJ and Tamura 3-p method.
All topologies obtained with ribosomal and mitochondrial markers were solid, well-supported and congruent, corroborating the phylogenetic relationship of F. nyanzae within the genus Fasciola. Moreover, the three phylogenies agree in the inclusion of the species Fs. jacksoni and Fs. magna in the genus Fascioloides and its distant relationship with the genus Fasciola.
Discussion
Fasciola nyanzae phenotypic and genotypic comparison
Fasciola nyanzae is a parasite specific of the bile ducts of the hippopotamus. No adult flukes were produced in experimental infections of calves, goats and rabbits (29), and it was never reported in cattle around areas where it is frequent in Uganda (47). Parasitological studies on hippopotamuses have reported the parasite several times in Uganda (48, 49), Kenya (50), South Africa (51), the southern part of Chad (52), Gabon (53), and in an artificial lake in Zimbabwe (30, 54). These findings indicate a very wide distribution throughout Africa, which overlaps with that of its specific freshwater lymnaeid snail vector species R. natalensis (29) (Figure 2).
The species markedly differs morphologically from F. hepatica and F. gigantica by its length, the great breadth of the shoulders, the large cone, and the long tapering body, almost entirely occupied by vitelline follicles, with the distance between the posterior testis and the posterior extremity of the body being longer than the remaining anterior part of the body (Figure 1). The maximum length of the adult stage infecting the hippopotamus has been noted to reach 69 mm (55), and up to 91 mm (29), but reached even 136.6 mm in the longest specimen from the South African reserves. The adult maximum length is only around 55 mm in F. gigantica and only 31 mm in F. hepatica (25, 56). However, egg size overlap: F. nyanzae eggs measure 150–190/70–110 μm in the uterus (50), 132–150/79–97 μm in bile, and 131–160/75–88 μm in specimens mounted in Canada balsam (29); F. gigantica eggs are of 130.3–182.8/74.0–123.6 μm in animals and 150.9–182.2/85.1–106.2 μm in humans; and F. hepatica eggs range 73.8–156.8/58.1–98.1 μm in animals and 100.6–162.2/65.9–104.6 μm in humans (5, 57).
In the 18S rDNA gene, the few mutations between the three species of Fasciola and the two species of Fascioloides agree with the conserved characteristics of this gene (26). The sequence of ITS-1 of F. nyanzae from hippopotamus in the South African reserves is identical to that obtained in snails and in one hippopotamus in an artificial lake of Zimbabwe (30).
The numerous mutations in the short cox1 fragments obtained from F. nyanzae in Zimbabwe (30, 54) represent a surprising intraspecific variability for specimens from only one place and are difficult to interpret in materials from an artificial lake. Therefore, these numerous mutations should be individually verified in the electropherograms and the geographical origin of the snail and hippopotamus hosts introduced in the artificial lake should be analyzed regarding the sequences in question.
In the phylogenetic tree obtained with the concatenated rDNA sequences, the Asian species Fasciolopsis buski proves to be phylogenetically very distant from the species of Fasciola and Fascioloides. This result agrees with the completely different characteristics of Fps. buski when compared to the other fasciolids, including the morpho-anatomy of its adult stage with non-ramified intestinal caeca (vs. ramified in the other fasciolids), the intestinal microhabitat of infection (vs. a liver microhabitat), the omnivore definitive host (vs. herbivores), and planorbids as snail vectors (vs. lymnaeids) (45). This indicates that the genus Fasciolopsis has no close evolutionary relationship with the genera Fasciola and Fascioloides and may consequently be ruled out when analyzing the direct derivation of the paleobiogeographical origins of F. hepatica and F. gigantica. The species Fps. buski has been therefore used as outgroup for the two phylogenetic reconstructions by means of the cox1 and nad1 genes to obtain better trees than with the more distant P. cervi.
Both sequence analyses and phylogenetic trees evidence (i) the close relationships of F. nyanzae with F. gigantica and F. hepatica and consequently its belonging to the genus Fasciola, and (ii) the distant relationships of Fasciola with the two species Fs. jacksoni and Fs. magna included in the genus Fascioloides.
Divergence estimates
Trematodes do not fossilize, but their evolution may be estimated considering the fossil record of their specific, co-evolving definitive hosts and snail vectors (58). Hippopotamid and ruminant hosts of the three Fasciola species have evolved between the Early Miocene and the Pleistocene (59–61), whereas lymnaeid snails are pronouncedly older, follow a slower evolution, and their oldest fossil record dates back to the Jurassic time period (62). When dating estimates of relative divergences in such past periods, the conserved nuclear rDNA 18S gene becomes appropriate because (i) it allowed for a reliable alignment of the sequences across the taxa analyzed, (ii) it follows a low mutation rate in both fasciolids and lymnaeids (63), (iii) it is a non-coding gene, (iv) general traits and length of the phases of the life cycle of Fasciola (64) and Fascioloides (22) are very similar and timely overlap, indicating a similar number of generations along time and therefore a similar evolutionary speed rate, and (v) snail vectors of Fasciola and Fascioloides belong to the same or similar species. The last two factors exclude the need to apply different evolutionary rates for the lineages of Fasciola and Fascioloides. As it has been highlighted, when a constancy rate is not rejected, the use of a strict clock is preferred because it has the fewest parameters and generally leads to the most precise divergence estimates (65). Moreover, given that only three species of Fasciola and two species of Fascioloides are used for the analysis, this molecular clock meets the premise of a simple model which captures the essential biological features of the data. Recent studies have shown how increasing the data set size lead to increasing estimate errors, even in data sets of moderate size (65). Additionally, the relative divergence estimates obtained for fasciolids proved to fit very well with the fossil records of the oldest representatives of the definitive host groups and their subsequent intercontinental migrations, as well as with available knowledge on paleobiogeographical changes of the landscape, mainly concerning interconnecting bridges between continents.
When applying the conventional molecular clock rate of 1.8 × 10−10 substitutions per site per year (1.8% per 100 my) for the evolution of the 18S gene obtained in many studies of different organism groups which count on well-dated fossil remains (66), the time elapsed between the appearance of F. gigantica and F. hepatica by derivation from F. nyanzae is estimated to be around 8.41 my. If the oldest hippopotamus fossil record in Asia, of 6.1–5.9 mya (67–69), is considered, the appearance of F. gigantica should be rolled back to at least around 14.5 mya. This fits the upper limit marked by the appearance of the first primitive hippopotamids, dated back to 21.0–15.9 mya in the Early Miocene in Africa (56, 60). When analyzing the different phases of the split of the bovids along the Miocene (61), these data suggest that the capture phenomenon from early hippopotamids to ancient bovids grazing in flooded plains and lowlands close to freshwater collections inhabited by hippopotamuses most probably occurred during the third split phase corresponding to the radiation of mainly Reduncinae and when several ancestors of Bovinae and Alcelaphinae were also present in Africa, at about 13.5 mya. These evolutionary estimates indicate a time frame more recent than the 19 mya divergence obtained according to sequence analysis of cathepsin L-like cysteine proteases (70). However, subsequent assessments have demonstrated that metabolic rates are not appropriate for the calibration of the molecular clock (71).
The 18S molecular clock estimates the divergence between Fs. magna of cervids and caribou in North America and Fs. jacksoni of the Asian elephant at about 19.77 mya. This fits well with the faunal exchange between the Palearctic and Nearctic regions during which ancestral odocoileine cervids entered America from Siberia via the Bering Strait in the late Miocene/early Pliocene (72).
The divergence between F. nyanzae and Fs. jacksoni is of 22.44 mya and that between F. nyanzae and Fs. magna of 25.48 mya, whereas that between F. gigantica and Fs. jacksoni dates back to 22.44 mya and that between F. gigantica and Fs. magna to 28.31 mya. This indicates that the divergence between ancestral Fasciola and Fascioloides rolls back to the Late Oligocene-Earliest Miocene, pronouncedly before the appearance of F. gigantica and F. hepatica. Therefore, a direct derivation of the current species of Fasciola from Fascioloides does not appear to be supported.
Additional molecular clock divergence estimations based on the rDNA 18S gene were analyzed on lymnaeids (52) to verify that the snail vectors of the three Fasciola species were there in the past to assure the respective fasciolid life cycles. The molecular clock furnishes a divergence of 61.1 my between G. truncatula and R. auricularia, of 39.0 my between R. natalensis and R. auricularia, and of 0.33 my between R. natalensis of Burkina Faso and R. natalensis of Egypt. These dating estimates agree with the very old history and slow evolutionary rates of Lymnaeidae, whose oldest fossil record dates back to around 208–146 mya (62). The divergence between Galba and Radix appears to be very old, although it occurred pronouncedly later then the Gondwana division leading to the separation of the landmasses of South America and Africa by the Atlantic Ocean according to a very long process lasting between around 130 and 85 mya (73), which explains the absence of Radix in the New World. The time of divergence between R. natalensis and R. auricularia indicates that the former, the only Radix species present in the whole sub-Saharan region, became isolated in Africa from all other Palaearctic Radix species even before the Oligocene. This is sufficient time to expand passively transported by watercourses and ancient animals and cover wide zones of this continent. Concerning the time needed for R. auricularia to expand from the sub-Saharan region up to Egypt, is should be considered that the Nile river flow is indiscernibly slow and almost inexistent and that consequently it is more logical to think that this snail species should have been passively transported by animals, hippopotamuses included, following the river basin northward.
Past and present distribution of hippopotamuses
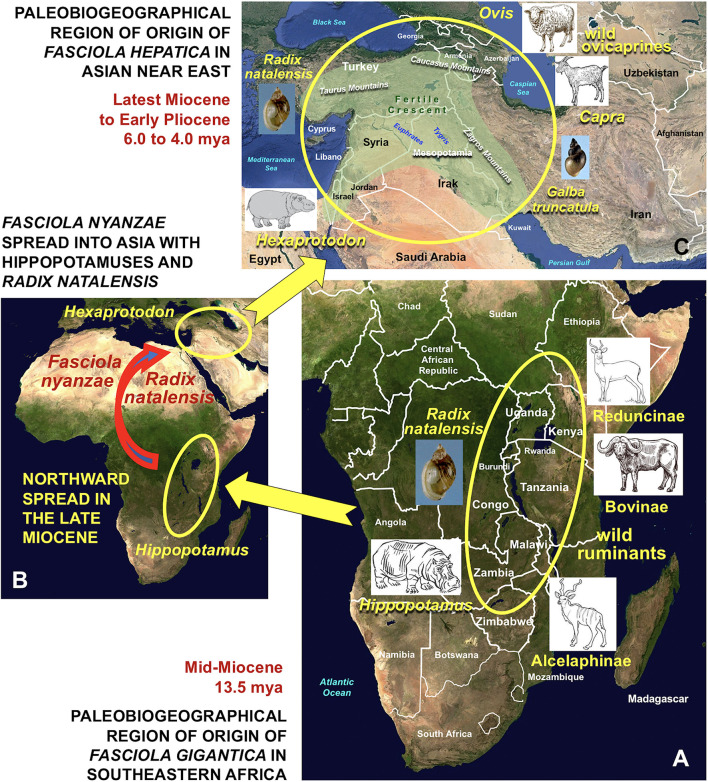
The African Hipppotamus amphibious is at present showing a very wide geographical distribution covering whole Africa southward from the Sahara desert (74), according to a markedly fragmented presence of populations linked to the peaceful type of water collections it inhabits. Hippopotamuses are amphibious herbivorous mammals which leave water to graze vegetation in the surroundings over night (75), a behavior which facilitates its infection by fasciolid flukes. The oldest fossils of hippopotamuses are dated back to the Early Miocene and indicate that these mammals reached their major diversity in sub-Saharan Africa afterwards during the Plio-Pleistocene (76). Molecular assessments suggest that their original area might have been in eastern Africa, around present-day Uganda, southwestern Kenya, eastern Congo, Tanzania, Zambia, and northern Zimbabwe (77) (Figure 2). Paleontological data indicate that hippopotamuses spread northwards to North Africa and Europe during the late Middle or Late Pleistocene (76).
This northward spread led hippopotamuses to expand throughout southern Asia giving rise to the Asian fossil lineage included in the genus Hexaprotodon (Figure 6), which eastwards further reached the Indian subcontinent and up to South East Asia (76). The oldest fossils of Asian Hexaprotodon were found in the Siwalik hills of the present-day state of Himachal Pradesh in northeastern India, neighboring northern Pakistan and were dated back to the latest Miocene to Early Pliocene, between 6.1 and 5.9 mya (67–69). The passage from northeastern Africa into westernmost Asia probably occurred along the Levantine corridor. This is suggested by hippopotamid fossil remains found in present-day Georgia around 1.40 mya, in the Israel-Libano area around 1.40–0.70 mya, and in Syria around 0.30–0.25 mya (76). The present H. amphibius was still present in Palestina in the Neolithic and even until very recently in the northern Nile river basin (78).
Figure 6.
Paleobiogeographical regions of origin of Fasciola gigantica and F. hepatica by direct evolutionary derivation from F. nyanzae infecting past hippopotamuses. (A) Origin of F. gigantica in southeastern Africa by definitive host capture phenomenon from African past hippopotamuses to wild ruminants, mainly ancestors of the bovid subfamilies Reduncinae, Bovinae and Alcelaphinae by keeping the same snail vector Radix natalensis in the Mid-Miocene, around 13.5 mya; (B) northward spread of F. nyanzae with African hippopotamuses and R. natalensis and introduction from Africa into the Asian Near East with Hexaprotodon in the Late Miocene, around 6.1 and 5.9 mya; (C) origin of F. hepatica in Near East Asia by definitive host capture phenomenon from Hexaprotodon to wild species of ovicaprines of the genera Ovis and Capra and vector capture event from R. natalensis to Galba truncatula in the Latest Miocene to Early Pliocene, around 6.0–4.0 mya. Countries and geographic names are only those noted in the text. Geographic background from composed satellite maps of orthographic projections by NASA (public domain) via Wikimedia Commons. Schema S. Mas-Coma.
In the African hippopotamuses, the orbits are elevated above the skull, and their nostrils and ears can be closed while diving, whereas eyes are non- or less-elevated in the Asian Hexaprotodon indicating a less amphibious, i.e., more terrestrial, life style (68).
Past and present distribution of Radix natalensis
The snail R. natalensis is the only autochthonous lymnaeid in Africa southward of the Sahara desert. In this very wide region, other lymnaeids reported are all introduced species, whether recently (Pseudosuccinae columella, R. rubiginosa, R. plicata, G. truncatula) or evolutionarily recently (G. mweruensis in isolated refugia).
This freshwater snail shows a wide geographical distribution throughout the aforementioned sub-Saharan Africa, which covers that of the present populations of hippopotamus (79) (Figure 2). Three additional aspects of the geographical distribution of R. natalensis should be highlighted:
(A) The findings of fossilized shells in Late Pleistocene-Holocene sites throughout the Sahara desert (80) (Figure 2) speak about the past presence of watercourses along this desert (81), and indicate that this snail was accompanying hippopotamuses during their northward spread during the late Middle or Late Pleistocene (76).
(B) The distribution of R. natalensis includes a northward arm extending along the Nile river basin up to the Nile Delta at the Mediterranean shore (79) (Figure 2).
(C) Additional populations have been found in Jordan found in Azraq oasis and the Jordan Valley (82), and in Kishda, Palestina (83) (Figure 2), which indicate that this snail also reached western Asia, most probably accompanying the aforementioned past spread of hippopotamuses along the Levantine corridor (76).
Radix natalensis is a freshwater snail which inhabits steady water habitats and swampy areas with slow-flowing streams and canals usually with abundant aquatic vegetation in warm lowlands. The adaptability of R. natalensis to varying conditions of temperature (84, 85) and dryness (86, 87) underlie the persistence of this radicine lymnaeid over time once it colonizes and establishes in a freshwater habitat. In this way, this snail finds its ideal habitat in the same type of water habitats as the hippopotamus. This explains the sharing of the same sites by hippopotamuses and R. natalensis. This coexistence allows the life cycle of F. nyanzae to be established and maintained, as indeed F. nyanzae was experimentally demonstrated to be transmitted by R. natalensis (29), and the larval stages of this fasciolid have also been found in naturally infected specimens of this radicine lymnaeid (30, 54).
Worth mentioning is the fact that R. natalensis also acts as the specific transmitter of F. gigantica in Africa. To date, populations of R. natalensis from different countries have always demonstrated to be experimentally susceptible to the infection and able to transmit the many geographical strains of African F. gigantica assayed (88–96). Moreover, in the Levantine corridor in westernmost Asia, where it was referred to as Lymnaea auricularia in initial reports (97), the R. natalensis population of the Azraq oasis in Jordan (Figure 2) also showed to transmit the local F. gigantica (98, 99).
Finally, it should be highlighted that R. natalensis from Egypt has experimentally demonstrated to also transmit F. hepatica from Europe (100). This means that R. natalensis is able to transmit the three species of Fasciola and might be interpreted as this lymnaeid species being a kind of “mother vector” for these trematodes.
Fasciola gigantica in the wild fauna in eastern Africa
Ungulates are hoofed mammals known to present their higher diversity in the savannah biome of sub-Saharan Africa. The analysis of population genetic data showed a concordance of the phylogeographic structuring of many ungulate species (77). The cradle of ungulate diversity appears in East Africa (101), where major biogeographic lineages of these herbivore mammals meet and overlap, including the hippopotamus.
Reports on F. gigantica infection in ungulates are numerous in several countries of eastern Africa (48, 102–105), including many different mammal species (Table 8). Five aspects should be highlighted:
Table 8.
Species of the wild fauna of Africa in which infection by Fasciola gigantica has been reported.
| Chad | Rwanda |
| *Alcelaphus buselaphus (Alcelaphinae) - Hartebeest *Kobus defassa (Reduncinae) - Waterbuck *Kobus kob (Reduncinae) – Kob Sudan *Damaliscus korrigum (Alcelaphinae) - Topi *Syncerus caffer (Bovinae) - buffalo Central African Republic *Syncerus caffer (Bovinae) - buffalo Congo *Alcelaphus buselaphus (Alcelaphinae) - Hartebeest *Kobus defassa (Reduncinae) - Waterbuck *Kobus kob (Reduncinae) - Kob Uganda *Alcelaphus buselaphus (Alcelaphinae) - Hartebeest *Kobus kob (Reduncinae) - Kob *Syncerus caffer (Bovinae) - buffalo *Taurotragus oryx (Bovinae) - Eland Kenya *Aepyceros melampus (Aepycerotinae) - Impala *Connochaetes taurinus (Alcelaphinae) - Wildebeest *Giraffa camelopardalis (Giraffidae) - Giraffe *Taurotragus oryx (Bovinae) - Eland |
Sylvicapra grimmia (Cephalophinae) - Common or Gray duiker Tanzania *Connochaetes taurinus (Alcelaphinae) - Wildebeest *Damaliscus korrigum (Alcelaphinae) - Topi *Syncerus caffer (Bovinae) - buffalo *Taurotragus oryx (Bovinae) - Eland Zambia * [Hippopotamus amphibius (Hippopotamdae) - hippopotamus]** Kobus leche (Reduncinae) - Kafue lechwe Kobus vardoni (Reduncinae) - Puku Zimbabwe *Aepyceros melampus (Aepycerotinae) - Impala *Connochaetes taurinus (Alcelaphinae) - Wildebeest Damaliscus lunatus (Alcelaphinae) - Tsessebe or sassaby *Giraffa camelopardalis (Giraffidae) - Giraffe *Hippotragus niger (Hippotraginae) - Sable antelope Redunca arundinum (Reduncinae) - Reedbuck Sylvicapra grimmia (Cephalophinae) - Gray duiker *Taurotragus oryx (Bovinae) - Eland Tragelaphus scriptus (Bovinae) - Bushbuck *Tragelaphus strepsiceros (Bovinae) - Kudu Swaziland *Aepyceros melampus (Aepycerotinae) - Impala |
Subfamilies are noted in the case of Bovidae.
*Ungulate species whose major biogeographic pattern inferred from phylogeographic data is found in eastern Africa coinciding with that of the hippopotamus.
**Finding in hippopotamus in Zambia initially reported as Fasciola sp. and later ascribed to F. gigantica by Round in 1968 (101) still remains a question mark.
(A) Although in most cases the coexistence with domestic ruminants indicates livestock to be the infection source for the sylvatic mammals due to the sharing of the same grazing areas, there have been reports in which it was specified that the infected wild animals had never been in contact with domestic ruminants, such as in cases of the Kob Kobus kob, the Hartebeest Alcelaphus buselaphus, and the buffalo Syncerus caffer in Uganda, or cases in which such a contact was noted to have been rare, as in cases of the buffalo in Uganda and Tanzania and the Eland Taurotragus oryx in Tanzania (48).
(B) Many of the wild ungulate species show major biogeographic patterns inferred from phylogeographic data which are found in eastern Africa coinciding with that of the hippopotamus (77).
(C) Several of the wild species found infected appear repeatedly in the different countries, such as the Hartebeest, Waterbuck, Kob, Topi, buffalo, Eland, Impala, and Wildebeest (Table 8), indicating a grazing behavior in appropriate areas for the infection by F. gigantica, i.e., where freshwater collections inhabited by R. natalensis are present.
(D) Very high prevalences were reported from the African buffalo (58%), kob K. kob (47%), and Hartebeest A. buselaphus (47%) in Uganda (103). as well as in the Kafue lechwe (52.5%) in Zambia and the Kudu (12.5%) in Zimbabwe (105).
(E) Infection intensities may reach high burdens in several of these wild animals, as it has been observed in the African buffalo (up to 66 flukes per host individual), kob K. kob (17 flukes) and Jackson's hartebeest A. buselaphus (21 flukes) in Uganda (103), and also in giraffe (41 flukes) from Kenya and Topi (30 flukes) in Tanzania (48).
It is evident that all necessary features for a host capture event of the large F. nyanzae from the big amphibious hippopotamus to smaller terrestrial ungulates were converging in eastern Africa in the past. This host capture phenomenon should have been the origin of speciation by adaptation to the new hosts giving rise to the smaller F. gigantica (Figure 6).
Fasciola hepatica in the wild fauna scenario of the Near East
The westernmost Asian region of the Near East is unique in presenting the geographic and timely convergence of the following mammal hosts, lymnaeid snail vectors, fasciolid flukes, appropriate altitude-varying milieu, and climate characteristics:
- F. gigantica and F. hepatica;
- small-sized ruminants as sheep and goats;
- large sized ruminants as taurine cattle, zebu and buffalo;
- equines as donkeys, mules and horses, also including onager donkeys in the past;
- omnivores as wild swine;
- Old World camelids as Bactrian camel and dromedary;
- R. natalensis, R. auricularia and G. truncatula;
- past existence of hippopotamuses presumably harboring F. nyanzae;
- warm flat lowlands and cool mountainous highlands.
It was in this region where the so-called Fertile Crescent emerged and the domestication of wild herbivores began around 12,000–10,000 years ago (106–109), marking the dawn of the Neolithic period. The Fertile Crescent was a fertile zone for agricultural use covering from present-day Syria, Lebanon, Jordan, Israel and Palestine in the so-called Levant and part of Anatolia (present-day Turkey) including the Taurus mountains, in the West, along the Tigris and Euphrates river basins in the Mesopotamia flatlands and northward reaching the Caucasus mountains, and up to the Zagros mountains in present-day Irak and Iran, in the East.
Archeological excavations in a cave in the western-central Zagros allowed for the finding of remains of bovids, cervids, suids and equines dating back to around 75,000 years BP (110). This indicates that potential hosts for fasciolids were present in this region in the past. Comparative data on infectivity, life span, egg shedding and immunity indicate that sheep and goats are the ideal hosts of F. hepatica (1, 53). Ovicaprines are mid-sized ruminants which should therefore be considered among the original hosts of the smaller F. hepatica. A definitive host capture phenomenon from a F. nyanzae lineage in the less amphibious southwest Asian Hexaprotodon hippopotamuses toward a more terrestrial ovicaprine/G. truncatula binome may be envisaged to have occurred in Near East Asia around the latest Miocene - earliest Pliocene period.
Species of the two genera Capra and Ovis diversified around 6.8–5.1 mya (61). The wild ancestor of the domesticated goat Capra hircus was the bezoar Capra aegagrus, which was widely distributed from the Taurus mountains and eastern Anatolia in the West, down to the Iranian plateau in the East, through the Zagros mountainous chain along present Irak and Iran (111, 112). The wild ancestor of the domesticated sheep Ovis aries was the mouflon Ovis gmelini (= urial sheep O. orientalis) (113, 114), whose distribution covered the region from the southern Levant through southeastern Anatolia and northern Syria, to the high Zagros mountain pastures and lowland plains of Irak and Iran (108, 109). Indeed, the biome inhabited by wild sheep are steep mountainous woods near tree lines, and they are known to migrate to lower altitudes in winter. Today, the wild bezoar goat Capra aegagrus and the smaller wild urial sheep O. orientalis are only found in Afghanistan and northern Pakistan further south. Close species as the Siberian ibex Capra ibex siberica and the wild argali sheep Ovis ammon show present ranges extending into the Hindu Kush.
The possibility for wild boars Sus scrofa to have played an intermediate evolutionary role in this capture phenomenon may not be ruled out. Indeed, Suidae are evolutionarily related to hippopotamids (78) and the wild boar is an animal living both in warmer lowlands and cooler highlands (115). Sus scrofa was present in the Asian Near East (116–118). Moreover, the wild boar in western Europe (119) and feral black pigs in Sicily (120) are known to be infected by F. hepatica. Additionally, the pig has recently demonstrated to play an important role of reservoir for F. hepatica, at least in South America (121).
In this Asian Near East region, initial domestication steps with goats and sheep occurred around 11,000 years BP, and pigs and cattle domestication in that area started around 10,500 and 10,000 years BP, respectively. These dating estimations were 9,000–8,500 years BP in Mesopotamia, 10,000–8,500 years BP in Anatolia, and 9,600-8,500 years BP in the Levant (108).
Three archeo-parasitological findings of F. hepatica eggs are worth mentioning.
(A) Five F. hepatica eggs were found in sepultures on the island of Cyprus, dated back to 8,000–7,000 years BC (122, 123). These findings are the oldest archeological record of F. hepatica, and indicate that it was infecting domesticated animals at the earliest beginning of the domestication process. Human migration from the mainland into Cyprus took place between 11,100 and 10,600 years BP and goat, sheep, cattle, and pig were introduced into the island a bit later, around 10,400 years BP (124). Fasciola hepatica was able to maintain its life cycle thanks to the past presence of G. truncatula on the island, dated back around 7,200–6,800 years BC (125), which indicates that this lymnaeid vector was present in the neighboring mainland of Near East Asia in the past. This lymnaeid continues to be present in Cyprus until today (126).
(B) Four F. hepatica eggs were detected in a latrine of an archeological site in Israel dated back 200–300 years AC (122). This finding speaks about the past presence of this fasciolid in the Levantine corridor.
(C) The first archeological report of F. hepatica eggs in whole Asia concerns the finding in coprolites from a donkey in an archaeological site at an altitude of 1,663 m a.s.l. in the Zanjan province, northwestern Iran, dated back to the Sassanid period, 224–651 AD. This donkey was the Persian onager Equus hemionus onager, the Asiatic Wild-ass native of Iran. This finding indicates that this fasciolid had already spread through the Zagros mountains eastward from the Fertile Crescent at that time. Onagers were present in that region and neighborhood of the Fertile Crescent long before the beginning of the domestication of the livestock reservoirs of Fasciola (127).
In the Iranian province of Guilan, neighboring Zanjan province, surveys on goats, sheep, cattle and buffaloes showed a relationship of liver fluke infection with altitude, with F. gigantica transmitted by R. auricularia in the lowlands besides the Caspian Sea and F. hepatica transmitted by G. truncatula in the highlands of the Talesh mountains (128). In this zone, vertical transhumance pastoralism was practiced since 6,500 years ago (129). Vertical altitudinal transhumance and horizontal transhumance nomadic pastoralism should have significantly contributed to the dispersal of both F. hepatica and F. gigantica in domestic ruminants and their respective lymnaeid vector species during the Neolithic. Additional to the ruminants, equines and camelids should also have contributed in such an expansion process in the Near East, as suggested by their infection in present times (130, 131).
From this paleobiogeographical Asian Near East region of origin (Figure 6), F. hepatica was able to spread westward into Europe, eastward into Asia and southward into Africa by means of human-guided movements of domestic ruminants, equines and camelids along the post-domestication period, and subsequently to the Americas and Oceania after transoceanic ship transport in the last 500 years (1).
Early introduction of Fasciola gigantica into Asia
Fasciola gigantica spread not only throughout its African continent of origin, but also Asia. Consequently, the question is posed on how, where and when was this fasciolid able to migrate out of Africa and reach the Asian continent, establish, and later expand up to the Far East, South East Asia and Pacific islands (1).
Paleontological data indicate that the eastern Mediterranean Levantine Corridor was the major route out for the African fauna into Eurasia, playing as a kind of bottleneck passage. This corridor is assumed to have been a northward extension of the East African Rift, with ecological and climatic characteristics similar to those of eastern Africa along the Plio-Pleistocene period. A high number of large mammal fossils of African origin have been reported from this Levantine Corridor (132). This corridor appears to have been the same route used by hippopotamuses and R. natalensis snails in their way out from the African continent.
This Levantine Corridor was, however, not always open. A land bridge connected Africa with Eurasia already in the Miocene according to the dramatic lowering of the level of the Mediterranean Sea. The fossil bovid record provides evidence for greater biological continuity between Africa and Eurasia in the late Miocene and earliest Pliocene than is found later in time (133). Although a great number of African faunal elements entered Eurasia around 18–17 mya (134), this was to early as F. gigantica was not yet originated. This bridge disappeared with the developing of the Red Sea and the flooding of the Mediterranean at the Miocene end, around 5 mya. During the subsequent Pliocene period of 5.33–2.58 mya, the barrier was maintained and the southern Levant was increasingly isolated. The quasi-isolation of this region finished with another regression of the Mediterranean toward the early Pleistocene (135). Other important faunal changes also occurred (i) at the beginning of the Pliocene around 5.5 mya, (ii) at the beginning of the Pleistocene around 2.0–1.8 mya, and (iii) at the early-middle Pleistocene boundary around 1.0 mya (134).
Among the African large mammals which used this corridor, there was a big assemblage of African bovids. The wide range of African animals indicates that there was no ecological selection on animals entering southwestern Asia (136). Three groups of these African bovids should be highlighted because of (i) being grazers, (ii) their ecological specializations linked to the presence of freshwater habitats, and (iii) showing the highest infection rates by F. gigantica in the wild fauna in Africa (Table 8), several even in the absence of or very rare contact with livestock. Indeed, bovid tribes and genera manifest evident relationships with given biome types which have been interpreted to underlie their original differentiation occurred along Miocene and Pliocene times (137):
(A) Reduncines are animals with a strong preference for flood plains and other areas close to water.
(B) Bovines of the cattle group live in floodplain grasslands and valleys within a short distance of water.
(C) Alcelaphalines live concentrated on greenbelts near water points in the dry season and disperse into dry savanna during the rainy season, preferring an open landscape with short grass, and they can also tolerate tree cover.
Among reduncines, the Plio-Pleistocene members of Asia belong to the extinct genus Sivacobus with evident relationships with the African crown kob cradle. Sivacobus is considered to have arisen from the same African ancestor of the kobs, which emigrated from Africa around 3.5 mya (138). Another fossilized species Dorcadoxa porrecticornis from the Siwaliks dated back 9.5–6.5 mya has also been considered to belong to the Kobus group because of its African relative D. aff. porrecticornis, which would mean an earlier entry of this African bovid group into Eurasia around the Late Miocene.
Among the bovines, the large buffalo Bos (Pelorovis) oldowayensis is considered the most significant element. The large buffalo Pelorovis sensu stricto is known to have been a common species in the African savannas in the Late Pliocene and Early Pleistocene, and has even been suggested to be the direct ancestor of the Middle Pleistocene Eurasian species Bos primigenius (132).
Among alcelaphalines, members of the genus Damalops, already extinct in Africa, have been found in Tajikistan dated to the Late Pliocene and were also present in the Siwaliks around 2.5 mya. Indeterminate alcelaphines were also recorded in southwestern Asia from the early Pleistocene onwards (136).
Hippotragines (in the Early to Middle Pleistocene) (136) and giraffids (in the Late Miocene and again later in the early Pleistocene) (134) were also included among the African immigrants, and may have also participated in the dispersal of F. gigantica once already in Asia, as they have been verified to be infected by this fasciolid in Africa (Table 8), although only secondarily given their less appropriate ecological preferences regarding the fluke life cycle.
It should, moreover, be considered that, additional to the aforementioned introduction of R. natalensis, polymorphic fossilized shells of R. auricularia indicate that this lymnaeid was widely present in the plain of Lower Mesopotamia during the Quaternary (139). Radix auricularia is currently found as a pronouncedly aquatic snail in permanent, still or slow flowing, and temporary collections of water of this plain (140), as well as in central Irak (141). Consequently, lymnaeid vectors able to assure the life cycle of F. gigantica were there.
Summing up, F. gigantica had many opportunities to emigrate from Africa into southwestern Asia and the possibility of more than an old introduction wave may even not be ruled out. All this indicates that F. gigantica should have been there transmitted by African-introduced R. natalensis and Asian-autochthonous R. auricularia in the lowlands of the Levant and Near East Asia at the time of the beginning of the animal domestication in the Fertile Crescent around 12,000–10,000 years ago (107–109). This should have allowed F. gigantica to benefit from herding and pastoralism for its spread throughout the Near East during the Neolithic and for its subsequent further expansion eastward into Asia by means of human-guided movements of the domestic herbivores including ruminants, equines and Old World camelids (1).
Conclusions
The sequences of the nuclear rDNA and mtDNA markers show evidence of a close relationship of F. gigantica and F. hepatica of worldwide distribution with F. nyanzae of sub-Saharan Africa. The direct evolutionary derivation of the two former from the latter species agrees with the strict specificity of F. nyanzae in hippopotamus (older origin leading to long-term coevolution specialization) and the pronouncedly low specificity of the two other fasciolids in herbivore mammals, including a few omnivores like pigs and humans (modern origin only enabling a short-term coevolution insufficient for specialization). Fasciolid adult stage size show a length cline, from the biggest F. nyanzae to the smallest F. hepatica, manifesting a parallelism with the size of their main definitive hosts, which agrees with the recognized influence of the biliary canal inner space on fasciolid fluke size. Molecular clock dating estimates define a chronological scenario which adequately fits the fossil picture and paleobiogeographical events in Africa and Asian Near East.
The multidisciplinary analysis for F. gigantica suggests southeastern Africa as its paleobiogeographical region of origin, with a core zone around present-day Uganda and neighboring countries. Its chronological period for appearance should have been the mid-Miocene, around 13.5 mya. The original host capture phenomenon leading to speciation should have involved from the primitive hippopotamuses to bovid ancestors of Reduncinae, Bovinae, and Alcelaphinae, characterized by grazing preferences in flooded plains and proximity to freshwater collections. African Radix natalensis was the original lymnaeid snail vector species, and warm lowlands surrounding peaceful freshwater collections inhabited by hippopotamuses should have been the original biome (Figure 6).
For F. hepatica, all data suggest the Near East of Asia to have been its paleobiogeographical region of origin, from the Levant and Mesopotamia to the mountains of the Taurus, the Caucasus and Zagros. This fasciolid should have appeared after a host capture phenomenon from primitive, less amphibious Hexaprotodon hippopotamuses to southwest Asian ovicaprines (Bovidae: Caprinae) in the period of the latest Miocene to Early Pliocene, around 6.0–4.0 mya and perhaps shortly afterwards. The wild bezoar Capra aegagrus and the wild mouflon Ovis gmelini, direct ancestors of the domesticated goat Capra hircus and the domesticated sheep Ovis aries, respectively, should be considered the most probable original definitive hosts of F. hepatica, although a possible evolutionary intermediate passage through the wild boar Sus scrofa should not be ruled out. The lymnaeid Galba truncatula may be considered its original snail vector species after another freshwater snail capture phenomenon from radicine lymnaeids. The original ecological habitat should have included cooler areas and mountainous foothills, where the aforementioned hippopotamuses and mid-sized mammals overlapped (Figure 6).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.
Ethics statement
The animal study was reviewed and approved by the Evaluation of Projects concerning Animal Research at University of Valencia (Organo Habilitado para la Evaluación de Proyectos de Experimentación Animal de la Universidad de Valencia) (A1263 915389140), strictly following the institution's guidelines based on Directive 2010/63/EU. Permission for animal research was additionally obtained from the Servicio de Sanidad y Bienestar Animal, Dirección General de Producción Agraria y Ganadería, Consellería de Presidencia y Agricultura, Pesca, Alimentación y Agua, Generalitat Valenciana, Valencia, Spain (No. 2015/VSC/PEA/00001 tipo 2). Animal ethics guidelines regarding animal care were strictly adhered to Parasitological studies on hippopotamuses of Private Nature Reserve 1, Hoedspruit, the Private Nature Reserve 3 (New camp dam), and the Private Nature Reserve 2 of the Mpumalanga Province of South Africa were performed within an official permission agreement (Agreement document No. 13582). Written informed consent was obtained from the owners for the participation of their animals in this study. In Sri Lanka, liver flukes were obtained by dissection of naturally deceased wild elephants in fieldwork collaboration with members of the Minneriya National Park and the Department of Veterinary Pathobiology, Faculty of Veterinary Medicine and Animal Science, University of Peradeniya, under the auspices of the national Department of Wildlife Conservation (DWC). No ethics approval nor consent was needed for snail collections done on public land.
Author contributions
MDB, AH, PA, WJL-P, MAV, and SM-C participated in the investigation. MDB, AH, WJL-P, and SM-C obtained the materials. AH and WJL-P coordinated local activities in South Africa. MDB, PA, MAV, and SM-C applied the methods. SM-C designed the study. MDB, WJL-P, and SM-C obtained and administered the project fundings. SM-C and MDB analyzed the results and wrote the manuscript. All authors read and approved the final manuscript.
Funding
Field work on hippopotamus carcasses in South Africa funded by Project No. 101054 of the South African Research Chairs Initiative (SARChI) of the Department of Science and Innovation and the National Research Foundation of South Africa. Field work for the obtaining of samples in other countries and laboratory research in Spain funded by CIBER de Enfermedades Infecciosas Project No. CB21/13/00056, CIBER - Consorcio Centro de Investigación Biomédica en Red (CB 2021), Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación, Madrid, Spain, and Unión Europea—NextGenerationEU; by the Red de Investigación de Centros de Enfermedades Tropicales—RICET (Project No. RD16/0027/0023 of the PN de I+D+I, ISCIII-Subdirección General de Redes y Centros de Investigación Cooperativa RETICS), Ministry of Health and Consumption, Madrid; by Projects Nos. 2016/099 and 2021/004 of the PROMETEO Program, Programa of Ayudas para Grupos de Investigación de Excelencia, Generalitat Valenciana, Valencia, Spain; and by Health Research Project No. PI16/00520, Subprograma Estatal de Generación de Conocimiento de la Acción Estratégica en Salud (AES) y Fondos FEDER, Plan Estatal de Investigación Científica y Técnica y de Innovación, ISCIII-MINECO, Madrid, Spain. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Studies of this article have been performed within the framework of the Worldwide Initiative of WHO against Human Fascioliasis (WHO Headquarters, Geneva, Switzerland). Special thanks are given to Dr. François Roux, Mpumalanga Tourism and Parks Agency, Lydenburg, Mpumalanga Province, South Africa, for his coordination of fieldwork, as well as the management teams of the Private Game Reserves for providing carcasses of hippopotamuses for research and allowing us to investigate health and parasitic disease in the populations of these animals. We also thank people who helped during the fieldwork, especially Pieter Olivier, Kris Bal, Hendrik Hattingh, Jean Roux, Moses M. Matla, Willem J. Smit, Katlego D. Kunutu, Makubu P. Mokgawa, and Nokukhanya Makuwa. The authors also acknowledge the facilities provided and the collaboration received from the organisms, institutions and centers, as well as all the numerous local collaborators of the other countries helping in the obtaining of samples. Collection of liver fluke materials has been possible thanks to the help of many scientists in field work during missions to different endemic countries: Joseph Jourdane (France) in Niger, Jérôme Boissier (France) in Cameroon, Roger Fons (France) in Corsica, Marc Desquesnes (France) in Burkina Faso, Filippo Curtale (Italy) in Egypt, Carlos Feliu (Spain) in Senegal, Alberto Martínez-Ortí (Spain) in Angola, Petr Horak (Czech Republic) in Eastern Europe, René Angles in Bolivia, José R. Espinoza and Pedro Ortiz in Peru, Roberto Mera y Sierra in Argentina, Gabriel Trueba in Ecuador, José Lino Zumaquero-Ríos in Mexico, Valeria Gayo in Uruguay, Luisa Carolina González in Venezuela, Gholamreza Mowlavi in Iran, Nino Iashvili in Georgia, Linda Chougar in Algeria, Jayanthe Rajapakse, and Vijitha Perera in Sri Lanka. Technical support for DNA sequencing was provided by the Servicio Central de Secuenciación para la Investigación Experimental (SCSIE) of the Universidad de Valencia (Dr. A. Martínez).
References
- 1.Mas-Coma S, Valero MA, Bargues MD. Fasciola, lymnaeids and human fascioliasis, with a global overview on disease transmission, epidemiology, evolutionary genetics, molecular epidemiology and control. Adv Parasitol. (2009) 69:41–146. 10.1016/S0065-308X(09)69002-3 [DOI] [PubMed] [Google Scholar]
- 2.Mas-Coma S, Cafrune MM, Funatsu IR, Mangold AJ, Angles R, Buchon P, et al. Fascioliasis in llama, Lama glama, in Andean endemic areas: experimental transmission capacity by the high altitude snail vector Galba truncatula and epidemiological analysis of its reservoir role. Animals. (2021) 11:2693. 10.3390/ani11092693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayward AD, Skuce PJ, McNeilly TN. The influence of liver fluke infection on production in sheep and cattle: a meta-analysis. Int J Parasitol. (2021) 51:913–24. 10.1016/j.ijpara.2021.02.006 [DOI] [PubMed] [Google Scholar]
- 4.Mas-Coma S, Bargues M, Valero MA. Fascioliasis and other plant-borne trematode zoonoses. Int J Parasitol. (2005) 35:1255–78. 10.1016/j.ijpara.2005.07.010 [DOI] [PubMed] [Google Scholar]
- 5.Mas-Coma S, Bargues MD, Valero MA. Diagnosis of human fascioliasis by stool and blood techniques: Update for the present global scenario. Parasitology. (2014) 141(Special Issue):1918–46. 10.1017/S0031182014000869 [DOI] [PubMed] [Google Scholar]
- 6.Chen MG, Mott KE. Progress in assessment of morbidity due to Fasciola hepatica infection: a review of recent literature. Trop Dis Bull. (1990) 87:R1–38. [Google Scholar]
- 7.Mas-Coma S, Agramunt VH, Valero MA. Neurological and ocular fascioliasis in humans. Adv Parasitol. (2014) 84:27–149. 10.1016/B978-0-12-800099-1.00002-8 [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez-Miguel J, Valero MA, Reguera-Gomez M, Mas-Bargues C, Bargues MD, Simon-Martin F, et al. Numerous Fasciola plasminogen-binding proteins may underlie blood-brain barrier leakage and explain neurological disorder complexity and heterogeneity in the acute and chronic phases of human fascioliasis. Parasitology. (2019) 146:284–98. 10.1017/S0031182018001464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lalor R, Cwiklinski K, Calvani NED, Dorey A, Hamon S, Corrales JL, et al. Pathogenicity and virulence of the liver flukes Fasciola hepatica and Fasciola gigantica that cause the zoonosis fasciolosis. Virulence. (2021) 12:2839–67. 10.1080/21505594.2021.1996520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rondelaud D, Dreyfuss G, Vignoles P. Clinical and biological abnormalities in patients after fasciolosis treatment. Méd Mal Infect. (2006) 36:466–8. 10.1016/j.medmal.2006.07.018 [DOI] [PubMed] [Google Scholar]
- 11.Dalton JP, Robinson MW, Mulcahy G, O'Neill SM, Donnelly S. Immunomodulatory molecules of Fasciola hepatica: candidates for both vaccine and immunotherapeutic development. Vet Parasitol. (2013) 195:272–85. 10.1016/j.vetpar.2013.04.008 [DOI] [PubMed] [Google Scholar]
- 12.Girones N, Valero MA, Garcia-Bodelon MA, Chico-Calero MI, Punzon C, Fresno M, et al. Immune supression in advanced chronic fascioliasis: an experimental study in a rat model. J Infect Dis. (2007) 195:1504–12. 10.1086/514822 [DOI] [PubMed] [Google Scholar]
- 13.Aldridge A, O'Neill SM. Fasciola hepatica tegumental antigens induce anergic like T cells via dendritic cells in a mannose receptor dependent manner. Eur J Immunol. (2016) 46:1180–92. 10.1002/eji.201545905 [DOI] [PubMed] [Google Scholar]
- 14.Esteban JG, Flores A, Aguirre C, Strauss W, Angles R, Mas-Coma S. Presence of very high prevalence and intensity of infection with Fasciola hepatica among Aymara children from the Northern Bolivian Altiplano. Acta Trop. (1997) 66:1–14. 10.1016/S0001-706X(97)00669-4 [DOI] [PubMed] [Google Scholar]
- 15.Esteban JG, Flores A, Angles R, Strauss W, Aguirre C, Mas-Coma S, et al. A population-based coprological study of human fascioliasis in a hyperendemic area of the Bolivian Altiplano. Trop Med Int Health. (1997) 2:695–9. 10.1046/j.1365-3156.1997.d01-356.x [DOI] [PubMed] [Google Scholar]
- 16.Angles R, Buchon P, Valero MA, Bargues MD, Mas-Coma S. One Health action against human fascioliasis in the Bolivian Altiplano: food, water, housing, behavioural traditions, social aspects, and livestock management linked to disease transmission and infection sources. Int J Environ Res Publ Health. (2022) 19:1120. 10.3390/ijerph19031120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.World Health Organization . Ending the neglect to attain the Sustainable Development Goals. A Road Map for Neglected Tropical Diseases 2021–2030. Geneva: Department of Control of Neglected Tropical Diseases, World Health Organization, WHO Headquarters; (2020). p. 1–47. [Google Scholar]
- 18.World Health Organization. Ending the Neglect to Attain the Sustainable Development Goals. One Health Companion Document to the Neglected Tropical Diseases Road Map 2021–2030. Draft for Public Consultation (updated 19 October 2021). Geneva: Department of Control of Neglected Tropical Diseases, World Health Organization, WHO Headquarters (2020). p. 1–23. Available online at: https://cdn.who.int/media/docs/default-source/ntds/rabies/online-public-consultation.-one-health-companion-document/draft-for-public-consultation-one-health-companion-document-for-ntd-road-map.pdf (accessed December 15, 2021).
- 19.Choi YJ, Fontenla S, Fischer PU, Le TH, Costabile A, Blair D, et al. Adaptive radiation of the flukes of the family Fasciolidae inferred from genome-wide comparisons of key species. Mol Biol Evol. (2020) 37:84–99. 10.1093/molbev/msz204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mas-Coma S, Bargues MD, Valero MA. Human fascioliasis infection sources, their diversity, incidence factors, analytical methods and prevention measures. Parasitology. (2018) 145(13, Special Issue):1665–99. 10.1017/S0031182018000914 [DOI] [PubMed] [Google Scholar]
- 21.Rajapakse RPVJ, Pham KLT, Karunathilake KJK, Lawton SP, Le TH. Characterization and phylogenetic properties of the complete mitochondrial genome of Fascioloides jacksoni (syn. Fasciola jacksoni) support the suggested intergeneric change from Fasciola to Fascioloides (Platyhelminthes: Trematoda: Plagiorchiida). Infect Genet Evol. (2020) 82:104281. 10.1016/j.meegid.2020.104281 [DOI] [PubMed] [Google Scholar]
- 22.Malcicka M. Life history and biology of Fascioloides magna (Trematoda) and its native and exotic hosts. Ecol Evol. (2015) 5:1381–97. 10.1002/ece3.1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bazsalovicsová E, Králová-Hromadová I, Štefka J, Minárik G, Bokorová S, Pybus M. Genetic interrelationships of North American populations of giant liver fluke Fascioloides magna. Parasit Vectors. (2015) 8:288. 10.1186/s13071-015-0895-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bargues MD, Gayo V, Sanchis J, Artigas P, Khoubbane M, BirrieI S, Mas-Coma S. DNA multigene characterization of Fasciola hepatica and Lymnaea neotropica and its fascioliasis transmission capacity in Uruguay, with historical correlation, human report review and infection risk analysis. PLoS Negl Trop Dis. (2017) 11:e0005352. 10.1371/journal.pntd.0005352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bargues MD, Valero MA, Trueba GA, Fornasini M, Villavicencio AF, Guaman R, et al. DNA multi-marker genotyping and CIAS morphometric phenotyping of Fasciola gigantica-sized flukes from Ecuador, with an analysis of the Radix absence in the New World and the evolutionary lymnaeid snail vector filter. Animals. (2021) 11:2495. 10.3390/ani11092495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mas-Coma S, Bargues MD. Populations, hybrids and the systematic concepts of species and subspecies in Chagas disease triatomine vectors inferred from nuclear ribosomal and mitochondrial DNA. Acta Trop. (2009) 110:112–36. 10.1016/j.actatropica.2008.10.013 [DOI] [PubMed] [Google Scholar]
- 27.Chougar L, Mas-Coma S, Artigas P, Harhoura K, Aissi M, Agramunt VH, et al. Genetically “pure” Fasciola gigantica discovered in Algeria: DNA multimarker characterization, trans-Saharan introduction from a Sahel origin and spreading risk into northwestern Maghreb countries. Transb Emerg Dis. (2020) 67:2190–205. 10.1111/tbed.13572 [DOI] [PubMed] [Google Scholar]
- 28.Valero MA, Mas-Coma S. Comparative infectivity of Fasciola hepatica metacercariae from isolates of the main and secondary reservoir animal host species in the Bolivian Altiplano high human endemic region. Folia Parasitol. (2000) 47:17–22. 10.14411/fp.2000.004 [DOI] [PubMed] [Google Scholar]
- 29.Dinnik JA, Dinnik NN. On the morphology and life history of Fasciola nyanzae (Leiper, 1910) from the hippopotamus. J Helminthol. (1961) RT Leiper supplement:53-62. 10.1017/S0022149X00017570 [DOI] [PubMed] [Google Scholar]
- 30.Schols R. Carolus H, Hammoud C, Muzarabani KC, Barson M, Huyse T. Invasive snails, parasite spillback, and potential parasite spillover drive parasitic diseases of Hippopotamus amphibius in artificial lakes of Zimbabwe. BMC Biol. (2021) 19:160. 10.1186/s12915-021-01093-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bildfell RJ. Whipps CW, Gillin CM, Kent ML. DNA-based Identification of a hepatic trematode in an Elk Calf. J Wildlife Dis. (2007) 43:762–9. 10.7589/0090-3558-43.4.762 [DOI] [PubMed] [Google Scholar]
- 32.Lee JK, Graham Rosser T, Cooley J. Pulmonary embolization of immature Fascioloides magna causing fatal hemothorax confirmed by molecular technique in a heifer in the United States. J Vet Diagn Inv. (2016) 28:584–8. 10.1177/1040638716660129 [DOI] [PubMed] [Google Scholar]
- 33.Kralova-Hromadova I, Špakulova M, Horačkova E, Turčekova L, Novobilský A, Beck R, et al. Sequence analysis of ribosomal and mitochondrial genes of the giant liver fluke Fascioloides magna (Trematoda: Fasciolidae): intraspecific variation and differentiation from Fasciola hepatica. J Parasitol. (2008) 94:58–67. 10.1645/GE-1324.1 [DOI] [PubMed] [Google Scholar]
- 34.Blair D, Barker SC. Affinities of the Gyliauchenidae: utility of the 18S rRNA gene for phylogenetic inference in the Digenea (Platyhelminthes). Int J Parasitol. (1993) 23:527–32. 10.1016/0020-7519(93)90042-W [DOI] [PubMed] [Google Scholar]
- 35.Le TH, Pham KLT, Doan HTT, Le TKX, Nguyen KT, Lawton SP. Description and phylogenetic analyses of ribosomal transcription units from species of Fasciolidae (Platyhelminthes: Digenea). J Helminthol. (2020) 94:e136. 10.1017/S0022149X20000164 [DOI] [PubMed] [Google Scholar]
- 36.Dao TTH, Nguyen TTG, Gabriël S, Bui KL, Dorny P, Le TH. Updated molecular phylogenetic data for Opisthorchis spp. (Trematoda: Opisthorchioidea) from ducks in Vietnam. Parasit Vectors. (2017) 10:575. 10.1186/s13071-017-2514-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mas-Coma S, Funatsu IR, Bargues MD. Fasciola hepatica and lymnaeid snails occurring at very high altitude in South America. Parasitology. (2001) 123:S115–27. 10.1017/S0031182001008034 [DOI] [PubMed] [Google Scholar]
- 38.Bargues MD, Artigas P, Mera y Sierra RL, Pointier JP, Mas-Coma S. Characterisation of Lymnaea cubensis, L. viatrix and L. neotropica n. sp., the main vectors of Fasciola hepatica in Latin America, by analysis of their ribosomal and mitochondrial DNA. Ann Trop Med Parasitol. (2007) 101:621–41. 10.1179/136485907X229077 [DOI] [PubMed] [Google Scholar]
- 39.Bargues MD, Mangold AJ, Muñoz-Antoli C, Pointier JP, Mas-Coma S. SSU rDNA characterization of lymnaeid snails transmitting human fascioliasis in South and Central America. J Parasitol. (1997) 83:1086–92. 10.2307/3284367 [DOI] [PubMed] [Google Scholar]
- 40.Kumar S, Stecher G, Li M, Knyaz C, Tamura K, MEGA X. Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 35:1547–9. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glez-Peña D, Gómez-Blanco D, Reboiro-Jato M, Fernandez-Riverola F, Posada D. ALTER: program-oriented conversion of DNA and protein alignments. Nucl Acids Res. (2010) 38:W14–8. 10.1093/nar/gkq321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bargues MD, Mas-Coma S. Reviewing lymnaeid vectors of fascioliasis by ribosomal DNA sequence analyses. J Helminthol. (2005) 79:257–67. 10.1079/JOH2005297 [DOI] [PubMed] [Google Scholar]
- 43.Lin CP, Danforth BN. How do insect nuclear and mitochondrial gene substitution patterns differ? Insights from Bayesian analyses of combined datasets. Mol Phyl Evol. (2004) 30:686–702. 10.1016/S1055-7903(03)00241-0 [DOI] [PubMed] [Google Scholar]
- 44.Ballard JWO, Rand DM. The population biology of mitochondrial DNA and its phylogenetic implications. Ann Rev Ecol Evol Syst. (2005) 36:621–42. 10.1146/annurev.ecolsys.36.091704.175513 [DOI] [Google Scholar]
- 45.Mas-Coma S, Valero MA, Bargues MD. Fasciolopsis buski and fasciolopsiasis. In: Smithers G. editor. Encyclopedia of Food Safety, Second Edition. Reference Collection in Food Sciences - via ScienceDirect, Elsevier Major Reference Works. Netherlands: Elsevier; (2022). [Google Scholar]
- 46.Ma J, Sun MM, He JJ, Liu GH Ai L, Chen MX, Zhu XQ. Fasciolopsis buski (Digenea: Fasciolidae) from China and India may represent distinct taxa based on mitochondrial and nuclear ribosomal DNA sequences. Parasit Vectors. (2017) 10:101. 10.1186/s13071-017-2039-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coyle TJ. The Epidemiology of Fasciola gigantica in Cattle in Uganda Protectorate (thesis). Royal College of Veterinary Surgeons, United Kingdom (1961). [Google Scholar]
- 48.Hammond JA. Infections with Fasciola spp. in wildlife in Africa. Trop Anim Health Prod. (1972) 4:1–13. 10.1007/BF02357089 [DOI] [PubMed] [Google Scholar]
- 49.Cowan DEF, Thurlbeck WM, Law RM. Some diseases of the hippopotamus in Uganda. Pathol Vet. (1967) 4:553–67. 10.1177/030098586700400604 [DOI] [PubMed] [Google Scholar]
- 50.Jackson HG. revision on the genus Fasciola with particular reference to F. gigantica (Cobbold) and F nyanzi (Leiper). Parasitology. (1921) 13:48–56. 10.1017/S0031182000012294 [DOI] [Google Scholar]
- 51.McCully RM, Van Niekerk J, Kruger SP. Observations on the pathology of bilharziasis and other parasitic infestations of Hippopotamus amphibius Linnaeus, 1758, from the Kruger National Park. Onderstepoort J Vet Res. (1967) 34:563–617. [PubMed] [Google Scholar]
- 52.Graber M. Helminthes parasites de certains animaux domestiques et sauvages du Tchad. Bull Epiz Dis Africa. (1969) 17:403–28. [PubMed] [Google Scholar]
- 53.Rietmann S, Walzer C. Parasitological examination of common hippopotamus (Hippopotamus amphibius) faeces in the Gamba Complex of Protected Area in Gabon. Wien Tierärztl Monatsschr. (2014) 101:66–73. [Google Scholar]
- 54.Carolus H, Muzarabani KC, Hammoud C, Schols R, Volckaert FA, Barson M, et al. A cascade of biological invasions and parasite spillback in man-made Lake Kariba. Sci Total Environ. (2019) 659:1283–92. 10.1016/j.scitotenv.2018.12.307 [DOI] [PubMed] [Google Scholar]
- 55.Leiper RT. The entozoa of the hippopotamus. Proc Zool Soc London. (1910) 233–51. 10.1111/j.1096-3642.1910.tb01894.x [DOI] [Google Scholar]
- 56.Periago MV, Valero MA, Panova M, Mas-Coma S. Phenotypic comparison of allopatric populations of Fasciola hepatica and Fasciola gigantica from European and African bovines using a computer image analysis system (CIAS). Parasitol Res. (2006) 99:368–78. 10.1007/s00436-006-0174-3 [DOI] [PubMed] [Google Scholar]
- 57.Valero MA, Perez-Crespo I, Periago MV, Khoubbane M, Mas-Coma S. Fluke egg characteristics for the diagnosis of human and animal fascioliasis by Fasciola hepatica and F. gigantica Acta Trop. (2009) 111:150–9. 10.1016/j.actatropica.2009.04.005 [DOI] [PubMed] [Google Scholar]
- 58.Mas-Coma S. (1992). Parasitología ecológica y evolutiva: aspectos aplicados. In: Sanmartín Durán ML. editor. Avances en Parasitología - Protozoología, No. 1. Santiago de Compostela: Servicio de Publicacións e Intercambio Científico, Universidade de Santiago; (1992). p. 43–65. [Google Scholar]
- 59.Orliac M, Boisserie JR, Lihoreau F, MacLatchy L. Early Miocene hippopotamids (Cetartiodactyla) constrain the phylogenetic and spatiotemporal settings of hippopotamid origin. Proc Natl Acad Sci USA. (2010) 107:11871–6. 10.1073/pnas.1001373107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lihoreau F, Boisserie JR, Manthi FK, Ducrocq S. Hippos stem from the longest sequence of terrestrial cetartiodactyl evolution in Africa. Nature Commun. (2015) 6:6264. 10.1038/ncomms7264 [DOI] [PubMed] [Google Scholar]
- 61.Hernandez Fernandez M, Vrba ES. A complete estimate of the phylogenetic relationships in Ruminantia: a dated species-level supertree of the extant ruminants. Biol Rev. (2005) 80:269–302. 10.1017/S1464793104006670 [DOI] [PubMed] [Google Scholar]
- 62.Remigio EA, Blair D. Molecular systematics of the freshwater snail family Lymnaeidae (Pulmonata: Basommatophora) utilising mitochondrial ribosomal DNA sequences. J Moll Stud. (1987) 63:173–85. 10.1093/mollus/63.2.173 [DOI] [Google Scholar]
- 63.Bargues MD, Mas-Coma S. Phylogenetic analysis of lymnaeid snails based on 18S rDNA sequences. Mol Biol Evol. (1997) 14:569–77. 10.1093/oxfordjournals.molbev.a025794 [DOI] [PubMed] [Google Scholar]
- 64.Mas-Coma S, Buchon P, Funatsu IR, Angles R, Artigas P, Valero MA, et al. Sheep and cattle reservoirs in the highest human fascioliasis hyperendemic area: experimental transmission capacity, field epidemiology, and control within a One Health initiative in Bolivia. Front Vet Sci. (2020) 7:583204. 10.3389/fvets.2020.583204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ho SYW, Duchene S. Molecular-clock methods for estimating evolutionary rates and timescales. Mol Ecol. (2014) 23:5947–65. 10.1111/mec.12953 [DOI] [PubMed] [Google Scholar]
- 66.Bargues MD, Marcilla A, Ramsey J, Dujardin JP, Schofield CJ, Mas-Coma S. Nuclear rDNA-based molecular clock of the evolution of Triatominae (Hemiptera: Reduviidae), vectors of Chagas disease. Mem Inst Oswaldo Cruz. (2000) 95:567–73. 10.1590/S0074-02762000000400020 [DOI] [PubMed] [Google Scholar]
- 67.Barry JC, Morgan ME, Flynn LJ, Pilbeam D, Behrensmeyer AK, Raza SM, et al. Faunal and environmental change in the late Miocene Siwaliks of northern Pakistan. Paleobiol Mem Memoir. (2002) 3:1–71. 10.1666/0094-8373(2002)28[1:FAECIT]2.0.CO;2 [DOI] [Google Scholar]
- 68.Boisserie JR. The phylogeny and taxonomy of Hippopotamidae (Mammalia: Artiodactyla): a review based on morphology and cladistic analysis. Zool J Linn Soc. (2005) 143:1–26. 10.1111/j.1096-3642.2004.00138.x [DOI] [Google Scholar]
- 69.Htike T, Takai M. Reevaluation of the phylogeny and taxonomy of the Asian fossil Hippopotamuses. Univ Res J. (2016) 8:171–97. [Google Scholar]
- 70.Irving JA, Spithill TW, Pike RN, Whisstock JC, Smooker PM. The evolution of enzyme specificity in Fasciola spp. J Mol Evol. (2003) 57:1–15. 10.1007/s00239-002-2434-x [DOI] [PubMed] [Google Scholar]
- 71.Lanfear R. Thomas JA, Welch JJ, Bromham L, Metabolic rate does not calibrate the molecular clock. Proc Natl Acad Sci USA. (2007) 104:15388–93. 10.1073/pnas.0703359104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heckeberg NS. The systematics of the Cervidae: a total evidence approach. PeerJ. (2020) 8:e8114. 10.7717/peerj.8114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seton M, Müller RD, Zahirovic S, Gaina C, Torsvik T, Shephard G, et al. Global continental and ocean basin reconstructions since 200 Ma. Earth Sci Rev. (2012) 113:212–70. 10.1016/j.earscirev.2012.03.002 [DOI] [Google Scholar]
- 74.Lewison R, Pluháček J. Hippopotamus amphibius. The International Union for Conservation of Nature and Natural Resources (IUCN) Red List of Threatened Species (2017). [Google Scholar]
- 75.Nowak RM. Walker's Mammals of the World. 6th ed. Baltimore, MD: The Johns Hopkins Press; (1999) 1-1936. [Google Scholar]
- 76.van der Made J, Sahnouni M, Boulaghraief K. Hippopotamus gorgops from El Kherba (Algeria) and the context of its biogeography. In: Sahnouni M, Semaw S, Garaizar JR. editors. Proceedings of the II Meeting of African History (Burgos 15-16 April 2015), Centro Nacional de Investigación sobre la Evolución Humana, Consorcio CENIEH. Burgos: Gráficas Eujos. (2017). p. 137–68. [Google Scholar]
- 77.Lorenzen ED, Heller R, Siegismund HR. Comparative phylogeography of African savannah ungulates. Mol Ecol. (2012) 21:3656–70. 10.1111/j.1365-294X.2012.05650.x [DOI] [PubMed] [Google Scholar]
- 78.Thenius E. Grundzüge der Faunen- und Verbreitungsgeschichte der Säugetiere. Stuttgart: Gustav Fischer Verlag; (1980). p. 1–375. [Google Scholar]
- 79.Brown D. Freshwater snails of Africa and their medical importance. First Edition. London: Taylor & Francis Ltd. (1994). p. i-x + 1-609. [Google Scholar]
- 80.Van Damme D. The Freshwater Mollusca of Northern Africa - Distribution, Biogeography and Paleoecology. Dordrecht: Dr. W. Junk Publishers; (1984). p. 1–164. [Google Scholar]
- 81.Drake NA, Blench RM, Armitage SJ, Bristow CS, White KH. Ancient watercourses and biogeography of the Sahara explain the peopling of the desert. Proc Natl Acad Sci USA. (2011) 108:458–62. 10.1073/pnas.1012231108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Amr ZS, Abu Baker M. Freshwater snails of Jordan. Denisia. (2004) 14:221–7. [Google Scholar]
- 83.Handal EN, Amr Z, Qumsiyeh MB. Some records of freshwater snail from the Occupied Palestinian territories. Jordan J Nat Hist. (2015) 2:23–9. [Google Scholar]
- 84.Dinnik JA, Dinnik NN. Effect of the seasonal variations of temperature on the development of Fasciola gigantica in the snail host in Kenya Highlands. Bull Epizootiol Dis Africa. (1963) 11:197–207. [PubMed] [Google Scholar]
- 85.Dinnik JA, Dinnik NN. The influence of temperature on the succession of redial and cercarial generations of Fasciola gigantica in a snail host. Parasitology. (1964) 54:59–65. 10.1017/S0031182000074321 [DOI] [PubMed] [Google Scholar]
- 86.Cridland CC. Resistance of Bulinus (Physopsis) globosus, Bulinus (Ph.) africanus, Biomphalaria pfeifferi and Lymnaea natalensis to experimental desiccation. Bull WHO. (1967) 36:507–13. [PMC free article] [PubMed] [Google Scholar]
- 87.Vassiliadès G. Capacité de résistance à la sécheresse de la limnée (Lymnaea natalensis), mollusque hôte intermédiaire de Fasciola gigantica, au Sénégal. Rev Elev Méd Vét Pays Trop. (1978) 31:57–62. 10.19182/remvt.8174 [DOI] [PubMed] [Google Scholar]
- 88.Porter A. The life history of the African sheep and cattle fluke Fasciola gigantica. South African J Sci. (1920) 17:126–30. [Google Scholar]
- 89.Fain A. Lymnaea (Radix) natalensis undussumae von Martens, transmetteur naturel de Fasciola gigantica Cobbold au Congo Beige. Reproduction expérimentale du cycle évolutif de cette douve. Ann Soc Beige Méd Trop. (1951) 31:531–9. [PubMed] [Google Scholar]
- 90.Kendall SB, Parfitt JW. Life-history of Fasciola gigantica Cobbold 1856. Nature. (1953) 171:1164–5. 10.1038/1711164a0 [DOI] [PubMed] [Google Scholar]
- 91.Dinnik JA, Dinnik NN. Observations on the succession of redial generations of Fasciola gigantica Cobbold in a snail host. Zeitschr Tropenm Parasitol. (1956) 7:397–419. [PubMed] [Google Scholar]
- 92.Kendall SB. Relationships between the species of Fasciola and the molluscan hosts. Adv Parasitol. (1965) 3:59–98. 10.1016/S0065-308X(08)60363-2 [DOI] [PubMed] [Google Scholar]
- 93.Rao CMP. On the comparative susceptibility of Lymnaea natalensis (Kraus) and L. rufescens (Gray) to infection with Fasciola gigantica (West African Strain) and the tissue responses in the snails. J Helminthol. (1966) 40:131–40. 10.1017/S0022149X00034143 [DOI] [PubMed] [Google Scholar]
- 94.Ogambo-Ongoma AH, Goodman JD. Fasciola gigantica Cobbold 1856 in the snail. J Parasitol. (1975) 62:33–8. 10.2307/3279037 [DOI] [PubMed] [Google Scholar]
- 95.Ibidapo CA. Cercarial infection of the lymnaeid snail Lymnaea natalensis (Krauss) in Ibadan, Nigeria. J Exp Appl Biol. (1989) 1:1–13. [Google Scholar]
- 96.Khoubbane M, Bargues MD, Mas-Coma S. Fascioliasis epidemiology in Egypt: lymnaeid susceptibility and their role in the transmission. In: Mas-Coma S, Bargues MD, Esteban JG, Valero MA. editors. Multidisciplinarity for Parasites, Vectors and Parasitic Diseases. II K.E. Mott Symposium on schistosomiasis and distomatoses. IX European Multicolloquium of Parasitology (EMOP 9) (Valencia, Spain, 18-23 July 2004), Valencia: J. Aguilar S. L., Programme and Abstracts (2004) Abstract No. 486:290. (2004). [Google Scholar]
- 97.Burch J, Amr Z. Freshwater snail fauna of Jordan. Walkerana. (1990) 11:27–58. [Google Scholar]
- 98.Saliba EK, Lutfy RG, Ismail NS. Fascioliasis in Azraq Oasis, Jordan. 2. Infection of Lymnaea auricularia snails with Fasciola gigantica Cobb., 1885 cercariae. Acta Parasitol Polon. (1978) 25:341–5. [Google Scholar]
- 99.Saliba EK, Othman MI. Further studies on natural infection of Lymnaea auricularia with larval trematodes and its susceptibility to infection with Fasciola gigantica Cobb. from Azraq, Jordan. Acta Parasitol Polon. (1980) 27:285–92. [Google Scholar]
- 100.Dar Y, Djuikwo Teukeng FF, Vignoles P, Dreyfuss G, Rondelaud D. Radix natalensis (Gastropoda: Lymnaeidae), a potential intermediate host of Fasciola hepatica in Egypt. Parasite. (2010) 17:251–6. 10.1051/parasite/2010173251 [DOI] [PubMed] [Google Scholar]
- 101.du Toit JT, Cumming DHM. Functional significance of ungulate diversity in African savannas and the ecological implications of the spread of pastoralism. Biodiv Conserv. (1999) 8:1643–61. 10.1023/A:1008959721342 [DOI] [Google Scholar]
- 102.Round MC. Check list of the helminth parasites of African mammals of the Orders Carnivora, Tubulidentata, Proboscidea, Hyracoidea, Artiodactyla and Perissodactyla, Vol. 38. St. Albans, Farnham Royal, Bucks: Commonwealth Agricultural Bureaux Commonwealth Bureau of Helminthology, Technical Communication. (1968) 38:i-vi + 1-252. [Google Scholar]
- 103.Bindernagel JA. Liver fluke Fasciola gigantica in African buffalo and antelopes in Uganda, East Africa. J Wildlife Dis. (1972) 8:315–7. 10.7589/0090-3558-8.4.315 [DOI] [PubMed] [Google Scholar]
- 104.Losos GJ. Infectious Tropical Diseases of Domestic Animals. Essex: Longman Scientific & Technical; (1986) p. i-xiii + 1-938. [Google Scholar]
- 105.Malatji MP, Pfukenyi DM, Mukaratirwa S. Fasciola species and their vertebrate and snail intermediate hosts in East and Southern Africa: a review. J Helminthol. (2019) 94:e63. 10.1017/S0022149X19000531 [DOI] [PubMed] [Google Scholar]
- 106.Mac Hugh DE, Bradley DG. Livestock genetic origins: goats buck the trend. Proc Natl Acad Sci USA. (2001) 98:5382–4. 10.1073/pnas.111163198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vigne JD. Les débuts néolithiques de l'élévage des bovidés et de l'exploitation laitière dans l'Ancien Monde. In: Poulain JP. Director, L'homme, le mangeur et l'animal Qui nourrit l'autre? Les Cahiers de l'OCHA, Observatoire CNIEL des Habitudes Alimentaires, Paris. (2007). p. 45–57. [Google Scholar]
- 108.Zeder MA. Domestication and early agriculture in the Mediterranean Basin: origins, diffusion, and impact. Proc Natl Acad Sci USA. (2008) 105:11597–604. 10.1073/pnas.0801317105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vigne JD, Balasse M, Gourichon L, Helmer D, Lesur J, Mashkour M, et al. Etat des connaissances archéologiques sur les débuts de l'élévage du mouton dans l'Ancien Monde. Ethozootechnie. (2011) 91:11–9. [Google Scholar]
- 110.Mashkour M, Monchot H, Trinkaus E, Reyss JL, Biglari F, Bailon S, et al. Carnivores and their prey in the Wezmeh Cave (Kermanshah, Iran): a late Pleistocene refuge in the Zagros. Int J Osteoarchaeol. (2009) 19:678–94. 10.1002/oa.997 [DOI] [Google Scholar]
- 111.Zeder MA, Hesse B. The initial domestication of goats (Capra hircus) in the Zagros mountains 10,000 years ago. Science. (2000) 287:2254–7. 10.1126/science.287.5461.2254 [DOI] [PubMed] [Google Scholar]
- 112.Naderi S, Rezaei HR, Pompanon F, Blum MGB, Negrini R, Naghash HR, et al. The goat domestication process inferred from large-scale mitochondrial DNA analysis of wild and domestic individuals. Proc Natl Acad Sci USA. (2008) 105:46:17659–64. 10.1073/pnas.0804782105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chessa B, Pereira F, Arnaud F, Amorim A, Goyache F, Mainland I, et al. Revealing the history of sheep domestication using Retrovirus integrations. Science. (2009) 324:532–6. 10.1126/science.1170587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Moradi MH, Phua SH, Hedayat N, Khodaei-Motlagh M, Razmkabir M. Haplotype and genetic diversity of mtDNA in indigenous Iranian sheep and an insight into the history of sheep domestication. J Agr Sci Technol. (2017) 19:591–601. [Google Scholar]
- 115.Janoska F, Farkas A, Marosan M, Fodor JT. Wild boar (Sus scrofa) home range and habitat use in two Rumanian habitats. Acta Silv Lign Hung. (2018) 14:51–63. 10.2478/aslh-2018-0003 [DOI] [Google Scholar]
- 116.Mashkour M. Boars and pigs: A view from the Iranian plateau. In: Lion B, Michel C. Editors. De la domestication au tabou: Le cas des suidés dans le Proche-Orient ancien. Travaux de la Maison René-Ginouvès. Paris: De Boccard; (2006). p. 155–67. [Google Scholar]
- 117.Larson G, Dobney K, Albarella U, Fang M, Matisoo-Smith E, Robins J, et al. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science. (2005) 307:1618–21. 10.1126/science.1106927 [DOI] [PubMed] [Google Scholar]
- 118.Larson G, Albarella U, Dobney K, Rowley-Conwy P, Schibler J, Tresset A, et al. Ancient DNA, pig domestication, and the spread of the Neolithic into Europe. Proc Natl Acad Sci USA. (2007) 104:15276–81. 10.1073/pnas.0703411104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mezo M, Gonzalez-Warleta M, Castro-Hermida JA, Manga-Gonzalez MY, Peixoto R, Mas-Coma S, et al. The wild boar (Sus scrofa Linnaeus, 1758) as secondary reservoir of Fasciola hepatica in Galicia (NW Spain). Vet Parasitol. (2013) 198:274–83. 10.1016/j.vetpar.2013.09.009 [DOI] [PubMed] [Google Scholar]
- 120.Capucchio MT, Catalano D, Di Marco V, Russo M, Aronica V, Tomaselli A, et al. Natural trematode infestation in feral Nebrodi Black pigs: Pathological investigations. Vet Parasitol. (2009) 159:37–42. 10.1016/j.vetpar.2008.10.017 [DOI] [PubMed] [Google Scholar]
- 121.Mas-Coma S, Funatsu IR, Angles R, Buchon P, Mas-Bargues C, Artigas P, et al. Domestic pig prioritized in one health action against fascioliasis in human endemic areas: Experimental assessment of transmission capacity and epidemiological evaluation of reservoir role. One Health. (2021) 13:100249. 10.1016/j.onehlt.2021.100249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Harter S. Implication de la Paléoparasitologie dans l'étude des populations anciennes de la vallée du Nil et du Proche-Orient: études de cas (PhD thesis). Université de Reims Champagne-Ardenne, Unité de Formation et de Recherche de Pharmacie, Ecole Doctorale “Sciences Exactes et Biologie,” Reims, France. (2003). 271 p. [Google Scholar]
- 123.Harter-Lailheugue S, Le Mort F, Vigne JD, Guilaine J, Le Brun A, Bouchet F. Premières données parasitologiques sur les populations humaines précéramiques chypriotes (VIIIe et VIIe millénaires av. J-C). Paléorient. (2005) 31:43–54. 10.3406/paleo.2005.5124 [DOI] [Google Scholar]
- 124.Vigne JD, Briois F, Zazzo A, Willcox G, Cucchi T, Thiébault S, et al. First wave of cultivators spread to Cyprus at least 10,600 y ago. Proc Natl Acad Sci USA. (2012) 109:8445–9. 10.1073/pnas.1201693109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ridout-Sharpe J. The Mollusca from some prehistorisc wells and ditches on the west coast of Cyprus. In: Ridout-Sharpe J, Editor. The Archaeo-Malacological Group Newsletter. (2007) 11:9–10. Available online at: https://paperzz.com/doc/9332937/issue-11-may-2007-archaeomalacology-working-group [Google Scholar]
- 126.Fischer W. Süsswassermollusken aus Zypern. Nachrichtenblatt der Ersten Vorarlberger Malakol Gesellsch. (1994) 2:47–8.34093076 [Google Scholar]
- 127.Askari Z, Mas-Coma S, Bouwman AS, Boenke N, Stöllner T, Aali A, et al. Fasciola hepatica eggs in paleofaeces of the Persian onager Equus hemionus onager, a donkey from Chehrabad archaeological site, dating back to the Sassanid Empire (224-651 AD), in ancient Iran. Infect Genet Evol. (2018) 62:233–43. 10.1016/j.meegid.2018.04.028 [DOI] [PubMed] [Google Scholar]
- 128.Ashrafi K, Valero MA, Peixoto RV, Artigas P, Panova M, Mas-Coma S. Distribution of Fasciola hepatica and F. gigantica in the endemic area of Guilan, Iran: relationships between zonal overlap and phenotypic traits. Infect Genet Evol. (2015) 31:95–109. 10.1016/j.meegid.2015.01.009 [DOI] [PubMed] [Google Scholar]
- 129.Ponel P, Andrieu-Ponel V, Djamali M, Lahijani H, Leydet M, Mashkour M. Fossil beetles as possible evidence for transhumance during the middle and late Holocene in the high mountains of Talysch (Talesh) in NW Iran? J Environ Archaeol. (2013) 18:201–10. 10.1179/1749631413Y.0000000007 [DOI] [Google Scholar]
- 130.Sazmand A, Joachim A. Parasitic diseases of camels in Iran (1931–2017) – a literature review. Parasite. (2017) 24:21. 10.1051/parasite/2017024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sazmand A, Bahari A, Papi S, Otranto D. Parasitic diseases of equids in Iran (1931–2020): a literature review. Parasit Vectors. (2020) 13:586. 10.1186/s13071-020-04472-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Martínez-Navarro B. Early Pleistocene faunas of Eurasia and hominin dispersals. In: Fleagle J, Shea J, Grine F, Baden A, Leakey R. Editors. Out of Africa I. Vertebrate Paleobiology and Paleoanthropology. Dordrecht: Springer; (2010). p. 207–24. [Google Scholar]
- 133.Bibi F. Mio-Pliocene faunal exchanges and African biogeography: the record of fossil bovids. PLoS ONE. (2011) 6:e16688. 10.1371/journal.pone.0016688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Koufos GD, Kostopoulos DS, Vlachou TD. Neogene/Quaternary mammalian migrations in Eastern Mediterranean. Belg J Zool. (2005) 135:181–90. [Google Scholar]
- 135.Belmaker M. Criteria for identifying the African origin of early Pleistocene mammalian fauna in Eurasia. Comp Rend Palevol. (2018) 17:262–75. 10.1016/j.crpv.2017.08.003 [DOI] [Google Scholar]
- 136.O'Regan HJ, Turner A, Bishop LC, Elton S, Lamb A. Hominins without fellow travellers? First appearances and inferred dispersals of Afro-Eurasian large-mammals in the Plio-Pleistocene. Quatern Sci Rev. (2011) 30:1343–52. 10.1016/j.quascirev.2009.11.028 [DOI] [Google Scholar]
- 137.Estes RD. The Behaviour Guide to African Mammals, Including Hoofed Mammals, Carnivores and Primates. Berkeley, CA: University of California Press; (1991). p. i-xxii + 1-611. [Google Scholar]
- 138.Gujarat I. and a revision of the Asian genus Sivacobus Pilgrim, 1939. J Vert Paleontol. (2014) 35:e943399. 10.1080/02724634.2014.943399 [DOI] [Google Scholar]
- 139.Plaziat JC, Younis, WR,. The Modern Environments of Molluscs in Southern Mesopotamia, Iraq: A Guide to Paleogeographical Reconstructions of Quaternary fluvial, Palustrine Marine Deposits. Carnets de Géologie, Notebooks on Geology, Brest. (2005). p. 1–18. Available online at: http://paleopolis.rediris.es/cg/05/A01/ (accessed December 16, 2021).
- 140.Al-Mashhadani HM. Morphology and ecology of lymnaeid snails of Iraq with special reference to fascioliasis. Trans Roy Soc Trop Med Hyg. (1974) 67:10–1. 10.1016/0035-9203(74)90231-4 [DOI] [PubMed] [Google Scholar]
- 141.Mohammad MK. The parasitic infection of the freshwater snails collected in central Iraq. Int J Curr Microbiol Appl Sci. (2015) 4:4755. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.